Abstract.
Purpose: The mean linear intercept (MLI) score is a common metric for quantification of injury in lung histopathology images. The automated estimation of the MLI score is a challenging task because it requires accurate segmentation of different biological components of the lung tissue. Therefore, the most widely used approaches for MLI quantification are based on manual/semi-automated assessment of lung histopathology images, which can be expensive and time-consuming. We describe a fully automated pipeline for MLI estimation, which is capable of producing results comparable to human raters.
Approach: We use a convolutional neural network based on U-Net architecture to segment the diagnostically relevant tissue segments in the whole slide images (WSI) of the mouse lung tissue. The proposed method extracts multiple field-of-view (FOV) images from the tissue segments and screen the FOV images, rejecting images based on presence of certain biological structures (i.e., blood vessels and bronchi). We used color slicing and region growing for segmentation of different biological structures in each FOV image.
Results: The proposed method was tested on ten WSIs from mice and compared against the scores provided by three human raters. In segmenting the relevant tissue segments, our method obtained a mean accuracy, Dice coefficient, and Hausdorff distance of 98.34%, 98.22%, and , respectively. Our proposed method yields a mean precision, recall, and -score of 93.37%, 83.47%, and 87.87%, respectively, in screening of FOV images. There was substantial agreement found between the proposed method and the manual scores (Fleiss Kappa score of 0.76). The mean difference between the calculated MLI score between the automated method and average rater’s score was ().
Conclusion: The proposed pipeline for automated calculation of the MLI score demonstrates high consistency and accuracy with human raters and can be a potential replacement for manual/semi-automated approaches in the field.
Keywords: image segmentation, histopathology, mean linear intercept, bronchopulmonary dysplasia
1. Introduction
Bronchopulmonary dysplasia (BPD) is the most common complication of preterm birth.1 BPD is a chronic lung disease, characterized by an arrest in alveolar and vascular growth within the lung. BPD is a multifactorial disease, caused by ventilator and oxygen therapy administered for acute respiratory failure, and is commonly associated with ante- and post-natal inflammation.2,3 Although, there is currently no effective treatment for BPD, ongoing investigations are in progress to better understand the pathophysiology of this disease. To discover new potential therapies, it is crucial that researchers quantify the lung injury phenotype in an accurate and efficient manner.
A common metric used to quantify lung injury is the mean linear intercept (MLI), which represents the mean distance between alveolar septa within the lung.4 Investigators using animal models to mimic neonatal chronic lung disease, often use the MLI as a parameter to describe the simplification of the lung architecture, characteristic of BPD. The conventional method for MLI quantification of lung tissue specimens usually includes the microscopic assessment of histopathological slides by an expert, which is inefficient, tedious, and time-consuming. Moreover, there is a lack of objective visual gold standards for structures found in microscopic views of tissues, which leads to inter- and intra- expert variation and reproducibility issues.5–7 In recent years, there has been a shift toward the development of automated methods for assessment of histopathology images to address the shortcomings in the conventional approaches.8–11 However, due to technical impediments such as object variability, varying straining, and artifacts, the development of robust and comprehensive methods for assessment of histopathology images remains a challenging task.
Automated estimation of the MLI score requires detailed and accurate segmentation of biological structures in lung histopathology images, which makes development of such approaches difficult. In recent years, few studies have been reported on lung histopathology image analysis with a focus of automating the MLI quantification process. However, these methods often have difficulty identifying non-alveolar structures (e.g., blood vessels and bronchi), which leads to underestimation of the MLI score in comparison to manual measurements.12 Moreover, the technical details provided for such algorithms are often limited. As a result, there are currently no accessible and reliable automated approach for MLI quantification in the literature and consequently, the most trusted methods remain manual/semi-automated techniques,13–15 which can be laborious, time-consuming, and subjective. In this work, our aim is to present a fully automated pipeline for estimation of the MLI score in histopathology images of mouse lung tissue. The main contributions of this paper are: (1) proposing an innovative approach for assessment of digitized histopathology slides to automate the estimation of the MLI scoring; (2) performing accurate segmentation of different biological structures in the histopathology images of mouse lung tissue; and (3) evaluation of the proposed method against human raters.
2. Materials and Methods
2.1. Histopathology Images of Mouse Lung Tissue
Our dataset comprises high-resolution whole slide images (WSIs) of 10 lung histopathology specimens of mice obtained from the Sinclair Centre for Regenerative Medicine (Ottawa Hospital Research Institute, Ottawa, Ontario). All animal experiments were conducted in accordance with protocols approved by the University of Ottawa Animal Care Committee. The lungs specimens were inflation fixed through the trachea with 10% buffered formalin, under 20-cm pressure, for 5 min. After the trachea was ligated, the lungs were immersion fixed in 10% buffered formalin for 48 h at room temperature and then immersed in 70% ethanol for 24 h at room temperature. The Louise Pelletier Histology Core Facility at the University of Ottawa paraffin-embedded, cut ( sections), mounted, and stained the lung tissue with hematoxylin and eosin (H&E).
The slides were scanned using an Aperio CS2 slide scanner (Leica), and high-resolution color images at magnification (i.e., per pixel) were obtained. In total, 10 WSIs were generated from two different experimental groups. The WSIs were randomly selected from different mice lung tissue while blinded to the experimental groups. This procedure was consistent across all animals. The first group contains five WSIs of healthy mouse lung tissue from mice that were housed in room air (RA) (RA group), which is used as the control group in this experiment. The second group consists of five WSIs of diseased mouse lung tissue from mice that were exposed to a high concentration of oxygen and lipopolysaccharide (LPS) ( group). The experimental group mimics the conditions that a preterm infant is exposed to (high concentration of oxygen and inflammation), which can lead to a lung injury phenotype seen in BPD. The WSIs generated from the group normally contain fewer and larger alveoli, in comparison to WSIs from the RA control group. This is expected to reflect in the MLI score by calculation of a higher value of MLI in the group in comparison to the RA control group, which have much more and smaller alveoli.
2.2. Conventional MLI Quantification
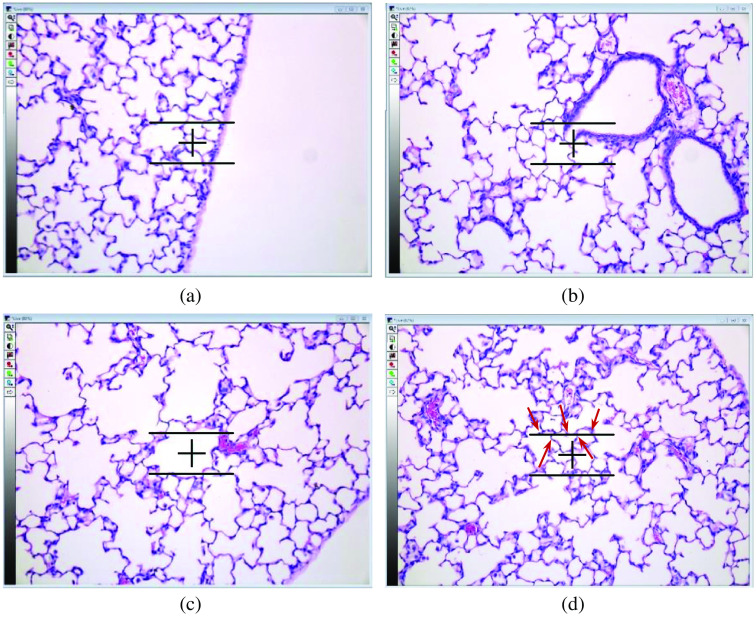
Conventionally, the MLI score is calculated using a semi-automated process.4 In this process, the microscope software (MetaMorph Software version 7.8, Molecular Devices, LLC) automatically presents the human rater with a field of view (FOV) from pre-defined grid points. Each FOV image is a sub-image from the input WSI, overlaid with guidelines in the middle of the image (see Fig. 1); the top horizontal guideline, of length , is used by a human rater in the MLI quantification procedure.
Fig. 1.
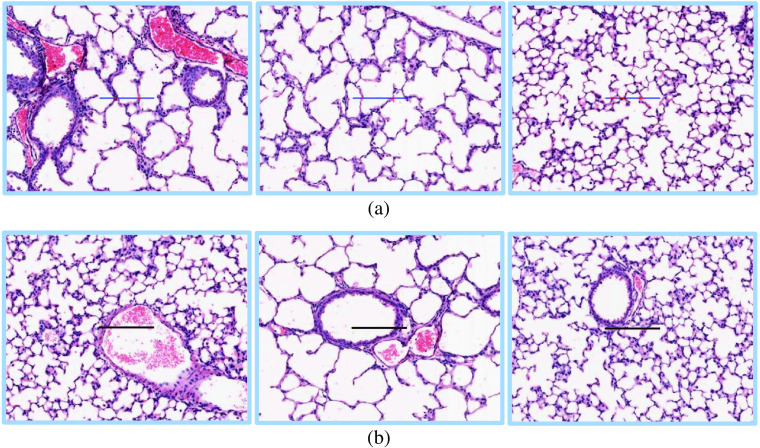
Examples of extracted fields of view (FOV) images using the image analysis software. (a) This FOV image is rejected because the guideline is partially outside of the section. (b) The FOV image is rejected due to intersection of the guideline with a bronchus. (c) The FOV image is rejected because the guideline intersects with a blood vessel. (d) An example of accepted FOV image with five intersections. The intersections of the guideline with the septa is shown using red arrows.
The human rater first decides whether the shown FOV image can be used for MLI quantification or not. The rejection of a FOV image is based on two criteria. First, if a part of the horizontal guideline is in the pleural space [i.e., outside the lung space; Fig. 1(a)], the FOV image is rejected; i.e., it is not used in the calculation of the MLI score. Second, if the horizontal line intersects a bronchus [Fig. 1(b)] or a vessel [Fig. 1(c)], the FOV image is rejected. If the FOV image is not rejected, the human rater counts the number of intersections. An intersection is when the horizontal guideline fully crosses over the septa, which is the alveolar border wall [Fig. 1(d)]. The MLI score is calculated as
| (1) |
where is the number of accepted FOV images, 155.34 refers to the length of the horizontal line in , and is the total number of intersections counted from all of the accepted FOV images. A minimum of 250 accepted FOV images (i.e., number of FOV images, not including those that were rejected), is desired for the computation of the MLI score.16
2.3. Automated Calculation of MLI Score
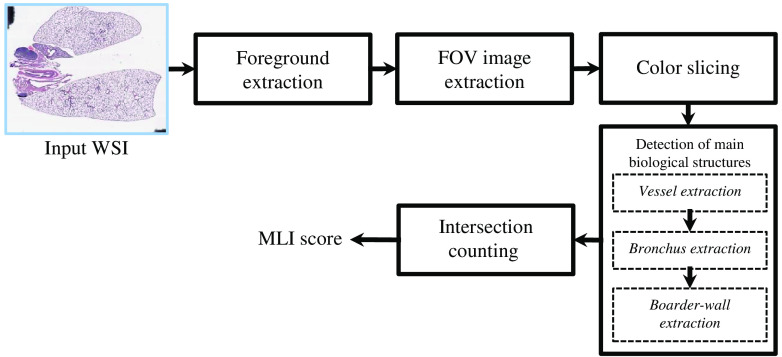
Our proposed pipeline for the automated estimation of the MLI score consists of five steps, which are shown in Fig. 2.
Fig. 2.
Block diagram of the proposed methodology. The abbreviations WSI, FOV, and MLI are referred to whole slide image, FOV, and MLI, respectively.
2.3.1. Foreground extraction
Each WSI may contain various imaging artifacts and undesired biological structures (see Input WSI in Fig. 2). As an initial step in our pipeline, we segment the lung space from the pleural space and undesired artifacts, which are considered the foreground and background, respectively. The foreground regions of interest (ROIs) were segmented using a convolutional neural network (CNN), based on the U-Net17 architecture.
Since the histopathology slides were scanned at magnification, images’ sizes are large (average size of ) and contain a high level of detail that is not required to segment the foreground ROI. As such, images were down-sampled by a factor of 10, greatly reducing the computational cost while still allowing for accurate ROI segmentation. From each WSI, image patches () were extracted using a sliding window, with 50% overlap in the horizontal and vertical directions. Data augmentation (90-deg rotations and image flipping) was employed to the training dataset to increase the classifier performance.18
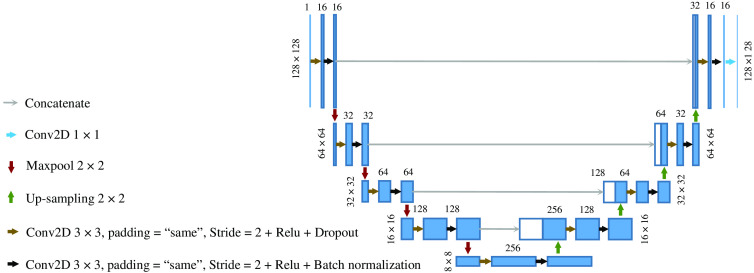
The CNN was trained from scratch, with four convolution layers in the contracting path and four transpose convolution layers in the expansive path. The complete architecture of the network is illustrated in Fig. 3. The CNN was trained using the Adam optimizer,19 binary cross-entropy loss function, and mean intersection over union evaluation metric. To account for the overfitting problem, we perform dropout by a factor of 0.5 at each layer and perform cross-validation at each epoch with the ratio of 10:1 (train on 90% of the training data and validation on the remaining 10% at each epoch). Batch normalization was applied to each layer to reduce the training time and prevent diverging gradients. We trained our model for 200 epochs with a batch size of 100. We used the Keras framework for algorithm development on a standard workstation with an Intel Core i7-3770 3.40 GHz CPU, 12 GB of installed RAM, and a single NVIDIA RTX 2060 with 6 GB memory. A total number of 2,161,649 trainable parameters were optimized in our segmentation model.
Fig. 3.
The CNN architecture. The blue and white boxes represent the multi-channel feature maps and copied feature maps, respectively. The size of the feature maps is indicated on top of each box and the size of each input layer is denoted on the left-hand side.
2.3.2. FOV image extraction
From the foreground region of each original high-resolution WSI, FOV images (sub-images of size ) were extracted using a sliding window, with a 50% and 75% overlap in the horizontal and vertical directions, respectively. For each FOV image, a horizontal guideline (thickness 1 pixel; length 312 pixels, which corresponds to ) was superimposed at the center of each image and used by human raters to count the intersections. In the automated process the guideline is only virtually superimposed, appearing only for visualization purposes. If the horizontal guideline does not fully reside within the foreground region, the FOV image is rejected.
2.3.3. Color slicing
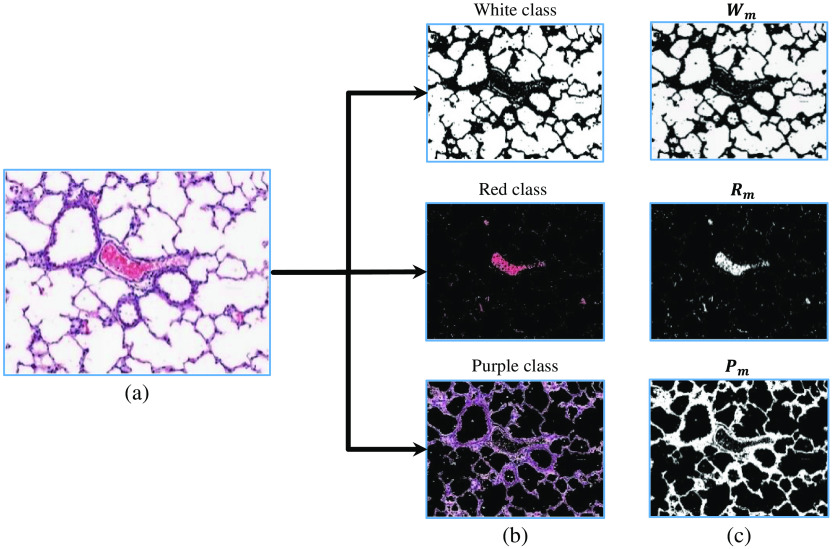
A number of factors can contribute to variations of the color content in histological images (e.g., histochemical staining time, amount of histology stain used) across different WSIs. We applied color normalization20 to the input WSIs to mitigate such variations. Next, pixels are classified, using a color slicing algorithm,21 into the three main categorizes of color in the lung images (see Fig. 4): (1) white, (2) red, and (3) purple.
Fig. 4.
The organization of extracted colors in a FOV image. (a) The original input FOV image. (b) The RGB representation of each color slice. (c) The visualization of each color binary mask, , , and .
The white pixels in each FOV image belong to two main sources: (1) white areas in the pleural space (i.e., background regions) and (2) white areas within the lung space. The white pixels located in the pleural space are segmented in the foreground extraction step. Therefore, only the white pixels corresponding to the lung region need to be identified. The binary mask denotes the white pixels within the lung space and is determined using Eq. (2):
| (2) |
In Eq. (2), , , and are the red, green, and blue channels in RGB color space, respectively. is a threshold whose value is determined using Otsu’s method22 at each fold. This threshold ensures that the extracted pixels have high intensities (close to white). The constant is a fixed threshold for all folds to ensure that the intensity difference between the R, G, and B channels is small. The value assigned to was 15, which was determined empirically. The mask is generated by the intersection of the and binary masks, and contains the white pixels within the lung space. The and are the corresponding number of rows and columns, respectively.
is the binary mask denoting the red pixels in each FOV image. These pixels usually represent the remaining blood cells in the lung tissue, which can indicate the presence of a vessel in the neighboring region. The binary mask is determined using Eq. (3):
| (3) |
In Eq. (3), the represents all of the pixels that are not in . The value of the threshold is determined using Otsu’s method22 at each fold.
The remaining pixels are assigned to the purple color binary mask , which represents several cellular compartments (e.g., pneumocytes cells, glands, and smooth muscle).
2.3.4. Detection of main biological structures
Each FOV image is segmented into three biological structures: (1) alveoli, (2) vessel, and (3) bronchi. As stated in Sec. 2.2, FOV images whose horizontal guideline intersects with vascular and bronchus regions should be rejected (i.e., excluded from MLI calculation). We first automatically segment the lumen region (LR) of vessels based on a region growing method.23 We then separate bronchi from alveoli, based on multiple morphological features extracted from each individual LR. Finally, the segmentation approach is completed by identification of the border wall of the vessel and bronchus (see Fig. 5).
Fig. 5.
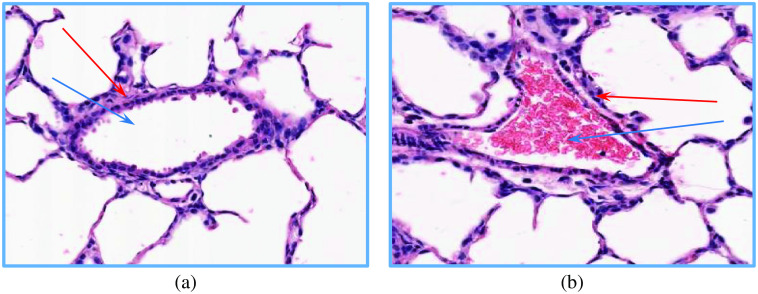
The visualization of (a) a bronchus and (b) vessel. The corresponding LR and border wall region of each structure is identified by blue and red arrows, respectively. As can be seen in (b), the LR of the vessel is densely covered by the red blood cells.
Vessel extraction
To identify different lung structures in each FOV image, it is desirable to detect all of the LRs using . However, due to high density of blood cells in the LR of the vessels, accurate extraction of the LR of vessels from is not feasible. Therefore, we develop an alternative approach, where we first extract candidate seed regions for each individual vessel in the FOV image and then use a region growing method to segment all of the LR of the vessels.
Since most vessel structures have a considerable amount of red blood cells within their LR [see Fig. 5(b)], it is possible to locate the candidate seed regions by detection of the areas in the red color binary mask , where the density of red pixels is relatively high. To detect these regions in , using a window, we iteratively sweep with 50% horizontal and vertical overlap. At each iteration, the local density of the red pixels is calculated. If the local density is higher than 0.9, all of the red pixels present in the window will be added to the binary mask .
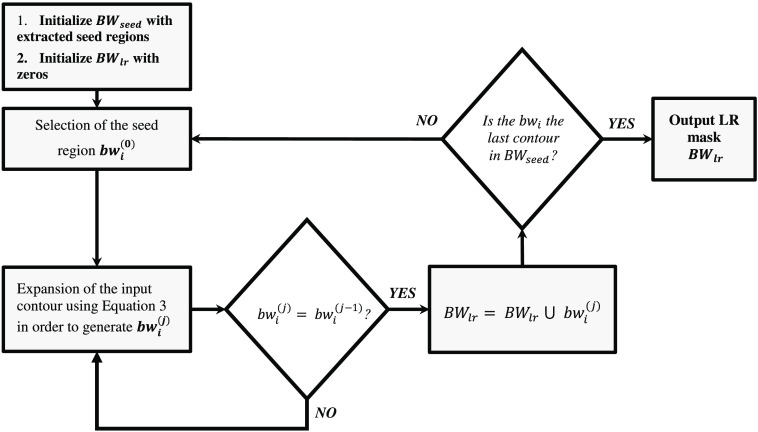
Figure 6 shows different steps in the LR segmentation method. To extract the LRs associated with each seed region in , we apply a region growing segmentation approach to each individual seed region. Let denote different seed regions in . In the proposed LR segmentation, the selected seed region is iteratively expanded using Eq. (4).
| (4) |
Fig. 6.
The block diagram of the region growing based LR segmentation. The notation refers to the ’th selected seed region and superscript () indicates that is expanded times.
Equation (4) shows the morphological dilation operation () of input binary mask by a circular structuring element with radius of two pixels (), followed by a bitwise AND operation of the resultant mask with . If the expanded binary mask does not change after applying Eq. (4) (), the algorithm quits the iterative expansion loop and adds the binary mask to the output LR binary mask [see Eq. (5)]. The operation is continued until all of the seed regions are processed by the algorithm.
| (5) |
Bronchus extraction
The LR of the remaining structures (bronchi and alveoli) are mostly white and can be directly extracted using . The next step is to separate the LR of the bronchi from the alveoli’s LR. Based on unique visual characteristics of the LR of the bronchi, multiple morphological and textural features are extracted from each individual LR, which is used for classification of the remaining structures. These features are briefly introduced in the following:
-
a.
Area of the LR: One of the salient visual features in classification of the remaining LRs is the area of the LR. In the room air condition (i.e., RA group), there is a considerable size difference between the LR area of bronchi compared to the LR of alveoli, and therefore the size feature plays an effective role in classification of bronchi versus alveoli. However, in experimental group, the alveoli structures are enlarged. As such, LR area may not be as reliable a feature for the group and the classifier may be more reliant on the other extracted features.
We define the area of an object as the overall comprising pixels in the LR of the object.
-
b.Shape of the LR: Based on our visual observation, the shape of the LR is another distinguishing feature to discern bronchi versus alveoli LR. For instance, the LR of bronchus structures are mostly ovular and do not contain many branches. The alveoli LR are often non-ovular and contain many sub-branched [see Fig. 7(a)]. We used Eq. (6) to quantify the circularity of the LR.
(6)
Fig. 7.
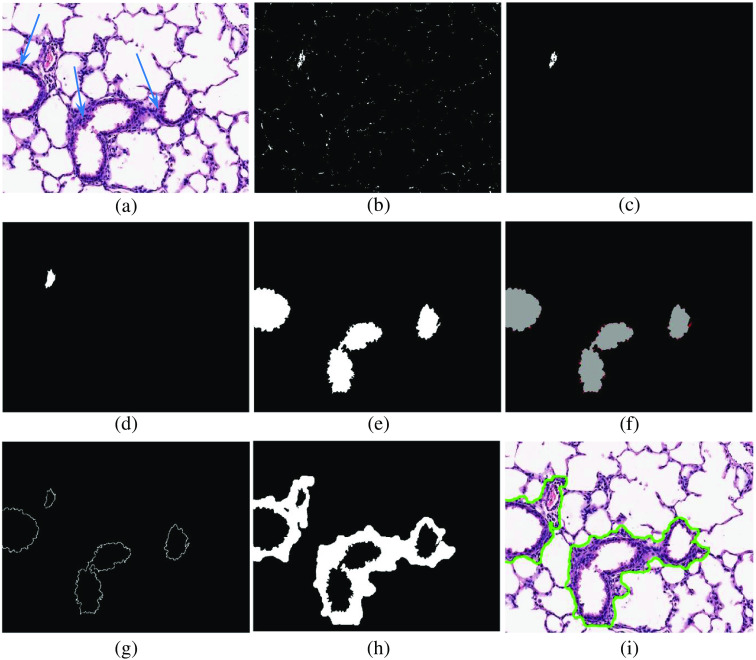
(a) A sample FOV image. The blue arrows indicate the border-wall region of the bronchi structures. (b) Red color binary mask . (c) The extracted vessel’s seed region. (d) The extracted vessel LR. (e) The extracted bronchi LR. (f) The visualization of the ripple pattern in bronchi structures. The ripple pattern is marked as red in this figure. (g) The corresponding seed region in border-wall extraction step. (h) The extracted border-wall regions of every vessel and bronchi. (i) The segmented bronchi and vessel in the FOV image.
In Eq. (6), and represent the functions that calculate the area and the perimeter of the input binary mask input contour , respectively. The value of is an integer between “0” and “1”, where 1 represents a circular contour.
-
c.Thickness of the boarder wall region: The majority of bronchus structures have a thick border wall region in comparison to other structures in lung tissue [see Fig. 7(a)]. Therefore, the thickness of the border-wall region of the remaining structures are extracted. To quantify the thickness of the boarder-wall region, the complete segmentation of the boarder-wall of each structure is not required. Instead, we expand the selected LR using Eq. (7) until the ratio of pixels in the expanded area become larger than 25%. The number of times that Eq. (7) will be used to expand the LR can be used as an indication of the border-wall region thickness.
(7)
Equation (7) shows the morphological dilation operation () of input binary mask by a circular structuring element with radius of one pixels (), followed by removing the pixels of the from the resultant binary mask ().
-
d.LR’s perimeter ripple pattern: Another important biological feature is the frequent detection of ripple shape patterns on the perimeter of the LR of bronchi [see Fig. 7(f)]. Considering the fact that this ripple pattern is more apparent in the LR of the bronchi, it can be a valuable discerning feature. For each LR, this feature is quantified by calculation of the ratio of the total area of the ripples over the area of the LR [Eq. (8)].
(8)
In Eq. (8), is the generated binary mask from the input LR, is the binary mask of the ripple pattern, represents the opening morphological operator with a circular structuring element with a radius of three, represents the function that calculates the area of the input binary mask, and is the ripple pattern feature.
After the feature extraction step, we classify the remaining structures as bronchi or alveoli using a decision tree based classifier. The classifier is trained on FOV images extracted from eight WSIs and will be tested on the remaining two WSIs at each fold. In our dataset, the population ratio of the bronchus samples to alveoli samples is . Therefore, we perform synthetic oversampling of minority class method (SMOTE)24 to address the class imbalance in our dataset. In this approach, the synthetic observations are created based on the existing minority observations. For each minority class observation, SMOTE calculates -nearest neighbors. Depending on the number required oversampled observations, one or more of the -nearest neighbors are selected to create the synthetic examples.
After identification of the LRs of the bronchi structures, the vessels, and the bronchi structures LR will be used in the next step for extraction of the boarder-wall region.
2.3.5. Border-wall extraction
As seen in Fig. 7(a), all of the biological structures in the FOV image are surrounded by a border-wall. From a biological standpoint, these border-wall regions are considered as a part of each structure in the lung tissue. Therefore, the border-wall corresponding to each vessel and bronchus LR, are extracted. Here, we use a region growing approach similar to that of the extraction of the vessels LR. First, we acquire the initial seed pixels using the surrounding neighboring pixels of each structure LR [see Fig. 7(g)]. The initial seed pixel is then iteratively expanded using Eq. (9) until the entire border-wall region of the selected LR is extracted.
| (9) |
In Eq. (9), is the binary mask containing the border-wall pixels, is the morphological dilation of the input binary mask by a circular structuring element with radius of two pixels, & indicates the arithmetic AND operation, is the purple color binary mask, and is the morphological opening operation by a circular structuring element with radius of .
To define the expansion stopping criteria, we take advantage of the fact that the border-wall surrounding each vessel and bronchus LR is relatively thicker than the alveoli’s border-wall (i.e., septa). As a result, it is possible to separate the border-wall of the vessel and bronchi from septa by measuring the thickness of the wall [see Fig. 7(h)]. The erosion criteria is implemented as the morphological opening term in Eq. (9), using a circular structuring element with radius of . The optimized value of the over our training data (extracted FOV images, which were used for training at each fold) was 16 pixels in average.
2.3.6. Intersection counting
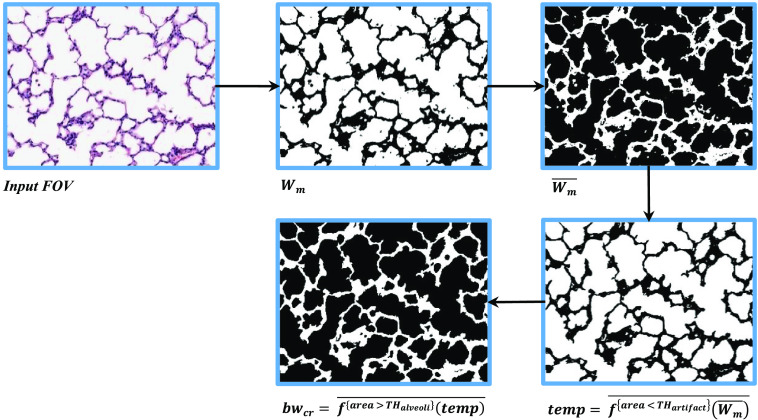
To calculate the MLI score, we are required to count the total number of intersections of the horizontal guideline with septa within the FOV image. First, we reject any FOV image, where the horizontal guideline touches a vessel or bronchus. Then, in the remaining FOV images, we segment the septa regions using Eq. (10). Figure 8 visualizes different steps in segmentation of septa region in a FOV image.
| (10) |
Fig. 8.
An example of septa segmentation in a FOV image.
In Eq. (10), is the estimated septa region, is an operator that removes the contours with area less than the threshold , and is an operator that removes the contours with area larger than the threshold . After extraction of septa regions, it is straightforward to count the total number of intersections in each FOV image.
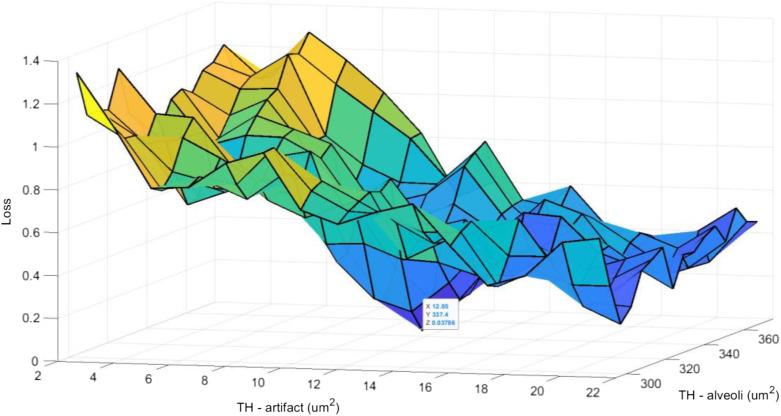
The values of and were determined using the grid search hyper-parameter optimization algorithm25 (see Fig. 9). To acquire the optimum values of the hyper-parameters at each fold, the algorithm was run multiple times (number of grid points used in the grid search) using different hyper-parameters values. The cost value [calculated using Eq. (11)] was observed to capture the hyper-parameters associated with the minimum cost values.
| (11) |
where, is the estimated intersections in the ’th FOV image, is the true number of intersections according to the manual assessment, and is the batch size.
Fig. 9.
An example of hyper-parameters grid search at each fold. At each fold, the values of and were determined over eight training WSIs and will be tested on two remaining WSIs in the test set. This procedure continues until all WSIs used as test set.
2.4. Evaluation of the Developed Method
The overall performance of the proposed method is affected by three main steps: (1) Foreground extraction, (2) detection of main biological structures, and (3) intersection counting. Performance metrics are provided for each step. We performed five-fold cross validation. Within each fold, we used eight of the WSIs from the dataset (four from RA and four from ) as the training dataset, and the remaining two WSIs were used as the test dataset in each stage of the proposed pipeline. This was repeated five times such that each WSI was used as the test set.
2.4.1. Foreground extraction
In the foreground extraction step, the training dataset (i.e., eight WSIs) was used for training and validation (90% training and 10% validation) of the CNN and the test dataset (i.e., remaining two WSIs) for testing. On average, 28,092 image patches were generated from the eight WSIs at each fold, which were assigned to the training and validation sets with a 9:1 ratio (i.e., 25,283 and 2809 for training and validation, respectively). The remaining two WSIs in each fold, used for the test dataset, had the average of 7023 image patches.
The proposed foreground extraction results were compared with the manual segmentation. Manual segmentation of the foreground was performed on each individual WSI image using ImageJ software26 by Sina Salsabili. The manual segmentation was used as the ground truth for our foreground extraction approach.
To quantitatively evaluate the performance of our segmentation method, we used both region-based and boundary-based metrics. For region-based, we computed the Dice similarity coefficient27 (DSC) and pixel-wise accuracy (AC).
| (12) |
| (13) |
where,
-
•
X: are the set of pixels within the ROI region in the manual segmentation.
-
•
Y: are the set of pixels within the ROI region in the CNN segmentation.
-
•
|·|: is the cardinality of the set.
-
•
True positive (TP): ROI region is correctly detected as ROI region (i.e., ).
-
•
True negative (TN): Non-ROI region is correctly detected as Non-ROI region (i.e., ).
-
•
False positive (FP): Non-ROI region is incorrectly detected as ROI region (i.e., ).
-
•
False negative (FN): ROI region is incorrectly detected as non-ROI region (i.e., ).
For boundary-based, the Hausdorff distance28 between the border of the segmented WSI and that of the ground truth was calculated.
2.4.2. Detection of main biological structures
For our dataset, we extracted 18,321 FOV images of size (9009 FOV images from the RA group and 9312 FOV images from the group). At training phase, we used the ground truth foreground extraction as the guideline for extraction of FOV images to generate the training data. At each fold, an average of 14,657 FOV images extracted from the eight WSIs, which were used for training of the later steps (i.e., bronchi versus alveoli classification) of our pipeline. At the testing phase, the CNN model was used to extract FOV images from the test dataset (i.e., the remaining two WSIs). An average of 3664 FOV images were used for testing at each fold.
Manual MLI scoring was conducted independently by co-authors Sina Salsabili and Shreyas Sreeraman (both are considered novice scorers, who were provided 5 h of training in Bernard Thebaud’s laboratory), and co-author Marissa Lithopoulos (verified by co-author Bernard Thebaud; and considered an expert scorer with three years of experience), referred to as rater 1, rater 2, and rater 3, respectively.
The detection of vessels and bronchi is necessary to properly reject FOV images, where the horizontal guideline touches one of these biological structures. In this step, the metrics in Eqs. (14)–(16) were utilized to evaluate the performance of the proposed method in detection of the accepted/rejected FOV images.
| (14) |
| (15) |
| (16) |
We compared the performance of the proposed method against the ground truth, which consists of manual scores for calculation of the MLI for each WSI by three human raters. Our ground truth includes the assessment of accepted/rejected FOV images and the number of intersections with alveoli wall in each accepted FOV image. To evaluate our method in detection of accepted/rejected FOV images, two evaluation approaches were conducted:
-
1.
Acquiring the ground truth for accepted/rejected FOV images based on the majority vote (i.e., correct if it agrees with at least two of the raters).
-
•
TP: an FOV image is rejected, and at least two raters rejected the FOV image.
-
•
FP: an FOV image is rejected, and at least two raters accepted the FOV image.
-
•
FN: an FOV image is accepted, and at least two raters rejected the FOV image.
-
•
-
2.
A less restrictive approach, where the automated method is deemed correct if it agrees with at least one of the raters.
-
•
TP: an FOV image is rejected, and at least one rater rejected the FOV image.
-
•
FP: an FOV image is rejected, and no rater rejected the FOV image.
-
•
FN: an FOV image is accepted, and no rater accepted the FOV image.
-
•
2.4.3. Intersection counting
To evaluate the reliability of agreement between the manually assessed FOV images and the algorithm-generated assessment, the Fleiss’ Kappa29 statistical measure is calculated in each study group (i.e., RA and ). The Fleiss’ Kappa score reflects the performance of our method for counting the number of intersections in accepted FOV images.
As the final step in the evaluation of the results, the MLI scores were calculated and compared to that of the manually assessed scores.
3. Results
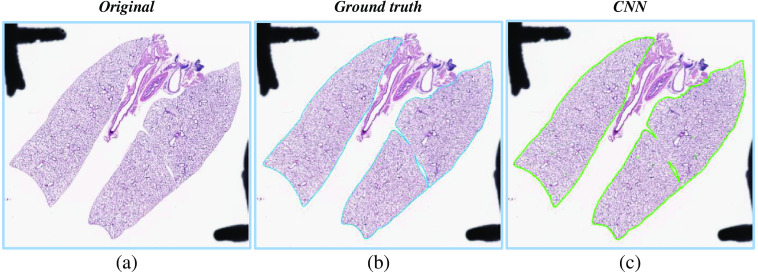
Figure 10 visualizes an example of foreground extraction over a histopathology WSI and the corresponding manual segmentation. Table 1 contains performance metrics for foreground extraction, which indicate that the proposed method performed well for all WSIs.
Fig. 10.
An example of the complete foreground extraction approach. (a) The original WSI. (b) The manually segmented ground truth mask. (c) The proposed method segmentation mask.
Table 1.
The evaluation of the foreground extraction step performance using pixel-wise accuracy (AC), Dice coefficient (DSC), and Hausdorff distance.
| Image | AC (%) | DSC (%) | Hausdorff distance () |
|---|---|---|---|
| Average score | 98.34 | 98.22 | 109.68 |
| Maximum | 98.95 | 98.58 | 182.85 |
| Minimum | 97.50 | 97.49 | 39.55 |
To evaluate the performance of the proposed method in detection of accepted FOV images, the precision, recall, and -score metrics are calculated for each fold for the RA group and the group. Table 2 gives the distribution of rejected and accepted FOV images in our dataset. In Table 3, the results for evaluation of our proposed method in detection of FOV images for different evaluation approaches are presented. As it is shown in Table 3, the RA group as compared to group has higher precision and lower recall in both evaluation approaches, whereas the -scores are similar. Examples of automated FOV image assessment using our proposed method are visualized in Fig. 11.
Table 2.
Distribution of extracted FOV images. The “completely inside pleural space” column represents the FOV images that contain no specific tissue compartments and therefore, are eliminated in the foreground extraction step. The remaining FOV images are used in detection of main biological structures step to identify the accepted FOV images.
| Fold # | FOV images in the dataset | Completely inside pleural space | Number of rejected FOV images | Number of accepted FOV images | |
|---|---|---|---|---|---|
| Intersection with pleural space | Intersection with vessel/bronchi | ||||
| 1 | 3219 | 1637 | 249 | 334 | 999 |
| 2 | 3024 | 1444 | 182 | 300 | 1098 |
| 3 | 4266 | 2666 | 185 | 276 | 1139 |
| 4 | 3780 | 2180 | 256 | 302 | 1042 |
| 5 | 4032 | 2432 | 227 | 291 | 1082 |
| Total | 18,321 | 10,359 | 1099 | 1503 | 5360 |
Table 3.
Evaluation of the proposed method performance in detection of FOV images. For evaluation approach (1), the decisions are correct if the automated method agrees with at least two out of three raters. For evaluation approach (2), the decisions are correct if the automated method agrees with at least one rater out of three raters. The results are presented for five different folds for each group. The abbreviation STD stands for standard deviation. In Table 3, the abbreviations RA and refers to the mice group housed in room air and mice group were exposed to a high concentration of oxygen and LPS, respectively.
| RA | Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold #1 | Fold #2 | Fold #3 | Fold #4 | Fold #5 | Mean ± STD | Fold #1 | Fold #2 | Fold #3 | Fold #4 | Fold #5 | Mean ± STD | Mean ± STD | ||
| Evaluation approach (1) | Precision (%) | 89.17 | 96.50 | 93.68 | 97.09 | 97.42 | 84.25 | 76.56 | 88.16 | 86.64 | 90.59 | |||
| Recall (%) | 68.81 | 60.79 | 69.26 | 76.50 | 72.14 | 88.17 | 72.32 | 83.40 | 82.72 | 81.00 | ||||
| -score (%) | 77.68 | 74.59 | 79.64 | 85.58 | 82.89 | 86.16 | 74.38 | 85.71 | 84.63 | 85.53 | ||||
| Evaluation approach (2) | Precision (%) | 93.75 | 97.90 | 94.74 | 98.91 | 98.97 | 90.41 | 80.22 | 93.47 | 92.67 | 92.68 | |||
| Recall (%) | 74.01 | 73.30 | 78.95 | 81.44 | 80.67 | 92.96 | 80.81 | 90.16 | 92.27 | 90.17 | ||||
| -score (%) | 82.72 | 83.83 | 86.12 | 89.33 | 88.89 | 91.67 | 80.51 | 91.78 | 92.47 | 91.41 | ||||
Fig. 11.
Visualization of the automated assessment results. (a) Accepted FOV images. The intersections with the septa are marked as red. The blue color indicates that there are no intersections. (b) Rejected FOV images.
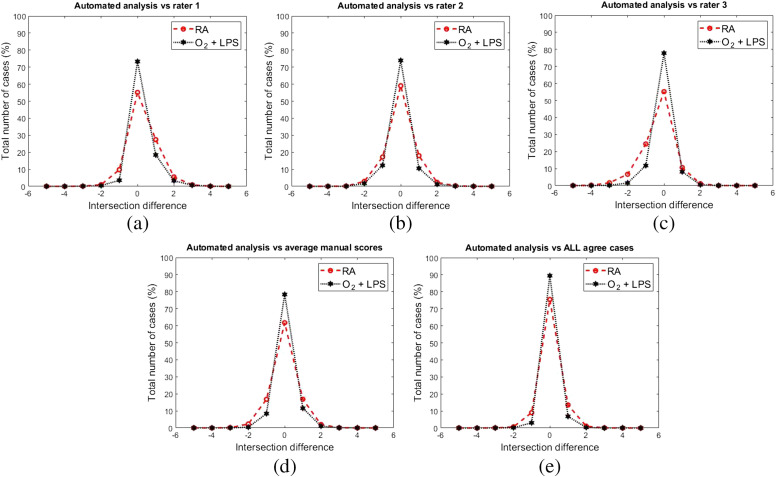
Another step in the determination of the overall performance is the accuracy by which our method can count the number of intersections in each accepted sample FOV image. Figure 12 shows the comparison between the proposed method accuracy in detection of the intersections in accepted FOV images against the manually generated scores by human raters. Rater 1 exibits a slight negative skew [Fig. 12(a)] and rater 3 a slight positive skew [Fig. 12(c)], rater 2 [Fig. 12(b)], and the rater’s average [Fig. 12(d)] distributions are quiet symetric. Figure 12(d) shows a comparison between the proposed method intersection counting scores and the rater’s average score (i.e., the arithmetic mean over the reported intersections by human raters and rounding it to the nearest whole number). In Fig. 12(e), the intersection difference between the proposed method and the manual scores is visulized, when all three raters were agreed on the number of intersections.
Fig. 12.
The evaluation of the proposed method in detection of the intersections against the human raters. Panels (a), (b), and (c) represent the intersection difference between the automated method and each individual raters scores. (d) Comparison between the generated intersections by the proposed method and the average number of intersections reported by human raters. (e) The comparison in instances that all human raters agree on number of intersections (e.g., all raters agree that there are five intersections with alveoli septa in the corresponding FOV image).
The calculated Fleiss’ Kappa scores are presented in Table 4, showing the reliability of agreement in counting the number of intersections in accepted FOV images between manual assessment by human raters and automated algorithm. In Table 4, the qualitative interpretation of scores’ agreement in “agreement assessment” section has been derived from Ref. 30.
Table 4.
The evaluation of the reliability of agreement in detection of intersections in accepted FOV images between the proposed method and the manual assessments using Fleiss’ Kappa score.
| RA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rater #1 | Rater #2 | Rater #3 | Rater’s average | All agree cases | Rater #1 | Rater #2 | Rater #3 | Rater’s average | All agree cases | Overall score | |
| Weighted percent agreement | 0.55 | 0.59 | 0.55 | 0.59 | 0.75 | 0.78 | 0.74 | 0.73 | 0.77 | 0.89 | 0.82 |
| Weighted percent chance agreement | 0.22 | 0.23 | 0.24 | 0.22 | 0.25 | 0.30 | 0.30 | 0.32 | 0.31 | 0.33 | 0.29 |
| Kappa score | 0.42 | 0.47 | 0.41 | 0.46 | 0.67 | 0.68 | 0.63 | 0.61 | 0.66 | 0.84 | 0.76 |
| Agreement assessment | Moderate | Moderate | Moderate | Moderate | Substantial | Substantial | Substantial | Substantial | Substantial | Perfect | Substantial |
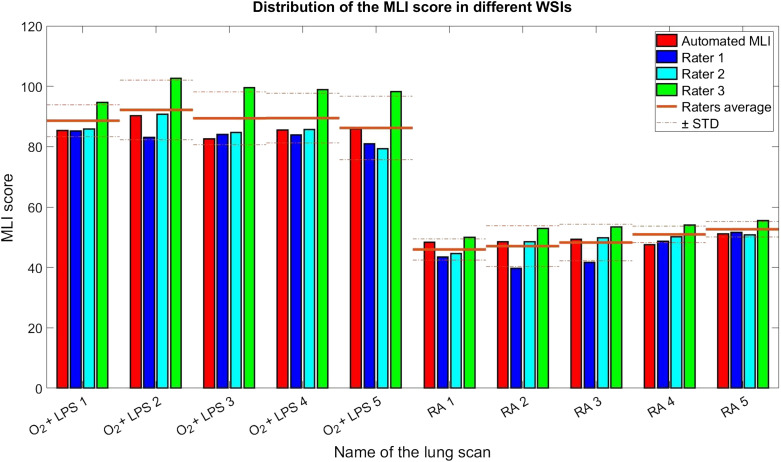
Figure 13 shows a comparison between calculated MLI scores by the three raters and the automatically generated MLI scores by the proposed method. As it can be seen in this figure, there is noticeable varibility in the MLI scores between individual raters and the automated MLI score falls within this variability. All raters and the automated method successfully discern the RA and conditions based on their MLI score.
Fig. 13.
The evaluation of the calculated MLI score by the proposed method against human raters.
4. Discussion
The current approaches in estimation of MLI score involves manual/semi-automated assessment of histopathology images,14,15,31–33 which can be time-consuming, subjective, and expensive. In recent years, there has been a growing interest in developing fully automated methods to encounter the inefficiencies of manual/semi-automated MLI scoring but none of these methods addressed the segmentation of lung main biological structures, which plays a vital role in estimation of MLI score. Jacob et al.31 proposed an automated approach for estimation of MLI score in histopathology images of mouse lung tissue. However, their proposed method was not applied to WSIs and the authors used a few selective images to provide their experimental results. Rieger-Fackeldey et al.34 proposed an automated method for estimation of MLI score to study the effects of hyperoxia in histopathology images of newborn mice lung tissue. The authors used a digital image analysis software (Image Pro Plus version 4.0) and a custom macro commands for automated investigations of alveolar morphological characteristics. However, they provided no technical details on their proposed method, nor how well it worked in comparison to manual assessments by human raters. Sallon et al.12 proposed an automated approach for estimation of MLI score in WSIs of mouse lung tissue. The authors used a thresholding approach and closed contour assessment based on size to classify the alveolar structures. However, their proposed algorithm was unable to distinguish between main lung biological structures (alveoli, bronchi, and blood vessels). To the best of our knowledge, our fully automated pipeline is the first approach capable of comprehensively account for main challenges involved with estimation of MLI score, including (1) taking a histopathology WSIs as input and extract the diagnostically relevant tissue compartments for extraction of FOV images, (2) screening of the FOV images, rejecting images based on presence of certain biological structures (bronchi and blood vessel).
To evaluate the performance of our proposed method, we compared our work against MLI scores from three human raters. Using ten high-resolution WSIs of mouse lung tissue, comprised of two distinct experimental groups (i.e., RA and ). We independently tested the performance of each step in our pipeline against manual assessment.
In extraction of the foreground regions, the proposed approach showed promising performance in removing the imaging artifacts and undesired biological components and identifying the ROIs, yielding 98.34%, 98.22%, and , AC, DSC, and Hausdorff distance, respectively.
We proposed two different approaches to evaluate the performance of our proposed method in detection of the main biological structures in histopathology images of mouse lung tissue. In evaluation approach (1), the proposed method was able to detect the rejected FOV images with mean precision, recall, and -score of 90.01%, 75.51%, and 81.68%, respectively. In evaluation approach (2), the proposed method was able to detect the rejected FOV images with mean precision, recall, and -score of 93.37%, 83.47%, and 87.87%, respectively. The higher performance metrics of evaluation approach (2), as compared to evaluation approach (1), can be an indication of the subjectivity involved with the process of calculating the manual MLI scores.
Two main observation can be drawn from the Table 3. First, the mean precision in RA group is higher in comparison to the mean precision in group (94.77% versus 85.24%). This indicates that the number of incorrectly rejected FOV images (i.e., FPs) are higher in group. This may be related to fact that the alveoli’s LR are enlarged in the group. As we used the area of the LR as a feature in our classification approach, this may result in more misclassifications of alveoli as bronchi (higher number of FPs). This may potentially explain the higher precision in RA group compared to group. Second, the mean recall in RA group is much lower in comparison to group (69.50% versus 81.52%). This indicates that the total number of incorrectly accepted FOV images (i.e., FNs) are much higher in RA group. In our dataset, we noticed that the density of remaining blood cells in RA group was lower in comparison to group. Since we used the presence of blood cells as a feature to identify blood vessels, the reduced amount of remaining red blood cells in the RA group results in increased number of FNs.
In classification of bronchi versus alveoli strucutres, as the alveoli structures are enlarged in group, there is a possibility of a bronchi misclassified as a bronchi as alveoli or vice versa. If an alveoli is misclassified as a bronchi, the FOV would be rejected and may result in underestimation of MLI. If a bronchi is misclassified as an alveoli, the FOV would not be successfully rejected and this may result in an overestimation in MLI, as bronchi tend to be larger than alveoli. These types of misclassifications do not appear to be happening with any noticeable frequency in our dataset. We speculate that the occasional misclassifications would also not have a large impact, as the MLI is computed across a large number of FOVs.
We also used Fleiss’ Kappa score to measure the reliability of agreement between the human raters and automatically generated intersections. The mean Fleiss’ Kappa scores were 0.46 and 0.66 for the RA group and the group, respectively. This shows that the proposed method has a slightly higher agreement with the group than the RA group against the average manual. We hypothesize two contributing factors for this difference: (1) The MLI scoring task in RA group is a more complex task compared to group. The Fleiss Kappa scores among human raters in RA group and group were 0.5949 and 0.6991, respectively. This indicates that the images in the RA group were inherently more difficult to analyze. (2) As it is mentioned in Sec. 2.3.6, a contributing factor in preparation of the septa region for automated intersection counting is the value of , which represent the minimum area of the alveoli’s LR. Using this threshold, the white objects that are smaller than are eliminated form the process of intersection counting. We optimized this value over both study groups (RA and ). As the alveoli’s LR are enlarged in the group, we expect that the calculated would result in more misclassifications of alveoli in RA group compared to the group, affecting the agreement between human raters and the proposed method.
In Table 4, we observe that the rate of agreement in both experimental groups is increased considerably, when all raters agree on the number intersections (Fleiss’ Kappa scores of 0.67 and 0.84 in RA group and group). This is also another indication that the subjectivity of the manual analysis can dramatically influence the results. The mean difference between the calculated MLI score between the automated method and average rater’s score was 2.33 (4.25%) with standard deviation of 4.13 (5.67%), which shows that our proposed method has the ability to accurately estimate the MLI scores with regards to manual scores calculated by human raters. A Student’s -test was performed to see if the average MLI scores of the human raters and the MLI scores from the proposed method could statistically differentiate the RA and groups (, with a Bonferroni correction). Results demosntrate a statistically signfiicant difference for both the human raters () and proposed method ().
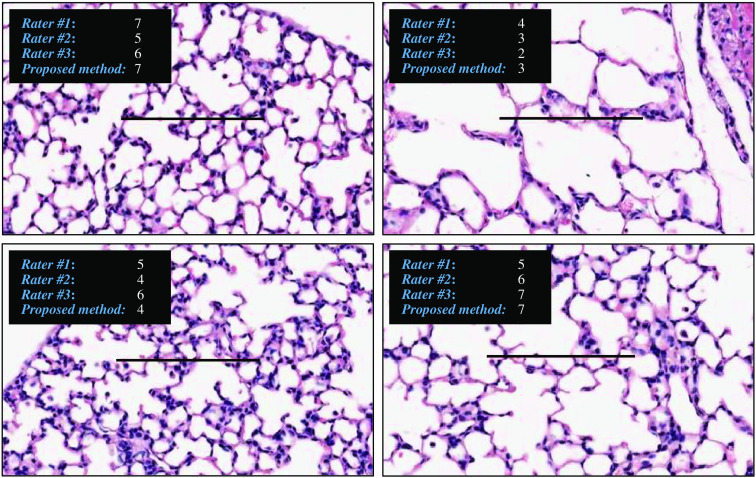
There is a strong agreement between the manual assessment and the proposed method, when the average intersections are calculated for all three raters [Fig. 12(d)]. Therefore, the proposed method agrees with the average raters’ score in 69.79% of cases and the agreement will include 96.77% of cases, if a maximum of one intersection difference is included in the calculations. The agreement between manual scores and the proposed method is stronger, when all raters counted the exact same number of intersections [see Fig. 12(e)]. In the cases where all raters agree on the number of intersections, the proposed method counts the exact same number of intersections with a probability of 83.84% or counts a maximum of one intersection difference with probability of 98.77%. Considering the fact that the proposed method is fully automated and completely reproducible, this result can be an indication of reproducibility issues in detection of intersections in FOV images by human raters. To demonstrate the subjectivity in the manual MLI scoring, a few examples of intersection counting are shown in Fig. 14. This discrepancy in their counting could imply the raters may have a particular bias in their MLI scoring. The results suggest the relative MLI scorings of the proposed method in comparison to each individual rater’s scores are constant (see Fig. 13). As a result, even if the automated method has a bias itself, it would be constant and highly reproducible.
Fig. 14.
The variability in intersection counting by human raters.
The processing time for foreground extraction within a single fold was and few minutes for CNN training and testing, respectively. This was performed on a standard workstation with an Intel Core i7-3770 3.40 GHz CPU, 12 GB of installed RAM, and a single NVIDIA RTX 2060 with 6 GB memory. The processing time required for the remaining steps of our proposed pipeline (i.e., color slicing, detection of the main biological structures, and intersection counting) was and for each fold for training and testing, respectively. The vast majority of this time is attributed to detection of main biological structures. The average time required to manually calculate the MLI score for each WSI was . In total, the time required for the proposed method and a human rater to score the entire dataset (10 WSIs) was and 100 h, respectively. Considering the fact that the algorithm can automatically run in the background 24/7 with no supervision, the manual scoring of the dataset by a human rater that may take up to several weeks for a human rater, can be achieved by the proposed method in less than two days.
It should be noted that optimizing processing time was not a focus of this work. Reduction of processing time could be easily achieved by leveraging parallel processing capabilities.
In our work, we faced various limitations and challenges that we were not able to address in our proposed pipeline. One of the limitations of our work was detection of the seed regions in segmentation of the blood vessels. We used the remaining blood cells in the perimeter of each vessel as an indication of existing blood vessel. Although, the majority of the blood vessels in our dataset had remaining blood cells in their perimeters, there were some cases with no blood cells present. As a result, in these cases our algorithm fails to correctly identify and segment the vessel regions. The complexity of blood vessels made it difficult for us to find a suitable handcrafted feature, which could be effective in identification of these structures. Another limitation was the way we have evaluated the performance of our proposed method in segmentation of biological structures. Although, the presented evaluation procedure can be a good indication of how detection error propagates into calculation of MLI scores, it does not evaluate the segmentation performance of our proposed approach in the detection of the lung structures. The main barrier is the unavailability of manual segmentation of the various biological structures of lung tissue in our dataset. Our future work is to develop a database of images with manual segmentation, which will also support the development of more advanced supervised learning segmentation algorithms, such as deep learning, that we anticipate will improve the overall segmentation performance.
In this work, we developed a pipeline to measure the changes in the lung architecture observed in mice with experimental bronchopulmonary dysplasia compared to control, healthy animals. The pathology of bronchopulmonary dysplasia in humans is more complex than in mice. In humans, the disease is variable within the lungs of one patient and there is more variability between patients. However, by accounting a wider variability in disease pathology, it is feasible that in the future, the current pipeline to be translated for use in human lung histopathology for bronchopulmonary dysplasia.
5. Conclusion
In this paper, we proposed a new pipeline for automating the estimation of the MLI score. The proposed method uses U-Net architecture for segmentation of diagnostically relevant tissue specimens, which yielded accurate results. Our proposed method utilized color image analysis and region growing for segmentation of the main biological structures (bronchi and vessels) in histopathology images of mouse lung tissue, which showed promising performance. The comparison between the automated method and the manual assessment showed substantial agreement in the calculation of the MLI score. The result demonstrated that the proposed method could replace the manual/semi-automated methods for calculating the MLI score.
Acknowledgments
This work was supported in part by the Ontario Trillium Scholarship (OTS), Natural Sciences and Engineering Research Council (NSERC) of Canada, and Canadian Institutes of Health Research (CIHR) Doctoral Award.
Biographies
Sina Salsabili is a PhD candidate at the Department of Systems and Computer Engineering at Carleton University. He works under supervision of Dr. Adrian D. C. Chan and Dr. Eranga Ukwatta. He received the Ontario Trillium Scholarship in 2016 from government of Canada. His PhD research focus on computer vision applications in automated analysis of histopathology images.
Marissa Lithopoulos is a PhD candidate at the Department of Cellular and Molecular Medicine at the University of Ottawa. She works under the supervision of Dr. Bernard Thébaud at the Ottawa Hospital Research Institute. Marissa is a recipient of a Canadian Institutes of Health Research Frederick Banting and Charles Best Doctoral Scholarship.
Shreyas Sreeraman is a second-year medical student at McMaster University in Hamilton, Ontario. He received his Bachelor of Health Sciences degree in 2019 from McMaster University. His interests include medical education, medical devices, and quality improvement.
Arul Vadivel received his PhD from the Council of Scientific and Industrial Research (CSIR) institutes, the Central Leather Research Institute (CLRI) in Chennai, India. He is currently working with Dr. Thébaud on the mechanisms of oxygen-induced lung injury and potential therapeutic strategies.
Bernard Thebaud is a professor of pediatrics at the University of Ottawa, a neonatologist at Children’s Hospital of Eastern Ontario and The Ottawa Hospital, and a senior scientist at the Ottawa Hospital Research Institute. His research is focused on the role of stem cells during normal and impaired lung development. His goal is to translate cell- and gene-based therapies into the clinic to improve the outcome of patients with life-threatening and debilitating lung diseases.
Adrian D. C. Chan is a professor at the Department of Systems and Computer Engineering. He is a biomedical engineering researcher with expertise in biomedical signal processing, biomedical image processing, assistive devices, and noninvasive sensor systems. He is a registered professional engineer, a senior member of the IEEE, member of the IEEE Engineering in Medicine and Biology Society, member of the Canadian Medical and Biological Engineering Society, member of the Biomedical Engineering Society, and 3M Teaching Fellow.
Eranga Ukwatta received his master’s and PhD degrees in electrical and computer engineering and biomedical engineering from Western University, Canada, in 2009 and 2013, respectively. From 2013–2015, he was a multicenter postdoctoral fellow with Johns Hopkins University and University of Toronto. He is currently an assistant professor with the School of Engineering, University of Guelph, Canada, and an adjunct professor in systems and computer engineering at Carleton University, Canada.
Disclosures
No conflicts of interest.
Contributor Information
Sina Salsabili, Email: sina.salsabili@carleton.ca.
Marissa Lithopoulos, Email: mlithopoulos@gmail.com.
Shreyas Sreeraman, Email: shreyas.sreeraman@gmail.com.
Arul Vadivel, Email: avadivel@ohri.ca.
Bernard Thébaud, Email: bthebaud@toh.ca.
Adrian D. C. Chan, Email: adrianchan@cunet.carleton.ca.
Eranga Ukwatta, Email: erangauk@gmail.com.
References
- 1.Thébaud B., et al. , “Bronchopulmonary dysplasia,” Nat. Rev. Dis. Prim. 5, 78 (2019). 10.1038/s41572-019-0127-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jobe A. H., Bancalari E., “Bronchopulmonary dysplasia,” Am. J. Respir. Crit. Care Med. 163(7), 1723–1729 (2001). 10.1164/ajrccm.163.7.2011060 [DOI] [PubMed] [Google Scholar]
- 3.Stoll B. J., et al. , “Trends in care practices, morbidity, and mortality of extremely preterm Neonates, 1993–2012,” J. Am. Med. Assoc. 314, 1039–1051 (2015). 10.1001/jama.2015.10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudsen L., et al. , “Assessment of air space size characteristics by intercept (chord) measurement: an accurate and efficient stereological approach,” J. Appl. Physiol. 108, 412–421 (2010). 10.1152/japplphysiol.01100.2009 [DOI] [PubMed] [Google Scholar]
- 5.Rizzardi A. E., et al. , “Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring,” Diagn. Pathol. 7, 42 (2012). 10.1186/1746-1596-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poggi S. H., et al. , “Variability in pathologists’ detection of placental meconium uptake,” Am. J. Perinatol. 26, 207–210 (2009). 10.1055/s-0028-1103030 [DOI] [PubMed] [Google Scholar]
- 7.Meyer J. S., et al. , “Erratum: breast carcinoma malignancy grading by Bloom–Richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index (Modern Pathology (2005) 18 (1067-1078 DOI: 10.1038/modpathol.3800388),” Mod. Pathol. 18, 1649 (2005). 10.1038/modpathol.3800479 [DOI] [PubMed] [Google Scholar]
- 8.Jothi J. A. A., Rajam V. M. A., “A survey on automated cancer diagnosis from histopathology images,” Artif. Intell. Rev. 48, 31–81 (2017). 10.1007/s10462-016-9494-6 [DOI] [Google Scholar]
- 9.Gurcan M. N., et al. , “Histopathological image analysis: a review,” IEEE Rev. Biomed. Eng. 2, 147–171 (2009). 10.1109/RBME.2009.2034865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alzubaidi A. K., et al. , “Computer aided diagnosis in digital pathology application: review and perspective approach in lung cancer classification,” in Annu. Conf. New Trends Inf. and Commun. Technol. Appl., (2017). 10.1109/NTICT.2017.7976109 [DOI] [Google Scholar]
- 11.Veta M., et al. , “Breast cancer histopathology image analysis: a review,” IEEE Trans. Biomed. Eng. 61, 1400–1411 (2014). 10.1109/TBME.2014.2303852 [DOI] [PubMed] [Google Scholar]
- 12.Sallon C., et al. , “Automated high-performance analysis of lung morphometry,” Am. J. Respir. Cell Mol. Biol. 53, 149–158 (2015). 10.1165/rcmb.2014-0469MA [DOI] [PubMed] [Google Scholar]
- 13.Crowley G., et al. , “C80-C Imaging methodology and application to lung disease: quantitative lung morphology: semiautomated method of mean chord length measurements,” Am. J. Respir. Crit Care Med. 195, A6510 (2017). 10.1164/ajrccmconference.2017.C80C [DOI] [Google Scholar]
- 14.Crowley G., et al. , “Quantitative lung morphology: semi-automated measurement of mean linear intercept,” BMC Pulm. Med. 19, 206 (2019). 10.1186/s12890-019-0915-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horai Y., et al. , “Quantitative analysis of histopathological findings using image processing software,” J. Toxicol. Pathol. 30, 351–358 (2017). 10.1293/tox.2017-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tschanz S. A., Burri P. H., Weibel E. R., “A simple tool for stereological assessment of digital images: the STEPanizer,” J. Microsc. 243, 47–59 (2011). 10.1111/j.1365-2818.2010.03481.x [DOI] [PubMed] [Google Scholar]
- 17.Ronneberger O., Fischer P., Brox T., “U-Net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 18.Mikołajczyk A., Grochowski M., “Data augmentation for improving deep learning in image classification problem,” in Int. Interdisciplinary PhD Workshop, (2018). 10.1109/IIPHDW.2018.8388338 [DOI] [Google Scholar]
- 19.Kingma D. P., Ba J. L., “Adam: a method for stochastic optimization,” in 3rd Int. Conf. Learn. Represent., ICLR 2015—Conf. Track Proc. (2015). [Google Scholar]
- 20.Macenko M., et al. , “A method for normalizing histology slides for quantitative analysis,” in Proc.—2009 IEEE Int. Symp. Biomed. Imaging: From Nano to Macro (2009). 10.1109/ISBI.2009.5193250 [DOI] [Google Scholar]
- 21.Salsabili S., et al. , “Automated segmentation of villi in histopathology images of placenta,” Comput. Biol. Med. 113, 103420 (2019). 10.1016/j.compbiomed.2019.103420 [DOI] [PubMed] [Google Scholar]
- 22.Otsu N., “Threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 23.Adams R., Bischof L., “Seeded region growing,” IEEE Trans Pattern Anal. Mach. Intell. 16, 641–647 (1994). 10.1109/34.295913 [DOI] [Google Scholar]
- 24.Chawla N. V., et al. , “SMOTE: synthetic minority over-sampling technique,” J. Artif. Intell. Res. 16, 321–357 (2002). 10.1613/jair.953 [DOI] [Google Scholar]
- 25.Jobe A. H., “The new bronchopulmonary dysplasia,” Curr. Opin. Pediatr. 23, 167–172 (2011). 10.1097/MOP.0b013e3283423e6b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasband W. S., “ImageJ,” U. S. National Institutes of Health, Bethesda, Maryland, 1997–2018 https://imagej.nih.gov/ij/. [Google Scholar]
- 27.Crum W. R., Camara O., Hill D. L. G., “Generalized overlap measures for evaluation and validation in medical image analysis,” IEEE Trans. Med. Imaging 25, 1451–1461 (2006). 10.1109/TMI.2006.880587 [DOI] [PubMed] [Google Scholar]
- 28.Beauchemin M., Thomson K. P. B., Edwards G., “On the Hausdorff distance used for the evaluation of segmentation results,” Can. J. Remote Sens. 24, 3–8 (1998). 10.1080/07038992.1998.10874685 [DOI] [Google Scholar]
- 29.Fleiss J. L., “Measuring nominal scale agreement among many raters,” Psychol. Bull. 76, 378–382 (1971). 10.1037/h0031619 [DOI] [Google Scholar]
- 30.Landis J. R., Koch G. G., “The measurement of observer agreement for categorical data,” Biometrics 33, 159–174 (1977). 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 31.Jacob R. E., et al. , “Comparison of two quantitative methods of discerning, airspace enlargement in smoke-exposed mice,” PLoS One 4, e6670 (2009). 10.1371/journal.pone.0006670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schittny J. C., Mund S. I., Stampanoni M., “Evidence and structural mechanism for late lung alveolarization,” Am. J. Physiol.–Lung Cell Mol. Physiol. 294, L246–L254 (2008). 10.1152/ajplung.00296.2007 [DOI] [PubMed] [Google Scholar]
- 33.Mund S. I., Stampanoni M., Schittny J. C., “Developmental alveolarization of the mouse lung,” Dev. Dyn. 237, 2108–2116 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Rieger-Fackeldey E., et al. , “Lung development alterations in newborn mice after recovery from exposure to sublethal hyperoxia,” Am. J. Pathol. 184, 1010–1016 (2014). 10.1016/j.ajpath.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]