Abstract
Increasingly, quantitative lung computed tomography (qCT)-derived metrics are providing novel insights into chronic inflammatory lung diseases, including chronic obstructive pulmonary disease, asthma, interstitial lung disease, and more. Metrics related to parenchymal, airway, and vascular anatomy together with various measures associated with lung function including regional parenchymal mechanics, air trapping associated with functional small airways disease, and dual-energy derived measures of perfused blood volume are offering the ability to characterize disease phenotypes associated with the chronic inflammatory pulmonary diseases. With the emergence of COVID-19, together with its widely varying degrees of severity, its rapid progression in some cases, and the potential for lengthy post-COVID-19 morbidity, there is a new role in applying well-established qCT-based metrics. Based on the utility of qCT tools in other lung diseases, previously validated supervised classical machine learning methods, and emerging unsupervised machine learning and deep-learning approaches, we are now able to provide desperately needed insight into the acute and the chronic phases of this inflammatory lung disease. The potential areas in which qCT imaging can be beneficial include improved accuracy of diagnosis, identification of clinically distinct phenotypes, improvement of disease prognosis, stratification of care, and early objective evaluation of intervention response. There is also a potential role for qCT in evaluating an increasing population of post-COVID-19 lung parenchymal changes such as fibrosis. In this work, we discuss the basis of various lung qCT methods, using case-examples to highlight their potential application as a tool for the exploration and characterization of COVID-19, and offer scanning protocols to serve as templates for imaging the lung such that these established qCT analyses have the best chance at yielding the much needed new insights.
Introduction
Coronavirus disease 2019 1 has taken the world by storm. The disease is caused by a novel strain of coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The virus primarily affects the respiratory system and manifests as pneumonia, which frequently can be seen on imaging, especially CT. 2,3 The disease started from mainland China in 2019, and by the end of January 2020 was declared as a global health emergency by the World Health Organization. 4,5 The disease has highlighted the lack of preparedness of health care to deal with a new and emerging pandemic. 6,7 The therapeutic options, long-term effects, and methods to follow up the disease are still being studied. Given the public health concerns with the disease, any step that can help understand disease pathogenesis, improve the diagnosis, prognostication, or management of the disease will be critical. Currently, there are significant challenges and questions pertaining to the diagnosis and management of the disease. Some of the challenges include the choice of test for screening and diagnosis of the disease (especially when the reverse transcriptase-polymerase chain reaction (RT-PCR) tests have not been readily available or were unreliable), assessment of disease severity, triage of patients for a hospital vs home care, appropriate prognostication of individual patients, and assessing early response to therapy. 8 A recent statement from the Fleischner Society 9 has highlighted that key roles for imaging are to identify patients at risk of disease progression and to help triage moderate-to-severe cases for hospital vs home care. With PCR availability and accuracy improving, at this stage of the pandemic (as of October 2020), tools are needed that allow better characterization of the lung disease phenotypes leading to an enhanced understanding of the lung pathology and variable patient prognosis. Hence, in this practice and policy paper, we outline a set of well-utilized set of quantitative lung CT (qCT) metrics that can be applied to COVID-19 if basic principles of qCT scanning are followed.
We would add that, as we seek to identify interventions, there is a need for tools to detect successes early in the course of the disease rather than waiting to treat late. In addition, early studies have demonstrated that imaging will likely play an important, retrospective role in understanding the unique phenotypes of this disease 10 as well as understanding the residual pathologies upon recovery. 11
Understanding of the role of CT imaging in the disease is evolving. Initial reports suggested that chest CT may be more sensitive than the current tests of choice, such as RT-PCR. 12 These reports suggested that CT should be used for screening in symptomatic or suspected patients, but such an observation may be related to a lack of technical expertise with RT-PCR. Additionally, CT imaging can be very non-specific with regard to COVID-19, with features overlapping with other pathologies, such as influenza, bacterial pneumonia, or inflammatory drug reactions. Factors currently inhibiting the use of CT are concerns, which include poor imaging room ventilation, relative lack of personal protective equipment, difficult logistics involved in patient transportation to the CT facility, and the need for lengthy room disinfection after testing. Hence, major radiology organizations worldwide (American College of Radiology [ACR], Society of Thoracic Radiology, and American Society of Emergency Radiology [STR and ASER], British Society of Thoracic Imaging [BSTI], The Royal Australian and New Zealand College of Radiologists [RANZCR]) have suggested that CT should not be used for screening or diagnosis if reliable RT-PCR testing is available. The role of CT should be reserved for hospitalized, symptomatic patients with specific clinical indications for CT, and a portable chest radiograph should be preferred in the suspected patients. 13–16 However, the clinical manifestations frequently overlap with other diseases for which CT imaging is usually performed (i.e. chest pain, shortness of breath, fever in immunocompromised patients, rule-out pulmonary embolism etc.), and both chest CT and radiographic imaging are being performed among COVID-19 suspected or confirmed cases. As putative interventions emerge, qCT can provide a better understanding of disease pathogenesis and an early objective assessment of outcomes, allowing for continuance of the intervention or early termination.
The methodology of qCT allows for automated, reproducible, and quantifiable metrics that can measure normal and diseased lungs on CT images. Quantitative lung CT metrics have been primarily studied and validated for chronic lung diseases and, compared to a conventional review of images or pulmonary functional tests (PFTs), have been shown to provide a better characterization of distribution and percent of the lung affected by the disease 17 ; disease prognostication 18 ; prediction of worse clinical outcomes and mortality 18 ;as well as objective monitoring of early response to therapy. 19,20 Quantitative lung CT applications in inflammatory lung disease have included, for instance, chronic obstructive pulmonary disease (COPD), 21–25 interstitial lung disease (ILD), 26–28 or lymphangioleiomyomatosis (LAM). 29,30 However, qCT metrics have not been as well studied in acute lung diseases. Pulmonary applications of qCT in association with COVID-19 are a novel application, allowing the following of rapidly changing pulmonary disease processes. Quantitative features that can be applied to lung CT images are based on lung density, texture, and airway or vascular mapping. 28,31,32 These advanced lung imaging tools have been developed using a consistent imaging protocol that has been validated and applied in various multicenter studies. 33 The use of these validated advanced lung imaging tools can help better quantify the disease burden and may help triage the patients. Additionally, as COVID-19 patients with comorbidities or baseline lung disease 34 have a poor prognosis and are at higher odds for severe disease, qCT can help by objectifying the characterization of the underlying disease phenotypes associated with, e.g. COPD, asthma, ILD and most certainly have a new role to play in the acute and potentially more chronic state of the lung associated with COVID-19. These tools can help with better patient prognostication and aggressive management, when needed. In this practice and policy document, we highlight the applications of advanced lung imaging tools as applied to COVID-19 disease and their potential role. With the desire for objective quantification comes the need for standardizing imaging protocols to ensure accuracy of the resulting metrics and to allow for harmonization of image findings across centers, 33 which in turn will allow for the application of emerging technologies such as deep learning. 35–38
Current status of artificial intelligence (AI) and machine learning-based tools
As with the understanding of the disease itself, the role of AI (e.g. classical ML or deep learning methodologies) for COVID-19 pneumonia is evolving by utilizing the quantitative information from CT images and submitting these to various computer-based processes. Radiomics is a methodology that extracts a large number of features from imaging data that would not otherwise be apparent to visual inspection. Radiomics uses a data-characterization algorithm to bin the features into clusters that are most relevant for a specific clinical context. Such processes yield new insights into distinct disease phenotypes, which in turn can provide for an understanding of underlying pathology. A recent position statement by RANZCR briefly addresses this topic and highlights that AI research and technological imaging advances for their added role in the clinical care of patients should be supported. 15 A few critical but very important points highlighted in the position statement are that quantitative image analysis tools should add value to visual image inspection by providing a better understanding of the disease rather than simply targeting a reduction in workload or turnaround times as those are currently not the limiting factors. Another vital step towards the usefulness of quantitative image analysis tools is generalizability to different population groups and integration into the existing clinical practices.
In the current stage of the pandemic (as of October 2020), there is increasing utilization of CT imaging in patients with COVID-19 pneumonia. Based on the experience with the use of quantitative analysis tools for multiple other lung pathologies, there is a potential for valuable applications in COVID-19 disease. The initial reports on applications of AI-based tools for COVID-19 disease are promising. 5,39–42 Some of the deep learning-based methods are focused on improving the sensitivity or diagnostic accuracy for COVID-19, which are new and have scant (if any) validation data for their generalized or wide-spread applicability. 5,39,43 In this work, we will focus on established qCT methodologies that have been validated and utilized in various multicenter studies in collaboration with our research laboratory over last two decades. 31,32,44–51 These methodologies have been combined with various statistical and other machine-learning analyses, imparting added insights into lung diseases. If appropriately used, these methods can provide direct insights into the disease severity, distribution, progression, and regression as well as identify underlying variations (sub-phenotypes such as parenchyma vs vasculature) in pathology, which may not be otherwise apparent. The development and testing of the generalizability of these tools require the use of standardized imaging protocols. By organizing such commonality of imaging practices, we have an opportunity to utilize this crisis to bring radiology fully into the emerging era of computer-assisted objective, quantitative image analysis. 52
Quantitative lung CT imaging tools
Density and histogram-based disease quantification
The attenuation values of voxels on the lung CT vary between −1000 Hounsfield units (HUs) (air), 0 HU (water), and +1800 HU (cortical bone) and is related to the tissue characteristics. Various lung diseases can be objectively measured using quantification of voxel attenuation. The image feature to be quantified is based on the type of abnormality on the CT image (low attenuation area [LAA] vs high attenuation area [HAA]). Because the lung is composed essentially of air and tissue with just two HU values, the HU value of a voxel allows one to determine the percent air and percent “tissue” (blood, tissue, and extracellular fluid). 53,54 As regional inflammation progresses, voxel “tissue” volume increases. Thus, short-term (weeks or months) longitudinal changes in tissue volume can serve as an index of progression or regression of inflammation, since it would not be expected that actual parenchymal volume would change acutely. Emphysema quantification is performed by measuring the LAAs. Initially, for emphysema quantification, a cut-off HU unit value of −910 HU was suggested (areas having an attenuation of −910 HU or less characterized as emphysema). 55 It has now been demonstrated that the cut-off value of −950 correlates better with the pathological finding of emphysema, 56,57 while −910 HU 58 captured emphysema at earlier stages of the pathology. Other studies suggest that a threshold of −960 or −970 HU can be used 59,60 ; the differentiation between these thresholds is mostly immaterial. 61 One needs to remain consistent, and −950 HU for advanced disease and −910 HU for early disease processes remain widely accepted. 62,63 CT has been used to identify the COPD phenotypes which were not identifiable by the diagnostic standard PFT. COPD phenotypes identifiable by CT are also demonstrated to have similar clinical symptoms and prognosis, 63,64 and the presence of emphysema-like voxels in subjects with normal PFTs has been found to be a predictor of future PFT changes. When quantified, the degree of emphysema is associated with poor exercise capacity, 63 have poor overall health-status, 65 and higher mortality. 66 Identification of COPD phenotypes have also allowed management of individuals based on patterns of disease that are only identifiable by CT. 67 While attenuation values of less than −950 HU helps to quantify emphysema, quantification of the lung density voxel histogram has been demonstrated to characterize fibrotic lung processes 68 and areas with attenuation between −250 and −600 (HAAs) has been used to identify fibrotic-like lung. 69 Because normal lung has some voxels below −950 HU and voxels between −250 and −600 HU, large population-based studies have been used to establish normative equations taking into account sex, age, race, scanner make and model etc. to provide a threshold above which one can consider an individual as having the presence of emphysema or fibrosis. 21,70 This has been extensively studied in the context of interstitial lung diseases, where the HAA corresponds to the areas of inflammation and fibrotic lung processes. 71–73 These HAA are also shown to correlate with worse patient health status and poor outcomes. 71,72,74,75 In the MESA Lung study, 73 subjects with HAA areas on CT were found to have higher serum inflammatory markers, poor respiratory function, and increased overall mortality. However, due to physiological changes in the lungs with inspiration and expiration, adherence to a standard imaging protocol is imperative to allow disease quantification and prevent errors due to variability in interpretation by the radiologists. Studies have shown that submaximal inspiration leads to an underestimation of emphysema 76 and overestimation of HAA.
In patients with COVID-19 pneumonia, bilateral patchy peripheral ground glass opacity (GGO) and consolidation are typical findings. 77 These areas are represented by HAAs, 78 which have been validated for quantification and utility in various studies on lung diseases, especially interstitial pulmonary fibrosis. HAA can be quantified using a density- or histogram-based technique with a threshold of −250 to −600 HU (Figure 1, middle column). As in other lung diseases, disease quantification may allow objective determination of disease severity, better patient prognostication, and can also help with patient management. This tool can be used for objective quantification of the disease and can also help in finding cut-offs for disease severity on imaging that can complement clinical assessment to triage patients for care strategies during the current pandemic and in the future. Some early studies using density-based disease quantification have shown promise. In a single-center study of 262 patients with COVID-19 infection, 79 the quantification of the compromised lung (defined as lung with HU between −500 and +100 HU) showed a significant correlation with the need for oxygenation support, intubation, and in-hospital death. In this study, it was also demonstrated that the %compromised lung between 6 and 23% was associated with oxygenation therapy, >23% was associated with the need for intubation, and the quantification of disease lung had a negative correlation with PaO2/FiO2 ratio on arterial blood gas (ABG) analysis. In another 79 patient study, 80 quantification of lung inflammation on CT (cut-off value >14.2%) correlated with decreased PaO2/FiO2 ratio based on ABG. In another study with 236 patients with COVID-19 infection, 81 automated density-based quantification of the well-aerated lung (WAL, defined as lung with HU between −950 and −700 HU) was performed. A higher percentage of WAL correlated with better clinical outcomes, and lower WAL correlated with significantly higher odds of ICU admission and death. When quantification of WAL from CT was added to the clinical prognostication markers, the diagnostic performance of the models significantly improved. These early feasibility studies highlight the potential of CT-based disease quantification for triage and prognostication of patients with COVID-19 pneumonia.
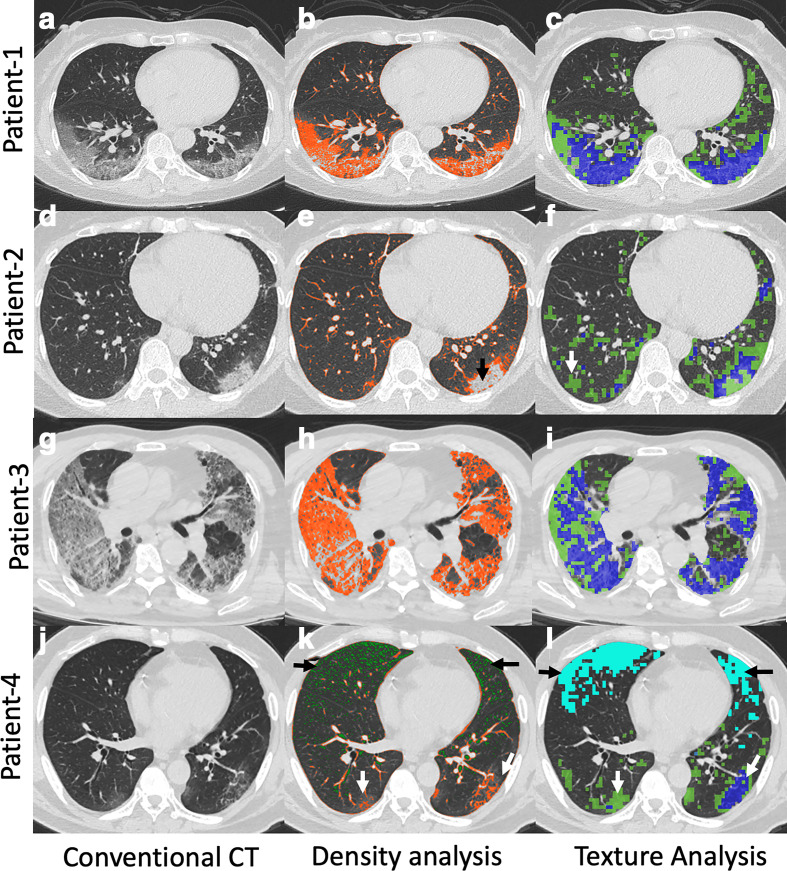
Figure 1.
Conventional CT (first row), lung density (second row), and 3D AMFM Texture (third row) quantification in four patients with COVID-19 pneumonia. Patient 1: 67-year-old female with conventional CT (a) demonstrating posterior bilateral lower lobe mixed GGO and interstitial thickening. Density analysis (b) demonstrating HAAs (orange color) which was quantified as 14%. 3D AMFM Texture analysis (c) demonstrating abnormal lung (green color = GGO; blue color = GGO with reticular thickening) which was quantified as 27% (GGO,16%; GGO with reticular thickening, 11%). Patient 2: 46-year-female with conventional CT (d) demonstrating left more than right high-attenuation posterior GGO. Density analysis (e) underestimated the disease (quantified as 8%) because of relatively high attenuation in the central portion of the abnormality (black arrow). However, 3D AMFM Texture analysis (f) quantified the entire extent of disease, characterized it as predominantly GGO (green, 15%) with intermixed areas of interstitial thickening (blue, 3%), and also highlighted certain areas in the right lung (white arrow) that did not have identifiable disease on conventional CT. Patient 3: Adult male patient with COVID-19 pneumonia, conventional CT (g) demonstrating extensive bilateral disease with intermixed areas of GGO and interstitial thickening. Density analysis (h) showing HAA (orange color) quantified as 46% and 3D AMFM Texture analysis (i) quantifying the amount of GGO (green color, 31%) and mixed GGO and interstitial thickening (Blue, 35%). Patient 4: Conventional CT (j) demonstrating bilateral lower lobe GGO intermixed with areas of interstitial thickening with disease more in left. No abnormality is appreciated in the anterior lung on conventional CT. Density analysis (k) showing HAA (orange, white arrows) quantified as 6% with low attenuation areas in the anterior lung (green color in K, black arrows, 5%). 3D AMFM Texture analysis (l) also demonstrated abnormality corresponding to a region of hyperlucency (light blue, black arrows, 10%) in the anterior lung with better quantification of disease (GGO 8%, GGO with reticular thickening 2%) in the posterior lungs as compared to density analysis (white arrows in L). hyperlucent areas, which visually do not appear to be emphysematous, (detected as a texture abnormality) are possibly due to microemboli-induced reduction in regional blood volume. Despite relatively less lung involvement, this patient had a protracted ICU course and died on Day 25. AMFM, adaptive multiplefeature method; GGO, ground glassopacity; HAA, high attenuation area.
Texture analysis
Computational methods for texture have been developed, and their utility has been analyzed in multiple diseases, including COPD, ILD, and lung cancer. CT-based texture assessment is automated and can help differentiate between normal and abnormal lung quantitatively. Furthermore, it can characterize and quantify the abnormality into various disease or imaging patterns. Texture-based methods make use of dozens of mathematical formulations of grayscale patterns within the CT image, and can be used in combination with various forms of supervised or unsupervised machine learning to reduce a regional multidimensional set of textures down to several textures serving to define a parenchymal classification. Some of the expected patterns which AI-based texture software can be trained to classify include: normal aerated lung, normal bronchovascular bundles, GGO, mixed ground-glass and reticular disease, consolidation, and honeycombing. Quantitative lung CT-based texture analysis can be obtained on a two-dimensional (2D) or a three-dimensional (3D, isotropic voxels) data set. An earlier 2D texture-based classification method applied to the lung was the adaptive multiple feature method (AMFM), which utilized observer assigned classifications to 2D regions of interest combined with a Bayesian classifier to convert combinations of texture types into a parenchymal descriptor. 17,82,83 This form of development of automated learning methodology is known as ‘supervised’ ML. Such a development relies on a standardized training set and consistent labeling of the normal and diseased lung. The 2D texture analysis was later extended to a more sensitive and specific 3D AMFM method. 84,85 CT texture-based analysis has been shown to identify and differentiate, e.g. normal lung, respiratory bronchiolitis, emphysema, and honeycombing. There is increasing evidence in support of CT texture-based methods for investigating clinical questions. Among studies of smoking subjects, 3D AMFM texture-based classification has, as an example, been shown to differentiate a non-smoker and a smoker lung from a pool of subjects with normal PFT. 85 For COPD evaluation, various multicenter research studies have utilized texture methodologies to objectively characterize parenchymal pathologies and correlate them with outcomes. 74,86–89 For interstitial lung diseases, texture-based quantification of imaging abnormalities has been shown to be useful to predict disease progression among patients with idiopathic pulmonary fibrosis. 18
In patients with COVID-19 pneumonia, texture analysis can differentiate normal from abnormal lung (providing a score representing the percentage of the normal lung) by characterizing abnormal lung as various imaging patterns such as GGO, mixed GGO/interstitial, and consolidation (Figure 1, right column). Of interest, a portion of this patient’s ventral lung was labeled by the AMFM as having “emphysema-like” features. Visually, the lung did not appear emphysematous, but the density measurements fell below −950 HU. It is possible that such regions (also observed in other COVID patients) may have a reduced regional blood pool related to emerging reports of microemboli. 90–92 Such examples suggest that CT texture analysis in COVID-19 patients may help quantify the imaging patterns and potentially explain low arterial oxygenation when visual assessments of disease extent fail. These observations may help identify distinct disease phenotypes (i.e. lung parenchymal disease vs vascular disease). As depicted in Figure 2, both texture analysis and density-based methods provide for an objective, quantitative assessment of progression or regression of disease. As can be seen in both Figure 1 and Figure 2, the density-based method misses the inclusion of dense consolidation with attenuations greater than −250 HU. However, if the threshold range is expanded to include the consolidation, normal bronchovascular bundles are included with the regions of pathology, leading to overestimation of the size of diseased regions. This is avoided by the use of the texture-based approach. As demonstrated in Figure 2, texture can be used to quantify changes in parenchymal involvement over time, offering a sensitive quantitative assessment of disease progression, regression, and, thus, response to therapy even when such changes are not apparent on subjective visual review of images. In this particular example, in addition to quantitation of texture types, the change in the non-air volume of the lung serves as a measure of changing inflammatory burden.
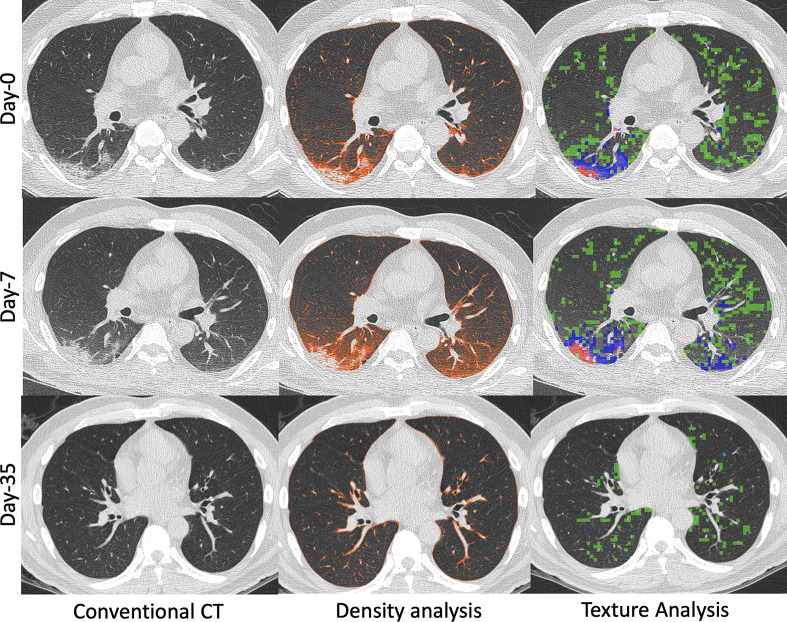
Figure 2.
55-year-old male with COVID-19 pneumonia that received imaging at presentation (Day 0), Day 7 of hospitalization, and Day 35 of hospitalization. Images generated at initial presentation (Top row, Day 0) along with computed density and texture analysis demonstrating predominantly posterior bilateral lower lobe disease (HAAs quantified as 13% on density analysis and abnormal lung quantified as 24% on texture analysis). The texture analysis characterized the disease as GGO (green, 21%), mixed GGO and interstitial (blue, 3.9%), and consolidation (red, 0.1%). Follow-up CT (Middle row, Day 7) obtained due to worsening shortness of breath demonstrated minimal disease progression on conventional CT images and density analysis and texture analysis provided objective assessments of disease worsening (HAA on density analysis quantified as 18% and abnormal lung quantified as 29% on texture analysis). Progression of imaging disease type was also quantified (GGO, green: 22.5%; mixed GGO and interstitial, blue: 6.3%; and consolidation, red: 0.2%). Another follow-up CT (Bottom row, Day 35) obtained after clinical improvement and prior to discharge from the hospital demonstrated complete resolution of disease on conventional images. The density and texture analysis identified some residual abnormalities (HAA 5% on density analysis and abnormal lung 6% on texture analysis). GGO, ground glassopacity; HAA, high attenuation area
Summary or topographic multiplanar reformatted views
While the review of conventional CT images (axial, sagittal, and coronal) is paramount for the diagnosis of lung injury, certain advanced image visualization techniques have the potential to help with the interpretation and improve the turn-around-time in challenging times like this. Topographic multiplanar reformatted (tMPR) 93 views, (VIDA Diagnostics, Coralville, Iowa) provide a comprehensive view of the airway tree and associated parenchyma in a single plane. These are developed using a tMPR technique that warps and flattens non-overlapping airways and their local environment. The warped parenchymal display can be further thickened into a maximum intensity projection (MIP). 94 The tMPR views are different from conventional MPR views (commonly generated at imaging workstations) as the tMPR view flattens the airway tree along with associated parenchyma, highlighting the association between affected parenchyma and the feeding bronchial path. This is not achieved with conventional MPR techniques. Unlike the MPR, the tMPR view requires a series of automated but complex processing steps, including airway segmentation and branching structure extraction. By capturing mid-coronal as well as left and right sagittal views this way, the tMPR views allow the review of typical bronchocenteric imaging patterns, 77 as well as the location of disease in one coronal and two sagittal snapshots (Figure 3). Additionally, this technique can be applied to the quantification of the disease presence within the individual sub lobar segments of the lungs (Figure 4), enabling objective metrics related to functional compromise and the means for follow-up, with important implications for subphenotyping and for the assessment of disease interventions. The technique thus has the potential to assess the correspondence of parenchymal vs airway pathology. Such displays, as has been seen with simple MIP or other sliding slab displays, 95 may also improve the interpretation efficiency, and can be useful for rapid evaluation of changes over time using serial scans for disease comparison. Unlike other thick or sliding slab views, the tMPR view lays out a significant portion of the airway tree and its associated parenchyma in a single flattened depiction. In Figure 4, note that all of the major segmental airways are present within a single coronal view. Similarly, the right or left sagittal oriented views demonstrate the full set of right or left segmental airways.
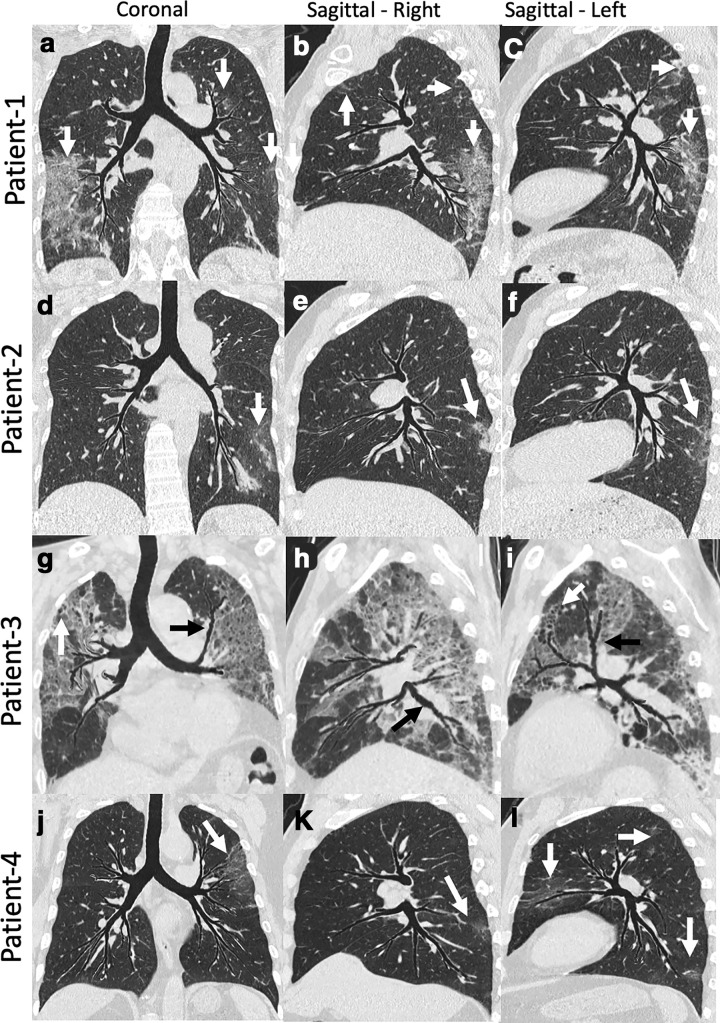
Figure 3.
tMPR view images of four patients with COVID-19 pneumonia. The tMPR view warps the airway tree such that most major subsegmental airway segments reside within a single flattened projection, and associated parenchyma and vasculature are similarly warped. Patient 1: Coronal (a) and Sagittal (b, c) images showing multifocal, predominantly peripheral bilateral ground glass opacification (white arrows). These images provide comprehensive visualization of airway and overall extent of disease. Patient 2: Coronal (d) and sagittal (e, f) images showing bilateral peripheral mixed ground glass opacity and reticular thickening (white arrows). The coronal image demonstrates left-sided disease, but the right sagittal image demonstrates posterior lower lobe disease as well. Patient 3: Coronal (g) and sagittal (h, i) images demonstrating extensive bilateral upper and lower lobe disease. The patient had a protracted course and the imaging was obtained later in the time course. Images show bilateral central bronchiectasis (black arrows) and distal bronchiolectasis (white arrow). Patient 4: Coronal (j) and sagittal (k, l) images demonstrate multifocal (white arrows) left-side-predominant disease. Review of these three images provide a good appreciation of localization and the relationship with the airway paths. tMPR, topographic multiplanar reformatted.
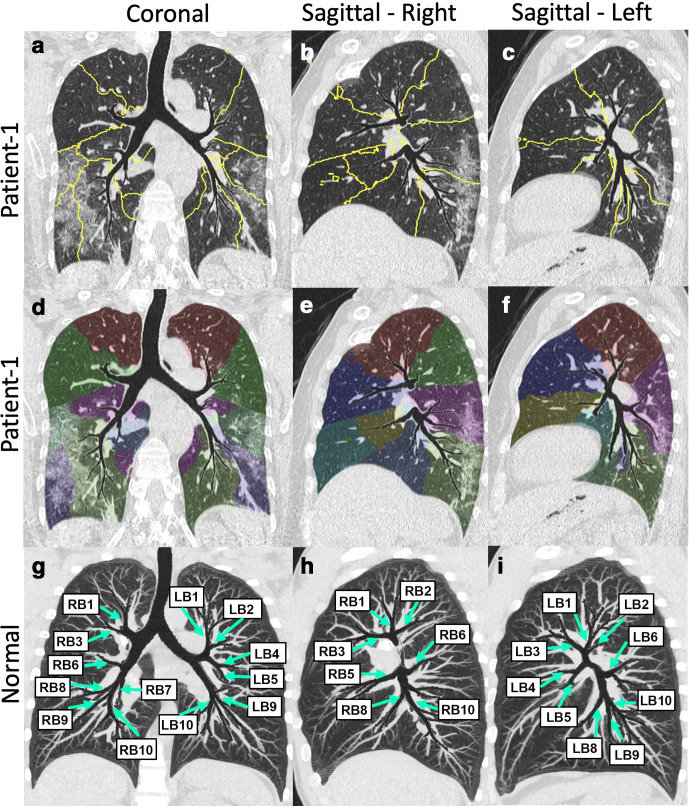
Figure 4.
AI-based delineation of sublobar segments based on regions of influence of the airway branches. Shown here are the tMPR views of Patient 1 shown in Figures 1 and 3. Top row coronal (a) and sagittal (c, d) images showing outlines of the sublobar segments within the tMPR views. Middle row again shows the coronal (d) and sagittal (e, f) oriented tMPR views highlighting the sublobar segments via use of transparent color coding. Bottom row shows tMPR view of a normal subject. The views are thickened by use of a maximum intensity projection with regions of the airways protected from overlapping structures. This serves to put the airways and vessel branching in context of each other. By labeling the airway segments, we emphasize that the full segmental tree has been included within a single, flattened view. AI, artificial intelligence;tMPR, topographic multiplanar reformatted.
Inspiratory and expiratory imaging for assessment of functional small airways disease (fSAD) representing regional air trapping
Due to the difficulties of breath-holding at low lung volumes, expiratory imaging may not be possible in acutely ill patients. However, it offers utility in the post-COVID-19 assessment of residual lung damage. Parametric response mapping (PRM) 96 and disease probability map (DPM) 97 utilize image registration to map inspiratory and expiratory images together. In emphysema, the PRM has been useful to identify the regions of air trapping on full expiration based on density thresholds which, may not be identified from an image obtained at full inspiration. The DPM also uses image matching between full inspiration and full expiration to identify hyperinflated regions and regions of fSAD. However, the warping function must be used to characterize regional volume changes together with the density threshold to identify regions of fSAD. In acute respiratory distress syndrome (ARDS) and other forms of acute lung injury requiring invasive mechanical ventilation, the PRM has been shown to help identify regions at increased risk for atelectrauma and ventilator-induced lung injury, based on voxels transitioning between high and low density during a breath. 98 Because of the highly contagious nature of COVID-19, and because of the respiratory difficulties of acutely ill COVID-19 patients, the use of only an inspiratory scan is preferred. However, PRM and DPM methodologies, used in the early stages of the mild-to-moderate disease or used to follow early interventional changes in the more normal lung regions, may still provide important quantitative information.
Suggested protocol and imaging guidelines
The development and validation of qCT rely on standardized imaging protocols, as various technical factors like voxel size, image kernel can affect various parameters. As the CT technology is evolving, and there are multiple vendors with varying nomenclature, having a single universal protocol is not possible. However, if there is a similar principle for setting protocols which derive similar images between different vendors, quantitative lung imaging, and refined imaging data collection for AI or deep learning across vendors becomes possible. The principles for low dose qCT include:
Coaching the patient to full inspiration (total lung capacity).
Non-contrast scanning.
Use of 3D capable thin slices (to accommodate current clinical practices trying to limit the number of slices, we recommend 1.25 mm sections with 1 mm slice increments).
A quantitative image reconstruction kernels (avoiding sharp kernels which alter the HU at edges of structures and accentuate noise). In the case of Siemens, this is a Qr 40 or B30f, and for GE and Philips, this is ‘standard reconstruction’ algorithm.
Use of more modern iterative reconstruction algorithms (ADMIRE or SAFIRE on Siemens, ASiR-V on GE, iDose on Philips).
Use of the most current dose modulation implementations of the various manufacturers.
A coached full expiration [residual volume (RV) scan, if taking advantage of PRM and DPM analysis]. This technique has the potential to be relevant for longitudinal studies in COVID-19 survivors but may not be possible in acutely ill patients.
Table 1 summarize suggested CT imaging protocols for a single source and a dual-source Siemens, GE, and Philips scanner, respectively, based upon slight modifications to protocols developed for the more recent phases of several ongoing NIH-sponsored longitudinal multicenter studies, including SPIROMICS, 99 MESA Lung, 100 and PrecISE. 101 Using the Siemens SOMATOM Force as the reference scanner, low-dose protocols have been established based upon the lowest dose, which does not interfere with the targeted airway and parenchymal metrics. 102,103 The protocol was developed so as to achieve a common image noise and spatial resolution across scanner types as judged by imaging a modified anthropomorphic phantom produced by Kyoto Kagaku 104 and inserts for assessment of density and modulation transfer functions produced by the Phantom Labs. 105 A protocol for a scanner vendor or model not mentioned in Table 1 can be made using the above-stated principles for qCT lung scanning. Protocols that allow for consistent qCT metrics have been detailed in our previous work, 106 but a minor difference is suggested in this protocol for COVID-19 to ensure integration with the existing clinical workflow. The slice thickness suggested is 1 mm (as compared to sub-mm), due to the technical challenges associated with a large number of CT images for clinical interpretation and storage requirements. The qCT metrics detailed in this work have been studied and validated on non-contrast images. Hence, if contrast-enhanced CT is performed for clinical indications (like rule out pulmonary embolism), the addition of low dose chest CT based on the suggested protocols will be useful for post-processing using qCT methods. Rapidly increasing literature highlights the clinical potential of qCT applications in COVID-19 pneumonia, and reiterates the need for standardization of lung CT imaging, such that these efforts can be applied to diverse populations, demographics, institutions, and scanner models.
Table 1.
CT Chest protocols for quantitative lung imaging
| Siemens Definition AS+ | Siemens FORCE | GE CT750 HD | GE Revolution | Philips iCT | Canon Genesis | Canon PrimeSP |
|
|---|---|---|---|---|---|---|---|
| Intravenous contrast | No | No | No | No | No | No | No |
| Detector configuration (number x mm) | 128 × 0.6 | 192 × 0.6 | 64 × 0.625 | 128 × 0.625 | 128 × 0.625 | 80 × 0.5 | 80 × 0.5 |
| SOV | – | – | Large body | Large body | – | Large | Large |
| Organ characteristic | Thorax Ensured by first selecting a Siemens default routine adult chest protocol |
Thorax Ensured by first selecting a Siemens default routine adult chest protocol |
– | – | – | – | – |
| Rotation time (s) | 0.5 | 0,025 | 0.5 | 0.5 | 0.5 | 0.275 | 0.35 |
| Scanner Pitch | 1.0 | 1.0 | 0.984 | 0.992 | 1.0 | Standard | Standard |
| kVp | 120 (Automated kV control – off) | 120 (Automated kV control – off) | 120 | 120 | 120 | 120 | 120 |
| Tube current modulation | Care dose: on | Care dose: on | AutomA & SmartmA: On | AutomA & SmartmA: On | DoseRight On | SureExposure On | SureExposure On |
| Quality reference mAs – Software v. 2012B, VA48, and VA50 | 42 | 36 | – | – | – | – | – |
| Quality reference mAs – (VB10 & VB20) | 29 | 25 | – | – | – | – | – |
| Max mA | – | – | 800 | – | 700 | 700 | |
| mA | – | – | – | SmartmA 30–500 | – | 20–700 | 20–700 |
| Noise index | – | – | 85 | 61 | – | 12.5 | 12.5 |
| DRI | – | – | – | – | 8 | – | |
| Slice thickness x interval (mm) | 1 × 0.5 | 1 × 0.5 | 1 × 0.5 | 1 × 0.5 | 1 × 0.5 | 1 × 0.5 | 1 × 0.5 |
| Reconstruction Kernel | Q30f | Qr40 | Standard | Standard | B (Standard) | Lung Standard | Lung Standard |
| Iterative algorithms | SAFIRE 5 | ADMIRE 5 | ASiR-V 60% | ASiR-V 60% | iDose 6 | AIDR3De | AIDR 3De |
| Reconstruction mode | – | – | Helical Plus | Helical Plus | – | – | |
| Additional filters | – | – | IQ Enhance OFF | IQ Enhance OFF | – | – | |
| Approximate dose (for a 30 cm scan, average-size adult) | CTDIvol: 2.46 mGy Effective dose: 1.65 mSv |
CTDIvol: 2.21 mGy Effective dose: 1.49 mSv |
CTDIvol 4.87 mGy Effective dose: 3.27 mSv |
CTDIvol: 3.38 mGy Effective dose: 2.27 mSv |
CTDIvol: 3.5 mGy Effective dose: 2.35 mSv |
CTDIvol: 2.4 mGy Effective dose: 1.61 mSv |
CTDIvol: 2.4 mGy Effective dose: 1.61 mSv |
CTDIvol, volume computed tomography dose index; DRI, dose right index ; SOV, scan field of view.
Potential of qCT imaging research for COVID-19
Based on extensive data on multiple lung diseases described, early feasibility studies on qCT metrics among COVID-19 pneumonia patients, and cases shown in this practice and policy document, the use of previously validated CT acquisition protocols and qCT imaging has great potential for research allowing a better understanding of the disease. Quantitative lung CT metrics may have a role in identifying individuals that are at a higher risk of lung disease progression as compared to individuals that are at lower-risk and hence can be managed by home care. Other potential roles for qCT metrics include better characterization of disease phenotype and how treatment may be optimized for individual patients. Delineation of bronchocenteric disease patterns and their quantification in the sublobar segments using tMPR or summary views may enhance diagnosis and disease quantification. Due to heterogeneous nature of the COVID-19 disease with varied lung affection among patients, there is an ongoing debate as to whether distinct clinical phenotypes of COVID-19 exist based on respiratory compliance 107 or CT imaging features, or if the lung disease is distributed along a continuum of severity similar to ARDS. 108,109 Given such wide distributions in the severity of the disease, voxel attenuation characteristics on CT imaging, ventilatory responsiveness, and time between onset and clinical presentation, qCT can play a significant role in assessing the response to treatment early and objectively, and help resolve such debates. Newly emerging radiomics methods, combined with supervised ML, has been considered a next step in line with qCT in the field of oncology, 110–114 whereby the growing list of qCT metrics yield disease insights. Beyond oncology, there are a growing number of pulmonary applications that employ ML methodologies to qCT-derived metrics to demonstrate patient clusters with common clinical characteristics. 50,115–123 Use of qCT metrics for featurization upstream of ML algorithms has the potential to enhance the positive impact of AI on medical imaging.
Summary
Imaging in COVID-19 pneumonia is challenging, but automated qCT can help beyond widely used qualitative subjective disease assessment. Quantitative lung imaging applications are fast and automated. They can help provide a reproducible objective assessment. Texture analysis can help to classify and quantify the radiological abnormality. Standardized lung CT acquisition is vital to apply qCT algorithms. Based on our experience in the role of qCT imaging in multiple other lung diseases, it is very likely that objective quantification of COVID-19 pneumonia and identification of its imaging phenotypes may have a role in diagnosis, patient triage, and predicting outcomes. The clustering of patients with common phenotypes and similar clinical outcomes may augment treatment decisions and validation of future clinical trials.
Footnotes
Acknowledgments: Authors would like to acknowledge VIDA Diagnostics, Coralville, Iowa, USA for their contribution of the tMPR view images.
Conflict of Interest: Eric Hoffman, PhD is a founder and shareholder of VIDA Diagnostics, a company commercializing lung image analysis software developed, in part, at the University of Iowa. Junfeng Guo, PhD is a shareholder of VIDA Diagnostics. Sam Peterson, PhD is an employee of VIDA Diagnostics. David Kaczka, MD, PhD has received a research grant from ZOLL Medical Corporation, and is a co-founder and shareholder of OscillaVent, Inc.
Funding: In part by NIH R01-HL-130883.
Contributors: PN, JG and EAH analyzed the cases for quantitative CT imaging. JKL, KBK, CHL and KMS provided the images and clinical data. All authors were responsible for manuscript writing and editing.
Contributor Information
Prashant Nagpal, Email: prashant-nagpal@uiowa.edu.
Junfeng Guo, Email: junfeng-guo@uiowa.edu.
Kyung Min Shin, Email: kyung-shin@uiowa.edu.
Jae-Kwang Lim, Email: limjaekwang@gmail.com.
Ki Beom Kim, Email: givemesunn@naver.com.
Alejandro P Comellas, Email: alejandro-comellas@uiowa.edu.
David W Kaczka, Email: david-kaczka@uiowa.edu.
Samuel Peterson, Email: speterson@vidalung.ai.
Chang Hyun Lee, Email: changhyun.lee.snuh@gmail.com.
Eric A Hoffman, Email: eric-hoffman@uiowa.edu.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–33. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. ATS Committee on Proficiency Standards for Clinical Pulmonary Function Laboratories . ATS statement: guidelines for the six-minute walk test. Am J Respir Crit Care Med 2002; 166: 111–7. doi: 10.1164/ajrccm.166.1.at1102 [DOI] [PubMed] [Google Scholar]
- 3. Li Y, Xia L, Disease C. COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol 2019; 2020: 1–7. [DOI] [PubMed] [Google Scholar]
- 4. WHO. Weekly epidemiological update - Coronavirus disease 2019 (COVID-19). 2020. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201005-weekly-epi-update-8.pdf.
- 5. Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. Ct imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020; 295: 202–7. doi: 10.1148/radiol.2020200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X, Zhang X, He J. Challenges to the system of reserve medical supplies for public health emergencies: reflections on the outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic in China. Biosci Trends 2020; 14: 3–8. doi: 10.5582/bst.2020.01043 [DOI] [PubMed] [Google Scholar]
- 7. Ranney ML, Griffeth V, Jha AK. Critical supply shortages - the need for ventilators and personal protective equipment during the Covid-19 pandemic. N Engl J Med 2020; 382: e41. doi: 10.1056/NEJMp2006141 [DOI] [PubMed] [Google Scholar]
- 8. Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 2020; 55: 105924. doi: 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology 2020; 296: 172–80. doi: 10.1148/radiol.2020201365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gattinoni L, Coppola S, Cressoni M, Busana M, Rossi S, Chiumello D. Covid-19 does not lead to a "Typical" acute respiratory distress syndrome. Am J Respir Crit Care Med 2020; 201: 1299–300. doi: 10.1164/rccm.202003-0817LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Dong C, Hu Y, Li C, Ren Q, Zhang X, et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology 2020; 296: E55–64. doi: 10.1148/radiol.2020200843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020; 296: E32–40. doi: 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. American College of Radiology (ACR). Recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. 2020. Available from: https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection.
- 14. Society of Thoracic Radiology. STR/ASER COVID-19 position statement. 2020. Available from: https://thoracicrad.org/wp-content/uploads/2020/03/STR-ASER-Position-Statement-1.pdf.
- 15. The Royal Australian and New Zealand College of Radiologists (RANZCR). COVID-19: position statements and guidance. 2020. Available from: https://www.ranzcr.com/our-work/coronavirus/position-statements-and-guidance.
- 16. British Society of Thoracic Imaging. COVID-19 BSTI statement and guidance. 2020. Available from: https://www.bsti.org.uk/standards-clinical-guidelines/clinical-guidelines/covid-19-bsti-statement-and-guidance/.
- 17. Uppaluri R, Hoffman EA, Sonka M, Hartley PG, Hunninghake GW, McLennan G. Computer recognition of regional lung disease patterns. Am J Respir Crit Care Med 1999; 160: 648–54. doi: 10.1164/ajrccm.160.2.9804094 [DOI] [PubMed] [Google Scholar]
- 18. Salisbury ML, Lynch DA, van Beek EJR, Kazerooni EA, Guo J, Xia M, et al. Idiopathic pulmonary fibrosis: the association between the adaptive multiple features method and fibrosis outcomes. Am J Respir Crit Care Med 2017; 195: 921–9. doi: 10.1164/rccm.201607-1385OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roth MD, Tseng C-H, Clements PJ, Furst DE, Tashkin DP, Goldin JG, et al. Predicting treatment outcomes and Responder subsets in scleroderma-related interstitial lung disease. Arthritis Rheum 2011; 63: 2797–808. doi: 10.1002/art.30438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sciurba FC, Ernst A, Herth FJF, Strange C, Criner GJ, Marquette CH, et al. A randomized study of endobronchial valves for advanced emphysema. N Engl J Med 2010; 363: 1233–44. doi: 10.1056/NEJMoa0900928 [DOI] [PubMed] [Google Scholar]
- 21. Hoffman EA, Ahmed FS, Baumhauer H, Budoff M, Carr JJ, Kronmal R, et al. Variation in the percent of emphysema-like lung in a healthy, nonsmoking multiethnic sample. The MESA lung study. Ann Am Thorac Soc 2014; 11: 898–907. doi: 10.1513/AnnalsATS.201310-364OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo Cascio CM, Quante M, Hoffman EA, Bertoni AG, Aaron CP, Schwartz JE, et al. Percent emphysema and daily motor activity levels in the general population: multi-ethnic study of atherosclerosis. Chest 2017; 151: 1039–50. doi: 10.1016/j.chest.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camiciottoli G, Bartolucci M, Maluccio NM, Moroni C, Mascalchi M, Giuntini C, et al. Spirometrically gated high-resolution CT findings in COPD: lung attenuation vs lung function and dyspnea severity. Chest 2006; 129: 558–64. doi: 10.1378/chest.129.3.558 [DOI] [PubMed] [Google Scholar]
- 24. Camiciottoli G, Bigazzi F, Bartolucci M, Cestelli L, Paoletti M, Diciotti S, et al. BODE-index, modified BODE-index and ADO-score in chronic obstructive pulmonary disease: relationship with COPD phenotypes and CT lung density changes. COPD 2012; 9: 297–304. doi: 10.3109/15412555.2012.661000 [DOI] [PubMed] [Google Scholar]
- 25. Andrianopoulos V, Celli BR, Franssen FME, Pinto-Plata VM, Calverley PMA, Vanfleteren LEGW, et al. Determinants of exercise-induced oxygen desaturation including pulmonary emphysema in COPD: results from the eclipse study. Respir Med 2016; 119: 87–95. doi: 10.1016/j.rmed.2016.08.023 [DOI] [PubMed] [Google Scholar]
- 26. Camiciottoli G, Orlandi I, Bartolucci M, Meoni E, Nacci F, Diciotti S, et al. Lung CT densitometry in systemic sclerosis: correlation with lung function, exercise testing, and quality of life. Chest 2007; 131: 672–81. doi: 10.1378/chest.06-1401 [DOI] [PubMed] [Google Scholar]
- 27. Ariani A, Aiello M, Silva M, Alfieri V, Bonati E, Lumetti F, et al. Quantitative CT indexes are significantly associated with exercise oxygen desaturation in interstitial lung disease related to systemic sclerosis. Clin Respir J 2017; 11: 983–9. doi: 10.1111/crj.12451 [DOI] [PubMed] [Google Scholar]
- 28. Wu X, Kim GH, Salisbury ML, Barber D, Bartholmai BJ, Brown KK, et al. Computed tomographic biomarkers in idiopathic pulmonary fibrosis. The future of quantitative analysis. Am J Respir Crit Care Med 2019; 199: 12–21. doi: 10.1164/rccm.201803-0444PP [DOI] [PubMed] [Google Scholar]
- 29. Schmithorst VJ, Altes TA, Young LR, et al. Automated algorithm for quantifying the extent of cystic change on volumetric chest CT: initial results in lymphangioleiomyomatosis. AJR Am J Roentgenol 2009; 192: 1037–44. [DOI] [PubMed] [Google Scholar]
- 30. Avila NA, Kelly JA, Dwyer AJ, Johnson DL, Jones EC, Moss J. Lymphangioleiomyomatosis: correlation of qualitative and quantitative thin-section CT with pulmonary function tests and assessment of dependence on pleurodesis. Radiology 2002; 223: 189–97. doi: 10.1148/radiol.2231010315 [DOI] [PubMed] [Google Scholar]
- 31. Hoffman EA, Lynch DA, Barr RG, van Beek EJR, Parraga G, .IWPFI Investigators . Pulmonary CT and MRI phenotypes that help explain chronic pulmonary obstruction disease pathophysiology and outcomes. J Magn Reson Imaging 2016; 43: 544–57. doi: 10.1002/jmri.25010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Castro M, Fain SB, Hoffman EA, Gierada DS, Erzurum SC, Wenzel S, et al. Lung imaging in asthmatic patients: the picture is clearer. J Allergy Clin Immunol 2011; 128: 467–78. doi: 10.1016/j.jaci.2011.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sieren JP, Newell JD, Barr RG, Bleecker ER, Burnette N, Carretta EE, et al. SPIROMICS protocol for multicenter quantitative computed tomography to phenotype the lungs. Am J Respir Crit Care Med 2016; 194: 794–806. doi: 10.1164/rccm.201506-1208PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guan W-J, Liang W-H, Zhao Y, Liang H-R, Chen Z-S, Li Y-M, et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J 2020; 55: 2000547. doi: 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 2020; 296: E65–71 Epub 2020 Mar 19. doi: 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walsh SLF, Humphries SM, Wells AU, Brown KK. Imaging research in fibrotic lung disease; applying deep learning to unsolved problems. Lancet Respir Med 2020; 8: 1144–53. doi: 10.1016/S2213-2600(20)30003-5 [DOI] [PubMed] [Google Scholar]
- 37. Agarwala S, Kale M, Kumar D, Swaroop R, Kumar A, Kumar Dhara A, et al. Deep learning for screening of interstitial lung disease patterns in high-resolution CT images. Clin Radiol 2020; 75: 481.e1–481.e8. doi: 10.1016/j.crad.2020.01.010 [DOI] [PubMed] [Google Scholar]
- 38. Gerard SE, Patton TJ, Christensen GE, Bayouth JE, Reinhardt JM. FissureNet: a deep learning approach for pulmonary fissure detection in CT images. IEEE Trans Med Imaging 2019; 38: 156–66. doi: 10.1109/TMI.2018.2858202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bai HX, Wang R, Xiong Z, Hsieh B, Chang K, Halsey K, et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology 2020; 296: E156–65. doi: 10.1148/radiol.2020201491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Y, Ma L, Yang F, Chen Y, Ma K, Yang J, et al. A collaborative online AI engine for CT-based COVID-19 diagnosis. medRxiv 2020; 0: 2020.05.10.20096073: 2020.05.10.20096073 19 May 2020. doi: 10.1101/2020.05.10.20096073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Belfiore MP, Urraro F, Grassi R, Giacobbe G, Patelli G, Cappabianca S, et al. Artificial intelligence to codify lung CT in Covid-19 patients. Radiol Med 2020; 125: 500–4. doi: 10.1007/s11547-020-01195-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaganti S, Grenier P, Balachandran A, Chabin G, Cohen S, Flohr T, et al. Automated quantification of CT patterns associated with COVID-19 from chest CT. Radiology: Artificial Intelligence 2020; 2: e200048. doi: 10.1148/ryai.2020200048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fang M, He B, Li L, Dong D, Yang X, Li C. Ct radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Science China Information Sciences 2020; 63: 172103: 172103. doi: 10.1007/s11432-020-2849-3 [DOI] [Google Scholar]
- 44. APPIL. Research projects. 2020. Available from: https://www.i-clic.uihc.uiowa.edu/research/projects/.
- 45. Hoffman EA, McLennan G. Assessment of the pulmonary structure-function relationship and clinical outcomes measures: quantitative volumetric CT of the lung. Acad Radiol 1997; 4: 758–76. doi: 10.1016/S1076-6332(97)80080-3 [DOI] [PubMed] [Google Scholar]
- 46. Hoffman EA, Clough AV, Christensen GE, Lin C-L, McLennan G, Reinhardt JM, et al. The comprehensive imaging-based analysis of the lung: a forum for team science. Acad Radiol 2004; 11: 1370–80. doi: 10.1016/j.acra.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 47. Hoffman EA, Simon BA, McLennan G. State of the art. A structural and functional assessment of the lung via multidetector-row computed tomography: phenotyping chronic obstructive pulmonary disease. Proc Am Thorac Soc 2006; 3: 519–32. doi: 10.1513/pats.200603-086MS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin C-L, Tawhai MH, Hoffman EA. Multiscale image-based modeling and simulation of gas flow and particle transport in the human lungs. Wiley Interdiscip Rev Syst Biol Med 2013; 5: 643–55. doi: 10.1002/wsbm.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trivedi A, Hall C, Hoffman EA, Woods JC, Gierada DS, Castro M. Using imaging as a biomarker for asthma. J Allergy Clin Immunol 2017; 139: 1–10. doi: 10.1016/j.jaci.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhatt SP, Washko GR, Hoffman EA, Newell JD, Bodduluri S, Diaz AA, et al. Imaging advances in chronic obstructive pulmonary disease. insights from the genetic epidemiology of chronic obstructive pulmonary disease (COPDGene) study. Am J Respir Crit Care Med 2019; 199: 286–301. doi: 10.1164/rccm.201807-1351SO [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoffman E, Olson L. Characteristics of respiratory system complexity captured via X-ray computed tomography: image acquisition, display, and analysis. Lung biology in health and disease 1998; 121: 325–78. [Google Scholar]
- 52. Kundu S, Elhalawani H, Gichoya JW, et al. How might AI and chest imaging help unravel COVID-19’s mysteries? Radiology 2020; 2: e200053. doi: 10.1148/ryai.2020200053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoffman EA, Newell JD. Lung mass as the complement to lung air content in quantitative CT of the COPD lung. Acad Radiol 2017; 24: 383–5. doi: 10.1016/j.acra.2017.01.016 [DOI] [PubMed] [Google Scholar]
- 54. Hoffman EA. Effect of body orientation on regional lung expansion: a computed tomographic approach. J Appl Physiol 1985; 59: 468–80. doi: 10.1152/jappl.1985.59.2.468 [DOI] [PubMed] [Google Scholar]
- 55. Muller NL, Staples CA, Miller RR, et al. "Density mask". An objective method to quantitate emphysema using computed tomography. Chest 1988; 94: 782–7. [DOI] [PubMed] [Google Scholar]
- 56. Gevenois PA, de Maertelaer V, De Vuyst P, Zanen J, Yernault JC. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. Am J Respir Crit Care Med 1995; 152: 653–7. doi: 10.1164/ajrccm.152.2.7633722 [DOI] [PubMed] [Google Scholar]
- 57. Coxson HO, Mayo JR, Behzad H, Moore BJ, Verburgt LM, Staples CA, et al. Measurement of lung expansion with computed tomography and comparison with quantitative histology. J Appl Physiol 1995; 79: 1525–30. doi: 10.1152/jappl.1995.79.5.1525 [DOI] [PubMed] [Google Scholar]
- 58. Hoffman EA, Jiang R, Baumhauer H, Brooks MA, Carr JJ, Detrano R, et al. Reproducibility and validity of lung density measures from cardiac CT Scans-The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study. Acad Radiol 2009; 16: 689–99. doi: 10.1016/j.acra.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Madani A, De Maertelaer V, Zanen J, Gevenois PA, et al. Pulmonary emphysema: radiation dose and section thickness at multidetector CT quantification-comparison with macroscopic and microscopic morphometry. Radiology 2007; 243: 250–7. doi: 10.1148/radiol.2431060194 [DOI] [PubMed] [Google Scholar]
- 60. Madani A, Van Muylem A, de Maertelaer V, Zanen J, Gevenois PA. Pulmonary emphysema: size distribution of emphysematous spaces on multidetector CT images--comparison with macroscopic and microscopic morphometry. Radiology 2008; 248: 1036–41. doi: 10.1148/radiol.2483071434 [DOI] [PubMed] [Google Scholar]
- 61. Hoffman EA, Barr RG. Drs Hoffman and Barr replies. Academic Radiology 2010; 17: 399–401. doi: 10.1016/j.acra.2009.12.002 20152729 [DOI] [Google Scholar]
- 62. Wang Z, Gu S, Leader JK, Kundu S, Tedrow JR, Sciurba FC, et al. Optimal threshold in CT quantification of emphysema. Eur Radiol 2013; 23: 975–84. doi: 10.1007/s00330-012-2683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Diaz AA, Bartholmai B, San José Estépar R, Ross J, Matsuoka S, Yamashiro T, et al. Relationship of emphysema and airway disease assessed by CT to exercise capacity in COPD. Respir Med 2010; 104: 1145–51. doi: 10.1016/j.rmed.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han MK, Agusti A, Calverley PM, Celli BR, Criner G, Curtis JL, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med 2010; 182: 598–604. doi: 10.1164/rccm.200912-1843CC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han MK, Bartholmai B, Liu LX, Murray S, Curtis JL, Sciurba FC, et al. Clinical significance of radiologic characterizations in COPD. COPD 2009; 6: 459–67. doi: 10.3109/15412550903341513 [DOI] [PubMed] [Google Scholar]
- 66. Martinez FJ, Foster G, Curtis JL, Criner G, Weinmann G, Fishman A, et al. Predictors of mortality in patients with emphysema and severe airflow obstruction. Am J Respir Crit Care Med 2006; 173: 1326–34. doi: 10.1164/rccm.200510-1677OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003; 348: 2059–73. doi: 10.1056/NEJMoa030287 [DOI] [PubMed] [Google Scholar]
- 68. Hartley PG, Galvin JR, Hunninghake GW, Merchant JA, Yagla SJ, Speakman SB, et al. High-resolution CT-derived measures of lung density are valid indexes of interstitial lung disease. J Appl Physiol 1994; 76: 271–7. doi: 10.1152/jappl.1994.76.1.271 [DOI] [PubMed] [Google Scholar]
- 69. Hoffman EA, Reinhardt JM, Sonka M, Simon BA, Guo J, Saba O, et al. Characterization of the interstitial lung diseases via density-based and texture-based analysis of computed tomography images of lung structure and function. Acad Radiol 2003; 10: 1104–18. doi: 10.1016/S1076-6332(03)00330-1 [DOI] [PubMed] [Google Scholar]
- 70. Easthausen I, Podolanczuk A, Hoffman E, Kawut S, Oelsner E, Kim JS, et al. Reference values for high attenuation areas on chest CT in a healthy, never-smoker, multi-ethnic sample: the MESA study. Respirology 2020; 25: 855–62. doi: 10.1111/resp.13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Podolanczuk AJ, Oelsner EC, Barr RG, Bernstein EJ, Hoffman EA, Easthausen IJ, et al. High-attenuation areas on chest computed tomography and clinical respiratory outcomes in community-dwelling adults. Am J Respir Crit Care Med 2017; 196: 1434–42. doi: 10.1164/rccm.201703-0555OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ash SY, Harmouche R, Vallejo DLL, Villalba JA, Ostridge K, Gunville R, et al. Densitometric and local histogram based analysis of computed tomography images in patients with idiopathic pulmonary fibrosis. Respir Res 2017; 18: 45. doi: 10.1186/s12931-017-0527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Podolanczuk AJ, Oelsner EC, Barr RG, Hoffman EA, Armstrong HF, Austin JHM, et al. High attenuation areas on chest computed tomography in community-dwelling adults: the MESA study. Eur Respir J 2016; 48: 1442–52. doi: 10.1183/13993003.00129-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jacob J, Bartholmai BJ, Rajagopalan S, Kokosi M, Nair A, Karwoski R, et al. Automated quantitative computed tomography versus visual computed tomography scoring in idiopathic pulmonary fibrosis: validation against pulmonary function. J Thorac Imaging 2016; 31: 304–11. doi: 10.1097/RTI.0000000000000220 [DOI] [PubMed] [Google Scholar]
- 75. Guisado-Vasco P, Silva M, Duarte-Millán MA, Sambataro G, Bertolazzi C, Pavone M, et al. Quantitative assessment of interstitial lung disease in Sjögren's syndrome. PLoS One 2019; 14: e0224772. doi: 10.1371/journal.pone.0224772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Madani A, Van Muylem A, Gevenois PA. Pulmonary emphysema: effect of lung volume on objective quantification at thin-section CT. Radiology 2010; 257: 260–8. doi: 10.1148/radiol.10091446 [DOI] [PubMed] [Google Scholar]
- 77. Nagpal P, Narayanasamy S, Vidholia A, Guo J, Shin KM, Lee CH, et al. Imaging of COVID-19 pneumonia: patterns, pathogenesis, and advances. Br J Radiol 2020; 93: 20200538. doi: 10.1259/bjr.20200538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, et al. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal 2019; 53: 197–207. doi: 10.1016/j.media.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lanza E, Muglia R, Bolengo I, Santonocito OG, Lisi C, Angelotti G, et al. Quantitative chest CT analysis in COVID-19 to predict the need for oxygenation support and intubation. Eur Radiol 2020; 30: 6770–8. doi: 10.1007/s00330-020-07013-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Turcato G, Panebianco L, Zaboli A, Scheurer C, Ausserhofer D, Wieser A, et al. Correlation between arterial blood gas and CT volumetry in patients with SARS-CoV-2 in the emergency department. Int J Infect Dis 2020; 97: 233–5. doi: 10.1016/j.ijid.2020.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Colombi D, Bodini FC, Petrini M, Maffi G, Morelli N, Milanese G, et al. Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology 2020; 296: E86–96. doi: 10.1148/radiol.2020201433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Uppaluri R, Hoffman EA, Sonka M, Hunninghake GW, McLennan G. Interstitial lung disease: a quantitative study using the adaptive multiple feature method. Am J Respir Crit Care Med 1999; 159: 519–25. doi: 10.1164/ajrccm.159.2.9707145 [DOI] [PubMed] [Google Scholar]
- 83. Uppaluri R, Mitsa T, Sonka M, Hoffman EA, McLennan G. Quantification of pulmonary emphysema from lung computed tomography images. Am J Respir Crit Care Med 1997; 156: 248–54. doi: 10.1164/ajrccm.156.1.9606093 [DOI] [PubMed] [Google Scholar]
- 84. Xu Y, van Beek EJR, Hwanjo Y, Guo J, McLennan G, Hoffman EA. Computer-aided classification of interstitial lung diseases via MDCT: 3D adaptive multiple feature method (3D AMFM). Acad Radiol 2006; 13: 969–78. doi: 10.1016/j.acra.2006.04.017 [DOI] [PubMed] [Google Scholar]
- 85. Xu Y, Sonka M, McLennan G, Guo J, Hoffman EA. MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE Trans Med Imaging 2006; 25: 464–75. doi: 10.1109/TMI.2006.870889 [DOI] [PubMed] [Google Scholar]
- 86. Romei C, Tavanti LM, Taliani A, De Liperi A, Karwoski R, Celi A, et al. Automated computed tomography analysis in the assessment of idiopathic pulmonary fibrosis severity and progression. Eur J Radiol 2020; 124: 108852. doi: 10.1016/j.ejrad.2020.108852 [DOI] [PubMed] [Google Scholar]
- 87. Jacob J, Bartholmai BJ, Rajagopalan S, van Moorsel CHM, van Es HW, van Beek FT, et al. Predicting outcomes in idiopathic pulmonary fibrosis using automated computed tomographic analysis. Am J Respir Crit Care Med 2018; 198: 767–76. doi: 10.1164/rccm.201711-2174OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim GHJ, Weigt SS, Belperio JA, Brown MS, Shi Y, Lai JH, et al. Prediction of idiopathic pulmonary fibrosis progression using early quantitative changes on CT imaging for a short term of clinical 18-24-month follow-ups. Eur Radiol 2020; 30: 726–34. doi: 10.1007/s00330-019-06402-6 [DOI] [PubMed] [Google Scholar]
- 89. Kim HG, Tashkin DP, Clements PJ, Li G, Brown MS, Elashoff R, et al. A computer-aided diagnosis system for quantitative scoring of extent of lung fibrosis in scleroderma patients. Clin Exp Rheumatol 2010; 28(5 Suppl 62): S26–35. [PMC free article] [PubMed] [Google Scholar]
- 90. Wichmann D, Sperhake J-P, Lütgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med 2020; 173: 268-277. doi: 10.7326/M20-2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Poissy J, Goutay J, Caplan M, Parmentier E, Duburcq T, Lassalle F, et al. Pulmonary embolism in patients with COVID-19: awareness of an increased prevalence. Circulation 2020; 142: 184–6. doi: 10.1161/CIRCULATIONAHA.120.047430 [DOI] [PubMed] [Google Scholar]
- 92. McGonagle D, O'Donnell JS, Sharif K, Emery P, Bridgewood C, et al. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. The Lancet Rheumatology 2020; 2: e437–45. doi: 10.1016/S2665-9913(20)30121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Peterson S. tMPR educational video. 2020. Available from: https://www.youtube.com/watch?v=BJ_Icy_djhw&feature=youtu.be.
- 94. Harris LD, Robb RA, Yuen TS, Ritman EL. Display and visualization of three-dimensional reconstructed anatomic morphology: experience with the thorax, heart, and coronary vasculature of dogs. J Comput Assist Tomogr 1979; 3: 439–46. doi: 10.1097/00004728-197908000-00002 [DOI] [PubMed] [Google Scholar]
- 95. Prokop M, Shin HO, Schanz A, Schaefer-Prokop CM. Use of maximum intensity projections in CT angiography: a basic review. Radiographics 1997; 17: 433–51. doi: 10.1148/radiographics.17.2.9084083 [DOI] [PubMed] [Google Scholar]
- 96. Galbán CJ, Han MK, Boes JL, Chughtai KA, Meyer CR, Johnson TD, et al. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med 2012; 18: 1711–5. doi: 10.1038/nm.2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kirby M, Yin Y, Tschirren J, Tan WC, Leipsic J, Hague CJ, et al. A novel method of estimating small airway disease using Inspiratory-to-Expiratory computed tomography. Respiration 2017; 94: 336–45. doi: 10.1159/000478865 [DOI] [PubMed] [Google Scholar]
- 98. Cereda M, Xin Y, Goffi A, Herrmann J, Kaczka DW, Kavanagh BP, et al. Imaging the injured lung: mechanisms of action and clinical use. Anesthesiology 2019; 131: 716–49. doi: 10.1097/ALN.0000000000002583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, et al. Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS. Thorax 2014; 69: 492–5. doi: 10.1136/thoraxjnl-2013-203897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Barr RG, Ahmed FS, Carr JJ, Hoffman EA, Jiang R, Kawut SM, et al. Subclinical atherosclerosis, airflow obstruction and emphysema: the MESA lung study. Eur Respir J 2012; 39: 846–54. doi: 10.1183/09031936.00165410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. PrecISE. Precision interventions for severe asthma: PrecISE network. 2020. Available from: https://preciseasthma.org/preciseweb/.
- 102. Hammond E, Sloan C, Newell JD, Sieren JP, Saylor M, Vidal C, et al. Comparison of low- and ultralow-dose computed tomography protocols for quantitative lung and airway assessment. Med Phys 2017; 44: 4747–57. doi: 10.1002/mp.12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Newell JD, Fuld MK, Allmendinger T, Sieren JP, Chan K-S, Guo J, et al. Very low-dose (0.15 mGy) chest CT protocols using the COPDGene 2 test object and a third-generation dual-source CT scanner with corresponding third-generation iterative reconstruction software. Invest Radiol 2015; 50: 40–5. doi: 10.1097/RLI.0000000000000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. KYOTO KAGAKU. Kyotokagaku product data. 2020. Available from: https://www.kyotokagaku.com/en/products_data/ph-1_01/.
- 105. The Phanom Laboratory. Products: CT - other phantoms. 2020. Available from: https://www.phantomlab.com/products.
- 106. Newell JD, Sieren J, Hoffman EA. Development of quantitative computed tomography lung protocols. J Thorac Imaging 2013; 28: 266–71. doi: 10.1097/RTI.0b013e31829f6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gattinoni L, Chiumello D, Caironi P, Busana M, Romitti F, Brazzi L, et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med 2020; 46: 1099–102. doi: 10.1007/s00134-020-06033-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. ARDS Definition Task Force, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin definition. JAMA 2012; 307: 2526–33. doi: 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 109. Bos LD, Paulus F, Vlaar APJ, et al. Subphenotyping ARDS in COVID-19 patients: consequences for ventilator management. Ann Am Thorac Soc 2020; 17: 1161–3. doi: 10.1513/AnnalsATS.202004-376RL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Avanzo M, Stancanello J, El Naqa I, Naqa E I. Beyond imaging: the promise of radiomics. Phys Med 2017; 38: 122–39. doi: 10.1016/j.ejmp.2017.05.071 [DOI] [PubMed] [Google Scholar]
- 111. Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol 2016; 61: R150–66. doi: 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Keek SA, Leijenaar RT, Jochems A, Woodruff HC. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol 2018; 91: 20170926. doi: 10.1259/bjr.20170926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Uthoff J, Nagpal P, Sanchez R, Gross TJ, Lee C, Sieren JC. Differentiation of non-small cell lung cancer and histoplasmosis pulmonary nodules: insights from radiomics model performance compared with clinician observers. Transl Lung Cancer Res 2019; 8: 979–88. doi: 10.21037/tlcr.2019.12.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Haghighi B, Choi S, Choi J, Hoffman EA, Comellas AP, Newell JD, et al. Imaging-Based clusters in former smokers of the COPD cohort associate with clinical characteristics: the subpopulations and intermediate outcome measures in COPD study (SPIROMICS). Respir Res 2019; 20: 153. doi: 10.1186/s12931-019-1121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Smith BM, Kirby M, Hoffman EA, Kronmal RA, Aaron SD, Allen NB, et al. Association of Dysanapsis with chronic obstructive pulmonary disease among older adults. JAMA 2020; 323: 2268–80. doi: 10.1001/jama.2020.6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Smith BM, Traboulsi H, Austin JHM, Manichaikul A, Hoffman EA, Bleecker ER, et al. Human airway branch variation and chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A 2018; 115: E974–81. doi: 10.1073/pnas.1715564115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Choi J, LeBlanc LJ, Choi S, Haghighi B, Hoffman EA, O'Shaughnessy P, et al. Differences in particle deposition between members of imaging-based asthma clusters. J Aerosol Med Pulm Drug Deliv 2019; 32: 213–23. doi: 10.1089/jamp.2018.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Haghighi B, Choi S, Choi J, Hoffman EA, Comellas AP, Newell JD, et al. Imaging-based clusters in current smokers of the COPD cohort associate with clinical characteristics: the subpopulations and intermediate outcome measures in COPD study (SPIROMICS). Respir Res 2018; 19: 178. doi: 10.1186/s12931-018-0888-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Choi S, Haghighi B, Choi J, Hoffman EA, Comellas AP, Newell JD, et al. Differentiation of quantitative CT imaging phenotypes in asthma versus COPD. BMJ Open Respir Res 2017; 4: e000252. doi: 10.1136/bmjresp-2017-000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Choi S, Hoffman EA, Wenzel SE, Castro M, Fain S, Jarjour N, et al. Quantitative computed tomographic imaging-based clustering differentiates asthmatic subgroups with distinctive clinical phenotypes. J Allergy Clin Immunol 2017; 140: 690–700. doi: 10.1016/j.jaci.2016.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hartley RA, Barker BL, Newby C, Pakkal M, Baldi S, Kajekar R, et al. Relationship between lung function and quantitative computed tomographic parameters of airway remodeling, air trapping, and emphysema in patients with asthma and chronic obstructive pulmonary disease: A single-center study. J Allergy Clin Immunol 2016; 137: 1413–22. doi: 10.1016/j.jaci.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gupta S, Hartley R, Khan UT, Singapuri A, Hargadon B, Monteiro W, et al. Quantitative computed tomography-derived clusters: redefining airway remodeling in asthmatic patients. J Allergy Clin Immunol 2014; 133: 729–38. doi: 10.1016/j.jaci.2013.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]