Abstract
The past decade has witnessed the blossom of two fields: nucleic acid therapeutics and cancer immunotherapy. Unlike traditional small molecule medicines or protein biologics, nucleic acid therapeutics have characteristic features such as storing genetic information, immunomodulation, and easy conformational recovery. Immunotherapy uses the patients’ own immune system to treat cancer. A variety of strategies have been developed for cancer immunotherapy including immune checkpoint blockade, adoptive cell transfer therapy, therapeutic vaccines, and oncolytic virotherapy. Interestingly, nucleic acid therapeutics have emerged as a pivotal class of regimen for cancer immunotherapy. Examples of such nucleic acid immunotherapeutics include immunostimulatory DNA/RNA, mRNA/plasmids that can be translated into immunotherapeutic proteins/peptides, and genome-editing nucleic acids. Like many other therapeutic nucleic acids, nucleic acid immunotherapeutics often require chemical modifications to protect them from enzymatic degradation and need drug delivery systems for optimal delivery to target tissues and cells and subcellular locations. In this review, we attempted to summarize recent advancement in the interfacial field of nucleic acid immunotherapeutics for cancer treatment.
Keywords: nucleic acid therapeutics, vaccine, adjuvant, cancer, immunotherapy
Graphical Abstarct
INTRODUCTION
Cancer immunotherapy harnesses the host immune system to treat cancer,1 inhibit the progression of primary tumors and metastatic tumor,2 and prevent tumor relapse via elicit antitumor immune memory.3,4 Current approaches to cancer immunotherapy include adoptive cell transfer therapy,5 immune checkpoint blockade,6,7 oncolytic virotherapy,8 and cancer therapeutic vaccines.9 Nucleic acid therapeutics hold great potential for all these immunotherapy approaches. Natural nucleic acids encode, transmit, and express genetic information, and noncoding nucleic acids can also modulate biological functions.10 Technology advancement has enabled the synthesis of virtually all forms of nucleic acids ranging from oligonucleotides and oligodeoxynucleotides to large mRNA, plasmids, and even whole chromosomes and genomes. The coupling of the versatile functionalities of nucleic acids with the capability to synthesize nucleic acids on demand offers virtually unlimited opportunities to develop functional nucleic acids including nucleic acid therapeutic.11-15 In the past few decades, cancer immunotherapy has emerged as another pivotal approach to cancer treatment. Cancer immunotherapy is an emerging field in which nucleic acid therapeutics hold tremendous potential. For instance, via RNA interference (RNAi)-mediated gene silencing, therapeutic interventions using small interfering RNA (siRNA) or small hairpin RNA (shRNA) that can inhibit the production of a pathological protein have been explored for cancer immunotherapy.16-20 In addition to siRNA/shRNA, other types of nucleic acid therapeutics such as antisense oligonucleotides, aptamers, immunostimulatory DNA/RNA, plasmid, mRNA, and more recently CRISPR/Cas9 gene editing systems have also been studied for cancer immunotherapy (Figure 1).21 The clinical translation of nucleic acid immunotherapeutics has faced unique challenges due to the unique physical chemical properties, pharmacological behaviors, and toxicology profiles. Nucleic acids are distinct from conventional small molecule medicines or peptides or proteins, in terms of chemistry and formulation, pharmacokinetics and pharmacodynamics, and pharmacology as well as adverse effects. For instance, nucleic acids, unless modified or formulated for protection from nucleases, are susceptible to enzymatic degradation.10,21 In the past few decades, hundreds of nucleic acid chemical modifications have been developed to address this challenge and promote the resistance of nucleic acids to enzymatic degradation. In addition, the intrinsic hydrophilicity and high negative electronic charge can often present multiple barriers to the effective delivery in vivo, and lead to fast clearance from the body and limited retention in the target tissues.22 Consequently, this can narrow the therapeutic windows for these nucleic acid therapeutics. By using chemical conjugates or drug delivery systems such as nanoparticles, nucleic acid therapeutics can be efficiently delivered to target tissues and cells and even subcellular locations.15,23 In this review, we will summarize multiple types of nucleic acid immunotherapeutics for cancer immunotherapy.
Figure 1.
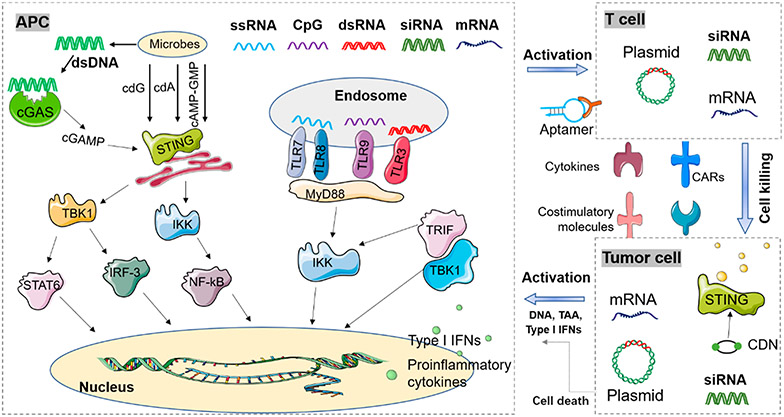
Schematic depiction of common nucleic acid therapeutics for cancer immunotherapy. Immunostimulatory (IS) nucleic acids of PAMPs are detected by pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) on the endosome membrane and cyclic GMP-AMP synthase (cGAS) in the cytosol that culminate in the production of type I interferons (IFNs) and proinflammatory cytokines, leading to the promotion of anticancer immune responses. Moreover, genetic carriers such as plasmids and mRNA can express functional RNA or protein/peptides that promote anticancer immune responses. Gene-regulating nucleic acids, such as siRNA/shRNA, gene activating nucleic acids, antisense oligonucleotides, and gene-editing nucleic acids, can regulate immune-related genes for the activation of anticancer immune responses. Other nucleic acids such as aptamers can function as agonists or antagonists against immune-related molecular targets so as to promote anticancer immune responses. For cancer immunotherapy, these nucleic acid immunotherapeutics can be engineered to function in a wide variety of cells including antigen-presenting cells (APCs), T cells or natural killing (NK) cells, and cancer cells. dsDNA, double-stranded DNA; dsRNA, double-stranded RNA; STING, stimulator of interferon genes; cGAS, cyclic GMP-AMP synthase; TBK1, TANK-binding kinase 1; Stat6, signal transducer and activator of transcription 6; IRF3, interferon regulatory factor 3; IKK, IκB kinase; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; TLR, Toll-like receptors; TRIF, TIR-domain-containing adapter-inducing interferon-β (TRIF); MHC, major histocompatibility complex; GM-CSF, granulocyte-macrophage colony-stimulating factor; CARs, chimeric antigen receptors; CDNs, cyclic dinucleotides; TAA, tumorassociated antigen.
■ VERSATILE NUCLEIC ACID THERAPEUTICS FOR CANCER IMMUNOTHERAPY
For the application in cancer therapy, there are versatile nucleic acid immunotherapeutics including immunostimulatory (IS) nucleic acids of pathogen-associated molecular patterns (PAMPs), plasmid, mRNA, immunomodulatory aptamers, and immunomodulatory gene regulation systems (Figure 1). IS nucleic acids of PAMPs can be recognized by the immune system as “foreign” or “danger” signals, thereby triggering innate immune responses.24 In addition, genetic carriers, such as plasmids and mRNA, can be engineered to transcribe RNA (for plasmids) or express proteins/peptides (for plasmids and mRNA), which subsequently activate anticancer immune responses for the immunotherapy of cancer.19,25 Moreover, nucleic acid genetic tools, such as gene-editing,26 gene silencing, or activating systems, can be leveraged to promote antitumor immune responses for cancer immunotherapy. Finally, nucleic acid aptamers have also been developed as agonists or antagonists of immune-related molecular targets for the purpose of immune activation that promotes the immunotherapeutic efficacy of cancer.27 Despite different target tissues and cells and even subcellular locations, the optimal immune activation and therapeutic efficacy of almost all these classes of nucleic acid immunotherapeutics require efficient drug delivery systems to prolong their bioavailability and overcome multiple biological barriers. In this section, we will discuss several common types of nucleic acid immunotherapeutics in terms of their properties, functionalities, and examples of delivery systems used for these therapeutics.
■ IMMUNOSTIMULATORY NUCLEIC ACIDS
PAMPs are independent immune modulators, which are regarded as “danger signals”, that are increasingly considered key components of many modern vaccines. PAMPs are highly conservative and distinct microbial molecules that bind to PRRs such as TLRs, retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and cytosolic cGAS expressed in the endosomes and the cytosol of innate immune cells (Figure 1).24,28 prrs, which can be activated by PAMPs, can upregulate the expression of proinflammatory cytokines, chemokines, type I IFNs, and costimulatory signaling molecules, all of which are important for the activation of innate and adaptive immunity for cancer immunotherapy.28
Double-stranded RNA (dsRNA) can be recognized as PAMPs including polyinosinic-polycytidylic acid (poly-I:C) and its derivatives such as poly-IC12U (Ampligen) and poly-ICLC (Hiltonol). These synthetic dsRNA can activate multiple elements of host defense in a pattern similar to that of viral infection. Nevertheless, an early clinical study of poly-I:C at exceptionally high dose (up to 75 mg/m2) revealed poor interferon induction, high toxicity, and no antitumor activity.29 To promote the therapeutic efficacy of poly-IC, derivatives have been developed to modulate the toxicity, interferon induction, and immunogenicity of poly-I:C.30 Poly-I:C and its derivatives can enhance and prolong antigen-specific immune responses when used with antigens.31 By activating the TLR3 and RLRs signaling pathways, poly-I:C can induce a strong IFN response accompanied by upregulated expression of immunostimulatory cytokines, chemokines, and costimulators.32-34 In poly-IC12U (Ampligen), the uracil and guanosine residues are mismatched to decrease its half-life in vivo, which was found to overcome some toxicity issues associated with the parent poly-I:C.35 As an immune adjuvant, poly-IC12U stimulates signal entirely through TLR3 and does not function through MDA-5.36,37 Poly-IC12U induces a lower expression of type I interferon than poly-I:C.36 Another poly-I:C derivative, poly-ICLC, which is electrostatic complexes of poly-I:C with cationic poly-L-lysine, shows obviously enhanced resistance to nucleolytic hydrolysis, which prolongs and enhances its activity in vivo.38,39 Similar to poly-I:C, poly-ICLC has an independent signal through TLR3 and MDA-5 that is localized in cell endosome and cytoplasm, respectively.40,41
CpG oligodeoxynucleotides (CpG ODN or CpG) are another class of commonly studied PAMP nucleic acid immunotherapeutics. CpG DNA is an unmethylated sequence containing CpG-dinucleotides that is more common in bacterial genomes than in vertebrate genomes, where the activity of CpG dinucleotides is generally inhibited by methylation at the CG sites.42 CpG stimulates immune cells via TLR9 signaling pathway.43 In early studies, immunostimulatory activity of bacterial DNA was reported to inhibit the growth of a variety of tumors in syngeneic animal tumors, enhance NK cell activity, and induce the production of type I IFNs in mouse spleen cells and human peripheral blood leukocytes.44 Further studies showed that bacterial DNA as well as synthetic ODN containing a central CpG can induce B cell proliferation and activate macrophages and DCs.45 DNA sequences with immunostimulatory activity were identified, with a generic structure of 5′-purine–purine-CpG-pyrimidinepyrimidine-3′ in the most immunostimulatory motifs.46 Activation of TLR9 by CpG subsequently triggers the activation of downstream signaling pathways involving IRAK, TRAF6, NF-kB, and MAP kinases, similar to immunestimulating components derived from other pathogens.47 Worth noting, due to the location of TLR9 on endosome membrane, the uptake and endosomal maturation is required for CpG DNA to exhibit immunostimulatory activity.48 One caveat though is that the expression pattern of TLR9 is different in humans and mice. Specifically for DCs, which are pivotal for antigen presentation, murine TLR9 is expressed in both plasmacytoid DCs (pDCs) and myeloid DCs (mDCs), whereas human TLR9 is only expressed in pDCs but not in mDCs.49 Such discrepancy will likely impact TLR9 agonists, such as CpG, to be translated based on preclinical studies in mice into clinical studies in humans. In summary, CpG can activate DCs, NK cells, and B cells through TLR9 signaling pathways to elicit immune responses that can be leveraged to promote the therapeutic efficacy of diseases such as cancer.
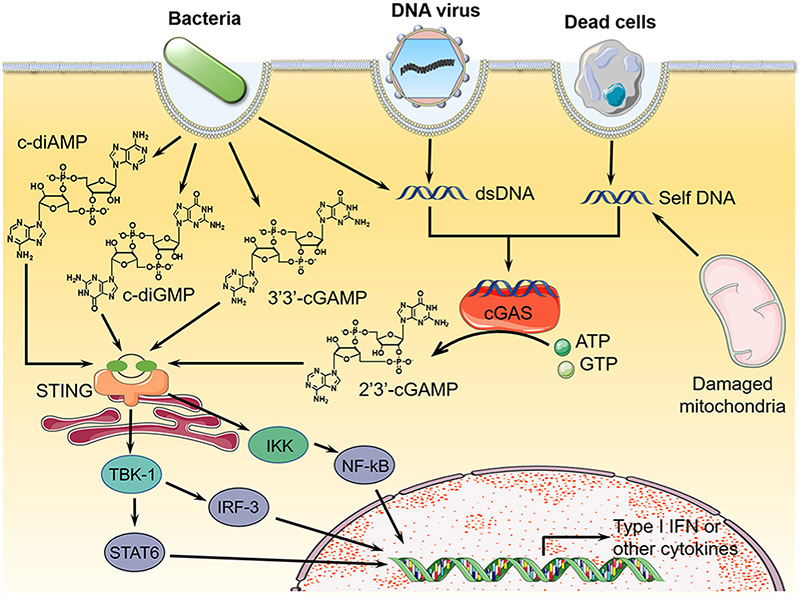
Cytosolic dsDNA, especially if long enough, make up another class of PAMP nucleic acids that stimulates cytosolic dsDNA sensor, cGAS, and subsequently elicit proinflammatory immune responses. For instance, dsDNA from bacteria, viruses, protozoa, and dead cells can introduce dsDNA into cytosol of eukaryotic cells.50 Specifically, cytosolic dsDNA can activate cGAS to synthesize 2’3′-cGAMP, which activates STING signaling pathway to promote type I IFN responses.51 Indeed, in addition to 2′3′-cGAMP, many other types of CDNs such as cyclic dimeric guanosine monophosphate (c-di-GMP or cdG), cyclic dimeric adenosine monophosphate (c-di-AMP, cdA), and 3′3′-cyclic GMP-AMP (3′3′-cGAMP), which can be secreted by bacteria,52 can activate the STING signaling pathway (Figure 2). The stimulation of innate immune responses by STING activation defends eukaryotic cells against the invasion from bacteria, DNA viruses, or eukaryotic pathogens,24,42,53,54 and defends bacteria against infection from phage.55 Biochemically, upon CDN binding to STING, a conformational change of STING leads to the formation of “closed pocket” to tightly bind to the CDN ligand.56 After activated STING was transferred from endoplasmic reticulum (ER) to the discrete foci in the cell cytosol, STING recruited TBK1 and IKK kinases, which in turn activated IRF-3, STAT6, and NF-kB. After transposition to the nucleus, these activated transcription factors bind to the corresponding promoters to induce the production of type I IFNs and cytokines.57 In summary, nucleic acid agonists that activate the cGAS-STING pathway hold great potential to elicit or augment proinflammatory responses that can be leveraged for the immunotherapy of cancer.
Figure 2.
Overview of nucleic acid immunotherapeutics that can activate the cGAS-STING signaling pathway. CDNs activate STING to produce type I IFNs that can be leveraged for cancer immunotherapy.52 Adapted with permission from ref 52. Copyright (2013) Elsevier Publishing Group.
Given the great potential of these nucleic acid therapeutics for cancer immunotherapy, a variety of drug delivery systems have been developed to promote the delivery of these therapeutics to target tissues, cells, and subcellular locations. For example, cationic liposomes,58 emulsion,59 and microspheres60 have been developed to deliver poly-I:C and elicit antitumor immune responses.61 Moreover, a series of CDN delivery systems including liposomes,62 polymeric nanoparticles,63 and inorganic materials64 has also been developed.50 Likewise, lipid nanoparticles (LNPs), including liposomes, ionizable lipids, and polymer–lipid nanoparticles, have been developed to deliver CpG to target cells.21 Meanwhile, we previously developed a DNA-inorganic hybrid nanoflower for the delivery of CpG alone,65 or in combination with synergistic immunostimulatory shRNA and tumor-specific neoantigen peptides.66
■ GENETIC NUCLEIC ACIDS FOR CANCER IMMUNOTHERAPY
Genetic DNA as Immunotherapeutics.
Gene therapy has made significant advancement for versatile applications including cancer immunotherapy. Plasmids are often used as genetic carriers.67 Synthetic plasmids typically possess one or more selective marker genes and one single synthetic polyclonal site sequence, which contains multiple restriction enzyme recognition site.67 For cancer immunotherapy, synthetic plasmids can be designed to encode tumor-specific antigens or tumor immunotherapeutic proteins or peptides (e.g., cytokines) that can elicit/augment antitumor immune responses in versatile target cells such as antigen-presenting cells, T cells, and tumor cells.68 One notable application of such immunotherapeutic plasmids is the genetic engineering of chimeric antigen receptor T cell (CAR-T cell). CAR-T cells are typically autologous T cells that are isolated from patients, then engineered ex vivo to express cancer cell-specific CAR and related immunostimulatory molecular signals, prior to proliferation and administration back into the donor patients for cancer immunotherapy.69 Moreover, plasmids that encode antitumor cytokines such as IL-2, IL-12, and GM-CSF, costimulatory molecules (B7.1 or B7.2), and MHC molecules have been found to enhances antitumor immune responses including tumor antigen-specific T cell responses [e.g., tyrosinase-related protein-1 (Trp1)70 and melanocyte-specific self-antigen (gp100)71 for melanoma].72 Plasmids have also been studied to express tumor antigens as tumor therapeutic vaccines. For instance, plasmids that express a model antigen ovalbumin (OVA) modulated antigen-specific Th1 immunity response and delayed the tumor growth of B16F10-OVA murine melanoma in syngeneic mice.73
Conventionally, plasmids are delivered via viral carriers, which can not only mediate effective transfection but also bear intrinsic safety concerns. While these viral vectors can mediate efficient transfection, they also encounter efficacy and safety concerns such as preexisting antiviral immunity and off-targeting or random gene integration and mutation.74 Nonviral gene delivery carriers, such as lipid nanoparticles and polymer nanoparticles, have been studied for plasmids delivery.75-77 For instance, poly(β-amino ester) nanoparticles78,79 have been developed with a lymphocyte-targeting ligand as nanocarriers that can efficiently load CAR-coding plasmids for the genetic engineering of CAR-T cells.80 Interestingly, in preclinical models, these nanoparticles programmed sufficient endogenous T cells in vivo for tumor immunotherapy, which hold the potential to use “off-the-shelf’ CAR-coding DNA nanoparticles for fast and economical CAR-T cell therapy of cancer.
mRNA Immunotherapeutics.
In addition to DNA plasmids, mRNA has recently garnered substantial enthusiasm for drug development including cancer immunotherapeutics.81,82 Traditional mRNA consists of a coding region (antigen translation) and noncoding flank [5′ and 3′ nontranslated regions (UTR) on either side of the coding region], a critical 5′ 7-methylguanosine triphosphate (m7G) cap, and a 3′ tail of poly(A) sequence.83 The 5′ m7G cap, 3′ poly(A) tail, and UTR are critical for the stability and translation of mRNA and mRNA therapeutics. mRNA therapeutics have several potential prominent advantages compared with DNA gene therapeutics.74,81,82,84 First, mRNA therapeutics are translated immediately when they are delivered to the cytoplasm, without the need of nucleus entry in the case of DNA gene therapeutics. Second, mRNA therapeutics avoid the risk of genomic integration for DNA gene therapeutics. Third, mRNA therapeutics are eventually degraded, and the expression from mRNA is transient, which bypass some of the long-term safety concerns over DNA gene therapeutics about the genotoxicity and long-term side effects.
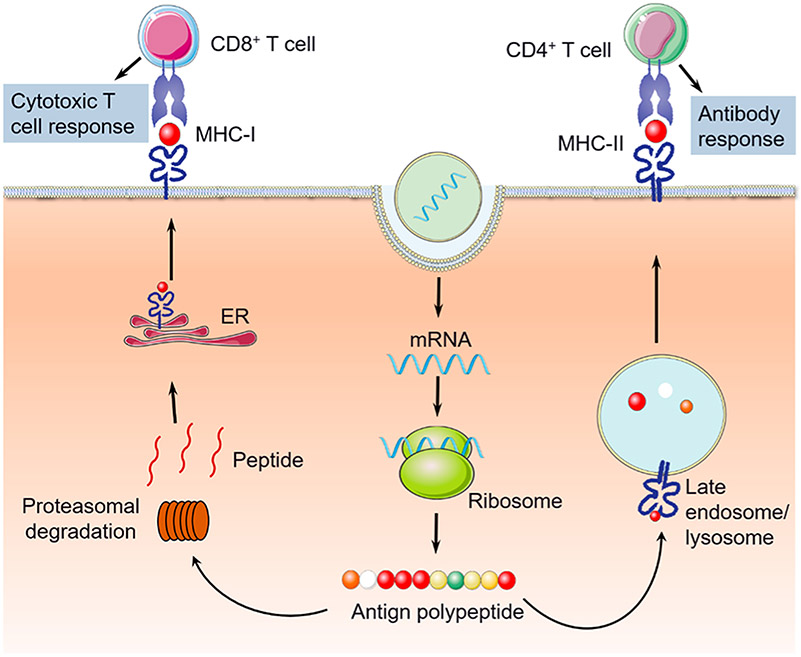
mRNA has been studied for versatile applications in cancer immunotherapy. One example is mRNA vaccines. Customized mRNA vaccines encoding cancer antigenic determinants (epitopes) can be delivered into the cytoplasm of APCs such as DCs, followed by antigen expression and presentation to B cells and T cells to stimulate antitumor adaptive immune responses.83 In some cases, the long antigens encoded by mRNA vaccines can be degraded by proteasomes to peptide epitopes, which bind with MHC-I or MHC-II molecules to form peptide-MHC complexes that are then transported and presented on APC cell surfaces. By designing MHC-I- or MHC-II-restricted antigens in mRNA vaccines, CD8+ T cell and CD4+ T cell response, respectively, can be elicited or augmented (Figure 3).85,86 This versatility is critical especially because both the CD8+ T cell population and the CD4+ T cell population are crucial in cancer immunotherapy. Further, the modularity of mRNA allows easy integration of multiepitope antigens and synergistic immunostimulatory signal peptides into one mRNA vector for the optimal immunostimulation of a broad spectrum of antitumor immune responses. Similarly, for the development of human mRNA vaccines, antigens that are able to bind with human leukocyte antigen (HLA) can be easily incorporated for immune modulation.87 Besides synthetic subunit antigen mRNA vaccine, tumor total RNA has also been studied to activate a full spectrum of tumor-specific antitumor immune responses for tumor immunotherapy.83,88 Overall, mRNA vaccines hold great potential for cancer immunotherapy.
Figure 3.
Schematic depiction of mRNA vaccines for cancer immunotherapy.86 Note that, by using MHC-I or MHC-II-restricted antigens that are translated from mRNA vaccines, both arms of CD8+ and CD4+ T cell responses, respectively, can be elicited or augmented. Adapted with permission from ref 86. Copyright (2013) Elsevier Publishing Group.
Like DNA gene therapeutics, the delivery of mRNA, including mRNA vaccines, into target tissues, targets cells, and their cytosol is also pivotal for the optimal immune modulation efficacy as well as the consequent therapeutic efficacy.89,90 A variety of nanoparticles have been engineered and tested in preclinical models and in the clinic to deliver mRNA immunotherapeutics. Nanocarriers can protect mRNAs from nuclease degradation and enhance delivery efficiency by facilitating cell uptake into APCs.89 Preclinical studies have indicated that mRNA-based nanovaccines, using drug delivery carriers such as cationic liposomes, can effectively deliver mRNA in vivo and trigger efficient antitumor immune responses.91,92 Further, by complexing with positively charged protamine, mRNA vaccines can induce cellular and humoral immune responses in both mice and human, resulting in the production of antigen-specific IgG antibodies and activation of antigen-specific T cell responses for cancer immunotherapy.93 Moreover, polymer nanoparticles based on cationic and pH-responsive polymer such as poly(b-amino ester) have been investigated to load ionic mRNA via electrostatic interaction, and the resulting nanoparticles improved transfection efficiency and therapeutic effects of mRNA therapeutics.94
Gene-Regulating Nucleic Acids as Immunotherapeutics.
Cancer occurrence is caused by a wide range of abnormal gene expressions, and normalizing the expression of such genes holds the potential for cancer therapy including immunotherapy. Nucleic acid approaches to gene regulation include gene downregulation using antisense oligonucleotides (ASOs), RNA interference (RNAi) using small interfering RNAs (siRNAs) or small hairpin RNA (shRNA), and gene upregulation using small activating RNA (saRNA). For example, siRNA, which is typically 21–23 nucleotides in length, can facilitate the degradation of target complementary mRNA or inhibit the corresponding protein translation.20 In addition to designing siRNA for selective target gene therapy, targeted siRNA delivery systems have also been delivered to increase the therapeutic efficacy. For example, Yu et al. developed a conjugate of CpG oligonucleotide (a TLR9 agonist) with signal transducer and activator of transcription-3 (STAT3) siRNA. The resulting CpG-siRNA conjugate promoted the delivery efficiency of siRNA to TLR9+ APCs, in which the expression of the immunosuppressive STAT3 is significantly inhibited to promote antitumor immune responses.95 In other examples, siRNAs that silence the expression of immune checkpoints CTLA-4 and programmed cell death-ligand 1 (PD-L1) were studied to enhance T cell-mediated antitumor immune response;96,97 siRNA against immunosuppressive cytokine TGF-β knocked down the expression of TGF-β, thereby changing the melanoma immune microenvironment by ameliorating the immunosuppression to promote melanoma immunotherapy.18 Like many other types of nucleic acid therapeutics, nucleic acid chemistry and pharmacoengineering principles have been incorporated to develop siRNA that enhance their biostability, target selectivity, bioavailability, and the penetration ability across tissue barriers and cell membrane as well as endosome membrane, while reducing their unwanted immunogenicity.98 A variety of drug delivery systems ranging from bioconjugates and nanoparticles to hydrogels have been studied for siRNA delivery for versatile biomedical application including cancer immunotherapy.99-102
Gene-Editing Nucleic Acid Immunotherapeutics.
Gene editing has made a historical breakthrough in the past decade. Multiple gene-editing technologies, such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and more recently clustering regularly spaced short palindromic repeats (CRISPR)-associated proteins (CRISPR-Cas), have been investigated for gene therapy of a wide variety of diseases including cancer.103 Particularly, CRISPR-Cas-based RNA-guided genome-editing has been revolutionizing fields such as biomedicine and biotechnology.104,105 For example, one of the CRISPR-Cas systems, CRISPR-Cas9, comprises two key components, a Cas9 as DNA endonuclease and a single-stranded RNA (sgRNA) that is used for site recognition based on Watson–Crick base pairing between sgRNA and target DNA.106,107 CRISPR-Cas9 can be used to engineer therapeutic immune cells by approaches such as building precisely genetically engineered CAR-T cells and knocking out immune checkpoints such as PD-1 and CTLA-4. The reduced expression level of such immune checkpoints would reinvigorate the otherwise exhausted antitumor immune cells, thereby promoting the antitumor immune responses and immune cells. For example, CRISPR/Cas9 was used to produce PD-1-deficient CD19-targeting CAR-T cells, thereby promoting the potency of CAR-T cells and enhancing the resulting therapeutic efficacy in PD-L1+ tumor.108,109 In 2017, the FDA approved Kymriah, a cellular gene therapy that uses CAR-T cells, to treat leukemia.110 Kymriah’s early success and the potential of CRISPR/Cas9 laid the foundation to further advance gene-editing therapy for cancer by further promoting T cell-mediated cancer cell killing and reducing adverse side effects.111
Currently, CRISPR-based gene editing systems have been often delivered using viral vectors such as adenovirus (AV),112,113 adeno-associated virus (AAV),114 and lentivirus (LV).115 Given the safety concerns such as preexisting antiviral immunity and off-targeting or random gene integration and mutation, as discussed above,74 alternative nonviral vectors have been investigated to address the above concerns for the delivery of CRISPR systems in various forms such as Cas-expressing DNA, Cas-expressing mRNA, and Cas ribonucleases (Figure 4).116-118
Figure 4.
Nonviral delivery of CRISPR-Cas9 system for gene editing. Cas9 ribonuclease can be delivered in the forms of Cas9-expressing DNA, Cas9-expressing mRNA, and Cas9 protein.116 sgRNA can be delivered by expression from sgRNA-coding DNA or as independent oligonucleotides together with Cas9-expressing mRNA or Cas9 proteins. Adapted with permission from ref 116. Copyright (2017) American Chemical Society Publishing Group.
Aptamer Immunotherapeutics.
Aptamers are single-stranded RNA or DNA oligonucleotides with high compatibility and specificity to the target molecules.119 Aptamers are typically screened by a method called systematic evolution of ligands by exponential enrichment (SELEX).119 Compared to monoclonal antibodies used for cancer immunotherapy,120 aptamers may have several advantages121 including fast aptamer screening to a variety of molecular and cellular targets, reproducible and programmable aptamer synthesis, site-specific chemical modifications that can promote the stability or pharmacology or functionalities, and recoverable conformations and functionalities upon experiencing some high temperature or denaturing conditions. Aptamers can be designed to activate costimulus receptors or block immunosuppressive signals for triggering specific antitumor immune responses.122 In the tumor microenvironment, the lack of costimulating ligands causes the exhaustion of T cells, thus compromising their efficacy to elicit or augment antitumor immune responses.122 Pastorhas et al. has developed multivalent RNA aptamers as agonists for costimulation receptor CD28. These aptamers improved the costimulatory signal, which promoted the proliferation of CD4+ T cells and CD8+ T cells in vitro.123 In another study, CTLA-4 aptamers were developed, which, in multivalence, bound to CTLA-4 immune checkpoint with high affinity and inhibited the immunosuppression efficacy of CTLA-4.124 Relative to monomers, the tetrameric CTLA-4 aptamer enhanced its immunomodulation and therapeutic efficacies in vitro and in vivo. In a similar manner, to block the immunosuppression of immune checkpoint PD-1, a PD-1 DNA aptamer was developed to block the binding of PD-1 with PD-L1. By reinvigorating T cells after PD-1 blockade, this PD-1 aptamer inhibited tumor growth and improved mouse survival rate in PD-L1-positive colon carcinoma in a syngeneic mouse model.125 In addition to serving as agonists or antagonists by aptamers per se, aptamers have been studied as targeting ligands for targeted delivery of molecular cargoes.126,127
■ CHEMICAL MODIFICATIONS FOR NUCLEIC ACID IMMUNOTHERAPEUTICS
The evolution of nucleic acid chemistry over the past few decades has resulted in versatile chemical modifications of nucleic acid therapeutics (Figure 5).128 Note that these modifications can often be programably and site-specifically incorporated into nucleic acids during automated synthesis of oligonucleotides or oligodeoxynucleotides or during enzymatic synthesis of large nucleic acids. Versatile nucleic acid modifications have been developed to improve the biostability, enhance tissue- or cell-level delivery efficiency, add functionalities, or tune the immunogenicity of nucleic acids. In this section, we will discuss some of these chemical modifications that can be critical for nucleic acid immunotherapeutics.
Figure 5.
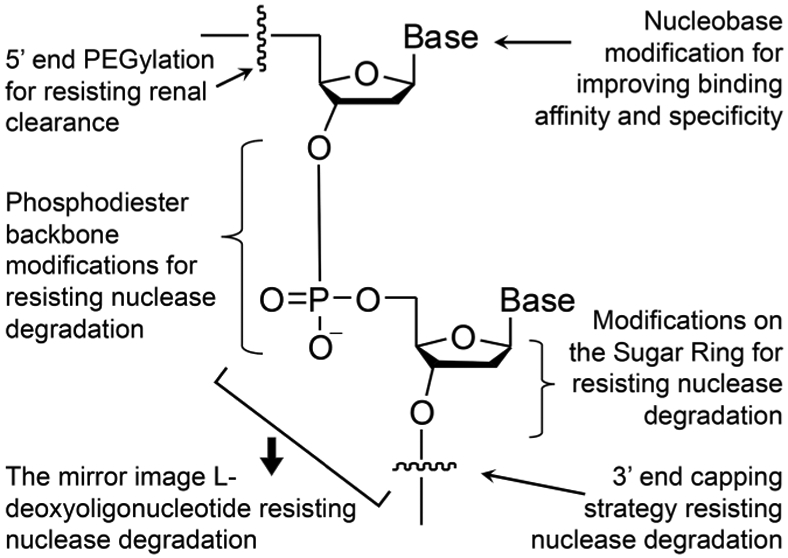
Versatile chemical modifications that can improve the biostability, pharmacokinetics and pharmacodynamics, and immunogenicity of nucleic acids therapeutics. These modifications can be at the 5′- or 3′-terminals, the phosphodiester linkage, on the sugar rings, or on the bases.128 Adapted with permission from ref 128. Copyright (2017) MDPI Publication.
Chemical Modifications to Overcome Nuclease Degradation.
Natural nucleic acids are often susceptible to nuclease degradation and hydrolysis. Chemical modifications of nucleic acids have been developed to confer protection from nuclease degradation. These modifications can be on the terminal ends, phosphate backbone, pentose sugar, or nucleotides. For instance, 3′-inverted thymidine increased the biostability of nucleic acids to resist 3′-exonuclease degradation in serum.129,130 Moreover, multiple types of modifications on the 2′-position of pentose sugar have been developed to increase nuclease resistance such as 2′-O-methyl (2′-OMe), 2′-amino (2′-NH2), or 2′-fluoro (2′-F).128 Locked nucleic acids (LNA), in which the 2′-O and 4′-C of ribonucleotide is linked, are also often used for resistance against nuclease degradation and thermal denaturation.131 Besides, the backbone of nucleic acids can also be engineered to increase nuclease resistance, and to reduce the negative charge of phosphodiester and weaken the electrostatic repulsion, the latter of which may to improve the penetration of the resulting nucleic acids through negatively charged cell membrane for cell uptake. Examples of such backbone modifications include peptide nucleic acids (PNAs), phosphorothioate (PS), tetramethyl phosphorodiamidate morpholino (PPMO), phosphoryl guanidine (Tmg), and triazole.15,132,133 In addition, the chiral transition of natural DNA in D-configuration to its mirror L-configuration may promote nuclease resistance as well as binding affinity.134 Finally, nucleic acid molecular engineering such as nucleic acid circularization and nanoengineering such as spherical nucleic acids have also proven able to increase the biostability of nucleic acids.135
Chemical Modifications to Reduce Immunogenicity of Nucleic Acids.
Nucleic acids may stimulate innate or adaptive immune responses, which are involved in the immune responses underlying many diseases such as autoimmune diseases and inflammation.136 While such immunogenicity could be leveraged for immune modulation, the immunogenicity of nucleic acids might also impair the efficacy of nucleic acid therapeutics in certain scenarios. For example, the immunogenicity of mRNA can be used to promote immune responses when used as immunostimulatory vaccines, whereas such immunogenicity often needs to be inhibited in the cases of mRNAs that express therapeutic proteins or peptide.137 For example, natural RNA can be edited by adenosine deaminase (ADAR1) to generate A-to-I mutation of RNA, which alleviates the immune responses elicited by excessive RNA.138 For synthetic mRNA generated by in vitro transcription (IVT),139 modifications such as pseudoacridine are often incorporated to reduce the immunogenicity of RNA and increase its stability and translational capacity.140 On the other hand, chemical modifications can also be developed to increase the immunogenicity of nucleic acids for applications such as immunostimulatory vaccines. An example is that a 3′-tripphosphate moiety in RNA can increase the immunogenicity of RNA motifs as RIG-1 agonists, which have shown the potential for the immunotherapy of diseases such as cancer.139
Chemical Modifications to Improve Pharmacokinetics and Pharmacodynamics of Nucleic Acids.
Nucleic acids are featured with high negative charge and high hydrophilicity, and many oligonucleotide therapeutics generally have small molecular sizes. All of these could be attributed to the fast clearance from the body and poor pharmacokinetics and pharmacodynamics. This often presents a challenge against the development of nucleic acid therapeutics that are required to have a therapeutically effective level over a relatively long period. To address this challenge, a variety of approaches, such as chemical modifications as well as nanoformulation, have been developed. PEGylation of nucleic acids, via conjugating nucleic acids with poly(ethylene glycol) (PEG), is commonly used to enlarge the molecular size of nucleic acids to slow down renal clearance and extend the in vivo half-life. For example, PEGylated MP7 DNA aptamer extended the in vivo half-life to block the interaction between PD-1 and PD-L1.125 Another strategy involves enabling nucleic acids to hitchhike endogenous molecular or cellular vehicles. For instance, a modification of cholesterol of oligonucleotides such as p40-targeting siRNA141 enables the conjugate to insert into cell membrane, which naturally contains of abundant cholesterol, thereby increasing the in vivo half-life. Lipidmodified oligonucleotides such as immunostimulatory CpG can also insert into cell membrane or interact with endogenous albumin to extend the in vivo half-life.142 Similar albuminbinding approaches using Evans blue derivatives or albuminbinding domain (ABD) peptides have also the ability to extend the half-life of oligonucleotides.143 The extended in vivo half-life of nucleic acid immunotherapeutics subsequently promoted the immune modulation and enhanced the cancer immunotherapeutic efficacy. In addition, drug delivery systems based on nanomaterials or macromaterials (e.g., hydrogel) have been developed for nucleic acid immunotherapeutics. Generally, these biomaterials can serve as a drug depot, protect nucleic acids from degradation, promote the delivery to target tissues and cells, or mediate efficient codelivery of multiple agents (e.g., vaccine adjuvants and antigens) for the optimal therapeutic efficacy.21,50,89,144
■ CONCLUSIONS
Nucleic acid therapeutics make up an emerging class of therapeutics that have characteristics compared to conventional small molecule medicines and monoclonal antibodies. The past few decades of advancement in this field have developed multiple unconventional therapeutic strategies, established a plethora of nucleic acid chemistry, and developed formulations and delivery systems for the efficient delivery of different types of nucleic acid therapeutics for a variety of therapeutic purposes including cancer immunotherapy. The field of cancer immunotherapy has also made remarkable progress in the past decades, resulting in the FDA approval of multiple immune checkpoint inhibitors and CAR-T cell therapy as well as cancer therapeutic vaccines. However, current immunotherapy is only effective in a small subset of cancer patients of a limited subtypes of cancer. Novel approaches that complement or synergize with current immunotherapy have the potential to broaden the population of cancer patients that can benefit from immunotherapy. To this end, nucleic acid therapeutics represent an attractive class of therapeutics due to their versatile functionalities. As discussed, examples of cancer immunotherapeutic nucleic acids include immunostimulatory nucleic acids, gene-expressing or gene-regulating or gene-editing nucleic acids, and aptamers. Of note, the advancement of these nucleic acid therapeutics, including those for cancer immunotherapy, has been fueled by the development of a plethora of nucleic acid chemistries that enhance their biostability, improve their in vivo pharmacokinetics and pharmacodynamics, increase their functionalities, or tune their immunogenicity. Further, the development of drug delivery systems for different types of nucleic acids has also facilitated the development of the field of nucleic acid therapeutics. Quite a few nucleic acid therapeutics have been approved by the US FDA for the treatment of noncancer diseases. Many clinical trials involving nucleic acid therapeutics have been investigating their therapeutic efficacy and safety for cancer immunotherapy. It is expected that relative to conventional immunotherapeutics, nucleic acid immunotherapeutics would face unique challenges and opportunities. Meanwhile, the past experiences of currently FDA approved nucleic acid therapeutics would facilitate the development of nucleic acid therapeutics for cancer immunotherapy. Built on the versatile unique functionalities as well as the established and evolving nucleic acid chemistry and formulation technologies, it is expected that nucleic acids hold great potential to further advance the field of cancer immunotherapy and benefit a broad population of cancer patients.
ACKNOWLEDGMENTS
G.Z. acknowledges partial support from VCU Center for Pharmaceutical Engineering and Sciences, NIH KL2 Scholarship and Endowment Fund from VCU C. Kenneth and Dianne Wright Center for Clinical and Translational Research (UL1TR002649), and American Cancer Society Institutional Research Grants (IRG-18-159-43). S.L. was supported by grants from National Natural Science Foundation of China (81772999) and Guangzhou People’s Livelihood Science and Technology Project (201903010006). Y.Z. and T.S. were supported by grants from National Science Foundation for Young Scientists of China (21804038). We thank Kush Shah for assistance with proofreading.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Tingting Shen, Molecular Sciences and Biomedicine Laboratory, State Key Laboratory for Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering and College of Biology, Collaborative Innovation Center for Molecular Engineering and Theranostics, Hunan University, Changsha 410082, China; Department of Pharmaceutics, Center for Pharmaceutical Engineering and Sciences-School of Pharmacy; Massey Cancer Center; Institute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, Virginia 23298, United States.
Yu Zhang, Department of Pharmaceutics, Center for Pharmaceutical Engineering and Sciences-School of Pharmacy; Massey Cancer Center; Institute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, Virginia 23298, United States; Department of Rehabilitation Medicine, Center for Translational Medicine, Precision Medicine Institute, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou 510080, China.
Shurong Zhou, Department of Pharmaceutics, Center for Pharmaceutical Engineering and Sciences-School of Pharmacy; Massey Cancer Center; Institute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, Virginia 23298, United States.
Shuibin Lin, Department of Rehabilitation Medicine, Center for Translational Medicine, Precision Medicine Institute, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou 510080, China.
Xiao-Bing Zhang, Molecular Sciences and Biomedicine Laboratory, State Key Laboratory for Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering and College of Biology, Collaborative Innovation Center for Molecular Engineering and Theranostics, Hunan University, Changsha 410082, China.
Guizhi Zhu, Department of Pharmaceutics, Center for Pharmaceutical Engineering and Sciences-School of Pharmacy; Massey Cancer Center; Institute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, Virginia 23298, United States.
REFERENCES
- (1).Schuster M; Nechansky A; Kircheis R Cancer immunotherapy. Biotechnol. J 2006, 1 (2), 138–147. [DOI] [PubMed] [Google Scholar]
- (2).Gabrilovich DI; Ishida T; Nadaf S; Ohm JE; Carbone DP Antibodies to Vascular Endothelial Growth Factor Enhance the Efficacy of Cancer Immunotherapy by Improving Endogenous Dendritic Cell Function. Clin. Cancer Res 1999, 5 (10), 2963–2970. [PubMed] [Google Scholar]
- (3).Kwon ED; Foster BA; Hurwitz AA; Madias C; Allison JP; Greenberg NM; Burg MB Elimination of residual metastatic prostate cancer after surgery and adjunctive cytotoxic T lymphocyteassociated antigen 4 (CTLA-4) blockade immunotherapy. Proc. Natl. Acad. Sci. U. S. A 1999, 96 (26), 15074–15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chen Q; Xu L; Liang C; Wang C; Peng R; Liu Z Photothermal therapy with immune-adjuvant nanoparticles together with checkpoint blockade for effective cancer immunotherapy. Nat. Commun 2016, 7, 13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yee C; Thompson JA; Byrd D; Riddell SR; Roche P; Celis E; Greenberg PD Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc. Natl. Acad. Sci. U. S. A 2002, 99 (25), 16168–16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mahoney KM; Freeman GJ; McDermott DF The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma. Clin. Ther 2015, 37 (4), 764–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hargadon KM; Johnson CE; Williams CJ Immune checkpoint blockade therapy for cancer: An overview of FDAapproved immune checkpoint inhibitors. Int. Immunopharmacol 2018, 62, 29–39. [DOI] [PubMed] [Google Scholar]
- (8).Wollmann G; Ozduman K; van den Pol AN Oncolytic virus therapy for glioblastoma multiforme: concepts and candidates. Cancer J. 2012, 18 (1), 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Finn OJ Cancer vaccines: between the idea and the reality. Nat. Rev. Immunol 2003, 3 (8), 630–641. [DOI] [PubMed] [Google Scholar]
- (10).Kuwahara M; Sugimoto N Molecular Evolution of Functional Nucleic Acids with Chemical Modifications. Molecules 2010, 15 (8), 5423–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sridharan K; Gogtay NJ Therapeutic nucleic acids: current clinical status. Br. J. Clin. Pharmacol 2016, 82 (3), 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Seth PP; Tanowitz M; Bennett CF Selective tissue targeting of synthetic nucleic acid drugs. J. Clin. Invest 2019, 129 (3), 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dirks RW; Tanke HJ Advances in fluorescent tracking of nucleic acids in living cells. BioTechniques 2006, 40 (4), 489–496. [DOI] [PubMed] [Google Scholar]
- (14).Mizukami S; Takikawa R; Sugihara F; Shirakawa M; Kikuchi K Dual-function probe to detect protease activity for fluorescence measurement and 19F MRI. Angew. Chem., Int. Ed 2009, 48 (20), 3641–3643. [DOI] [PubMed] [Google Scholar]
- (15).Chen C; Yang Z; Tang X Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy. Med. Res. Rev 2018, 38 (3), 829–869. [DOI] [PubMed] [Google Scholar]
- (16).Manuel ER; Blache CA; Paquette R; Kaltcheva TI; Ishizaki H; Ellenhorn JD; Hensel M; Metelitsa L; Diamond DJ Enhancement of cancer vaccine therapy by systemic delivery of a tumor-targeting Salmonella-based STAT3 shRNA suppresses the growth of established melanoma tumors. Cancer Res. 2011, 71 (12), 4183–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang Z; Rao DD; Senzer N; Nemunaitis J RNA interference and cancer therapy. Pharm. Res 2011, 28 (12), 2983–2995. [DOI] [PubMed] [Google Scholar]
- (18).Xu Z; Wang Y; Zhang L; Huang L Nanoparticle-Delivered Transforming Growth Factorβ siRNA Enhances Vaccination against Advanced Melanoma by Modifying Tumor Microenvironment. ACS Nano 2014, 8 (4), 3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li CX; Parker A; Menocal E; Xiang S; Borodyansky L; Fruehauf JH Delivery of RNA interference. Cell Cycle 2006, 5 (18), 2103–2109. [DOI] [PubMed] [Google Scholar]
- (20).Sharma VK; Rungta P; Prasad AK Nucleic acid therapeutics: basic concepts and recent developments. RSC Adv. 2014, 4 (32), 16618–16631. [Google Scholar]
- (21).Mukalel AJ; Riley RS; Zhang R; Mitchell MJ Nanoparticles for nucleic acid delivery: Applications in cancer immunotherapy. Cancer Lett. 2019, 458, 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Juliano RL The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44 (14), 6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Rodriguez-Gascon A; del Pozo-Rodriguez A; Solinis MA Development of nucleic acid vaccines: use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomed 2014, 9 (1), 1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Iurescia S; Fioretti D; Rinaldi M Nucleic Acid Sensing Machinery: Targeting Innate Immune System for Cancer Therapy. Recent Pat. Anti-Cancer Drug Discovery 2018, 13 (1), 2–17. [DOI] [PubMed] [Google Scholar]
- (25).Liu MA A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7 (2), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Naldini L Gene therapy returns to centre stage. Nature 2015, 526 (7573), 351–360. [DOI] [PubMed] [Google Scholar]
- (27).Soldevilla MM; Villanueva H; Pastor F Aptamers: A Feasible Technology in Cancer Immunotherapy. J. Immunol. Res 2016, 2016, 1083738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tan X; Sun L; Chen J; Chen ZJ Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol 2018, 72, 447–478. [DOI] [PubMed] [Google Scholar]
- (29).Robinson RA; DeVita VT; Levy HB; Baron S; Hubbard SP; Levine AS Levine, A Phase I–II Trial of Multiple-Dose Polyriboinosinic-Polyribocytidylic Acid in Patients With Leukemia or Solid Tumors. J. Natl. Cancer Inst 1976, 57 (3), 599–602. [DOI] [PubMed] [Google Scholar]
- (30).Carter WA; Pitha PM; Marshall LW; Tazawa I; Tazawa S; Ts’O POP Structural requirements of the rIn·rCn complex for Induction of human interferon. J. Mol. Biol 1972, 70 (3), 567–587. [DOI] [PubMed] [Google Scholar]
- (31).Martins KA; Bavari S; Salazar AM Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev. Vaccines 2015, 14 (3), 447–59. [DOI] [PubMed] [Google Scholar]
- (32).Wang Y; Cella M; Gilfillan S; Colonna M Cutting edge: polyinosinic:polycytidylic acid boosts the generation of memory CD8 T cells through melanoma differentiation-associated protein 5 expressed in stromal cells. J. Immunol 2010, 184 (6), 2751–2755. [DOI] [PubMed] [Google Scholar]
- (33).Alexopoulou L; Holt AC; Medzhitov R; Flavell RA Recognition of double-stranded RNA and activation of NF-kB by Toll-like receptor 3. Nature 2001, 413, 732–738. [DOI] [PubMed] [Google Scholar]
- (34).Kato H; Takeuchi O; Mikamo-Satoh E; Hirai R; Kawai T; Matsushita K; Hiiragi A; Dermody TS; Fujita T; Akira S Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med 2008, 205 (7), 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Stahl-Hennig C; Eisenblatter M; Jasny E; Rzehak T; Tenner-Racz K; Trumpfheller C; Salazar AM; Uberla K; Nieto K; Kleinschmidt J; Schulte R; Gissmann L; Muller M; Sacher A; Racz P; Steinman RM; Uguccioni M; Ignatius R Synthetic double-stranded RNAs are adjuvants for the induction of T helper 1 and humoral immune responses to human papillomavirus in rhesus macaques. PLoS Pathog. 2009, 5 (4), No. e1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gowen BB; Wong MH; Jung KH; Sanders AB; Mitchell WM; Alexopoulou L; Flavell RA; Sidwell RW TLR3 is essential for the induction of protective immunity against Punta Toro Virus infection by the double-stranded RNA (dsRNA), poly(I:C12U), but not Poly(I:C): differential recognition of synthetic dsRNA molecules. J. Immunol 2007, 178 (8), 5200–5208. [DOI] [PubMed] [Google Scholar]
- (37).Trumpfheller C; Caskey M; Nchinda G; Longhi MP; Mizenina O; Huang Y; Schlesinger SJ; Colonna M; Steinman RM The microbial mimic poly IC induces durable and protective CD4+ T cell immunity together with a dendritic cell targeted vaccine. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (7), 2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Levy HB; Baer G; Baron S; Buckler CE; Gibbs CJ; Iadarola MJ; London WT; Rice J A Modified Polyriboinosinic-Polyribocytidylic Acid Complex That Induces Interferon in Primates. J. Infect. Dis 1975, 132 (4), 434–439. [DOI] [PubMed] [Google Scholar]
- (39).Sammons ML; Stephen EL; Levy HB; Baron S; Hilmas DE Interferon Induction in Cynomolgus and Rhesus Monkeys After Repeated Doses of a Modified Polyriboinosinic- Polyribocytidylic Acid Complex. Antimicrob. Agents Chemother 1977, 11 (1), 80–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schreiner B; Voss J; Wischhusen J; Dombrowski Y; Steinle A; Lochmuller H; Dalakas M; Melms A; Wiendl H Expression of toll-like receptors by human muscle cells in vitro and in vivo: TLR3 is highly expressed in inflammatory and HIV myopathies, mediates IL-8 release, and upregulation of NKG2D-ligands. FASEB J. 2006, 20 (1), 118–120. [DOI] [PubMed] [Google Scholar]
- (41).Lech M; Avila-Ferrufino A; Skuginna V; Susanti HE; Anders HJ Quantitative expression of RIG-like helicase, NOD-like receptor and inflammasome-related mRNAs in humans and mice. Int. Immunol 2010, 22 (9), 717–728. [DOI] [PubMed] [Google Scholar]
- (42).de Jong SD; Basha G; Wilson KD; Kazem M; Cullis P; Jefferies W; Tam Y The immunostimulatory activity of unmethylated and methylated CpG oligodeoxynucleotide is dependent on their ability to colocalize with TLR9 in late endosomes. J. Immunol 2010, 184 (11), 6092–6102. [DOI] [PubMed] [Google Scholar]
- (43).Akira S; Hemmi H Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett 2003, 85 (2), 85–95. [DOI] [PubMed] [Google Scholar]
- (44).Tokunaga T; Yamamoto H; Shimada S; Abe H; Fukuda T; Fujisawa Y; Furutani Y; Yano O; Kataoka T; Sudo T; Makiguchi N; Suganuma T Antitumor Activity of Deoxyribonucleic Acid Fraction From Mycobacterium bovis BCG. I. Isolation, Physicochemical Characterization, and Antitumor Activity1. J. Natl. Cancer Inst 1984, 72 (4), 955–962. [PubMed] [Google Scholar]
- (45).Bode C; Zhao G; Steinhagen F; Kinjo T; Klinman DM CpG DNA as a vaccine adjuvant. Expert Rev. Vaccines 2011, 10 (4), 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Krieg AM; Yi A-K; Matson S; Waldschmidt TJ; Bishop GA; Teasdale R; Koretzky GA; Klinman DM CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374 (6522), 546–549. [DOI] [PubMed] [Google Scholar]
- (47).Kawasaki T; Kawai T Toll-like receptor signaling pathways. Front. Immunol 2014, 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bauer S; Kirschning CJ; Hacker H; Redecke V; Hausmann S; Akira S; Wagner H; Lipford GB Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. U. S. A 2001, 98 (16), 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wagner H The immunobiology of the TLR9 subfamily. Trends Immunol. 2004, 25 (7), 381–386. [DOI] [PubMed] [Google Scholar]
- (50).Su T; Zhang Y; Valerie K; Wang X-Y; Lin S; Zhu G STING activation in cancer immunotherapy. Theranostics 2019, 9 (25), 7759–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Diner EJ; Burdette DL; Wilson SC; Monroe KM; Kellenberger CA; Hyodo M; Hayakawa Y; Hammond MC; Vance RE The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013, 3 (5), 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Danilchanka O; Mekalanos JJ Cyclic dinucleotides and the innate immune response. Cell 2013, 154 (5), 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Holm CK; Jensen SB; Jakobsen MR; Cheshenko N; Horan KA; Moeller HB; Gonzalez-Dosal R; Rasmussen SB; Christensen MH; Yarovinsky TO; Rixon FJ; Herold BC; Fitzgerald KA; Paludan SR Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol 2012, 13 (8), 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sharma S; DeOliveira RB; Kalantari P; Parroche P; Goutagny N; Jiang Z; Chan J; Bartholomeu DC; Lauw F; Hall JP; Barber GN; Gazzinelli RT; Fitzgerald KA; Golenbock DT Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity 2011, 35 (2), 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Cohen D; Melamed S; Millman A; Shulman G; Oppenheimer-Shaanan Y; Kacen A; Doron S; Amitai G; Sorek R Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 2019, 574 (7780), 691–695. [DOI] [PubMed] [Google Scholar]
- (56).Gao P; Ascano M; Zillinger T; Wang W; Dai P; Serganov AA; Gaffney BL; Shuman S; Jones RA; Deng L; Hartmann G; Barchet W; Tuschl T; Patel DJ Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 2013, 154 (4), 748–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Cavlar T; Ablasser A; Hornung V Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol. Cell Biol 2012, 90 (5), 474–482. [DOI] [PubMed] [Google Scholar]
- (58).Zaks K; Jordan M; Guth A; Sellins K; Kedl R; Izzo A; Bosio C; Dow S Efficient immunization and cross-priming by vaccine adjuvants containing TLR3 or TLR9 agonists complexed to cationic liposomes. J. Immunol 2006, 176 (12), 7335–7345. [DOI] [PubMed] [Google Scholar]
- (59).Jin B; Wang RY; Qiu Q; Sugauchi F; Grandinetti T; Alter HJ; Shih JW Induction of potent cellular immune response in mice by hepatitis C virus NS3 protein with double-stranded RNA. Immunology 2007, 122 (1), 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hafner AM; Corthesy B; Textor M; Merkle HP Tuning the immune response of dendritic cells to surface-assembled poly(I:C) on microspheres through synergistic interactions between phagocytic and TLR3 signaling. Biomaterials 2011, 32 (10), 2651–2661. [DOI] [PubMed] [Google Scholar]
- (61).Colapicchioni V; Palchetti S; Pozzi D; Marini ES; Riccioli A; Ziparo E; Papi M; Amenitsch H; Caracciolob G Killing cancer cells with nanotechnology: novel poly(I:C) loaded liposome-silica hybrid nanoparticles. J. Mater. Chem. B 2015, 3 (37), 7408–7416. [DOI] [PubMed] [Google Scholar]
- (62).Hanson MC; Crespo MP; Abraham W; Moynihan KD; Szeto GL; Chen SH; Melo MB; Mueller S; Irvine DJ Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J. Clin. Invest 2015, 125 (6), 2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Wilson DR; Sen R; Sunshine JC; Pardoll DM; Green JJ; Kim YJ Biodegradable STING agonist nanoparticles for enhanced cancer immunotherapy. Nanomedicine 2018, 14 (2), 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).An M; Yu C; Xi J; Reyes J; Mao G; Wei WZ; Liu H Induction of necrotic cell death and activation of STING in the tumor microenvironment via cationic silica nanoparticles leading to enhanced antitumor immunity. Nanoscale 2018, 10 (19), 9311–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Zhang L; Zhu G; Mei L; Wu C; Qiu L; Cui C; Liu Y; Teng IT; Tan W Self-Assembled DNA Immunonanoflowers as Multivalent CpG Nanoagents. ACS Appl. Mater. Interfaces 2015, 7 (43), 24069–24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Zhu G; Mei L; Vishwasrao HD; Jacobson O; Wang Z; Liu Y; Yung BC; Fu X; Jin A; Niu G; Wang Q; Zhang F; Shroff H; Chen X Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy. Nat. Commun 2017, 8 (1), 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Al Doghaither H; Gull M Plasmids as Genetic Tools and Their Applications in Ecology and Evolution. Plasmid 2019, 1–15. [Google Scholar]
- (68).Lowe DB; Shearer MH; Jumper CA; Kennedy R C. Towards progress on DNA vaccines for cancer. Cell. Mol. Life Sci 2007, 64 (18), 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Curran KJ; Pegram HJ; Brentjens RJ Chimeric antigen receptors for T cell immunotherapy: current understanding and future directions. J. Gene Med 2012, 14 (6), 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Bowne WB; Wolchok JD; Hawkins WG; Srinivasan R; Gregor P; Blachere NE; Moroi Y; Engelhorn ME; Houghton AN; Lewis JJ Injection of DNA encoding granulocyte-macrophage colony stimulating factor recruits dendritic cells for immune adjuvant effects. Cytokines Cell. Mol. Ther 1999, 5 (4), 217–225. [PubMed] [Google Scholar]
- (71).Ferrone CR; Perales MA; Goldberg SM; Somberg CJ; Hirschhorn-Cymerman D; Gregor PD; Turk MJ; Ramirez-Montagut T; Gold JS; Houghton AN; Wolchok JD Adjuvanticity of plasmid DNA encoding cytokines fused to immunoglobulin Fc domains. Clin. Cancer Res 2006, 12 (18), 5511–5519. [DOI] [PubMed] [Google Scholar]
- (72).Gorschluter M; Ziske C; Glasmacher A; Schmidt-Wolf IGH Current Clinical and Laboratory Strategies to Augment the Efficacy of Immunotherapy in Multiple Myeloma. Clin. Cancer Res 2001, 7 (8), 2195–2204. [PubMed] [Google Scholar]
- (73).Lambricht L; Vanvarenberg K; De Beuckelaer A; Van Hoecke L; Grooten J; Ucakar B; Lipnik P; Sanders NN; Lienenklaus S; Preat V; Vandermeulen G Coadministration of a Plasmid Encoding HIV-1 Gag Enhances the Efficacy of Cancer DNA Vaccines. Mol. Ther 2016, 24 (9), 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-viral vectors for gene-based therapy. Nat. Rev. Genet 2014, 15 (8), 541–555. [DOI] [PubMed] [Google Scholar]
- (75).Tolstyka ZP; Phillips H; Cortez M; Wu Y; Ingle N; Bell JB; Hackett PB; Reineke TM Trehalose-Based Block Copolycations Promote Polyplex Stabilization for Lyophilization and in Vivo pDNA Delivery. ACS Biomater. Sci. Eng 2016, 2 (1), 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Olden BR; Cheng Y; Yu JL; Pun SH Cationic polymers for non-viral gene delivery to human T cells. J. Controlled Release 2018, 282, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Feng G; Chen H; Li J; Huang Q; Gupte MJ; Liu H; Song Y; Ge Z Gene therapy for nucleus pulposus regeneration by heme oxygenase-1 plasmid DNA carried by mixed polyplex micelles with thermo-responsive heterogeneous coronas. Biomaterials 2015, 52, 1–13. [DOI] [PubMed] [Google Scholar]
- (78).Guerrero-Cázares H; Tzeng SY; Young NP; Abutaleb AO; Quiňones-Hinojosa A; Green JJ Biodegradable Polymeric Nanoparticles Show High Efficacy and Specificity at DNA Delivery to Human Glioblastoma in Vitro and in Vivo. ACS Nano 2014, 8 (5), 5141–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Mangraviti A; Tzeng SY; Kozielski KL; Wang Y; Jin Y; Gullotti D; Pedone M; Buaron N; Liu A; Wilson DR; Hansen SK; Rodriguez FJ; Gao G-D; DiMeco F; Brem H; Olivi A; Tyler B; Green JJ Polymeric Nanoparticles for Nonviral Gene Therapy Extend Brain Tumor Survival in Vivo. ACS Nano 2015, 9 (2), 1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Smith TT; Stephan SB; Moffett HF; McKnight LE; Ji W; Reiman D; Bonagofski E; Wohlfahrt ME; Pillai SPS; Stephan MT In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 2017, 12 (8), 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Hajj KA; Whitehead KA Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater 2017, 2 (10), 17056. [Google Scholar]
- (82).Sahin U; Kariko K; Tureci O mRNA-based therapeuticsdeveloping a new class of drugs. Nat. Rev. Drug Discovery 2014, 13 (10), 759–780. [DOI] [PubMed] [Google Scholar]
- (83).Pardi N; Hogan MJ; Porter FW; Weissman D mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discovery 2018, 17 (4), 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Weissman D mRNA transcript therapy. Expert Rev. Vaccines 2015, 14 (2), 265–281. [DOI] [PubMed] [Google Scholar]
- (85).McNamara MA; Nair SK; Holl EK RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res 2015, 2015, 794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Pollard C; De Koker S; Saelens X; Vanham G; Grooten J Challenges and advances towards the rational design of mRNA vaccines. Trends Mol. Med 2013, 19 (12), 705–713. [DOI] [PubMed] [Google Scholar]
- (87).Van Nuffel AM; Benteyn D; Wilgenhof S; Pierret L; Corthals J; Heirman C; van der Bruggen P; Coulie PG; Neyns B; Thielemans K; Bonehill A Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients. Mol. Ther 2012, 20 (5), 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Sayour EJ; Grippin A; De Leon G; Stover B; Rahman M; Karachi A; Wummer B; Moore G; Castillo-Caro P; Fredenburg K; Sarkisian MR; Huang J; Deleyrolle LP; Sahay B; Carrera-Justiz S; Mendez-Gomez HR; Mitchell DA Personalized Tumor RNA Loaded Lipid-Nanoparticles Prime the Systemic and Intratumoral Milieu for Response to Cancer Immunotherapy. Nano Lett. 2018, 18 (10), 6195–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Zhang Y; Lin S; Wang XY; Zhu G Nanovaccines for cancer immunotherapy. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol 2019, 11 (5), No. e1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Steitz J; Britten CM; Wolfel T; Tuting T Effective induction of anti-melanoma immunity following genetic vaccination with synthetic mRNA coding for the fusion protein EGFP.TRP2. Cancer Immunol. Immunother 2006, 55 (3), 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Wasungu L; Hoekstra D Cationic lipids, lipoplexes and intracellular delivery of genes. J. Controlled Release 2006, 116 (2), 255–264. [DOI] [PubMed] [Google Scholar]
- (92).Kranz LM; Diken M; Haas H; Kreiter S; Loquai C; Reuter KC; Meng M; Fritz D; Vascotto F; Hefesha H; Grunwitz C; Vormehr M; Husemann Y; Selmi A; Kuhn AN; Buck J; Derhovanessian E; Rae R; Attig S; Diekmann J; Jabulowsky RA; Heesch S; Hassel J; Langguth P; Grabbe S; Huber C; Tureci O; Sahin U Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534 (7607), 396–401. [DOI] [PubMed] [Google Scholar]
- (93).Hoerr I; Obst R; Rammensee H-G; Jung G In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur. J. Immunol 2000, 30, 1–7. [DOI] [PubMed] [Google Scholar]
- (94).Su X; Fricke J; Kavanagh D; Irvine DJ In vitro and in vivo mRNA delivery using lipid-enveloped pH-responsive polymer nanoparticles. Mol. Pharmaceutics 2011, 8 (3), 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Kortylewski M; Swiderski P; Herrmann A; Wang L; Kowolik C; Kujawski M; Lee H; Scuto A; Liu Y; Yang C; Deng J; Soifer HS; Raubitschek A; Forman S; Rossi JJ; Pardoll DM; Jove R; Yu H In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat. Biotechnol 2009, 27 (10), 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Li SY; Liu Y; Xu CF; Shen S; Sun R; Du XJ; Xia JX; Zhu YH; Wang J Restoring anti-tumor functions of T cells via nanoparticle-mediated immune checkpoint modulation. J. Controlled Release 2016, 231, 17–28. [DOI] [PubMed] [Google Scholar]
- (97).Wang D; Wang T; Liu J; Yu H; Jiao S; Feng B; Zhou F; Fu Y; Yin Q; Zhang P; Zhang Z; Zhou Z; Li Y Acid-Activatable Versatile Micelleplexes for PD-L1 Blockade-Enhanced Cancer Photodynamic Immunotherapy. Nano Lett. 2016, 16 (9), 5503–5513. [DOI] [PubMed] [Google Scholar]
- (98).Tatiparti K; Sau S; Kashaw SK; Iyer AK siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials 2017, 7 (4), E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Warashina S; Nakamura T; Sato Y; Fujiwara Y; Hyodo M; Hatakeyama H; Harashima H A lipid nanoparticle for the efficient delivery of siRNA to dendritic cells. J. Controlled Release 2016, 225, 183–191. [DOI] [PubMed] [Google Scholar]
- (100).Lee SJ; Huh MS; Lee SY; Min S; Lee S; Koo H; Chu J-U; Lee KE; Jeon H; Choi Y; Choi K; Byun Y; Jeong SY; Park K; Kim K; Kwon IC Tumor-Homing Poly-siRNA/Glycol Chitosan Self-Cross-Linked Nanoparticles for Systemic siRNA Delivery in Cancer Treatment. Angew. Chem., Int. Ed 2012, 51 (29), 7203–7207. [DOI] [PubMed] [Google Scholar]
- (101).Neagoe IB; Braicu C; Matea C; Bele C; Florin G; Gabriel K; Veronica C; Irimie A Efficient siRNA delivery system using carboxilated single-wall carbon nanotubes in cancer treatment. J. Biomed. Nanotechnol 2012, 8 (4), 567–574. [DOI] [PubMed] [Google Scholar]
- (102).Pridgen EM; Langer R; Farokhzad OC Biodegradable, polymeric nanoparticle delivery systems for cancer therapy. Nanomedicine 2007, 2 (5), 669–680. [DOI] [PubMed] [Google Scholar]
- (103).Cox DB; Platt RJ; Zhang F Therapeutic genome editing: prospects and challenges. Nat. Med 2015, 21 (2), 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Corrigan-Curay J; O’Reilly M; Kohn DB; Cannon PM; Bao G; Bushman FD; Carroll D; Cathomen T; Joung JK; Roth D; Sadelain M; Scharenberg AM; von Kalle C; Zhang F; Jambou R; Rosenthal E; Hassani M; Singh A; Porteus MH Genome editing technologies: defining a path to clinic. Mol. Ther 2015, 23 (5), 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Kang Y; Chu C; Wang F; Niu Y CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis. Models & Mech 2019, 12 (10), No. dmm039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Sternberg SH; LaFrance B; Kaplan M; Doudna JA Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 2015, 527 (7576), 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Singh D; Sternberg SH; Fei J; Doudna JA; Ha T Realtime observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun 2016, 7, 12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Rupp LJ; Schumann K; Roybal KT; Gate RE; Ye CJ; Lim WA; Marson A CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep 2017, 7 (1), 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Ren J; Liu X; Fang C; Jiang S; June CH; Zhao Y Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res 2017, 23 (9), 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Prasad V Immunotherapy: Tisagenlecleucel - the first approved CAR-T-cell therapy: implications for payers and policy makers. Nat. Rev. Clin. Oncol 2018, 15 (1), 11–12. [DOI] [PubMed] [Google Scholar]
- (111).Hsu PD; Lander ES; Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157 (6), 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Chakraborty S; Ji H; Kabadi AM; Gersbach CA; Christoforou N; Leong KW A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep. 2014, 3 (6), 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Ding Q; Strong A; Patel KM; Ng SL; Gosis BS; Regan SN; Cowan CA; Rader DJ; Musunuru K Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res 2014, 115 (5), 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Ran FA; Cong L; Yan WX; Scott DA; Gootenberg JS; Kriz AJ; Zetsche B; Shalem O; Wu X; Makarova KS; Koonin EV; Sharp PA; Zhang F In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520 (7546), 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Wang T; Wei JJ; Sabatini DM; Lander ES Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2014, 343 (6166), 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Wang HX; Li M; Lee CM; Chakraborty S; Kim HW; Bao G; Leong KW CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev 2017, 117 (15), 9874–9906. [DOI] [PubMed] [Google Scholar]
- (117).Chen G; Abdeen AA; Wang Y; Shahi PK; Robertson S; Xie R; Suzuki M; Pattnaik BR; Saha K; Gong S A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol 2019, 14 (10), 974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Nguyen DN; Roth TL; Li PJ; Chen PA; Apathy R; Mamedov MR; Vo LT; Tobin VR; Goodman D; Shifrut E; Bluestone JA; Puck JM; Szoka FC; Marson A Polymerstabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020, 38, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Tan W; Donovan MJ; Jiang J Aptamers from cell-based selection for bioanalytical applications. Chem. Rev 2013, 113 (4), 2842–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Pardoll DM The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12 (4), 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Ma H; Liu J; Ali MM; Mahmood MA; Labanieh L; Lu M; Iqbal SM; Zhang Q; Zhao W; Wan Y Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev 2015, 44 (5), 1240–1256. [DOI] [PubMed] [Google Scholar]
- (122).Soldevilla MM; Villanueva H; Pastor F Aptamers as a Promising Therapeutic Tool for Cancer Immunotherapy. In Immunotherapy: Myths, Reality, Ideas, Future 2017, 129–150. [Google Scholar]
- (123).Pastor F; Soldevilla MM; Villanueva H; Kolonias D; Inoges S; de Cerio AL; Kandzia R; Klimyuk V; Gleba Y; Gilboa E; Bendandi M CD28 aptamers as powerful immune response modulators. Mol. Ther.–Nucleic Acids 2013, 2, No. e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Santulli-Marotto S; Nair SK; Rusconi C; Sullenger B; Gilboa E Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003, 63, 7483–7489. [PubMed] [Google Scholar]
- (125).Prodeus A; Abdul-Wahid A; Fischer NW; Huang EH; Cydzik M; Gariepy J Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Mol. Ther.–Nucleic Acids 2015, 4, No. e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Meyer C; Hahn U; Rentmeister A Cell-specific aptamers as emerging therapeutics. J. Nucleic Acids 2011, 2011, 904750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Bagalkot V; Farokhzad OC; Langer R; Jon S An aptamer-doxorubicin physical conjugate as a novel targeted drugdelivery platform. Angew. Chem. Int. Ed 2006, 45 (48), 8149–8152. [DOI] [PubMed] [Google Scholar]
- (128).Ni S; Yao H; Wang L; Lu J; Jiang F; Lu A; Zhang G Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci 2017, 18 (8), 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Shum KT; Tanner JA Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. ChemBioChem 2008, 9 (18), 3037–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Dougan H; Lyster DM; Vo CV; Stafford A; Weitz JI; Hobbs JB Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Biol 2000, 27, 289–297. [DOI] [PubMed] [Google Scholar]
- (131).Shi H; He X; Cui W; Wang K; Deng K; Li D; Xu F Locked nucleic acid/DNA chimeric aptamer probe for tumor diagnosis with improved serum stability and extended imaging window in vivo. Anal. Chim. Acta 2014, 812, 138–144. [DOI] [PubMed] [Google Scholar]
- (132).Abeydeera ND; Egli M; Cox N; Mercier K; Conde JN; Pallan PS; Mizurini DM; Sierant M; Hibti FE; Hassell T; Wang T; Liu FW; Liu HM; Martinez C; Sood AK; Lybrand TP; Frydman C; Monteiro RQ; Gomer RH; Nawrot B; Yang X Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016, 44 (17), 8052–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Varizhuk AM; Tsvetkov VB; Tatarinova ON; Kaluzhny DN; Florentiev VL; Timofeev EN; Shchyolkina AK; Borisova OF; Smirnov IP; Grokhovsky SL; Aseychev AV; Pozmogova GE Synthesis, characterization and in vitro activity of thrombin-binding DNA aptamers with triazole internucleotide linkages. Eur. J. Med. Chem 2013, 67, 90–97. [DOI] [PubMed] [Google Scholar]
- (134).Hoellenriegel J; Zboralski D; Maasch C; Rosin NY; Wierda WG; Keating MJ; Kruschinski A; Burger JA The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123 (7), 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Soldevilla MM; Villanueva H; Casares N; Lasarte JJ; Bendandi M; Inoges S; Cerio A. L.-D. d.; Pastor F MRP1-CD28 bi-specific oligonucleotide aptamers: target costimulation to drugresistant melanoma cancer stem cells. Oncotarget 2016, 7 (17), 23182–23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Theofilopoulos AN; Kono DH; Baccala R The multiple pathways to autoimmunity. Nat. Immunol 2017, 18 (7), 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Durbin AF; Wang C; Marcotrigiano J; Gehrke L RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7 (5), No. e00833–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).O’Connell MA; Mannion NM; Keegan LP The Epitranscriptome and Innate Immunity. PLoS Genet. 2015, 11 (12), No. e1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Hornung V; Ellegast J; Kim S; Brzozka K; Jung A; Kato H; Poeck H; Akira S; Conzelmann KK; Schlee M; Endres S; Hartmann G 5’-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314 (5801), 994–997. [DOI] [PubMed] [Google Scholar]
- (140).Kariko K; Muramatsu H; Welsh FA; Ludwig J; Kato H; Akira S; Weissman D Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther 2008, 16 (11), 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Bruck J; Pascolo S; Fuchs K; Kellerer C; Glocova I; Geisel J; Dengler K; Yazdi AS; Rocken M; Ghoreschi K Cholesterol Modification of p40-Specific Small Interfering RNA Enables Therapeutic Targeting of Dendritic Cells. J. Immunol 2015, 195 (5), 2216–2223. [DOI] [PubMed] [Google Scholar]
- (142).Yu C; An M; Li M; Liu H Immunostimulatory Properties of Lipid Modified CpG Oligonucleotides. Mol. Pharmaceutics 2017, 14 (8), 2815–2823. [DOI] [PubMed] [Google Scholar]
- (143).Zhu G; Lynn GM; Jacobson O; Chen K; Liu Y; Zhang H; Ma Y; Zhang F; Tian R; Ni Q; Cheng S; Wang Z; Lu N; Yung BC; Wang Z; Lang L; Fu X; Jin A; Weiss ID; Vishwasrao H; Niu G; Shroff H; Klinman DM; Seder RA; Chen X Albumin/vaccine nanocomplexes that assemble in vivo for combination cancer immunotherapy. Nat. Commun 2017, 8 (1), 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Fliervoet LAL; Engbersen JFJ; Schiffelers RM; Hennink WE; Vermonden T Polymers and hydrogels for local nucleic acid delivery. J. Mater. Chem. B 2018, 6 (36), 5651–5670. [DOI] [PubMed] [Google Scholar]