Abstract
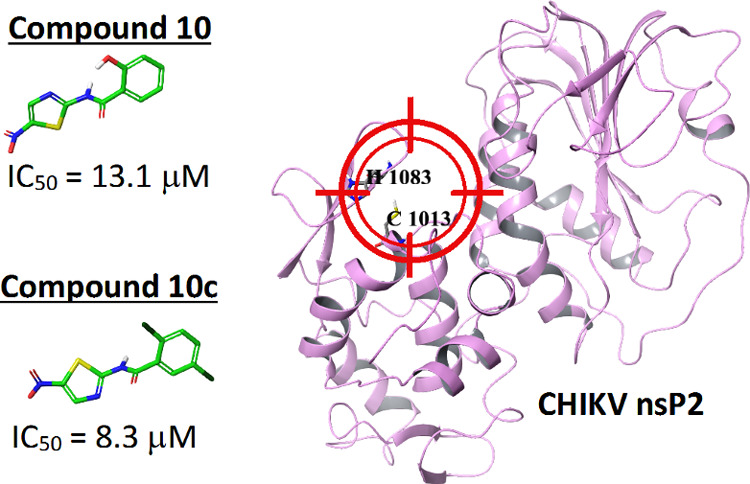
Chikungunya fever results from an infection with Chikungunya virus (CHIKV, genus Alphavirus) that is prevalent in tropical regions and is spreading fast to temperate climates with documented outbreaks in Europe and the Americas. Currently, there are no available vaccines or antiviral drugs for prevention or treatment of Chikungunya fever. The nonstructural proteins (nsPs) of CHIKV responsible for virus replication are promising targets for the development of new antivirals. This study was attempted to find out new potential inhibitors of CHIKV nsP2 protease using the ligand-based drug design. Two compounds 10 and 10c, identified by molecular docking, showed antiviral activity against CHIKV with IC50 of 13.1 and 8.3 μM, respectively. Both compounds demonstrated the ability to inhibit the activity of nsP2 in a cell-free assay, and the impact of compound 10 on virus replication was confirmed by western blot. The molecular dynamics study of the interactions of compounds 10 and 10c with CHIKV nsP2 showed that a possible mechanism of action of these compounds is the blocking of the active site and the catalytic dyad of nsP2.
Introduction
Chikungunya virus (CHIKV) is an Alphavirus of the Togaviridae family, one of the arthropod-borne member of genus Alphavirus, which is transmitted by the Aedes aegypti and Aedes albopictus mosquitos. Infection with this virus causes Chikungunya fever, whose major symptoms are an acute febrile illness with arthralgia, myalgia, rash, lymphopenia, thrombocytopenia, and gastrointestinal symptoms.1−4 CHIKV infection is rarely fatal, but in 60% of cases, the disease can progress to a chronic stage, which can seriously disable a patient for a long time.4,5 Currently, there are no approved vaccines or specific antiviral drugs, and the treatment of CHIKV infection is mostly based on the relief of symptoms.6
In the last decade, many research groups have focused on the identification of novel inhibitors of CHIKV replication to develop clinical candidate drugs for the treatment of CHIKV infections, as described in the recent review articles.7,8 For example, the compounds consisting of benzofuran, pyrrolopyridine, and thiazole carboxamide derivatives,9 harringtonine, that is, cephalotaxine alkaloid,10 a natural compound derivative (ID1452-2),11 and compounds similar to ribavirin12 were identified as possible CHIKV inhibitors using a cell-based high-throughput screening assay. Potential drug candidates were also searched using a computer-aided drug design (CADD). In this way, hydrazine derivatives,13 various carboxamide, acrylamide, and rhodanine analogues,14 plant-derived secondary metabolites,15 hesperetin,16 designed peptidomimetics,17 FDA-approved drugs and known cysteine protease inhibitors,18 and flavonoids19 were identified as possible anti-CHIKV agents. Besides extensive search from libraries, a smaller specific group of compounds was also tested in a virus-cell-based assay, for example, phenothiazines,20 synthesized triazolopyrimidines21 and thiazolidone derivatives,22 plant-derived tigliane-type diterpenoids,23 jatropane esters,24 phorbol esters,25 aplysiatoxins,26 and new daphnane diterpenoid orthoesters and their chlorinated analogues.27
In the current work, nsP2, one of the nonstructural proteins (nsPs) of CHIKV, was used as a target for molecular design. All nsPs, which are components of CHIKV RNA replicase, are translated from the virus RNA genome in the form of a P1234 polyprotein precursor. Active forms of the replicase enzymes are generated using the autoproteolytic activity of nsP2, making the enzyme indispensable for virus infection.14,28 The aim of our current study was to find out potential CHIKV nsP2 protease inhibitors using the ligand-based approach, molecular docking, and molecular dynamics (MD) simulations. In this work, two potential inhibitors of CHIKV nsP2 protease were identified. The thorough verification of the effects of identified compounds on protease activity of CHIKV nsP2 and replication of CHIKV was carried out using both CADD methods, cell-based and cell-free assays.
Results and Discussion
Molecular Modeling
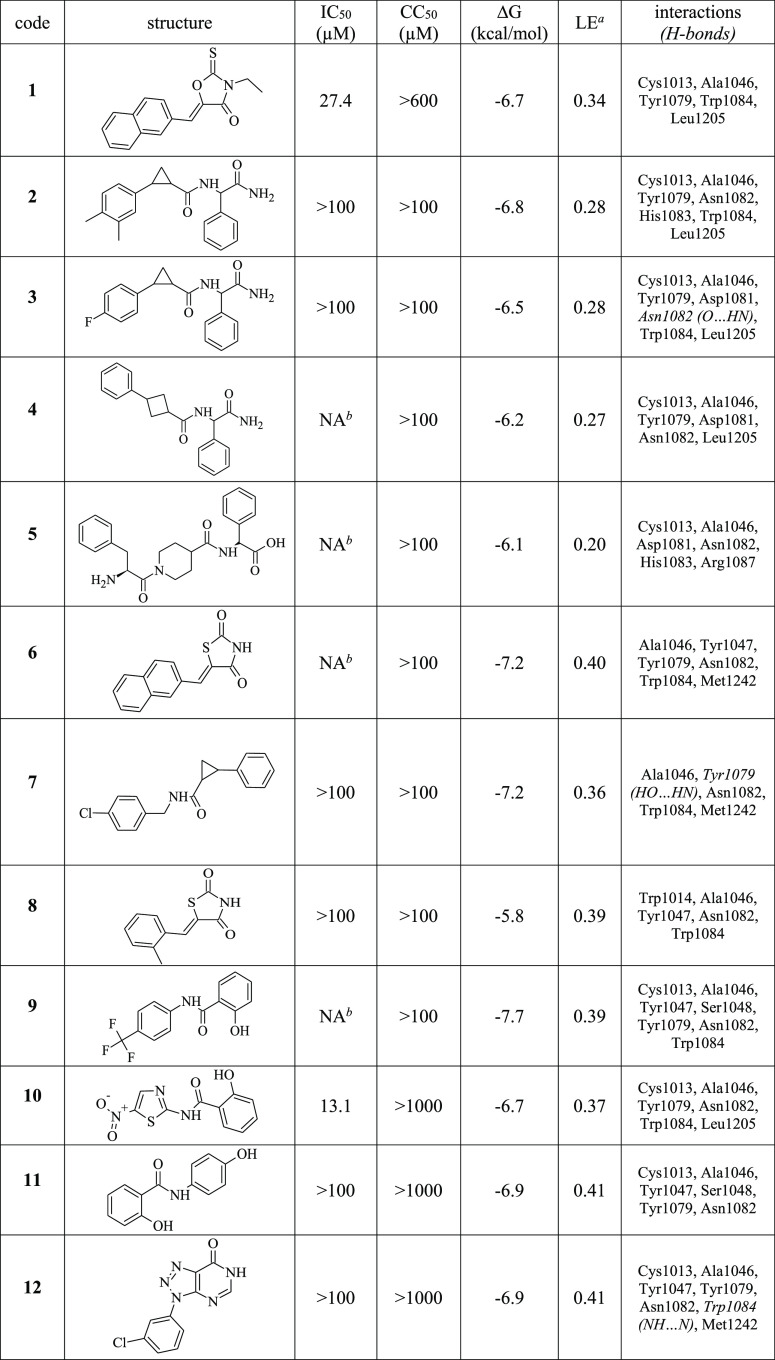
A virtual screening of the previously described CHIKV inhibitors for their potential to act as inhibitors of CHIKV nsP2 protease was performed. CHIKV inhibitors with an IC50 of up to 15 μM were selected from the review article by da Silva-Júnior et al.29 Thereafter, the selected compounds (Table S1) were docked to the potential active site of CHIKV nsP2 using AutoDock Vina 1.1.2.30 CHIKV inhibitors with ligand efficiency greater than 0.27 were selected as templates for further search of commercially available compounds using the Tanimoto similarity coefficient (≥60%) from MolPort31 database. The search resulted in 96 compounds that were also docked to the active site of CHIKV nsP2. Based on the molecular docking results, 12 compounds with ligand efficiency greater than or equal to 0.27 were selected for biological study. The calculated energies for the 12 selected compounds were in the range of −5.8 to −7.7 kcal/mol (Table 1). The binding modes of the compounds involve hydrophobic contacts with amino acid residues Cys1013 and Trp1084 (Figure 1), which are important for the activity of CHIKV nsP2 protease (here and in the rest of the text, the residue numbers correspond to these in P1234 polyprotein of CHIKV).32 The binding mode of compound 1 was found to be different from that of compounds 10, 10b, and 10c: the benzene ring of compound 1 is located below the 1H-indole ring of the Trp1084 residue of the target protein, but the benzene rings of compounds 10, 10b, and 10c are located below the benzene ring of the Tyr1079 residue of the protein. In addition, the thiazole rings of compounds 10, 10b, and 10c are oriented parallel to the 1H-indole ring of the Trp1084 residue. It should be noted that compounds 10, 10b, and 10c are derivatives of nitazoxanide, a known CHIKV inhibitor that inhibits the attachment and entry of the CHIKV into the cell.33 According to the molecular docking results, the binding mode of nitazoxanide is slightly different compared to the binding modes of compounds 10, 10b, and 10c. The substitution of the hydroxyl group by an acetyl group results in a small displacement of nitazoxanide in the active site of CHIKV nsP2 relative to compound 10 and, therefore, to the loss of a hydrophobic contact with the amino acid residue Cys1013 of the catalytic dyad (Figure 1e). Probably, the acetyl group of nitazoxanide is a steric hindrance for optimal binding to the active site of CHIKV nsP2 that can be crucial for the ability of nitazoxanide to inhibit protease activity.
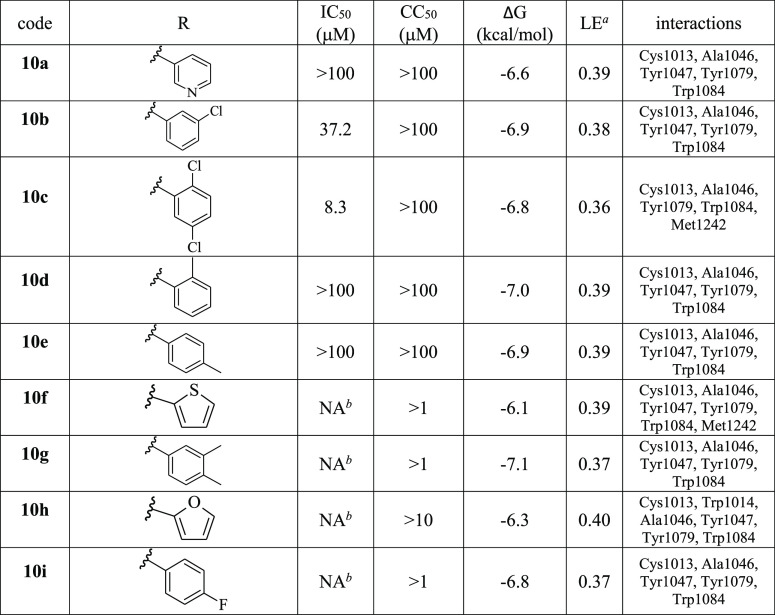
Table 1. Antiviral Activity of Compounds 1–12 against CHIKV-NanoLuc in BHK-21 Cells and Calculated Binding Energies, Ligand Efficiencies, and Interactions.
LE: ligand efficiency, that is, ΔG/N (heavy atoms).
NA: not active, that is, no activity at the maximum nontoxic concentration.
Figure 1.
Calculated binding modes of compounds 1 (a), 10 (b), 10b (c), 10c (d), and nitazoxanide (e) in the active site of CHIKV nsP2 (PDB ID: 3TRK).
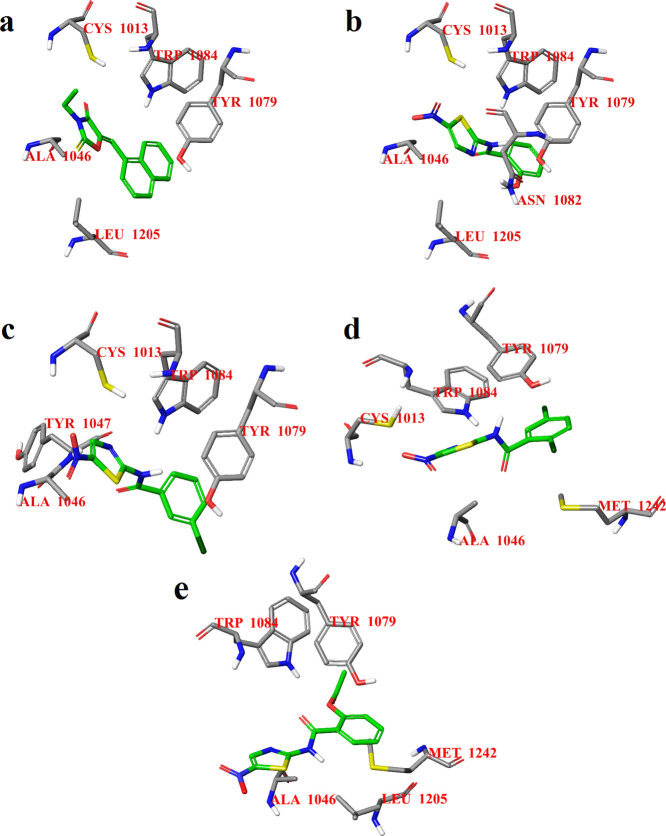
The docking calculations were followed by MD simulations using the Desmond package34 in the active site of CHIKV nsP2. The root mean square deviation (rmsd) of the atomic position behavior is notably small for all four active compounds, but compound 10c has a smaller rmsd that shows the stability of the ligand binding with CHIKV nsP2 (Figure S1). The MD modeling also confirms the location of the binding modes of all compounds (Figure 2). In the case of the compounds with the highest activity, 10 and 10c, there is a significant hydrogen bond between the side chain of Trp1084 and the carbonyl oxygen atom of the corresponding ligands (Figures 2 and S2). All potential inhibitors form a stacking (π–π) interaction with the 1H-indole ring of Trp1084 or the benzene ring of Tyr1079 (Figure 2), especially this contact is very strong for compound 10c (Figure 2d). Analysis of the MD trajectories shows that all compounds have a hydrogen bond with Cys1013 of the catalytic dyad of CHIKV nsP2 (Figure S2); however, this bond is short-term and probably does not play a significant role in the activity of the compound. However, the presence of a stable hydrogen bond with the side chain of the amino acid residue of Trp1084 is characteristic of compounds with higher activity. Thus, the hydrogen bonding of a ligand to the side chain of Trp1084 can be important for both CHIKV nsP2 protease activity and antiviral activity of potential inhibitors. It should be noted, according to the MD results (Figure S2), that all selected compounds form short-term contacts with amino acid residues of the loop between the β7 strand and α9 helix, which with the loop between β1 and β2 strands are closing the access to the active site.35 However, the duration of these contacts is so short that it can be assumed that the main mechanism of action of these compounds is the blocking of the active site and the catalytic dyad of nsP2 protease. Probably, the binding of the identified compounds to the catalytic dyad of CHIKV nsP2 prevents the binding of the substrate and, thus, prevents the stabilization of the thiolate–imidazolium ion pair required for the nsP2-activated state.36
Figure 2.
2D summary diagram of the MD-calculated contacts between CHIKV nsP2 (PDB ID: 3TRK) and compounds 1 (a), 10 (b), 10b (c), and 10c (d). Interactions that occur more than 10% of the simulation time are shown.
Enzymatic Assay
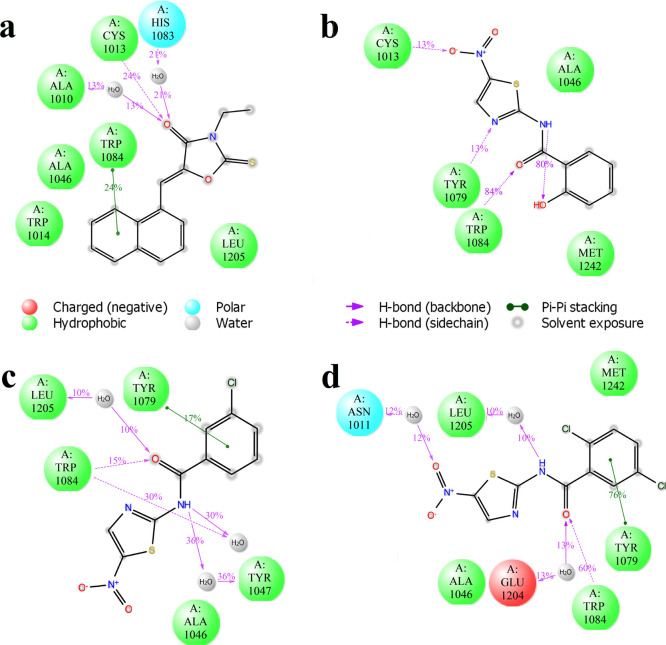
The ability of the selected compounds to inhibit the protease activity of the purified recombinant CHIKV nsP2 was analyzed using the cell-free assay. Among the selected potential inhibitors, compounds 10, 10b, and 10c were found to inhibit the protease activity of nsP2 (Figure 3a). Compound 10 had the strongest inhibitory effect, as at its presence, the least amount of cleavage product was formed. Compounds 10b and 10c had a smaller inhibitory effect than compound 10, and there was more cleavage product than in the presence of compound 10, but still less product compared to the noninhibitor control sample. Compound 1 had no effect on the protease activity of nsP2 in enzymatic assay.
Figure 3.
(a) Effects of compounds 1, 10, 10b, and 10c (indicated on the top) used at a concentration of 1 mM on the ability of CHIKV nsP2 to cleave a recombinant protein substrate. Names of the proteins are indicated on the right, and molecular masses of marker bands are indicated on the left. (b) Western blot analysis. BHK-21 cells infected with CHIKV-NanoLuc (MOI 10) were treated with increasing concentrations of compound 10. Cell lysates were collected 6 h post infection and run on 10% SDS-PAGE, and proteins were transferred onto the PVDF membrane. CHIKV proteins were detected using the respective rabbit primary antibodies and secondary anti-rabbit IRDye680-conjugated fluorescent antibodies. Loading control—β-actin—was detected using the primary mouse and secondary anti-mouse IRDye800-conjugated antibody. Names of the proteins are indicated on the left, and molecular masses of marker bands are indicated on the right. Neg: BHK-21 cells treated with 1% DMSO; Pos: infected BHK-21 cells treated with 1% DMSO (no inhibitor).
Cell Assay
The selected compounds were initially tested at three concentrations (1, 10, and 100 μM) for their potential to inhibit replication of CHIKV-NanoLuc in BHK-21 cells. Compounds 1 and 10 were very potent inhibitors; subsequent experiments revealed that their IC50 were 27.4 and 13.1 μM, respectively. Other compounds had very low antiviral activity or no activity at their maximum nontoxic concentrations (Table 1). Compound 10c, the dichloro-substituted in the benzene ring, was found to be the most potent among the derivatives of compound 10 (Table 2). The metasubstituted derivative 10b had a lower activity than compounds 10 and 10c. Probably, the substitution in the benzene ring is very important for the antiviral activity. All active compounds showed no cytotoxic effect at their active concentrations. The antiviral activity of compound 10 was also confirmed using a different assay system (CellTox Green assay) in another cell line (retinal pigment epithelium (RPE) cells). In RPE cells, compound 10 had an IC50 of 7.3 μM, about twofold lower than in BHK-21 cells, and CC50 was ≥30 μM. The antiviral activity of compound 10 was also evaluated in BHK-21 cells infected at a high MOI (multiplicity of infection) of 10. Using western blotting, it was found that at a concentration 10 times higher than IC50, the virus replication was completely inhibited, as it is evident from the lack of ns-protein and capsid protein expression (Figure 3b). This experiment demonstrated that the prominent inhibitory effect starts at a concentration of 20 μM and is associated by reduction of synthesis of ns-proteins and capsid protein. These data are consistent with the proposed mechanism of compound 10: inhibiting protease activity of nsP2 is expected to result in an inhibition of the formation of viral replication complexes and therefore to reduce synthesis of all viral RNAs and products of their translation. It is possible that compound 1, which was predicted to bind to nsP2 (Figures 1a, 2a, Table 1), inhibited virus replication in infected cells but failed to inhibit its protease activity in the cell-free assay (Figure 3a) and may also act via binding to nsP2 possibly disturbing other functions of this multifunctional protein.
Table 2. Antiviral Activity of Analogues of Compound 10 against CHIKV-NanoLuc in BHK-21 Cells and Calculated Binding Energies, Ligand Efficiencies, and Interactions.
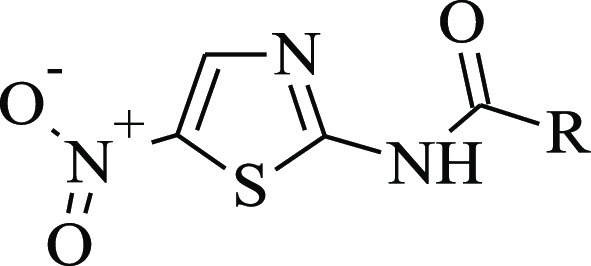
LE: ligand efficiency, that is, ΔG/N (heavy atoms).
NA: not active, that is, no activity at the maximum nontoxic concentration.
Conclusions
In this work, new potential CHIKV nsP2 protease inhibitors were searched using the ligand-based drug design approach. Two potential nsP2 CHIKV inhibitors were identified based on the virtual screening of the previously described CHIKV inhibitors. It is worth noting that compounds 10 and 10c are derivatives of nitazoxanide, which inhibits the attachment and entry of the CHIKV into the cell.33 In our work, it was shown that CHIKV nsP2, which plays an important role not only in the replication of the virus but also in the pathogenesis of the viral infection,37 is a possible target for the identified compounds 10 and 10c. Thus, these compounds are of great interest for further development of the efficient and targeted CHIKV inhibitors.
Methods
Molecular Modeling
The crystal structure of CHIKV nsP2 protease was obtained from Protein Data Bank (PDB ID: 3TRK).38 The structural model was measured by X-ray diffraction with a resolution of 2.40 Å. Protein preparation was carried out using Schrödinger’s Protein Preparation Wizard of Maestro 10.7.39 Water molecules were removed from the crystal structure. The two-dimensional chemical structures of ligands were obtained from MolPort31 database. Ligand structures were prepared for further molecular docking procedure using LigPrep with the OPLS_2005 force field from the Schrödinger Suite.40 All possible states such as generation and ionization states were enumerated for each ligand using Epik at a pH of 7.0 ± 2. Stereoisomers were determined from the three-dimensional structures. PDB files for the molecular docking procedure were created from lowest-energy conformers for each ligand. The docking studies were carried out using AutoDock Vina 1.1.2.30 The potential active site of CHIKV nsP2 was based on the work by Jadav et al.22 with key residues Cys1013, His1083, and Trp1084 (numeration based on residues in P1234 of CHIKV). The active site was surrounded with grid-box-sized 20 × 20 × 20 points with a spacing of 1.000 Å. The docking settings of AutoDock Vina 1.1.230 were used in their default values, namely, one CPU to use, the number of output poses is 9, and exhaustiveness is 8. The MD simulations were carried out using the Desmond simulation package of Schrödinger LLC.34 The NPT ensemble with the temperature 300 K and a pressure of 1 bar was applied in all runs. The simulation lengths were 50 ns with a relaxation time of 1 ps. The force field OPLS_200541 was used for each simulation. The long-range electrostatic interactions were calculated using the particle mesh Ewald method.42 The cutoff radius in Coulomb interactions was 9.0 Å. The water molecules were described using the simple point charge model (SPC).43 The Martyna–Tuckerman–Klein chain coupling scheme44 with a coupling constant of 2.0 ps was used for the pressure control, and the Nosé-Hoover chain coupling scheme44 was used for the temperature control. The nonbonded forces were calculated using an RESPA integrator where the short-range forces were updated every step and the long-range forces were updated every three steps. The trajectories were saved at 50.0 ps intervals for further analysis. The behavior and interactions between the ligands and protein were analyzed using the Simulation Interaction Diagram tool implemented in the Desmond MD package.
Compounds
Compounds for experimental study were purchased from MolPort, Inc.31 10 mM stocks of compounds were prepared by dissolving compounds in sterile dimethyl sulfoxide (DMSO) (Sigma, USA) and stored at −20 °C until further use. (1) (5Z)-3-ethyl-5-(naphthalen-1-ylmethylidene)-2-sulfanylidene-1,3-oxazolidin-4-one; ChemBridge Corp., cat. no. 7374012, purity: 90%; (2) 2-{[2-(3,4-dimethylphenyl)cyclopropyl]formamido}-2-phenylacetamide; Enamine, Ltd., cat. no. Z898678328, purity: >90%; (3) 2-{[2-(3,4-dimethylphenyl)cyclopropyl]formamido}-2-phenylacetamide; Enamine, Ltd., cat. no. Z225701352, purity: >90%; (4) 2-phenyl-2-[(3-phenylcyclobutyl)formamido]acetamide; Enamine, Ltd., cat. no. Z875424876, purity: >90%; (5) (2S)-1-(4-{[(S)-carboxy(phenyl)methyl]carbamoyl}piperidin-1-yl)-1-oxo-3-phenylpropan-2-aminium chloride; IBScreen NP, cat. no. STOCK1N-56416, purity: 90%; (6) (5E)-5-(naphthalen-2-ylmethylidene)-1,3-thiazolidine-2,4-dione; Vitas-M Laboratory, Ltd., cat. no. STK244409, purity: >90%; (7) N-[(4-chlorophenyl)methyl]-2-phenylcyclopropane-1-carboxamide; Vitas-M Laboratory, Ltd., cat. no. STK440795, purity: >90%; (8) (5E)-5-[(2-methylphenyl)methylidene]-1,3-thiazolidine-2,4-dione; Vitas-M Laboratory, Ltd., cat. no. STK038906, purity: >90%; (9) 2-hydroxy-N-[4-(trifluoromethyl)phenyl]benzamide; Alinda Chemical, Ltd., cat. no. IBS-L0127348, purity: 90%; (10) 2-hydroxy-N-(5-nitro-1,3-thiazol-2-yl)benzamide; TargetMol, cat. no. T2279, purity: 99%; (11) 2-hydroxy-N-(4-hydroxyphenyl)benzamide; TargetMol, cat. no. T0353, purity: 99%; (12) sodium 3-(3-chlorophenyl)-7-oxo-6H-[1,2,3]triazolo[4,5-d]pyrimidin-6-ide; Life Chemicals, Inc., cat. no. F2199-0574, purity: >90%; (10a) N-(5-nitro-1,3-thiazol-2-yl)pyridine-3-carboxamide; ChemBridge Corp., cat. no. 5530525, purity: 90%; (10b) 3-chloro-N-(5-nitro-1,3-thiazol-2-yl)benzamide; ChemDiv, Inc., cat. no. 1786-0083, purity: >90%; (10c) 2,5-dichloro-N-(5-nitro-1,3-thiazol-2-yl)benzamide; Vitas-M Laboratory, Ltd., cat. no. STK059294, purity: >90%; (10d) 2-methyl-N-(5-nitro-1,3-thiazol-2-yl)benzamide; Vitas-M Laboratory, Ltd., cat. no. STL356377, purity: >90%; (10e) 4-methyl-N-(5-nitro-1,3-thiazol-2-yl)benzamide; Vitas-M Laboratory, Ltd., cat. no. STK060782, purity: >90%; (10f) N-(5-nitro-1,3-thiazol-2-yl)thiophene-2-carboxamide; Vitas-M Laboratory, Ltd., cat. no. STK071581, purity: >90%; (10g) 3,4-dimethyl-N-(5-nitro-1,3-thiazol-2-yl)benzamide; Vitas-M Laboratory, Ltd., cat. no. STK072231, purity: >90%; (10h) N-(5-nitro-1,3-thiazol-2-yl)furan-2-carboxamide; BIONET—Key Organics, Ltd., cat. no. 7N-023, purity: 90%; (10i) 4-fluoro-N-(5-nitro-1,3-thiazol-2-yl)benzamide; BIONET—Key Organics, Ltd., cat. no. 6N-020, purity: 90%.
Enzymatic Assay
Full-length recombinant CHIKV nsP2 was used as the protease. The recombinant protein substrate contained 15 amino acid residues corresponding to P10 to P′5 residues of the nsP2 cleavage site located between nsP1 and nsP2 regions of P1234, which was placed between enhanced green fluorescent protein and thioredoxin. The recombinant proteins were expressed and purified, as described in detail earlier.45,46 Protease inhibition assay was carried out at 30 °C for 1.5 h in 10 μL volume in protease assay buffer (20 mM HEPES [pH 7.2] and 2 mM dithiothreitol). The CHIKV nsP2 final concentration was 1.4 μM, the protease substrate’s final concentration was 6 μM, the inhibitor’s final concentration was 1 mM, and 10% DMSO was used as a solvent control. The maximally allowed concentration of DMSO was described in our previous work.14 Protease inhibition assay reaction products (5.5 μL) were analyzed by 10% SDS-PAGE and Coomassie blue staining. The experiment was carried out three times with very similar results.
Cells and Viruses
Baby hamster kidney (BHK-21) cells (ATCC CCL-10) were grown in Glasgow’s minimal essential medium (GMEM; PAN Biotech) containing 7.5% fetal bovine serum (FBS), 2% tryptose phosphate broth, 20 mM HEPES, and 1% dilution of penicillin/streptomycin stock. RPE cells (ATCC CRL-4000) were grown in Dulbecco’s modified Eagle medium: nutrient mixture F-12 (DMEM/F12) containing 10% FBS, 1% penicillin/streptomycin stock, and 0.25% sodium bicarbonate. Both cell cultures were maintained at 37 °C in a 5% CO2 atmosphere. The CHIKV-NanoLuc virus was obtained from the icDNA clone pICRES1, representing the LR2006OPY1 strain belonging to the East/Central/South African genotype.47 The virus stocks were stored at −80 °C. All virus experiments were conducted in accordance with the guidelines of the national authorities using appropriate biosafety laboratories under appropriate safety approvals.
Cytotoxicity Assay in BHK-21 Cells
Cells were plated in 96-well plates containing a complete growth medium and cultured overnight. The cells were then treated with compounds at the indicated concentrations for specified times. After drug treatment, cell viability was measured using the MTT assay. Briefly, 10 μL of MTT solution (5 mg/mL) was added to each well and incubated at 37 °C for 4 h. After removing the medium, the formed crystals produced were dissolved in 100 μL of DMSO. The optical density of the obtained solution was measured at 540 nm. All experiments were performed in triplicates.
Antiviral Activity in BHK-21 Cells
BHK-21 cells were seeded on 24-well tissue culture plates (Thermo Fisher Scientific) at a density 2 × 105 cells/well in 400 μL of GMEM (PAN Biotech) and were allowed to adhere overnight. Next, BHK-21 cells were infected with CHIKV-NanoLuc at an MOI of 0.001 PFU (plaque forming units)/cell in a virus growth medium (100 μL/well) containing GMEM, 0.2% BSA, 1% penicillin/streptomycin stock, and compounds at final concentrations ranging from 0.1 to 200 μM. At 1 h post infection, the complete growth medium (300 μL/well) containing compounds at final concentrations ranging from 0.1 to 200 μM was added. At 16 h post infection, the medium was discarded, cells were lysed, and nanoluciferase activity was measured using a Renilla luciferase assay system (Promega, Madison, WI, USA). Percent inhibition was calculated by comparing values obtained from compound-treated wells with those from infected wells treated with 1% DMSO as a solvent control. The assay was carried out in three parallels. IC50 calculation was performed using GraphPad Prism version 8.0 for Windows, GraphPad Software, La Jolla California, USA.48 The calculated IC50 graphs are presented in Figure S3.
Western Blot Analysis of Infected Cells Treated with Compound 10
BHK-21 cells were seeded on six-well tissue culture plates (Thermo Fisher Scientific) at a density 1 × 106 cells/well in 2 mL of GMEM (PAN Biotech). Cells were infected with CHIKV-NanoLuc at an MOI of 10 in the presence of compound 10 at concentrations of 10, 20, 50, 100, and 200 μM and 200 μL of GMEM containing 0.2% BSA. Control cells were mock infected under the same condition with the presence of 1% DMSO as a solvent control. At 1 h post infection, the complete growth medium (300 μL/well) was added, containing the same concentration of compound 10 or DMSO. Cells were incubated at 37 °C for 6 h, and then, cells were lysed in 100 μL of SDS sample buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 0.2% bromophenol blue) and denatured at 100 °C for 8 min. Proteins were separated by SDS-PAGE in 10% gels and transferred onto poly(vinylidene difluoride) (PVDF) membranes. The CHIKV nsP1, nsP2, nsP3, and capsid protein were detected using the corresponding rabbit polyclonal antibodies (all generated in-house); β-actin (sc-47778; Santa Cruz Biotechnology) was used as a loading control. The membranes were incubated with the appropriate secondary antibodies conjugated to fluorescent infrared dyes (LI-COR), and the signals were visualized with the LI-COR Odyssey Fc imaging system.
Cytotoxicity and Antiviral Activity of Compounds in RPE Cells
RPE cells were seeded on 24-well tissue culture plates (Thermo Fisher Scientific) at a density of 5 × 104 cells/well in 100 μL of DMEM/F-12 (Corning) and were allowed to adhere overnight. Confluent cells were infected with CHIKV-NanoLuc at an MOI of 2 in the virus growth medium DMEM-F12 containing 0.2% BSA, 2 mM l-glutamine, 0.348% sodium bicarbonate, and 1 μg/mL l-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin (TPCK)-trypsin (Sigma-Aldrich). Compound 10 was added to the cells in threefold dilution at seven different concentrations starting from 30 μM. No compound was added to the control wells. At 48 h post infection, the medium was replaced with the virus growth medium containing the CellTox Green Dye reagent (1:2000 dilution in the assay well, Promega, Madison, WI, USA). Fluorescence was measured using the Synergy M microplate reader (BioTek, USA); then, cells were lysed with Renilla lysis buffer in twofold dilution containing the Renilla luciferase assay substrate (1:66), and luminescence was measured using the Synergy M microplate reader (BioTek, USA). IC50 calculation was carried out with GraphPad Prism version 8.0 for Windows, GraphPad Software, La Jolla California, USA.48 The calculated IC50 graph is presented in Figure S3c.
Acknowledgments
Current work was financially supported by the EU European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering (project no. 2014-2020.4.01.15-0013), Estonia.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c06191.
Table of calculated binding energies, ligand efficiencies, and interactions of starting compounds selected from the literature; MD-calculated contacts and rmsd of the atomic positions of compounds 1, 10, 10b, and 10c with CHIKV nsP2; determination of IC50 of compounds 1, 10, 10b, and 10c (PDF)
Author Contributions
L.I. performed molecular docking and MD simulations; L.I., J.T.-T., and M.K. analyzed modeling results. L.I., K.R., and E.Ž. performed biological experiments; L.I., K.R., E.Ž., A.M., and M.K. analyzed experimental data. A.M. and M.K. coordinated the project. All authors participated in the preparation of the manuscript and approved the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Pialoux G.; Gaüzère B.-A.; Jauréguiberry S.; Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. 10.1016/s1473-3099(07)70107-x. [DOI] [PubMed] [Google Scholar]
- Borgherini G.; Poubeau P.; Staikowsky F.; Lory M.; Moullec N. L.; Becquart J. P.; Wengling C.; Paganin F.; Paganin F. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin. Infect. Dis. 2007, 44, 1401–1407. 10.1086/517537. [DOI] [PubMed] [Google Scholar]
- Chusri S.; Siripaitoon P.; Silpapojakul K.; Hortiwakul T.; Charernmak B.; Chinnawirotpisan P.; Nisalak A.; Thaisomboonsuk B.; Klungthong C.; Gibbons R. V.; Jarman R. G. Kinetics of Chikungunya Infections during an Outbreak in Southern Thailand, 2008-2009. Am. J. Trop. Med. Hyg. 2014, 90, 410–417. 10.4269/ajtmh.12-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc T.; Lecuit M. Chikungunya virus pathogenesis: from bedside to bench. Antiviral Res. 2015, 121, 120–131. 10.1016/j.antiviral.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Simon F.; Javelle E.; Oliver M.; Leparc-Goffart I.; Marimoutou C. Chikungunya virus infection. Curr. Infect. Dis. Rep. 2011, 13, 218–228. 10.1007/s11908-011-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T.; Couderc T.; Ng L. F. P.; Hallengärd D.; Powers A.; Lecuit M.; Esteban M.; Merits A.; Roques P.; Liljeström P. Therapeutics and vaccines against chikungunya virus. Vector Borne Zoonotic Dis. 2015, 15, 250–257. 10.1089/vbz.2014.1681. [DOI] [PubMed] [Google Scholar]
- Jadav S. S.; Sinha B. N.; Hilgenfeld R.; Jayaprakash V. Computer-Aided Structure Based Drug Design Approaches for the Discovery of new Anti-CHIKV Agents. Curr. Comput.-Aided Drug Des. 2017, 13, 346–361. 10.2174/1573409913666170309145308. [DOI] [PubMed] [Google Scholar]
- Subudhi B.; Chattopadhyay S.; Mishra P.; Kumar A. Current Strategies for Inhibition of Chikungunya Infection. Viruses 2018, 10, 235. 10.3390/v10050235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D. J. M.; Bonotto R. M.; Gomes R. G. B.; da Silva C. T.; No J. H.; Lombardot B.; Schwartz O.; Hansen M. A. E.; Freitas-Junior L. H.; Freitas-Junior L. H. Identification of Novel Compounds Inhibiting Chikungunya Virus-Induced Cell Death by High Throughput Screening of a Kinase Inhibitor Library. PLoS Neglected Trop. Dis. 2013, 7, e2471 10.1371/journal.pntd.0002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P.; Thiruchelvan M.; Lee R. C. H.; Chen H.; Chen K. C.; Ng M. L.; Chu J. J. H. Inhibition of Chikungunya Virus Replication by Harringtonine, a Novel Antiviral That Suppresses Viral Protein Expression. Antimicrob. Agents Chemother. 2013, 57, 155–167. 10.1128/aac.01467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Hourani M.; Lupan A.; Desprès P.; Thoret S.; Pamlard O.; Dubois J.; Guillou C.; Tangy F.; Vidalain P.-O.; Munier-Lehmann H. A Phenotypic Assay to Identify Chikungunya Virus Inhibitors Targeting the Nonstructural Protein nsP2. J. Biomol. Screen 2013, 18, 172–179. 10.1177/1087057112460091. [DOI] [PubMed] [Google Scholar]
- Mudhasani R.; Kota K. P.; Retterer C.; Tran J. P.; Tritsch S. R.; Zamani R.; Whitehouse C. A.; Bavari S. High-Content Image-Based Screening of a Signal Transduction Pathway Inhibitor Small-Molecule Library against Highly Pathogenic RNA Viruses. J. Biomol. Screen 2015, 20, 141–152. 10.1177/1087057114556253. [DOI] [PubMed] [Google Scholar]
- Bassetto M.; De Burghgraeve T.; Delang L.; Massarotti A.; Coluccia A.; Zonta N.; Gatti V.; Colombano G.; Sorba G.; Silvestri R.; Tron G. C.; Neyts J.; Leyssen P.; Brancale A. Computer-aided identification, design and synthesis of a novel series of compounds with selective antiviral activity against chikungunya virus. Antiviral Res. 2013, 98, 12–18. 10.1016/j.antiviral.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Das P. K.; Puusepp L.; Varghese F. S.; Utt A.; Ahola T.; Kananovich D. G.; Lopp M.; Merits A.; Karelson M. Design and Validation of Novel Chikungunya Virus Protease Inhibitors. Antimicrob. Agents Chemother. 2016, 60, 7382–7395. 10.1128/AAC.01421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byler K. G.; Collins J. T.; Ogungbe I. V.; Setzer W. N. Alphavirus protease inhibitors from natural sources: A homology modeling and molecular docking investigation. Comput. Biol. Chem. 2016, 64, 163–184. 10.1016/j.compbiolchem.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Oo A.; Hassandarvish P.; Chin S. P.; Lee V. S.; Abu Bakar S.; Zandi K. In silico study on anti-Chikungunya virus activity of hesperetin. PeerJ 2016, 4, e2602 10.7717/peerj.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindwal S.; Kesari P.; Singh H.; Kumar P.; Tomar S. Conformer and pharmacophore based identification of peptidomimetic inhibitors of chikungunya virus nsP2 protease. J. Biomol. Struct. Dyn. 2017, 35, 3522–3539. 10.1080/07391102.2016.1261046. [DOI] [PubMed] [Google Scholar]
- Kumar P.; Kumar D.; Giri R. Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors. Pathogens 2019, 8, 128. 10.3390/pathogens8030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain W.; Amir A.; Rasool N. Computer-aided study of selective flavonoids against chikungunya virus replication using molecular docking and DFT-based approach. Struct. Chem. 2020, 31, 1363–1374. 10.1007/s11224-020-01507-x. [DOI] [Google Scholar]
- Pohjala L.; Utt A.; Varjak M.; Lulla A.; Merits A.; Ahola T.; Tammela P. Inhibitors of Alphavirus Entry and Replication Identified with a Stable Chikungunya Replicon Cell Line and Virus-Based Assays. PLoS One 2011, 6, e28923 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante A.; Canela M.-D.; Delang L.; Priego E.-M.; Camarasa M.-J.; Querat G.; Neyts J.; Leyssen P.; Pérez-Pérez M.-J. Identification of [1,2,3]Triazolo[4,5-d]pyrimidin-7(6H)-ones as Novel Inhibitors of Chikungunya Virus Replication. J. Med. Chem. 2014, 57, 4000–4008. 10.1021/jm401844c. [DOI] [PubMed] [Google Scholar]
- Jadav S. S.; Sinha B. N.; Hilgenfeld R.; Pastorino B.; de Lamballerie X.; Jayaprakash V. Thiazolidone derivatives as inhibitors of chikungunya virus. Eur. J. Med. Chem. 2015, 89, 172–178. 10.1016/j.ejmech.2014.10.042. [DOI] [PubMed] [Google Scholar]
- Bourjot M.; Delang L.; Nguyen V. H.; Neyts J.; Guéritte F.; Leyssen P.; Litaudon M. Prostratin and 12-O-Tetradecanoylphorbol 13-Acetate Are Potent and Selective Inhibitors of Chikungunya Virus Replication. J. Nat. Prod. 2012, 75, 2183–2187. 10.1021/np300637t. [DOI] [PubMed] [Google Scholar]
- Nothias-Scaglia L.-F.; Retailleau P.; Paolini J.; Pannecouque C.; Neyts J.; Dumontet V.; Roussi F.; Leyssen P.; Costa J.; Litaudon M. Jatrophane Diterpenes as Inhibitors of Chikungunya Virus Replication: Structure-Activity Relationship and Discovery of a Potent Lead. J. Nat. Prod. 2014, 77, 1505–1512. 10.1021/np500271u. [DOI] [PubMed] [Google Scholar]
- Corlay N.; Delang L.; Girard-Valenciennes E.; Neyts J.; Clerc P.; Smadja J.; Guéritte F.; Leyssen P.; Litaudon M. Tigliane diterpenes from Croton mauritianus as inhibitors of chikungunya virus replication. Fitoterapia 2014, 97, 87–91. 10.1016/j.fitote.2014.05.015. [DOI] [PubMed] [Google Scholar]
- Gupta D.; Kaur P.; Leong S.; Tan L.; Prinsep M.; Chu J. Anti-Chikungunya Viral Activities of Aplysiatoxin-Related Compounds from the Marine Cyanobacterium Trichodesmium erythraeum. Mar. Drugs 2014, 12, 115–127. 10.3390/md12010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourjot M.; Leyssen P.; Neyts J.; Dumontet V.; Litaudon M. Trigocherrierin A, a Potent Inhibitor of Chikungunya Virus Replication. Molecules 2014, 19, 3617–3627. 10.3390/molecules19033617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla A.; Lulla V.; Tints K.; Ahola T.; Merits A. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J. Virol. 2006, 80, 5413–5422. 10.1128/jvi.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva-Júnior E. F.; Leoncini G. O.; Rodrigues É. E. S.; Aquino T. M.; Araújo-Júnior J. X. The medicinal chemistry of Chikungunya virus. Bioorg. Med. Chem. 2017, 25, 4219–4244. 10.1016/j.bmc.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.molport.com/shop/index; MolPort: Lacplesa iela 41, Riga, LV-1011, Latvia: (accessed 2021-01-10).
- Rausalu K.; Utt A.; Quirin T.; Varghese F. S.; Zusinaite E.; Das P. K.; Ahola T.; Merits A. Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease’s active site cysteine residue. Sci. Rep. 2016, 6, 37124. 10.1038/srep37124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-M.; Lu J.-W.; Lin C.-C.; Chin Y.-F.; Wu T.-Y.; Lin L.-I.; Lai Z.-Z.; Kuo S.-C.; Ho Y.-J. Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Res. 2016, 135, 81–90. 10.1016/j.antiviral.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bowers K. J.; Chow D. E.; Xu H.; Dror R. O.; Eastwood M. P.; Gregersen B. A.; Klepeis J. L.; Kolossvary I.; Moraes M. A.; Sacerdoti F. D.; Salmon J. K.; Shan Y.; Shaw D. E.. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, 2006; p 43.; b Schrödinger Release 2020-4: Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, 2020; Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020.
- Narwal M.; Singh H.; Pratap S.; Malik A.; Kuhn R. J.; Kumar P.; Tomar S. Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site. Int. J. Biol. Macromol. 2018, 116, 451–462. 10.1016/j.ijbiomac.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Russo A. T.; Malmstrom R. D.; White M. A.; Watowich S. J. Structural basis for substrate specificity of alphavirus nsP2 proteases. J. Mol. Graphics Modell. 2010, 29, 46–53. 10.1016/j.jmgm.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fros J. J.; van der Maten E.; Vlak J. M.; Pijlman G. P. The C-terminal domain of chikungunya virus nsP2 independently governs viral RNA replication, cytopathicity, and inhibition of interferon signaling. J. Virol. 2013, 87, 10394–10400. 10.1128/jvi.00884-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Protein Data Bank . Research Collaboratory for Structural Bioinformatics. https://www.rcsb.org/structure/3TRK (accessed 2021-02-11).; b Cheung J.; Franklin M.; Mancia F.; Rudolph M.; Cassidy M.; Gary E.; Burshteyn F.; Love J.. Structure of the Chikungunya virus nsP2 protease. Unpublished work. [Google Scholar]
- a Sastry G. M.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]; Schrödinger Release 2020-4: Schrödinger Suite 2020-4 Protein Preparation Wizard, Epik; Schrödinger, LLC: New York, NY, 2020; Impact; Schrödinger, LLC: New York, NY, 2020; Prime; Schrödinger, LLC: New York, NY, 2020.
- Schrödinger Release 2020-4: LigPrep; Schrödinger, LLC: New York, NY, 2020.
- Banks J. L.; Beard H. S.; Cao Y.; Cho A. E.; Damm W.; Farid R.; Felts A. K.; Halgren T. A.; Mainz D. T.; Maple J. R.; Murphy R.; Philipp D. M.; Repasky M. P.; Zhang L. Y.; Berne B. J.; Friesner R. A.; Gallicchio E.; Levy R. M. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukmaji A. Y.; Board J. A. Jr. Ewald summation techniques in perspective: a survey. Comput. Phys. Commun. 1996, 95, 73–92. 10.1016/0010-4655(96)00016-1. [DOI] [Google Scholar]
- Zielkiewicz J. Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J. Chem. Phys. 2005, 123, 104501. 10.1063/1.2018637. [DOI] [PubMed] [Google Scholar]
- Martyna G. J.; Klein M. L.; Tuckerman M. Nosé-Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635. 10.1063/1.463940. [DOI] [Google Scholar]
- Das P. K.; Merits A.; Lulla A. Functional Cross-talk between Distant Domains of Chikungunya Virus Non-structural Protein 2 Is Decisive for Its RNA-modulating Activity. J. Biol. Chem. 2014, 289, 5635–5653. 10.1074/jbc.m113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utt A.; Das P. K.; Varjak M.; Lulla V.; Lulla A.; Merits A. Mutations Conferring a Noncytotoxic Phenotype on Chikungunya Virus Replicons Compromise Enzymatic Properties of Nonstructural Protein 2. J. Virol. 2015, 89, 3145–3162. 10.1128/jvi.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utt A.; Quirin T.; Saul S.; Hellström K.; Ahola T.; Merits A. Versatile Trans-Replication Systems for Chikungunya Virus Allow Functional Analysis and Tagging of Every Replicase Protein. PLoS One 2016, 11, e0151616 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- www.graphpad.com: La Jolla California USA: (accessed 2020-09-19).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.