Abstract
Objective
SARS‐CoV‐2 has caused a worldwide pandemic of COVID‐19. The existence of prolonged SARS‐CoV‐2 positivity (PP) has further increased the burden on the health system. Since T cells are vital for viral control, we aimed to evaluate the characteristics of T‐cell responses associated with PP.
Methods
We established a PP cohort and two age‐ and sex‐matched control cohorts: a regular clinical recovery (CR) cohort and a healthy donor (HD) cohort. The mean time for RNA negativity conversion in the PP cohort was markedly longer than that in the CR cohort (66.2 vs 25.3 days), while the time from illness onset to sampling was not significantly different. T‐cell responses in the PP cohort were assayed, analysed and compared with those in the CR and HD cohorts by flow cytometry and ELISpot analysis of peripheral blood mononuclear cells.
Results
Compared with the CR cohort, the proliferation, activation and functional potential of CD8+ and CD4+ T cells in the PP cohort were not significantly different. However, the frequencies and counts of Teff and Tem in CD8+ but not in CD4+ T cells of the PP cohort were prominently lower. Moreover, a weaker SARS‐CoV‐2 N protein‐specific IFN‐γ+ T‐cell response and a higher frequency of Tregs were detected in the PP cohort.
Conclusion
Suppressed CD8+ T‐cell differentiation is associated with PP and may be an indicator for the prediction of prolonged SARS‐CoV‐2 positivity in COVID‐19 patients. The association between suppressed CD8+ T‐cell differentiation and elevated Tregs warrants studies in the future.
Keywords: COVID‐19, differentiation, prolonged SARS‐CoV‐2 positivity, T‐cell response
In this study, the characteristics of T‐cell responses of peripheral blood mononuclear cells in a COVID‐19 cohort with prolonged SARS‐CoV‐2‐positive (Pht P) were comparatively assayed and analysed with those in clinically recovered (CR) and healthy donor (HD) cohorts by FACS and ELISpot analysis of peripheral blood mononuclear cells. The results showed that the frequencies of Teff and Tem in CD8+ but not in CD4+ of the PP cohort were prominently lower than those of the CR cohort. Moreover, lowered SARS‐CoV‐2 N protein‐specific IFN‐γ+ T‐cell response and higher frequency of Tregs were detected in the PP cohort.

Introduction
SARS‐CoV‐2 has caused a worldwide pandemic of COVID‐19 that poses a global threat to humans and society. As of 16 January 2021, there were over 93.4 million confirmed infections globally, with more than 2 million deaths. The estimated duration time from SARS‐CoV‐2 infection to the loss of virus RNA detection for nasopharyngeal swab samples in approximately 95% of mild cases and severe cases was 45.6 and 48.9 days, respectively, 1 which was longer than that of MERS‐CoV and pandemic influenza A/H1N1 2009 infection. 2 , 3 In the remaining 5% of the SARS‐CoV‐2‐infected population, the duration of SARS‐CoV‐2 positivity was estimated to be up to 90 days. 1 A recent study showed that the median viral RNA carrying duration of long‐term carriers was 92 days after the first admission, and the longest carrying duration was up to 118 days. Moreover, infectious SARS‐CoV‐2 could be isolated from sputum, where high levels of viral RNA were found. Virus sequencing suggested that these patients persistently carried SARS‐CoV‐2 and were not re‐infected. 4 Although the proportion of prolonged SARS‐CoV‐2‐positive individuals is relatively low in the whole COVID‐19 population, the phenomenon further suggests a complex relationship between SARS‐CoV‐2 infection and disease outcomes, in addition to the range of clinical manifestations from asymptomatic or mild infection to severe COVID‐19 that requires hospitalisation. A markedly prolonged duration of viral positivity in COVID‐19 patients obviously requires more medical treatments and should be given more attention. It is of great significance to identify factors associated with prolonged SARS‐CoV‐2 positivity.
Case reports have suggested that immunosuppression may prolong the duration of SARS‐CoV‐2 positivity. 5 , 6 Similar to other respiratory viral infections, adaptive immune responses, particularly T‐cell responses, play a prominent role in SARS‐CoV‐2 infection. 7 , 8 , 9 , 10 A growing body of studies has shown that a more robust clonal expansion of CD8+ T cells is detected in mild but not severe COVID‐19 patients, 11 convalescent COVID‐19 patients usually mount robust T‐cell responses against SARS‐CoV‐2, 12 , 13 and robust T‐cell responses against SARS‐CoV‐2 are essential for viral clearance and disease resolution. 9 Li et al. 4 demonstrated similar SARS‐CoV‐2‐specific binding and neutralising antibody profiles between long‐term carriers and recovered patients. Together, these results suggest a key role for T cells in SARS‐CoV‐2 clearance. However, most studies have only focused on SARS‐CoV‐2‐specific T‐cell responses; thus, there is a scarcity of information on the composition, phenotype and functional potential of whole T‐cell populations in SARS‐CoV‐2‐infected individuals, let alone in prolonged viral‐positive patients.
Early in the COVID‐19 crisis in Wuhan during March and April 2020, we found that a moderate number of COVID‐19 inpatients could not be discharged because of SARS‐CoV‐2 positivity in Wuhan Jinyintan Hospital, even though some of them showed no symptoms. Hence we also recruited a clinical recovery (CR) cohort, in whom the time for viral RNA negativity conversion was less than 42 days, and a SARS‐CoV‐2‐naive healthy donor (HD) cohort. Since measurements of immune responses in blood appear to provide a reasonable reflection of those in the lung in the context of SARS‐CoV‐2 infection, 14 T cells in peripheral blood of the PP cohort were analysed and compared with the age‐ and sex‐matched CR cohort and HD cohort.
Results
Establishment of the study cohorts
Prolonged SARS‐CoV‐2‐positive (PP) individuals were recruited and selected from among 70 hospitalised COVID‐19 patients whose duration from illness onset to sampling for the present study was at least 49 days, aged 20–80 years, at Wuhan Jinyintan Hospital in March and April 2020. The patients in the PP cohort were discharged upon two consecutive negative respiratory reverse transcription–polymerase chain reaction (RT‐PCR) results. These patients were followed for 3 months, and virus positivity was not re‐detected. The duration from illness onset to the first of two consecutive negative RNA tests was designated as the time for RNA negativity conversion (TRN). Patients with co‐infections, such as Aspergillus fumigatus (three patients), a TRN < 49 days (13 patients), SARS‐CoV‐2 RBD‐specific IgM and IgG dual negativity (two patients) or severe chronic problems, such as HIV, HCV infection, cerebrovascular disease, kidney disease or tumor (four patients), were excluded (Figure 1).
Figure 1.
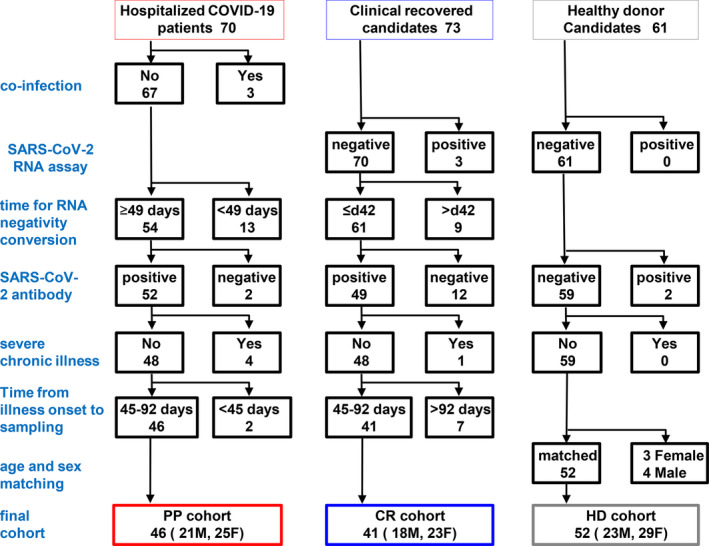
Establishment of the prolonged SARS‐CoV‐2 positivity cohort (PP), the COVID‐19 clinically recovered cohort (CR) and the SARS‐CoV‐2‐naive healthy donor cohort (HD). The PP, CR and HD cohorts were recruited and selected from hospitalised COVID‐19 patients, clinically recovered persons from COVID‐19 patients and HD, respectively. The exclusion standards were as follows: co‐infection, time for RNA negativity conversion, SARS‐CoV‐2 RBD‐specific antibody response and severe chronic problem. Patients in whom the time from illness onset to sampling was less than 45 days or more than 92 days were also excluded. To ensure that the PP and HD cohorts were age‐ and sex‐matched, three young female and four young male healthy individuals were further excluded from the HD cohort.
Seventy‐three individuals who had clinically recovered from COVID‐19 (CR) were recruited and selected during their physical re‐examination in Wuhan Jinyintan Hospital after discharge in April 2020. Individuals with suspected re‐detected virus‐positive (three patients), a TRN more than 42 days (nine patients), SARS‐CoV‐2 RBD‐specific IgM and IgG dual negativity (12 patients) or severe chronic problems (one patient) were excluded (Figure 1).
To ensure that there was a comparable duration from onset to our sampling for the T‐cell assay between the CR individuals and the PP individuals, patients in whom time from illness onset to sampling (TIS) for the T‐cell assay was < 45 days or > 92 days were excluded. After the stringent exclusion process, a PP cohort of 46 individuals (mean TRN, 66.2 days; 21 males and 25 females; mean age, 54.7 years; mean TIS, 67.5 days) and a CR cohort of 41 individuals (mean TRN, 25.3 days; 18 males and 23 females; mean age, 50.6 years; mean TIS, 71.0 days) were established (Figure 1). A significant difference existed only in the TRN between the PP and CR cohorts, but no other significant differences were found in regard to age, sex or TIS (Supplementary figure 1a and b). Additionally, the total number of lymphocytes in the blood of the PP cohort was comparable to that of the HD and CR cohorts (Supplementary figure 1c). The proportions of comorbidities, including diabetes and hypertension (4/47 and 11/47), in the PP cohort were not significantly different from those in the CR cohort (5/41 and 6/41).
A SARS‐CoV‐2‐naïve HD cohort was established by recruiting and selecting 61 healthy individuals who were undergoing a regular physical examination at Wuhan Jinyintan Hospital. The individuals were confirmed to not have an ongoing or past SARS‐CoV‐2 infection based on a SARS‐CoV‐2 nucleic acid assay of nasopharyngeal swab samples and SARS‐CoV‐2 RBD‐specific IgM and IgG assays of plasma (two individuals excluded). To ensure that the cohort was age‐ and sex‐matched with the PP and HD cohorts, 3 young female and 4 young male individuals were further excluded. Thus, an HD cohort of 52 individuals (23 males and 29 females; mean age, 50.4 years) was established to serve as a healthy control group in the study (Figure 1).
The clinical information for the HD, PP and CR cohorts is listed in Supplementary table 1. For the detection of SARS‐CoV‐2‐specific T‐cell responses in Figure 5, a sub‐PP cohort of 16 PP individuals, a sub‐CR cohort of 20 CR individuals and a sub‐HD cohort of 18 HD individuals from the cohorts above were established.
Figure 5.
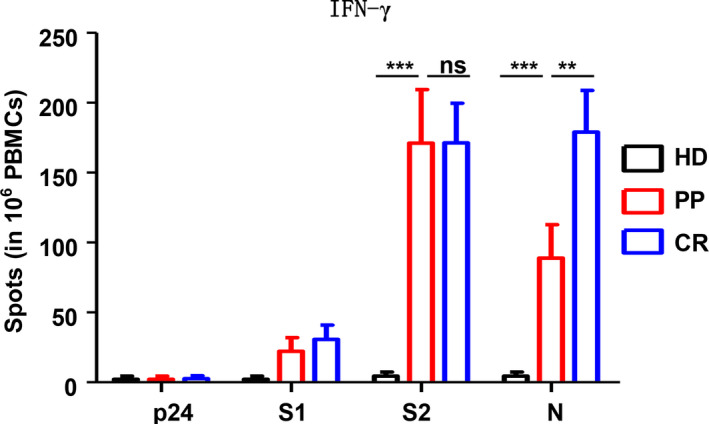
SARS‐CoV‐2‐specific T‐cell responses in the peripheral blood of the HD, PP and CR cohorts. IFN‐γ‐secreting cells were detected by ELISpot following stimulation with the SARS‐CoV‐2 N, S1 and S2 proteins or the irrelevant HIV‐1 p24 protein. The data were obtained from a sub‐PP cohort of 16 individuals, a sub‐CR cohort of 20 individuals and a sub‐HD cohort of 18 individuals. Data were analysed with two‐way ANOVA. ns, non‐significant; **P < 0.01; ***P < 0.001.
The antibody profiles, including total IgM‐ and IgG‐ and SARS‐CoV‐2‐specific antibodies, of the PP and CR cohorts were not significantly different
Before examining the differences in T‐cell responses in the PP and CR cohorts, we first analysed the antibody profiles of the three cohorts. As shown in Figure 2a and b, the total IgM level in the PP and CR cohorts was lower than that in the HD cohort, whereas the total IgG level in the PP and CR cohorts was greater than that in the HD cohort. As expected, the titres of SARS‐CoV‐2 S1, S2, N and RBD protein‐specific antibodies against the PP and CR cohorts were remarkably higher than those in the HD cohort. However, when considering the total IgM‐ and IgG‐ or SARS‐CoV‐2‐specific antibodies, there were no significant differences between the PP and CR cohorts. These results were in agreement with reports from Li et al. 4
Figure 2.
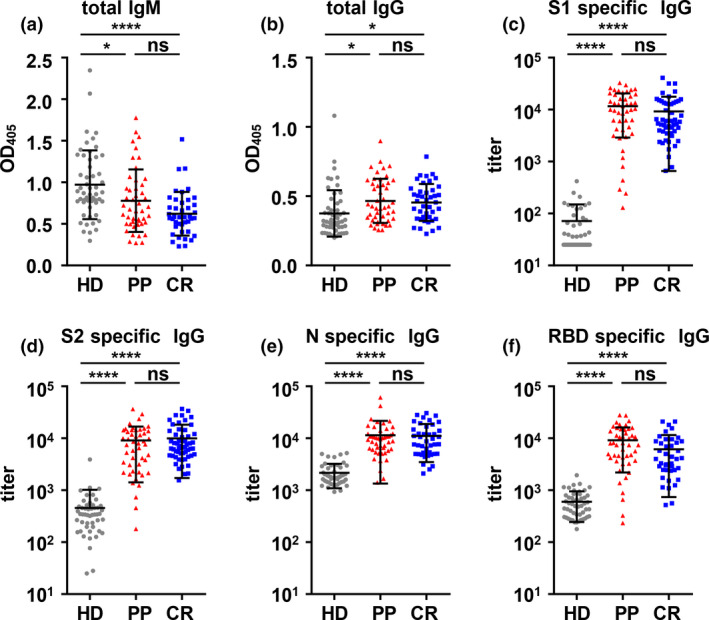
Total IgM and IgG and SARS‐CoV‐2‐specific antibody profiles in the plasma of the PP, CR and HD cohorts. (a, b) Total IgM (a) and IgG (b) in plasma were assayed by ELISA. A dilution of 1:25 600 was used for IgM and 1:512 000 for IgG. (c–f) SARS‐CoV‐2 S1‐specific (c), S2‐specific (d), N‐specific (e) and RBD‐specific (f) IgG in plasma. *P < 0.05; ****P < 0.0001
CD8+ Teff and Tem in the PP cohorts were lower than those in the CR cohort
We then assayed and analysed the composition, differentiation, proliferation, activation and function of CD8+ and CD4+ T cells in the PP cohort, the CR cohort and the HD cohort by flow cytometry. The gating strategies for the total, the differentiated subsets, IFN‐γ+, IL‐4+ and IL‐17A+ of CD8+ and CD4+ T cells are shown in Supplementary figures 2a, Figure 3a and 4a, and Supplementary figure 2c, respectively. The percentages and absolute numbers of CD8+ T cells and CD4+ T cells in the peripheral blood mononuclear cells (PBMCs) of the PP cohort were both comparable to those of the CR and HD cohorts (Supplementary figure 2b, Figure 3c and 4c).
Figure 3.
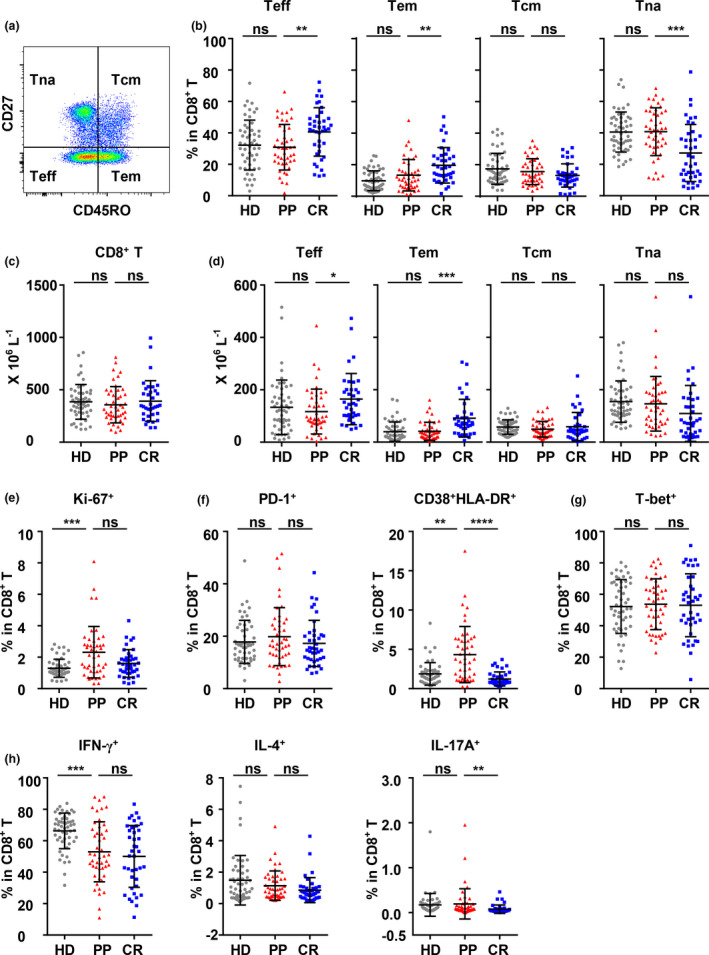
Differentiation, proliferation, activation and function of CD8+ T cells in peripheral blood of the HD, PP and CR cohorts. (a) Gating strategy and the frequencies of the effector T‐cell (Teff, CD45RO−CD27−), effector memory T‐cell (Tem, CD45RO+CD27−), central memory T‐cell (Tcm, CD45RO+CD27+) and naïve T‐cell (Tna, CD45RO−CD27+) subsets in CD8+ T cells of PBMCs. (b, e–g) Frequencies of Ki‐67+, PD‐1+, CD38+HLA‐DR+ and T‐bet+ CD8+ T cells. (c, d) The counts of total (c) and the Teff, Tem, Tcm and Tna subsets of CD8+ T cells (d). (h) To analyse the function of CD8+ T cells, PBMCs were stimulated with PMA/ionomycin for 4.5 h in the presence of BFA and monensin. The production of IFN‐γ, IL‐4 and IL‐17A by CD8+ T cells was analysed by intracellular staining. The data were obtained from 46 PP individuals, 41 CR individuals and 52 HD individuals. Data were analysed with one‐way ANOVA followed by Dunnett's multiple comparison test to compare the PP cohort with the HD and CR cohorts. ns, non‐significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Figure 4.
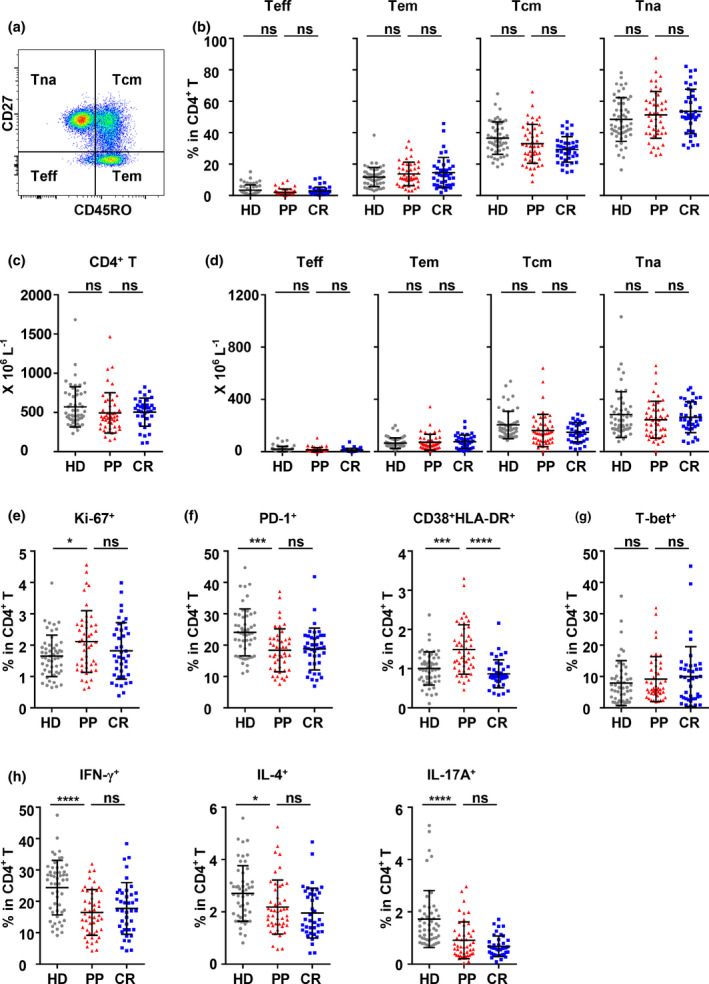
Differentiation, proliferation, activation and function of CD4+ T cells in peripheral blood of the HD, PP and CR cohorts. (a) Gating strategy and the frequencies of the effector T‐cell (Teff, CD45RO−CD27−), effector memory T‐cell (Tem, CD45RO+CD27−), central memory T‐cell (Tcm, CD45RO+CD27+) and naïve T‐cell (Tna, CD45RO−CD27+) subsets in CD4+ T cells of PBMCs. (b, e–g) Frequencies of Ki‐67+, PD‐1+, CD38+HLA‐DR+ and T‐bet+ CD4+ T cells. (c, d) The absolute counts of total (c) and the Teff, Tem, Tcm and Tna subsets of CD4+ T cells (d). (h) To analyse the function of CD4+ T cells, PBMCs were stimulated with PMA/ionomycin for 4.5 h in the presence of BFA and monensin. The production of IFN‐γ, IL‐4 and IL‐17A by CD4+ T cells was analysed by intracellular staining. The data were obtained from 46 PP individuals, 41 CR individuals and 52 HD individuals. Data were analysed with one‐way ANOVA followed by Dunnett's multiple comparison test to compare the PP cohort with the HD and CR cohorts. ns, non‐significant; *P < 0.05; ***P < 0.001; ****P < 0.0001.
Most acute viral infections in humans induce the differentiation of both CD8+ T cells and CD4+ T cells into effector subsets to exert their antiviral functions, as does SARS‐CoV‐2 infection. In our previous studies, significant and gradual increases in effector T cells (CD45RO−CD27−, Teff) in both CD8+ and CD4+ T cells were detected in acute asymptomatic SARS‐CoV‐2 infection, and increases in Teff and effector memory T cells (CD45RO+CD27−, Tem) were also detected in the CR cohort. 15 , 16 However, we found that the frequencies and counts of CD8+ Teff and Tem in the PP cohort were much lower than those in the CR cohort, while all three differentiated subsets, Teff, Tem, central memory T cells (CD45RO+CD27+, Tcm) and naïve T cells (Tna) in the PP cohort were comparable to those in the HD cohort (Figure 3b and d).
In terms of proliferation, the frequency of Ki‐67‐expressing CD8+ T cells in the PP cohort was higher than that in the HD cohort but similar to that in the CR cohort (Figure 3e), though the frequencies of Ki‐67‐expressing CD8+ Teff, Tem and Tcm cell subsets in the PP cohort showed different patterns compared to those in either the HD or CR cohort (Supplementary figure 3a), indicating that the prominent proliferation of CD8+ T cells in the PP cohort was induced by SARS‐CoV‐2 infection at the assay time point. In terms of activation, the frequency of PD‐1‐expressing CD8+ T cells in the PP cohort showed no difference from that in the HD or CR cohort, but the frequency of CD38+HLA‐DR+ CD8+ T cells in the PP cohort was much higher than that in the HD and CR cohorts (Figure 3f and Supplementary figure 3b). Regarding functional potential, the frequency of T‐bet+ CD8+ T cells in the PP cohort was comparable to that in the CR or HD cohort (Figure 3g). However, during polyclonal stimulation, the frequency of IFN‐γ+ CD8+ T cells (Tc1) in the PP cohort was similar to that in the CR cohort but much lower than that in the HD cohort (Figure 3h). Meanwhile, the frequency of IL‐4+ CD8+ T cells (Tc2) in the PP cohort appeared similar to that in the CR or HD cohort, and the frequency of IL‐17+ CD8+ T cells (Tc17) in the PP cohort was significantly higher than that in CR cohort but comparable with that in the HD cohort (Figure 3h).
The data above suggested that the antiviral effector subsets CD8+ Teff and Tem were prominently suppressed in the PP cohort, though the proliferation and activation of CD8+ T cells in the PP cohort appeared to be stimulated by viral infection, as indicated by Ki‐67 and CD38 HLA‐DR expression, respectively. As a result of the potential function of CD8+ T cells, Tc1 in the PP cohort were repressed to a level similar to those in the CR cohort, but Tc2 and Tc17 in the PP cohort did not appear to be repressed, if the HD cohort was taken as the normal non‐repressed control.
The differentiation, proliferation, activation and functional potential of CD4+ T cells in the PP cohort showed no significant differences from those in the CR cohort
In the context of CD4+ T cells, the frequencies and counts of Teff, Tem, Tcm and Tna in CD4+ T cells of the PP cohort were not significantly different from those of the HD or CR cohort (Figure 4b and d). The frequency of Ki‐67‐expressing CD4+ T cells in the PP cohort was higher than that in the HD cohort and the CR cohort, although the difference was not significant (Figure 4e and Supplementary figure 4a). In terms of activation, the frequency of PD‐1+ CD4+ T cells in the PP cohort was remarkably lower than that in the HD cohort but similar to that in the CR cohort (Figure 4f). Specifically, the frequencies of PD‐1+ CD4+ Tcm and Tna in the PP cohort were much lower than those in the CR cohort (Supplementary figure 4b). In contrast, the frequency of CD38+HLA‐DR+ CD4+ T cells in the PP cohort was much higher than that in the HD and CR cohorts (Figure 4f). The functional analysis of CD4+ T cells under polyclonal stimulation showed that the frequency of IFN‐γ+, IL‐4+ and IL‐17+ CD4+ T cells in the PP cohort was notably lower than that in the HD cohort but comparable with that in the CR cohort (Figure 4h). However, the frequency of T‐bet+ CD4+ T cells was not significantly different between the PP and CR cohorts and between the PP and HD cohorts (Figure 4g). The data indicated that no specific pattern of CD4+ T‐cell differentiation was observed after SARS‐CoV‐2 infection in either the PP or CR cohort compared to the HD cohort, though the proliferation of CD4+ T cells might be stimulated. On the other hand, the functions of Th1, Th2 and Th17 cells in the PP cohort and the CR cohort were all reduced compared to those in the HD cohort, which suggested that the functional status of CD4+ T cells was repressed by SARS‐CoV‐2 infection to a significant extent regardless of TRN. It was noted that CD4+ T cells in the PP cohort might be activated for a longer duration, as indicated by the higher frequency of CD38+HLA‐DR+ CD4+ T cells in the PP cohort than that in the CR cohort (Figure 4f).
SARS‐CoV‐2 N protein‐specific IFN‐γ+ T‐cell response in the PP cohort was significantly weaker than that in the CR cohort
Since a significant difference in the general differentiation of CD8+ Teff and Tem was found between the PP and CR cohorts, we examined whether the SARS‐CoV‐2‐specific effector T‐cell responses between the PP and CR cohorts were different. The noncytolytic viral clearance mediated by IFN‐γ that favors the generation of an antiviral microenvironment is also an important antiviral mechanism of effector T cells in addition to cytolytic T cells. Therefore, an ELISpot assay was performed with freshly isolated PBMCs stimulated ex vivo by both surface and non‐surface SARS‐CoV‐2 proteins, including S1, S2 and N. As shown in Figure 5, SARS‐CoV‐2‐specific IFN‐γ+‐expressing T‐cell responses were detected in both the PP and CR cohorts but not in the HD cohort. In terms of antigen specificity, significant S2‐ and N‐specific but only marginal S1‐specific T‐cell responses were detected in both the PP and CR cohorts compared to the irrelevant protein HIV‐1 p24‐negative control. However, it is worth noting that the N‐specific IFN‐γ+ T‐cell response in the PP cohort was significantly weaker than that in the CR cohort, while the S2‐specific IFN‐γ+ T‐cell response in the PP cohort was similar to that in the CR cohort. These data suggested that the SARS‐CoV‐2‐specific T‐cell response in the PP cohort might also be suppressed to a certain extent, which was especially shown by the N‐specific IFN‐γ+ T‐cell response.
The frequencies of total Tregs and CTLA4+ aTregs in the PP cohort were significantly higher than those in the CR cohort
Since regulatory T cells (Tregs) usually play an essential role in compromised viral control, the composition of regulatory T cells (CD127−CD25+, Tregs) in CD4+ T cells of the PP cohort was further analysed and compared with those of the CR and HD cohorts. The frequency of Tregs in CD4+ T cells of the PP cohort was significantly higher than that of the CR cohort and much higher than that of the HD cohort (Figure 6a and b). In Tregs, the frequencies of activated Tregs (CD45RA−FoxP3hi, aTregs) and resting Tregs (CD45RA+FoxP3lo, rTregs) in the PP cohort were not different from those in the CR cohort (Figure 6b). However, the frequency of CTLA4+ aTregs in the PP cohort was significantly higher than that in the CR cohort (Figure 6c), suggesting the higher inhibitory activity of activated Tregs in the PP cohort.
Figure 6.
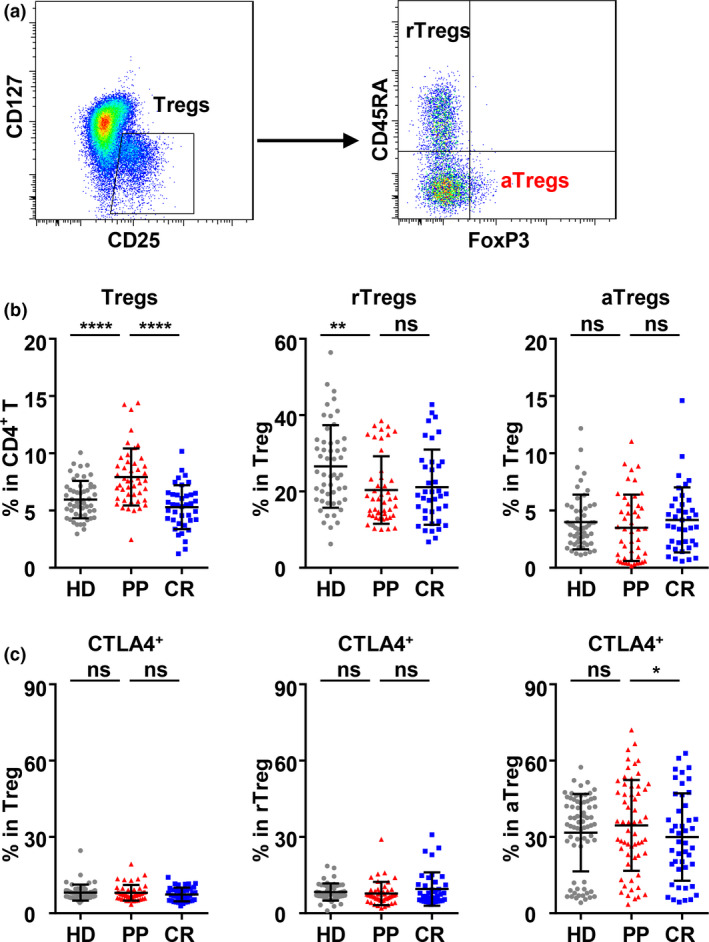
The frequency of Tregs, Treg subsets and CTLA4 expression on Tregs in the peripheral blood of the HD, PP and CR cohorts. (a) Gating strategy of CD25+CD127− Tregs in CD3+CD8−CD4+ T cells and Treg subsets: activated Treg cells (aTregs, CD45RA−Foxp3hi) and resting Treg cells (rTregs, CD45RA+Foxp3lo). (b) The frequency of Tregs in CD4+ T cells and aTregs and rTregs in Tregs. (c) The frequencies of CTLA4+ cells in Tregs and their subsets: rTregs and aTregs. The data were obtained from 46 PP individuals, 41 CR individuals and 52 HD individuals. Data were analysed with one‐way ANOVA followed by Dunnett's multiple comparison test to compare the PP cohort with the HD and CR cohorts. ns, non‐significant; *P < 0.05; **P < 0.01; ****P < 0.0001.
Discussion
Since T‐cell responses play key roles in both viral control and pathogenesis, a growing number of studies on SARS‐CoV‐2‐specific T‐cell responses have been conducted on COVID‐19 patients and individuals who have recovered. 7 , 8 , 10 , 11 , 12 , 17 , 18 , 19 However, alterations in either the phenotype or function potential of the whole T‐cell population have not been widely studied in COVID‐19 patients or convalescent populations, not to mention in the prolonged SARS‐CoV‐2‐positive population.
In the present study, we established a prolonged SARS‐CoV‐2‐positive (PP) cohort of hospitalised patients whose TRN was significantly longer than that of regular clinically recovered individuals. We tried to detect and analyse both SARS‐CoV‐2‐specific and non‐specific antibodies and T‐cell responses in the prolonged SARS‐CoV‐2‐positive PP cohort and in the regularly clinically recovered CR cohort and a HD cohort. We found that the antibody profiles in the PP cohort were not significantly different from those in the CR cohort. However, profound suppression was detected not only in the SARS‐CoV‐2 N protein‐specific IFN‐γ+ T‐cell response but also in the general CD8+ Teff and Tem subsets of the PP cohort. Moreover, a significant increase in Tregs was detected in the PP cohort.
Previous studies have suggested that sex, age and comorbidities including diabetes and hypertension are some of the most common underlying conditions associated with worse clinical outcomes of COVID‐19. 7 , 11 Li et al. 4 also reported that prolonged SARS‐CoV‐2‐positive patients were mostly elderly people. In the process of establishing our cohort, these potential influencing factors, including TIS (Supplementary figure 1b), were all screened as stringently as possible. Notably, lymphopenia, a conventional feature of SARS‐CoV‐2 infection reported to affect CD4+ and CD8+ T cells, was reversed to a normal cell frequency and count at the sampling time point for the PP cohort (Supplementary figures 1c, 2b and Figures 3c, 4c). Moreover, the antibody profiles, including total IgM‐ and IgG‐ and SARS‐CoV‐2‐specific antibodies, of the PP and CR cohort were not significantly different (Figure 2). In addition, the neutralising antibody profiles were similar between long‐term carriers and recovered patients, as reported by a recent work. 4 Therefore, the detected suppression of CD8+ Teff and Tem, the low level of the N‐specific IFN‐γ‐producing T‐cell response and the increase in Tregs (Figures 3b and d, 5 and 6) were most likely to be associated with prolonged SARS‐CoV‐2 positivity, although it is not clear whether these phenotypic alterations are the causes or consequences of delayed viral clearance.
In the course of acute virus infection, T cells usually differentiate into effector cells to exert their antiviral functions. CD8+ T cells are the primary effector cells and receive help from CD4+ T cells. We observed significant increases in Teff and Tem in CD8+ T cells in a follow‐up case study of an asymptomatic SARS‐CoV‐2 patient. 15 Although effector cells declined after the elimination of infection, the increase in Teff and Tem subsets in CD8+ T cells of the CR cohort was observed in our previous study 16 and in this study, suggesting that the general differentiation of CD8+ T cells occurred after SARS‐CoV‐2 infection. A growing number of studies have confirmed that there are broad and strong SARS‐CoV‐2‐specific CD8+ T‐cell responses with a central and effector memory phenotype in recovered subjects. 12 , 13 , 20 Odak et al. 21 also showed that the differentiation of effector/memory T cells is linked to recovery from COVID‐19, and a more robust clonal expansion of CD8+ T cells in peripheral blood may be associated with milder disease or recovery. Our results from the PP cohort further suggested that the potential suppression of CD8+ effector T cells was likely to be linked to prolonged SARS‐CoV‐2 positivity.
The suppression of CD8+ Teff and Tem in the PP cohort may result from differentiation arrest in the activation stage or the quick waning of differentiated CD8+ T subsets. T‐cell differentiation is a tightly regulated process, and the success of differentiation is dependent on efficient and sustained signals such as those provided by professional antigen‐presenting cells, the duration of antigenic stimulation, the help from CD4 cells and the type of cytokine milieu. 22 , 23 , 24 Moreover, the components of viruses also have a strong potential to interfere with the process of Teff differentiation. In patients with acute infection, IL‐2 deficiency during T‐cell activation and circulating HCV core protein may play a central role in inhibiting effector T‐cell differentiation. 25 , 26 Aberrant cytokine production 8 , 18 or delayed or defective type I interferon responses 27 can also potentially skew T‐cell differentiation. Whether and how cytokines or proteins from SARS‐CoV‐2 play roles in the suppressed differentiation of CD8+ effector T cells in the PP population warrant further study.
There is evidence of the association of SARS‐CoV‐2‐specific CD4+ and CD8+ T cells with milder disease, suggesting roles for SARS‐CoV‐2‐specific T‐cell responses in protective immunity in COVID‐19. 28 In agreement with the generally suppressed differentiation of CD8+ Teff and Tem observed with flow cytometry, we found that the SARS‐CoV‐2‐specific IFN‐γ+ T‐cell response in the PP cohort was significantly weaker than that in the CR cohort (Figure 5). Since PBMCs but not purified CD4+ or CD8+ T cells were used in the ELISpot assay, it could not be determined whether the weaker IFN‐γ+ T‐cell response was derived from CD8+ or CD4+ T cells. Nevertheless, it was suggested that a weaker N‐specific T‐cell response was likely to be associated with delayed viral clearance in the PP cohort. On the other hand, it is also suggested that the N‐specific T‐cell response might play an important role in the clearance of SARS‐CoV‐2 infection. Given that SARS‐CoV‐1‐specific memory T cells persist in recovered SARS‐CoV‐1 patients for up to 6 years post‐infection, 29 , 30 vaccines that promote a robust SARS‐CoV‐2‐specific T‐cell response might provide long‐term protection. In our present study, compared with S1, which contains the RBD region, S2 and N induced a more robust SARS‐CoV‐2‐specific T‐cell response. Therefore, S2 and N might be more appropriate targets for the design of T‐cell‐based SARS‐CoV‐2 vaccines.
Tregs play an indispensable role in the maintenance of self‐tolerance and immune homeostasis, possibly at the cost of compromised viral control. 31 , 32 In patients with chronic hepatitis C, the frequency of Tregs was reported to be high, and these cells can suppress virus‐specific CD8+ T cells, while the depletion of Tregs from peripheral blood resulted in the recovery of CD8+ T cells. 33 ‘CTLA4 is a key Treg suppressive molecule as illustrated by its germline deletion or its specific loss in Treg cells, either of which leads to fatal autoimmunity similar to that seen in scurfy mice’. 34 CTLA4 expressed by Tregs can modulate CD80 and CD86 expression by DCs and thereby inhibit the activation of effector T cells. 35 In this study, the frequencies of total Tregs and CTLA4+ aTregs in the PP cohort were both higher than those in the CR cohort, suggesting the involvement of Tregs in the suppression of general and SARS‐CoV‐2‐specific Teff and Tem differentiation. However, we cannot exclude the possibility that prolonged SARS‐CoV‐2 virus positivity, at least of viral RNA, enhanced the induction of Tregs and the CTLA4+ aTreg subset.
In summary, this cohort study found that suppressed CD8+ T‐cell differentiation was likely to be associated with prolonged SARS‐CoV‐2 positivity, suggesting that CD8+ effector T cells are vital for eliminating SARS‐CoV‐2 and that profoundly suppressed CD8+ T‐cell differentiation might be an indicator for the prediction of prolonged SARS‐CoV‐2 positivity in COVID‐19 patients. Whether promoting the SARS‐CoV‐2‐specific T‐cell response and/or inhibiting Tregs could reduce the time of hospitalisation warrants further study in the future.
Methods
Clinical laboratory measurements
Clinical laboratory measurements were performed at Wuhan Jinyintan Hospital in April 2020. Nasopharyngeal swab samples were collected on the day of peripheral blood collection in the CR and HD cohorts and were tested by qRT‐PCR to amplify and assay the E gene, RdRp gene and N gene of SARS‐CoV‐2, as described in our previous study. 36
Lymphocyte response evaluation by flow cytometry
Plasma and cell pellets were separated from fresh peripheral blood from PP patients, clinically recovered donors and HDs. Plasma was used to detect SARS‐CoV‐2‐binding IgM or IgG. PBMCs were separated from cell pellets after resuspension with PBS by density gradient centrifugation and resuspended in complete RPMI 1640 medium containing 10% FBS, 1% penicillin and 1% streptomycin. PBMCs were then divided for 3 analysis panels: panel 1 for T‐cell differentiation, proliferation and activation detection; panel 2 for Treg detection; and panel 3 for cell cytokine production detection. PBMCs in panel 1 and panel 2 were directly used for staining, while PBMCs in panel 3 were stimulated with 200 ng mL−1 PMA (Beyotime, Shanghai, China) and 2.5 μm ionomycin (Beyotime) in the presence of 1 μm monensin (BioLegend, San Diego, CA, USA) and 2.5 μg mL−1 brefeldin A at 37°C and 5% CO2 for 4.5 h before staining. PBMCs were stained with a dead cell discrimination marker (Fixable Viability Dye eFluor™ 506, FVD; eBioscience, San Diego, CA, USA) and surface‐staining antibodies in PBS at 4°C for 30 min. After washing with PBS, cells were fixed with fixation/permeabilisation buffer (eBioscience) at 4°C overnight and then stained with the respective panel of intracellular markers in permeabilisation buffer at 4°C for 30 min. For intracellular cytokine staining, PE/Cy7‐conjugated anti‐human IL‐4, PerCP/Cy5.5‐conjugated anti‐human IL‐17A and PE/Dazzle™ 594‐conjugated anti‐human IFN‐γ were used at 1:250 dilutions. The antibodies used are listed in Supplementary table 2. A BD LSRFortessa flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) was used to assess stained cells, and data were analysed using FlowJo V7.0 (Becton Dickinson).
Enzyme‐linked immunospot assay (ELISpot)
IFN‐γ (UC Tech, Utrecht, Netherlands)‐secreting cells were detected by ELISpot following the manufacturer’s instructions. Briefly, PBMCs were seeded into pre‐coated ELISpot plates and stimulated with 2 μg mL−1 SARS‐CoV‐2 N protein (kindly provided by Professor Ningshao Xia at Xiamen University), S1 and S2 (Sino Biological, Beijing, China) or the irrelevant HIV‐1 p24 protein (homemade) for 45 h at 37°C and 5% CO2. After cell lysis, biotin‐conjugated mAbs were applied prior to adding streptavidin‐HRP, and spots were then developed with an AEC colouring system. The number of spots was counted by an automatic ELISpot reader (AID, Strassberg Germany). Cells stimulated with 10 μg mL−1 phytohaemagglutinin or medium alone were used as positive or negative controls, respectively.
Measurement of total IgM and IgG and SARS‐CoV‐2‐specific IgG in plasma
Total IgM and IgG and SARS‐CoV‐2‐specific IgG in plasma were measured by ELISA. Briefly, each well of flat‐bottom 96‐well ELISA plates (Greiner Bio‐One, Frickenhausen Germany) was coated with 100 μL of 3 μg mL−1 unlabelled goat anti‐human IgM or IgG polyclonal antibodies (Southern Biotechnology, Birmingham, AL, USA) or SARS‐CoV‐2 S1 (Sino Biological), S2 (Sino Biological), N (a gift from Professor Ningshao Xia of Xiamen University) or RBD proteins (aa319‐527, expressed by Sf9 cells) and incubated at 4°C overnight. Plates were blocked with 1% bovine serum albumin in PBS for 2 h, and 100 μL of serially diluted plasma was subsequently added in blocking buffer. Alkaline phosphatase‐labelled goat anti‐human IgM or IgG polyclonal antibodies (Southern Biotechnology) and p‐nitrophenyl phosphate substrate (Sigma, St. Louis, MO, USA) were used for detection. The absorbance was read by a microplate reader (Thermo Labsystems Waltham, MA, USA) at 405 nm. For total IgM and IgG detection, preliminary experiments were carried out with the serial dilution of a small number of plasma samples to obtain the optimal dilutions. Finally, a dilution of 1:25 600 was used for IgM and of 1:512 000 for IgG. Therefore, the result for total IgM and IgG is expressed as the absorbance value at 405 nm. For SARS‐CoV‐2‐specific IgG evaluation, reciprocal values of the lowest dilution with an OD reading that was two times the background were determined as the titre.
Statistics
Data analysis was performed with InStat, version 7.0 (GraphPad Software, La Jolla, CA, USA), and the data are presented as the mean ± SD. Except in the indicated settings, statistical analysis was performed using one‐way ANOVA followed by Dunnett's multiple comparison test to compare the PP cohort with the HD and CR cohorts. For the data in Supplementary figure 1a and b, a t‐test was used to compare differences between two groups to establish and validate the PP and CR cohorts. Two‐way ANOVA was used to analyse the ELISpot results in Figure 5. P‐values < 0.05 were considered significant.
Study approval
This study was reviewed and approved by the Medical Ethical Committee of Wuhan Jinyintan Hospital (Approval Number KY‐2020‐47.01). Written informed consent was obtained from COVID‐19 patients, recovered COVID‐19 individuals and HDs.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
Jingyi Yang: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Visualization; Writing‐original draft; Writing‐review & editing. Maohua Zhong: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Visualization; Writing‐original draft; Writing‐review & editing. Ke Hong: Investigation; Resources; Writing‐review & editing. Qingyu Yang: Investigation; Resources; Writing‐review & editing. Ejuan Zhang: Investigation; Methodology; Visualization; Writing‐review & editing. Dihan Zhou: Investigation; Writing‐review & editing. Jianbo Xia: Investigation; Writing‐review & editing. Yao‐Qing Chen: Investigation; Writing‐review & editing. Mingbo Sun: Funding acquisition; Writing‐review & editing. Bali Zhao: Investigation; Writing‐review & editing. Jie Xiang: Investigation; Writing‐review & editing. Ying Liu: Investigation; Writing‐review & editing. Yang Han: Investigation; Writing‐review & editing. Mengxin Xu: Investigation; Writing‐review & editing. Xi Zhou: Conceptualization; Writing‐review & editing. Chaolin Huang: Conceptualization; Project administration; Resources; Supervision; Validation; Writing‐review & editing. You Shang: Conceptualization; Project administration; Resources; Supervision; Validation; Writing‐review & editing. Huimin Yan: Conceptualization; Project administration; Supervision; Validation; Writing‐original draft; Writing‐review & editing.
Supporting information
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary table 1
Supplementary table 2
Acknowledgments
We thank the patients, the individuals who recovered from COVID‐19 and HDs involved in this study and the staff at Wuhan Jinyintan Hospital. This work was supported by the Kunming Science and Technology Department under grant 2020‐1‐N‐037.
Contributor Information
Chaolin Huang, Email: 88071718@qq.com.
You Shang, Email: you_shanghust@163.com.
Huimin Yan, Email: hmyan@wh.iov.cn.
References
- 1. Sun J, Xiao J, Sun R et al. Prolonged persistence of SARS‐CoV‐2 RNA in body fluids. Emerg Infect Dis 2020; 26: 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oh MD, Park WB, Choe PG et al. Viral load kinetics of MERS coronavirus infection. N Engl J Med 2016; 375: 1303–1305. [DOI] [PubMed] [Google Scholar]
- 3. Na S, Chong YP, Kim MN et al. Duration of viral shedding in patients admitted to hospital with pandemic influenza A/H1N1 2009 infection. J Med Virol 2011; 83: 5–9. [DOI] [PubMed] [Google Scholar]
- 4. Li Q, Zheng XS, Shen XR et al. Prolonged shedding of severe acute respiratory syndrome coronavirus 2 in patients with COVID‐19. Emerg Microbes Infect 2020; 9: 2571–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Decker A, Welzel M, Laubner K et al. Prolonged SARS‐CoV‐2 shedding and mild course of COVID‐19 in a patient after recent heart transplantation. Am J Transplant 2020; 20: 3239–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Man Z, Jing Z, Huibo S, Bin L, Fanjun Z. Viral shedding prolongation in a kidney transplant patient with COVID‐19 pneumonia. Am J Transplant 2020; 20: 2626–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z, John Wherry E. T cell responses in patients with COVID‐19. Nat Rev Immunol 2020; 20: 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathew D, Giles JR, Baxter AE et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science 2020; 369: 1203.12–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun J, Zhuang Z, Zheng J et al. Generation of a broadly useful model for COVID‐19 pathogenesis, vaccination, and treatment. Cell 2020; 182: 734–743.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilk AJ, Rustagi A, Zhao NQ et al. A single‐cell atlas of the peripheral immune response in patients with severe COVID‐19. Nat Med 2020; 26: 1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wen W, Su W, Tang H et al. Immune cell profiling of COVID‐19 patients in the recovery stage by single‐cell sequencing. Cell Discov 2020; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ni L, Ye F, Cheng ML et al. Detection of SARS‐CoV‐2‐specific humoral and cellular immunity in COVID‐19 convalescent individuals. Immunity 2020; 52: 971–977.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng Y, Mentzer AJ, Liu G et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol 2020; 21: 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sette A, Crotty S. Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell 2021. S0092‐8674(21)00007‐6. 10.1016/j.cell.2021.1001.1007. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang J, Zhang E, Zhong M et al. Longitudinal characteristics of T cell responses in asymptomatic SARS‐CoV‐2 infection. Virol Sin 2020; 35: 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J, Zhong M, Zhang E et al. Broad phenotypic alterations and potential dysfunctions of lymphocytes in COVID‐19 recovered individuals. medRxiv 2020. 10.1101/2020.1107.1101.20144030v20144031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen G, Wu D, Guo W et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020; 130: 2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giamarellos‐Bourboulis EJ, Netea MG, Rovina N et al. Complex immune dysregulation in COVID‐19 patients with severe respiratory failure. Cell Host Microbe 2020; 27: 992–1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thevarajan I, Nguyen THO, Koutsakos M et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med 2020; 26: 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grifoni A, Weiskopf D, Ramirez SI et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell 2020; 181: 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Odak I, Barros‐Martins J, Bosnjak B et al. Reappearance of effector T cells is associated with recovery from COVID‐19. EBioMedicine 2020; 57: 102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manjunath N, Shankar P, Wan J et al. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J Clin Invest 2001; 108: 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaech SM, Wherry EJ, Ahmed R. Effector and memory T‐cell differentiation: implications for vaccine development. Nat Rev Immunol 2002; 2: 251–262. [DOI] [PubMed] [Google Scholar]
- 24. Lanzavecchia A, Sallusto F. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science 2000; 290: 92–97. [DOI] [PubMed] [Google Scholar]
- 25. Accapezzato D, Francavilla V, Rawson P et al. Subversion of effector CD8+ T cell differentiation in acute hepatitis C virus infection: the role of the virus. Eur J Immunol 2004; 34: 438–446. [DOI] [PubMed] [Google Scholar]
- 26. Francavilla V, Accapezzato D, De Salvo M et al. Subversion of effector CD8+ T cell differentiation in acute hepatitis C virus infection: exploring the immunological mechanisms. Eur J Immunol 2004; 34: 427–437. [DOI] [PubMed] [Google Scholar]
- 27. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell 2020; 181: 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rydyznski Moderbacher C, Ramirez SI, Dan JM et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell 2020; 183: 996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan YY, Huang ZT, Li L et al. Characterization of SARS‐CoV‐specific memory T cells from recovered individuals 4 years after infection. Arch Virol 2009; 154: 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang F, Quan Y, Xin ZT et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six‐year follow‐up study. J Immunol 2011; 186: 7264–7268. [DOI] [PubMed] [Google Scholar]
- 31. Arpaia N, Green JA, Moltedo B et al. A distinct function of regulatory T cells in tissue protection. Cell 2015; 162: 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan Z, Zhou Z, Liu Y et al. Regulatory T cells and T helper 17 cells in viral infection. Scand J Immunol 2020; 91: e12873. [DOI] [PubMed] [Google Scholar]
- 33. Chigbu DI, Loonawat R, Sehgal M, Patel D, Jain P. Hepatitis C virus infection: host‐virus interaction and mechanisms of viral persistence. Cells 2019; 8: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity 2019; 50: 302–316. [DOI] [PubMed] [Google Scholar]
- 35. Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 2010; 10: 490–500. [DOI] [PubMed] [Google Scholar]
- 36. Cao B, Wang Y, Wen D et al. A trial of Lopinavir‐Ritonavir in adults hospitalized with severe Covid‐19. N Engl J Med 2020; 382: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary table 1
Supplementary table 2


