Abstract
The last step of cholesterol biosynthesis is the conversion of 7-dehydrocholesterol (7-DHC) into cholesterol, a reaction catalyzed by dehydrocholesterol reductase 7 (DHCR7). Investigation of the effect of Dhcr7 single-allele mutations on the metabolism of aripiprazole (ARI) and cariprazine (CAR) in maternally exposed transgenic pups revealed that ARI, CAR, and their active metabolites were decreased in the liver and brain of Dhcr7+/–. This difference in the drug and metabolite levels resulted in an increased turnover of ARI and CAR in tissues from Dhcr7+/– animals, indicating an enhanced metabolism, which was at least partially due to increased levels of Cyp2d6 in the liver of Dhcr7+/– mice. Finally, experiments with both WT and DHCR7+/– human fibroblasts revealed lower drug levels in DHCR7+/– heterozygous cells. Our findings have potential clinical implications, as DHCR7 heterozygosity is present in 1–3% in the human population, and these individuals might have reduced therapeutic levels of Cyp2d6-metabolized medications and are putatively more susceptible to unwanted side effects.
Introduction
Cholesterol biosynthesis is a complex process that makes cholesterol from acetyl-CoA.1 The last step of the pathway consists of the conversion of 7-dehydrocholesterol (7-DHC) into cholesterol, a step catalyzed by the enzyme dehydrocholesterol reductase 7 (DHCR7).1 It is estimated that 1–3% of the human population carries single-allele mutations in the DHCR7 gene.2 However, despite the high prevalence of DHCR7 heterozygous carriers in the human population, very little is known about the biological consequences of these single-allele mutations. It is known that Dhcr7+/– mice have higher 7-DHC levels than Dhcr7+/+ animals in all tissues and circulation.3−5 Similarly, 7-DHC is elevated in fibroblasts from DHCR7+/– when compared to DHCR7+/+, suggesting that animal observations translate to human physiology.6Dhcr7 heterozygosity has effects beyond biochemistry. Behavioral studies comparing the number and duration of ultrasonic vocalizations (USVs) between WT and Dhcr7+/– mice found that heterozygous animals made fewer and shorter USV calls than the WT animals.7
Many FDA-approved pharmaceuticals have a side effect of DHCR7 enzymatic activity inhibition.8−10 Aripiprazole (ARI) and cariprazine (CAR), two atypical antipsychotics, are among the most potent DHCR7 inhibitors, strongly elevating 7-DHC levels.10 This chemical inhibition of the DHCR7 enzyme can have profound consequences, as 7-DHC-derived oxysterols are cytotoxic. A 2016 review by Boland and Tatonetti demonstrates that first-trimester exposure to DHCR7 inhibitors results in outcomes similar to those of known teratogens and that DHCR7 activity should be considered during drug development and prenatal toxicity assessment.11 A set of recent studies revealed that Dhcr7+/– animals and DHCR7+/– human fibroblasts respond differently to inhibitors of the DHCR7 enzyme.3,4,6Dhcr7 heterozygosity by itself leads only to a small increase in 7-DHC levels,5 but a combination of both inhibitory insults (genetic and chemical) results in a robust elevation in 7-DHC. When subjected to the same dose of Dhcr7 inhibitors, heterozygous animals (or cells) have much higher 7-DHC elevation than those with a WT genotype, indicating a drug × gene interaction and a higher vulnerability of the Dhcr7+/– genotype to the inhibitors.3,4,6
This drug × gene interaction appears to be of particular importance during development. We recently evaluated sterol levels in the brains of offspring maternally exposed to ARI and CAR. We found that offsprings’ brains with a Dhcr7+/– genotype, when maternally exposed to CAR or ARI, have 7-DHC levels comparable to those detected in mild cases of the Smith–Lemli–Optiz Syndrome (SLOS), a neurodevelopmental disorder caused by mutations in both DHCR7 alleles.3,4 In addition, we observed that levels of CAR and its metabolites were significantly lower in Dhcr7+/– pups compared to their WT littermates, which suggests that WT and Dhcr7+/– mice may metabolize the drugs differently.
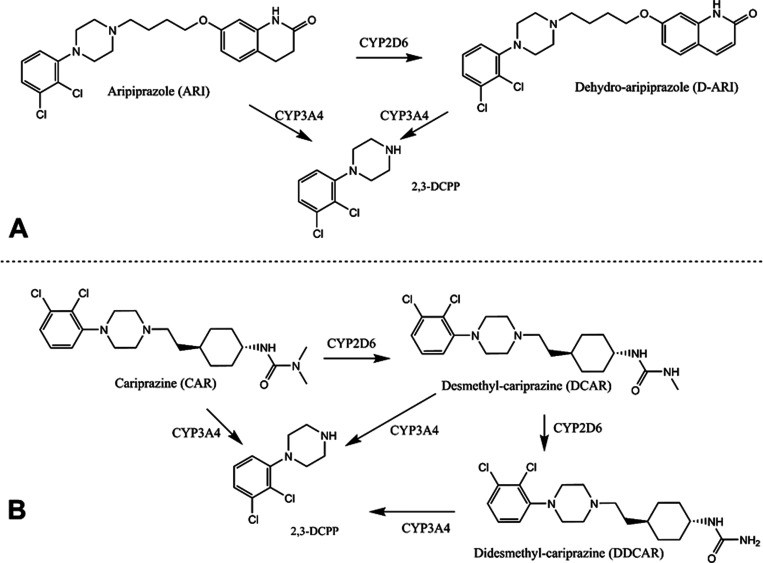
The metabolism and clearance of antipsychotics is complex and the entire mechanism is not fully understood. They are primarily cleared by hepatic metabolism with enzymes in the cytochrome P450 superfamily (CYPs).12,13 Most CYPs are abundantly expressed in the liver, with some isoforms present in other organs.13,14 ARI and CAR are cleared by multiple CYP isoforms (Figure 1).12,15−18 ARI is converted by CYP2D6 into D-ARI, which then undergoes N-dealkylation by CYP3A4 to generate 2,3-dichlorophenyl piperazine (2,3-DCPP).18,19 Similarly, CAR is converted into desmethyl-CAR (DCAR) and didesmethyl-CAR (DDCAR) by CYP2D6 and CYP3A4 or undergoes N-dealkylation to generate 2,3-DCPP.16
Figure 1.
Simplified metabolism of ARI and CAR. (A) CYP2D6 catalyzes ARI’s dehydrogenation to generate D-ARI. Both ARI and D-ARI can undergo N-dealkylation by CYP3A4 to generate 2,3-DCPP. (B) CYP2D6 catalyzes the conversion of CAR into DCAR, which is then converted into DDCAR. CAR, DCAR, and DDCAR can undergo N-dealkylation to generate 2,3-DCPP.
In this current study, we investigated the influence of Dhcr7 heterozygosity on ARI, CAR, and their metabolites. The study was performed using WT and Dhcr7+/– human fibroblasts and transgenic animal models and assessed the expression of an ARI and CAR metabolizing enzyme.
Results
Dhcr7+/– Heterozygosity Influences Drug Levels across Multiple Tissues
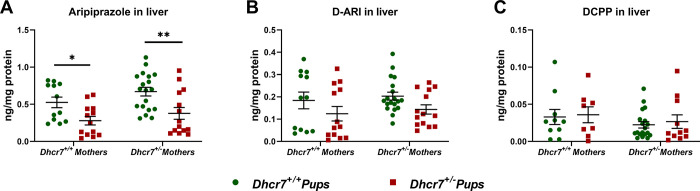
In order to investigate the effect of a Dhcr7 heterozygosity on drug metabolism, we assessed the levels of ARI, CAR, and their active metabolites, across different tissues of WT and Dhcr7+/– mice. WT and Dhcr7+/– female mice were injected with ARI from E12 to E19 and the offspring tissue was collected at birth (P0). The levels of ARI and its metabolites (D-ARI and 2,3-DCPP) were measured in the liver of P0 animals (Figure 2). The analysis of medication levels with regards to embryonic genotype alone revealed that ARI levels in the livers of Dhcr7+/– pups were 52% lower than those of WT pups (Dhcr7+/– pups: 0.33 ± 0.05 ng/mg protein vs WT pups: 0.68 ± 0.06; p < 0.0001) (Figure 2A). D-ARI levels followed the same pattern as ARI and were found decreased in Dhcr7+/– pups compared to their WT littermates (Dhcr7+/– pups: 0.13 ± 0.02 ng/mg protein vs WT pups: 0.21 ± 0.02; p = 0.0095) (Figure 2B). A two-way ANOVA analysis accounting for the contribution of both genotypes is presented in Table 1. No significant differences were observed in 2,3-DCPP between WT and Dhcr7+/– pups (Figure 2C). Importantly, maternal genotype had no significant effect on the medication levels in the liver of pups, suggesting that the effect is primarily driven by the pup genotype.
Figure 2.
Dhcr7+/– heterozygosity decreases ARI levels in the liver. Levels of ARI and its metabolites were determined in maternally exposed pups at an age of P0. Samples were grouped taking into account both the maternal and newborn mouse genotypes. WT and Dhcr7+/– pups are depicted in green or red, respectively. ARI (A), D-ARI (B), and 2,3-DCPP (C) were determined using LC–MS/MS. Bars correspond to the mean ± SEM; statistical significance: *p < 0.05; **p < 0.01. A two-way ANOVA analysis is presented in Table 1. Note the lower levels of ARI and D-ARI in Dhcr7+/– pups.
Table 1. ANOVA Analysis of ARI Levels in P0 Liversa.
| # | Comparison | ARI | D-ARI | 2,3-DCPP |
| 1 | embryonic genotype: Dhcr7+/+vs Dhcr7+/– | 0.0002 | 0.0286 | 0.6585 |
| 2 | maternal genotype: Dhcr7+/+vs Dhcr7+/– | 0.0788 | 0.4764 | 0.2262 |
| 3 | two-way interaction: maternal Dhcr7 vs embryonic Dhcr7 genotypes | 0.7311 | 0.9987 | 0.9333 |
Rows #1–2 denote statistical significance for single variables; #3 reports probability for the two interacting factors; values highlighted in bold denote p < 0.05. No statistical difference was observed between male and female animals.
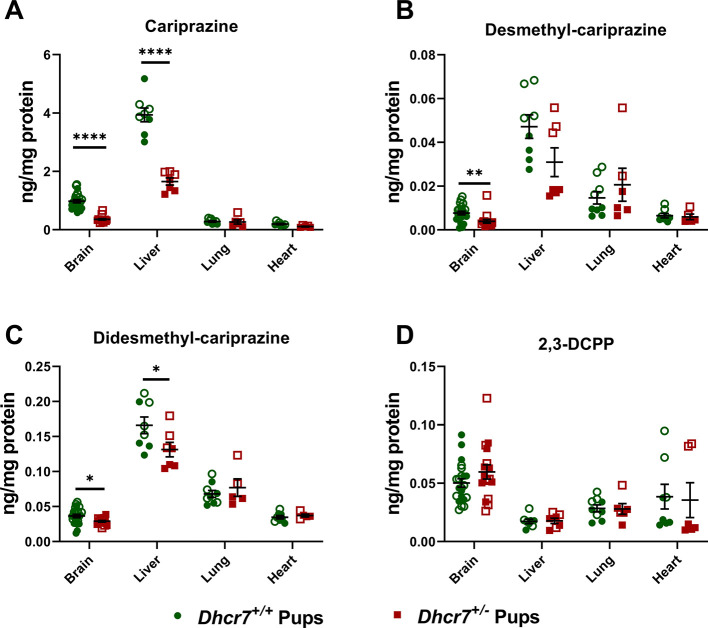
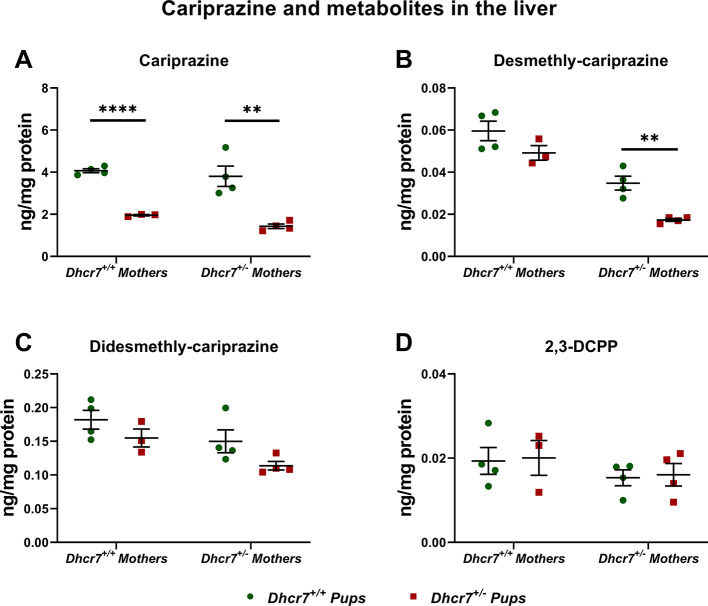
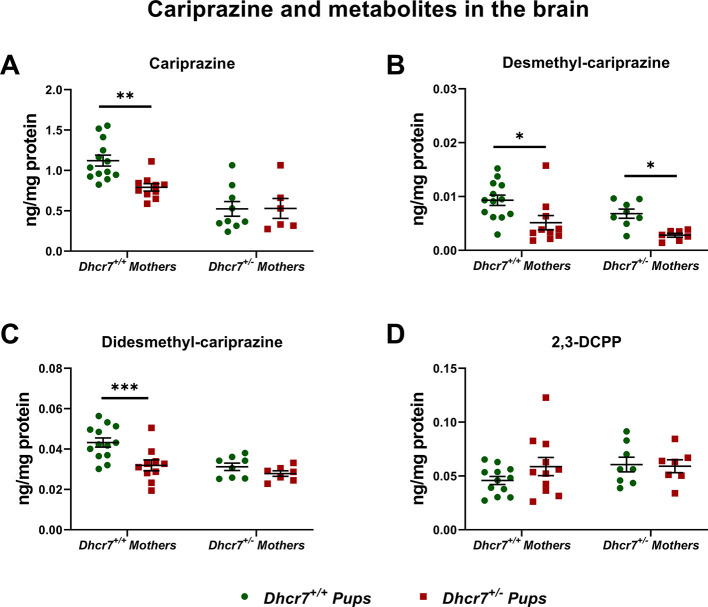
Next, we were interested if this finding is also observed in response to CAR exposure and if this effect can be seen across multiple tissues. Developmental exposure (E12-E19) to 0.2 mg/kg CAR was followed by tissue harvest at birth and parent drug/metabolites (DCAR, DDCAR, and 2,3-DCPP) were measured in the liver, heart, lungs, and brain of offspring with the Dhcr7+/– and Dhcr7+/+ genotypes (Figure 3). Highest levels of CAR and its metabolites were observed in the liver, followed by the brain, lungs, and heart. Importantly, CAR levels were significantly lower in Dhcr7+/– pups when compared to their WT littermates in both the liver and brain (liver: Dhcr7+/– pups: 1.65 ± 0.12 ng/mg protein vs WT pups: 3.94 ± 0.23; p < 0.0001; brain: Dhcr7+/– pups: 0.36 ± 0.03 ng/mg protein vs WT pups: 0.98 ± 0.06; p < 0.0001) (Figure 3A). DCAR and DDCAR followed the same pattern, with lower levels detected in Dhcr7+/– pups compared to their WT littermates across all four investigated tissues, although only DDCAR levels reached significant difference between the two groups (Figure 3B,C, respectively). Levels of 2,3-DCPP were not significantly different between the two genotypes (Figure 3D).
Figure 3.
Dhcr7+/– heterozygosity decreases CAR levels in the liver and brain. The levels of CAR and metabolites were determined at P0 in the brain, liver, lung, and heart. Samples were grouped taking into account the pups’ genotype. WT and Dhcr7+/– pups are depicted in green and red, respectively. CAR (A), DCAR (B), DDCAR (C), and 2,3-DCPP (D) were determined using LC–MS/MS. Opened symbols denote pups from WT mothers and filled symbols denote pups from Dhcr7+/– mothers. Bars correspond to the mean ± SEM; statistical significance: *p < 0.05; ****p < 0.0001. Note the decreased levels of CAR and DDCAR in both the liver and brain and the different CAR levels across the four investigated tissues.
To address the origin of these changes, we selected the two tissues with the highest drug levels (brain and liver) and reanalyzed the samples taking into account both maternal and embryonic genotypes. The liver data are depicted in Figure 4, and a two-way ANOVA analysis of our findings is presented in Table 2. The embryonic Dhcr7+/– genotype had a significant effect on CAR, DCAR, and DDCAR levels, with lower levels in heterozygous pups compared to their WT littermates. No effect on 2,3-DCPP was observed. Similarly, the levels of DCAR and DDCAR were also significantly decreased in pups born to Dhcr7+/– mothers, indicating that the maternal genotype also contributes to the decreased drug levels in the liver of heterozygous animals. Similar results were observed in the brain tissue, where the CAR, DCAR, and DDCAR levels were affected by both maternal and embryonic Dhcr7+/– genotypes (Figure 5 and Table 3). Furthermore, an analysis of the maternal serum revealed lower levels of CAR and its metabolites in the serum of Dhcr7+/– mothers when compared to their WT counterparts (Figure S1).
Figure 4.
Levels of CAR and its metabolites in the liver depend on both maternal and embryonic genotypes. The levels of CAR and metabolites were determined in the livers of P0 pups. The data are presented taking into account both maternal and embryonic Dhcr7 genotypes. A two-way ANOVA analysis is presented in Table 2. Each symbol corresponds to a single pup. Bars correspond to the mean ± SEM; statistical significance: **p < 0.01; ****p < 0.0001. Note that for CAR, DCAR, and DDCAR, there is a summation effect between the maternal and pup Dhcr7 genotype, with the Dhcr7+/– pups born to Dhcr7+/– mothers showing the lowest levels.
Table 2. ANOVA Analysis of CAR Levels in P0 Liversa.
| # | Comparison | CAR | DCAR | DDCAR | 2,3-DCPP |
|---|---|---|---|---|---|
| 1 | embryonic genotype: Dhcr7+/+vs Dhcr7+/– | <0.0001 | 0.0017 | 0.0380 | 0.8120 |
| 2 | maternal genotype: Dhcr7+/+vs Dhcr7+/– | 0.1782 | <0.0001 | 0.0199 | 0.2030 |
| 3 | two-way interaction: maternal Dhcr7 vs embryonic Dhcr7 genotypes | 0.6421 | 0.3202 | 0.7388 | 0.9998 |
Rows #1–2 denote statistical significance for single variables; #3 reports probability for the two interacting factors; values highlighted in bold denote p < 0.05. No statistical difference was observed between male and female animals.
Figure 5.
Levels of CAR and its metabolites in the brain depend on both maternal and embryonic genotypes. The levels of CAR and metabolites were determined in the brains of P0 pups. The data are presented taking into account both the maternal and embryonic Dhcr7 genotypes. A two-way ANOVA analysis is presented in Table 3. Each symbol corresponds to a single pup. Bars correspond to the mean ± SEM; statistical significance: *p < 0.05; **p < 0.01; ***p < 0.001. Note that for CAR, DCAR, and DDCAR, there is a summation effect between the maternal and pup Dhcr7 genotype, with the Dhcr7+/– pups born to Dhcr7+/– mothers showing the lowest levels.
Table 3. ANOVA Analysis of CAR Levels in P0 Brainsa.
| # | Comparison | CAR | DCAR | DDCAR | 2,3-DCPP |
|---|---|---|---|---|---|
| 1 | embryonic genotype: Dhcr7+/+vs Dhcr7+/– | 0.0525 | 0.0005 | 0.0039 | 0.3980 |
| 2 | maternal genotype: Dhcr7+/+vs Dhcr7+/– | <0.0001 | 0.0304 | 0.0018 | 0.2655 |
| 3 | two-way interaction: maternal Dhcr7 vs embryonic Dhcr7 genotypes | 0.0452 | 0.9414 | 0.1026 | 0.2885 |
Rows #1–2 denote statistical significance for single variables; #3 reports probability for the two interacting factors; values highlighted in bold denote p < 0.05. No statistical difference was observed between male and female animals.
Dhcr7+/– Heterozygosity Increases Drug Turnover
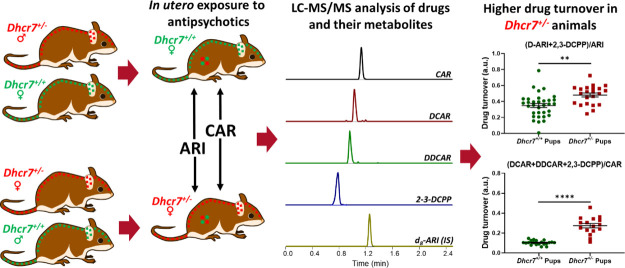
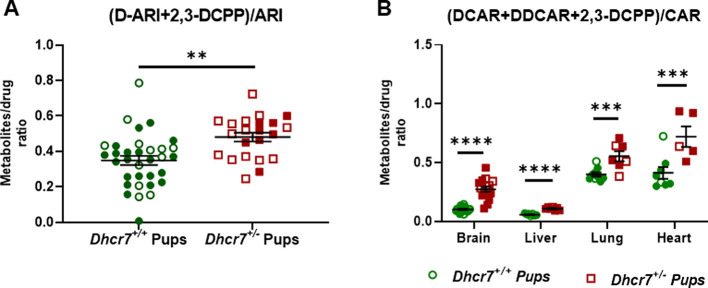
Using data in Figures 2 and 3, we assessed drug turnover in the four investigated organs of both WT and Dhcr7+/–P0 mice by calculating the ratio of metabolites over the parent drug (Figure 6). While the overall drug metabolites’ levels were decreased by the pup Dhcr7+/– genotype, the metabolite/parent drug ratio was increased in the same samples. ARI turnover (D-ARI+2,3-DCPP)/ARI revealed a 40% higher turnover ratio in the liver of Dhcr7+/– animals when compared to their WT littermates, suggesting that the drug is metabolized at a higher rate in heterozygous animals (Dhcr7+/– pups: 0.21 ± 0.02 ng/mg protein vs WT pups: 0.13 ± 0.02; p = 0.0011) (Figure 6A). Similar results were obtained for CAR (DCAR + DDCAR + 2,3-DCPP)/CAR) with significantly higher turnover rates observed in Dhcr7+/– pups when compared to their WT littermates (Figure 6B). Drug turnover values for ARI and CAR grouped according to both maternal and embryonic genotypes are presented in Figures S2 and S3, respectively. Tables S1 and S2 depict the two-way ANOVA analysis assessing the contribution of both maternal and embryonic genotypes to the altered drug turnover. There are two possible explanations for these findings. First, the drug is metabolized independently across each tissue, and the higher turnover reflects tissue-specific metabolism. Alternatively, the turnover difference observed between WT and Dhcr7+/– animals is a result of hepatic activity, where the drug is converted into the metabolites in the liver and distributed by the systemic circulation to all tissues.
Figure 6.
Dhcr7+/– mice metabolize ARI and CAR faster than WT mice. ARI (A) and CAR turnovers (B) were calculated by determining the ratio of drug metabolites over the parent drug. WT and Dhcr7+/– pups are depicted in green and red, respectively. Opened symbols denote pups from WT mothers and filled symbols denote pups from Dhcr7+/– mothers. Bars correspond to the mean ± SEM; statistical significance: **p < 0.01; ***p < 0.0001. Note that the parent drug/metabolite ratio was increased in Dhcr7+/– pups across all tissues.
Cyp2d6 Protein Expression is Increased in the Liver of Dhcr7+/– Animals
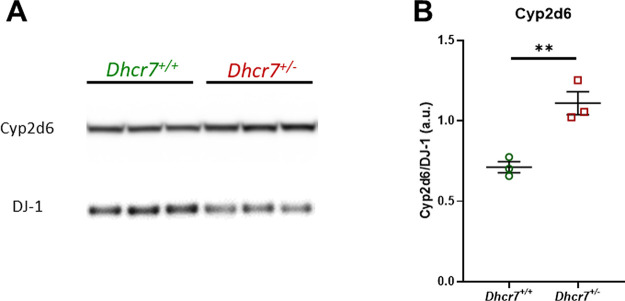
Cyp2d6 is a critical enzyme in the metabolism of both ARI and CAR (Figure 1).12,16,17 In order to determine if the increased metabolism of ARI and CAR was due to the increased level of the Cyp2d6 enzyme, Cyp2d6 protein expression was compared in WT and Dhcr7+/– livers of adult female mice (Figure 7). Three animals from each genotype were used for the western blot analysis. Cyp2d6 levels were normalized to the housekeeping protein DJ-1. We found that normalized Cyp2d6 levels were ∼40% higher in Dhcr7+/– animals when compared to WT (protein levels (a.u.): WT: 0.71 ± 0.06 versus Dhcr7+/–: 1.11 ± 0.07, p = 0.0075). Livers from adult male mice showed a similar pattern (Figure S4).
Figure 7.
Cyp2d6 protein expression is increased in the liver of adult Dhcr7+/– mice. (A) Western blot for Cyp2d6. Expression of housekeeping protein DJ-1 was used as a loading control. Each lane corresponds to the liver samples from different animals. (B) Optical density quantification of bands, with Cyp2d6 normalized to DJ-1. The values on panel B are shown as averages ± SEM of three biological replicates. **p < 0.01.
Human Fibroblasts with a DHCR7+/– Genotype Have Lower Drug Levels than WT Cells
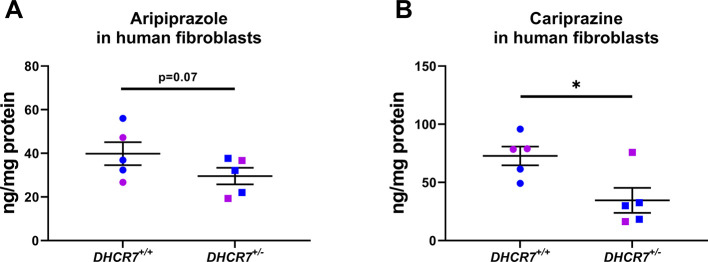
In order to gather an insight into the translational aspects of our transgenic animal observations, next, we treated five pairs of sex- and age-matched WT and DHCR7+/– human fibroblasts with ARI and CAR (Figure 8). After five days of treatment, ARI levels were lower in DHCR7+/– cells in comparison with WT (DHCR7+/– cells: 29.6 ± 3.8 ng/mg protein vs WT cells: 39.9 ± 5.3; p = 0.0757), although this difference did not reach statistical significance (Figure 8A). In contrast, CAR levels in DHCR7+/– cells were significantly lower than those detected in WT cells (DHCR7+/– cells: 34.6 ± 10.8 ng/mg protein vs WT cells: 72.8 ± 8.0; p = 0.0217) (Figure 8B). No sex or age differences were observed in the experiments with either ARI or CAR, but due to a limited sample size and statistical power, these findings cannot be considered conclusive.
Figure 8.
Human fibroblasts with a DHCR7+/– genotype have lower drug levels than WT cells. Cells were treated for five days with either ARI (A) or CAR (B) and the drug levels were determined by LC–MS/MS. Drug metabolites were below the limit of quantitation and could not be determined. Blue and pink symbols denote cells from male and female human donors, respectively. Each symbol corresponds to cell cultures derived from a different individual. Bars correspond to the mean ± SEM; statistical significance: *p < 0.05.
Discussion
Our findings can be summarized as follows: (1) animals with a Dhcr7+/– genotype have lower drug levels than WT controls when exposed to the same levels of DHCR7 inhibiting medications; (2) drug turnover is higher in heterozygous animals, suggesting a faster metabolism of both ARI and CAR; (3) the differential drug and metabolite levels can be observed across multiple tissues, primarily in the liver and brain; (4) the embryonic Dhcr7+/– genotype is the primary contributing factor to the final drug levels and turnover observed in the offspring; (5) a critical enzyme in ARI and CAR metabolism, Cyp2d6, is expressed at higher levels in Dhcr7+/– animals; and (6) ARI- and CAR-exposed human fibroblasts from heterozygous individuals have lower drug levels than controls.
Brain cholesterol synthesis is critical for neurodevelopment. Our studies were designed to investigate the effects of a Dhcr7 heterozygosity on drug metabolism in utero at a time when intrinsic sterol synthesis starts in the developing brain. Interestingly, many medications that inhibit the DHCR7 enzyme are commonly prescribed to pregnant women, underscoring the public health relevance of our findings.20−22 Many medications with a DHCR7-inhibiting side effect have been associated with a wide range of negative pregnancy outcomes, including spontaneous abortions, intrauterine death, and major or minor fetal malformations.11
Our results suggest that the different metabolism and turnover of ARI and CAR is a stable effect of the DHCR7 genotype and does not depend on the age or pregnancy: the findings were consistent across multiple tissues, human and mouse models, and at least two different ages (P0 and adult pregnant females).
The altered drug levels and turnover observed between WT and Dhcr7+/– mice can be at least partially explained by higher Cyp2d6 levels detected in the heterozygous animals. This enzyme is responsible for the dehydrogenation of ARI into dehydro-ARI as well as the demethylation of CAR into DCAR and DDCAR. Therefore, more enzyme translates into a faster metabolism, leading to lower drug levels and higher turnover. Importantly, the effect of maternal and embryonic Dhcr7+/– genotypes appears to be summative, as heterozygous pups born to heterozygous mothers had the least amount of drugs and the highest drug turnover rate.
It is estimated that CYP2D6 constitutes ∼3% of the total hepatic CYP metabolism and is responsible for the metabolism of ∼20% of the drugs.23,24 Antipsychotics, antidepressants, beta-blockers, antiarrhythmics, and several opioids are all metabolized by CYP2D6.17,25−28 The above presented findings, using ARI and CAR, could be looked upon as a proof-of-concept study: it is plausible that our observations with ARI and CAR could be extrapolated to multiple DHCR7-inhibiting drugs cleared by CYP2D6. This raises the question if in DHCR7+/– individuals, the dosage of CYP2D6-metabolized, DHCR7-inhibiting medications should be adjusted.
However, should this adjustment be warranted at all for the 1–3% of the human population who carry single-allele DHCR7? Perhaps not. Increasing the dose might result in a more appropriate therapeutic level across the tissues but is also likely to increase the side effects, as higher medication doses will result in increased DHCR7 inhibition. This would further elevate unwanted 7-DHC levels which are already elevated at the baseline in DHCR7+/– individuals. In turn, as 7-DHC is the most reactive known sterol, it would spontaneously give rise to many oxysterols.29−33 These 7-DHC-derived oxysterols are known to be toxic, and their further elevation by drugs in DHCR7+/– individuals might not be a wise course of action.
Finally, it is interesting to consider our observations as a protective mechanism of the body against a toxic insult. Elevation of 7-DHC and 7-DHC-derived oxysterols disrupts cellular homeostasis and the body responds by increasing clearance to minimize the toxic side effects. Should this be true, the feedback mechanism by which a defensive increased CYP2D6 expression might occur should be further investigated.
Materials and Methods
Chemicals
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Co (St. Louis, MO). HPLC-grade solvents were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). CAR was obtained from Sigma-Aldrich and dissolved in 0.9% saline solution for the experiments. ARI and d8-ARI were obtained from Sigma-Aldrich (St. Louis, MO).
Mice Studies
Adult male and female B6.129P2(Cg)-Dhcr7tm1Gst/J stock # 007453 mice were purchased from Jackson Laboratories. Mice homozygous for the Dhcr7Ex8 allele lack the exon 8 coding sequence and flanking splice acceptor site of the targeted gene, resulting in the truncated DHCR7 mutation most frequently observed in SLOS patients (IVS8-1G > C). Homozygous mice die shortly after birth. Dhcr7+/– mice are well, fertile, and indistinguishable from control, wild-type mice. Mice were housed under a 12 h light–dark cycle at constant temperature (25 °C) and humidity with ad libitum access to food (Teklad LM-485 Mouse/Rat Irradiated Diet 7012) and water in Comparative Medicine at the UNMC, Omaha, NE. The time-pregnant female mice received i/p injections of vehicle (VEH) or CAR (0.2 mg/kg) from E12 to E19. 18 WT and thirteen Dhcr7+/– mothers were used in our study. WT mothers were mated with Dhcr7+/– fathers and Dhcr7+/– mothers were mated with WT fathers, as described previously.3,4 Time-pregnant female mice received i/p injections of VEH or ARI (5 mg/kg) from E12 to E19. Half of each genotype group was injected with VEH and the other half with CAR. Similarly, eight WT and seven Dhcr7+/– mothers were used in our study and half of each genotype was injected with VEH and the other half with ARI. The mouse colony was monitored three times a day and all newborn pups (P0) were collected for dissection shortly after birth. All pups were born naturally; thus, we had no access to placental tissue. Adult female mice were killed after pups’ delivery. Both ARI and CAR doses were determined based on bioequivalency, as described previously, according to the formula of the animal equivalent dose (AED in mg/kg) = human dose (mg/kg) × Km ratio.34 The E12-E19 exposure window was chosen based on the time point when de novo cholesterol biosynthesis starts in the mouse brain.35 After dissection, frozen tissue samples were sonicated in ice-cold PBS containing butylated hydroxytoluene (BHT) and triphenylphosphine (PPh3). The aliquots of homogenized tissue were used for drug extraction and protein measurements. The protein was measured using BCA assay (Pierce). All procedures were performed in accordance with the Guide for the Humane Use and Care of Laboratory Animals. The use of mice in this study was approved by the Institutional Animal Care and Use Committee of UNMC.
Human Fibroblasts
All WT and DHCR7+/– human fibroblasts were described previously.6 All cultured human fibroblasts used were passages 5–18. All cells were subcultured once a week, and the culture medium was changed every two days. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose (25 mM), 1 mM l-glutamine, 10% fetal bovine serum, and penicillin/streptomycin at 37 °C and 5% CO2. For the drug exposure experiments, human fibroblasts were cultured in DMEM with 25 mM glucose, 1 mM l-glutamine, 10% delipidated fetal bovine serum (FBS), and penicillin/streptomycin. Medium was changed every 2 days during the course of the treatment. At the endpoint, cells were collected in ice-cold PBS and stored at −80 °C for further analyses.
LC–MS/MS (SRM) Analyses
Drug levels were determined as described previously.3,4 CAR and ARI levels were acquired in an Acquity UPLC system coupled to a Thermo Scientific TSQ Quantis mass spectrometer using an ESI source in the positive ion mode. A total of 5 μL of each sample was injected onto the column (Phenomenex Luna Omega C18, 1.6 μm, 100 Å, 2.1 mm × 50 mm) using water (0.1% v/v acetic acid) (solvent A) and acetonitrile (0.1% v/v acetic acid) (solvent B) as the mobile phase. The gradient was: 10–40% B for 0.5 min; 40–95% B for 0.4 min; 95% B for 1.5 min; 95–10% B for 0.1 min; and 10% B for 0.5 min. CAR and its metabolites were analyzed by selective reaction monitoring (SRM) using the following transitions: CAR 427 → 382, desmethyl-CAR (DCAR) 413 → 382, didesmethyl-CAR (DDCAR) 399 → 382 and 2,3-DCPP 230 → 187. The SRM for the internal standard (d8-ARI) was set to 456 → 293 and response factors were determined to accurately determine the drug levels. Similarly, ARI and its metabolites were analyzed by SRM using the following transitions: ARI 448 → 285, dehydroaripiprazole 446 → 285, 2,3-DCPP 230 → 187. The SRM for the internal standard (d8-ARI) was set to 456 → 293. Final drug levels are reported as ng/mg of protein.
Western Blot Analyses
Liver samples were homogenized by sonication in ice-cold RIPA lysis buffer (VWR International, Radnor, PA) plus phosphatase inhibitors (Sigma-Aldrich) and protease inhibitors (Thermo Fisher Scientific) and incubated on ice for 30 min. To clear the lysates, the samples were spun at 14,000 g at 4 °C for 5 min to pellet the debris. The protein concentration of the supernatant was quantified using the Bio-Rad BCA assay. Equal amounts of protein from each sample were mixed with the reducing reagent and loading buffer and heated to 70 °C for 10 min. Proteins were separated on NuPAGE 4–12% bis–tris protein gels (Thermo Fisher Scientific). Prestained protein ladder was used to evaluate the molecular weight. The Bio-Rad Mini Trans-Blot Electrophoretic Transfer Cell was used for the electrophoretic transfer using the polyvinylidene difluoride membranes (Immobilon-P PVDF Membrane, Sigma-Aldrich) and transfer buffer (25 mM Tris, 192 mM glycine and 20% (v/v) methanol (pH 8.3)). Following transfer, PVDF membranes were blocked in 5% milk in TBS (50 mM Tris-Cl, 150 mM NaCl, pH 7.5) with 0.05% Igepal (Spectrum Chemical, New Brunswick, NJ) and incubated in primary antibody overnight at +4 °C and secondary antibodies at room temperature for 1 h. Membranes were probed with the following primary antibodies: Cyp2d6 (Cell Signaling) and DJ-1 (Cell Signaling). Western blots were developed using Azure’s Radiance Substrate, imaged on Azure C300 with the cSeries Capture Software and saved as TIFF images (Azure Biosystems). The TIFF images were analyzed and quantified with AzureSpot.
Statistical Analyses
Statistical analyses were performed using Graphpad Prism 9 for Windows. Unpaired two-tailed t-tests were performed for individual comparisons between two groups. The Welch’s correction was employed when the variance between the two groups was significantly different. Two-way ANOVA analyses were performed to assess the contributions of the maternal and embryonic genotypes to drug metabolism and to test for any potential interactions between these variables. The p values for statistically significant differences are highlighted in the figure legends..
Acknowledgments
This work was supported by The National Institutes of Health NIMH MH110636 (K.M.), MN067234 (K.M.), and NICHD HD064727 (Z.K.). The authors also would like to thank the human fibroblast donors.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c05817.
Levels of CAR and its metabolites depend on maternal genotype; Dhcr7+/– mice metabolize ARI faster than WT mice; Dhcr7+/– mice metabolize CAR faster than WT mice; Cyp2d6 protein expression is increased in the liver of adult male Dhcr7+/– mice; ANOVA analysis of the ARI turnover in P0 livers; and ANOVA analysis of the CAR turnover in different tissues of P0 mice (PDF)
Author Contributions
Experimental design and research concept: T.C.G.-M., Z.K., and K.M.; cell culture experiments: T.C.G.-M. and Z.K.; animal injections: A.A. and Z.K.; mouse dissections, sample preparation, and drug measurement: T.C.G.-M., Z.K., L.A., and A.A.; protein measurements: A.A., L.A., and Z.K.; mouse colony maintenance: A.A.; LC–MS/MS analyses: T.C.G.-M. and L.A.; western blot analyses: T.C.G.-M.; statistical analysis: T.C.G.-M. and K.M.; funding: K.M.; draft of the manuscript: T.C.G.-M., Z.K., and K.M., and the final version was approved by all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Nes W. D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451. 10.1021/cr200021m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross J. L.; Iben J.; Simpson C. L.; Thurm A.; Swedo S.; Tierney E.; Bailey-Wilson J. E.; Biesecker L. G.; Porter F. D.; Wassif C. A. Determination of the allelic frequency in Smith-Lemli-Opitz syndrome by analysis of massively parallel sequencing data sets. Clin. Genet. 2015, 87, 570–575. 10.1111/cge.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Allen L. B.; Anderson A.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Maternal aripiprazole exposure interacts with 7-dehydrocholesterol reductase mutations and alters embryonic neurodevelopment. Mol. Psychiatr. 2019, 24, 491–500. 10.1038/s41380-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Anderson A.; Allen L. B.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Maternal cariprazine exposure inhibits embryonic and postnatal brain cholesterol biosynthesis. Mol. Psychiatr. 2020, 25, 2685–2694. 10.1038/s41380-020-0801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Xu L.; Lamberson C.; Haas D.; Korade Z.; Porter N. A. A highly sensitive method for analysis of 7-dehydrocholesterol for the study of Smith-Lemli-Opitz syndrome. J. Lipid Res. 2014, 55, 329–337. 10.1194/jlr.d043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Genaro-Mattos T. C.; Tallman K. A.; Liu W.; Garbett K. A.; Koczok K.; Balogh I.; Mirnics K.; Porter N. A. Vulnerability of DHCR7+/– mutation carriers to aripiprazole and trazodone exposure. J. Lipid Res. 2017, 58, 2139–2146. 10.1194/jlr.m079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif N. F.; Korade Z.; Porter N. A.; Harrison F. E. Oxidative stress, serotonergic changes and decreased ultrasonic vocalizations in a mouse model of Smith-Lemli-Opitz syndrome. Gene Brain Behav. 2017, 16, 619–626. 10.1111/gbb.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Ž.; Liu W.; Warren E. B.; Armstrong K.; Porter N. A.; Konradi C. Effect of psychotropic drug treatment on sterol metabolism. Schizophr. Res. 2017, 187, 74–81. 10.1016/j.schres.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Kim H.-Y. H.; Tallman K. A.; Liu W.; Koczok K.; Balogh I.; Xu L.; Mirnics K.; Porter N. A. The Effect of Small Molecules on Sterol Homeostasis: Measuring 7-Dehydrocholesterol in Dhcr7-Deficient Neuro2a Cells and Human Fibroblasts. J. Med. Chem. 2016, 59, 1102–1115. 10.1021/acs.jmedchem.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y. H.; Korade Z.; Tallman K. A.; Liu W.; Weaver C. D.; Mirnics K.; Porter N. A. Inhibitors of 7-Dehydrocholesterol Reductase: Screening of a Collection of Pharmacologically Active Compounds in Neuro2a Cells. Chem. Res. Toxicol. 2016, 29, 892–900. 10.1021/acs.chemrestox.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M. R.; Tatonetti N. P. Investigation of 7-dehydrocholesterol reductase pathway to elucidate off-target prenatal effects of pharmaceuticals: a systematic review. Pharmacogenomics J. 2016, 16, 411–429. 10.1038/tpj.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccia S. Pharmacokinetics and metabolism update for some recent antipsychotics. Expert Opin. Drug Metab. Toxicol. 2011, 7, 829–846. 10.1517/17425255.2011.575061. [DOI] [PubMed] [Google Scholar]
- Urichuk L.; Prior T.; Dursun S.; Baker G. Metabolism of atypical antipsychotics: involvement of cytochrome p450 enzymes and relevance for drug-drug interactions. Curr. Drug Metab. 2008, 9, 410–418. 10.2174/138920008784746373. [DOI] [PubMed] [Google Scholar]
- Renaud H. J.; Cui J. Y.; Khan M.; Klaassen C. D. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol. Sci. 2011, 124, 261–277. 10.1093/toxsci/kfr240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccia S.; Invernizzi R. W.; Nobili A.; Pasina L. A new generation of antipsychotics: pharmacology and clinical utility of cariprazine in schizophrenia. Ther. Clin. Risk Manage. 2013, 9, 319–328. 10.2147/tcrm.s35137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citrome L. Cariprazine: chemistry, pharmacodynamics, pharmacokinetics, and metabolism, clinical efficacy, safety, and tolerability. Expert Opin. Drug Metab. Toxicol. 2013, 9, 193–206. 10.1517/17425255.2013.759211. [DOI] [PubMed] [Google Scholar]
- Dean L.Aripiprazole Therapy and CYP2D6 Genotype. In Medical Genetics Summaries; Pratt V. M., McLeod H. L., Rubinstein W. S., Scott S. A., Dean L. C., Kattman B. L., Malheiro A. J., Eds.; National Center for Biotechnology Information: Bethesda, 2012. [PubMed] [Google Scholar]
- Kubo M.; Koue T.; Maune H.; Fukuda T.; Azuma J. Pharmacokinetics of aripiprazole, a new antipsychotic, following oral dosing in healthy adult Japanese volunteers: influence of CYP2D6 polymorphism. Drug Metab. Pharmacokinet. 2007, 22, 358–366. 10.2133/dmpk.22.358. [DOI] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Tallman K. A.; Allen L. B.; Anderson A.; Mirnics K.; Korade Z.; Porter N. A. Dichlorophenyl piperazines, including a recently-approved atypical antipsychotic, are potent inhibitors of DHCR7, the last enzyme in cholesterol biosynthesis. Toxicol. Appl. Pharmacol. 2018, 349, 21–28. 10.1016/j.taap.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile S.; Tofani S.; Bellantuono C. Aripiprazole and Pregnancy. J. Clin. Psychopharmacol. 2011, 31, 531–532. 10.1097/jcp.0b013e318222bc65. [DOI] [PubMed] [Google Scholar]
- Iqbal M. M.; Aneja A.; Rahman A.; Megna J.; Freemont W.; Shiplo M.; Nihilani N.; Lee K. The potential risks of commonly prescribed antipsychotics: during pregnancy and lactation. Psychiatry 2005, 2, 36–44. [PMC free article] [PubMed] [Google Scholar]
- Lutz U. C.; Hiemke C.; Wiatr G.; Farger G.; Arand J.; Wildgruber D. Aripiprazole in Pregnancy and Lactation. J. Clin. Psychopharmacol. 2010, 30, 204–205. 10.1097/jcp.0b013e3181d27c7d. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Antona C.; Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene 2006, 25, 1679–1691. 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- Tirona R. G.; Kim R. B.. Introduction to Clinical Pharmacology. In Clinical and Translational Science: Principles of Human Research; Robertson D., Williams G. H., Eds.; Academic Press: London, 2017; pp 365–388. [Google Scholar]
- Bertilsson L.; Dahl M.-L.; Dalén P.; Al-Shurbaji A. Molecular genetics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br. J. Clin. Pharmacol. 2002, 53, 111–122. 10.1046/j.0306-5251.2001.01548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic M. M.; Smith R. L.; Haslemo T.; Molden E.; Ingelman-Sundberg M. Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: a retrospective, cohort study. Lancet Psychiatr. 2019, 6, 418–426. 10.1016/s2215-0366(19)30088-4. [DOI] [PubMed] [Google Scholar]
- Dean L.Propafenone Therapy and CYP2D6 Genotype. In Medical Genetics Summaries; Pratt V. M., McLeod H. L., Rubinstein W. S., Scott S. A., Dean L. C., Kattman B. L., Malheiro A. J., Eds.; National Center for Biotechnology Information: Bethesda, 2012. [PubMed] [Google Scholar]
- Brandl E. J.; Tiwari A. K.; Zhou X.; Deluce J.; Kennedy J. L.; Müller D. J.; Richter M. A. Influence of CYP2D6 and CYP2C19 gene variants on antidepressant response in obsessive-compulsive disorder. Pharmacogenomics J. 2014, 14, 176–181. 10.1038/tpj.2013.12. [DOI] [PubMed] [Google Scholar]
- Xu L.; Davis T. A.; Porter N. A. Rate constants for peroxidation of polyunsaturated fatty acids and sterols in solution and in liposomes. J. Am. Chem. Soc. 2009, 131, 13037–13044. 10.1021/ja9029076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Mirnics K.; Bowman A. B.; Liu W.; Da J.; Porter N. A.; Korade Z. DHCEO accumulation is a critical mediator of pathophysiology in a Smith-Lemli-Opitz syndrome model. Neurobiol. Dis. 2012, 45, 923–929. 10.1016/j.nbd.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Xu L.; Mirnics K.; Porter N. A. Lipid biomarkers of oxidative stress in a genetic mouse model of Smith-Lemli-Opitz syndrome. J. Inherit. Metab. Dis. 2013, 36, 113–122. 10.1007/s10545-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Xu L.; Porter N. A. Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev. 2011, 111, 5944–5972. 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- Korade Z.; Xu L.; Shelton R.; Porter N. A. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. J. Lipid Res. 2010, 51, 3259–3269. 10.1194/jlr.m009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A.; Jacob S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tint G. S.; Yu H.; Shang Q.; Xu G.; Patel S. B. The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. J. Lipid Res. 2006, 47, 1535–1541. 10.1194/jlr.m600141-jlr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.