Abstract
Epigenetic regulation has profound influence on stem cell fate during normal development in maintenance of physiologic tissue homeostasis. Here we report diminished ten-eleven translocation (TET) methylcytosine dioxygenase expression and loss of the DNA hydroxymethylation mark, 5-hydroxymethylcytosine (5-hmC), in keratinocyte stem cells (KSCs) and transit-amplifying cells (TACs) in human psoriasis and in imiquimod-induced murine psoriasis. Loss of 5-hmC was associated with dysregulated KSC kinetics, resulting in accumulation of nestin and FABP5-expressing TACs to produce classic psoriatic epidermal architecture. Moreover, 5-hmC loss was accompanied by diminished TET1 and TET2 mRNA expression. Genome-wide mapping of epidermal 5-hmC in murine psoriasis revealed loci-specific loss of 5-hmC in genes regulating stem cell homeostasis, including MBD1, RTN1, STRN4, PRKD2, AKT1, MAPKAP2, as well as those associated with RAR and Wnt/β-catenin signaling pathways. In vitro restoration of TET expression by ascorbic acid was accomplished in cultured human KSCs to show similar [Ca++]-induced differentiation, resulting in increased 5-hmC levels and reduced nestin expression. To our knowledge, an epigenetic deficiency in psoriasis with relevance to stem cell dysregulation has not been previously reported. This observation raises the possibility that epigenetic modifiers that impact on the TET-5-hmC pathway may be a relevant approach of heretofore unappreciated therapeutic utility.
INTRODUCTION
Psoriasis is a chronic autoimmune skin disease that affects more than 125 million people worldwide (Griffiths et al., 2017). Patients with psoriasis suffer from physical symptoms, psychological effects, and social consequences that result in costs of around $135 billion annually in the United States alone (Brezinski, Dhillon, & Armstrong, 2015). Despite decades of research, the etiology and pathogenesis of psoriasis remains unclear. One histopathological hallmark of the psoriatic epidermis is the downward and centrifugal expansion of epidermal rete ridges, the tips of which contain a subset of basal cells with stem-like characteristics (Lavker & Sun, 1982; Whitaker-Menezes et al., 2003; Naik et al., 2017; Watt, 2001). Upon activation, keratinocyte stem cells (KSCs) divide asymmetrically to produce suprabasal transit amplifying cells (TACs) that then normally mature with ascent toward the epidermal surface, and such asymmetrical divisions have been documented to be increased in psoriasis (Charruyer et al., 2017). We therefore reasoned that aberrant stem cell activation and anomalous TAC accumulation within pathologically expanding epidermal rete might account for the psoriatic cytoarchitecture. This hypothesis was fueled by recognition that certain mediators of epidermal dysfunction, such as RAC1 (Ras-related C3 botulinum toxin substrate 1) that drive pathological interactions between keratinocytes and immune cells in psoriasis are also essential for KSC maintenance and homeostasis (Winge et al., 2016; Benitah, 2005). Moreover, IL-17, a critical cytokine effector in psoriasis, was recently implicated in KSC activation and proliferation via Steap4-p63 inductive pathway (Wu et al., 2015). Finally, previous work by us and others has implicated loss of the hydroxymethylation mark, 5-hmC (5-hydroxymethylcytosine) in regulation of normal and neoplastic skin stem cell function, activation and proliferation (Lian et al., 2012; Saldanha et al., 2015), and accordingly we also sought to determine whether this epigenetic pathway was relevant to psoriasis.
DNA methyltransferases (DNMTs) are known to be essential in preserving the self-renewing capacity of stem cells by maintaining DNA methylation patterns (Sen, Reuter, Webster, Zhu, & Khavari, 2010). Genome-wide studies have established aberrant DNA methylation sites that correlate altered gene expression with major histopathological features of psoriasis (Chandra, Senapati, Roy, Chatterjee, & Chatterjee, 2018; Zhou et al., 2016). Furthermore, restoration to a more normal methylation status at specific loci was seen in patients who experienced improvement of their psoriasis from anti-TNF-α therapy or UVB phototherapy (Gu, Nylander, Coates, Fahraeus, & Nylander, 2015; Roberson et al., 2012). Studies have shown that the DNA methylation profile can be altered by the Ten-eleven translocation (TET) dioxygenase family that catalyzes the conversion of genomic 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) (Ito et al., 2011). 5-hmC is a stable epigenetic modification that acts as an intermediate in the DNA demethylation pathway and can regulate gene expression (An, Rao, & Ko, 2017). The three known mammalian TET isozymes (TET1, TET2, and TET3) require Fe2+ and 2-oxoglutarate for their activity. Ascorbic acid is required to reduce oxidized iron species back to catalytically active Fe2+ in order to maintain the full enzymatic capacities of TET (Cimmino et al., 2017; Gustafson et al., 2015; Sant et al., 2018). 5-hmC levels can modulate genome-wide transcription patterns by recruiting different sets of binding proteins in comparison to 5-mC (Gustafson et al., 2015). There is a dynamic balance between 5-mC and 5-hmC during stem cell differentiation where knockdown of TET enzymes and loss of 5-hmC in embryonic stem cells has been shown to downregulate pluripotency-related genes (Ficz et al., 2011). In summary, DNA hydroxymethylation is a key player in maintaining stem cell homoeostasis that has been relatively unstudied in psoriasis.
Here, we investigated DNA hydroxymethylation in KSCs and TACs during psoriasis pathogenesis. Our findings indicate a relationship between TET-mediated DNA hydroxymethylation and keratinocyte stem cell participation in psoriasis pathogenesis, and suggest an innovative approach to psoriasis therapy involving targeting of epigenetic pathways that may restore more physiologic stem cell function.
RESULTS
KSC activation and TAC accumulation are associated with loss of 5-hmC in human psoriasis
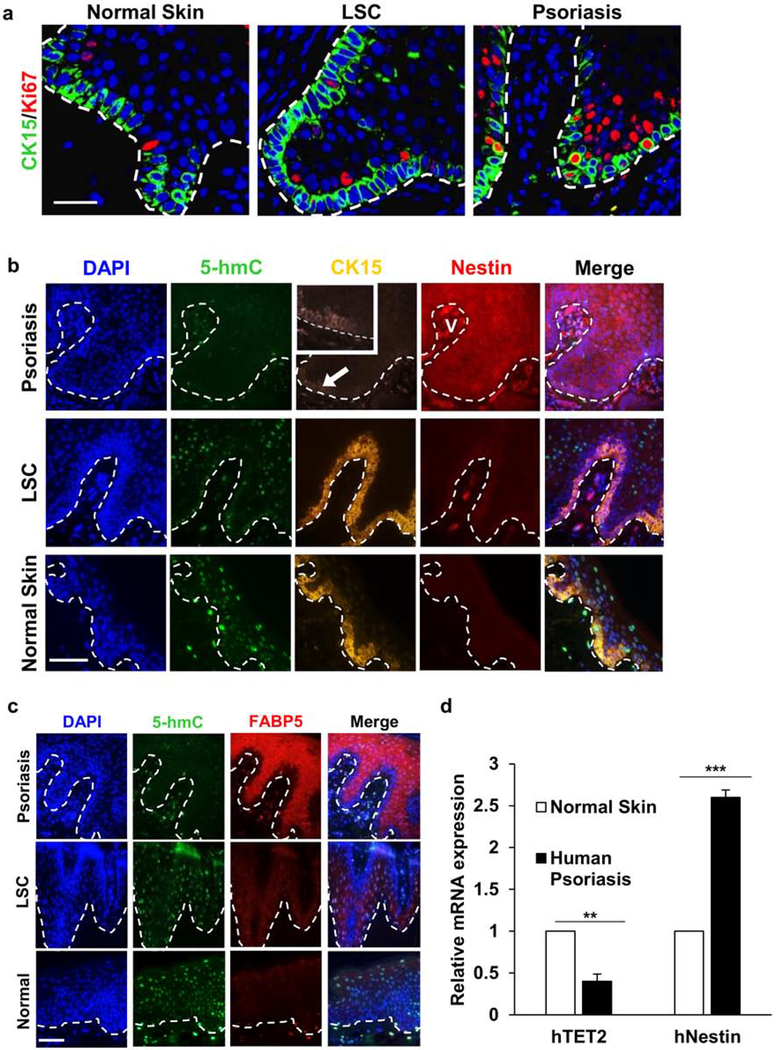
Multiplex immunofluorescence (IF) was performed for KSC and TAC biomarkers on 13 samples of clinical psoriasis, 10 samples of a dermatitis that also exhibits epidermal thickening (lichen simplex chronicus, LSC), and 10 samples of normal skin. Early human psoriasis that lacked well-developed and uniform downward extension and expansion of rete ridges, but no cases of LSC or normal skin, showed cells at the tips of rete ridges that were double-labeled for the stem cell marker, CK15, and activation marker, Ki67, that indicates entry into the cell cycle (Fig. 1a). With lesion evolution and consistent with previous studies, we observed a decrease in CK15-labeled KSCs in the basal layer of the progressively acanthotic psoriatic epidermis (Fig. 1b) (Jia et al., 2016) in contrast to LSC, where CK-15-positive KSCs appeared to be increased in number. The pathological hyperproliferation of keratinocytes in psoriasis has been shown to result from increased asymmetrical divisions of KSCs (Jia et al., 2016), as opposed to symmetrical stem cell divisions that produce identical progeny. Because asymmetrical divisions result in KSC self-renewal as well as the genesis of TACs, we evaluated the latter using antibodies to FABP5 and nestin (Watarai et al., 2013). Dramatic expansion of the TAC compartment was seen only in psoriasis, while LSC and normal skin failed to show similar findings (Fig. 1b, c). Moreover, the suprabasal layer of psoriatic epidermis, but not of LSC and normal skin, consistently exhibited increased expression of Ki67 in keeping with sustained proliferation within the TAC niche (Fig. 1a). Importantly, the psoriatic TAC region defined by FABP5 and nestin immunoreactivity exhibited a differential loss of 5-hmC when compared to disease and normal skin controls, suggesting the possibility of altered epigenetic regulation. The overall 5-hmC levels of KSCs and TACs were significantly decreased in psoriasis samples (0.43 ± 0.23, n = 10) versus LSC (2.73 ± 0.14, n = 10) and normal skin specimens (3.17 ± 0.11, n = 10) by semi-quantitative grading (p < 0.0001). In addition, quantitative RT-PCR of human psoriatic epidermis confirmed upregulation of nestin gene expression and downregulation of TET2, which mediates 5-hmC levels (Fig. 1d and Table S1).
Figure 1. KSCs in early psoriasis, LSC, and normal skin.
(a) Normally quiescent basal CK15+ KSCs become Ki67+ as CK-/Ki67+ TAC niche expands in psoriasis, but not in LSC (bar = 50 μm for all panels; broken line = dermal-epidermal junction). (b) Unlike LSC and normal skin, psoriatic lesions show 5-hmC loss (note dermal controls), decreased CK15 (arrow), and nestin upregulation (nestin+ vessel, V; bar = 100 μm for all b&c panels). (c) FABP5 is increased and 5-hmC is decreased in suprabasal TACs in psoriasis (opposed to LSC and normal skin). (d) Quantitative real-time PCR of human psoriatic epidermis showing decreased TET2 expression and increased nestin expression. ** P<0.01, ***P<0.001. Data = mean ± SEM of biological replicates (n=3).
The imiquimod-induced mouse psoriasis model confirms KSC/TAC dysregulation and links decreased TET-mediated DNA hydroxymethylation in KSC pathways
The imiquimod (IMQ)-induced psoriasis mouse model is widely used for correlative studies with human psoriasis (van der Fits et al., 2009). Psoriatic mouse back and ear skin after six consecutive days of topical imiquimod application revealed erythematous, scaling lesions that histologically showed epidermal acanthosis, parakeratosis, hypogranulosis, and prominence of superficial dermal microvessels (Fig. 2a), as demonstrated previously and in keeping with changes seen in human lesions (Boehncke & Schön, 2015; Jia et al., 2016). 5-hmC levels in IMQ-treated specimens were significantly lower (1.67 ± 0.12, n = 6) versus untreated control samples (3.78 ± 0.11, n = 6) by semi-quantitative grading (p < 0.0001). Immunohistochemical (IHC) multiplex labeling approaches performed instead of IF to enable higher magnification and resolution imaging of the normally thin murine epidermis confirmed CK15 depletion and expansion of the suprabasal FABP5/nestin-positive, 5-hmC-negative TAC niche (Fig. 2a).
Figure 2. Significant decrease of TET-mediated 5-hmC in IMQ-induced mouse psoriasis.
(a) Compared to untreated epidermis, psoriatic epidermis exhibits decreased 5-hmC. Nestin is upregulated in psoriatic lesions (S = sebaceous lobule). CK15-positive cells (encircled) express low 5-hmC but are undetectable in psoriatic epidermis (insets show CK15 and 5-hmC in hair follicle bulge region). FABP5 detected in sebaceous lobules (arrows) but not in control epidermis, whereas expression is present in psoriatic suprabasal keratinocyte cytoplasm and nuclei. Scale bar = 500 μm for HE. Scale bar = 100 μm for IHC. Downregulation of TET1 and TET2 (b) as well as upregulation of nestin (b) and IL-17A (c) gene expression is detected by RT-PCR in the IMQ-treated epidermis. ** P<0.01, ***P<0.001. Data shows mean ± SEM of n=3.
Quantitative RT-PCR was next performed on mouse epithelium removed from the dermal layer to prevent skewing of results due to cells extraneous to the epidermal layer. A significant decrease in predominantly TET1 over TET2 expression and increased nestin expression in IMQ-treated lesional epidermis compared to untreated epidermis was observed (Fig. 2b). IL-17RA gene expression was significantly upregulated with concomitant significant decrease in TET2 gene expression (Fig. 2c). Overall, these findings suggested that loss of TET expression and reduced 5-hmC levels in human and experimentally-induced psoriatic lesions could be related to stem cell dysregulation that correlates with acquisition of the psoriatic epidermal phenotype.
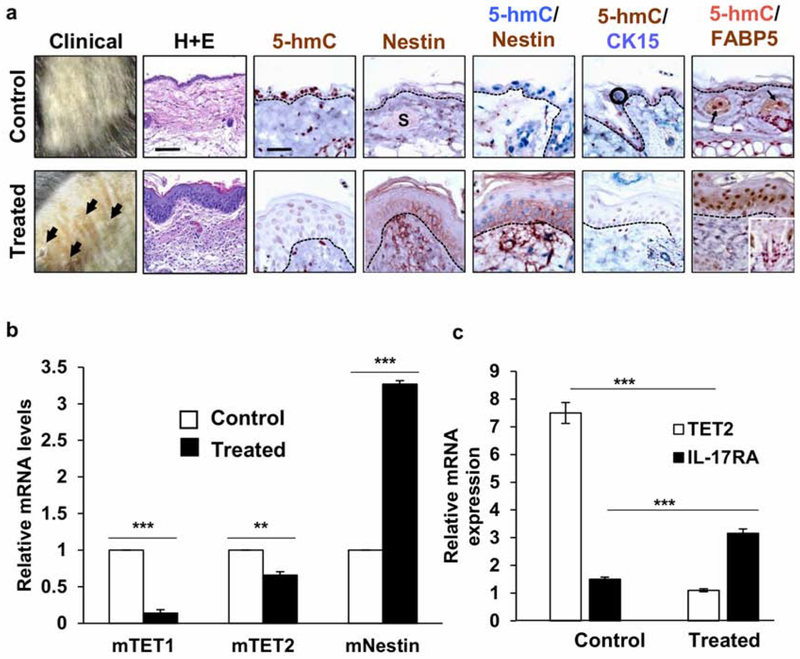
To further characterize the potential consequences of DNA hydroxymethylation in psoriasis, we conducted genomic-wide mapping studies of 5-hmC levels comparing IMQ-induced epidermal lesions with untreated non-lesional epidermis. We used a barcoded hydroxymethylated DNA immunoprecipitation (hMeDIP) method coupled with deep sequencing (hMeDIP-seq) for quantitative comparison of 5-hmC tag density at specific genome loci. Analysis of the tag densities showed lower 5-hmC levels in both CpG islands and genebodies in IMQ-treated epidermal samples (Fig. 3a; data pooled from 2 biological replicates). We looked further into loci-specific changes of 5-hmC and found a total of 364 differentially hydroxymethylated regions (2 fold change, MaxTags ≥ 50). Within these regions, 321 genes showed significantly lower 5-hmC levels and 67 genes had significantly higher 5-hmC levels in the IMQ-treated epidermis compared to control (Table S2). Gene ontology (GO) analysis was performed on genes differentially 5-hydroxymethylated in untreated versus IMQ-treated skin (Fig. 3b). This analysis calculates a p-value based on enrichment of hydroxymethylated genes in a particular GO annotation (log10 (p-value)) reported as an enrichment score for downregulated genes. GO analysis of genes downregulated (decreased activity) in IMQ-treated skin indicated a decrease in chromosome condensation, regulation of canonical Wnt signaling pathway, and regulation of cell differentiation (enrichment scores −11.19, −4.04, and −1.86 respectively) (Fig. 3b). Ingenuity Pathway Analysis (IPA) revealed that the differentially hydroxymethylated genes were closely associated with pathways involved in stem cell homeostasis, such as RAR activation (P = 2.80x10−3) and Wnt/β-catenin signaling (P = 5.28x10−3) (Fig. 3c, Fig. S1) (Lee et al., 2009; Veltri, Lang, & Lien, 2018). Akt1, a key player in both the RAR and Wnt/β-catenin pathways, showed loss of 5-hmC at the promoter region of the gene (Fig. 3d, Fig. S1a, b). Furthermore, MAPKAPK2, which is downstream of Akt1 in the RAR pathway, also showed loss of 5-hmC in the promoter region (Fig. 3d, Fig. S1a). Top 10 most and least 5-hydroxymethylated genes in IMQ-treated lesional epidermis are summarized in Figure 3e. IPA analysis also generated network diagrams showing that some of these differentially hydroxymethylated genes are involved in the NFκB, ERK1/2, and PI3K pathways (Fig. S2). Additional Integrative Genomic Viewer (IGV) diagrams of 5-hmC levels in other genes that have been shown to be important in stem cell regulation are included in the supplemental figures (Fig. S3). Thus, genome-wide mapping of 5-hmC revealed that loss of hydroxymethylation at certain gene loci had potential to influence pathways that govern stem cell homeostasis resulting in epidermal dysmaturation upon exposure to proliferative stimuli (summarized in schematic, Fig. 5).
Figure 3. Genome-wide mapping of 5-hmC in epithelial samples from IMQ-treated skin VS untreated skin in mice.
(a) Data obtained from 2 biological replicates (n=2). Loss of 5-hmC is seen in both CpG islands and genebodies in IMQ-treated epidermis compared to untreated epidermis. (b) GO terms of downregulated pathways found in IMQ-treated skin. (c) Canonical pathway analysis of 377 differentially hydroxymethylated genes. The top 10 statistically significant canonical pathways identified by IPA shown here with genes identified by our study listed to the right. (d) hMeDIP results showing peaks correlating with 5-hmC levels at the promoter regions of Akt1 and MAPKAPK2 genes. (e) Top upregulated and downregulated genes identified in IMQ-treated lesional epidermis compared to non-lesional epidermis.
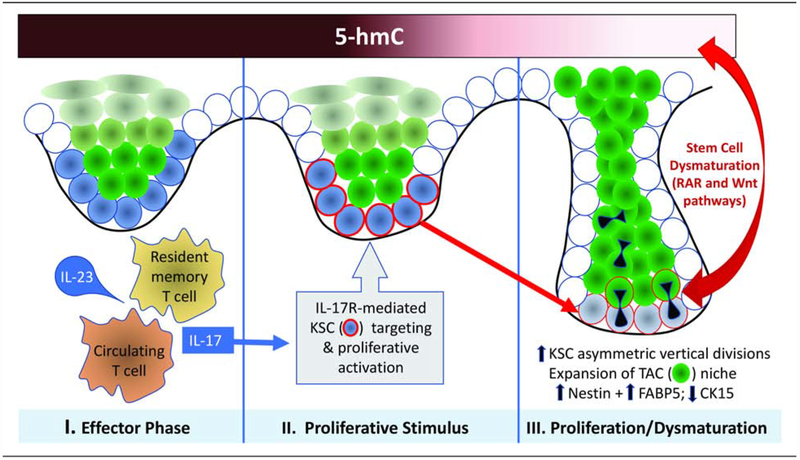
Figure 5. Schematic diagram of posited evolutionary phases of psoriasis initiation and progression.
The Effector Phase (I) involves generation of IL-23-responsive circulating and/or resident memory Th17 cells. Ligation of IL-17 with its receptor (II) provides a Proliferative Stimulus that targets KSC niche (blue cells), resulting in KSC activation and proliferation (acquisition of red rim; Wu et al., 2015). Proliferation is accompanied in psoriasis by dysmaturation signals that affect KSC kinetics and differentiation via loss of DNA hydroxymethylation mark, 5-hmC, that affects the stem cell-associated RAR and Wnt pathways. This Proliferation/Dysmaturation phase (III) involves increased vertical, asymmetric KSC divisions and accumulation of nestin/FABP5-positive TACs (green cells), resulting in downward and lateral expansion of the rete ridge/KSC/TAC niche to produce characteristic psoriatic epidermal architecture.
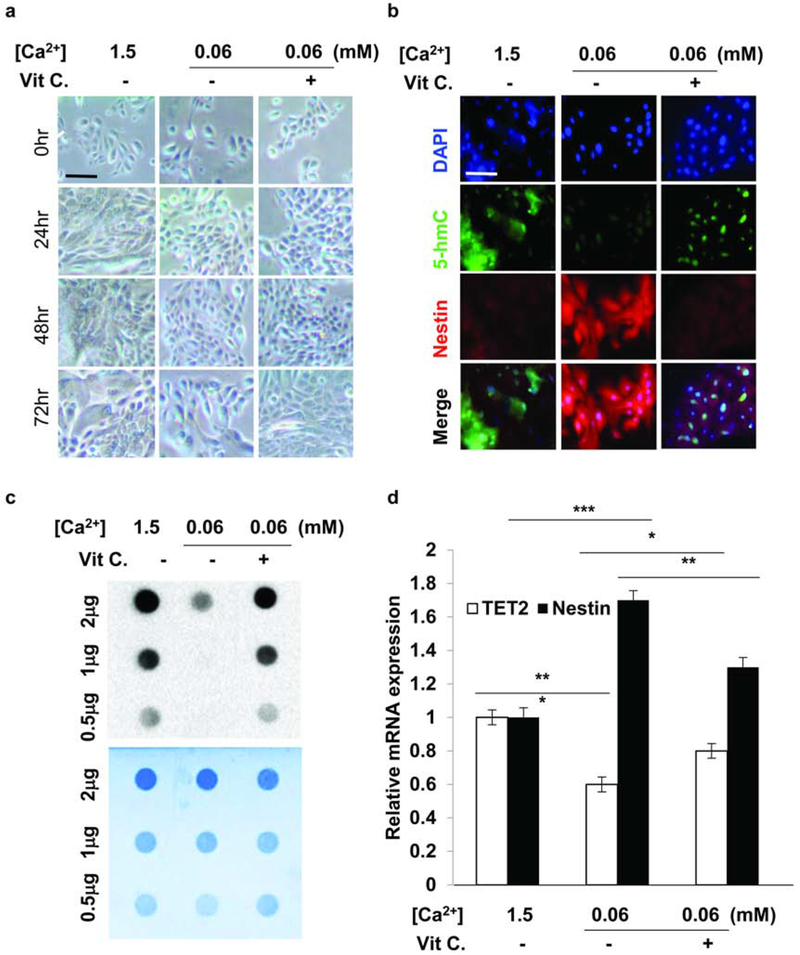
Modulation of TET-mediated 5-hmC levels via ascorbic acid alters KSC/TAC equilibrium in vitro
Our data suggested that loss of 5-hmC in psoriasis may result in KSC proliferative disequilibrium, thus contributing to the classical psoriatic epidermal phenotype. Therefore, we next sought to determine whether modulation of TET-mediated 5-hmC levels is capable of influencing KSC/TAC dynamics in an in vitro human keratinocyte stem cell culture system where differentiation may be modulated by calcium concentration (Hennings et al., 1980). At normal calcium concentrations (1.5mM), normal KSC cultured over 72 hours exhibited squamoid features consistent with differentiation with associated 5-hmC expression and little to no nestin expression (Fig. 4a, b), whereas normal KSCs cultured in low calcium medium (0.06mM) showed a more basaloid phenotype with low to no 5-hmC levels and increased numbers of nestin-positive cells (Fig. 4a, b). The low calcium conditions therefore replicated an apparent delay in differentiation associated with increased KSC-derived, nestin-expressing TACs similar to that seen in human and murine psoriatic epidermis. The loss of 5-hmC in the low calcium KSC group was confirmed by immune dot-blot (Fig. 4c). To modulate 5-hmC levels, we used ascorbic acid treatment previously shown to increase TET2-dependent DNA 5-hydroxymethylation in retinal pigment epithelial cells and melanoma cells (Gustafson et al., 2015; Sant et al., 2018). Treatment of KSCs in low calcium medium with ascorbic acid (200 μM) for 48 hours resulted in an increase in squamoid differentiation by 72 hours (Fig. 4a) accompanied by enhanced 5-hmC and diminished to no nestin staining (Fig. 4b). These findings were confirmed with quantitative RT-PCR showing decreased nestin expression with restored TET2 expression upon ascorbic acid treatment of low calcium KSC cultures (Fig. 4d). These initial in vitro experiments support our findings that 5-hmC influences downstream genes that play a role in maintaining KSC homeostasis.
Figure 4. Human KSC proliferation assay in vitro suggests 5-hmC regulation.
(a) Human KSCs cultured in normal calcium (1.5mmol/L) exhibited differentiation, while lack of calcium-induced differentiation of KSCs cultured in low calcium (0.06mmol/L) even after 72 hours incubation was observed. Scale bar = 100 μm. (b) IF studies showed that the low 5-hmC levels and high nestin expression in low calcium KSC cultures could be rescued with 200μmol/L ascorbic acid treatment. Scale bar = 100 μm. (c) Dot blot showing restoration of 5-hmC levels in low calcium KSC cultures after vitamin C treatment. (d) RT-PCR showed elevated TET2 expression and reduced nestin expression after ascorbic acid treatment. * P<0.05, ** P<0.01, ***P<0.001. Data are shown as mean ± SEM of three biological replicates (n=3).
DISCUSSION
In this study, we demonstrated significant loss of 5-hmC in KSCs and TACs in psoriasis. Depletion of CK15-expressing KSCs and associated expansion of the nestin and FABP5-labeled TAC compartment in both human and murine psoriasis suggested that decreased 5-hmC levels may alter gene expression important in regulating stem cell homeostasis and responses to proliferative stimuli. Quantitative RT-PCR of isolated IMQ-treated mouse epidermis showed diminished TET1 and TET2 gene expression that correlated with loss of 5-hmC detected by IHC and IF, and hMeDIP-seq indicated that gene pathways involved in stem cell maintenance are preferentially affected by 5-hmC perturbations. Finally, the ability to rescue TET2/5-hmC and aberrant nestin expression in vitro with an epigenetic regulator suggests that such approaches may hold promise for therapeutic restoration of stem cell dynamics in psoriasis in vivo.
Loss of 5-hmC in psoriatic lesions correlated with an increase in nestin expression in both basal and suprabasal epidermis. This is consistent with our previous findings of negative regulation of nestin expression via TET2 in human melanoma (Gomes et al., 2016). Furthermore, nestin expression has been shown to correlate directly with neural stem cell proliferation and inversely with cellular differentiation (Cui et al., 2012). Until now, the downstream pathways affected by 5-hmC/nestin switching have remained unexplored. Recent developments of hMeDIP-seq technology now permit study of the genome-wide 5-hmC landscape in the isolated epidermis of IMQ-induced murine psoriasis. Interestingly, analysis of the genomic data revealed loci-specific modulation of DNA hydroxymethylation of genes involved in pathways related to stem cell homeostasis and related keratinocyte differentiation and proliferation, including the RAR and Wnt/β-catenin pathways (Lee et al., 2009; Veltri et al., 2018).
In vitro human KSC calcium-induced differentiation assays now provide further support for the potential importance of 5-hmC in regulating stem cell dysequilibrium in psoriasis. Upon restoration of TET expression in low calcium cultures via ascorbic acid treatment, human KSCs exhibited higher levels of 5-hmC and switched to a more normal differentiation status, as assessed cytologically and via nestin expression. Ascorbic acid derivatives have been shown to ameliorate psoriasis (Kitahata K, et al. 2018), and our results raise the possibility of that increase of TET-mediated DNA hydroxymethylation by ascorbic acid is an underlying mechanism in psoriasis treatment by ascorbic acid derivatives. Thus, loss of 5-hmC in psoriasis appears to at least in part function in modulating gene expression important for maintenance of stem cell behavior during psoriasis pathogenesis. DNA methylation profiling in psoriasis has previously indicated that hypermethylation in lesional skin with altered expression of differentially methylated genes contributes to the psoriatic phenotype (P. Zhang, Su, Chen, Zhao, & Lu, 2010; Zhou et al., 2016; Chandra et al., 2018). Successful treatment of psoriasis with phototherapy not only significantly reduced the histopathological features in psoriasis, but also reverted DNA methylation to a more normal pattern (Gu et al., 2015; Ozkanli et al., 2015). Importantly, studies have shown that most 5-hmC in the genome arises from pre-existing 5-mC, which stresses the dynamic balance between methylation and hydroxymethylation required to maintain normal gene expression (Ficz et al., 2011).
Our hMeDIP-seq data not only revealed loss of 5-hmC in CpG islands and genebodies, but also identified differentially hydroxymethylated genes in the epidermis including Akt1 and MAPKAPK2 that play an important role in maintaining stem cell homeostasis (Schwermann et al., 2009; Segrelles et al., 2014; M. Zhang & Zhang, 2019). Decreased hydroxymethylation and resultant modulation of Akt1 expression could lead to aberrant regulation of stem cell activation and proliferation (Segrelles et al., 2014; M. Zhang & Zhang, 2019). Downstream of Akt1, MAPKAPK2 signaling has been shown to influence stem cell self-renewal capacity as well as inflammatory responses affecting cutaneous wound healing (Schwermann et al., 2009; Thuraisingam et al., 2010). Both Akt1 and MAPKAP2 are involved in the RAR activation pathway, which plays a major role in regulating keratinocyte proliferation (Gericke et al., 2013; Lee et al., 2009). These data now provide a foundation for future studies to dissect how TET-mediated 5-hmC regulates transcriptional and proteomic pathways in psoriasis.
Epigenetic maintenance by TET2 also has been found to be crucial for self-renewal and differentiation of hematopoietic stem cells (Kohli & Zhang, 2013). In our study, we found loss of 5-hmC in psoriatic lesional epidermis possibly due to decreased TET1 and TET2 expression. Diminished TET expression could explain the loss of KSC self-renewal capabilities as more stem cells undergo asymmetric division to become FABP5-labeled TACs (Jia et al., 2016). Our GO analysis of hMeDIP-seq data supports this hypothesis by showing downregulation of genes involved in controlling the canonical Wnt signaling pathway and cell differentiation. Studies have shown the importance of Wnt signaling in tissue homeostasis as well as aberrant regulation of KSCs in psoriatic lesions (Gudjonsson et al., 2010; Veltri et al., 2018; Yanfei Zhang et al., 2015). Wnt5a, which regulates epidermal differentiation in adult skin, was identified as a gene whose expression did not return to baseline even after successful treatment and resolution of the epidermal abnormalities in psoriasis. Residual Wnt5a expression therefore may contribute to the recurrent nature of psoriasis (Suárez-Fariñas, Fuentes-Duculan, Lowes, & Krueger, 2011). In aggregate, these findings along with our hMeDIP-seq analysis indicate that differentially hydroxymethylated genes in the IMQ-treated epidermis may disrupt stem cell homeostasis that can lead to a psoriatic disease state. Despite potential limitations of using the IMQ-induced psoriasis model to replicate the precise pathogenesis of human psoriasis (Hawkes, Gudjonsson, & Ward, 2017), further studies in TET enzyme knockout mice may provide valuable insights into TET function and its downstream regulation.
Loss of 5-hmC is also seen in other hyperproliferative diseases in cancer biology, including hematopoietic cancers and melanoma where re-introduction of TET activity leads to pathogenic phenotype rescue (An et al., 2017; Lian et al., 2012). By manipulating TET expression in vitro, as previously accomplished in melanoma, we were able to epigenetically reprogram KSCs by increasing levels of 5-hmC (Gustafson et al., 2015). Efforts are presently underway to deploy similar approaches in in vivo models in order to assess the effects of 5-hmC restoration on experimentally-induced psoriatic lesions. Nonetheless, these early findings demonstrate the potential of 5-hmC as a targetable epigenetic anomaly in psoriasis.
In summary, this study establishes the 5-hmC landscape in psoriatic KSCs and TACs and sets the stage for future functional studies of 5-hmC regulation in psoriasis. As summarized in Figure 5, we hypothesize that IL-23/IL-17 pathways known to be capable of inducing epidermal proliferation via KSC targeting, in concert with loss of 5-hmC in epigenetically-regulated genes affecting KSC/TAC homeostasis, results in KSC/TAC niche expansion and the characteristic cytoarchitecture of the psoriatic phenotype. The potential key role of hydroxymethylation in psoriasis pathogenesis could lead to promising epigenetic-based therapies. In addition, recent recognition of similarities between psoriasis and other IL-17-associated dermatitides such as atopic dermatitis (Krueger & Guttman-Yassky, 2017) now paves the way for broader inquiry into the potential role of anomalies in the epigenome in chronic skin conditions characterized by aberrant epidermal proliferation/maturation.
MATERIALS AND METHODS
Human Tissue Samples and Cells
All human tissue samples were derived from the Pathology Archives of the Brigham and Women’s Hospital with full IRB approval. Patient consent for experiments was not required because de-identified pathological specimens of human samples is considered to be discarded material by our institution, and thus the studies were exempt. Human keratinocyte stem cells were provided by the Cutaneous Biology Research Center at Massachusetts General Hospital.
Immunofluorescence Staining
Immunofluorescence (IF) studies were performed on paraffin-embedded sections of formalin-fixed tissue. Tissue sections cut at 5 micron intervals were deparaffinized, rehydrated, and heated with Target Antigen Retrieval Solution (Dako, Agilent Technologies, Santa Clara, CA) in a pressure cooker. Sections were blocked with 10% animal serum for 30 min prior to incubation with primary antibodies. Cell culture samples were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and permeabilized using 0.5% TritonX-100 (Sigma-Aldrich, St. Louis, MO). Cell culture stains for 5-hmC requires additional incubation with 4 N HCl for 30 min. Samples were incubated at 4°C overnight with the following primary antibodies: rabbit IgG-anti-5hmC (1:2000; Active Motif, Carlsbad, CA), mouse IgG2a-anti-CK15 (1:100; Thermo Fisher Scientific, Waltham, MA), mouse IgG1-anti-nestin (1:800; Millipore, Burlington, MA), rabbit IgG-anti-FABP5 (1:600; Cell Signaling Technology, Danvers, MA), or rabbit IgG-anti-Ki67 (1:250; Novus Biologicals, Littleton, CO). Sections were then blocked with 0.1% Sudan Black (Abcam, Cambridge, UK) for 10 min. The secondary antibodies used were: AF488 goat-anti-rabbit (1:2000; Invitrogen, Carlsbad, CA), AF546 goat-anti-mouse IgG1 (1:2000, Vector Laboratories, Burlingame, CA), goat-anti-rabbit Biotin (1:2000, Thermo Fisher Scientific), or rat-anti-mouse IgG1 Biotin (1:100, eBioscience). Biotinylated secondary antibodies were processed with Strep AF647 Biotin (1:1000, Thermo Fisher Scientific). Cell nuclei were labeled with DAPI (Invitrogen).
Immunohistochemistry Staining
Immunohistochemistry (IHC) studies were performed on paraffin-embedded sections of formalin-fixed mouse tissue. Tissue sections were deparaffinized, rehydrated, and blocked with 3% hydrogen peroxide. Heat-induced antigen retrieval was completed using Target Antigen Retrieval solution (Dako, Carpinteria, CA) in a pressure cooker for 45 min. Sections were blocked with 10% animal serum for 30 min prior to incubation with primary antibodies. Sections were incubated at 4°C overnight with primary antibodies: rabbit IgG-anti-5hmC (1:2000; Active Motif), mouse IgG2a-anti-CK15 (1:100; Thermo Fisher Scientific), mouse IgG1-anti-nestin (1:800; Millipore), or rabbit IgG-anti-FABP5 (1:600; Cell Signaling Technology). Simultaneous staining was performed for 5-hmC/CK15 and 5-hmC/nestin double labeling, while sequential staining was performed for 5-hmC/FABP5 double labeling. Sections were incubated with the following secondary antibodies for an hour: horse-anti-mouse HRP (Vector Laboratories), goat-anti-rabbit HRP (Vector Laboratories), mouse polymer HRP (EnVision, Agilent Technologies), horse-anti-mouse AP (Vector Laboratories), or donkey-anti-rabbit AP (Abcam). They were then processed by one of the following: DAB + Chromagen (Dako), Vector Red Alkaline Phosphatase (Vector Laboratories), Vector NovaRed (Vector Laboratories), Vector Blue Alkaline Phosphatase (Vector Laboratories), and ImmPRESS-AP anti-mouse IgG polymer detection kit (Vector Laboratories). Sections were counterstained with Gill’s Hematoxylin (Fisher Scientific, Kalamazoo, MI) and treated with Defining Solution (Fisher Scientific, Kalamazoo) and Bluing Solution (American MasterTech Scientific, McKinney, TX). Hematoxylin and eosin stains were done by Harvard Medical Area Core Management System, Rodent Histopathology.
Mice and Epidermal Isolation
C57BL/6J mice (female) were treated under defined conditions in accordance with institutional guidelines. Experiments were performed according to approved experimental protocols. Mice at 6 weeks of age received a topical dose of 62.5 μg of Imiquimod (Aldara) 5% cream (Aldara, 3M Pharmaceuticals, St. Paul, MN) daily onto the shaved back and right ear for 6 consecutive days. This translates to a daily dose of 3.125 mg of the active compound. Control mouse skin was similarly treated with Vaseline as a control vehicle cream. Punch biopsies of mouse skin were used for IHC studies. Isolation of the epidermis from shaved biopsies was also performed in order to collect samples for RT-PCR and hMeDIP-seq. This was accomplished by soaking the samples in 0.2% dispase in 4°C for 6 hours and manually separating epidermis from dermis.
hMeDIP-seq
Genomic DNA from the epithelium of mouse models was purified, sonicated, and ligated to Illumina barcode adapters before hMeDIP-seq (hydroxymethylated DNA immunoprecipitation followed by high throughput sequencing). Adaptor-ligated DNA (5 μg) was denatured and incubated with 3 μL of 5-hmC antibody (Active Motif) at 4°C overnight. Antibody-DNA complexes were captured by protein A/G beads. The immunoprecipitated DNA was purified and sequenced using the Illumina platform followed by standard Illumina protocols (Xu et al., 2011b). Read sequences were aligned to the mouse genome (mm10) using ELAND v2 in the CASAVA (Illumina, v1.6) package. MACS2 was used to identify differentially hydroxymethylated genes (Yong Zhang et al., 2008). Ingenuity Pathway Analysis (Krämer, Green, Pollard, & Tugendreich, 2014) was used for network analysis and Gene Ontology (Ashburner et al., 2000; The Gene Ontology Consortium, 2017) for pathway analysis.
Human Keratinocyte Stem Cell Culture
Human keratinocyte stem cells taken from neonatal foreskin were given by the Cutaneous Biology Research Center at Massachusetts General Hospital. 5.0 × 104 cells were plated in 6 cm cell culture dishes. Cells were cultured in EpiLife Calcium-Free MEPICF cell culture medium (Cascade Biologics, Carlsbad, CA) and incubated at 37°C, 5% CO2 for 24 hours prior to addition of calcium. Cultures were treated with either 1.5 mM Ca2 or 0.06 mM Ca2+. After 24 hours of incubation with calcium, some cultures were treated with 200 μmol/L ascorbic acid using L-Ascorbic Acid (Thermo Fisher Scientific) for 48 hours. The cultures were then harvested for immunofluorescence studies or RT-PCR.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the LEO Foundation to C.G.L, G.F.M, M.H.F; the Harvard Stem Cell Institute to C.G.L, G.F.M, M.H.F, N.Y.F.; the Department of Pathology, Brigham and Women’s Hospital to C.G.L, G.F.M; DOD PPCRP Idea Award with Special Focus to C.G.L., G.F.M.; NIH/NEI grants 1RO1EY025794 and R24EY028767, VA BLR&D 1I01BX000516 and VA RR&D 1I01RX000989 Merit Review Awards, DOD translational team science award to N.Y.F.; and NIH/NIBIB grant EB016652-05 to C.A.A.L.
Dr. Feng Li and Christine W. Yuan contribute equally to this study.
Dr. Christine G. Lian and Dr. George F. Murphy serve as co-corresponding/senior authors.
Abbreviations:
- 5-hmC
5-hydroxymethylcytosine
- CK15
cytokeratin 15
- GO
gene ontology
- FABP5
fatty acid binding protein 5
- hMeDIP
hydroxymethylated DNA immunoprecipitation
- IMQ
imiquimod
- IPA
ingenuity pathway analysis
- KSC
keratinocyte stem cell
- LSC
lichen simplex chronicus
- TAC
transit amplifying cell
- TDC
terminally differentiated cell
- TET
ten-eleven translocation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY STATEMENT
The 5-hmC-seq datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138921, hosted at NCBI’s Gene Expression Omnibus and are accessible through GEO series Accession Number GSE138921.
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, three figures, and two tables.
REFERENCES
- An J, Rao A, & Ko M TET family dioxygenases and DNA demethylation in stem cells and cancers. Experimental & Molecular Medicine. 2017; 49: e323–e323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000; 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitah SA Stem cell depletion through epidermal deletion of Rac1. Science. 2005; 309: 933–935 [DOI] [PubMed] [Google Scholar]
- Boehncke W-H, & Schön MP Psoriasis. The Lancet. 2015; 386: 983–994 [DOI] [PubMed] [Google Scholar]
- Brezinski EA, Dhillon JS, & Armstrong AW Economic burden of psoriasis in the United States: a systematic review. JAMA Dermatology. 2015; 151: 651. [DOI] [PubMed] [Google Scholar]
- Chandra A, Senapati S, Roy S, Chatterjee G, & Chatterjee R Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clinical Epigenetics. 2018; 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charruyer A, Fong S, Vitcov GG, Sklar S, Tabernik L, Taneja M, et al. Brief report: interleukin-17A-dependent asymmetric stem cell divisions are increased in human psoriasis: a mechanism underlying benign hyperproliferation: IL17A and stem cell division in hyperproliferation. Stem Cells. 2017; 35: 2001–2007 [DOI] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017; 170: 1079–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, et al. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progeny cells by targeting Nestin. BMC Neuroscience. 2012; 13: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011; 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Gericke J, Ittensohn J, Mihály J, Álvarez S, Álvarez R, Töröcsik D, et al. Regulation of retinoid-mediated signaling involved in skin homeostasis by RAR and RXR agonists/antagonists in mouse skin. PLoS ONE. 2013; 8: e62643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CBF, Zechin KG, Xu S, Stelini RF, Nishimoto IN, Zhan Q, et al. TET2 negatively regulates nestin expression in human melanoma. The American Journal of Pathology. 2016; 186: 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths CEM, van der Walt JM, Ashcroft DM, Flohr C, Naldi L, Nijsten T, et al. The global state of psoriasis disease epidemiology: a workshop report. The British Journal of Dermatology, 2017; 177: e4–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Nylander E, Coates PJ, Fahraeus R, & Nylander K Correlation between reversal of DNA methylation and clinical symptoms in psoriatic epidermis following narrow-band UVB phototherapy. Journal of Investigative Dermatology, 2015; 135: 2077–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson JE, Johnston A, Stoll SW, Riblett MB, Xing X, Kochkodan JJ, et al. Evidence for altered Wnt signaling in psoriatic skin. Journal of Investigative Dermatology, 2010; 130: 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson CB, Yang C, Dickson KM, Shao H, Van Booven D, Harbour JW, et al. Epigenetic reprogramming of melanoma cells by ascorbic acid treatment. Clinical Epigenetics. 2015; 7: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, & Krueger JG Atopic dermatitis and psoriasis: two different immune diseases or one spectrum? Current Opinion in Immunology. 2017; 48: 68–73. [DOI] [PubMed] [Google Scholar]
- Hawkes JE, Gudjonsson JE, & Ward NL The snowballing literature on imiquimod-induced skin inflammation in mice: A Critical Appraisal. Journal of Investigative Dermatology. 2017; 137: 546–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, & Yuspa SH Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980; 19: 245–254 [DOI] [PubMed] [Google Scholar]
- Hsu Y-C, Li L, and Fuchs E Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014; 157: 935–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H-Y, Shi Y, Luo L-F, Jiang G, Zhou Q, Xu S-Z, and Lei T-C Asymmetric stem-cell division ensures sustained keratinocyte hyperproliferation in psoriatic skin lesions. International Journal of Molecular Medicine. 2016; 37: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahata K, Matsuo K, Hara Y, Naganuma T, Oiso N, Kawada A., et al. Ascorbic acid derivative DDH-1 ameliorates psoriasis-like skin lesions in mice by suppressing inflammatory cytokine expression. J Pharmacol Sci. 2018; 138: 284–288 [DOI] [PubMed] [Google Scholar]
- Kohli RM and Zhang Y TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013; 502: 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, & Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014; 30: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker R, and Sun T Heterogeneity in epidermal basal keratinocytes: morphological and functional correlations. Science. 1982; 215: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Lee D-D, Stojadinovic O, Krzyzanowska A, Vouthounis C, Blumenberg M, & Tomic-Canic M Retinoid-responsive transcriptional changes in epidermal keratinocytes. Journal of Cellular Physiology. 2009; 220: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012; 150: 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Gomez NC, Alaverdyan K, Sendoel A, Yuan S, et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 2017; 550: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkanli S, Zemheri E, Karadag AS, Akbulak O, Zenginkinet T, Zindanci I, et al. A comparative study of histopathological findings in skin biopsies from patients with psoriasis before and after treatment with acitretin, methotrexate and phototherapy. Cutaneous and Ocular Toxicology. 2015; 34: 276–281 [DOI] [PubMed] [Google Scholar]
- Roberson EDO, Liu Y, Ryan C, Joyce CE, Duan S, Cao L, et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. Journal of Investigative Dermatology. 2012; 132: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha S, Royston K, Udayakumar N, & Tollefsbol T Epigenetic regulation of epidermal stem cell biomarkers and their role in wound healing. International Journal of Molecular Sciences. 2015; 17: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha G, Joshi K, Lawes K, Bamford M, Moosa F, Teo KW, and Pringle JH 5-Hydroxymethylcytosine is an independent predictor of survival in malignant melanoma. Modern Pathology. 2016; 30: 60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant DW, Camarena V, Mustafi S, Li Y, Wilkes Z, Van Booven D, et al. Ascorbate suppresses VEGF expression in retinal pigment epithelial cells. Investigative Opthalmology & Visual Science. 2018; 59: 3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwermann J, Rathinam C, Schubert M, Schumacher S, Noyan F, Koseki H, et al. MAPKAP kinase MK2 maintains self-renewal capacity of haematopoietic stem cells. The EMBO Journal. 2009; 28: 1392–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrelles C, García-Escudero R, Garín MI, Aranda JF, Hernández P, Ariza JM, et al. Akt signaling leads to stem cell activation and promotes tumor development in epidermis: Akt activation of HF-SCs. Stem Cells. 2014; 32, 1917–1928. 10.1002/stem.1669 [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, & Khavari PA DNMT1 maintains progeny function in self-renewing somatic tissue. Nature. 2010; 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Fuentes-Duculan J, Lowes MA, & Krueger JG Resolved psoriasis lesions retain expression of a subset of disease-related genes. Journal of Investigative Dermatology. 2011; 131: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Research. 2017; 45: D331–D338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuraisingam T, Xu YZ, Eadie K, Heravi M, Guiot M-C, Greemberg R, et al. MAPKAPK-2 signaling is critical for cutaneous wound healing. Journal of Investigative Dermatology. 2010; 130: 278–286 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Mourits S, Voerman JSA, Kant M, Boon L, Laman JD, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. The Journal of Immunology. 2009; 182: 5836–5845 [DOI] [PubMed] [Google Scholar]
- Veltri A, Lang C, & Lien W-H Concise Review: Wnt signaling pathways in skin development and epidermal stem cells: Wnt signaling in skin development and stem cells. Stem Cells. 2018; 36: 22–35 [DOI] [PubMed] [Google Scholar]
- Wang X, Liu X, Duan X, Zhu K, Zhang S, Gan L, et al. Ten-eleven translocation-2 regulates DNA hydroxymethylation status and psoriasiform dermatitis progression in mice. Acta Dermato Venereologica. 2008; 98: 585–593 [DOI] [PubMed] [Google Scholar]
- Watarai A, Amoh Y, Maejima H, Hamada Y, and Katsuoka K Nestin expression is increased in the suprabasal epidermal layer in psoriasis vulgaris. Acta Dermato Venereologica. 2013; 93: 39–43 [DOI] [PubMed] [Google Scholar]
- Watt FM Stem cell fate and patterning in mammalian epidermis. Current Opinion in Genetics & Development. 2001; 11: 410–417 [DOI] [PubMed] [Google Scholar]
- Weatherhead SC, Farr PM, Jamieson D, Hallinan JS, Lloyd JJ, Wipat A, and Reynolds NJ Keratinocyte apoptosis in epidermal remodeling and clearance of psoriasis induced by UV radiation. Journal of Investigative Dermatology. 2011; 131: 1916–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker-Menezes D, Jones SC, Friedman TM, Korngold R, & Murphy GF An epithelial target site in experimental graft-versus-host disease and cytokine-mediated cytotoxicity is defined by cytokeratin 15 expression. Biology of Blood and Marrow Transplantation. 2003; 9: 559–570 [DOI] [PubMed] [Google Scholar]
- Whitbread LA, & Powell BC Expression of the intermediate filament keratin gene,K15,in the basal cell layers of epithelia and the hair follicle. Experimental Cell Research. 1998; 244: 448–459 [DOI] [PubMed] [Google Scholar]
- Winge MC, Ohyama B, Dey CN, Boxer LM, Li W, Ehsani-Chimeh N, et al. RAC1 activation drives pathologic interactions between the epidermis and immune cells. Journal of Clinical Investigation. 2016; 126: 2661–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen X, Zhao J, Martin B, Zepp JA, Ko JS, et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. The Journal of Experimental Medicine. 2015; 212: 1571–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular Cell. 2011b; 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, and Zhang X The role of PI3K/AKT/FOXO signaling in psoriasis. Archives of Dermatological Research. 2019; 311: 83–91 [DOI] [PubMed] [Google Scholar]
- Zhang P, Su Y, Chen H, Zhao M, & Lu Q Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. Journal of Dermatological Science. 2010; 60: 40–42 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tu C, Zhang D, Zheng Y, Peng Z, Feng Y, et al. Wnt/β-Catenin and Wnt5a/Ca2+ pathways regulate proliferation and apoptosis of keratinocytes in psoriasis lesions. Cellular Physiology and Biochemistry. 2015; 36: 1890–1902 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology. 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Wang W, Shen C, Li H, Zuo X, Zheng X, et al. Epigenome-wide association analysis identified nine skin DNA methylation loci for psoriasis. Journal of Investigative Dermatology. 2016; 136: 779–787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.