Version Changes
Revised. Amendments from Version 1
Amendments were made based on reviewer comments to improve the clarity and flow of the review. Previously undescribed acronyms have been described, and redundant references and points in figure legend removed.
Abstract
Mitochondrial vitality is critical to cellular function, with mitochondrial dysfunction linked to a growing number of human diseases. Tissue and cellular heterogeneity, in terms of genetics, dynamics and function means that increasingly mitochondrial research is conducted at the single cell level. Whilst there are several technologies that are currently available for single-cell analysis, each with their advantages, they cannot be easily adapted to study mitochondria with subcellular resolution. Here we review the current techniques and strategies for mitochondrial isolation, critically discussing each technology’s limitations for future mitochondrial research. Finally, we highlight and discuss the recent breakthroughs in sub-cellular isolation techniques, with a particular focus on nanotechnologies that enable the isolation of mitochondria from subcellular compartments. This allows isolation of mitochondria with unprecedented spatial precision with minimal disruption to mitochondria and their immediate cellular environment.
Keywords: Mitochondria, mtDNA, mitochondrial isolation, heterogeneity, subcellular, nanoprobes, nanobiopsy, nanotweezers.
Introduction
Mitochondrial genetics
The mitochondrial genome consists of multiple double-stranded, circular DNA molecules ( Anderson et al., 1981; Andrews et al., 1999). In healthy individuals the default state of the mitochondrial genome is that of homoplasmy, where only wild-type mitochondrial DNA (mtDNA) exists. Low levels of mtDNA heterogeneity as a result of de novo mutations, termed heteroplasmy, is not deleterious since the polyploid nature of the mitochondrial genome buffers low levels of heteroplasmy ( Payne et al., 2013). However, clonal expansion of mutant mtDNA above a threshold level, results in biochemical defects and disease ( Payne et al., 2013; Rossignol et al., 2003). Over a life-course it is believed that clonal expansion of mtDNA point mutations occurs through random genetic drift in both germline and mitotic somatic cells ( Greaves et al., 2014); as well as in post-mitotic cells, facilitated by relaxed mtDNA replication, which can explain the long-time required for a threshold level to be reached as observed in age-related disease ( Elson et al., 2001). In comparison, it is suggested that some degree of selection influences clonal expansion of mtDNA deletions, acting in conjunction or independently of random genetic drift ( Lawless et al., 2020). Asides from random genetic drift, two alternative theories of clonal expansion of mtDNA deletions are the ‘negative feedback loop’ and ‘perinuclear niche’ theories ( Kowald & Kirkwood, 2014; Vincent et al., 2018). The non-uniform nature of mtDNA deletions suggests deletion of a single locus, leading to impaired feedback of polymerase γ activity, is unlikely to be solely responsible for clonal expansion of mtDNA deletions ( Damas et al., 2014), whilst the contribution of retrograde signalling within the ‘perinculear theory’ requires further investigation ( Lawless et al., 2020). To elucidate the mechanism of clonal expansion of mtDNA mutations, further investigation of mtDNA heterogeneity at the tissue, single-cell and subcellular level is necessary ( Lawless et al., 2020).
Mitochondrial disease and heterogeneity
Clonally expanded mtDNA mutations cause mitochondrial disease in one of two forms: primary, due to inherited mtDNA mutations and acquired, due to nuclear DNA mutations that lead to mtDNA mutation formation throughout life, as a result of impaired mtDNA maintenance ( Alston et al., 2017; Trifunovic et al., 2004). Primary mitochondrial disease, caused by inherited mtDNA mutations, has a variable age of onset and has a range of different presentations ( Gorman et al., 2016). In contrast, acquired mitochondrial disease, resulting from clonal expansion of sporadic mtDNA mutations, typically begins in adult life and causes progressive external ophthalmoplegia often with additional myopathic and neurodegenerative symptoms ( Alston et al., 2017); or neurodegeneration associated with advanced age ( Bender et al., 2006; Trifunovic et al., 2004).
Mitochondrial disease often expresses mosaicism in oxidative phosphorylation (OXPHOS) deficiency at the organ and tissue level, caused by cellular heterogeneity in mtDNA ( Ahmed et al., 2018). Mitochondrial heterogeneity exists as either genetic or non-genetic heterogeneity ( Aryaman et al., 2019). Sources of genetic heterogeneity associated with mitochondrial disease include mtDNA copy number, which is depleted in OXPHOS deficient skeletal muscle fibres ( Lehmann et al., 2019) and in Parkinson’s disease (PD) substantia nigra (SN) neurons but elevated in aged control SN neurons, possibly indicative of a neuroprotective mechanism that is overwhelmed in PD ( Chen et al., 2020; Lehmann et al., 2019). Genetic heterogeneity is also observed in elevated intercellular heteroplasmy, corresponding with oxidative phosphorylation (OXPHOS) deficiency between skeletal muscle fibres, colonic crypt cells and PD SN neurons ( Lehmann et al., 2019; Greaves et al., 2014 and Bender et al., 2006) but also localised intracellular heteroplasmy within skeletal muscle fibres and PD SN neurons ( Reeve et al., 2018; Vincent et al., 2018).
Non-genetic heterogeneity is observed in morphology, membrane potential and dynamics ( Kuznetsov & Margreiter, 2009). Mitochondrial heterogeneity can be physiological, due to different subcellular roles of mitochondrial subpopulations. In skeletal muscle, intermyofibrillar mitochondria have a more complex structure, likely to facilitate the contractile movement of muscle fibres ( Kuznetsov & Margreiter, 2009; Vincent et al., 2019). In contrast, subsarcolemmal mitochondria are more punctate mitochondria and have a lower capacity for OXPHOS ( Lai et al., 2019), but are likely to be involved in mito-nuclear signalling ( Paszkiewicz et al., 2017). It is also suggested synaptic mitochondria subpopulations have different proteomic profiles compared with non-synaptic mitochondria, notably in complex I expression, resulting in a smaller, punctate mitochondrial morphology and may modulate ATP supply for synaptic transmission and Ca 2+ buffering ( Graham et al., 2017). This proteomic heterogeneity, however, may also make synaptic mitochondria more vulnerable to OXPHOS deficiency and increased synaptic mitochondria density may serve as a compensatory mechanism, as observed in neurodegenerative disease ( Graham et al., 2017; Reeve et al., 2018). Membrane potential (Ψm) also expresses intercellular heterogeneity based on specific cell functions, such as in pancreatic β cells in response to elevated glucose and ATPase activity. Highly metabolic cells also tend to have a higher Ψm and likewise cellular regions with variable metabolic demands express localised Ψm heterogeneity ( Wikstrom et al., 2009). In neurodegeneration, it has been proposed that dysfunctional, depolarised mitochondria are selectively transported towards the cell body for degradation, whilst healthy mitochondria populate areas of greatest ATP demand ( Miller & Sheetz, 2004). Genetic heterogeneity is suggested to impact on non-genetic heterogeneity and that mtDNA might locally modulate cristae remodelling and respiratory chain complex structure ( Busch et al., 2014), equally non-genetic factors may affect mtDNA heterogeneity through mitochondrial dynamics and Ψm depolarisation modulating selective degradation of mitochondria harbouring high levels of heteroplasmy ( Wikstrom et al., 2009). Precise sampling of mitochondria from within subcellular localisations, to supplement existing data from studies at the single-cell and tissue level, would provide insight not only into mtDNA heterogeneity but also how this influences heterogeneity in mitochondrial structure, function and dynamics in health and disease.
Studying mitochondrial subpopulations
There is still much we do not understand about mitochondrial heterogeneity. Investigating mitochondrial heterogeneity at the inter- and intracellular level, will provide insight into other key outstanding questions within the mitochondrial field ( Aryaman et al., 2019; Lawless et al., 2020). What is the physiological role of intracellular heterogeneity? What differences exist between inherited and acquired mtDNA mutations? What interplay exists between genetic and non-genetic heterogeneity? Historically, mitochondrial fractionation techniques were used and remain popular due to being well characterised and reliable; despite being laborious, resource intensive and potentially damaging to mitochondria ( Liao et al., 2020). When selecting a method of mitochondrial isolation, in addition to mitochondrial purity, function and yield; efficiency, downstream analyses and sample should be considered. Whilst cell lines are more readily manipulatable and enable longitudinal studies ( Lawless et al., 2020), disease burden is better characterised in tissue, which also has greater clinical relevance ( Ahmed et al., 2018; Chen et al., 2020; Grady et al., 2018; Spinazzi et al., 2012). In tissue, some foci of deficiency measure as little as 10µm in diameter ( Vincent et al., 2018) and accurate sampling of individual foci would be challenging using current techniques. The purpose of this review is to critically discuss the advancement and application of different techniques for mitochondrial isolation. This review will also highlight the recent development of micro- and nanoscale techniques, allowing isolation of mitochondria at the cellular and subcellular level.
Macroscale mitochondrial isolation
Macroscale mitochondrial isolation entails the release of mitochondria from organs or large tissue samples through physical disruption. This is achieved through manual homogenisation, although low reproducibility and the degree of skill required affect mitochondrial integrity and yield of mitochondria ( Lai et al., 2019). Subcellular fractionation, through homogenisation followed by centrifugation, is a traditional and well-established means of reliably acquiring a high-yield mitochondrial fraction. To overcome some of the noted limitations of centrifugation-based methods of mitochondrial isolation, a number of alternative macroscale techniques have been developed for use with, or instead of, differential or density gradient centrifugation ( Gauthier & Lazure, 2014).
Centrifugation
Differential centrifugation (DC) and density gradient centrifugation (DGC) are used to fractionate organelles, including mitochondria, based on mass and sedimentary characteristics. Cell or tissue homogenate is centrifuged at increasing velocities and, in DGC, through a series of increasingly dense media bands ( Liao et al., 2020). DGC produces a purer fraction but a lower mitochondrial yield, relative to DC, though yield and mitochondrial integrity may be improved using detergents and reducing the homogenisation and centrifugation speed ( Lai et al., 2019). These methods of ‘mitochondrial fractionation’ are both reliable, acquire highly functional mitochondria and are well characterised in the literature but have a high resource and time cost. Dependent on the nature of the study, using DC alone may be preferable in obtaining a higher yield despite sacrificing mitochondrial purity. Centrifugation methods do remain the most popular methods of large-scale mitochondrial isolation however post-centrifugation methods are seeing an increase in usage, such as affinity purification ( Liao et al., 2020).
Affinity purification
Affinity purification (AP) captures mitochondria, through the adhesion of mitochondrial surface proteins ( Ru et al., 2012), to magnetic bead-conjugated antibodies which are retained within a magnetic field. In addition to favourable yield and purity, AP allows isolation of mitochondrial subpopulations, even without centrifugation ( Ahier et al., 2018; Hornig-Do et al., 2009; Hubbard et al., 2019) and has a low sample requirement and run time ( Ru et al., 2012). AP does have a high reagent cost and is less well established than DC or DGC but with continued development it is suggested that AP could succeed DGC as the preeminent method of large-scale mitochondrial isolation and AP is particularly advantageous for isolating mitochondrial subpopulations from homogenate ( Afanasyeva et al., 2018; Hubbard et al., 2019).
Flourescence activated organelle sorting
Following detection of a fluorescent label, including dyes or reporter genes ( Ashley et al., 2005; Cohen & Fox, 2001; Floros et al., 2018), fluorescence activated organelle sorting (FAOS) is able to isolate mitochondria from other organelles or mitochondrial subpopulations by assigning mitochondria, separated into individual droplets, a specific charge ( Cavelier et al., 2000). FAOS enables isolation of a higher yield of functional, purified mitochondrial subpopulations reducing the starting material requirement ( Daniele et al., 2016; Gauthier & Lazure, 2014). A consideration when using FAOS is dye cytotoxicity, however, the increasing number of available dyes should allow substitution of potentially toxic dyes ( Barteneva et al., 2014); nevertheless, fluorescent dyes may still lead to mitochondrial aggregation and overestimation of mtDNA copy number ( Pflugradt et al., 2011). FAOS is best utilised for high-throughput and high-yield mitochondrial isolation from larger samples ( Poe et al., 2006).
Electrophoresis and field-flow fractionation
Capillary electrophoresis (CE), free-flow electrophoresis (FFE) and field-flow fractionation (FFF) techniques also separate subpopulations of organelles based on sedimentary characteristics. Samples are carried along a channel by a laminar flow and in CE and FFE, organelles are separated based on their isoelectric point, through application of an electric field ( Poe et al., 2006; Zischka et al., 2006), whilst FFF utilises a perpendicular cross flow to separate particles based on size and mass ( Yang et al., 2015). FFE is shown to be more time efficient and able to produce a purer but smaller yield, however, equipment cost was higher compared with DGC ( Hartwig et al., 2009). There is consensus in the literature that FFE is optimally used in conjunction with DGC and for use in studies where high-throughput and mitochondrial fraction purity are more desirable than a higher mitochondrial yield ( Islinger et al., 2018; Zischka et al., 2006). Compared with FFE, CE-LIF (capillary electrophoresis -laser-induced fluorescence) has a lower throughput but requires less starting material, and has a better signal-to-noise ratio compared with FAOS ( Kostal et al., 2009; Poe et al., 2006). A limitation of CE-LIF is mitochondria retention within the fine capillary tube, which can impact on yield and, like FFE, is best used when purity is preferable to high yield ( Poe et al., 2010). FFF has the highest working range of the techniques reviewed and does not require mitochondrial labelling ( Kang et al., 2008; Reschiglian et al., 2005; Yang et al., 2015). Whilst isolation of mitochondrial subpopulations was demonstrated with FFF, isolated fractions were highly contaminated and with no direct comparison of yield relative to DGC, it is difficult to determine the practicability of FFF for mitochondrial isolation. Unlike FFE and CE, FFF is best used when rapid acquisition of a high mitochondrial yield is required at the expense of purity ( Kang et al., 2008).
Overview of macroscale mitochondrial isolation techniques
Macroscale methods, incorporating centrifugation, remain the gold standard for high throughput, large-scale mitochondrial isolation but are limited by high time and resource demands, coupled with limited separation of mitochondrial subpopulations ( Lai et al., 2019; Liao et al., 2020) ( Table 1). Combining DGC or DC with post-fractionation purification, isolation of purified mitochondrial subpopulations is possible but at the risk of increasing the run time, sacrificing purity or impacting on mitochondrial vitality or function. Additionally, these techniques cannot provide information on cellular provenance or allow longitudinal studies, due to the necessity of cell lysis. Whilst macroscale techniques excel at investigating mitochondrial heterogenity at the tissue level, to study patterns of mitochondrial heterogeneity and dysfunction at the cellular and sub-cellular level, low-throughput analysis of smaller samples is likely to be more appropriate ( Picard et al., 2011).
Table 1. Summary of mitochondrial isolation methods (Macroscale).
A table highlighting the relative benefits and disadvantages of different ‘macroscale’ mitochondrial methods of mitochondrial isolation, incorporating either density gradient centrifugation (DGC), differential centrifugation (DC) or none. Techniques compared relative to the performance of DGC alone*: ++ = higher; + = slightly higher or similar; - = lower; 2D-PAGE = two-dimensional polyacrylamide gel electrophoresis; AP = affinity purification; CE = capillary electrophoresis; FAOS = fluorescence activated organelle sorting; FFE = free flow electrophoresis; FFF = field-flow fractionation; LC-MS = liquid chromatography mass spectrometry; SEM = Scanning Electron Microscopy; TEM = transmission electron microscopy.
| Isolation
Method |
Prior
Fractionation |
Mitochondrial
Yield * |
Mitochondrial
Purity * |
Starting
Material * |
Expense * | Throughput * | Subcellular
Spatial Precision |
Serial
Sampling |
Confirmation | General
Comments |
References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | DC
(or none) |
++
(++) |
+
(++) |
-
(-) |
++
(++) |
++
(++) |
No | No | Western blotting,
immunofluorescence, qPCR |
High
selectivity; sonication may impact mitochondria viability; not suitable for larger samples |
Hornig-Do
et al. (2009); Ahier et al. (2018) and Hubbard et al. (2019) |
| FAOS | DC | ++ | ++ | ++ | + | ++ | No | No | qPCR, NGS | High
selectivity; shear damage and dyes may impact mitochondrial viability |
Daniele
et al.
(2016) |
| FFE | DC
(and DGC) |
+
(-) |
++
(++) |
+
(+) |
++
(++) |
++
(++) |
No | No | Western blotting,
TEM |
High
mitochondrial viability; high selectivity |
Zischka
et al.
(2006) |
| FFF | DC | ++ | - | + | ++ | ++ | No | No | Western blotting,
SEM, 2D-PAGE, LC-MS |
High
mitochondrial viability; high selectivity; high working range |
Kang
et al.
(2008) and Yang et al. (2015) |
| CE | DC | - | ++ | - | ++ | ++ | No | No | qPCR | High
selectivity; predisposed to clogging |
Poe
et al.
(2006) and Poe et al. (2010) |
Microscale and nanoscale mitochondrial isolation
Whilst mitochondrial heterogeneity does exist at the single and subcellular level, until relatively recently there has been a distinct lack of techniques capable of effective subcellular manipulation. Previous methods enabling mechanical penetration of cells relied on micropipettes which are inclined to cause physical trauma to the cell and lack the spatio-temporal control required for subcellular sampling. Use of microfluidics and nanoprobes offer a less damaging and more controlled means of sampling of organelles such as mitochondria ( Kang et al., 2016) ( Table 2 and Table 3).
Table 2. Summary of mitochondrial isolation methods (Microscale).
A table highlighting the relative benefits and disadvantages of different ‘microscale’ mitochondrial methods of mitochondrial isolation, incorporating either density gradient centrifugation (DGC), differential centrifugation (DC) or none. Techniques compared relative to the performance of DGC alone*: ++ = higher; + = slightly higher or similar; - = lower; ddPCR = digital droplet PCR; LCM = laser capture microdissection; NGS = next generation sequencing; OT = optical tweezers; SEM = Scanning Electron Microscopy.
| Isolation
Method |
Prior
Fractionation |
Mitochondrial
Yield * |
Mitochondrial
Purity * |
Starting
Material * |
Expense * | Throughput * | Subcellular
Spatial Precision |
Serial
Sampling |
Confirmation | General
Comments |
References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Micro-AP | DC | - | ++ | - | - | ++ | No | No | fluorescence
microscopy |
High
mitochondrial viability; low run time; predisposed to clogging |
Kayo
et al.
(2013) and Banik et al. (2016) |
| Micro-FFE | DC | - | ++ | - | - | ++ | No | No | electrophoretic
profiling |
High
mitochondrial viability; low run time; predisposed to clogging |
Kostal
et al.
(2009) |
| Micro-FFF | DC | - | ++ | - | - | ++ | No | No | fluorescence
microscopy |
High
mitochondrial viability; low run time |
Lu
et al.
(2004) |
| LCM | None | - | ++ | - | ++ | - | Yes | No | fluorescence
microscopy, ddPCR, qPCR |
Targets a
single cell; may cause laser induced mtDNA damage |
Vincent
et al. (2018) and Trifunov et al. (2018) |
| OT | None | - | ++ | - | ++ | - | Yes | No | fluorescence
microscopy; SEM |
Targets a
single cell; may cause laser induced mtDNA damage |
König
et al.
(2005) and Jeffries et al. (2007) |
Table 3. Summary of mitochondrial isolation methods (Nanoscale).
A table highlighting the relative benefits and disadvantages of different methods of nanoscale mitochondrial isolation, incorporating either density gradient centrifugation (DGC), differential centrifugation (DC) or none. Techniques compared relative to the performance of DGC alone *: ++ = higher; + = slightly higher or similar; - = lower; nFAMS = Nano-Fluorescence Activated Mitochondrial Sorting; NGS = next generation sequencing; NT = nanotweezers MS = Mass spectrometry.
| Isolation
Method |
Prior
Fractionation |
Mitochondrial
Yield * |
Mitochondrial
Purity * |
Starting
Material * |
Expense * | Throughput * | Subcellular
Spatial Precision |
Fluorescent
labelling |
Serial
Sampling |
Confirmation | General
Comments |
References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nFAMS | DGC | ++ | ++ | - | + | ++ | No | Yes | No | fluorescence
microscopy, MS, qPCR |
High
selectivity; shear damage may impact mitochondrial viability; dyes may induce aggregation |
MacDonald
et al. (2019) |
| Nanobiopsy | None | - | ++ | - | ++ | - | Yes | Yes | Yes | fluorescence
microscopy, NGS, SICM |
Maintains
cellular environment and viability; limited selectivity |
Actis
et al.
(2014a) |
| NT | None | - | ++ | - | ++ | - | Yes | Yes | Yes | fluorescence
microscopy |
Maintains
cell viability and natural environment; limited selectivity |
Nadappuram
et al. (2019) |
Microfluidics
Microfluidic analogues of macro-techniques, including micro-AP, micro-FFF, and micro-FFE, have been developed for the isolation of mitochondria ( Kostal et al., 2009; Kayo et al., 2013; Lu et al., 2004). Microfluidic isolation methods also exist that trap mitochondria through geometric traps, such as the nanohole array ( Kumar et al., 2015). Not only are microfluidic assays more resource efficient, they are less disruptive to the physiological organelle environment, cause less physical trauma to cells and organelles and provide more representative results ( Kang et al., 2016; Moutaux et al., 2018). Whilst ‘ Mito-magento’, a micro-AP variant, forgoes the need for immunoprecipitation and minimises the risk of clogging, which can impact on yield, nanoparticle toxicity to cells and mitochondria is not ruled out ( Banik et al., 2016; Tesauro et al., 2017; Yarjanli et al., 2017). Microfluidic techniques have a much lower yield and do not allow insight into the subcellular location of mitochondrial subpopulations or facilitate longitudinal analysis. Microfluidic techniques are best suited to high-throughput isolation of pure and highly functional mitochondrial fractions from a limited amount of starting material.
Nano-fluorescence activated mitochondrial sorting
FAOS is generally optimised to isolate mitochondria from larger samples ( Daniele et al., 2016). Nano-fluorescence activated mitochondrial sorting (nFAMS) utilises a microfluidic chip device with confocal microscopy. Application of a small voltage allows sorting of mitochondrial subpopulations with minimal impact to viability whilst achieving a high throughput, yield and purity ( MacDonald et al., 2019; Schiro et al., 2012). It is noted single molecule PCR may be more applicable to enrichment of smaller mtDNA concentrations, obtained using nFAMS and similar formats, to avoid PCR errors ( Kraytsberg et al., 2008; MacDonald et al., 2019). Respiratory function assays did show that isolated mitochondria were highly functional, however, like the other techniques discussed so far, obligatory cell lysis also means that obtaining information on cell provenance and serial sampling are not possible with nFAMS ( MacDonald et al., 2019).
Laser-capture microdissection and optical tweezers
Laser capture microdissection (LCM) allows for single-cell isolation by cutting around a delinated cell using a ‘train’ of sequential nanosecond pulses from a nitrogen laser. Translocation and capture is either gravity assisted or through laser pulse induced propulsion into a collection vessel ( Vogel et al., 2007). LCM was used to isolate mitochondria from single cells and from subcellular foci, to investigate the spread of clonal expansion of mtDNA mutations a ( Trifunov et al., 2018; Vincent et al., 2018).
Optical tweezers (OT) are capable of simultaneously lysing cells, whilst trapping subcellular organelles within a focused laser beam, with nanoscale precision ( König et al., 2005; Reiner et al., 2010). LCM and OT are exciting techniques for isolating mitochondria within a specific cell, demonstrating not only the means to retain cellular provenance but also demonstrating cellular and subcellular precision ( König et al., 2005; Vincent et al., 2018). However, laser induced damage to mitochondrial and mtDNA damage when targeting smaller areas limits the effectiveness of LCM and OT in serial sampling and isolating mitochondria from smaller subcellular foci ( Botchway et al., 2010; Vincent et al., 2018). Alongside flow cytometry based methods, LCM and OT are gold standard techniques used for mitochondrial isolation for understanding mitochondrial heterogeneity at the single-cell level.
Nanoprobe based techniques
Nanotechnologies show great promise to enable mitochondrial isolation at the subcellular level ( Laforge et al., 2007). Whilst micromanipulators incorporating micropipettes have been shown to successfully isolate and transplant nuclei ( Hyslop et al., 2016), a challenge for this technology has been the ability to sample smaller subcellular structures ( Jeffries et al., 2007). Nanoscale probes can allow the manipulation of biomolecules and organelles in a non-invasive manner with greater spatial precision compared to their microscale counterparts. Nanoprobes cause minimal perturbation to cell viability or their natural cellular environment, allowing for longitudinal sampling of mitochondria and enabling the comparison of mitochondrial heterogeneity between and within foci of deficiency ( Actis, 2018). Four key methods for nanomanipulation of cytoplasmic contents have been developed: nanobiopsy, where cellular contents are aspirated within a nanopipette using an applied voltage; fluid-force microscopy (FluidFM), where an atomic force microscope (AFM) probe, containing a nanofluidic channel, enables pressure-driven sampling from living cells; dielectrophoretic nanoprobes, where an applied high-frequency AC is used to trap and manipulate organelles and biomolecules within living cells; and nanoelectroporation, where cells are grown on an array of channels and a transient electrical current causes temporary pore formation in the cellular membrane, allowing sampling from the cytoplasm ( Ino et al., 2018). So far only nanobiopsy and nanotweezers (NT), which harnesses dielectrophoresis, have been demonstrated to isolate mitochondria ( Actis et al., 2014a; Nadappuram et al., 2019).
Nanobiopsy. Nanopipettes are nanoprobes that are readily and reproducibly fabricated from glass capillaries using a laser puller ( Laforge et al., 2007). Nanopipettes can be integrated within nanomanipulators to comprise a scanning ion conductance microscope (SICM), which allows for the high resolution topographical mapping of living cells and tissues ( Novak et al., 2009). The nanobiopsy approach relies on electrowetting, within a nanopipette, to extract cytoplasmic material from living cells. Electrowetting is a physical effect where the wetting, or interaction of a liquid with another surface, across two interfaces, can be controlled via an externally applied voltage. If a nanopipette is filled with an organic solution and immersed in an aqueous solution a liquid-liquid interface is formed at the nanopipette tip ( Laforge et al., 2007). Depending on the voltage applied, aqueous solution can be drawn or expelled from the nanopipette ( Dale & Unwin, 2008). Electrowetting within the cytoplasm of a living cell allows cytosol and organelles to be aspirated ( Actis et al., 2014a; Figure 1). Successful aspiration of mitochondria has been reported using nanobiopsy. This was demonstrated by the presence of MitoTracker green, a mitochondrial matrix specific fluorescent dye, in the nanopipette tip, a reduction in fluorescence in the biopsied area and successful sequencing of mtDNA acquired from the biopsy ( Actis et al., 2014a). Dependent on the target cell and the pipette geometry, fine tuning of the electrowetting settings is necessary ( Actis et al., 2014b; Seger et al., 2012). The precise volume aspirated or expelled can be determined, and therefore controlled, by measuring the nanopipette resistance during nanobiopsy. A drawback of nanobiopsy is that the use of an organic solvent within the nanopipette does not allow the acquisition of topographical images because of the low signal-noise ratio. To be able to properly take advantage of the fine placement of the nanopipette, a double barrel pipette - filled with aqueous and organic solutions respectively, would be required so that scanning and electrowetting could be performed simultaneously ( Seger et al., 2012). For biopsies taken from tissue, a possible alternative could be either the pre- or post-biopsy topographical scanning of the area of interest. Nanobiopsy is highly optimised for the highly-precise isolation of small mitochondrial subpopulations either from subcellular foci or for longitudinal sampling of mitochondria from the same cell. Whilst this has been demonstrated in cultured cells this has yet to be demonstrated in tissue.
Figure 1. Mitochondrial nanobiopsy.
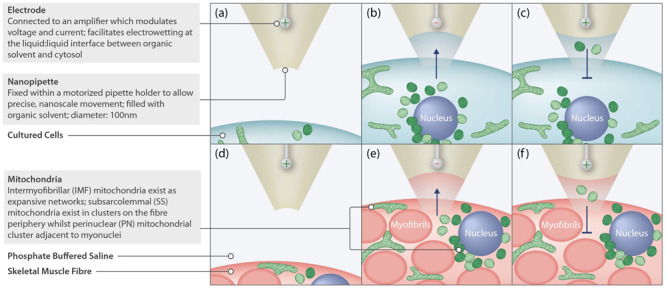
Nanobiopsy is carried out using a nanopipette, filled with organic solvent, that is incorporated into a Scanning Ion Conductance Microscope (SICM) system which consists of an amplifier, that modulates and detects extremely small changes in current, as well as piezo motors that allow for highly precise movement of the nanopipette to and within the area of interest. This can be used with cultured cells ( a– c) or in tissue ( d– f). ( a, d) To function, the SICM requires the nanopipette to be immersed in an aqueous solution. Whilst being lowered, the SICM system constantly measures the current through the nanopipette tip. If the current magnitudes drops below a predetermined threshold, this will result in negative feedback causing the nanopipette to stop typically within 1µm of the cell of interest. At this point the nanopipette be lowered at high speed to penetrate the cell membrane, whilst a small positive voltage (200mV) is applied to prevent premature aspiration of the cytosolic components. ( b, e) Once within the cell and at the area of interest, application of a small voltage (-200mV) allows for the aspiration of mitochondria within the cytosol as a result of the phenomenon of electrowetting. ( c, f) After successful aspiration of mitochondria, reapplication of a positive voltage (100mV) prevents further aspiration but allows retention of the collected sample. The sample can then be transferred to a collection vessel where the sample can be expelled at a higher voltage (1V).
Nanotweezers. Dielectrophoretic nanotweezers (DENT) were first introduced as an adaption of AFM, where by employing dielectrophoresis, mRNA could be selectively targeted and captured with minimal impact to cell viability. DENT utilises a nanoprobe consisting of a closely distanced silicon core and a metal alloy layer which serve as inner and outer electrodes ( Nawarathna et al., 2009). Application of an AC between the inner and outer electrodes results in an electrical field sufficient to prompt polarisable subcellular molecules to become induced dipoles that are trapped at the probe tip ( Ramos et al., 1999). A dielectrophoretic force can be intensified by increasing the voltage or decreasing the distance between the two electrodes, since the force is proportionate to V 2d −3 ( V being voltage; d being distance). An inter-electrode distance of <20nm enabled authors to achieve a dielectrophoretic force sufficient to capture a single mitochondrion at a physiologically viable ionic strength, with a NT probe incorporated into a micromanipulator system. Successful acquisition of healthy mitochondria was shown through a reduction in localised tetramethylrhodamine methyl ester fluorescent dye post-acquisition ( Nadappuram et al., 2019; Figure 2). Like nanobiopsy, NT are capable of sampling subcellular biomolecules with high spatial resolution with minimal impact on cell and organelle viability. A potential disadvantage of the NT format could be the manner in which samples are retained at the end of the nanotweezers probe, similar to a holding pipette. Without protection from the pipette tip, the sample would be exposed and prone to physical trauma or loss when being transferred from the cell to collection vessel ( Imura et al., 2002; Nadappuram et al., 2019). Loss of nucleic acid samples can be circumvented through use of specific oligonucleotide probes at the tip of the nanotweezers, to hybridised and trap mRNA or DNA isolated through dielectrophoresis and leading to the suggestion that this technique has a greater propensity for mRNA isolation or even cell-free mtDNA, though nanotweezers still offer great promise for mitochondrial isolation ( Nadappuram et al., 2019; Nawarathna et al., 2009).
Figure 2. Nanotweezers.
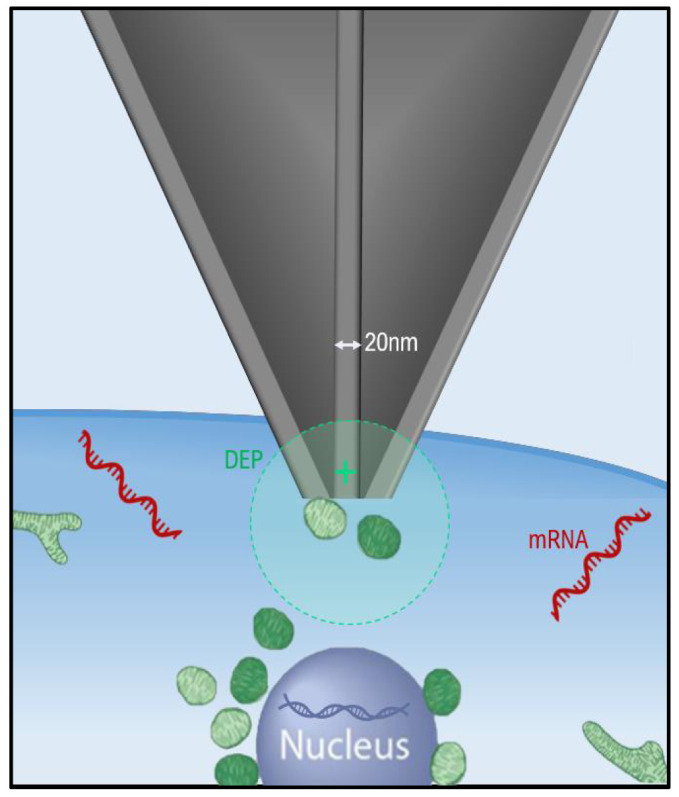
The nanotweezers tip is inserted into a cell and then application of an alternating current (A.C.) results in a localised electric field that traps biomolecules at the probe tip through dielectrophoretic (DEP). This DEP force is strong enough to capture nucleic acids and mitochondria in solution. Once captured the molecules can then we withdrawn from out of the cell of interest with the removal of the nanotweezers and transferred to a collection vessel. The applied DEP force is reversible and turning off the A.C. results in the release of the captured biomolecules.
Fluid-Force Microscopy. FluidFM is another adaptation of AFM. AFM works using a cantilever, which moves relative to the changes in depth of a given surface either through direct contact, tapping or oscillating just above the surface ( Verbiest & Rost, 2016). The movement of the cantilever is measured by a laser which produces a 3-D image of the surface and negative feedback control of a piezo motor allows adjustment of the probe position ( Guillaume-Gentil et al., 2016; Uehara et al., 2004). The unique feature of FluidFM is that it utilizes a hollow AFM tip coupled with a pressure regulator that allows the manipulation of liquid in and around cells with high precision. During FluidFM, the AFM tip is lowered into the cell and held with a predetermined force (~550nN) to allow capture of cellular contents by applying negative pressure (-800mbar) ( Guillaume-Gentil et al., 2016). Whilst Guillaume-Gentil and colleagues did test whether mitochondria could be isolated using FluidFM, a negligible reduction in MitoTracker Green likely indicates that this was unsuccessful. Authors concede that lack of success could have been because of probe aperture size was insufficient to capture mitochondria within an interconnected network. Like nanobiopsy and NT, FluidFM minimally impacts on cell viability. Also, like nanobiopsy, AFM allows precise placement of the AFM probe in a cellular localisation, though not with the same precision afforded by SICM. A ‘snap’ phenomenon, caused by the intermolecular forces acting on the AFM probe as it approaches the cellular membrane, can cause loss of control of the probe, reducing sampling accuracy ( Uehara et al., 2004; Verbiest & Rost, 2016). FluidFM also prioritises high sampling resolution above yield and should successful isolation of mitochondria be demonstrated using Fluid FM, it would fit into a similar niche to nanobiopsy and NT isolating mitochondria within subcellular foci.
Overview of micro- and nanoscale mitochondrial isolation techniques
Microfluidic techniques are promising, being high-throughput and the microfluidic devices themselves are inexpensive. However, adapting existing platforms for microfluidics is less cost effective and has resulted in slower development and adoption of microfluidic techniques ( Sonker et al., 2017). Of the more developed micro- and nanoscale techniques, flow cytometry based methods appear quicker, higher throughput and demonstrate the greatest yield, though may impair mitochondrial viability ( Pflugradt et al., 2011). Whilst LCM and OT can potentially result in photodamage to mtDNA, careful consideration of cutting parameters can negate this ( Keloth et al., 2018). Some macro- and microscale techniques may be able to separate mitochondrial subpopulations but lack the spatio-temporal range to isolate and contextualise mitochondrial subpopulations in their natural cellular environment, which affects our ability to fully understand the cellular conditions that contribute to mitochondrial heterogeneity and dysfunction ( Lanza & Nair, 2009; Moutaux et al., 2018).
Nanoprobes allow a means of isolating mitochondria with superior sampling specificity whilst being minimally invasive to the cell – preserving the viability of live cells and the native environment of the mitochondria ( Actis et al., 2014a; Nadappuram et al., 2019). It is suggested that nanobiopsy lacks specificty in the cytoplasmic contents sampled ( Actis et al., 2014a; Nadappuram et al., 2019). However, nanoscale adjustment of the nanopipette position above the area of interest, using an SICM system guided by topographical scanning in addition to optical confirmation alone, enables highly precise movement and mitochondrial capture allowing for a superior sampling specificity using nanobiopsy ( Seger et al., 2012). ‘Microbiospy’ of mitochondria in vivo has been demonstrated using a micropipette incorporated into a micromanipulator ( Morris et al., 2017). Whilst the data from this study is certainly valuable, the method was limited by poor control of aspirated volumes and the inability to contextualise labelled mitochondria without a secondary mode of visualising the location of the pipette relative to labelled mitochondria. This indeed is a common limitation of both NT and other micromanipulator based methods of mitochondrial isolation. Mitochondria can be as large 2µm in diameter ( Imura et al., 2002). Incorporation of micropipettes within an SICM system may enable the positioning of the pipette to the mitochondrial sub-population of interest with the same nanoscale level of precision, achieved through the combination of topographical scanning and optical visualisaton, but the larger pore size would guard against sampling bias favouring smaller mitochondrial subpopulations.
NT preferentially captures polarisable molecules. Whilst this enables a degree of selectivity, this may also lead to increased contamination, especially from non-specific capture of nucleic acids ( Nadappuram et al., 2019). Use of a ‘physiologically safe’ current, stated as being less than the total current of all ion channels in a given cell, also prevents direct damage to the sampled mitochondria ( Laforge et al., 2007). Whilst not necessarily a disadvantage, nanobiopsy and NT both require samples to be immersed in ionic solution ( Actis, 2018). FluidFM, conversely, can be adapted to faciliate high-throughput sampling by connecting the AFM probe to a microchannel and can be carried out in or out of solution ( Kang et al., 2016; Meister et al., 2009). Whilst as of yet, there has only been a modest number of examples demonstrating isolation of mitochondria using nanotechnologies, pioneering proof-of-principle study data does make the future development of nanotechnologies for mitochondrial isolation an exciting prospect. Whilst sampling of mitochondria, from within foci of deficiency in tissue, has yet to be demonstrated this provides the next challenge for nanotechnologies going forward.
Concluding remarks
Mitochondrial isolation can be organised into three general categories based on their sampling resolution: macroscale, microscale and nanoscale. Macroscale and microfluidic techniques are most useful for respective large or small-scale studies investigating mitochondrial heterogeneity, or a particular mechanism in mitochondrial dysfunction, at the tissue and organ level ( Brand & Nicholls, 2011); LCM, OT and nanoprobe based techniques, however, are designed to acertain information on mitochondrial heterogeneity at the cellular and subceullar level. The latter will ultimately help us to understand how mitochondrial dysfunction may originate and spread ( Picard et al., 2011; Vincent & Picard, 2018). Whilst mitochondrial isolation has only been demonstrated using nanobiopsy and NT, asides FluidFM, other nanoprobe techniques have potential utility for mitochondrial isolation. The “mille-feuille” probe, like its namesake, contains alternating aqueous and organic phase layers, allowing continuous sampling through nano-electrophoresis ( Ito et al., 2017). Nanoneedles and nanostraws offer the potential for longitudinal analysis of mitochondria from cells, cultured directly on top of these nanostructures, and sampling is achieved through laser-induced poration or electroporation of the cell membrane and high throughput isolation with a similar level of precision ( Cao et al., 2019; Chiappini et al., 2015).
The usefulness of nanoprobe techniques is not limited to isolating mitochondria. The ability to aspirate and inject using nanoprobes has great therapeutic potential, including the injection of mitochondria at precise cellular locations for the purposes of experimentation but also the eventual possibility for use in mitochondrial targeted therapies, building upon existing mitochondrial transfer techniques whilst avoiding ‘mitochondrial carryover’ ( Hudson et al., 2019; Hyslop et al., 2016; Seger et al., 2012). Mitochondrial fractionationation based techniques are still the most extensively used, but other technologies are catching up in terms of popularity and development. This is in part driven by the need to better understand mitochondrial physiology and pathophysiology with subcellular precision ( Picard et al., 2011), but also coincides with the transition of cellular biology from era of ‘single-cell omics’ to that of ‘subceullar omics’. No single technique is all-encompassing, in aiding our understanding of mechanisms of mitochondrial dysfunction, instead each has objective superiority. Nanoprobe based technologies are a great addition to the arsenal of mitochondrial isolation methods and their continued development is opening up an entirely new avenue of research into the spread of mitochondrial dysfunction at the subcellular level. This will ultimately help us better understand the bigger picture of mitochondrial heterogeneity and dysfunction.
Data availability
Underlying data
No data are associated with this article.
Funding Statement
This work was supported by the Wellcome Turst through a Sir Henry Wellcome Postdoctoral Fellowship to AV [215888, <a href=https://doi.org/10.35802/215888>https://doi.org/10.35802/215888</a>] and a Wellcome Trust Centre grant to the Wellcome Centre for Mitochondrial Research [203105]. This work was also supported by the Medical Research Council [OSR/0200/2018/BURY] and the Michael J. Fox Foundation for Parkinson’s Research [ID-15643].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- Actis P: Sampling from Single Cells. Small Methods. 2018;2(3):1700300. 10.1002/smtd.201700300 [DOI] [Google Scholar]
- Actis P, Maalouf MM, Kim HJ, et al. : Compartmental genomics in living cells revealed by single-cell nanobiopsy. ACS nano. 2014a;8(1):546–553. 10.1021/nn405097u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Actis P, Tokar S, Clausmeyer J, et al. : Electrochemical nanoprobes for single-cell analysis. ACS Nano. 2014b;8(1):875–884. 10.1021/nn405612q [DOI] [PubMed] [Google Scholar]
- Afanasyeva MA, Ustiugova AS, Golyshev SA, et al. : Isolation of Large Amounts of Highly Pure Mitochondria for "Omics" Studies. Biochemistry (Mosc). 2018;83(1):76–85. 10.1134/S0006297918010108 [DOI] [PubMed] [Google Scholar]
- Ahier A, Dai CY, Tweedie A, et al. : Affinity purification of cell-specific mitochondria from whole animals resolves patterns of genetic mosaicism. Nat Cell Biol. 2018;20(3):352–360. 10.1038/s41556-017-0023-x [DOI] [PubMed] [Google Scholar]
- Ahmed ST, Craven L, Russell OM, et al. : Diagnosis and Treatment of Mitochondrial Myopathies. Neurotherapeutics. 2018;15(4):943–953. 10.1007/s13311-018-00674-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alston CL, Rocha MC, Lax NZ, et al. : The genetics and pathology of mitochondrial disease. J Pathol. 2017;241(2):236–250. 10.1002/path.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, et al. : Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, et al. : Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Aryaman J, Johnston IG, Jones NS: Mitochondrial Heterogeneity. Front Genet. 2019;9:718. 10.3389/fgene.2018.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley N, Harris D, Poulton J: Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp Cell Res. 2005;303(2):432–446. 10.1016/j.yexcr.2004.10.013 [DOI] [PubMed] [Google Scholar]
- Banik B, Askins BW, Dhar S: Mito-magneto: a tool for nanoparticle mediated mitochondria isolation. Nanoscale. 2016;8(47):19581–19591. 10.1039/c6nr05882e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barteneva NS, Ponomarev ED, Tsytsykova A, et al. : Mitochondrial staining allows robust elimination of apoptotic and damaged cells during cell sorting . J Histochem Cytochem. 2014;62(4):265–75. 10.1369/0022155413520404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, et al. : High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38(5):515–517. 10.1038/ng1769 [DOI] [PubMed] [Google Scholar]
- Botchway SW, Reynolds P, Parker AW, et al. : Use of near infrared femtosecond lasers as sub-micron radiation microbeam for cell DNA damage and repair studies. Mutat Res. 2010;704(1–3):38–44. 10.1016/j.mrrev.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG: Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435(2):297–312. 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KB, Kowald A, Spelbrink JN: Quality matters: how does mitochondrial network dynamics and quality control impact on mtDNA integrity? Philos Trans R Soc Lond B Biol Sci. 2014;369(1646):20130442. 10.1098/rstb.2013.0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Zhou X, Zeng Y: Microfluidic Exponential Rolling Circle Amplification for Sensitive microRNA Detection Directly from Biological Samples. Sens Actuators B Chem. 2019;279:447–457. 10.1016/j.snb.2018.09.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelier L, Johannisson A, Gyllensten U: Analysis of mtDNA copy number and composition of single mitochondrial particles using flow cytometry and PCR. Exp Cell Res. 2000;259(1):79–85. 10.1006/excr.2000.4949 [DOI] [PubMed] [Google Scholar]
- Chen C, Vincent A, Blain A, et al. : Investigation of mitochondrial biogenesis defects in single substantia nigra neurons using post-mortem human tissues. Neurobiol Dis. 2020;134:104631. 10.1016/j.nbd.2019.104631 [DOI] [PubMed] [Google Scholar]
- Chiappini C, De Rosa E, Martinez J, et al. : Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization. Nat Mater. 2015;14(5):532–539. 10.1038/nmat4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JS, Fox TD: Expression of green fluorescent protein from a recoded gene inserted into Saccharomyces cerevisiae mitochondrial DNA. Mitochondrion. 2001;1(2):181–189. 10.1016/s1567-7249(01)00012-5 [DOI] [PubMed] [Google Scholar]
- Dale SEC, Unwin PR: Polarised liquid/liquid micro-interfaces move during charge transfer. Electrochem commun. 2008;10(5):723–726. 10.1016/j.elecom.2008.02.023 [DOI] [Google Scholar]
- Damas J, Samuels DC, Carneiro J, et al. : Mitochondrial DNA rearrangements in health and disease--a comprehensive study. Hum Mutat. 2014;35(1):1–14. 10.1002/humu.22452 [DOI] [PubMed] [Google Scholar]
- Daniele JR, Heydari K, Arriaga EA, et al. : Identification and Characterization of Mitochondrial Subtypes in Caenorhabditis elegans via Analysis of Individual Mitochondria by Flow Cytometry. Anal Chem. 2016;88(12):6309–6316. 10.1021/acs.analchem.6b00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Samuels DC, Turnbull DM, et al. : Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68(3):802–806. 10.1086/318801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floros VI, Pyle A, Dietmann S, et al. : Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat Cell Biol. 2018;20(2):144–151. 10.1038/s41556-017-0017-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier DJ, Lazure C: Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev Proteomics. 2014;5(4):603–617. 10.1586/14789450.5.4.603 [DOI] [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, Dimauro S, et al. : Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- Grady JP, Pickett SJ, Ng YS, et al. : mtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol Med. 2018;10(6):e8262. 10.15252/emmm.201708262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LC, Eaton SL, Brunton PJ, et al. : Proteomic profiling of neuronal mitochondria reveals modulators of synaptic architecture. Mol Neurodegener. 2017;12(1):77. 10.1186/s13024-017-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Nooteboom M, Elson JL, et al. : Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing. PLOS Genetics. 2014;10(9):e1004620. 10.1371/journal.pgen.1004620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume-gentil O, Grindberg RV, Kooger R, et al. : Tunable Single-Cell Extraction for Molecular Analyses. Cell. 2016;166(2):506–516. 10.1016/j.cell.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Hartwig S, Feckler C, Lehr S, et al. : A critical comparison between two classical and a kit-based method for mitochondria isolation. Proteomics. 2009;9(11):3209–3214. 10.1002/pmic.200800344 [DOI] [PubMed] [Google Scholar]
- Hornig-do HT, Günther G, Bust M, et al. : Isolation of functional pure mitochondria by superparamagnetic microbeads. Anal Biochem. 2009;389(1):1–5. 10.1016/j.ab.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Hubbard WB, Harwood CL, Prajapati P, et al. : Fractionated mitochondrial magnetic separation for isolation of synaptic mitochondria from brain tissue. Sci Rep. 2019;9(1):9656. 10.1038/s41598-019-45568-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Takeda Y, Herbert M: Reversion after replacement of mitochondrial DNA. Nature. 2019;574(7778):E8–E11. 10.1038/s41586-019-1623-3 [DOI] [PubMed] [Google Scholar]
- Hyslop LA, Blakeley P, Craven L, et al. : Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature. 2016;534(7607):383–6. 10.1038/nature18303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura F, Nakada A, Egashira Y, et al. : Development of nano-surgery system for cell organelles. Proceedings of the 41st SICE Annual Conference. SICE. 2002;5:3236–3241. 10.1109/SICE.2002.1195633 [DOI] [Google Scholar]
- Ino K, Nashimoto Y, Taira N, et al. : Intracellular Electrochemical Sensing. Electroanalysis. 2018;30(10):2195–2209. 10.1002/elan.201800410 [DOI] [Google Scholar]
- Islinger M, Wildgruber R, Völkl A: Preparative free-flow electrophoresis, a versatile technology complementing gradient centrifugation in the isolation of highly purified cell organelles. Electrophoresis. 2018;39(18):2288–2299. 10.1002/elps.201800187 [DOI] [PubMed] [Google Scholar]
- Ito H, Tanaka M, Zhou Y, et al. : Continuous collection and simultaneous detection of picoliter volume of nucleic acid samples using a mille-feuille probe. Anal Bioanal Chem. 2017;409(4):961–969. 10.1007/s00216-016-0006-y [DOI] [PubMed] [Google Scholar]
- Jeffries GDM, Edgar JS, Zhao Y, et al. : Using polarization-shaped optical vortex traps for single-cell nanosurgery. Nano Lett. 2007;7(2):415–420. 10.1021/nl0626784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Oh S, Reschiglian P, et al. : Separation of mitochondria by flow field-flow fractionation for proteomic analysis. Analyst. 2008;133(4):505–15. 10.1039/b716851a [DOI] [PubMed] [Google Scholar]
- Kang W, Mcnaughton RL, Espinosa HD: Micro- and Nanoscale Technologies for Delivery into Adherent Cells. Trends Biotechnol. 2016;34(8):665–678. 10.1016/j.tibtech.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayo S, Bahnemann J, Klauser M, et al. : A microfluidic device for immuno-affinity-based separation of mitochondria from cell culture. Lab Chip. 2013;13(22):4467–4475. 10.1039/c3lc50739d [DOI] [PubMed] [Google Scholar]
- Keloth A, Anderson O, Risbridger D, et al. : Single Cell Isolation Using Optical Tweezers. Micromachines (Basel). 2018;9(9):434. 10.3390/mi9090434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König K, Riemann I, Stracke F, et al. : Nanoprocessing with nanojoule near-infrared femtosecond laser pulses. Medical Laser Application. 2005;20(3):169–184. 10.1016/j.mla.2005.07.009 [DOI] [Google Scholar]
- Kostal V, Fonslow BR, Arriaga EA, et al. : Fast determination of mitochondria electrophoretic mobility using micro free-flow electrophoresis. Anal Chem. 2009;81(22):9267–9273. 10.1021/ac901508x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowald A, Kirkwood TBL: Transcription could be the key to the selection advantage of mitochondrial deletion mutants in aging. Proc Natl Acad Sci U S A. 2014;111(8):2972–2977. 10.1073/pnas.1314970111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Nicholas A, Caro P, et al. : Single molecule PCR in mtDNA mutational analysis: Genuine mutations vs. damage bypass-derived artifacts. Methods. 2008;46(4):269–273. 10.1016/j.ymeth.2008.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Wolken GG, Wittenberg NJ, et al. : Nanohole Array-Directed Trapping of Mammalian Mitochondria Enabling Single Organelle Analysis. Anal Chem. 2015;87(24):11973–11977. 10.1021/acs.analchem.5b03604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov A, Margreiter R: Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci. 2009;10(4):1911–1929. 10.3390/ijms10041911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforge FO, Carpino J, Rotenberg SA, et al. : Electrochemical attosyringe. Proc Natl Acad Sci U S A. 2007;104(29):11895–11900. 10.1073/pnas.0705102104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai N, Kummitha CM, Rosca MG, et al. : Isolation of mitochondrial subpopulations from skeletal muscle: Optimizing recovery and preserving integrity. Acta Physiol (Oxf). 2019;225(2):e13182. 10.1111/apha.13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza IR, Nair KS: Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–372. 10.1016/S0076-6879(09)05020-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Greaves L, Reeve AK, et al. : The rise and rise of mitochondrial DNA mutations. Open Biol. 2020;10(5):200061. 10.1098/rsob.200061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann D, Tuppen HAL, Campbell GE, et al. : Understanding mitochondrial DNA maintenance disorders at the single muscle fibre level. Nucleic Acids Res. 2019;47(14):7430–7443. 10.1093/nar/gkz472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Bergamini C, Fato R, et al. : Isolation of mitochondria from cells and tissues. Methods Cell Biol. 2020;155:3–31. 10.1016/bs.mcb.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gaudet S, Schmidt MA, et al. : A Microfabricated Device for Subcellular Organelle Sorting. Anal Chem. 2004;76(19):5705–5712. 10.1021/ac049794g [DOI] [PubMed] [Google Scholar]
- MacDonald JA, Bothun AM, Annis SN, et al. : A nanoscale, multi-parametric flow cytometry-based platform to study mitochondrial heterogeneity and mitochondrial DNA dynamics. Commun Biol. 2019;2:258. 10.1038/s42003-019-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Gabi M, Behr P, et al. : FluidFM: Combining Atomic Force Microscopy and Nanofluidics in a Universal Liquid Delivery System for Single Cell Applications and Beyond. Nano Lett. 2009;9(6):2501–2507. 10.1021/nl901384x [DOI] [PubMed] [Google Scholar]
- Miller KE, Sheetz MP: Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117(Pt 13):2791–2804. 10.1242/jcs.01130 [DOI] [PubMed] [Google Scholar]
- Moutaux E, Christaller W, Scaramuzzino C, et al. : Neuronal network maturation differently affects secretory vesicles and mitochondria transport in axons. Sci Rep. 2018;8(1):13429. 10.1038/s41598-018-31759-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J, Na YJ, Zhu H, et al. : Pervasive within-Mitochondrion Single-Nucleotide Variant Heteroplasmy as Revealed by Single-Mitochondrion Sequencing. Cell Rep. 2017;21(10):2706–2713. 10.1016/j.celrep.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadappuram BP, Cadinu P, Barik A, et al. : Nanoscale tweezers for single-cell biopsies. Nat Nanotechnol. 2019;14(1):80–88. 10.1038/s41565-018-0315-8 [DOI] [PubMed] [Google Scholar]
- Nawarathna D, Turan T, Wickramasinghe HK: Selective probing of mRNA expression levels within a living cell. Appl Phys Lett. 2009;95(8):83117–83117. 10.1063/1.3213343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Li C, Shevchuk AI, et al. : Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat Methods. 2009;6(4):279–281. 10.1038/nmeth.1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkiewicz G, Gualberto JM, Benamar A, et al. : Arabidopsis Seed Mitochondria Are Bioenergetically Active Immediately upon Imbibition and Specialize via Biogenesis in Preparation for Autotrophic Growth. Plant Cell. 2017;29(1):109–128. 10.1105/tpc.16.00700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne BA, Wilson IJ, Yu-Wai-Man P, et al. : Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22(2):384–90. 10.1093/hmg/dds435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugradt R, Schmidt U, Landenberger B, et al. : A novel and effective separation method for single mitochondria analysis. Mitochondrion. 2011;11(2):308–314. 10.1016/j.mito.2010.12.009 [DOI] [PubMed] [Google Scholar]
- Picard M, Taivassalo T, Gouspillou G, et al. : Mitochondria: isolation, structure and function. J Physiol. 2011;589(Pt 18): 4413–21. 10.1113/jphysiol.2011.212712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe BG, 3rd, DUFFY CF, Greminger MA, et al. : Detection of heteroplasmy in individual mitochondrial particles. Anal Bioanal Chem. 2010;397(8):3397–3407. 10.1007/s00216-010-3751-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe BG, Navratil M, Arriaga EA: Analysis of subcellular sized particles: Capillary electrophoresis with post-column laser-induced fluorescence detection versus flow cytometry. J Chromatogr A. 2006;1137(2):249–255. 10.1016/j.chroma.2006.10.011 [DOI] [PubMed] [Google Scholar]
- Ramos A, Morgan H, Green NG, et al. : The role of electrohydrodynamic forces in the dielectrophoretic manipulation and separation of particles. J Electrostat. 1999;47:71–81. 10.1016/S0304-3886(99)00031-5 [DOI] [Google Scholar]
- Reiner JE, Kishore RB, Levin BC, et al. : Detection of Heteroplasmic Mitochondrial DNA in Single Mitochondria. PLoS One. 2010;5(12):e14359. 10.1371/journal.pone.0014359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Grady JP, Cosgrave EM, et al. : Mitochondrial dysfunction within the synapses of substantia nigraneurons in Parkinson’s disease. NPJ Parkinsons Dis. 2018;4:9. 10.1038/s41531-018-0044-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschiglian P, Zattoni A, Roda B, et al. : Field-flow fractionation and biotechnology. Trends Biotechnol. 2005;23(9):475–483. 10.1016/j.tibtech.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, et al. : Mitochondrial threshold effects. Biochem J. 2003;370(Pt 3):751–762. 10.1042/BJ20021594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru Y, Yin L, Sun H, et al. : A micropreparation of mitochondria from cells using magnetic beads with immunoaffinity. Anal Biochem. 2012;421(1):219–226. 10.1016/j.ab.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Schiro PG, Gadd JC, Yen GS, et al. : High-Throughput Fluorescence-Activated Nanoscale Subcellular Sorter with Single-Molecule Sensitivity. J Phys Chem B. 2012;116(35):10490–10495. 10.1021/jp3019233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger RA, Actis P, Penfold C, et al. : Voltage controlled nano-injection system for single-cell surgery. Nanoscale. 2012;4(19):5843–5846. 10.1039/c2nr31700a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonker M, Sahore V, Woolley AT: Recent advances in microfluidic sample preparation and separation techniques for molecular biomarker analysis: A critical review. Anal Chim Acta. 2017;986:1–11. 10.1016/j.aca.2017.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzi M, Casarin A, Pertegato V, et al. : Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc. 2012;7(6):1235–46. 10.1038/nprot.2012.058 [DOI] [PubMed] [Google Scholar]
- Tesauro C, Ferrando B, Ma X, et al. : Isolation of functional mitochondria by inertial microfluidics – a new method to sort intracellular organelles from a small scale biological sample. RSC Adv. 2017;7:23735–23741. 10.1039/C7RA03384B [DOI] [Google Scholar]
- Trifunov S, Pyle A, Valentino ML, et al. : Clonal expansion of mtDNA deletions: different disease models assessed by digital droplet PCR in single muscle cells. Sci Rep. 2018;8(1):11682. 10.1038/s41598-018-30143-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, et al. : Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Uehara H, Osada T, Ikai A: Quantitative measurement of mRNA at different loci within an individual living cell. Ultramicroscopy. 2004;100(3–4):197–201. 10.1016/j.ultramic.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Verbiest GJ, Rost MJ: Resonance frequencies of AFM cantilevers in contact with a surface. Ultramicroscopy. 2016;171:70–76. 10.1016/j.ultramic.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Vincent AE, Picard M: Multilevel heterogeneity of mitochondrial respiratory chain deficiency. J Pathol. 2018;246(3):261–265. 10.1002/path.5146 [DOI] [PubMed] [Google Scholar]
- Vincent AE, Rosa HS, Pabis K, et al. : Subcellular origin of mitochondrial DNA deletions in human skeletal muscle. Ann Neurol. 2018;84(2):289–301. 10.1002/ana.25288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, White K, Davey T, et al. : Quantitative 3D Mapping of the Human Skeletal Muscle Mitochondrial Network. Cell Rep. 2019;27(1):321. 10.1016/j.celrep.2019.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Lorenz K, Horneffer V, et al. : Mechanisms of Laser-Induced Dissection and Transport of Histologic Specimens. Biophys J. 2007;93(12):4481–4500. 10.1529/biophysj.106.102277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom JD, Twig G, Shirihai OS: What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int J Biochem Cell Biol. 2009;41(10):1914–1927. 10.1016/j.biocel.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Yang JS, Lee JY, Moon MH: High Speed Size Sorting of Subcellular Organelles by Flow Field-Flow Fractionation. Anal Chem. 2015;87(12):6342–6348. 10.1021/acs.analchem.5b01207 [DOI] [PubMed] [Google Scholar]
- Yarjanli Z, Ghaedi K, Esmaeili A, et al. : Iron oxide nanoparticles may damage to the neural tissue through iron accumulation, oxidative stress, and protein aggregation. BMC Neurosci. 2017;18(1):51. 10.1186/s12868-017-0369-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zischka H, Braun RJ, Marantidis EP, et al. : Differential Analysis of Saccharomyces cerevisiae Mitochondria by Free Flow Electrophoresis. Mol Cell Proteomics. 2006;5(11):2185–2200. 10.1074/mcp.T600018-MCP200 [DOI] [PubMed] [Google Scholar]


