Abstract
Considerable attention has been recently given to possible transmission of SARS-CoV-2 via water media. This review addresses this issue and examines the fate of coronaviruses (CoVs) in water systems, with particular attention to the recently available information on the novel SARS-CoV-2. The methods for the determination of viable virus particles and quantification of CoVs and, in particular, of SARS-CoV-2 in water and wastewater are discussed with particular regard to the methods of concentration and to the emerging methods of detection. The analysis of the environmental stability of CoVs, with particular regard of SARS-CoV-2, and the efficacy of the disinfection methods are extensively reviewed as well. This information provides a broad view of the state-of-the-art for researchers involved in the investigation of CoVs in aquatic systems, and poses the basis for further analyses and discussions on the risk associated to the presence of SARS-CoV-2 in water media. The examined data indicates that detection of the virus in wastewater and natural water bodies provides a potentially powerful tool for quantitative microbiological risk assessment (QMRA) and for wastewater-based epidemiology (WBE) for the evaluation of the level of circulation of the virus in a population. Assays of the viable virions in water media provide information on the integrity, capability of replication (in suitable host species) and on the potential infectivity. Challenges and critical issues relevant to the detection of coronaviruses in different water matrixes with both direct and surrogate methods as well as in the implementation of epidemiological tools are presented and critically discussed.
Abbreviations: µPAD, microfluidic paper analytic device; BSL, Biosafety Level; CCoV, canine coronavirus; CDC, Centers for Disease Control and Prevention; CoV, coronavirus; COVID-19, CoronaVirus Disease 19; CPE, cytopathic effects; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; ddPCR, droplet digital polymerase chain reaction; DMEM, Dulbecco minimal essential medium; dPCR, digital polymerase chain reaction; E, envelope protein; EPA, United States Environmental Protection Agency; FBS, fetal bovine serum; FET, field-effect transistor; FIPV, feline infectious peritonitis virus; ICC-MS, integrated cell culture-mass spectrometry; M, membrane protein; MBRs, membrane bioreactors; MEM, minimal essential medium; MERS, Middle East respiratory syndrome; MHV, murine hepatitis virus; MPAD, Multiplex Paired-Antibody Amplified Detection; MS, mass spectrometry; MWCO, molecular weight cut-off; N, nucleocapsid protein; NoV, noroviruses; OSHA, United States Occupational Safety and Health Administration; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PEG, poly(ethylene glycol); PS, polystyrene; QMRA, quantitative microbiological risk assessment; qPCR, quantitative polymerase chain reaction; RdRp, RNA-dependent RNA polymerase; RIVM, Rijksinstituut voor Volksgezondheid en Milieu; RNA, ribonucleic acid; rRT-LAMP, real-time reverse transcription loop mediated isothermal amplification; RT-ddPCR, reverse transcription droplet digital polymerase chain reaction; RT-LAMP, reverse transcription loop mediated isothermal amplification; RT-PCR, reverse transcription polymerase chain reaction; S, spike protein; SARI, Sorveglianza Ambientale Reflue in Italia; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; TCID50, tissue culture infectious dose-50; TGEV, transmissible gastroenteritis virus; UV, ultraviolet radiation; VIRADEL, virus adsorption-elution; WBE, wastewater-based epidemiology; WHO, World Health Organization; WWTP, wastewater treatment plant
Keywords: SARS-CoV-2, Coronavirus, Hazards, Viable virus, Infectivity, Detection, Concentration, Persistence, Disinfection, Epidemiology, Environment, Wastewater, Drinking water
Graphical Abstract
1. Introduction
A novel human coronavirus (CoV), named severe acute respiratory syndrome coronavirus-2, SARS-CoV-2, was identified in Wuhan, China, in December 2019. SARS-CoV-2 has caused an outbreak of respiratory illness called COVID-19 (Holshue et al., 2020, Mallapaty, 2020, Wang et al., 2020a, Wrapp et al., 2020, Yang et al., 2020, Zhang et al., 2020b). SARS-CoV-2 is a member of the Coronaviridae family which comprises enveloped and single-stranded ribonucleic acid (RNA) viruses with sizes ranging from 60 to 220 nm (Corpuz et al., 2020). The viral protein capsid, enclosing the viral RNA, is covered by a lipid bilayer membrane that contains proteins or glycoproteins and crown-like spikes on the surface. CoVs can infect birds, rodents, felines, canids, chiropters, and other mammals, including human beings (Cui et al., 2019, Decaro and Lorusso, 2020, La Rosa et al., 2012, Robinson et al., 2016, Wigginton et al., 2015). The initial animal-to-person transfer of CoV, referred to as the zoonotic transmission, appears to have occurred via a natural genetic mutation enabling the virus to infect humans (Andersen et al., 2020, Cui et al., 2019, Zhou et al., 2020b). Subsequently, the person-to-person transfer occurred by inhaling infected aerosols and respiratory droplets (Asadi et al., 2020a, Bourouiba, 2020, Cevik et al., 2020, Fears et al., 2020, Gralton et al., 2013, Morawska and Cao, 2020, Senatore et al., 2021, Somsen et al., 2020, Stadnytskyi et al., 2020, van Doremalen et al., 2020). However, other potential routes of SARS-CoV-2 transmission have been suggested. This include transmission via fomites (van Doremalen et al., 2020), ocular surfaces (Lu et al., 2020a) and the fecal–oral route (Gupta et al., 2020, Gwenzi, 2020, Lodder and de Roda Husman, 2020, Wu et al., 2020a, Wu et al., 2020c, Yeo et al., 2020).
CoVs were first identified in the mid-1960s and up to now seven human CoVs have been reported. Three of them, notably SARS-CoV, Middle East respiratory syndrome-CoV (MERS-CoV), and SARS-CoV-2 have emerged recently. The containment of human mortality associated with CoVs transmission, as well as of the number of people requiring hospitalization and the consequent saturation of hospital facilities, forces the application of strict isolation measures in the countries affected by the infection.
During epidemics, the high rate of transmission of human CoVs has principally occurred via the transfer of infected respiratory droplets (Asadi et al., 2020a, Bourouiba, 2020, Cevik et al., 2020, Fears et al., 2020, Gralton et al., 2013, Morawska and Cao, 2020, Senatore et al., 2021, Somsen et al., 2020, Stadnytskyi et al., 2020, van Doremalen et al., 2020). Before the emergence of SARS-CoV (below indicated as SARS-CoV-1 in order to avoid confusion with the SARS-CoV-2) in 2002 in China, CoVs were considered as exclusively respiratory pathogens. However, SARS-CoV-1 can affect the human enteric tract as well (Chan et al., 2004, Jevšnik et al., 2013, Lai et al., 2005, Leung et al., 2003, Peiris et al., 2003, Poon et al., 2004, Wang, 2005a). Aqueous media, in particular wastewater, can be directly contaminated with CoVs via infected feces. Aqueous media have been thus considered as a potential transmission carrier (Gundy et al., 2008, Lodder and de Roda Husman, 2020, Wigginton et al., 2015, Ye et al., 2016). CoVs have been reported to persist in aqueous media from few hours to few weeks, though their viability and infectivity strongly depend on several factors (vide infra).
Given that it is difficult to evaluate the impact of waterborne viral infections, the role of these infections is often underestimated as was the case during the SARS epidemic in Hong Kong in 2003. Bioaerosols, generated from the aeration in sewer pipelines and not subject to specific disinfection treatments, were identified to be source of the SARS-CoV-1 spreading in Amoy Gardens, a private housing estate in Hong Kong (Hung, 2003, McKinney et al., 2006, WHO, 2003). The promiscuous sharing of ponds by ducks, pigs and humans was indicated as origin and hot spots of resurgence of the influenza A virus subtype H5N1: an enveloped virus with spike-like proteins on surface similar to those of CoVs (Matsui, 2005). The spillover transmission of H5N1 virus to humans occurred from ducks, the original host species for the virus, through pigs as an intermediate species (Li et al., 2004, Matsui, 2005). Animals have also been proven to be reservoirs for the novel SARS-CoV-2. In fact, the virus has been found to be efficiently replicant in cats and ferrets and comparatively and poorly replicant in dogs, pigs, chickens and ducks (Shi et al., 2020).
SARS-CoV-2 has been rapidly spreading worldwide (Holshue et al., 2020, Mallapaty, 2020, Wang et al., 2020a, Wrapp et al., 2020, Yang et al., 2020). In addition to respiratory dysfunctions, the virus induces severe enteric symptoms and has been detected in the feces of infected patients (Holshue et al., 2020, Wu et al., 2020d, Yeo et al., 2020, Zhang et al., 2020b). Hence, the digestive system has been recognized as a potential route of infection (Holshue et al., 2020, Wang et al., 2020d, Zhang et al., 2020b) and the virus RNA ended up in wastewater. Urine and feces of patients affected by COVID-19 have been proven infectious, by assessing the viable SARS-CoV-2 virus particles in suitable host cells (vide infra) (Xiao et al., 2020). However, in spite of the high concentrations of the RNA of the virus worldwide found in wastewaters (see Table 1) and the potential concerns associate, (Adelodun et al., 2020, Amoah et al., 2020, Arslan et al., 2020, Bhowmick et al., 2020, Bilal et al., 2020, Bogler et al., 2020, Carducci et al., 2020, Carraturo et al., 2020, Collivignarelli et al., 2020, El Baz and Imziln, 2020, Foladori et al., 2020, Gwenzi, 2020, Jones et al., 2020, Paleologos et al., 2020, Kitajima et al., 2020, La Rosa et al., 2020a, Mandal et al., 2020, Nghiem et al., 2020, Shutler et al., 2020) recent investigations on the infectivity have indicated a scarce persistence of the virus in these water media (Bivins et al., 2020, Ge et al., 2020, Rimoldi et al., 2020a, Rimoldi et al., 2020b, Wang et al., 2020c, Westhaus et al., 2021).
Table 1.
Quantification and assay of viable f SARS-CoV-2 in wastewater and sludge.
| Reference; Location | Sample type; Sampling mode; Volume; Storage temperature | Sample pre-treatment | Concentration method | Treatment for PCR inhibitors | RNA extraction |
Detection/Quantization |
Concentration in genome copies/L |
Viability; (infectious samples/total samples) | |
|---|---|---|---|---|---|---|---|---|---|
| Influent | Effluent | ||||||||
| (Medema et al., 2020a,b); The Netherlands | Municipal wastewater; Composite sampling (24 h); 250 mL; 4 °C | Centrifugation | Centrifugal filtration (Centricon Plus-70, MWCO of 100 kDa) | n.r. | RNeasy PowerMicrobe (Qiagen) and Biomerieux Nulisens Kit (Biomerieux) in combination with semi-automated KingFisher mL purification system (Thermo Scientific) | RT-qPCR; Indirect evaluation by F-specific RNA phages assay; | 2.60 103–2.2 106 | a | a |
| (Wu et al., 2020b); Massachusetts, USA | Municipal wastewater; Grab and composite sampling (24 h); n.r.; 4 °C | Pasteurization (60 °C, 90 min) and filtration (0.2 μm pore size) | PEG 8000/NaCl precipitation, centrifugal filtration; (10 kDa Amicon ultra centrifugal filter) | n.r. | TRIzol; RNeasy kit (Qiagen) | RT-qPCR | 100–103 | a | a |
| (Nemudryi et al., 2020a,b); Montana, USA | Municipal wastewater; Composite sampling (24 h); 500 mL; n.r. | Filtration (5 μm and 0.45 μm pore size) | Centrifugal filtration (Corning Spin-X UF, MWCO of 100 kDa) | n.r. | RNeasy Mini Kit (Qiagen) | RT-qPCR (one-step) TaqPath™ 1-Step RT-qPCR Master Mix, CG (ThermoFisher) RT-PCR SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific); Q5 High-Fidelity DNA Polymerase (New England Biolabs) | > 104 | a | a |
| (Ahmed et al., 2020a); Australia | Municipal wastewater; Composite sampling (24 h) and grab sampling; 100–200 mL; 4 °C | Filtration (0.45 μm pore size) |
|
n.r. | RNeasy PowerWater Kit; RNeasy PowerMicrobiome (Qiagen) | RT-qPCR (one-step) iTaq™ Universal Probes One-Step Reaction Mix (Bio-Rad) | 1.90 101–1.2 102 | a | a |
| (Wurtzer et al., 2020); Paris, France | Municipal wastewater; n.r.; 11 mL; n.r. | – | Ultracentrifugation | PCR Inhibitor removal resin (Zymo Research) | PowerFecal Pro kit with a QIAsymphony automated extractor (Qiagen) | RT-qPCR | 5.00 104–3.00 106 | a | a |
| (Wang et al., 2020c); China | Hospital sewage; n.r.; n.r.; n.r. | – | – | n.r. | SARS-CoV-2 nucleic acid detection kit (Shanghai Berger Medical Technology Co., China) | RT-PCR | a | a | Test in Vero-E6 cells; (0/6) |
| (Randazzo et al., 2020b,c); Region of Murcia, Spain | Municipal wastewater; Grab sampling 200 mL; 4 °C | pH Adjustment at 6; | Precipitation with AlCl3, centrifugation; elution with beef extract (3%, pH 7.4), centrifugation and resuspension in PBS | n.r. | Nucleospin RNA virus Kit (Macherey-Nagel) | PrimeScript™ One Step RT-PCR Kit; RT-qPCR diagnostic panel assays (US CDC, 2019-nCoV RUO Kit and the positive control 2019-nCoV_N_Positive Control by Integrated DNA Technologies). | 1.48 105–3.90 105 | Secondary Effluent: 2.51 105 Tertiary Effluent: No detection | a |
| (La Rosa et al., 2020b); Milan and Rome, Italy | Municipal wastewater; Grab sampling; 250 mL; −20 °C | Pasteurization (57 °C, 30 min) | PEG-Dextran precipitation | One step PCR Inhibitor removal kit (Zymo Research) | NucliSENS miniMAG semi-automated extraction system (bioMerieux,) | RT-PCR SuperScript III/IV Reverse Transcriptase (ThermoFisher Scientific); | a | a | a |
| Kit Platinum™ SuperFi™ | |||||||||
| Green PCR Master Mix, Thermo), | |||||||||
| (Rimoldi et al., 2020a,b); Milan, Italy | Municipal wastewater; Grab sampling; 500 mL; Specific temperature not reported (samples were under refrigeration) | Filtration (0.7 μm and 0.2 μm nominal pore size) | Not carried out | n.r. | QIAMP VIRAL RNA mini kit (Qiagen, Hilden, Germany) | Real-Time RT-PCR | a | a | Test in Vero E6 cells; |
| Influent: (0/8) | |||||||||
| Effluent: (0/4) | |||||||||
| (Bar Or et al., 2020); Israel | Municipal wastewater; Composite sampling (24 h); 0.25–1 L; -20 °C or −80 °C | Centrifugation |
|
n.r. | RNA extraction kit (RNeasy mini kit- QIAGEN and EasyMAG-bioMerieux, France) | RT-qPCR | Only Cycle Threshold (Ct) numbers were given (i.e. number of cycles required for the fluorescent signal to cross the threshold) | Only Cycle Threshold (Ct) numbers were given (i.e. number of cycles required for the fluorescent signal to cross the threshold) | a |
| (Haramoto et al., 2020); Japan | Municipal wastewater; Grab sampling; Influent: 200 mL Secondary Effluent: 5000 mL; n.r. | – | Electronegative membrane-vortex method; | n.r. | RNeasy PowerWater Kit (Qiagen) | RT-qPCR Nested PCR | Not detected | 2.4 103 | a |
| Adsorption direct RNA extraction method | |||||||||
| (Zhang et al., 2020a); China | Municipal wastewater; Grab sampling; 2.0 L; 4 °C | Centrifugation | PEG 6000/NaCl precipitation | n.r. | EZ1 virus Mini kit (Qiagen, Germany) | RT-qPCR | Not detected (After primary disinfection tank before septic tank) | 0.5 103 to 18.7 103 (After septic tank with disinfection with sodium hypochlorite) | a |
| (Feng et al., 2021); Hangzhou, China | Medical wastewater (From Isolation Facility for COVID-19 Patients); Grab Sampling; 15 mL; n.r. | n.r. | n.r. | n.r. | Roche MagNA Pure LC 2.0 | RT-qPCR | Septic Tank Influent: 5.89 105 Wastewater Inlet Pipe: 1.660 106 | Septic Tank Effluent: 3.092 106 | a |
| Sewage Treatment Effluent: | |||||||||
| Below detection limit | |||||||||
| Sewage Treatment Inlet Pipe: 3.63 105 | |||||||||
| (Westhaus et al., 2021); Germany | Municipal wastewater; Composite sampling (24 h); 45 mL; −18 °C | Centrifugation (4700g for 30 min) | Centrifugal ultrafiltration | n.r. | NucleoSpin RNA Virus kit | RT-qPCR; Test in Caco-2 (human epithelial cell line from colon adenocarcinoma) | Influent (Aqueous Phase): 3.00 103–2.00 104 | Effluent (Aqueous Phase): 2.70 103 to 3.70 104 | Influent: (0) Effluent: 0 |
| After Tertiary Treat.: 0 (Total number of samples not reported) | |||||||||
| Effluent (Solid Phase): 1.30 104 | |||||||||
| Influent (Solid Phase): 2.50 104 | |||||||||
| (La Rosa et al., 2020b, La Rosa et al., 2021); Milan, Turin and Bologna, Italy | Municipal wastewater; Composite sampling (24 h); 250 mL; −20 °C | Pasteurization (56 °C, 30 min) | PEG-Dextran precipitation | OneStep PCR Inhibitor Removal | NucliSENS miniMAG semi-automated extraction system with magnetic silica (bioMerieux) | Nested RT-PCR; RT-qPCR | Below limit of detection to 5.60 104 | a | a |
| Kit (Zymo Research) | |||||||||
| (Trottier et al., 2020); Montpellier, France | Municipal wastewater; Composite sampling (24 h); n.r.; 4 °C | Filtration (40 μm cell strainer) | Centrifugal ultrafiltration (Vivaspin 50 kDa MWCO filter membrane) | n.r. | NucleoSpin RNA Virus | RT-qPCR TaqPath One-Step RT-qPCR, CG master mix (ThermoFisher Scientific) | Not detected to 8.0 104 (value estimated from graph in manuscript) | a | a |
| kit (Macherey-Nagel) | |||||||||
| (Kumar et al., 2020); India | Municipal WWTP (receiving effluent from hospital treating COVID-19 patients); Composite sampling; 50 mL; 4 °C | Centrifugation and filtration (0.22 μm mixed cellulose esters syringe filter) | PEG 9000/NaCl precipitation | n.r. | NucleoSpin RNA Virus | RT-PCR (Quantity of SARS-CoV-2 gene copies was approximated using obtained Cycle threshold (Ct) values) | 5.60 101–3.50 102 | Not detected | a |
| kit (Macherey-Nagel) | |||||||||
| (Sherchan et al., 2020); Louisiana, USA | Municipal wastewater; Grab and composite sampling (24 h); 1 L; −80 °C | – | Ultrafiltration; Adsorption-elution with electronegative membrane | n.r. | ZR Viral RNA Kit (Zymo Research) | RT-qPCR | 3.10 103–7.50 103 | Not detected | a |
| (Gonzalez et al., 2020); Virginia, USA | Municipal wastewater; Grab and composite sampling (24 h); 1 L; n.r. | – | InnovaPrep Concentrating Pipette Select; | n.r. | NucliSENS easyMag (bioMerieux) | Reverse transcription droplet digital PCR (RT-ddPCR) | 102–105 | a | a |
| Electronegative filtration with mixed cellulose ester HA filters | |||||||||
| (Randazzo et al., 2020a); Valencia, Spain | Metropolitan wastewater; | – | Aluminum-driven flocculation | n.r. | Nucleospin RNA virus Kit (Macherey-Nagel) | RT-qPCR | 1.66 105–9.77 105 | Not detected | a |
| Grab sampling; 200 mL; 4 °C | |||||||||
| (Mlejnkova et al., 2020); Czech Republic | Municipal wastewater; | – | Direct flocculation with beef-extract solution | n.r. | NucliSENS miniMAG system (bioMérieux) | RT-qPCR | a | a | a |
| Composite sampling; 500 mL; 2–8 °C | |||||||||
| (Arora et al., 2020); India | Municipal wastewater; n.r.; n.r.; n.r. | Inactivation at 60 °C | PEG/NaCl precipitation | n.r. | Allplex™ 2019-nCoV 197 Assay kit | RT-PCR | a | a | a |
| (Ahmed et al., 2020b); Australia | Aircraft wastewater and cruise ship wastewater (from membrane bioreactor influent and effluent); | – | Adsorption–extraction with electronegative membrane; Ultrafiltration with Amicon® Ultra-15 centrifugal filter unit (30 kDa) | n.r. | RNeasy PowerWater Kit and RNeasy PowerMicrobiome Kit (Qiagen) | RT-qPCR; RT-ddPCR | Aircraft: Below Limit of Detection to 2.72 103 | Cruise ship effluent: Below Limit of Detection to 9.45 102 | a |
| Cruise ship influent: | |||||||||
| Below Limit of Detection to 8.80 103 | |||||||||
| Grab sampling; 1 L; 4 °C | |||||||||
| (Fongaro et al., 2020); Santa Catalina, Brazil | Urban sewage; n.r.; 200 mL; −80 °C | – | Glycine buffer method | n.r. | QIAampR Viral RNA Mini kit (Qiagen) | RT-qPCR | 2.95 105–5.01 106 | a | a |
| (Sharif et al., 2020); Pakistan | Raw sewage; | – | Two-phase separation with polymer, dextran, and PEG | n.r. | Spin star viral nucleic acid kit 1.0 (ADT Biotech) | RT-qPCR | Only Cq (quantification cycle) values were reported | a | a |
| Grab sampling; 1 L; n.r. | |||||||||
| (Curtis et al., 2020); Virginia, USA | Municipal wastewater; | – | Electronegative filtration | n.r. | NucliSENS easyMag (bioMerieux) | RT-ddPCR | Grab samples: 5.80 103–1.38 104 | a | a |
| Grab and composite sampling (24 h); 1 L; 4 °C | |||||||||
| Composite samples: 2.50 102–1.11 104 | |||||||||
| (Prado et al., 2020); Rio de Janeiro, Brazil | Municipal and hospital wastewater; | Pasteurization at 60 °C for 90 min | Ultracentrifugation | n.r. | QIAamp Viral RNA Mini kit and | RT-qPCR | Only Ct (cycle threshold) numbers were reported | a | a |
| QIAcubeautomated system (Qiagen) | |||||||||
| Composite sampling (10 h); n.r.; n.r. | |||||||||
| (Ampuero et al., 2020); Santiago, Chile | Municipal wastewater; | – | Ultracentrifugation | n.r. | QIAamp Viral RNA Mini kit (Qiagen) | RT-qPCR | Not detected to 4.81 106 | Not detected to 2.67 105 | a |
| Composite sampling (24 h); n.r.; n.r. | |||||||||
| (Green et al., 2020); New York; USA | Municipal wastewater; | – | Ultracentrifugation through a sucrose cushion | n.r. | AllPrep® PowerViral® DNA/RNA Kit (Qiagen) | Multiplex RT-qPCR | Below limit of quantitation to 1.12 105 | a | a |
| Composite sampling (24 h); 1.9 L; 4 °C | |||||||||
| (Manupati et al., 2020); Hyderabad, India | Municipal wastewater; | Filtration through blotting paper and 0.22 μm filter | Filtration with 30 kDa Amicon® Ultra-15 | n.r. | QIAamp Viral RNA isolation kit (Qiagen) | RT-qPCR | 3.08 104–2.66 105 | Not detected | a |
| Grab sampling; n.r.; 4 °C | |||||||||
| (Miyani et al., 2020); Detroit, Michigan, USA | Municipal wastewater; | – | Adsorption elution with Nanoceram filter | n.r. | Viral RNA kit (Qiagen) | Two-step RT-qPCR | 1.24 104– 4.33 105 | a | a |
| Grab sampling; 28–80 L; 4 °C | |||||||||
| (Yaqub et al., 2020); Lahore, Pakistan | Municipal wastewater; | Vortex mixing | Centrifugation | n.r. | Hero 32 Extraction System | RT-qPCR | Not detected to 3.55 107 | a | a |
| Grab sampling; n.r.; 4 °C | |||||||||
| (Zhao et al., 2020); Wuhan, China | Municipal and hospital wastewater and sludge; n.r.; n.r.; n.r | Centrifugation | PEG/NaCl precipitation | n.r. | Direct-zol RNA Kit (Zymoresearch) | RT-qPCR | Municipal wastewater, influent: 7.40 103 | Municipal wastewater, secondary treatment effluent: 5.30 103 to 1.00 104 | a |
| Hospital wastewater, influent: 3.80 103–9.30 103 | |||||||||
| Hospital wastewater, sludge: 1.40 104 | |||||||||
| (Hong et al., 2020); Jeddah, Saudi Arabia | Hospital wastewater; | – | Adsorption-elution with Millipore HA membrane | QIAmp Viral RNA kit (ThermoFisher Scientific) | RT-qPCR | Underground Septic Tank (untreated wastewater): 1.74 102–1.33 103 | Biologically Activated Tank(partially treated wastewater): 8.11 101 to 1.12 103 | a | |
| Grab sample; 1 L; 4 °C | |||||||||
| (Chavarria-Miró et al., 2020); Barcelona, Spain | Municipal wastewater; | – | PEG 6000 precipitation | n.r. | NucliSENS miniMAG extraction system (bioMérieux) | One-step RT-qPCR | Below limit of detection to less than 105 | a | a |
| Composite sampling (24 h); 800 mL; n.r. | |||||||||
| (Crits-Christoph et al., 2020); San Francisco, California | Municipal wastewater; | Pasteurization at 60 °C for 90 min; | Ultrafiltration with Amicon | n.r. | Qiagen AllPrep DNA/RNA Mini Kit; | RT-qPCR; | 1.07 103–1.02 106 | a | a |
| Genome Sequencing | |||||||||
| Ultra-15 100 kDa CentrifugalFilter | |||||||||
| Composite sampling (24 h); 1 L; n,r, | Direct RNA extraction with silica columns | ||||||||
| Filtration through 0.22 μm filters | |||||||||
| (Hata et al., 2020); Ishikawa and Toyama, Japan | Municipal wastewater; | Centrifugation | PEG 8000/NaCl precipitation | n.r. | QIAamp viral RNA mini kit (Qiagen) | RT-qPCR; RT-nested PCR | 1.20 104–4.40 104 | a | a |
| Grab sampling; 100 mL; n.r. | |||||||||
| (Neault et al., 2020); Ontario and Quebec, Canada | Municipal wastewater and | Influent: Settled for 1 h to separate influent filtrate and influent solids | PEG precipitation | n.r. | SARS-CoV-2 Protein Extraction using radioimmunoprecipitation assay (RIPA), urea and triton X-100 lysis buffers | Multiplex Paired-antibody Amplified Detection (MPAD) | Proteins in place of viral RNA are measured; | a | a |
| Wastewater Primary Sludge; n.r.; n.r.; Primary sludge at −80 °C | |||||||||
| Cycle threshold numbers are reported | |||||||||
| (D'Aoust et al., 2020); Ontario and Quebec, Canada | Municipal wastewater post-grit chamber solids (PGS) | PGS samples: settled for 1 h at 4 °C | PGS supernatant: Filtration with mixed cellulose ester filter | n.r. | RNeasy PowerMicrobiome Kit Qiagen) | RT-qPCR; | PGS (RT-qPCR): | a | |
| RT-ddPCR | 1.42 106– | ||||||||
| 1.93 106 | |||||||||
| PGS solids: PEG precipitation and centrifugation | PGS (RT-ddPCR): | ||||||||
| 1.24 106– | |||||||||
| 1.42 106 | |||||||||
| PGS: Composite sampling (24 h); 250 mL; 4 °C | PCS: PEG and centrifugation | PCS (RT-qPCR): | |||||||
| 1.10 106– | |||||||||
| 1.51 106 | |||||||||
| PCS (RT-ddPCR): | |||||||||
| 2.74 105– | |||||||||
| 3.93 105 | |||||||||
| Primary clarifier sludge (PCS); | |||||||||
| Grab and composite sampling (24 h); 250 mL; 4 °C | |||||||||
| (Alpaslan Kocamemi et al., 2020); Turkey | Municipal wastewater sludge; | Centrifugation; filtration (0.45 and 0.2 μm nominal pore size); pH Adjustment at 7.0–7.2 | PEG 8000/centrifugation | n.r. | Roche MagNA pure LC total nucleic acid isolation kit using Roche MagNA pure LC system (Penzberg, Germany) | RT-qPCR | Primary Sludge: | a | |
| 1.25 104 | |||||||||
| Waste Activated Sludge: | |||||||||
| Grab sampling; n.r.; n.r. | 1.17 104– | ||||||||
| 4.02 104 | |||||||||
| (Peccia et al., 2020b); Connecticut, USA | Municipal wastewater sludge; | Not carried out | Not carried out (Direct addition of sludge to RNA extraction kit) | n.r. | RNeasey PowerSoil Total RNA kit, Qiagen | RT-qPCR | Primary Sludge: | a | |
| 1.7 106– | |||||||||
| 4.6 × 108 | |||||||||
| Grab sampling; 2.5 mL; −80 °C | |||||||||
| (Balboa et al., 2020); Ourense, Spain | Municipal wastewater and sludge; | – | Wastewater samples: Ultrafiltration | n.r. | STARMag 96 × 4 Universal Cartridge Kit (Seegene) | One-step multiplex RT-qPCR | Wastewater Influent: Below 7.5 x 103 to less than 4.0 × 104 | ||
| Sludge samples: PEG 8000/NaCl precipitation | |||||||||
| Wastewater Effluent: Not detected to less than 1.0 × 104 | |||||||||
| Wastewater; Composite sampling (24 h); 250 mL; 4 °C | |||||||||
| Primary Sludge: Not detected to less than 4.0 × 104 | |||||||||
| Activated Sludge: Not detected to less than 1.0 × 104 | |||||||||
| Thickened Sludge: Not detected to less than 2.0 × 104 | |||||||||
| Digested Sludge: Not detected | |||||||||
| (Ge et al., 2020); China | Hospital Sewage from preprocessing disinfection equipment and final disinfection pool | Centrifugation (12,000 × g for 5 min at 4 °C) | – | n.r. | MagNA Pure LC 2.0 (Roche, Basel, Switzerland) | RT-qPCR | – | Virus culture in Vero E6 cell line inlet (0/14) outlet (0/14) | |
Note: n.r. Not reported.
No available quantitative or viable virions data on the virus in wastewater or sludge.
Conversely, data on the occurrence of SARS-CoV-2 traces in wastewater can be used by epidemiologists and government authorities for evaluation of the extent of circulation of the virus in the population associate to a water sanitation network. At the moment of writing of these manuscript, several countries, such as the Netherlands, Australia, Finland, France, Italy, Portugal and Spain have activated national wastewater surveillance programs. Governmental, academic, health and research organizations are combining their efforts in Italy, a country strongly affected by the epidemics of SARS-CoV-2, for the monitoring of the presence of the virus in wastewater (project SARI, Sorveglianza Ambientale Reflue in Italia). An ambitious program is taking place in The Netherlands by the National Institute of Public Health and the Environment (Rijksinstituut voor Volksgezondheid en Milieu, RIVM), involving the daily monitoring of over 300 existing plants in the country (RIVM, 2020).
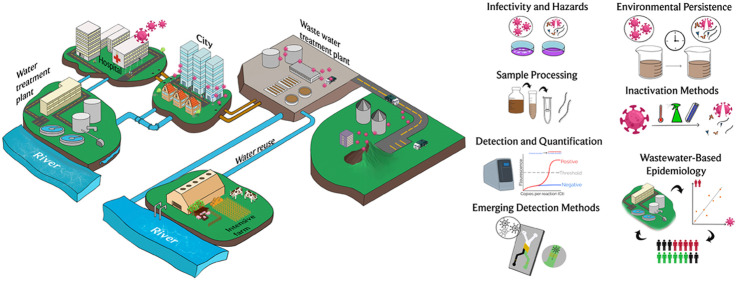
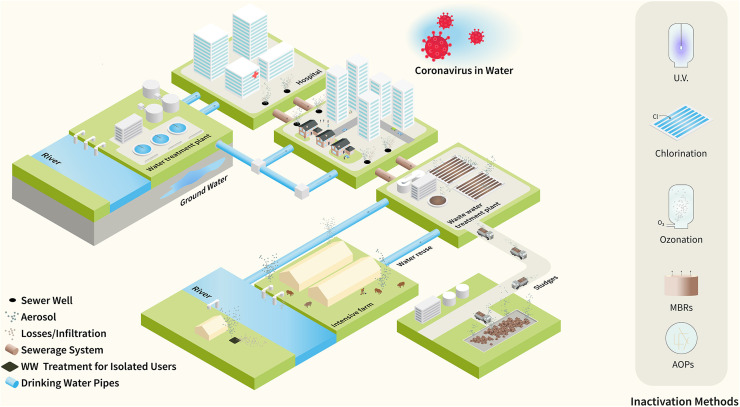
The presence of the virus in wastewater constitutes a potential hazard, due to its possible propagation through this medium, and its detection provides a powerful tool in the hands of the scientific, health and government communities, as an epidemiological indicator of the spread of the virus, including the number of asymptomatic infections. In this context, it is important the knowledge of the state-of-the-art on the methods for detection, quantification and determination of infectivity of the virus in aqueous matrices. The detection of the virus in wastewater is not directly correlated to the infectivity. Therefore, particular attention has been dedicated in the manuscript to the method for determination of the viable SARS-CoV-2 virions and the estimation of the potential risks associated. The current consolidated methods for molecular detection of the virus in wastewater, based on the amplification of viral genome, suffer in the case of the assay of the SARS-CoV-2 of some limitations, which were identified and discussed in this manuscript. The necessity to concentrate the SARS-CoV-2 traces present in wastewater, the absence of a robust and validated protocol for sample processing and the open challenges in this field are also critically analyzed and discussed. In addition, new methodologies and technologies, based on the detection of the SARS-CoV-2 RNA as well as of the proteins and other viral vestiges in wastewater are emerging and here examined. The development of these technologies could allow a more rapid and effective implementation of wastewater-based epidemiology. Fundamental is the exact knowledge on behavior and persistence of the SARS-CoV-2 in all aqueous media. This information combined with the knowledge on the efficacy of the methods for treatment of the water media for the disinfection allows establishing real risks and the correct strategy for control the spread of virus through the water environment (see Fig. 1).
Fig. 1.
Synoptic view of the topics discussed in the review.
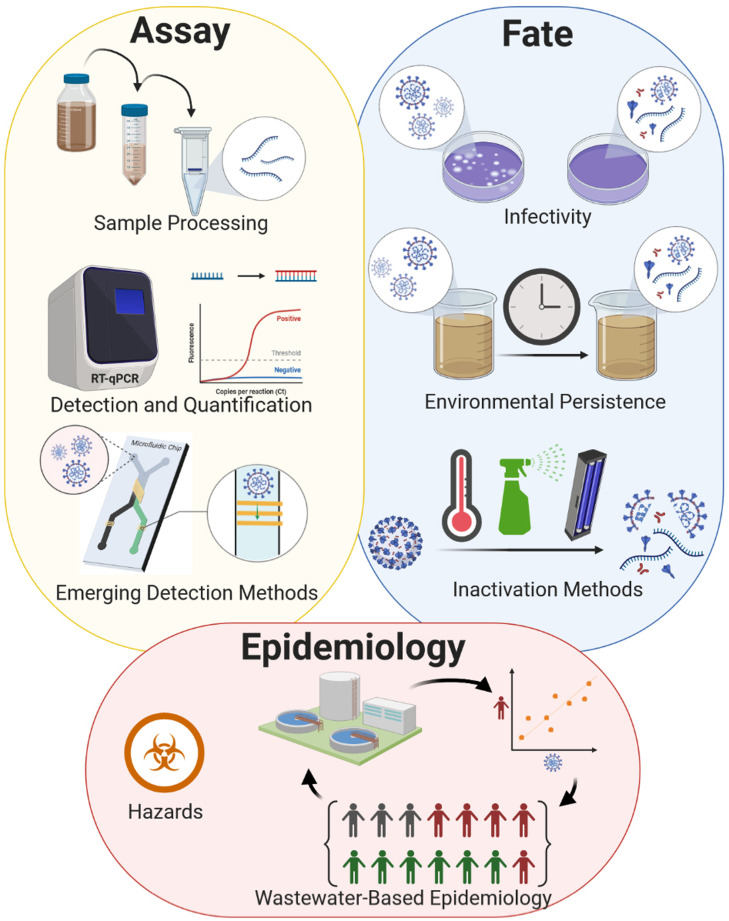
2. Assay of viable virions and quantification of coronaviruses in water media
The detection of pathogens, particularly viruses, in water media and wastewater plays a pivotal role for developing mitigation measures and health and safety plans (Xagoraraki and O’Brien, 2020; Naddeo, 2020) Viruses can be considered as supramolecular assemblies of biological small- and macro-molecules. They cannot reproduce by themselves and require host living cells for the replication. In fact, the debate of the living or non-living nature of viruses is old and still open (Gortner, 1938). Therefore, the determination of viral infectivity and concentration in a contaminated sample must be done using the culture in a suitable host organism and molecular techniques detecting the genetic material (DNA or RNA) or proteins of the virus.
The test for determination of the number of viable virus particles provides information on its state of integrity or inactivation by determining its ability to replicate when cultured in a suitable host species. The determination of the period during which the virus results viable, and thus capable of replication in determined conditions, provides information on its stability and persistence. However, detection of viable viruses in a sample does not necessarily implies infectivity: the route of transmission of the virus through that medium can be ineffective for the initiation of the actual infection in humans or other species targeted by the virus.
Assay of bacteria, that are aquatic host microorganisms for viruses, have been proposed as a simple and rapid indicator for the determination of the fate of viruses in water. In fact, the assay of bacteria in water media does not requires the “complex” sample pre-treatments and the molecular methods necessary for virus quantification (vide infra). For example, coliphages, a class of the bacteriophage viruses infecting the bacteria Escherichia coli, have been proposed as a viral indicator for enteric viruses in wastewater. Although the fate of coliphages had a good correlation with that of enteric viruses in wastewater, estimations of Escherichia coli as a bacterial indicator associable to a viral indicator have yielded inadequate results (Worley-Morse et al., 2019). Thus, results of the estimation of virus occurrence and the assessment of the efficacy of inactivation procedures for viruses based on a bacterial indicator, in place of dedicated viral indicators, require a comparison of data related to fate ad behavior between viruses and bacteria. Considered that CoVs do not infect bacteria or other aquatic microorganisms, it is impossible to quantify their presence using other indicators. In addition, the stability of these viruses in water media significantly differs from that of other viruses, typically infecting these environments, invalidating also the association of other viral indicators to CoV persistence. However, the fate of CoVs in water and wastewater and the efficacy of their inactivation methods can be estimated by analysing the fate of other enveloped viruses with higher resistance in wastewater.
2.1. Determination of viable coronavirus virions in water media
Detection of CoV RNA molecules in a sample does not necessarily implies infectivity of that specimen. RNA molecules or their fragments can be detected from damaged, and thus inactive, viral particles. Unaltered virus particles, thus potentially infective, can be quantified with molecular techniques (e.g. using quantitative polymerase chain reaction (qPCR; see Section 2.2.) targeting specific gene sequences or other molecular sections of the virus susceptible to fragmentation, or by enumeration of enlarged gene sequences (Ho et al., 2016, Polston et al., 2014, Wu et al., 2019). Capsid-integrity PCR is a recent technique that quantifies intact virions by excluding the genomic material coming from damaged viruses from the molecular analysis. RNA or DNA molecules coming from fragmented viruses are covalently functionalized with specific molecular markers before the molecular analysis (i.e. PCR) that, in this way, quantifies only the genomic material coming from intact virions (Leifels et al., 2021, Leifels et al., 2020).
The infectivity of a virus can be determined by plaque assay, a quantitative method of measuring infectious viruses by quantifying the plaques formed in cell cultures upon infection with serial dilutions of the virus specimen (Baer and Kehn-Hall, 2014) or by tissue culture infectious dose-50 (TCID50), which detects the presence or absence of cytopathic effects (CPEs) in cells infected with serial dilutions of a virus specimen.
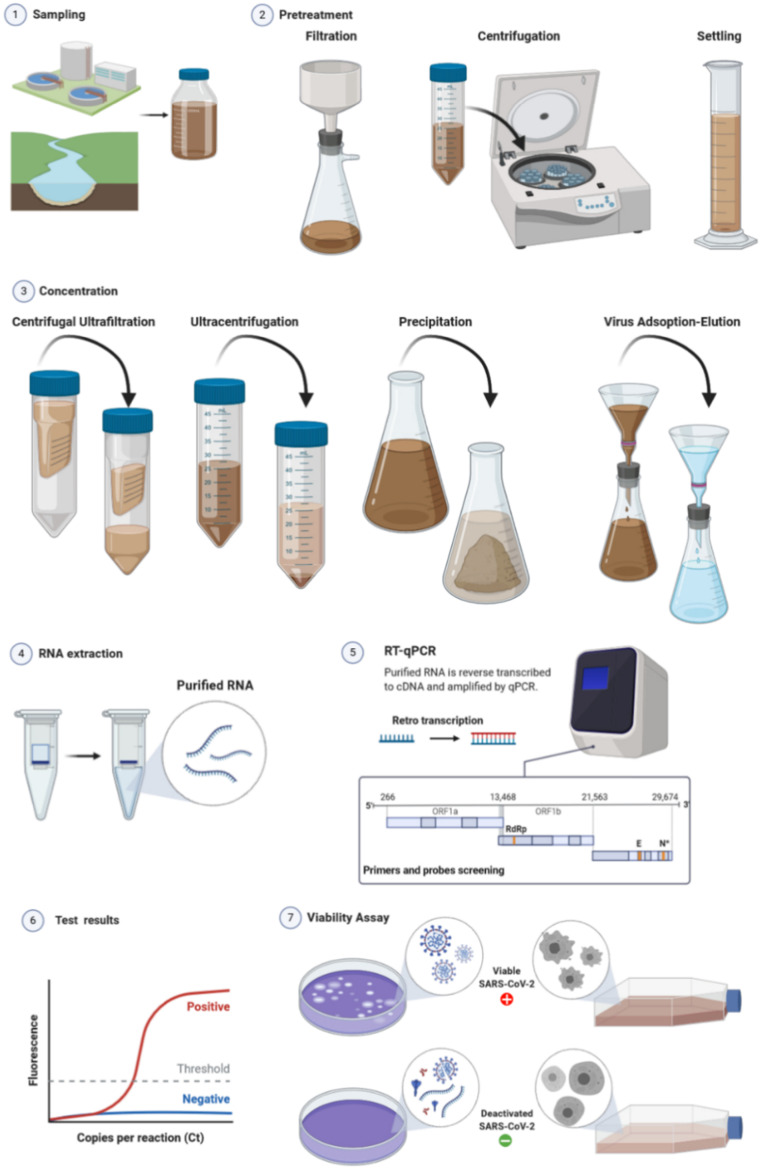
The host cells typically used for in vitro culture of coronaviruses, in particular for SARS-CoV-2, include Vero E6, Vero CCL-81, HUH 7.0, 293T, A549, EFKB3 and Caco-2 cell lines (Harcourt et al., 2020, Hoehl et al., 2020). The host cell line is cultured in Dulbecco minimal essential medium (DMEM) supplemented with heat-inactivated fetal bovine serum (FBS), antibiotics and antimycotics (Harcourt et al., 2020). After incubation with the potentially infected specimen, viable viruses induce CPEs on the host cells, determining the formation of visually discernible plaques or morphological modification of the cells (see Fig. 2). The standard plaque assay for SARS-CoV-2 (Harcourt et al., 2020), was based on previous protocols established for SARS-CoV-1 and MERS-CoV (Josset et al., 2013, Sims et al., 2013). Considering that other pathogens, particularly those present in wastewater, can induce CPE on CoV host cells, confirmatory tests via nucleic acid extraction, amplification and sequencing (vide infra) are necessary in case of positive results. Laboratories with qualified Biosafety Level 3 (BSL-3) are required for the handling of those viruses.
Fig. 2.
General scheme of wastewater analysis to detect occurrence and viable viruses.
The number of infectious virus particles can be calculated by tissue culture infectious dose (TCID50) assay. The virus sample is serially diluted and added to cells, selected to show cytopathic effect, placed in a multiwell plate. The infection with the virus results in cell morphological changes or mortality, and each well is classified as infected or not infected. Automated readings, colorimetric or fluorometric, are also possible. The calculation of the TCID50 is thus done identifying the dilution at which 50% of the wells show a CPE. The Spearman–Karber or the Reed–Muench methods are typically applied, and the viral loading is expressed as TCID50 per volume of sample (Ramakrishnan, 2016, Reed and Muench, 1938, Spearman, 1908).
Although the SARS-CoV-2 RNA has been detected worldwide in wastewater (see Table 1), results of viral culture experiments, used for the determination of the viable virions, have up to now been negative (Ge et al., 2020, Rimoldi et al., 2020a, Rimoldi et al., 2020b, Wang et al., 2020c, Westhaus et al., 2021). Wang et al. (2020c) analysed, without finding evidence of viable SARS-CoV-2, both the inlets and outlet sewages of pre-processing disinfection pool, as well as the final outlet of the sewage disinfection pool, of an hospital in China hosting COVID-19 cases. Similar negative results were found by Ge et al. analysing sewage samples from insolation wards during hospitalization of COVID-19 patients in the Hospital of Zhejiang University, China. The samples which were collected from the inlets and outlets of pre-processing disinfection equipment, were found positive for SARS-CoV-2 RNA detection and negative to the assay of viable virus particles carried out in Vero E6 cell line (Ge et al., 2020). Rimoldi et al., 2020a, Rimoldi et al., 2020b analysed influents and effluents of municipal WWTPs and water of river used for the discharge of the final effluents, during the emergence of COVID-19 epidemics in March 2020 in Milan, Italy. The infectivity of wastewater, both untreated and treated in WWTP, was investigated by Westhaus et al. (2021) culturing the specimen in Caco-2 cells in Germany. CPEs for this cell line can be observed by optical microscopy as morphological modifications of the Caco-2 cells (Hoehl et al., 2020). CPEs were not observed in any wastewater sample analyzed in this study (Westhaus et al., 2021). These findings provide important information on the real risk associable with the presence of SARS-CoV-2 in wastewater, indicating a very limited stability of this virus in this kind of aquatic medium. However, the possibility of false-negative results in the test of viable CoVs cannot be ruled out. As matter of facts, for SARS-CoV-1, which is the most proximal virus to SARS-CoV-2 (Andersen et al., 2020), 5-log of virus titer reduction was determined to occurs within 2 and 14 days in wastewater at 20 and 4 °C, respectively (see Table 3) (Wang et al., 2005b, Wang et al., 2005c). A similar persistence was determined for SARS-CoV-2, 1- and 2-log of virus titer reduction were observed respectively within the ranges of 1.4–3.3 and 2.9–6.5 days in frozen and thawed wastewater at room temperature (see Table 3) (Bivins et al., 2020).
Table 3.
Environmental persistence of SARS-CoV-2 and other representative CoVs at room temperature.
| CoV | Environment | Log10reduction | Time | Reference |
|---|---|---|---|---|
| E229 | Dechlorinated and filtered tap water | 3-log | 10 d at 23 °C (> 130 d at 4 °C) | (Gundy et al., 2008) |
| Primary wastewater | > 2.0-log | 2–4 d | ||
| Secondary wastewater | > 2.9-log | 2–4 d | ||
| PBS | n.r. | ≥ 6 d | (Sizun et al., 2000) | |
| MEM | 4-log | 9 d | (Rabenau et al., 2005) | |
| MEM + FBS (10%) | 4-log | 9 d | ||
| Plastics (PS) | 4-log | 72 h | ||
| FIPV | Dechlorinated and filtered tap water | 3-log | 10 d at 23 °C (> 130 d at 4 °C) | (Gundy et al., 2008) |
| Primary wastewater | > 3.1-log | 2–4 d | ||
| Secondary wastewater | > 3.7-log | 2–4 d | ||
| TGEV | Reagent grade water | 2-log | 22 d at 25 °C (> 49 d at 4 °C) | (Casanova et al., 2009) |
| Lake water | 2-log | 13 d at 25 °C | ||
| Pasteurized settled sewage | 2-log | 9 d at 25 °C | ||
| MHV | Reagent grade water | 2-log | 17 d at 25 °C (> 49 d at 4 °C) | (Casanova et al., 2009) |
| Lake water | 2-log | 10 d at 25 °C | ||
| Pasteurized settled sewage | 2-log | 7 d at 25 °C | ||
| Wastewater | 1-log | 13 h at 25 °C; 36 h at 10 °C | (Ye et al., 2016) | |
| Pasteurized wastewater | 1-log | 18 h at 25 °C; 149 h at 10 °C | ||
| OC43 | PBS | n.r. | ≥ 6 d | (Sizun et al., 2000) |
| MERS-CoV | Plastic | 6-log | 72 h (20–40% RH) | (van Doremalen et al., 2013) |
| Steel | 6-log | 72 h (20–40% RH) | ||
| SARS-CoV-1 | Cell culture media | 5-log | 60 h | (Duan et al., 2003) |
| Autoclaved water | 5-log | 72 h | ||
| Serum | 5-log | 72 h | ||
| Sputum | 5-log | 96 h | ||
| Feces | 5-log | 96 h | ||
| Urine | 5-log | 72 h | ||
| Glass | 5-log | 60 h | ||
| Mosaic | 5-log | 60 h | ||
| Metal | 5-log | 72 h | ||
| Plastics | 5-log | 60 h | ||
| Cloth | 5-log | 72 h | ||
| Filter paper | 5-log | 72 h | ||
| Autoclaved soil | 5-log | < 6 h | ||
| Dechlorinated tap water | 5-log | 2 d (≥ 14 d at 4 °C) | (Wang et al., 2005b) | |
| Domestic sewage (centrifuged) | 5-log | 2 d (≥ 14 d at 4 °C) | ||
| Hospital sewage (centrifuged and treated with sodium thiosulfate) | 5-log | 2 d (≥ 14 d at 4 °C) | ||
| PBS | 5-log | ≥ 14 d | ||
| Stool | 5-log | 3 d (≥ 17 d at 4 °C) | ||
| Urine | 5-log | 17 d | ||
| MEM | ~ 1-log | > 9 d | (Rabenau et al., 2005) | |
| MEM + FBS(10%) | ~ 1-log | > 9 d | ||
| Plastics (PS) | > 4-log | 9 d | ||
| Plastics | 2.7-log | 72 h | (van Doremalen et al., 2020) | |
| Stainless steel | 3.0-log | 72 h | ||
| Copper | ~ 3.0-log | 4 h | ||
| Cardboard | ~ 3.0-log | 24 h | ||
| SARS-CoV-2 | Plastics | 3.1-log | 72 h | (van Doremalen et al., 2020) |
| Stainless steel | 3.1-log | 72 h | ||
| Copper | ~ 3.0-log | 4 h | ||
| Cardboard | ~ 3.0-log | 24 h | ||
| Virus transport medium | 5.34-log | 5 min (70 °C) | (Chin et al., 2020) | |
| 6.65-log | 30 min (56 °C) | |||
| 6.57-log | 2 days (37 °C) | |||
| 6.51-log | 14 days (22 °C) | |||
| 0.47-log | 14 d (4 °C) | |||
| Paper | 4.76-log | 3 h | ||
| Banknote | 6.05-log | 4 d | ||
| Wood | 5.66-log | 2 d | ||
| Cloth | 4.84-log | 2 d | ||
| Respiratory mask | 2.99-log | 7 d | ||
| Glass | 5.83-log | 4 d | ||
| Stainless steel | 5.80-log | 7 d | ||
| Plastics | 5.81-log | 7 d | ||
| Tap water | 1-log | 1.8–2.2 d | (Bivins et al., 2020) | |
| 2-log | 3.6–4.4 d | |||
| Municipal wastewater | – | – | (Rimoldi et al., 2020a, Rimoldi et al., 2020b; Westhaus et al., 2021) | |
| WWTP effluent | – | – | ||
| River (contaminated with wastewater) | – | – | (Rimoldi et al., 2020a, Rimoldi et al., 2020b) | |
| Hospital wastewater | – | – | (Ge et al., 2020; Wang et al., 2020c) | |
| Wastewater (frozen and thawed) | 1-log | 1.4–3.3 d | (Bivins et al., 2020) | |
| 2-log | 2.9–6.5 d |
2.2. Concentration and detection methods of coronaviruses in water media
During an epidemic event, the loading of a virus in wastewater can be correlated with the contents in human stool or urine samples (Ye et al., 2016). Molecular methods based on polymerase chain reaction (PCR) are typically used for the quantitation of viruses in human samples and water media. A small sample of DNA can be rapidly reproduced to billions of copies by PCR: a large enough amount to be studied in detail by scientists for qualitative and quantitative analyses. CoVs present their genetic material in the form of RNA, therefore the retro-transcription (RT) in the form DNA is mandatory prior to PCR. At present, the analytical methods for CoVs in samples of human origin include the following molecular techniques for amplifying nucleic acids: i) reverse transcription polymerase chain reaction (RT-PCR), ii) real-time RT-PCR (rRT-PCR), iii) reverse transcription loop mediated isothermal amplification (RT-LAMP), and iv) real-time RT-LAMP (rRT-LAMP) (Bofill-Mas and Rusiñol, 2020, Corpuz et al., 2020). Analytical kits have been developed for the rapid and reliable assay of CoVs from biological human samples. Currently, detection kits based on RT-PCR are commercially available for SARS-CoV-1, MERS-CoV (Zhang et al., 2020c), and the novel SARS-CoV-2 virus (WHO, 2020a). Some of those analytical methods were applied also for detection of CoVs in water and wastewater samples (see Tables 1 and 2).
Table 2.
Different assays used for detection of SARS-CoV-2 RNA in wastewater samples.
| Method of detection | Primer | Target gene | Limit of detection | Reference |
|---|---|---|---|---|
| RT-qPCR | N_Sarbeco | N-gene | 8.3 copies/reaction | (Corman et al., 2020) |
| 3 copies/µL RNA template | (Ahmed et al., 2020b) | |||
| RT-qPCR | CDC_N1 | N-gene | 5 copies/reaction | (Lu et al., 2020b) |
| 1 copy/µL RNA template | (Ahmed et al., 2020b) | |||
| RT-ddPCR | 14.6 copies/reaction | (Gonzalez et al., 2020) | ||
| RT-qPCR | CDC_N2 | N-gene | 5 copies/reaction | (Lu et al., 2020b) |
| 2 copies/µL RNA template | (Ahmed et al., 2020b) | |||
| RT-ddPCR | 2 copies/reaction | (Gonzalez et al., 2020) | ||
| RT-qPCR | CDC_N3 | N-gene | 5 copies/reaction | (Lu et al., 2020b) |
| RT-ddPCR | 2.18 copies/reaction | (Gonzalez et al., 2020) | ||
| RT-qPCR | NIID_2019-nCOV_N | N-gene | 4 copies/ µL RNA template | (Ahmed et al., 2020b) |
| RT-qPCR | E_Sarbeco | E-gene | 3.9 copies/reaction | (Corman et al., 2020) |
| 5 copies/µL RNA template | (Ahmed et al., 2020b) | |||
| RT-qPCR | RdRP_SARSr | RdRp-gene | 3.6 copies/reaction | (Corman et al., 2020) |
| 316 gene equivalents/reaction | (Nalla et al., 2020) | |||
| > 500 copies/reaction | (Vogels et al., 2020) | |||
| 200 copies/reaction | (Westhaus et al., 2021) | |||
| First PCR; | Name | ORF1ab | 0.41 copies/µL RNA (LOD50 in pure RNA samples) | (La Rosa et al., 2020b, La Rosa et al., 2021) |
| Nested PCR | NIID_WH-1; | |||
| 2274 – CO | 1.46 copies/µL RNA (LOD50 in sewage samples) | |||
| 2275 – CO | ||||
| 2276 – CO | ||||
| 2277 – CO | ||||
| RT-qPCR | M-gene | 200 copies/reaction | (Westhaus et al., 2021) |
Typically, the abovementioned molecular techniques employed to ascertain the occurrence of viruses in unconcentrated wastewater and surface water (Corpuz et al., 2020) yield results below the detection limit of the methods. Consequently, it is mandatory to concentrate a large volume of water sample before the analysis (Ahmed et al., 2020d, Bofill-Mas and Rusiñol, 2020, Corpuz et al., 2020, Rusiñol et al., 2020, Ye et al., 2016). Concentration methods of CoVs in water media have been addressed in ongoing research (Ahmed et al., 2020d). These studies were recently reviewed by La Rosa et al. (2020a) and Corpuz et al. (2020).
Water samples can be pre-treated and concentrated using various processes, such as centrifugation/ultracentrifugation, virus adsorption-elution (VIRADEL), membrane (electropositive or electronegative) filtration, centrifugal ultrafiltration or precipitation with suitable coagulating agents (see Table 1 and Fig. 2) (Ahmed et al., 2020d, Bofill-Mas and Rusiñol, 2020, Corpuz et al., 2020, La Rosa et al., 2020a). Despite the extensive efforts dedicated to the development of methodologies for virus concentration, currently there is no consensus on the reliable application of these enrichment techniques for water samples. In fact, when these pre-treatments are applied a variable number of viral particles can be lost, depending on the nature of the specimen, the targeted virus and the method of concentration adopted (Ahmed et al., 2020d, Bofill-Mas and Rusiñol, 2020, Corpuz et al., 2020).
Most of the available methods for quantifying viruses in water samples are optimized for non-enveloped viruses, in particular for enteric viruses (e.g., adenoviruses, polioviruses, enteroviruses, noroviruses, and rotaviruses) (Corpuz et al., 2020). Compared to “simple” non-enveloped viruses, enveloped ones such as CoVs are more sensitive to sample pretreatment and are less stable in wastewater (see Table 3). In fact, SARS-CoV-1 has been shown to persist (5-log of virus titer reduction) only up to 2 days at room temperature, both in domestic and hospital sewages and for more than 14 days at 4 °C (see Table 3) (Wang et al., 2005b, Wang et al., 2005c) However, it should be noted that those tests of virus persistence were carried out in wastewater centrifuged at 6000 rpm for 30 min for the removal of suspended particles and bacteria, whereas the hospital wastewater was treated with sodium thiosulfate for the inactivation of disinfectants prior to seeding of the virus (Wang et al., 2005b). Therefore, the currently observed (scarce) resistance of SARS-CoV-1 in wastewater could be even overestimated.
CoVs tend to be adsorbed onto particles and debrides present in surface water and wastewater (Chaudhry et al., 2015, Gundy et al., 2008). Therefore, dedicated procedures for virus desorption from suspended solids in water should be implemented for quantitative measurements (Alpaslan Kocamemi et al., 2020, Balboa et al., 2020, Peccia et al., 2020b, Westhaus et al., 2021).
CoVs, like the other enveloped viruses, are sensitive to pH variations, with an optimal stability at slightly acidic pH range of 6.0–6.5 c.a. (Sattar et al., 2009). Nevertheless, SARS-CoV-2 have been demonstrated to exhibit sufficiently high stability in a wide range of pH values (Chin et al., 2020). This information should be taken into account in the case of application of VIRADEL techniques for the recovery of CoVs from wastewater, where the virus is captured by filtration on glass wool or with electropositive or electronegative membranes at strongly alkaline or acidic pHs, respectively, and then eluted from the filter. The acidification of the retained viruses with acidic solutions leads to the protonation of the capsid or of the envelope, depending on the virus typology, which assumes a positive charge and can be absorbed onto electronegative membranes. On the contrary, treatment with alkaline solutions causes the viruses to become negatively charged, therefore electropositive filters should be used for the recovery of the virus. After absorption onto filter the virus can be recovered by elution with buffered solutions eventually containing suitable agents for the desorption of the virus.
Few studies have examined the effect of the concentration method of wastewater on the efficiency of recovery of CoVs. To the best of our knowledge, only one study reported the recovery efficiency of SARS-CoV-1, the most proximal virus to SARS-CoV-2. Wang et al. (2005b) used a wastewater sample seeded with the virus and obtained ca. 1% of virus recovery by applying electropositive filtration for viral particle concentration in the sample. The recovery of bovine CoV (Abd-Elmaksoud et al., 2014, Collomb et al., 1986), transmissible gastroenteritis virus (TGEV) (Blanco et al., 2019) and murine hepatitis virus (MHV) (Ahmed et al., 2020d, Ye et al., 2016) have also been investigated. Virus adsorption on glass (Abd-Elmaksoud et al., 2014, Blanco et al., 2019, Collomb et al., 1986) or on silica gel compounded with aluminum hydroxide (Wang et al., 2005a), followed by elution with neutral (Wang et al., 2005a) or alkaline (Abd-Elmaksoud et al., 2014, Blanco et al., 2019, Collomb et al., 1986) buffers were adopted for concentration of the abovementioned CoVs. Poly(ethylene glycol) (PEG) precipitation, ultracentrifugation and ultrafiltration were compared by Ye et al. (2016) in the context of the recovery efficiency of MHV. Ultrafiltration was demonstrated the most efficient method with 25.1% of recovery of MHV. Blanco et al. (2019) applied virus adsorption onto glass wool with subsequent elution with alkaline buffer and PEG precipitation and explored on the recovery of TGEV, effects of pH, contact time and composition of eluent. 42.7% of adsorption degree and the complete removal of the virus from the glass wool adsorbent was obtained by overnight elution with glycine/beef extract buffer at pH of 11.0 in presence of TWEEN® 80 (0.3%). Ahmed et al. (2020a) adopted both a direct RNA extraction from electronegative membranes and ultrafiltration for the concentration of the SARS-CoV-2 virus in wastewater samples. However, the latter study did not evaluated the performance of the concentration methods using seeded samples. In another study, Ahmed et al. (2020d) compared percentage recoveries of MHV from raw wastewater using different concentration methods. MHV is a Betacoronavirus with envelope that well represents the behavior of SARS-CoV-2 in wastewater. MHV was used as a surrogate virus for SARS-CoV-2 due to the biosafety risks associated with handling the latter in the laboratory. The concentration methods that were examined in that study involved adsorption to electronegative membrane, ultrafiltration, PEG precipitation, and centrifugation (Fig. 1). Results of the study revealed that the more efficient methods were those involving the adsorption of the virus onto an electronegative membrane (without pre-acidification step) and subsequent direct RNA extraction from the filter. The relatively higher MHV recovery efficiencies were: adsorption to the electronegative membrane without pre-treatment: 60.5 ± 22.2%; adsorption to the electronegative membrane with the addition of MgCl2: 65.7 ± 23.8%. The high recovery efficiencies were mainly attributed to the extraction of viruses from both the liquid and solid fractions of the wastewater samples. The investigation reported by Ahmed et al. (2020d) also revealed that a pre-acidification step (pH 4.0) significantly reduced the recovery of MHV from the samples. A previous study also showed that enveloped viruses, such as MHV, tend to be adsorbed to the solid fraction of the wastewater (Ye et al., 2016). In the latter study, it was shown that a higher percentage of enveloped viruses (MHV: 26%, ϕ6: 22%), compared to non-enveloped viruses (MS2: 6%, T3: < 5%), was adsorbed to wastewater solids at equilibrium.
Those considerations should be taken into account during the sampling and storage of wastewater samples, as well as during the treatment for the detection, quantization and particularly in the determination of the viable virus particles. The development of a standard method for concentration of wastewater contaminated with SARS-CoV-2 should be based on this information. Currently, several procedures have been applied with success for the detection of the SARS-CoV-2 in wastewater, although scarce information is available on the actual efficacy of the recovery of this virus (Table 1).
Sample pre-treatments include conventional filtration, centrifugation or settling for removal of raw suspended solids and debrides. As discussed above, these treatments could affect the quantification of the virus by excluding from the molecular detection viral particles adsorbed onto particulate. Pasteurization at 56–60 °C for 60–90 min has been also applied for the inactivation of viruses and other pathogens for safe handling of the sample. Centrifugal ultrafiltration has been extensively applied for the concentration of the wastewater by using centrifugal filters with different molecular weight cut-offs (MWCO) (Ahmed et al., 2020a, Ahmed et al., 2020b, Balboa et al., 2020, Bar Or et al., 2020, Crits-Christoph et al., 2020, Manupati et al., 2020, Medema et al., 2020a, Medema et al., 2020b, Nemudryi et al., 2020a, Nemudryi et al., 2020b, Sherchan et al., 2020, Trottier et al., 2020, Westhaus et al., 2021, Wu et al., 2020b). Ultracentrifugation is also a convenient method for the rapid concentration of wastewater samples (Ampuero et al., 2020, Green et al., 2020, Prado et al., 2020, Wurtzer et al., 2020, Yaqub et al., 2020). The direct flocculation (Mlejnkova et al., 2020) or the treatment with PEG (Alpaslan Kocamemi et al., 2020, Bar Or et al., 2020, Chavarria-Miró et al., 2020, D'Aoust et al., 2020, Neault et al., 2020) in combination with sodium chloride (Arora et al., 2020, Balboa et al., 2020, Hata et al., 2020, Kumar et al., 2020, Wu et al., 2020b, Zhang et al., 2020a, Zhao et al., 2020) or dextran (La Rosa et al., 2020b, La Rosa et al., 2020c, La Rosa et al., 2021, Sharif et al., 2020) have been also adopted for the precipitation of the virus. These latter techniques were optimized for the precipitation of proteins and particularly utilized for the recovery of non-enveloped viruses with protein capsid exposed, such as enteric viruses (Corpuz et al., 2020). However, CoVs presents spike proteins on the viral envelope therefore this method of wastewater concentration works well also for this kind of viruses. VIRADEL methods were also largely employed for concentration of SARS-CoV-2 in wastewater, principally by using electronegative charged membranes (Ahmed et al., 2020a, Ahmed et al., 2020b, Curtis et al., 2020, Haramoto et al., 2020, Hong et al., 2020, Miyani et al., 2020, Sherchan et al., 2020). In absence of treatment for the concentration of the samples cannot be obtained reliable quantitative information on the virus concentration in wastewater (Table 1).
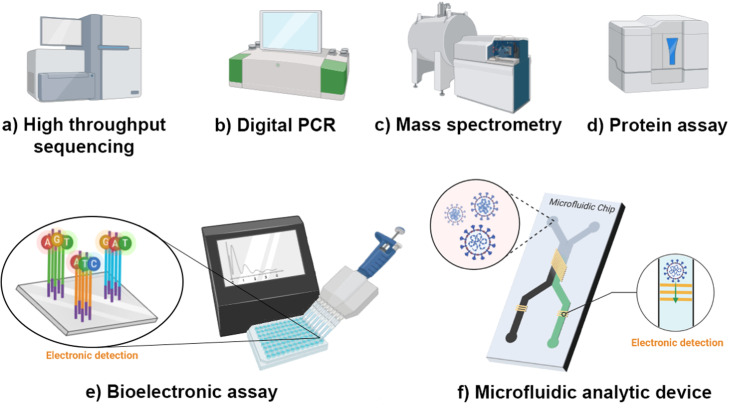
The application of the recent high-throughput sequencing methods will allow fast and reliable determination of viral parameters in water matrices (Bofill-Mas and Rusiñol, 2020). The genomic material of the viruses is often overshadowed by the host and bacterial genomes (Nieuwenhuijse and Koopmans, 2017). Therefore, automating the processing for analysing and interpreting viral genomic data is being investigated with great interest by virologists, environmental engineers, and bioinformatics.
2.3. Emerging and alternative methods for SARS-CoV-2 detection
2.3.1. RNA-based emerging and alternative methods
Previous studies of SARS-CoV-2 in wastewater focused on detection and quantification of the viral RNA through RT-PCR and RT-qPCR methods. The study of the different strains of the virus present in the wastewater and in the corresponding community is also important. High throughput sequencing has been used to study different strains of the virus to monitor its mutation (Feng et al., 2020). In a recent study by Crits-Christoph et al. (2020), high-throughput sequencing was used to study the different SARS-CoV-2 genotypes circulating in wastewater systems in California.
Another emerging approach in the detection and quantification of the SARS-CoV-2 RNA is digital PCR (dPCR), which is identified to be less affected by PCR inhibitors (Sidstedt et al., 2020). This is an advantage when detecting viruses from wastewater, which is a matrix that contains several possible PCR inhibitors. It is also noted that the dPCR has a lower limit of detection and has been reported to be more sensitive than qPCR (Ahmed et al., 2020b, Barceló, 2020, Falzone et al., 2020, Suo et al., 2020). The detection limit of dPCR is reported to be 10 times lower compared to that of the RT-qPCR (Barceló, 2020). Suo et al. (2020) showed that the limit of detection of an optimized droplet digital PCR (ddPCR) is 500 (maximum) times lower than the RT-qPCR in analytes with low-level of SARS-CoV-2 load (Suo et al., 2020). This may be favorable for detecting even low concentrations of SARS-CoV-2 in influents of wastewater in areas with low COVID-19 prevalence and in treated wastewater. The dPCR has been previously applied for clinical diagnostics, and its advantages show its potential as tool for detecting and quantifying SARS-CoV-2 in wastewater.
The ddPCR, an improvement of the RT-PCR, has been reported to be useful in quantification of very low target concentrations of nucleic acids in samples that are contaminated (Taylor et al., 2017). Most of the studies that quantified SARS-CoV-2 in wastewater samples utilized the RT-qPCR method. However, few studies have so far utilized the ddPCR method to not only detect but also quantify the SARS-CoV-2 genome in wastewater (Ahmed et al., 2020b, Gonzalez et al., 2020, Zhou et al., 2020a)]. RT-ddPCR’s main advantages over the RT-qPCR include i) the direct absolute quantification without the reliance on a calibration curve, and ii) reduced effects of PCR inhibitors (Deiana et al., 2020, Kuypers and Jerome, 2017). However, it also has its disadvantages, notably low reaction mixture volume and smaller dynamic range (Kuypers and Jerome, 2017). A one-step ddPCR assay was found by Graham et al. (2021) to be more sensitive than RT-qPCR in detecting and quantifying SARS-CoV-2 in primary settled solids from wastewater treatment plants, due to the reduced effect of PCR inhibitors.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based assays for SARS-CoV-2 RNA have also been recently developed for clinical applications (Broughton et al., 2020, Hou et al., 2020). Broughton et al. (2020) reported a limit of detection of a CRISPR-based assay at 10 copies/µL input, which is higher than that of the US Centers for Disease Control and Prevention (CDC) based on RT-qPCR assay (1–3.2 copies/µL input) (Broughton et al., 2020). However, in these recent studies by Broughton et al. and Hou et al., the total assay times for the CRISPR-based assays were reported to be shorter: 40–45 min compared to 4 h spent in the RT-qPCR assay. There is no published study that has yet reported the use of CRISPR-based assays for detection of SARS-CoV-2 in wastewater. However, its sensitivity and rapidity can be advantageous for wastewater-based epidemiological studies, where real-time and accurate information about SARS-CoV-2 circulating in wastewater is important.
It is to be noted that, at the time of writing of this manuscript, there was no established standard assay for the detection of SARS-CoV-2 in environmental samples, such as in wastewater and surface waters (La Rosa et al., 2020b). Recent studies that detected SARS-CoV-2 RNA in wastewaters utilized assays that target different regions of the SARS-CoV-2 genome, which include the following: i) N-gene encoding nucleocapsid protein, ii) S-gene encoding spike protein, iii) E-gene encoding envelope protein, iv) ORF1ab (La Rosa et al., 2020b, La Rosa et al., 2021), v) RNA-dependent RNA polymerase (RdRP), and vi) M-gene encoding membrane protein. Table 2 shows the different assays used in the studies on the detection of SARS-CoV-2 in wastewater and the corresponding limits of detection. The study of Ahmed et al. (2020a) showed that the N_Sarbeco assay was more sensitive than the NIID_2019-nCOV_N in detecting the SARS-CoV-2 RNA in wastewater (Ahmed et al., 2020a). In the study of Medema et al. (2020b), the CDC_N1 and CDC_N3 assays were shown to be more sensitive than CDC_N2. This is in agreement with the results of the study of Ahmed et al. (2020b), where CDC_N1 assay was found the most sensitive (limit of detection: 1 copy/µL RNA template) compared to other assays targeting the N-gene. In this study, it was also shown that the least sensitive was the E_Sarbeco assay (limit of detection: 5 copies/µL RNA template) (Ahmed et al., 2020b). La Rosa et al. (2020b) noted the low sensitivity of the RNA-dependent RNA polymerase (RdRp) assay, which led to no positive results in the RT-qPCR in their study. However, the authors in the latter study noted that the RdRP assay displayed a higher sensitivity than the S-gene assay and that it was able to reduce PCR inhibitor concentrations to below the acceptable levels (median at 29 %). In the study of Westhaus et al. (2021), it was shown that the assay targeting the RdRP gene (compared to assays targeting the N, E, and M genes) had the highest specificity to SARS-CoV-2 RNA since no signals were detected for CoV-229E and SARS-CoV-1 (Westhaus et al., 2021).
RT-ddPCR’s main advantages over the RT-qPCR include: i) the direct absolute quantification without the reliance on a calibration curve, and ii) the less effect of PCR inhibitors (Deiana et al., 2020, Kuypers and Jerome, 2017). However, it also has its disadvantages, notably low reaction mixture volume and smaller dynamic range (Kuypers and Jerome, 2017). Multiplex RT-qPCR and dPCR assays have also been used in clinical samples for detection and quantification of SARS-CoV-2 (da Silva Queiroz et al., 2021, de Kock et al., 2020).
These multiplex assays could be useful in using different well-established target SARS-CoV-2 genes; however, they have not yet been used in environmental samples.
2.3.2. Non-RNA-based emerging and alternative methods
Most of the recent studies on the presence and abundance of SARS-CoV-2 in wastewater samples have been performed through detection and quantification of the viral RNA. However, measurements of the SARS-CoV-2 RNA in wastewater have their own limitations. This includes the stability of the SARS-CoV-2 RNA in the wastewater at varying conditions such as higher temperature as discussed in Section 2.1 and will be discussed in Section 3. Detection of low levels of RNA in some wastewater samples is also a challenge, which means that significant levels of amplification are needed for the PCR-based methods (Neault et al., 2020).
Barceló suggested that the detection of biomolecules, which have already been used in clinical diagnostics, can be used to study the presence of SARS-CoV-2 in wastewater aside from detection of the viral RNA (Barceló, 2020). Among these biomolecules are the proteins of SARS-CoV-2, which may provide information that is supplementary to those obtained from detection and measurement of the viral RNA (see Fig. 3). An advantage of the detection of the viral proteins over the viral RNA is the reduction of the number false positive tests from the process of amplification since proteins could not be directly amplified (Feng et al., 2020). Neault et al. (2020) studied the presence of SARS-CoV-2 in samples from primary sludge and PEG-precipitated influent wastewater solids by detecting the structural proteins of the virus. In the latter study, the detection and quantification of proteins was achieved thanks to higher stability and higher number of gene copies of the proteins than the viral RNA. Immunoblot analysis was applied to detect the SARS-CoV-2 structural proteins, which include nucleocapsid (N), spike (S), membrane (M) and envelope (E) proteins. Quantification of the SARS-CoV-2 proteins was also conducted through Multiplex Paired-Antibody Amplified Detection (MPAD), an immunological method linked with PCR. The four structural proteins were detected in the primary sludge and influent solids samples. Results of the study also showed that the proteins measured by MPAD produced higher signals (64–208-fold in primary sludge samples, and 20–128-fold in influent solids samples) than the ones generated by viral RNA measured by RT-qPCR. In recent studies on the detection of SARS-CoV-2 in clinical samples, the target proteins were the N and S structural proteins (Feng et al., 2020). The N protein was chosen due to its higher abundance during infections (Ihling et al., 2020, Nikolaev et al., 2020a). On the other hand, the S protein is chosen in other studies due to its specificity compared to the other SARS-CoV-2 structural proteins (Mavrikou et al., 2020, Seo et al., 2020). It was noted that a longer S protein was observed to be encoded by SARS-CoV-2 compared to the other coronaviruses (Caruana et al., 2020). Thus, the S protein of SARS-CoV-2 potentially enhances the specific detection of the virus and its variants (Seo et al., 2020).
Fig. 3.
Emerging methods for SARS-CoV-2 detection.
However, Feng et al. (2020) noted that the content of proteins in a specimen is very low, the detection could be challenging and thus would require very sensitive methods. One of these methods that can be used is mass spectrometry (MS). This technique has recently been applied to detect SARS-CoV-2 proteins for clinical applications. Peptides that are unique to the SARS‐CoV‐2 N protein, which is identified as the most abundant protein in the virion, were identified using recently-developed MS-based assays applied to clinical samples (Ihling et al., 2020, Nikolaev et al., 2020b). The use of MS-based methods to detect SARS-CoV-2 proteins could be extended for wastewater samples. The detection of infectious viruses in wastewater samples using proteins has been previously carried out by Ye et al. (2019). This was accomplished using an integrated cell culture-mass spectrometry (ICC-MS), in which strain-specific viral peptides of reovirus were identified. Noteworthy, in this method, mass spectrometry eliminated the step of primer design, which is characteristic of PCR-based methods. Although reovirus was the only virus studied/detected, the ICC-MS could be potentially used for detection of SARS-CoV-2 proteins in wastewater. The use of mass-spectrometry could also be potentially used for quantification of SARS-CoV-2 proteins in wastewater samples (Feng et al., 2020). Barceló (2020) suggested that environmental proteomics could be a tool complementary to PCR-based methods for use in WBE (Barceló, 2020). Mass spectrometry for the characterization and quantitation of the SARS-CoV-2 viral proteins might be able to provide more insight on the fraction of the SARS-CoV-2 load that is capable of infecting cell culture. As discussed, SARS-CoV-2 has the structural proteins N, S, M, and E. SARS-CoV-2 infection is mediated by the S protein, which plays key roles in the attachment, fusion, and entry of the virus into host cells (Duan et al., 2020). Being able to obtain measurements of the N, S, M, and E proteins in different samples in the future could potentially deliver more information on the relationship between viral load and infectivity. However, there are still challenges in this approach because proteins cannot be directly amplified and consequently, direct detection of viral proteins still lack of sensitivity with the current analytical instruments (Feng et al., 2020).
The detection of SARS-CoV-2 proteins using biosensors suitable for clinical applications has also been recently explored (Mavrikou et al., 2020, Seo et al., 2020). Mavrikou et al. (2020) developed a biosensor that detects S proteins of SARS-CoV-2, which is reported to be more specific to the virus than other structural proteins and is responsible for the binding of the virus to the human cellular receptor. In the latter study, the binding of the SARS-CoV-2 S protein to a membrane-bound antibody, specific to this protein, produced changes in the bioelectric properties of the engineered cell membrane. A device based on the bioelectric recognition assay measured the changes in the properties of the membrane cell in real-time. Another biosensor that relies on the principle of the binding of SARS-CoV-2 S protein to a specific antibody was developed by Seo et al. (2020). A field-effect transistor (FET)-based biosensor with graphene sheets was coated with antibodies specific to the S protein. These studies showed that immunological methods can be used to detect SARS-CoV-2 rapidly and accurately, even without sample pretreatment. The latter studies showed the use of detection of SARS-CoV-2 proteins, particularly N and S proteins, for clinical diagnosis. This could be explored further for use in detection of SARS-CoV-2 in wastewater as demonstrated by Neault et al. (2020). However, there are also challenges presented by the detection of SARS-CoV-2 proteins such as cross-reactivity, specifically for N protein, and the availability of antibodies specific to the proteins (Feng et al., 2020). Another approach on molecular diagnosis of SARS-CoV-2 is the use of aptamers instead of antibodies to target the SARS-CoV-2 proteins (Feng et al., 2020, Song et al., 2020). The noted advantages of using aptamers for detection of coronaviruses’ proteins include the following: i) smaller size of aptamers permits more efficient binding on the surface of the virus, and ii) stability and facile synthesis of aptamers (Cho et al., 2011, Song et al., 2020). Song et al. (2020) have identified aptamers as probes that target the receptor-binding domain of the SARS-CoV-2 proteins. However, the use of these aptamers for detection of SARS-CoV-2 proteins are identified to be complementary to other diagnostic methods and studies are still on the “proof-of concept” stage (Chen et al., 2020, Song et al., 2020). This approach in detection still has to be further explored for use in the detection of SARS-CoV-2 in wastewater matrices.
As discussed, biosensors can provide rapid information about viruses and can complement other diagnostic methods, such as PCR-based methods. However, SARS-CoV-2 proteins quantification has been applied principally to clinical samples, which have different properties from wastewater matrices. However, biosensors have been studied for the detection of viruses in environmental samples. Limited number of studies have explored its use on the detection of viruses in the complex wastewater matrix. A study by Chung et al. reported the detection of noroviruses (NoV) in tap water and reclaimed wastewater using a microfluidic paper analytic device (µPAD) and quantified the viruses using a smart phone-based fluorescence microscope with a dedicated application software. The µPAD detection was realized using the principle of antigen-antibody binding instead of the detection of the viral RNA (Chung et al., 2019). The latter study shows the potential of paper-based sensors for detection and quantification of SARS-CoV-2 from wastewater. It is to be noted that these biosensors are meant to be complementary tools to other detection and quantification methods.
3. Environmental stability of coronaviruses
The understanding of the environmental persistence of pathogens and the effectiveness of applicable disinfection methods make it possible to assess the hazards associated to a contamination.
Aerosol stability of CoVs and in particular of SARS-CoV-2 has been extensively investigated and recently reviewed (Arslan et al., 2020, El Baz and Imziln, 2020, Schuit et al., 2020, Tang et al., 2020, van Doremalen et al., 2020, Wang et al., 2020b). The spontaneous inactivation of coronaviruses in the environment depends on several factors, such as temperature, relative humidity and, in water media, on pH, level of particulate, organic matter, chemicals, and of antagonistic microorganisms (Casanova et al., 2009, Gundy et al., 2008, Lai et al., 2005, Wang et al., 2005b, Wigginton et al., 2015, Ye et al., 2016). Noteworthy is the case of meat plants where suitable conditions favored the spreading of the SARS-CoV-2. Low temperature, very high or very low relative humidity conditions, combined with the large use of water and the dense production of aerosols resulted in the generation of hot-spots for SARS-CoV-2 diffusion in meet plants (Middleton et al., 2020).
Table 3 summarizes the persistence of representative CoVs at room temperature in various media under different conditions determined by plaque assay or TCID50 technique.
3.1. Persistence of CoVs on inanimate surfaces
Inanimate surfaces have been indicated to be potential sources of CoV contamination, both directly via fomite transmission and indirectly, via water media which came into contact with the infected surface (Peyrony et al., 2020). The aerosolization of fomites has also been demonstrated to be effective in the spreading of viruses such as the Influenza A virus (Asadi et al., 2020b). In this context, it is thus important to ascertain the persistence of CoVs on various surfaces and the adequacy of disinfection tools used to treat them (Bhardwaj and Agrawal, 2020). Human CoVs can remain infectious on inanimate surfaces from 2 h up to even several weeks, depending on the environmental conditions (see Table 3) (Casanova et al., 2009, Chan et al., 2011, Chan et al., 2004, Chin et al., 2020, Duan et al., 2003, Geller et al., 2012, Gundy et al., 2008, Lai et al., 2005, Rabenau et al., 2005, Sattar et al., 2009, Sizun et al., 2000, van Doremalen et al., 2020, van Doremalen et al., 2013, Wang et al., 2005b, Ye et al., 2016).
The infectivity of SARS-CoV-1 on porous and non-porous surfaces, such as those of cloth, filter paper, glass, mosaic, metal and plastic, persisted for 60–72 (5-log reduction of virus titer), as has been assessed by analysing the CPE of the infected surfaces in Vero cells (Duan et al., 2003). Up to 9 days were necessary for a 5-log reduction of the infectivity of the virus on plastic surfaces (polystyrene of Petri dishes) (Rabenau et al., 2005). A limited persistence of infectivity, less than 6 h, was found in autoclaved soil. The sterilization was applied to avoid the effect of concurrently present microorganisms on the inactivation of the virus (Duan et al., 2003). A similar stability was observed for MERS-CoV: 72 h were sufficient for 6-log of virus titer reduction on plastics and steel at 20 °C (van Doremalen et al., 2013). SARS-CoV-1 and SARS-CoV-2 showed similar stability when compared under the same environmental conditions (van Doremalen et al., 2020). A significant reduction of infectivity, of c.a. 3 log, was found for SARS-CoV-2 after 72 h on plastic and steel by TCID50 assay (van Doremalen et al., 2020). Under the same conditions, the persistence of SARS-CoV-1 was similar (see Table 3). A previous study showed that the stability of SARS-CoV-1 strongly depended on temperature and humidity conditions. At 22–25 °C and relative humidity of about 40–50%, the virus infectivity persisted up to 4 weeks on a laboratory multi-well plate made of plastics: 1-log loss of titer was observed in 5 days and a progressive reduction of the titer of 5-log was found in 4 weeks (Chan et al., 2011). The SARS-CoV-2 titer reduces by c.a. 3-log in 4 h on copper surfaces and in 24 h on cardboard, whereas SARS-CoV-1 requires 8 h on the same surfaces (van Doremalen et al., 2020). Another recent study on the environmental persistence of infectivity of SARS-CoV2 showed a virus titer reduction of 4.7-log in 3 h on printing and tissue papers, while from 2 up to 7 days were necessary to achieve 3–6-log of virus titer reduction both on porous and non-porous surfaces, such as banknote, wood, cloth, respiratory mask, glass, stainless steel, and plastics (Chin et al., 2020).
3.2. Persistence of CoVs in water media
Human beings are continuously in contact with the water for their daily activities, such as drinking, personal hygiene, washing, irrigation, food production, and recreational purposes (see Fig. 4). Used water, including stormwater and runoffs, is ultimately collected as wastewater. Feces, urine, or vomit infected with pathogens enter the human sewage; this can then affect the urban water cycle. The knowledge of the persistence of those microorganisms in water media allows to define accurately the levels of hazards for humans and environment. As matter of fact, the generation of bioaerosols in aeration basins in wastewater treatment plants constitutes a risk of infection in particular of enteric viruses (Mirskaya and Agranovski, 2018). Studies have demonstrated that field workers are exposed to infections from bioaerosols (Masclaux et al., 2014). On February 5, 2020, the U.S. Occupational Safety and Health Administration (OSHA) released a wastewater worker guidance, which stated that the current disinfection techniques used in WWTPs, such as oxidation with hypochlorite or peracetic acid and inactivation by ultraviolet (UV) irradiation, were sufficient to protect the health of wastewater plant operators and the public (OSHA, 2020, WEF, 2020). This recommendation was based on CoV disinfection data obtained from the healthcare settings, which conforms to OSHA’s position on the susceptibility of CoV to disinfection. WHO and United States Environmental Protection Agency (EPA) have not raised particular concerns on the current procedures of water sanitation with regard to the SARS-COV-2 (EPA. Coronavirus and Drinking Water and Wastewater, 2020, WHO, 2020b).
Fig. 4.
Integrated water cycle.
Representative CoVs, such as human CoV 229E (cause of a common cold) and the feline infectious peritonitis virus (FIPV), were examined determining the T99.9, i.e. the time required for the virus titer to decrease of 3-log (99.9%). That virus titer reduction was observed in dechlorinated and filtered tap water in 10 days at 23 °C and in 130 days at 4 °C. The inactivation of those CoVs occurred faster in wastewater with a T99.9 of only 2–4 days. Under the same conditions, a non-enveloped virus, such as the poliovirus 1, was more stable than CoV 229E and FIPV (Gundy et al., 2008). CoV 229E presented a similar stability with 5-log of titer reduction within 9 days when suspended in minimum essential medium (MEM) containing antibiotics (penicillin and streptomycin), both in the presence and absence of 10% of fetal bovine serum (FBS). In a dried form, deposed on a polystyrene petri dish, 72 h were needed (Rabenau et al., 2005).
The assay of viable virions of two surrogate CoVs, TGEV and MHV, was evaluated in reagent grade water, lake water, and pasteurized settled sewage by determining the T99, i.e the time required for a reduction in virus titer of 99%, in turn corresponding to 2 log) (Casanova et al., 2009). Thus, 2-log of virus titer reduction were observed at 25 °C for TGEV and MHV in reagent grade water respectively in 22 and 17 days, whereas at 4 °C no significant reduction of the infectious titer for both viruses was observed for 49 days. In a lake water sample at 25 °C, the same infectivity reduction was observed for TGEV and MHV respectively in 13 and 10 days. At 4 °C, 1-log of titer reduction of TGEV was achieved in 14 days, whereas the MHV resulted in comparison more stable. In pasteurized settled sewage at 25 °C, 9 and 7 days were necessary for 2-log of TGEV and MHV titer reduction, respectively.
The spontaneous inactivation of SARS-CoV-1 in water media, feces, and urine was investigated in vitro (Wang et al., 2005b, Wang et al., 2005c). The virus titer was found reduced of 5-log after 2 days at 20 °C in dechlorinated tap water, domestic sewage, or hospital sewage; after 3 days in feces and 17 days in urine. Reducing the temperature to 4 °C the infectivity of SARS-CoV-2 persisted for over 14 days in those media. Wet human specimens (blood serum, sputum, stool and urine) and biological media for cell and virus culture preserve quite well the infectivity of SARS-CoV-1 (Duan et al., 2003, Rabenau et al., 2005, Wang et al., 2005b, Wang et al., 2005c) and that of other human CoVs (see Table 3). The virus titer was reduced by 5-log in serum and sputum in 96 h and in urine in 72 h (Duan et al., 2003). A slow rate of reduction of the titer at room temperature for SARS-CoV-1 of 0.5-log reduction over 9 days was observed in serum-free cell culture medium, whereas under the same conditions, CoV E229 lost its infectivity completely. Phosphate-buffered saline (PBS) solution at pH 7.4 was proven to assure good stability to CoVs. E229 and OC43 persisted in this medium over 3 days (Sizun et al., 2000), while SARS-CoV-1 over 14 days were necessary for 5-log of virus titer reduction (Wang et al., 2005b, Wang et al., 2005c). Another more recent study on the environmental persistence of infectivity of SARS-CoV2 demonstrated that the virus is highly stable at 4 °C in virus transport medium: 0.7-log of titer reduction was observed after 14 day (Chin et al., 2020).
CoVs can survive for 2–4 days in sewage and wastewater at room temperature and for a more prolonged time at lower temperatures like that of the winter (Casanova et al., 2009, Duan et al., 2003, Gundy et al., 2008, Wang et al., 2005b, Wang et al., 2005c, Ye et al., 2016). The novel SARS-CoV-2 was detected and found infective in human stool samples (Cheung et al., 2020, Tian et al., 2020, Wang et al., 2020d) and detected worldwide in wastewater and surface water receiving wastewater (see Table 1). The RNA of the virus was found in sewage during and even before the emergence of COVID-19 cases (La Rosa et al., 2021, Medema et al., 2020b). However tests for the determination viable viruses in WWTP influent and effluent wastewater (Rimoldi et al., 2020a, Rimoldi et al., 2020b, Westhaus et al., 2021), as well as in hospital wastewater (Ge et al., 2020, Wang et al., 2020c) and in river receiving contaminated wastewater, (Rimoldi et al., 2020a, Rimoldi et al., 2020b) confirmed the limited resistance of the virus in such water media. The results indicate that the latter water medium is particularly aggressive for SARS-CoV-2 (see paragraph 2.1, Tables 1 and 3 and for further explanations paragraphs 4.4, 4.5 and 4.6). Infecting wastewater with SARS-CoV-2, Bivins et al. (2020) determined a persistence of 1.4–3.3 days for 1-log and of 2.9–6.5 days for 2-log of inactivation of the virus in this media. However, in the same study, a similar resistance was observed for the virus in tap water. In fact, at room temperature the spontaneous reduction of the virus titer of 1-log and 2-log occurred within 1.8–2.2 and 3.6–4.4 days, respectively (see Table 3). This further confirms the limited persistence of this virus in water media.
4. Methods for inactivation of coronaviruses
Lipid-enveloped viruses with high hydrophobicity, such as CoVs, are less stable in water when compared to non-enveloped viruses. In principle, the performance of water sanitation methods for CoVs can be evaluated based on the inactivation data of non-enveloped viruses with higher resistance. Unfortunately, the evaluation of infectivity based solely on determination of viable CoVs in wastewaters is considerably simplified.
The current guidelines for wastewater sanitation include the following main options ( Table 4): 1) thermal treatment, 2) UV irradiation, 3) chemical disinfection, 4) holding wastewater for a prolonged time; 5) sedimentation; 6) membrane filtration; and 7) attenuation in subsurface (WHO, 2018).
Table 4.
Inactivation of SARS-CoV-2 and other representative CoVs.
| Method | CoV | Substrate | Specification | Log10reduction | Time | References |
|---|---|---|---|---|---|---|
| Thermal | SARS-CoV-1 | Culture medium | 56 °C | 6 log | 90 min | (Duan et al., 2003) |
| 67 °C | 6 log | |||||
| 60 min | ||||||
| MEM | 60 °C | 5 log | 30 min | (Rabenau et al., 2005) | ||
| MEM + FBS(20%) | 60 °C | 1.93 log | 30 min | |||
| Plastics (PS) | 60 °C | 5 log | 30 min | |||
| MEM + FBS(10%) | 56 °C | 7 log | 30 min | (Kariwa et al., 2006) | ||
| MERS-CoV | Plastic | 20 °C | 5 log | 72 h | (van Doremalen et al., 2013) | |
| 30 °C | 5 log | 24 h | ||||
| Steel | 20 °C | 5 log | 72 h | |||
| 30 °C | 5 log | 24 h | ||||
| SARS-CoV-2 | Culture medium | 37 °C | 3 log | 2 days | (Chin et al., 2020) | |
| 56 °C | 6 log | 30 min | ||||
| 70 °C | 6 log | 5 min | ||||
| Wastewater | 50 °C | 1 log | 14–17 min | (Bivins et al., 2020) | ||
| 50 °C | 2 log | |||||
| 70 °C | 1 log | 28–34 min | ||||
| 70 °C | 2 log | |||||
| 1.8–2.9 min | ||||||
| 3.7–5.7 min | ||||||
| UV exposure | SARS-CoV-1 | Culture medium | UV-C (254 nm; 4.0 mW/cm2), | 4 log | 40 min | (Darnell and Taylor, 2006) |
| UV-A (365 nm; 2.1 mW/cm2) | 2 log | |||||
| 30 min | ||||||
| UV-C (260 nm; 90 μW/cm2) | 6 log | 60 min | (Duan et al., 2003) | |||
| UV 134 μW/cm2 | 6 log | 60 min | (Kariwa et al., 2006) | |||
| SARS-CoV-2 | Culture medium | UV-C (254 nm; 2.2 mW/cm2) | 1 log | 0.01 s | (Sabino et al., 2020) | |
| 0.016 mJ/cm2 | ||||||
| 0.706 mJ/cm2 | 2 log | 0.32 s | ||||
| 6.556 mJ/cm2 | 3 log | 2.98 s | ||||
| 31.880 mJ/cm2 | 4 log | 14.49 s | ||||
| 108.714 mJ/cm2 | 5 log | 49.42 s | ||||
| Chemical disinfection | SARS-CoV-1 | Culture medium | Povidone-iodine, Isodine® solution, Isodine Scrub®, Isodine Palm®, Isodine Gargle® and Isodine Nodo Fresh® | 6 log | 1 min | (Kariwa et al., 2006) |
| FBS/MEM | 2-Propanol (100%) | 3.31 log | 30 s | (Rabenau et al., 2005) | ||
| 2-propanol (70%) | 3.31 log | 30 s | ||||
| Desderman (78% Ethanol) | 5.01 log | 30 s | ||||
| Sterillium (45% 2-propanol,30% 1-propanol) | 2.78 log | 30 s | ||||
| Wine vinegar | 3.0 log | 60 s | ||||
| Formaldehyde (0.7%) | 3.01 log | 120 s | ||||
| Formaldehyde (1.0%) | 3.01 log | 120 s | ||||
| Glutardialdehyde (0.5%) | 4.01 log | 120 s | ||||
| Incidin plus (2%) | 1.68 log | 120 s | ||||
| Wastewater | Chlorine (10 mg/L) | 1.6 log | 30 min | (Wang et al., 2005b) | ||
| Chlorine dioxide (40 mg/L) | 1.75 log | |||||
| 30 min | ||||||
| CoV E229 | PBS | Sodium hypochlorite (0.05%), iodine solution (0.075%), soap (1%), ethanol (70%) | 1 log | 5 min | (Sizun et al., 2000) | |
| Faeces | Glutaraldehyde (2%), sodium hypochlorite (0.005%), povidone‐iodine (1%), ethanol (70%), chloramine T (0.3%), dimethyl-benzyl-ammonium chloride (0.04%), chlorhexidine (0.05%) | 3 log | 1 min | (Sattar et al., 2009) | ||
| OC43 | PBS | Sodium hypochlorite (0.5%), povidone‐iodine (0.075%), soap (1%), ethanol (70%) | 1 log | 5 min | (Sizun et al., 2000) | |
| SARS-CoV-2 | Culture medium | Sodium hypochlorite (1:49), sodium hypochlorite (1:99), ethanol (70%), iodine solution (7.5%), chloroxylenol (0.05%), chlorhexidine (0.05%), benzalkonium chloride (0.1%);Soap (1:49) | 7–8 | log, 5 min | (Chin et al., 2020) | |
| 7–8 log | ||||||
| > 5 min | ||||||
| Wastewater holding | SARS-CoV-1 | Wastewater | 22 °C | Not measured (Only detection after holding time was documented) | 2 days | (Wang et al., 2005b, Wang et al., 2005c) |
| 4 °C | ||||||
| 14 days | ||||||
| Hospital wastewater | Spontaneous inactivation (total residual chlorine was 0–1.0 mg/L or 3.0–12.5 mg/L; free residual chlorine was 0–0.5 mg/L or 1.5–5.0 mg/L) | n.r. | – | (Wang et al., 2005c) | ||
| SARS-CoV-2 | Wastewater | 1 log | 1.4–3.3 d | (Bivins et al., 2020) | ||
| 2 log | ||||||
| 2.9–6.5 d |
The performance of these disinfection methods in regard to CoVs and particular regard to SARS-CoV-2, from drinking water to wastewater effluent, is discussed below.
4.1. Thermal inactivation
Temperature is considered to be one of the most influential parameters for enveloped virus inactivation (Darnell and Taylor, 2006, Duan et al., 2003, John and Rose, 2005, Kariwa et al., 2006, Lamarre and Talbot, 1989, Rabenau et al., 2005). Membrane and capsid proteins are sensitive to heat-induced denaturation. Aqueous foods, such as milk and fruit juice, as well as drinking water, can be treated in pasteurization processes at moderate (56–65 °C for 30 min) or high temperature (80–135 °C for 1–4 s), in order to destroy or deactivate microorganisms, enzymes and viruses. The disinfection of SARS-CoV-1 (−6-log) was achieved at 56 °C, 67 °C, and 75 °C, respectively within 90, 30, and 30 min (see Table 4) (Duan et al., 2003). Similar results were reported on thermal inactivation of SARS-CoV-1 in vitro in media for cell or virus culture (Darnell and Taylor, 2006, Kariwa et al., 2006, Rabenau et al., 2005). In another study, a non-human CoV, the canine coronavirus (CCoV), was identified to be infectious even at 56 °C; however, it was inactivated at temperatures higher than 65 °C (Pratelli, 2008). MERS-CoV was found sensible to temperature variations: the virus titer was reduced by 5-log in 72 h at 20 °C and in 24 h at 30 °C, both on plastics and steel (van Doremalen et al., 2013).
The debate concerning the effects of environment temperature on the rate of transmission of SARS-CoV-2 is still open (Auler et al., 2020, Tobías and Molina, 2020). Nevertheless, the disinfecting effect of heat on the virus is undoubted. The thermal effect on the infectivity of SARS-CoV2 in virus culture media was investigated at 56 and 70 °C, demonstrating that treatments of 30 min and 5 min were respectively sufficient for achieving 6-log of virus deactivation (Chin et al., 2020). In wastewater 2-log of virus removal were obtained at 50 °C in 28–34 min, while at 70 °C were found sufficient 3.7–5.7 min (Bivins et al., 2020).
4.2. Inactivation by ultraviolet radiation
The inactivation of microorganisms by ultraviolet (UV) radiation is currently extensively applied for disinfection of surfaces, drinking water, as well as in tertiary treatments of wastewater for the abatement of the loading of highly resistant species. A relatively large number of the viruses, typically enteric ones, such as noroviruses, rotaviruses, reoviruses, sapoviruses, astroviruses, enteroviruses, adenoviruses and JC viruses, persist in the effluent of full-scale WWTPs with less than 2-log of loading removal (Qiu et al., 2018). Unlike the abovementioned viruses, enveloped viruses are more vulnerable to UV exposure. A prolonged exposure time of 40–60 min was necessary to achieve a satisfactory degree of inactivation for SARS-CoV-1 in vitro (Darnell and Taylor, 2006, Duan et al., 2003, Kariwa et al., 2006). Kariwa et al. showed 5-log of SARS-CoV-1 reduction at 134 μW/cm2 in 15 min and further 1-log after additional 45 min.
SARS-CoV-2 is susceptible to sunlight (Nicastro et al., 2020, Ratnesar-Shumate et al., 2020, Tang et al., 2021) and rapidly inactivated by UV-C (λ = 100–280 nm) (Bianco et al., 2020, Heilingloh et al., 2020, Sabino et al., 2020), UV-B (λ = 280–315 nm) and UV-A (λ = 315–400 nm) radiation (Nicastro et al., 2020, Ratnesar-Shumate et al., 2020). Sabino et al. (2020) accurately determined in vitro the kinetics and the light fluence for SARS-CoV-2 inactivation by UV-C at wavelength of 254 nm and light intensity of 2.2 mW/cm2. The removal of 3-log of viruses was observed in less than 3 s of irradiation, whereas for 5-log of inactivation were necessary almost 50 s (see Table 4).
4.3. Chemical disinfection
Conventional antiseptics and disinfectants, such as halogenated compounds (chlorine, sodium hypochlorite, chloramine and povidone-iodine), alcohols (ethanol and 2-propanol), aldehydes (formaldehyde and glutaraldehyde), quaternary ammonium compounds, phenolic compounds, and other decontaminating agents, have been found to be effective for the disinfection of the surfaces contaminated by 229E and SARS- and MERS- CoVs. The agents quenched the infectivity within a short exposure time of 1 min (see Table 4) (Eggers et al., 2015, Geller et al., 2012, Kampf et al., 2020, Kumar et al., 2015, Lai et al., 2005, Rabenau et al., 2005, Sattar et al., 2009). A significant SARS-CoV-1 titer reduction of 1.6–5-log was observed in vitro testing for 0.5–2 min using disinfectants, such as 2-propanol, formaldehyde, glutaraldehyde, Desderman, Sterillium, and Incidin plus (Rabenau et al., 2005) and of 6-log by using for 1 min commercially available disinfectant as povidone-iodine, isodine® solution, Isodine Scrub®, Isodine Palm®, Isodine Gargle® and Isodine Nodo Fresh® (Kariwa et al., 2006). Conventional disinfectants, such as sodium hypochlorite (1:99), ethanol (70%), iodine solution (7.5%), chloroxylenol (0.05%), chlorhexidine (0.05%) and benzalkonium chloride (0.1%), were found effective in the deactivation of SARS-CoV-2 in 5 min of treatment, with exception of the soap that required greater time for the removal of 7–8-log (Table 4).
The effect of pH on the stability of CoVs has been investigated for CoV 229E (Lamarre and Talbot, 1989), MHV (Sturman et al., 1990), TGEV (Pocock and Garwes, 1975), and the CCoV (Pratelli, 2008). Due to the lipid acidic envelope, CoVs were found to be sensitive to the variation of pH and the greater virus stability was found at slightly acidic pH levels of 6–6.5.(Geller et al., 2012) On the contrary, SARS-CoV-2 was found highly stable in a wide range of pH values (3–10) at room temperature (Chin et al., 2020).
Based on the data obtained from other viral indicators, the use of chlorine for water disinfection is the most effective and economical solution for this problem. Chlorine effectively inactivates the virus by destroying the viral envelope or capsid (Thurman and Gerba, 1988). In particular, free chlorine has been proven to affect directly the proteins present in the viral envelope, rather than the less reactive lipidic material and the RNA core (Ye et al., 2018). Chlorine can also react with the ammonia present in wastewater to form combined chlorine (e.g., chloramines). These compounds have the capability to disinfect; however, during disinfection, they are less active and behave differently from free chlorine. It is thus important for every wastewater treatment facility to examine the chlorine species and their relative abundance during the disinfection process.
Studies of the treatment of municipal water and wastewater using chlorine and its derivatives have reported significant inactivation efficiencies for SARS-CoV-1. In hospital wastewater, domestic sewage, and dechlorinated tap water, the virus remained active only for 2 days. Moreover, SARS-CoV-1 became more susceptible to disinfectants compared to E. coli and f2 phage. For SARS-CoV-1 inactivation, free chlorine was more relatively more effective compared to chlorine dioxide. Free residual chlorine exceeding 0.5 mg/L or chlorine dioxide exceeding 2.19 mg/L in wastewater ensured the complete inactivation of SARS-CoV (Wang et al., 2005b). Furthermore, extreme pH levels (pH > 12 or pH < 3), formalin, and glutaraldehyde were determined to inactivate SARS-CoV-1 quite well (Darnell et al., 2004). Peracetic acid has been determined to have the ability to destroy some non-enveloped viruses (e.g., norovirus), which are known to be more resistant to chemical agents compared to enveloped viruses (Dunkin et al., 2017).
The documented presence of SARS-CoV-2 RNA in wastewater influents and sludge, as well as in effluents released from WWTPs have raised the concern of the personnel of the treatment plants and necessitated examination of this issue by the scientific community involved in the field of water sanitation (Adelodun et al., 2020, Amoah et al., 2020, Arslan et al., 2020, Bhowmick et al., 2020, Bilal et al., 2020, Bogler et al., 2020, Carducci et al., 2020, Carraturo et al., 2020, Collivignarelli et al., 2020, El Baz and Imziln, 2020, Foladori et al., 2020, Gwenzi, 2020, Jones et al., 2020; Paleologos et al., 2020; Kitajima et al., 2020, La Rosa et al., 2020a, Mandal et al., 2020, Naddeo and Liu, 2020, Nghiem et al., 2020, Shutler et al., 2020, Silverman and Boehm, 2020, Zaneti et al., 2021).
These studies have shown that the virus tends to exhibit limited resistance in wastewater (Bivins et al., 2020). In general, investigations on viable SARS-CoV-2 in real influents and effluents, both from domestic and hospital sewages, have resulted in negative results (Ge et al., 2020, Rimoldi et al., 2020a, Rimoldi et al., 2020b, Wang et al., 2020c, Westhaus et al., 2021) albeit the RNA molecules of the virus have been shown to be highly persistent, even up to 50 days in wastewater at room temperature (Bivins et al., 2020). Concerns have also been raised due to the ecological risks associated to an excessive use of disinfectants for wastewater, such as the sodium hypochlorite that produces high levels of disinfection by-product residuals (Zhang et al., 2020a).
4.4. Effects of wastewater holding
Solvents, detergents and disinfectants, normally present in wastewater, can compromise the viral envelope of CoVs (see Table 4). In addition, during the biological treatment stage in WWTPs, the presence of antagonistic microorganisms can enhance the inactivation rates of many viruses (John and Rose, 2005).
As mentioned above, the assay of bacteriophages provides a suitable indicator for the fate of enveloped viruses in sewages (Adcock et al., 2009, Worley-Morse et al., 2019, Ye et al., 2016). The spontaneous inactivation of bacteriophage Φ6 was considered as a potential model for the survival and inactivation of enveloped human viruses. This virus undergoes inactivation of 5-log in 2 and 6 days at 22 and 30 °C, respectively. Longer holding times, for precaution, should be adopted at lower temperatures (Casanova and Weaver, 2015). Even for a more aggressively enveloped virus, such as Ebola, WHO recommended holding wastewater in a reservoir for one week prior to further handling or transport. Holding wastewater helped attenuate the viral activity (WHO, 2015).
Without disinfection treatments, SARS-CoV-1 have been shown to preserve its infectivity in municipal and hospital wastewater up to 2 days at 22 °C and for over 14 days at 4 °C (Wang et al., 2005b). A reduced persistence of the virus was observed in hospital wastewater where is typically present a high content of disinfectants (Wang et al., 2005b, Wang et al., 2005c). Sewage from two hospitals with SARS patients in Beijing, China was examined by Wang et al. (2005c). Electropositive filter media were used to concentrate the virus in the sample, whereas cell culture and RT-PCR techniques were utilized to verify viable virions and detect the amount of virus. The viral RNA was detected even 8 days after disinfection with chlorine, though the virus itself was inactive. SARS-CoV-1 was inactivated by the high concentration of disinfectants used in hospitals. The total residual chlorine was in the range of 0–1.0 mg/L and 3.0–12.5 mg/L, and the free residual chlorine was 0–0.5 mg/L and 1.5–5.0 mg/L, respectively for the two hospitals studied. Similar resistance was observed for SARS-CoV-2 in wastewater at room temperature, where up to 6.5 days were necessary for an attenuation of 2-log of the titer of viable virus (Bivins et al., 2020). On the contrary for a reduction of 2-log the RNA content of the virus in wastewater was found necessary over than 50 days.
Notwithstanding the limited resistance of the SARS-CoV-2 in sewages, fecal-to-oral route and aerosolization of these media could concur in the spreading of the virus. The aerosolizations of infected urines and feces from sewage pipelines, as well as during the washing of urinals and toilettes, have been indicated as potential routes of SARS-CoV-2 transmission (Ding et al., 2020, El Baz and Imziln, 2020, Ji-Xiang et al., 2020, McDermott et al., 2020, Patel, 2020, Yun-yun et al., 2020).
As matter of fact, this route of transmission was found effective for the spreading of SARS-CoV-1 in 2003 in Amoy Gardens, a private housing estate in Hong Kong (Hung, 2003, McKinney et al., 2006, WHO, 2003).
4.5. Sedimentation and inactivation in bioreactors
Suspended solids and particulate organic matter in both water and wastewater provide contribute to the physical protection of viruses, which can prolong the infectivity of CoVs (Chaudhry et al., 2015, Gundy et al., 2008). When the virus is adsorbed on the porous surface of the particulate, probably, it is sterically protected from the attack of antagonistic microorganisms. In fact, the high levels of suspended solids and organic matter in primary wastewater guarantee prolonged viral infectivity with respect to secondary wastewater effluents. However, the removal of suspended solids together with the adsorbed viruses by sedimentation ensures the reduction of infectivity. As a reference, CoVs inactivation in filtered tap water resulted greater than that in unfiltered samples (Gundy et al., 2008).
Membrane bioreactors (MBRs) have an important role in the removal of particulate matter, including viruses. Enveloped viruses can be effectively inactivated in MBRs (Bodzek et al., 2019, Chaudhry et al., 2015, Jumat et al., 2017, Lv et al., 2006, Simmons et al., 2011). Suspended solids and viruses can be retained using membrane filtration in the presence of antagonist microorganisms and adverse physicochemical conditions (e.g., aeration and chemical dosing) in the MBRs; this retention leads to the efficient inactivation of enveloped viruses, such as CoVs (Bodzek et al., 2019, Chaudhry et al., 2015, Lv et al., 2006; Naddeo et al., 2020).
5. SARS-CoV-2 in wastewater as an epidemiological tool
5.1. Wastewater-based epidemiology of SARS-CoV-2: current status
The concentration of substances stable in wastewater, excreted by humans or associable to their activities, can be used to back-estimate their initial occurrence in the serviced population. This constitutes the basis of the concept of the wastewater-based epidemiology (WBE). WBE has been utilized as an instrument for the real-time generation of information on the consumption and abuse of drugs, both legal and illegal, in a population (Lorenzo and Picó, 2019, Polo et al., 2020). WBE approach can be extended to other challenging purposes, such as the determination of the level of exposure of a population to some chemical and biological agents, such as pesticides, pollutants and pathogens, or even for gaining information on the incidence of specific diseases, such as diabetes, allergies and cancer. A consistent epidemiological surveillance in WWTPs, even in the time of inconspicuous infections, constitutes a sensitive tool for monitoring pathogen circulations in the public. This approach has been proposed both for bacteria (Diemert and Yan, 2019) as well as for viruses (Berchenko et al., 2017).
Viruses, such as SARS-CoV-2, cannot replicate in wastewater, thus their concentrations reflect the number of infected subjects in a population. Generally, there is a short time (from a few hours to a few days) during which the water is detained in the sanitation network. For this, the molecular vestiges of SARS-CoV-2 (RNA and proteins) in the wastewater arriving at a WWTP provide a snapshot of the number of people infected in the population associated with the sanitation system.
The potential utility of the WBE for SARS-CoV-2 monitoring has been demonstrated in a number of studies. The presence of RNA traces of SARS-CoV-2 has been detected in sewage in Amersfoort, the Netherlands before the emergence of cases with the symptoms of COVID-19 on March 5, 2020 (Medema et al., 2020a). Wu et al. (2020c) first correlated the SARS-CoV-2 titers in raw wastewater with the number of confirmed COVID-19 cases in Massachusetts, USA. These results showed that the actual concentration of SARS-CoV-2 genomes in wastewater was higher than the calculated concentration from the number of positive cases and from the rate of shedding of the virus in the fecal matter of the virus-positive patients. Wurtzer et al., (2020) also conducted a time-course survey of the concentration of SARS-CoV-2 genomes and correlated the quantitative data with the progress of the pandemic in Paris, France. The reported results showed that SARS-CoV-2 genomes were already detected in wastewater prior to the exponential growth of positive COVID-19 cases (Wurtzer et al., 2020). Similar results were reported in the study by La Rosa et al. (2020b), where the SARS-CoV-2 genome was detected in wastewater influents in Milan located in Lombardy region in Italy during February 2020, a period when there was still a limited number of cases in the region. A subsequent in-depth investigation by La Rosa et al., 2020c, La Rosa et al., 2021 on the detection of the SARS-CoV-2 in wastewater samples collected between October 2019 and February 2020, previously the first reported COVID-19 case in Italy detected in February 21, 2020, showed that the virus was likely to be circulating in Italy since December 2019. These investigations indicate that the wastewater-based epidemiology (WBE) approach can be effectively applied for the early revelation of the circulation of CoVs, as well as for the detection of asymptomatic infected shedders.
Randazzo et al., 2020c, Randazzo et al., 2020b found that the genome of SARS-CoV-2 was detected in wastewaters from some municipalities in the Region of Murcia (Spain) 12–16 days prior to the first confirmed COVID-19 case. Aside from using raw wastewater, Peccia et al., 2020a, Peccia et al., 2020b proposed the use of wastewater sludge as a leading indicator of community outbreak dynamics. As already mentioned, enveloped viruses such as SARS-CoV-2 tend to be adsorbed on or otherwise retained by colloidal and high molecular weight fractions of effluent organic matter, notably abundant in wastewater. Kitamura et al. (2021) showed that SARS-CoV-2 RNA concentrations in the solid fraction were higher than in the corresponding supernatant of wastewater samples. The concentration in the solid fraction ranged from 1.6 × 102 to 1.3 × 104 gene copies/L, while very low concentrations or mostly no detection of the SARS-CoV-2 RNA were observed in the supernatant. Similar results were obtained by Graham et al. (2021), where the SARS-CoV-2 N1 target and SARS-CoV-2 N2 target concentrations, on a per mass basis, were higher (N1: ~ 100, N2: ~ 1000 times higher) in the primary settled solids than in the corresponding influent samples of wastewater treatment plants. As discussed in Section 2.2, enveloped viruses were shown to be adsorbed to wastewater solids in equilibrium. The tendency of the SARS-CoV-2 to be adsorbed to the solid fraction leads to the accumulation of the virus in the raw sludge generated in wastewater treatment systems, making sludge suitable for monitoring SARS-CoV-2 shed by infected persons in the corresponding community. Peccia et al. (2020a) noted that the detection of significant SARS-CoV-2 concentrations in primary sludge preceded hospitalization reports by 1–4 days and test reports by approximately 1 week. The study suggested that this advanced information can be helpful in communities that experience delays in reporting of SARS-CoV-2 tests (in suspected cases) results. Aside from the quantitative data, another important information that can be gleaned from the presence of SARS-CoV-2 RNA in wastewater is the strains of the virus that is circulating in the population. Phylogenetic analysis done by Nemudryi et al., 2020a, Nemudryi et al., 2020b, helped determine the dominant strains of SARS-CoV-2 present in the wastewater, which reflects strain circulating in Bozeman, Montana (USA). The virus strains were found to be closely related to those circulating in California, USA and Victoria, Australia. Rimoldi et al., 2020a, Rimoldi et al., 2020b sequenced a SARS-CoV-2 genome isolated from wastewater in Milan and found that the strain of the virus is of the same origin as those strains dominantly found in Europe. Crits-Cristoph et al. (2020) demonstrated that genome sequencing of SARS-CoV-2 from wastewater can provide evidence of viral strains introduced from outside a region before they are detected through local patient-based sequencing. These studies show that sequencing of the SARS-CoV-2 virus RNA in wastewater can show the pattern of spread of the disease regionally and globally.
The discussed studies demonstrate the potential of WBE-based surveillance of SARS-CoV-2 in wastewater or sludge to determine the level of circulation of viruses in the corresponding community, and the pattern of spread of the disease among communities. WBE also has the potential to act as an early warning tool that predicts the emergence or re-emergence of a disease outbreak. The detection of higher viral concentration in wastewater samples as compared to the expected value based on the number of confirmed positive cases may also indicate that undetected asymptomatic cases are prevalent in the affected area. A study recently measured the SARS-CoV-2 concentration in the wastewaters of a commercial passenger aircraft and a cruise ship. The latter study suggested that data on the virus RNA concentration in wastewater systems can be used not only to determine the viral load of communities but also as an additional tool for prioritization of clinical testing and contact tracing (Ahmed et al., 2020b). Thus, the knowledge of the concentrations of SARS-CoV-2 in wastewater/sanitation systems may be utilized as a complementary tool to help in determining decisions related to public health.
However, the estimation of the number of infected persons in a community from the viral titers in wastewater involves uncertainties from the assumptions in the calculations. This includes the uncertainties in the used rate of viral shedding in the feces of infected persons. Zhang et al., (2020a) reported a maximum SARS-CoV-2 shedding rate of 105.8 copies/mL fecal sample of infected patients. On the other hand, Jeong et al. (2020) reported a range of 100.85 to 101.49 SARS-CoV-2 genome copies/mL of stool samples from COVID-19 positive patients (Jeong et al., 2020). It can be noted that not all stool samples from COVID-19 patients are positive for SARS-CoV-2, indicating that not all infected persons shed the virus in their feces (Wang et al., 2020c). The use of information on SARS-CoV-2 viral load in untreated wastewater should also consider the flow rate since variations due to rainfall or other weather conditions can significantly impact the resulting viral titers (Sims and Kasprzyk-Hordern, 2020). The collection of grey and rainwater together with human stools and urine in wastewater sanitation network affects the concentration of viruses and, in turn, the potential association of the virus concentration with the number of infected persons for epidemiological analyses.
These uncertainties must be accounted for in WBE-based models used to correlate the SARS-CoV-2 viral loads in wastewater or in sludge and the prevalence of COVD-19 cases in corresponding communities. This will ensure that accurate information from WBE, combined with clinical testing data, can be used as a tool to help determine approaches needed to manage the pandemic.
5.2. Wastewater-based epidemiology of SARS-CoV-2: limits, challenges and perspectives
The precision of the correlation between the number of infected people and the content of viral RNA in wastewater is influenced by numerous factors. First of these factors is the correct determination of the viral content in wastewater, as extensively discussed in the previous sections of this article. In turn, the viral genome content in wastewater can be influenced by external factors, such as dilution due to the conveyance of rainwater or by seasonal and periodic variations of grey and industrial water in the influx. The knowledge of the degree of dilution as a result of rainfall and a periodic monitoring of the wastewater flow, followed by a statistical analysis and computer modelling, would allow to minimize these effects. Alternatively, it is possible to normalize the viral concentration data with respect to an "internal standard" in wastewater. Such standard can be a chemical compound, resistant in wastewater, whose modality of release is known and, for this reason, it can act as an indicative analyte that can be related to the concentration of the virus. For this purpose, some molecules present in foods, drugs, cosmetics and personal care products, whose consumption by the population is well predictable, can be used. Caffeine, nicotine, creatinine, cholesterol, coprostanol, cortisol, 5-hydroxyindoleacetic acid (metabolite of the serotonin) and androstenedione have been proposed as endogenous and exogenous biomarkers for those applications (Polo et al., 2020, Sims and Kasprzyk-Hordern, 2020).
The detection of SARS-CoV-2 in wastewater could be even anticipated. Gallardo-Escárate et al. (2020) have associated modifications of the microbiome in wastewater, preceding the SARS-CoV-2 detection, to the early stage of gastrointestinal manifestations in COVID-19 cases. The microbiome in wastewater could be applied as an indicator for SARS-CoV-2 surveillance.
Other significant sources of uncertainty on the viral load in wastewater are the titer losses produced during sample acquisition and processing steps (Ahmed et al., 2020a). Notwithstanding the RNA of SARS-CoV-2 was found to be persistent in this media with a known decay rate (Ahmed et al., 2020c, Bivins et al., 2020), a study by Ahmed et al. (2020c) showed that storage temperature can affect the final determination of the loading of the virus in the sample. Hence, the storage of the sample at 4 °C and the subsequent rapid analysis was suggested in order to limit side effects of the decomposition of the virus. These aspects should be considered also during the phase of sampling and transport to the laboratory for analysis. WWTPs are often equipped with automated samplers that allow the withdrawal of aliquots at predetermined periods for the obtainment of reproducible composite specimens with predetermined characteristics. Unfortunately, this WWTP equipment is less typically refrigerated with resulting limitation of the sample stability. In fact, Hart and Halden (2020), showed that seasonal changes in temperature is also a source of uncertainty in the detection and quantification of SARS-CoV-2 RNA in wastewater, which subsequently affects the calculation of the viral load in the corresponding community.
The most difficult factor to address in the context of WWTP-based epidemiological observations is to determine the number of infected people associable to the wastewater (Peccia et al., 2020a). In an infected population, in addition to the confirmed COVID-19 cases, i.e. persons with symptoms of the disease, there is an unknown number of asymptomatic infected people. Efforts have been made by the countries affected by the pandemic to identify the active cases, but few investigations have established the exact level of circulation of the virus in a population, as performed by Lavezzo et al., 2020b, Lavezzo et al., 2020a via extensive investigation of nasopharyngeal swabs of the population exposed to SARS-CoV-2 in the municipality of Vo’, a small town in Veneto, Italy. The assessment of this parameter is complicated by the number of asymptomatic cases infected by the virus, largely excluded from the counting of the infected persons. The concentration of SARS-CoV-2 RNA in sewage samples therefore has the potential to reflect the actual circulation of the virus in the population since even the viruses shed by undocumented asymptomatic individuals are measured. Investigations on hospital sewages can help to overcome the issue of the association of the number of infect people with the SARS-CoV-2 concentration in wastewater. The number of COVID-19 cases in a hospital can be known with certainty and thus associable to the viral load in the relate sewage. However, this correlation needs further refinement for real epidemiological applications in a population wider than that of the number of cases in a hospital. However, the virus excretion from asymptomatic infect people differs from than of COVID-19 active cases and complicates the projection of such indicator in a “real” population different from that of a hospital (Cardillo et al., 2021, Hasanoglu et al., 2020).
6. Final remarks
-
1.
All coronaviruses, including the recently emerged SARS-CoV-2, present a limited persistence in water media: 2–5 days in tap water and 2–6 days in (frozen and thawed) wastewater were found sufficient for 2-log reduction of SARS-CoV-2 titer.
-
2.
The assay of viable virus particles provides crucial information on the potential infectivity of a contaminated specimen. Several studies focused the attention exclusively on the presence of molecular vestiges of virus and excluded the investigation on the number of viable virus particles in contaminated wastewater; that resulted in the increasing perception of the level of risk associate. On the contrary, only a few studies have examined the infectivity (in terms of viable virions) of SARS-CoV-2 in influents and effluents of municipal WWTPs and in hospital wastewater. The examination of the infectivity of wastewater contaminated with SARS-CoV-2 needs to be further investigated.
-
3.
An increasing number of studies have demonstrated that coronaviruses are transmitted primarily via airborne pathways and the hazards associated to the possible water-mediated transmission of the SARS-CoV-2 appear to be of low epidemiological significance.
-
4.
Transmission of SARS-CoV-2 could be however possible via fomites, fecal-oral route and aerosolization of infected sewages from urinals, toilets and sewage pipeline. Therefore, the precautionary alerts raised by the scientific community since the detection of SARS-CoV-2 in water media and in particular in wastewater still deserve attention and further detailed examinations.
-
5.
SARS-CoV-2 is characterized by a fragile structure and is susceptible to conventional disinfection methods that have been shown to be highly effective for its inactivation. Ca. 5 min of exposure to sodium hypochlorite (1%), ethanol (70%), iodine (7.5%), soap solution and other common disinfectants were sufficient for affording 7–8-log of SARS-CoV-2 titer reduction. Thermal inactivation, like the pasteurization process, is effective in SARS-CoV-2 deactivation: 30 min at 56 or 5 min at 70 °C were sufficient for achieving the complete depletion of the infectivity. SARS-CoV-2 is susceptible to sunlight and rapidly inactivated by UV radiation. UV-C with wavelength of 254 nm and intensity of 2.2 mW/cm2 affords 3-log of SARS-CoV-2 titer reduction in less than 3 s of irradiation.
-
6.
Excessive utilization of disinfectants has been associated with environmental and human health issues, therefore for SARS-CoV-2 disinfection conventional doses of disinfectants are recommended for sanitation and for wastewater treatment.
-
7.
Detection and quantification of virus in wastewater constitutes a powerful tool for the wastewater-based epidemiological applications and for the preventive identification of hotspots of virus resurgence. Unlike other pathologies, in case of SARS-CoV-2 outbreak the rapid resurgence of the virus needs a consistent routine of online detection (preferably based on a high frequency of sampling combined with a rapid detection method) which is necessary for the implementation of WBE and generation of reliable data with which to track the spread of viruses.
-
8.
Further studies are necessary to enhance the sensitivity and the rapidity in the detection of SARS-CoV-2, to improve the efficiency of sample preparation, virus concentration and molecular techniques of amplification of the viral genomic material.
-
9.
The development of a consistent, well documented and tested protocol for SARS-CoV-2 quantification in wastewater is needed. Various and dissimilar methods of virus concentration and detection have been applied worldwide. For the sake of comparison of the data and for prospective WBE applications, it is necessary to adopt a robust quantitative and widely available analytical method.
-
10.
Sampling (e.g. representative site, type of sampling, storage of samples) and processing (e.g. concentration and quantification methods) affect the measure of the concentration of coronaviruses included SARS-CoV-2 in wastewater. This is necessary to exploit and implement the significant potential of the WBE as an effective tool of surveillance. 24 h composite sampling and replicated analyses are recommended for robust and consistent results.
-
11.
Alternative analytical methods, based on technologies different from the molecular amplification of the SARS-CoV-2 RNA, are currently emerging. These methods could improve the rapidity, the cost-efficacy relation and extend the number of rapidly processable samples.
-
12.
The WBE-derived correlations between levels of contagion in a population and virus loading in the wastewater produced by the exposed population should be refined and made more accurate. Virus concentrations in wastewater are susceptible to dilution effects; thus, the results must be normalized with respect to a specific population, opportune bio-markers and networking conditions by using numerical models.
-
13.
The progress that can be reached during this pandemics in the WBE-based monitoring can be extended to other applications and monitoring of pathologies and diseases, such as diabetes, obesity and hypertension.
-
14.
An accurate knowledge of the behavior of virus in the environment can be implemented in a precise and informative model that can also use WBE-derived data. Such model can help to develop technological approaches and practical policies needed to mitigate the consequences on public health and economic caused by the ongoing virus epidemics or other potentially possible outbreaks.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors would like express sincere gratitude to: i) the University of Salerno (FARB Grants: ORSA11328, 300393FRB18NADDE and 300393FRB17NADDE); ii) the Inter-University Centre for Prediction and Prevention of Relevant Hazards (Centro Universitario per la Previsione e Prevenzione Grandi Rischi, C.U.G.RI.); iii) Italian National Institute of Health (Istituto Superiore di Sanità) for the Italian Environmental Surveillance of Wastewaters Project (Sorveglianza Ambientale Reflue in Italia, SARI); iv) the National Research Foundation of Korea (Grant no. 2019R1H1A2080148); v) the Center for Membranes and Advanced Water Technology (CMAT, Abu Dhabi, UAE).
Editor: Prof. Debora Rodriguez
References
- Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food Environ. Virol. 2014;6:253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adcock N.J., Rice E.W., Sivaganesan M., Brown J.D., Stallknecht D.E., Swayne D.E. The use of bacteriophages of the family Cystoviridae as surrogates for H5N1 highly pathogenic avian influenza viruses in persistence and inactivation studies. J. Environ. Sci. Health Part A. 2009;44:1362–1366. doi: 10.1080/10934520903217054. [DOI] [PubMed] [Google Scholar]
- Adelodun B., Ajibade F.O., Ibrahim R.G., Bakare H.O., Choi K.-S. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: any sustainable preventive measures to curtail the scourge in low-income countries? Sci. Total Environ. 2020;742 doi: 10.1016/j.scitotenv.2020.140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27:27. doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan Kocamemi, B., Kurt, H., Sait, A., Sarac, F., Saatci, A.M., Pakdemirli B., 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv. 〈10.1101/2020.05.12.20099358〉.
- Amoah I.D., Kumari S., Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020;143 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampuero, M., Valenzuela, S., Valiente-Echeverria, F., Soto-Rifo, R., Barriga, G.P., Chnaiderman, J., et al., 2020. SARS-CoV-2 Detection in Sewage in Santiago, Chile – Preliminary Results. medRxiv. 〈10.1101/2020.07.02.20145177〉.
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, S., Nag, A., Sethi, J., Rajvanshi, J., Saxena, S., Shrivastava, S.K., et al., 2020. Sewage Surveillance for the Presence of SARS-CoV-2 Genome as A Useful Wastewater Based Epidemiology (WBE) Tracking Tool in India. medRxiv. 〈10.1101/2020.06.18.20135277〉. [DOI] [PubMed]
- Arslan M., Xu B., Gamal El-Din M. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi S., Bouvier N., Wexler A.S., Ristenpart W.D. The coronavirus pandemic and aerosols: does COVID-19 transmit via expiratory particles? Aerosol Sci. Technol. 2020;54:635–638. doi: 10.1080/02786826.2020.1749229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi S., Gaaloul ben Hnia N., Barre R.S., Wexler A.S., Ristenpart W.D., Bouvier N.M. Influenza A virus is transmissible via aerosolized fomites. Nat. Commun. 2020;11:4062. doi: 10.1038/s41467-020-17888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auler A.C., Cássaro F.A.M., da Silva V.O., Pires L.F. Evidence that high temperatures and intermediate relative humidity might favor the spread of COVID-19 in tropical climate: a case study for the most affected Brazilian cities. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.139090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer A., Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J. Vis. Exp. 2014 doi: 10.3791/52065. e52065-e52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa, S., Mauricio-Iglesias, M., Rodríguez, S., Martínez-Lamas, L., Vasallo, F.J., Regueiro, B., et al., 2020. The fate of SARS-CoV-2 in Wastewater Treatment Plants Points Out the Sludge Line as A Suitable Spot for Incidence Monitoring. medRxiv. 〈10.1101/2020.05.25.20112706〉. [DOI] [PMC free article] [PubMed]
- Bar Or, I., Yaniv, K., Shagan, M., Ozer, E., Erster, O., Mendelson, E., et al., 2020. Regressing SARS-CoV-2 Sewage Measurements onto COVID-19 Burden in the Population: A Proof-of-Concept for Quantitative Environmental Surveillance. medRxiv. 〈10.1101/2020.04.26.20073569〉. [DOI] [PMC free article] [PubMed]
- Barceló D. Wastewater-based epidemiology to monitor COVID-19 outbreak: present and future diagnostic methods to be in your radar. Case Stud. Chem. Environ. Eng. 2020;2 doi: 10.1016/j.cscee.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchenko Y., Manor Y., Freedman L.S., Kaliner E., Grotto I., Mendelson E., Huppert A. Estimation of polio infection prevalence from environmental surveillance data. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf6786. [DOI] [PubMed] [Google Scholar]
- Bhardwaj R., Agrawal A. Tailoring surface wettability to reduce chances of infection of COVID-19 by a respiratory droplet and to improve the effectiveness of personal protection equipment. Phys. Fluids. 2020;32 doi: 10.1063/5.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick G.D., Dhar D., Nath D., Ghangrekar M.M., Banerjee R., Das S., Chatterjee J. Coronavirus disease 2019 (COVID-19) outbreak: some serious consequences with urban and rural water cycle. npj Clean Water. 2020;3:32. doi: 10.1038/s41545-020-0079-1. [DOI] [Google Scholar]
- Bianco, A., Biasin, M., Pareschi, G., Cavalleri, A., Cavatorta, C., Fenizia, C., et al., 2020. UV-C Irradiation Is Highly Effective in Inactivating and Inhibiting SARS-CoV-2 Replication. medRxiv. 〈10.1101/2020.06.05.20123463〉. [DOI] [PMC free article] [PubMed]
- Bilal M., Nazir M.S., Rasheed T., Parra-Saldivar R., Iqbal H.M.N. Water matrices as potential source of SARS-CoV-2 transmission – an overview from environmental perspective. Case Stud. Chem. Environ. Eng. 2020;2 doi: 10.1016/j.cscee.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodzek M., Konieczny K., Ra M. Membranes in water and wastewater disinfection: review. Arch. Environ. Prot. 2019;45:3–18. [Google Scholar]
- Bofill-Mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A., Packman A., Furman A., Gross A., Kushmaro A., Ronen A., Dagot C., Hill C., Vaizel-Ohayon D., Morgenroth E., Bertuzzo E., Wells G., Kiperwas H.R., Horn H., Negev I., Zucker I., Bar-Or I., Moran-Gilad J., Balcazar J.L., Bibby K., Elimelech M., Weisbrod N., Nir O., Sued O., Gillor O., Alvarez P.J., Crameri S., Arnon S., Walker S., Yaron S., Nguyen T.H., Berchenko Y., Hu Y., Ronen Z., Bar-Zeev E. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020;3:981–990. doi: 10.1038/s41893-020-00605-2. [DOI] [Google Scholar]
- Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA. 2020;323:1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo L., Martinis C.d., Viscardi M., Esposito C., Sannino E., Lucibelli G., et al. SARS-CoV-2 quantitative real time PCR and viral loads analysis among asymptomatic and symptomatic patients: an observational study on an outbreak in two nursing facilities in campania region (Southern Italy) Res. Sq. 2021 doi: 10.21203/rs.3.rs-139370/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., Cerullo V., Libralato G., Galdiero E., Guida M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana G., Croxatto A., Coste A.T., Opota O., Lamoth F., Jaton K., Greub G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 2020;26:1178–1182. doi: 10.1016/j.cmi.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. doi: 10.1021/acs.estlett.5b00029. [DOI] [Google Scholar]
- Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- Chan K.H., Poon L.L.L.M., Cheng V.C.C., Guan Y., Hung I.F.N., Kong J., Yam L.Y.C., Seto W.H., Yuen K.Y., Peiris J.S.M. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 2004;10:294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Malik Peiris J.S., Lam S.Y., Poon L.L.M., Yuen K.Y., Seto W.H. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011;2011:1–7. doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry R.M., Nelson K.L., Drewes J.E. Mechanisms of pathogenic virus removal in a full-scale membrane bioreactor. Environ. Sci. Technol. 2015;49:2815–2822. doi: 10.1021/es505332n. [DOI] [PubMed] [Google Scholar]
- Chavarria-Miró, G., Anfruns-Estrada, E., Guix, S., Paraira, M., Galofré, B., Sáanchez, G., et al., 2020. Sentinel Surveillance of SARS-CoV-2 in Wastewater Anticipates the Occurrence of COVID-19 Cases. medRxiv. 〈10.1101/2020.06.13.20129627〉.
- Chen Z., Wu Q., Chen J., Ni X., Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 2020;35:351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-J., Woo H.-M., Kim K.-S., Oh J.-W., Jeong Y.-J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Breshears L.E., Perea S., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.Y. Smartphone-based paper microfluidic particulometry of norovirus from environmental water samples at the single copy level. ACS Omega. 2019;4:11180–11188. doi: 10.1021/acsomega.9b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collomb J., Laporte J., Vautherot J.F., Schwartzbrod L. Research on coronaviruses in water. I. Adsorption and elution of the coronavirus on glass powder. Virologie. 1986;37:95–105. [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph, A., Kantor, R.S., Olm, M.R., Whitney, O.N., Al-Shayeb, B., Lou, Y.C., et al., 2020. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. medRxiv. 〈10.1101/2020.09.13.20193805〉. [DOI] [PMC free article] [PubMed]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, K., Keeling, D., Yetka, K., Larson, A., Gonzalez, R., 2020. Wastewater SARS-CoV-2 Concentration and Loading Variability from Grab and 24-Hour Composite Samples. medRxiv. 〈10.1101/2020.07.10.20150607〉.
- da Silva Queiroz J.A., de Cássia Pontello Rampazzo R., da Silva Filho E.B., Oliveira G.S., da Costa Oliveira S., Botelho Souza L.F., et al. Development of a quantitative one-step multiplex RT-qPCR assay for the detection of SARS-CoV-2 in a biological matrix. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust, P.M., Mercier, E., Montpetit, D., Jia, J.-J., Alexandrov, I., Neault, N., et al., 2020. Quantitative Analysis of SARS-CoV-2 RNA from Wastewater Solids in Communities with Low COVID-19 Incidence and Prevalence. medRxiv. 〈10.1101/2020.08.11.20173062〉. [DOI] [PMC free article] [PubMed]
- Darnell M.E.R., Taylor D.R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion. 2006;46:1770–1777. doi: 10.1111/j.1537-2995.2006.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods. 2004;121:85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020;244 doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiana M., Mori A., Piubelli C., Scarso S., Favarato M., Pomari E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020;10:18764. doi: 10.1038/s41598-020-75958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemert S., Yan T. Clinically unreported Salmonellosis outbreak detected via comparative genomic analysis of municipal wastewater Salmonella isolates. Appl. Environ. Microbiol. 2019;85:e00139–19. doi: 10.1128/aem.00139-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z., Qian, H., Xu, B., Huang, Y., Miao, T., Yen, H.-L., et al., 2020. Toilets Dominate Environmental Detection of SARS-CoV-2 Virus in A Hospital. medRxiv. 〈10.1101/2020.04.03.20052175〉. [DOI] [PMC free article] [PubMed]
- Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.576622. 576622-576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S.-M., Zhao X.-S., Wen R.-F., Huang J.-J., Pi G.-H., Zhang S.-X., et al. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed. Environ. Sci. BES. 2003;16:246–255. [PubMed] [Google Scholar]
- Dunkin N., Weng S., Schwab K.J., McQuarrie J., Bell K., Jacangelo J.G. Comparative inactivation of murine norovirus and MS2 bacteriophage by peracetic acid and monochloramine in municipal secondary wastewater effluent. Environ. Sci. Technol. 2017;51:2972–2981. doi: 10.1021/acs.est.6b05529. [DOI] [PubMed] [Google Scholar]
- Eggers M., Eickmann M., Zorn J. Rapid and effective virucidal activity of povidone-iodine products against Middle East respiratory syndrome coronavirus (MERS-CoV) and modified vaccinia virus Ankara (MVA) Infect. Dis. Ther. 2015;4:491–501. doi: 10.1007/s40121-015-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baz S., Imziln B. Can aerosols and wastewater be considered as potential transmissional sources of COVID-19 to humans? Eur. J. Environ. Public Health. 2020;4 [Google Scholar]
- EPA, 2020. Coronavirus and Drinking Water and Wastewater. 〈https://www.epa.gov/coronavirus/coronavirus-and-drinking-water-and-wastewater〉.
- Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C., Scalia G., Libra M., Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. 957-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears A.C., Klimstra W.B., Duprex P., Hartman A., Weaver S.C., Plante K.S., Mirchandani D., Plante J.A., Aguilar P.V., Fernández D., Nalca A., Totura A., Dyer D., Kearney B., Lackemeyer M., Bohannon J.K., Johnson R., Garry R.F., Reed D.S., Roy C.J. Persistence of severe acute respiratory syndrome coronavirus 2 in aerosol suspensions. Emerg. Infect. Dis. 2020;26:2168–2171. doi: 10.3201/eid2609.201806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Xu K., Gu S., Zheng S., Zou Q., Xu Y., Yu L., Lou F., Yu F., Jin T., Li Y., Sheng J., Yen H.L., Zhong Z., Wei J., Chen Y. Multi-route transmission potential of SARS-CoV-2 in healthcare facilities. J. Hazard. Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro, G., Hermes Stoco, P., Sobral Marques Souza, D., Grisard, E.C., Magri, M.E., Rogovski, P., et al., 2020. SARS-CoV-2 in Human Sewage in Santa Catalina, Brazil, November 2019. medRxiv. 〈10.1101/2020.06.26.20140731〉. [DOI] [PMC free article] [PubMed]
- Gallardo-Escárate C., Valenzuela-Muñoz V., Núñez-Acuña G., Valenzuela-Miranda D., Benaventel B.P., Sáez-Vera C., et al. The wastewater microbiome: a novel insight for COVID-19 surveillance. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.142867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T., Lu Y., Zheng S., Zhuo L., Yu L., Ni Z., Zhou Y., Ni L., Qu T., Zhong Z. Evaluation of disinfection procedures in a designated hospital for COVID-19. Am. J. Infect. Control. 2020 doi: 10.1016/j.ajic.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller C., Varbanov M., Duval R.E. Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses. 2012;4:3044–3068. doi: 10.3390/v4113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortner R.A. Viruses—living or non-living? Science. 1938;87:529–530. doi: 10.1126/science.87.2267.529. [DOI] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gralton J., Tovey E.R., McLaws M.-L., Rawlinson W.D. Respiratory virus RNA is detectable in airborne and droplet particles. J. Med. Virol. 2013;85:2151–2159. doi: 10.1002/jmv.23698. [DOI] [PubMed] [Google Scholar]
- Green, H., Wilder, M., Middleton, F.A., Collins, M., Fenty, A., Gentile, K., et al., 2020. Quantification of SARS-CoV-2 and Cross-Assembly Phage (crAssphage) from Wastewater to Monitor Coronavirus Transmission within Communities. medRxiv. 〈10.1101/2020.05.21.20109181〉.
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2008;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces – a rapid review. Colorectal Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwenzi W. Leaving no stone unturned in light of the COVID-19 faecal-oral hypothesis? A water, sanitation and hygiene (WASH) perspective targeting low-income countries. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanoglu I., Korukluoglu G., Asilturk D., Cosgun Y., Kalem A.K., Altas A.B., Kayaaslan B., Eser F., Kuzucu E.A., Guner R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection. 2020;49:117–126. doi: 10.1007/s15010-020-01548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata, A., Honda, R., Hara-Yamamura, H., Meuchi, Y., 2020. Detection of SARS-CoV-2 in Wastewater in Japan by Multiple Molecular Assays-Implication for Wastewater-Based Epidemiology (WBE). medRxiv. 〈10.1101/2020.06.09.20126417〉.
- Heilingloh C.S., Aufderhorst U.W., Schipper L., Dittmer U., Witzke O., Yang D., Zheng X., Sutter K., Trilling M., Alt M., Steinmann E., Krawczyk A. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control. 2020;48:1273–1275. doi: 10.1016/j.ajic.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Seidel M., Niessner R., Eggers J., Tiehm A. Long amplicon (LA)-qPCR for the discrimination of infectious and noninfectious phix174 bacteriophages after UV inactivation. Water Res. 2016;103:141–148. doi: 10.1016/j.watres.2016.07.032. [DOI] [PubMed] [Google Scholar]
- Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., Neumann P., Schork J., Tiarks-Jungk P., Walczok A., Eickmann M., Vehreschild M.J.G.T., Kann G., Wolf T., Gottschalk R., Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N. Engl. J. Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. 929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, P., Rachmadi, A.T., Mantilla-Calderon, D., Alkahtani, M., Bashwari, Y.M., Al Qarni, H., et al., 2020. How Early Into the Outbreak Can Surveillance of SARS-CoV-2 in Wastewater Tell Us? medRxiv. 〈10.1101/2020.08.19.20177667〉.
- Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLOS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.S. The SARS epidemic in Hong Kong: what lessons have we learned? J. R. Soc. Med. 2003;96:374–378. doi: 10.1258/jrsm.96.8.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihling C., Tänzler D., Hagemann S., Kehlen A., Hüttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- Jeong H.W., Kim S.-M., Kim H.-S., Kim Y.-I., Kim J.H., Cho J.Y., Kim S., Kang H., Kim S.G., Park S.J., Kim E.H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevšnik M., Steyer A., Zrim T., Pokorn M., Mrvič T., Grosek Š., Strle F., Lusa L., Petrovec M. Detection of human coronaviruses in simultaneously collected stool samples and nasopharyngeal swabs from hospitalized children with acute gastroenteritis. Virol. J. 2013;10:46. doi: 10.1186/1743-422X-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji-Xiang Wang, Li Yun-Yun, Liu Xiang-Dong, Cao Xiang. Virus transmission from urinals. Phys. Fluids. 2020;32 doi: 10.1063/5.0021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John D.E., Rose J.B. Review of factors affecting microbial survival in groundwater. Environ. Sci. Technol. 2005;39:7345–7356. doi: 10.1021/es047995w. [DOI] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4 doi: 10.1128/mBio.00165-13. e00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumat M.R., Hasan N.A., Subramanian P., Heberling C., Colwell R.R., Hong P.-Y. Membrane bioreactor-based wastewater treatment plant in Saudi Arabia: reduction of viral diversity, load, and infectious capacity. Water. 2017;9:534. [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariwa H., Fujii N., Takashima I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology. 2006;212(Suppl. 1):S119–S123. doi: 10.1159/000089211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kock R., Baselmans M., Scharnhorst V., Deiman B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2020 doi: 10.1007/s10096-020-04076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Mazur S., Ork B.L., Postnikova E., Hensley L.E., Jahrling P.B., Johnson R., Holbrook M.R. Inactivation and safety testing of Middle East respiratory syndrome coronavirus. J. Virol. Methods. 2015;223:13–18. doi: 10.1016/j.jviromet.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers J., Jerome K.R. Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 2017;55:1621–1628. doi: 10.1128/jcm.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., della Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanità. 2012;48:397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods – a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, G., Mancini, P., Bonanno Ferraro, G., Veneri, C., Iaconelli, M., Bonadonna, L., et al., 2020c. SARS-CoV-2 Has Been Circulating in Northern Italy Since December 2019: Evidence from Environmental Monitoring. medRxiv. 〈10.1101/2020.06.25.20140061〉. [DOI] [PMC free article] [PubMed]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since december 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.Y.Y., Cheng P.K.C., Lim W.W.L. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005;41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre A., Talbot P.J. Effect of pH and temperature on the infectivity of human coronavirus 229E. Can. J. Microbiol. 1989;35:972–974. doi: 10.1139/m89-160. [DOI] [PubMed] [Google Scholar]
- Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., Rossi L., Manganelli R., Loregian A., Navarin N., Abate D., Sciro M., Merigliano S., De Canale E., Vanuzzo M.C., Besutti V., Saluzzo F., Onelia F., Pacenti M., Parisi S.G., Carretta G., Donato D., Flor L., Cocchio S., Masi G., Sperduti A., Cattarino L., Salvador R., Nicoletti M., Caldart F., Castelli G., Nieddu E., Labella B., Fava L., Drigo M., Gaythorpe K.A.M., Brazzale A.R., Toppo S., Trevisan M., Baldo V., Donnelly C.A., Ferguson N.M., Dorigatti I., Crisanti A. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzo, E., Franchin, E., Ciavarella, C., Cuomo-Dannenburg, G., Barzon, L., Del Vecchio, C., et al., 2020b. Suppression of COVID-19 Outbreak in the Municipality of Vo, Italy. medRxiv. 〈10.1101/2020.04.17.20053157〉.
- Leifels M., Cheng D., Sozzi E., Shoults D.C., Wuertz S., Mongkolsuk S., Sirikanchana K. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications – a systematic review. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2020.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifels, M., Dan, C., Sozzi, E., Shoults, D.C., Wuertz, S., Mongkolsuk, S., et al., 2020. Capsid Integrity Quantitative PCR to Determine Virus Infectivity in Environmental and Food Applications; A Systematic Review. medRxiv. 〈10.1101/2020.05.08.20095364〉. [DOI] [PMC free article] [PubMed]
- Leung W.K., To K., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection1. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.S., Guan Y., Wang J., Smith G.J.D., Xu K.M., Duan L., Rahardjo A.P., Puthavathana P., Buranathai C., Nguyen T.D., Estoepangestie A.T.S., Chaisingh A., Auewarakul P., Long H.T., Hanh N.T.H., Webby R.J., Poon L.L.M., Chen H., Shortridge K.F., Yuen K.Y., Webster R.G., Peiris J.S.M. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health. 2019;9:77–84. doi: 10.1016/j.coesh.2019.05.007. [DOI] [Google Scholar]
- Lu C.-w., Liu X.-f., Jia Z.-f. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W., Zheng X., Yang M., Zhang Y., Liu Y., Liu J. Virus removal performance and mechanism of a submerged membrane bioreactor. Process Biochem. 2006;41:299–304. doi: 10.1016/j.procbio.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. Why does the coronavirus spread so easily between people? Nature. 2020 doi: 10.1038/d41586-020-00660-x. [DOI] [PubMed] [Google Scholar]
- Mandal P., Gupta A.K., Dubey B.K. A review on presence, survival, disinfection/removal methods of coronavirus in wastewater and progress of wastewater-based epidemiology. J. Environ. Chem. Eng. 2020;8 doi: 10.1016/j.jece.2020.104317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manupati, H., Kiran, U., Kuncha, S.K., Kopperi, H., Gokulan, C.G., Mohan, S.V., Mishra, R.K., et al., 2020. Comprehensive Surveillance of SARS-CoV-2 Spread Using Wastewater-Based Epidemiology Studies. medRxiv. 〈10.1101/2020.08.18.20177428〉. [DOI] [PMC free article] [PubMed]
- Masclaux F.G., Hotz P., Gashi D., Savova-Bianchi D., Oppliger A. Assessment of airborne virus contamination in wastewater treatment plants. Environ. Res. 2014;133:260–265. doi: 10.1016/j.envres.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Matsui S. Protecting human and ecological health under viral threats in Asia. Water Sci. Technol. 2005;51:91–97. doi: 10.2166/wst.2005.0234. [DOI] [PubMed] [Google Scholar]
- Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sens. Actuators B Chem. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott C.V., Alicic R.Z., Harden N., Cox E.J., Scanlan J.M. Put a lid on it: are faecal bio-aerosols a route of transmission for SARS-CoV-2? J. Hosp. Infect. 2020;105:397–398. doi: 10.1016/j.jhin.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health. 2006;68:26–30. [PubMed] [Google Scholar]
- Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., Brouwer, A., 2020a. Presence of SARS-Coronavirus-2 in Sewage. medRxiv. 〈10.1101/2020.03.29.20045880〉. [DOI] [PubMed]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Middleton J., Reintjes R., Lopes H. Meat plants—a new front line in the covid-19 pandemic. BMJ. 2020;370:m2716. doi: 10.1136/bmj.m2716. [DOI] [PubMed] [Google Scholar]
- Mirskaya E., Agranovski I.E. Sources and mechanisms of bioaerosol generation in occupational environments. Crit. Rev. Microbiol. 2018;44:739–758. doi: 10.1080/1040841X.2018.1508125. [DOI] [PubMed] [Google Scholar]
- Miyani B., Fonoll X., Norton J., Mehrotra A., Xagoraraki I. SARS-CoV-2 in detroit wastewater. J. Environ. Eng. 2020;146 doi: 10.1061/(ASCE)EE.1943-7870.0001830. [DOI] [Google Scholar]
- Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. IJERPH. 2020;17:5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V. Development of environmental biotechnology and control of emerging biological contaminants: the grand challenge for a sustainable future. Water Environ. Res. 2020;92:1246–1248. doi: 10.1002/wer.1439. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- Naddeo V., Secondes M.F.N., Borea L., Hasan S.W., Ballesteros Jr. F., Belgiorno V. Removal of contaminants of emerging concern from real wastewater by an innovative hybrid membrane process - UltraSound, Adsorption, and Membrane ultrafiltration (USAMe®) Ultrason. Sonochem. 2020;68 doi: 10.1016/j.ultsonch.2020.105237. [DOI] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58:e00557–20. doi: 10.1128/jcm.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neault, N., Baig, A.T., Graber, T.E., D'Aoust, P.M., Mercier, E., Alexandrov, I., et al., 2020. SARS-CoV-2 Protein in Wastewater Mirrors COVID-19 Prevalence. medRxiv. 〈10.1101/2020.09.01.20185280〉.
- Nemudryi, A., Nemudraia, A., Surya, K., Wiegand, T., Buyukyoruk, M., Wilkinson, R., et al., 2020a. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. medRxiv. 〈10.1101/2020.04.15.20066746〉. [DOI] [PMC free article] [PubMed]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem L.D., Morgan B., Donner E., Short M.D. The COVID-19 pandemic: considerations for the waste and wastewater services sector. Case Stud. Chem. Environ. Eng. 2020;1 doi: 10.1016/j.cscee.2020.100006. 100006-100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro, F., Sironi, G., Antonello, E., Bianco, A., Biasin, M., Brucato, J.R., et al., 2020. Modulation of COVID-19 Epidemiology by UV-B and -A Photons from the Sun. medRxiv. 〈10.1101/2020.06.03.20121392〉.
- Nieuwenhuijse D.F., Koopmans M.P.G. Metagenomic sequencing for surveillance of food- and waterborne viral diseases. Front. Microbiol. 2017;8:8. doi: 10.3389/fmicb.2017.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev E.N., Indeykina M.I., Brzhozovskiy A.G., Bugrova A.E., Kononikhin A.S., Starodubtseva N.L., Petrotchenko E.V., Kovalev G.I., Borchers C.H., Sukhikh G.T. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J. Proteome Res. 2020;19:4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
- Nikolaev E.N., Indeykina M.I., Brzhozovskiy A.G., Bugrova A.E., Kononikhin A.S., Starodubtseva N.L., Petrotchenko E.V., Kovalev G.I., Borchers C.H., Sukhikh G.T. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J. Proteome Res. 2020;19:4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
- OSHA, 2020. Standards and Directives for COVID-19. United States of America. Occupational Safety and Health Administration. 〈https://www.osha.gov/SLTC/covid-19/standards.html〉.
- Paleologos E.K., O’Kelly B.C., Tang Chao-Sheng, Cornell Ken, Rodríguez-Chueca J., Abuel-Naga Hossam, et al. Post COVID-19 water and wastewater management to protect public health and geoenvironment. Environ. Geotech. 2020:1–14. doi: 10.1680/jenge.20.00067. [DOI] [Google Scholar]
- Patel J. Viability of SARS-CoV-2 in faecal bio-aerosols. Colorectal Dis. 2020;22:1022. doi: 10.1111/codi.15181. 1022-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia, J., Zulli, A., Brackney, D.E., Grubaugh, N.D., Kaplan, E.H., Casanovas-Massana, A., et al., 2020b. SARS-CoV-2 RNA Concentrations in Primary Municipal Sewage Sludge As a Leading Indicator of COVID-19 Outbreak Dynamics. medRxiv. 〈10.1101/2020.05.19.20105999〉.
- Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M., Cheng V., Chan K., Tsang D., Yung R., Ng T., Yuen K. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrony O., Ellouze S., Fontaine J.-P., Thegat-Le Cam M., Salmona M., Feghoul L., Mahjoub N., Mercier-Delarue S., Gabassi A., Delaugerre C., Le Goff J., Achili Y., Ades L., Aguinaga L., Archer G., Benattia A., Bercot B., Bergeron A., Bertinchamp R., Bondeelle L., Bouaziz J.D., Bouda D., Boutboul D., Brindel Berthon I., Brugnet E., Caillat Zucman S., Cassonnet S., Celli Lebras K., Chabert J., Chaix M.L., Chevret S., Clément M., Davoine C., De Castro N., De Kerviler E., De Margerie-Mellon C., Depret F., Denis B., Djaghout L., Dupin C., Farge-Blancel D., Fauvaux C., Fenaux H., Feredj E., Feyeux D., Fremeaux-Bacchi V., Galicier L., Garestier J., Harel S., Jegu A.L., Kozakiewicz E., Lebel M Baye A., Le Guen P., Lengline E., Liegeon G., Lorillon G., Madelaine Chambrin I., Martin de Frémont G., Maylin S., Mehlman C., Meunier M., Molina J.M., Morin F., Oksenhendler E., Peffault de la Tour R., Plaud B., Rouveau M., Saussereau J., Schnepf N., Soret J., Tazi A., Tremorin M.T. Surfaces and equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the emergency department at a university hospital. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock D.H., Garwes D.J. The influence of pH on the growth and stability of transmissible gastroenteritis virus in vitro. Arch. Virol. 1975;49:239–247. doi: 10.1007/bf01317542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polston P.M., Rodríguez R.A., Seo K., Kim M., Ko G., Sobsey M.D. Field evaluation of an improved cell line for the detection of human adenoviruses in environmental samples. J. Virol. Methods. 2014;205:68–74. doi: 10.1016/j.jviromet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Poon L.L.M., Chan K.H., Wong O.K., Cheung T.K.W., Ng I., Zheng B., Seto W.H., Yuen K.Y., Guan Y., Peiris J.S.M. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Maranhão A.G., Siqueira M.M., Miagostovich M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115:115. doi: 10.1590/0074-02760200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A. Canine coronavirus inactivation with physical and chemical agents. Vet. J. 2008;177:71–79. doi: 10.1016/j.tvjl.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Rabenau H.F., Cinatl J., Morgenstern B., Bauer G., Preiser W., Doerr H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005;194:1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85. doi: 10.5501/wjv.v5.i2.85. 85-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, W., Truchado, P., Ferrando, E.C., Simon, P., Allende, A., Sanchez, G., 2020c. SARS-CoV-2 RNA Titers in Wastewater Anticipated COVID-19 Occurrence in A Low Prevalence Area. medRxiv. 〈10.1101/2020.04.22.20075200〉. [DOI] [PMC free article] [PubMed]
- Ratnesar-Shumate S., Williams G., Green B., Krause M., Holland B., Wood S., Bohannon J., Boydston J., Freeburger D., Hooper I., Beck K., Yeager J., Altamura L.A., Biryukov J., Yolitz J., Schuit M., Wahl V., Hevey M., Dabisch P. Simulated sunlight rapidly inactivates SARS-CoV-2 on surfaces. J. Infect. Dis. 2020;222:214–222. doi: 10.1093/infdis/jiaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi, S.G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., et al., 2020b. Presence and Vitality of SARS-CoV-2 Virus in Wastewaters and Rivers. medRxiv. 〈10.1101/2020.05.01.20086009〉. [DOI] [PMC free article] [PubMed]
- RIVM, 2020. Coronavirus Monitoring in Sewage Research. RIVM. 〈https://www.rivm.nl/en/covid-19/sewage〉.
- Robinson, C., Loeffelholz, M.J., Pinsky, B.A., 2016. Respiratory Viruses. Ch. 19. Respiratory Viruses. Clinical Virology Manual, pp. 255–276. 〈10.1128/9781555819156〉.
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino C.P., Sellera F.P., Sales-Medina D.F., Machado R.R.G., Durigon E.L., Freitas-Junior L.H., Ribeiro M.S. UV-C (254 nm) lethal doses for SARS-CoV-2. Photodiagn. Photodyn. Ther. 2020;32 doi: 10.1016/j.pdpdt.2020.101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar S.A., Springthorpe V.S., Karim Y., Loro P. Chemical disinfection of non-porous inanimate surfaces experimentally contaminated with four human pathogenic viruses. Epidemiol. Infect. 2009;102:493–505. doi: 10.1017/S0950268800030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuit M., Ratnesar-Shumate S., Yolitz J., Williams G., Weaver W., Green B., Miller D., Krause M., Beck K., Wood S., Holland B., Bohannon J., Freeburger D., Hooper I., Biryukov J., Altamura L.A., Wahl V., Hevey M., Dabisch P. Airborne SARS-CoV-2 is rapidly inactivated by simulated sunlight. J. Infect. Dis. 2020;222:564–571. doi: 10.1093/infdis/jiaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore V., Zarra T., Buonerba A., Choo K.-H., Hasan S.W., Korshin G., Li C.W., Ksibi M., Belgiorno V., Naddeo V. Indoor versus outdoor transmission of SARS-COV-2: environmental factors in virus spread and underestimated sources of risk. Eur-Mediterr. J. Environ. Integr. 2021;6:30. doi: 10.1007/s41207-021-00243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sharif, S., Ikram, A., Khurshid, A., Salman, M., Mehmood, N., Arshad, Y., et al., 2020. Detection of SARS-Coronavirus-2 in Wastewater, Using the Existing Environmental Surveillance Network: An Epidemiological Gateway to an Early Warning for COVID-19 in Communities. medRxiv. 〈10.1101/2020.06.03.20121426〉.
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Wen, Z., Zhong, G., Yang, H., Wang, C., Liu R., et al., 2020. Susceptibility of Ferrets, Cats, Dogs, and Different Domestic Animals to SARS-Coronavirus-2. bioRxiv. 〈10.1101/2020.03.30.015347〉. [DOI] [PMC free article] [PubMed]
- Shutler, J., Zaraska, K., Holding, T.M., Machnik, M., Uppuluri, K., Ashton, I., et al., 2020. Risk of SARS-CoV-2 Infection from Contaminated Water Systems. medRxiv. 〈10.1101/2020.06.17.20133504〉.
- Sidstedt M., Rådström P., Hedman J. PCR inhibition in qPCR, dPCR and MPS—mechanisms and solutions. Anal. Bioanal. Chem. 2020;412:2009–2023. doi: 10.1007/s00216-020-02490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Simmons F.J., Kuo D.H.W., Xagoraraki I. Removal of human enteric viruses by a full-scale membrane bioreactor during municipal wastewater processing. Water Res. 2011;45:2739–2750. doi: 10.1016/j.watres.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schafer A., Matzke M.M., Webb-Robertson B.J.M., Chang J., Luna M.L., Long C.E., Shukla A.K., Bankhead A.R., Burkett S.E., Zornetzer G., Tseng C.T.K., Metz T.O., Pickles R., McWeeney S., Smith R.D., Katze M.G., Waters K.M., Baric R.S. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87:3885–3902. doi: 10.1128/jvi.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizun J., Yu M.W.N., Talbot P.J. Survival of human coronaviruses 229E and OC43 in suspension and after drying onsurfaces: a possible source ofhospital-acquired infections. J. Hosp. Infect. 2000;46:55–60. doi: 10.1053/jhin.2000.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsen G.A., van Rijn C., Kooij S., Bem R.A., Bonn D. Small droplet aerosols in poorly ventilated spaces and SARS-CoV-2 transmission. Lancet Respir. Med. 2020;8:658–659. doi: 10.1016/S2213-2600(20)30245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- Spearman C. The method of ‘right and wrong cases’ (‘constant stimuli’) without Gauss's formulae. Br. J. Psychol. 1908;2:227–242. doi: 10.1111/j.2044-8295.1908.tb00176.x. 1904-1920. [DOI] [Google Scholar]
- Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. 2020;117:11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Ricard C.S., Holmes K.V. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37 degrees C correlates with virus aggregation and virus-induced cell fusion. J. Virol. 1990;64:3042–3050. doi: 10.1128/jvi.64.6.3042-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Liu M., Ren B., Wu Z., Yu X., Peng C., Tian J. Sunlight ultraviolet radiation dose is negatively correlated with the percent positive of SARS-CoV-2 and four other common human coronaviruses in the U.S. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Mao Y., Jones R.M., Tan Q., Ji J.S., Li N., Shen J., Lv Y., Pan L., Ding P., Wang X., Wang Y., MacIntyre C.R., Shi X. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ. Int. 2020;144 doi: 10.1016/j.envint.2020.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Laperriere G., Germain H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7:2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R.B., Gerba C.P. In: Laskin A.I., editor. Vol. 33. Academic Press; 1988. Molecular mechanisms of viral inactivation by water disinfectants; pp. 75–105. (Advances in Applied Microbiology). [DOI] [PubMed] [Google Scholar]
- Tian Y., Rong L., Nian W., He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobías A., Molina T. Is temperature reducing the transmission of COVID-19? Environ. Res. 2020;186 doi: 10.1016/j.envres.2020.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020;10 doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Munster V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eurosurveillance. 2013;18:20590. doi: 10.2807/1560-7917.ES2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-X., Cao X., Chen Y.-P. An air distribution optimization of hospital wards for minimizing cross-infection. J. Clean. Prod. 2020 doi: 10.1016/j.jclepro.2020.123431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005;11:4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B., Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W., zhu X., Gu C., Yin J., Wei W., Yao W., Liu C., Li J., Ou G., Wang M., Fang T., Wang G., Qiu Y., Wu H., Chao F., Li J. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People's Liberation Army. Water Sci. Technol. 2005;52:213–221. doi: 10.2166/wst.2005.0266. [DOI] [PubMed] [Google Scholar]
- WEF, 2020. Water Environment Federation, The Water Professional's Guide to COVID-19. 〈https://www.wef.org/news-hub/wef-news/thewater-professionals-guide-to-the-2019-novel-coronavirus/〉.
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2018. Guidelines on Sanitation and Health. [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus Disease (COVID-19) Technical Guidance: Laboratory Testing for 2019-nCoV in Humans.〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance〉 [Google Scholar]
- WHO, 2020b. Water, Sanitation, Hygiene and Waste Management for COVID-19. 〈https://www.who.int/publications-detail/water-sanitation-hygiene-and-waste-management-for-covid-19〉.
- WHO, 2003. WHO Environmental Health Team Reports on Amoy Gardens. 〈https://www.info.gov.hk/gia/general/200305/16/P200305160114_print.htm〉.
- WHO, 2015. World Health Organization. Manual for the Care and Management of Patients in Ebola Care Units/Community Care Centres Interim Emergency Guidance.
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/C5EW00125K. [DOI] [Google Scholar]
- Worley-Morse T., Mann M., Khunjar W., Olabode L., Gonzalez R. Evaluating the fate of bacterial indicators, viral indicators, and viruses in water resource recovery facilities. Water Environ. Res. 2019;91:830–842. doi: 10.1002/wer.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. eabb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zeng T., Guo W.-J., Bai Y.-Y., Pang D.-W., Zhang Z.-L. Digital single virus immunoassay for ultrasensitive multiplex avian influenza virus detection based on fluorescent magnetic multifunctional nanospheres. ACS Appl. Mater. Interfaces. 2019;11:5762–5770. doi: 10.1021/acsami.8b18898. [DOI] [PubMed] [Google Scholar]
- Wu, F., Xiao, A., Zhang, J., Gu, X., Lee, W.L., Kauffman, K., et al., 2020a. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. medRxiv. 〈10.1101/2020.04.05.20051540〉. [DOI] [PMC free article] [PubMed]
- Wu, F., Xiao, A., Zhang, J., Moniz, K., Endo, N., Armas, F., et al., 2020b SARS-CoV-2 Titers in Wastewater Foreshadow Dynamics and Clinical Presentation of New COVID-19 Cases. medRxiv. 〈10.1101/2020.06.15.20117747〉. [DOI] [PMC free article] [PubMed]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–20. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30083-2. 10.1016/S2468-1253(20)30083-2〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer, S., Marechal, V., Mouchel, J.-M., Maday, Y., Teyssou, R., Richard, E., et al., 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics through Viral Genome Quantification in Paris Wastewaters. medRxiv. 〈10.1101/2020.04.12.20062679〉. [DOI] [PMC free article] [PubMed]
- Xagoraraki I., O’Brien E. In: Women in Water Quality: Investigations by Prominent Female Engineers. O’Bannon D.J., editor. Springer International Publishing; Cham: 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [DOI] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqub, T., Nawaz, M., Shabbir, M.Z., Ali, MA, Altaf, I., Raza, S., et al., 2020. A Longitudinal Survey for Genome-Based Identification of SARS-CoV-2 in Sewage Water in Selected Lockdown Areas of Lahore City, Pakistan; A Potential Approach for Future Smart Lockdown Strategy. medRxiv. 〈10.1101/2020.07.31.20165126〉. [DOI] [PMC free article] [PubMed]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Ye Y., Chang P.H., Hartert J., Wigginton K.R. Reactivity of enveloped virus genome, proteins, and lipids with free chlorine and UV254. Environ. Sci. Technol. 2018;52:7698–7708. doi: 10.1021/acs.est.8b00824. [DOI] [PubMed] [Google Scholar]
- Ye Y., Zhao L., Imperiale M.J., Wigginton K.R. Integrated cell culture-mass spectrometry method for infectious human virus monitoring. Environ. Sci. Technol. Lett. 2019;6:407–412. doi: 10.1021/acs.estlett.9b00226. [DOI] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun-yun Li, Ji-Xiang Wang, Xi Chen. Can a toilet promote virus transmission? From a fluid dynamics perspective. Phys. Fluids. 2020;32 doi: 10.1063/5.0013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneti R.N., Girardi V., Spilki F.R., Mena K., Westphalen A.P.C., da Costa Colares E.R., Pozzebon A.G., Etchepare R.G. Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kang, Z., Gong, H., Xu, D., Wang, J., Li, Z., et al., 2020b. The Digestive System Is A Potential Route of 2019-nCov Infection: A Bioinformatics Analysis Based on Single-Cell Transcriptomes. bioRxiv. 〈10.1101/2020.01.30.927806〉.
- Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F., Du L., Jiang S. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92:408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L., Atoni, E., Du, Y., Zhang, H., Donde, O., Huang, D., et al., 2020. First Study on Surveillance of SARS-CoV-2 RNA in Wastewater Systems and Related Environments in Wuhan: Post-Lockdown. medRxiv. 〈10.1101/2020.08.19.20172924〉. [DOI] [PMC free article] [PubMed]
- Zhou J.B., Kong W.H., Wang S., Long Y.B., Dong L.H., He Z.Y., Liu M.Q. Potential transmission risk of SARS-CoV-2 through medical wastewater in COVID-19 outbreak cities. Res. Sq. 2020 doi: 10.21203/rs.3.rs-3743/v1. [DOI] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]