Abstract
The pandemic spread of Coronavirus Disease 2019 (COVID-19) is still ongoing since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is identified as the etiologic pathogen late December 2019. After over six-month spread of COVID-19, SARS-CoV-2 causes critical threats to global public health and economy. The investigations on evolution and genotyping on genetic variations are of great importance, therefore, the present study characterized the molecular variation of SARS-CoV-2 by analyzing 4230 complete genome sequences from the worldwide samples collected during the first 6-month pandemic. Phylogenetic tree analysis with Neighbor-Joining and Maximum-Parsimony methods indicated that the haplotypes of SARS-CoV-2 genome sequences were classified into four clades with the unique nucleotide and amino acid changes: T27879C (ORF8 L84S) in clade 1 (25.34%), A23138G (spike D614G) in clade 2 (63.54%), G10818T (nsp6 L37F), C14540T (nsp12 T442I), and G25879T (ORF3a V251F) in clade 3 (2.58%), and miscellaneous changes in clade 4 (8.54%). Interestingly, subclade 2B with the amino acid changes at nsp2 T85I, Spike D614G, and ORF3a Q57H was firstly reported on March 4, 2020 in United States of America, becoming the most frequent sub-haplogroup in the world (36.21%) and America (45.81%). Subclade 1C with the amino acid changes at nsp13 P504L and ORF8 L84S was becoming the second most frequent sub-haplogroup in the world (19.91%) and America (26.29%). Subclade 2A with the amino acid changes in Spike D614G and Nucleocapsid R203K and G204R was highly prevalent in Asia (18.82%) and Europe (29.72%). The study highlights the notable clades and sub-clades with unique mutations, revealing the genetic and geographical relevant post the six-month outbreak of COVID-19. This study thoroughly observed the genetic feature of SARS-CoV-2 haplotyping, providing an epidemiological trend of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Genetic variation, Phylogenetic tree, Clade, Epidemiological trend
1. Introduction
The ongoing outbreak of Coronavirus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global public health emergency. The emerged outbreak of SARS-CoV-2 rapidly spreads to the globe and manifests a wide spectrum of diseases which range from mild to severe symptoms such as pneumonia, acute respiratory distress syndrome, and even death. Since the first report in Wuhan city of Hubei Province in December 2019, there are more than 15 million confirmed cases in over 200 countries or territories and including over 640,000 deaths as of 26 July 2020 (WHO 2020). The COVID-19 ongoing pandemic is concern worldwide about its high contagiously. How quickly the virus could potentially raise its genetic variability is the valuable information to fill gaps in knowledge about the new COVID-19 over the world.
SARS-CoV-2, like other species include SARS-CoV, MERS-CoV, and bat SARS-related coronavirus, an enveloped single positive-stranded RNA virus, belonging to the Betacoronavirus genus which has been identified in human and animals. The whole-genome analysis comparisons showed that SARS-CoV-2 is the closest lineage with BatCoV RaTG13 (Zhou et al., 2020) and novel pangolin coronavirus (Lam et al., 2020). SARS-CoV-2 genome varies from 29.8 kb to 29.9 kb in size and encodes for ORF1ab (nsp1-nsp16: 256–21,545), ORF2 (spike protein: 21553–25,374), ORF3a (25383–26,210), ORF4 (envelope protein: 26235–26,462), ORF5 (membrane protein: 26513–27,181), ORF6 (27192–27,377), ORF7a (27384–27,749), ORF7b (27746–27,877), ORF8 (27884–28,294), ORF9 (nucleocapsid protein: 28264–29,523), and ORF10 (29548–29,664)(Yoshimoto, 2020). Besides, some regions are not encoded for viral protein, which is related to the non-canonical translational strategy employed by SARS-CoV-2. Recently, data sharing combined with the development of genetic sequence analysis tools have shed light on the pattern of global spread, the diversity in genetics, and dynamics of subtype evolution.
SARS-CoV-2 ORF1a/b is the large polyproteins (pp1a and pp1b) with the replicates consisting of non-structural proteins (nsps) 1–16, which perform numerous roles in the replication and virus assembly processes. Due to selective pressure, ORF1a/b displays a mutation rate of up to 29.47%, particularly in nsp2, nsp3 and nsp12, becoming one of the mutation hot spots (Pachetti et al., 2020; Ren et al., 2020). Spike (S) protein (ORF2) is the most important structural protein at the surface of the virion, responsible for virus infection via attaching onto the host receptor angiotensin-converting enzyme 2 (ACE2), a CLEC4M/DC-SIGNR receptor on the host cell surface. S protein is also a class I viral fusion protein mediating the fusion of the virion with cellular membranes. Thus, it is a known target for the host immune system and is commonly used for vaccine development (Walls et al., 2020; Wang et al., 2020). The changes and the variations in amino acid residues associated with spike protein–cell receptor interface are more vulnerable to viral infectivity. Recently, the spike variant D614G has emerged, as the most prevalent clade at multiple geographic levels that represented up to 78% (Korber et al., 2020; Ogawa et al., 2020). SARS-CoV-2 ORF3a, a transmembrane protein with a 72% homology to SARS-CoV ORF3a (Issa et al., 2020), forms the homotetrameric potassium sensitive ion channels that plays an important role in virus release. SARS-CoV-2 ORF3a shows a lower apoptotic activity than SARS-CoV ORF3a, as suggested to correlate with mild or even asymptomatic during early stages and follow by the high spread of the virus (Ren et al., 2020). It is noted that both the ORF3a and ORF8 highly divergent with interferon antagonist ORF3a and inflammasome activator ORF8b in SARS-CoV. Thus, based on the number of exported sequences during the early stages of human-to-human transmission, SARS-CoVs isolated from early patients contains the full-length ORF8. During its emergence, ORF8 partially deleted (−29 nt) and it may be reflects adaptation to humans (Chan et al., 2020). Interestingly, the novel coronavirus has a single ORF8 protein different from SARS-CoV which contains two ORF8s (ORF8a and ORF8b). The ORF8 in SARS-CoV-2 lacks any known functional domain or motif such as VLVVL (75-79aa) which activate the NLRP3 inflammasomes or bind to the IRF domain (IAD) region of interferon regulatory factor 3 (IRF3) then inactivates interferon signaling. ORF8 is one of the most relevant genes that have previously obvious genetic change during human-to-human transmission of the SARS-CoV epidemic (Muth et al., 2018; Yoshimoto, 2020).
Natural selection in coronaviruses could occur during each replication cycle and can act upon favorable mutations (Lauring and Andino, 2010). Since the availability of the first genomic sequence published, several studies have researched on the rapid genetic evolution of this novel virus through a phylogenetic tree tracking the geographical spread of the virus (Parlikar et al., 2020; Phan, 2020; van Dorp et al., 2020). The persistence of the pandemic may enable the accumulation of relevant mutations that are worth monitoring in the population even as drugs and vaccines are developed. After over six-month spread of COVID-19, SARS-CoV-2 presents critical threats to global public health and economy. The investigations on evolution and genotyping on genetic variations are of great importance. Therefore, the present study aims to characterize the molecular variation of SARS-CoV-2 by analyzing 4230 complete genome sequences in the open database, highlighting the notable clades and subclades (haplotypes) and the relevance of genetic and geographical patterns post the six-month outbreak of COVID-19. This study thoroughly observed the biological features of SARS-CoV-2 haplotypes, providing an epidemiological trend of COVID-19.
2. Material and methods
2.1. Sequence data and quality control
To construct a reference sequence dataset, 5801 complete full-length genome sequences (>29,000 bp) of SARS-CoV-2 in human host from December 2019 to 15 June 2020 were initially selected and downloaded through the website of Virus Pathogen Resource database (www.viprbrc.org). After removing the duplicate sequences and the sequences with a high variant with gaps or high numbers of mismatched over 20 nucleotides with ‘N' or other ambiguous IUPAC code, 4230 complete genomes were used for comparative analysis and geographic grouping (Supplementary Table 1, Fig. 1 ).
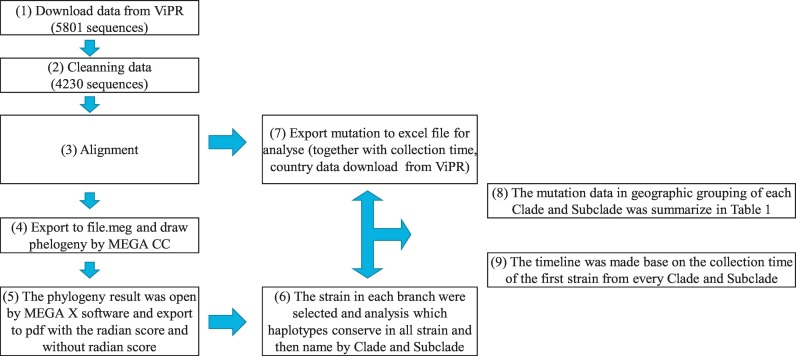
Fig. 1.
Data pipeline flowchart. ViPR was the available resource to provide the bioinformatics resource for applying to generate the data in this study. (1) In the first step we download the raw data with 5801 sequences. (2) By reducing the low-quality sequence, cleaning dataset was derived and contained 4230 sequences. (3) The cleaning process was in general a process of deletion, with the alignment of retained sequences and (4) exported the file for analyzing in (5), (6), (7). With the completion of these step, we are now ready to calculate the mutation and compare the data in geography and time (8), (9).
2.2. Sequence alignment and Phylogenetic tree
Assemblies were aligned against the Wuhan-Hu-1 strain NC_045512.2 as reference using a multiple sequence alignment program MAFFT (Multiple Alignment using Fast Fourier Transform) (Katoh and Standley, 2013). The upstream of nsp1 (start Condon ATG in ORF1ab) and downstream of ORF10 (from stop codon TAG) were deleted to keep the coding-regions. The alignment data were export into mega file (file.meg) for phylogenetic tree-building. Subsequently, the phylogenetic trees were constructed based on amino acid substitution type using the Neighbor-Joining (NJ) method with Maximum Composite Likelihood (MCL) distance, and Maximum Parsimony (MP) method with the Subtree-Pruning-Regrafting (SPR) algorithm, by MEGA CC (Kumar et al., 2018). To present the evolutionary history of the taxa analyzed, the bootstrap consensus tree was inferred using the NJ method with 1000 replicates (Felsenstein, 1985; Saitou and Nei, 1987). The proportion of associated taxa clustered together in the bootstrap test were shown next to the branches. The replicate tree Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The evolutionary distances were computed using the Poisson correction method, as in the units of the number of amino acid substitutions per site. The evolutionary history was inferred using the MP method with the SPR algorithm to compare with the phylogeny constructed by MCL distance. The tree was drawn to scale, with branch lengths calculated using the average pathway method, as in the units of the number of changes over the whole sequence. To observe variant sites using the phylogenetic circle map, we selected all variant clades from the phylogenetic map, then align with the Wuhan-Hu-1 strain (NC-045512.2) and analyzed by MEGA to check the mutations contain in each clade and subclade. After that, we exported the mutation mega file to an excel file for analysis. We also used MEGA CC to generate other calculation-specific results file (bootstrap consensus tree, csv files…) to analysis.
3. Results
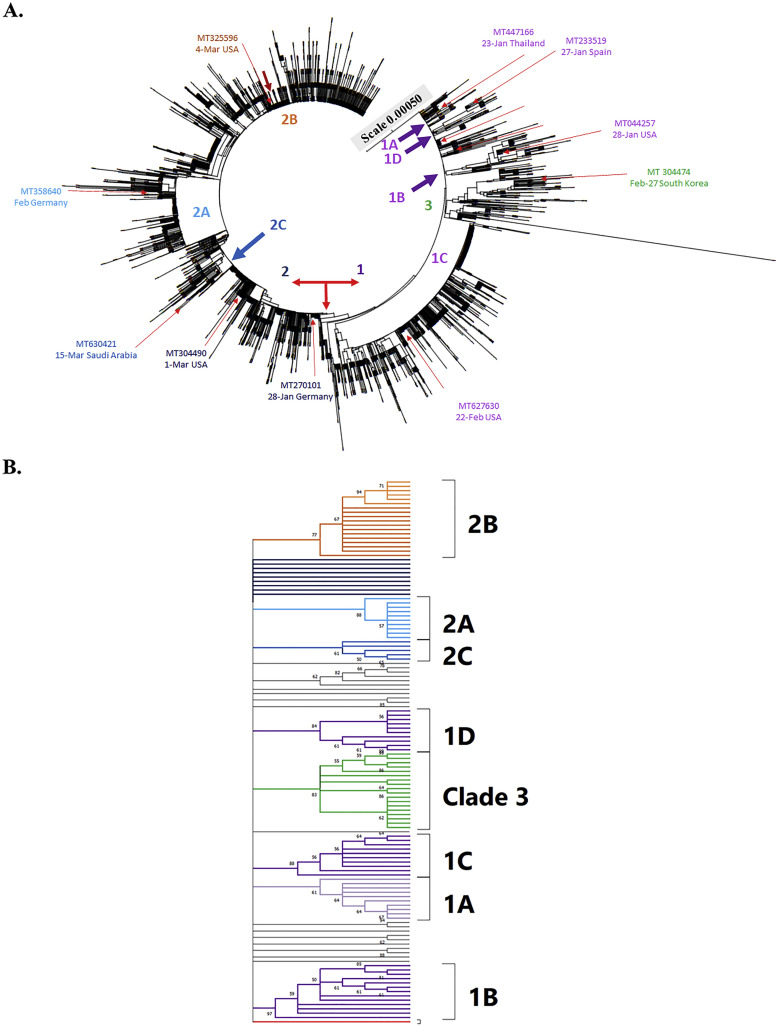
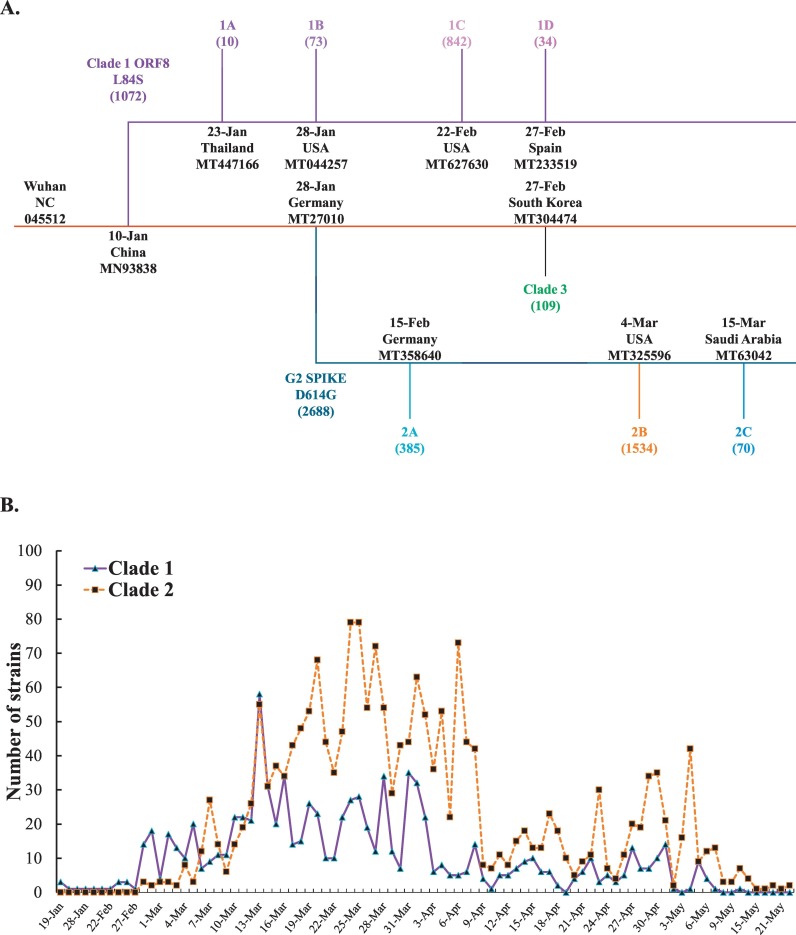
To investigate the genome diversity of SARS-CoV-2 variants after 6-month pandemic, 4230 complete genome sequences deposited in the Virus Pathogen Resource database were selected, which includes 3089 America variants (73.02%), 461 Oceania variants (10.89%), 441 Asia variants (10.43%), 212 Europe variants (5.01%), and 28 Africa variants (0.66%) collected during the period from December 2019 to June 2020. The phylogenetic trees were constructed based on amino acid substitution type by NJ and MP methods, and then rooted at the reference sequence of Wuhan-Hu-1 strain (NC_045512.2) (Fig. 2, Fig. 3, Fig. 4 , Supplemental Figs. 1–3, and Table 1 ). The phylogenetic analysis of genome sequences from 4230 SARS-CoV-2 variants by the NJ method indicated four haplotypes with the unique nucleotide variation and amino acid changes: T27879C (ORF8 L84S) in clade 1 (1072 variants, 25.34%), A23138G (spike D614G) in clade 2 (2688 variants, 63.54%), G10818T (nsp6 L37F), C14540T (nsp12 T442I), and G25879T (ORF3a V251F) in clade 3 (109 variants, 2.58%), and miscellaneous changes in clade 4 (361 variants, 8.54%) (Fig. 2A, Table 1). Interestingly, clade 2 with the mutation Spike D614G was the most predominant and prevalent haplotype among global variants.
Fig. 2.
Phylogeny analysis of 4230 SARS-CoV-2 genome sequences from the variants in worldwide. The tree was constructed based on amino acid substitution rate using Maximum Composite Likelihood distance by Neighbor-Joining tree from MEGA CC, which branch length in the units of the number of base substitutions per site was shown (A). The first strain by date and country from each clade and subclade was collected that showed in the same color with each group (A). Phylogenetic analysis of 126 genome sequences from each clade and subclade with 1000 bootstrap replicates was generated using the neighbor-joining method with Mega X software (B). The color mark for each clade and subclade was consistent with the color labeled in Fig. 2A. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
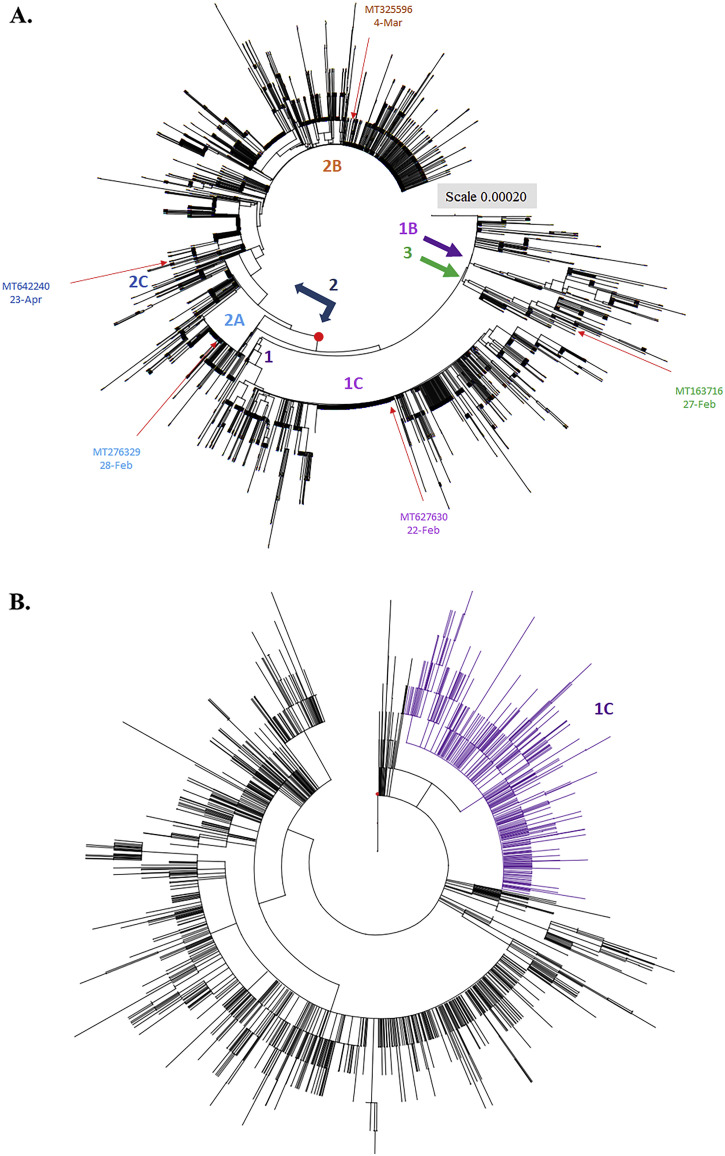
Fig. 3.
Phylogenetic tree of 3089 SARS-CoV-2 genome sequences from America variants. Phylogeny of SARS-CoV-2 lineages in worldwide construct with Maximum Composite Likelihood distance by Neighbor-Joining tree from MEGA CC (A). The evolutionary history was inferred using the Maximum Parsimony method (B). The first strain by date and country from each Clade and Subclade was collected that showed in the same color with each group in Fig. 2A. The topology was marked in violet contained the sequences conserved in haplotype 1C (812 sequences) generated in Maximum Composite Likelihood distance method.
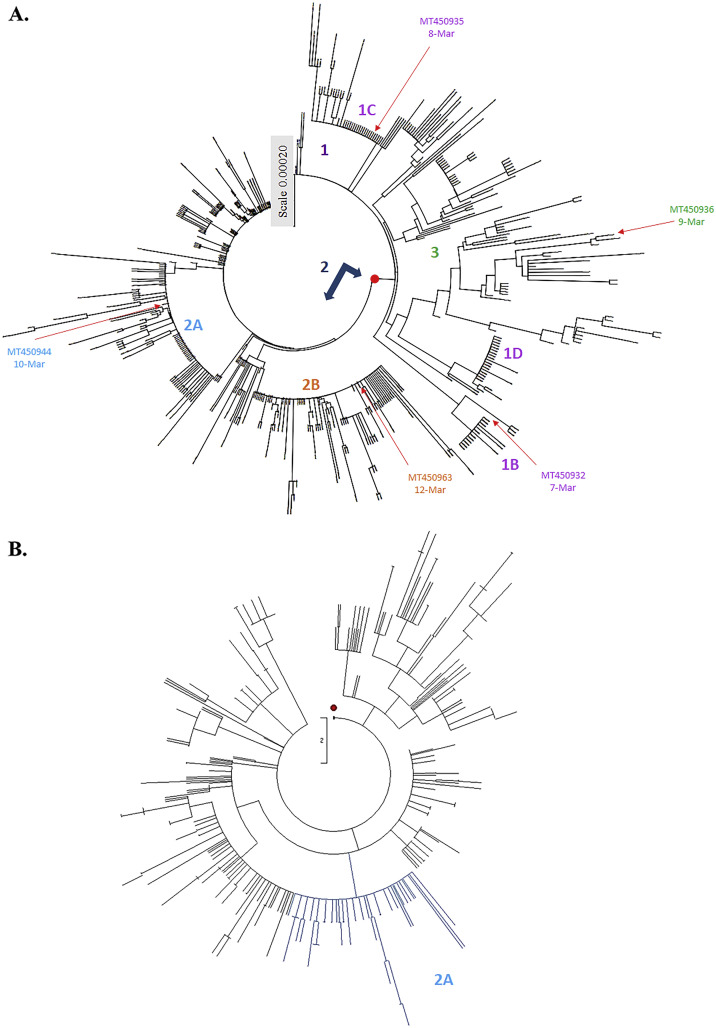
Fig. 4.
Phylogenetic tree of 460 SARS-CoV-2 genome sequences from Oceania variants. Phylogeny of SARS-CoV-2 lineages in Oceania construct with Maximum Composite Likelihood distance by Neighbor-Joining tree from MEGA CC (A). The evolutionary history was inferred using the Maximum Parsimony method for analysis in (B). The first strain by date and country from each clade and subclade was collected that showed in the same color with each group in Fig. 2A. Subclade 2A was marked by the light blue color, which contains 78 Oceania variants that maintained the same topology by Neighbor-Joining data in Fig. 4A.
Table 1.
Haplotypes and sub-haplogroups of 4230 SARS-CoV-2 genome sequences from different geographic areas.
| Nucleotide change | Amino acid change | Global |
America |
Europe |
Asia |
Africa |
Oceania |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (4230) |
(3089) |
(212) |
(441) |
(28) |
(460) |
|||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |||
| Clade 1 | T27879C | ORF8 L84 S | 1072 | 25.34 | 909 | 29.43 | 9 | 4.25 | 70 | 15.87 | 2 | 7.14 | 82 | 17.83 |
| 1A | T27879C | ORF8 L84S | 10 | 0.24 | 10 | 2.27 | ||||||||
| G19869T | nsp15 V172L | |||||||||||||
| G23782A | S A829T | |||||||||||||
| 1B | T27879C | ORF8 L84S | 73 | 1.73 | 57 | 1.85 | 16 | 3.48 | ||||||
| T225A | nsp1 D75E | |||||||||||||
| C2912T | nsp3 P153L | |||||||||||||
| G27812C | ORF8 V62L | |||||||||||||
| 1C | T27879C | ORF8 L84 S | 842 | 19.91 | 812 | 26.29 | 30 | 6.52 | ||||||
| C17482T | nsp13 P504L | |||||||||||||
| 1D | T27879C | ORF8 L84S | 34 | 0.8 | 3 | 0.1 | 7 | 3.3 | 1 | 0.23 | 23 | 5.0 | ||
| T9212A | nsp4 F308Y | |||||||||||||
| C14540T | nsp12 T455I | |||||||||||||
| G25714T | ORF3a G196V | |||||||||||||
| C28598T | N S197L | |||||||||||||
| Others in clade 1 | 113 | 2.67 | 37 | 1.19 | 2 | 0.94 | 59 | 13.38 | 2 | 7.14 | 13 | 2.83 | ||
| Clade 2 | A23138G | S D614G | 2688 | 63.54 | 1946 | 63.0 | 167 | 78.77 | 275 | 62.36 | 25 | 89.29 | 275 | 59.78 |
| 2A | A23138G | S D614G | 385 | 9.1 | 159 | 5.15 | 63 | 29.72 | 83 | 18.82 | 2 | 6.9 | 78 | 16.96 |
| G28616A G28617A G28618C |
N R203K, G204R | |||||||||||||
| 2B | A23138G | S D614G | 1532 | 36.21 | 1415 | 45.81 | 26 | 12.26 | 3 | 0.68 | 1 | 3.57 | 89 | 19.35 |
| G25298T | ORF3a Q57H | |||||||||||||
| C794T | nsp2 T85I | |||||||||||||
| 2C | A23138G | S D614G | 70 | 1.65 | 5 | 0.16 | 65 | 14.74 | ||||||
| G25298T | ORF3a Q57H | |||||||||||||
| C28589T | N S194L | |||||||||||||
| Others in clade 2 | 699 | 16.52 | 367 | 11.88 | 78 | 36.79 | 124 | 28.12 | 22 | 78.57 | 108 | 23.48 | ||
| Clade 3 | G10818T | nsp6 L37F | 109 | 2.58 | 54 | 1.75 | 16 | 7.55 | 4 | 0.91 | 1 | 3.57 | 34 | 7.39 |
| C14540T | nsp12 T442I | |||||||||||||
| G25879T | ORF3a V251F | |||||||||||||
| Clade 4 | A23138 | S D614 | 361 | 8.54 | 180 | 5.83 | 20 | 16.98 | 92 | 20.86 | 0 | 0 | 69 | 15 |
| T27879 | ORF8 L84 | |||||||||||||
Sub-haplogroups (subclades) in clades 1 and 2 has been clearly clustered with a relatively high bootstrap value (> 60% support) (Fig. 2B). In addition, the haplotypes and sub-haplogroups clustered by the MP method were consistent with the genome variation grouping by the NJ method (Figs. 3B–4B vs. 3A-4A, Supplemental Figs. 1B—3B vs. 1A-3A). Comparing the phylogenetic trees constructed by NJ and MP methods, the same topology for sub-clade 2A was identified, as marked in the light blue color in Figs. 3B, 4B, and Supplemental Figs. 1B, 2B, and 3B. Sub-clade 1C with the amino acid changes at nsp13 P504L and ORF8 L84S was the largest sub-haplogroup in clade 1 among all genome sequences from global (19.91%), America (26.29%) and Oceania (6.52%), respectively (Fig. 2, Fig. 3, Fig. 4, Table 1). Interestingly, sub-haplogroup 1A variants with the amino acid changes at nsp15 V172L and ORF8 L84S were observed only in Thailand (Supplemental Fig. 1, Table 1). Sub-clade 1B with the amino acid changes at nsp1 D75E, nsp3 P153L, and ORF8 V62L and L84S was reported in USA (57/73) and Australia (16/73). Sub-clade 1D was dominantly prevalent in Australia (23/34). Therefore, the result indicated that the prevalence of subclades in haplotype 1 showed the unique geographic pattern.
Among sub-haplogroups in clade 2, subclade 2A with the amino acid changes in Spike D614G and Nucleocapsid R203K and G204R was highly prevalent in Asia (18.82%) and Europe (29.72%), respectively (Supplemental Fig. 2, Fig. 3, Table 1). Subclade 2B with the amino acid changes at nsp2 T85I, Spike D614G, and ORF3a Q57H was firstly reported on March 4, 2020 in USA, becoming the most frequent sub-clade in the world (36.21%) and America (45.81%), respectively (Fig. 2, Fig. 3, Table 1). Sub-clade 2A with the amino acid changes at Spike D614G and Nucleocapsid R203K and G204R was the second most frequent sub-clade among haplotype 2, and appeared in all continents. Most subclade 2C variants (92.86%) with the amino acid changes at Spike D614G, ORF3a Q57H, and Nucleocapsid S194L was prevalent in Asia counties, including India (61/70), Bangladesh (3/70), and Saudi Aribia (1/65) (Supplemental Fig. 1, Table 1). The result demonstrated that sub-haplogroups 2A and 2B spread worldwide, as the dominant variants in the global pandemic.
Analysis of the daily new SARS-CoV-2 genome sequences indicated that the first clade 1 variant containing the amino acid change at ORF8 L84S was sequenced from the isolate (2019-nCoV_HKU-SZ-002a_2020, GenBank: MN938384) collected in January 10, 2020 in China, and then clade 1 variants were spread in all continents with approximately 85% (n = 909) in America, just around 7% in Asia (n = 70), and Oceania (n = 82), respectively (Fig. 2, Fig. 5A, Table 1). Sub-haplogroup analysis indicated the sequences of sub-clades 1A, 1B, 1C, and 1D among haplotype 1 variants were first identified on January 23, 2020 in Thailand (GenBank: MT447166), January 28, 2020 in USA (GenBank: MT044257), February 22, 2020 in USA (GenBank: MT627630), and February 28, 2020 in Spain (GenBank: MT233519), respectively (Fig. 2, Fig. 5A). In addition, the sequence of haplotype clade 2 was first reported in the variant with the change at Spike D614G on January 28, 2020 in Germany (GenBank: MT270101). Haplotype clade 2 variants gained prevalence in all over the world with 2688 variants sequenced (63.54% of 4230 variants) (Table 1). Subclade 2A variant with the changes at Spike D614G and nucleocapsid R203K and G204R (GenBank: MT358640) was firstly sequenced and reported on February 15 in Germany. The first subclade 2B variant with the changes at Spike D614G, ORF3a Q57H, and nsp2 T85I (GenBank: MT325596) was documented in USA on March 4 (Figs. 2 and 8A). Moreover, subclade 2C variant with the changes at Spike D614G, ORF3a Q57H, and nucleocapsid S194L (GenBank: MT630421) was first reported in Saudi Arabia on March 15. In addition, the haplotype clade 3 variant was first sequenced and reported on February 27, 2020 in South Korea (GenBank: MT304474). The prevalence of clade 3 variants was lower than clades 1 and 2 (Table 1). Recently, only 2 variants with the changes at Spike D614G (haplotype 1) and ORF8 L84S (haplotype 2) were detected in USA on March 24. However, such hybrid variants were infrequent.
Fig. 5.
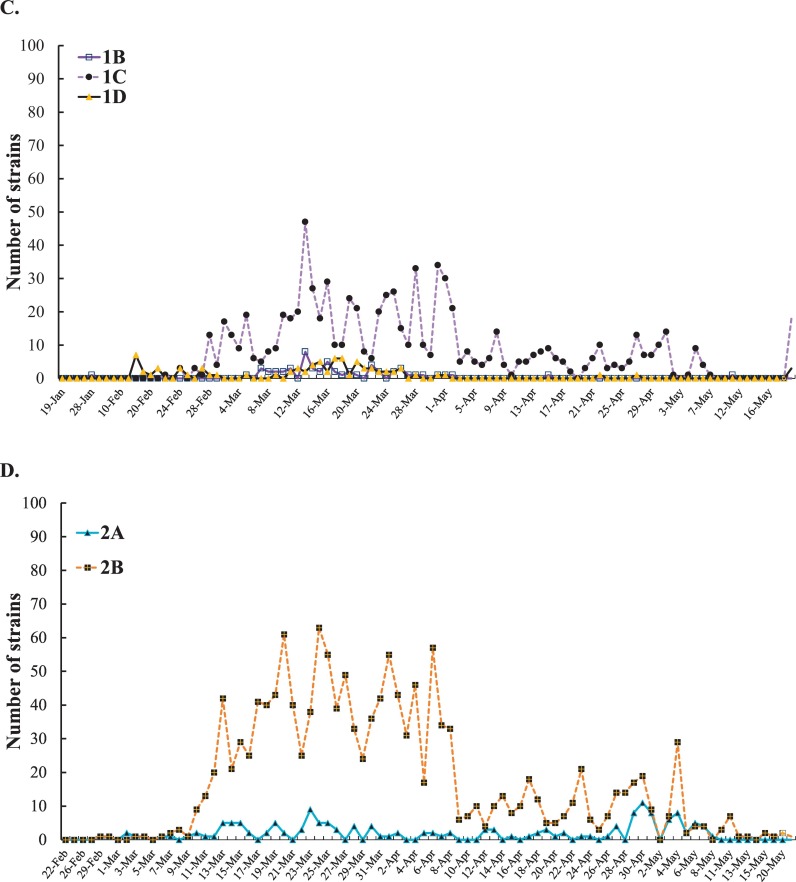
Analysis of the daily new SARS-CoV-2 genome sequences, haplotypes and sub-haplogroups. The information about the first variant in each clade and sub-clade was shown in Fig. A. The number of clades 1 and 2 in America variants was reported each day (B). The number of each sub-clade among haplotype 1 America variants was reported each day (C). Among haplotype 2 America variants, the number of each sub-clade was reported each day (D).
For the further investigation on the distribution of the variants post six-month spread, the frequency of each clade and sub-clade in America was counted since the countries in America had no implementation of strict social distancing and lockdown of communities with outbreaks in the early outbreak. Haplotype 2 variants showed a higher prevalence than haplotype 1 variants since March 2020 (Fig. 5B). Thus, the currently predominant haplotype in America was haplotype 2 variants accounted for about 63% among 3089 America sequenced-analysis post six-months spread. Among haplotype 1 variants, sub-haplogroups 1A, 1B, and 1D had a slow-spreading rate. Sub-haplogroup 1C variants appeared in late February 2020, and increased rapidly and remained for several months (Fig. 5C). Remarkably, among the subclades in clade 2, sub-haplogroup 2B variants (Spike D614G, ORF3a Q57H, and nsp2 T85I) distributed quickly, and then dominated in this continent (Fig. 5D).
4. Discussion
This study analyzed 4230 complete genome sequences of SARS-CoV-2 for grouping the specific mutants spreading in the world and determining how the virus accumulated the synonymous mutation after 6-month pandemic. Phylogenetic tree analysis highlighted that clade 2 with the amino acid change at Spike D614G was the most predominant and prevalent haplotype of SARS-CoV-2 variants since the first variant appeared in Germany lately on February (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Supplemental Figs. 1–3, Table 1). The variants with Spike G614 were over 60% to 78% (Korber et al., 2020; Mercatelli and Giorgi, 2020; Nguyen et al., 2020), which Spike G614 could be the main immunogenic for diagnostic reagents, vaccines, and antibody-based therapeutics against SARS-CoV-2. For the functions of SARS-CoV-2-endcoded accessory proteins, previous studies have reported that ORF3a can efficiently induce apoptosis in cell and influence on viral uptake and release(Issa et al., 2020; Ren et al., 2020).
Subclade 2B with the amino acid changes at nsp2 T85I, Spike D614G, and ORF3a Q57H was firstly reported on March 4, 2020 in United States of America, becoming the most frequent sub-haplogroup in the world (36.21%) and America (45.79%). SARS-CoV nsp2 was found to bind to two host proteins prohibitin 1 and prohibitin 2 that were known to play roles in cell cycle progression, cell migration, cellular differentiation, apoptosis, and mitochondrial biogenesis (Cornillez-Ty et al., 2009). Since Spike, ORF3a, and nsp2 were linked to virulence, infectivity, ion channel formation, and virus release, the mutations Spike D614G, ORF3a Q57H, and nsp2 T85I within Subclade 2B variants might alter the viral transmission and pathogenesis, which could correlate with the dominant spread of COVID-19 in USA.
Subclade 2A identified based on distance and maximum-likelihood methods from NJ phylogeny that contains the haplotype Spike D614G, and Nucleocapsid R203K, G204R were more frequent in Europe, Asia, and Africa areas than Subclade 2B (Figs. 2A–4A, Supplemental Figs. 1A-3A). In addition, Subclade 2C variants with the mutations Spike D614G, ORF3a Q57H, and Nucleocapsid S194L chiefly appeared in Asia area. Beside the amino acid substitutions occurred at SARS-CoV-2 nucleocapsid such as S194L, R203K and G204R in this study, Nucleocapsid RG103KR mutation was aforementioned as one genotype of current common variants in the global (Mercatelli and Giorgi, 2020). SARS-CoV-2 Nucleocapsid protein was primarily expressed during the early stages infection, playing a crucial role in viral RNA transcription and viral replication (Chan et al., 2020). SARS-CoV Nucleocapsid protein has been illustrated to upregulate the expression of the proinflammatory factor COX2, affect a variety of basic cellular processes, inflammatory responses and notably inhibit the innate immune responses in the host cell (Zeng et al., 2020). Of note, amino acid substation at SARS-CoV-2 Nucleocapsid protein within Subclades 2A and 2C might alter a significance in immune response compared to another.
Different with SARS-CoV that has two ORF8 proteins (ORF8a and ORF8b), SARS-CoV-2 has a single ORF8 protein without a VLVVL motif that triggered the intracellular stress pathways and enacted NOD-like receptor family pyrin region containing −3 (NLRP3). Notably, Subclade 1C variants with the mutations ORF8 L84S and Helicase P504L was over 79% (842 out of 1072) within Clade 1. SARS-CoV-2 Helicase (nsp13) shared 99.8% identity to SARS-CoV nsp13, processing the helicase activity and 5′-triphosphatase activity. Subclade 1C variants could have the unique feature of splicing, nuclear export, translation, and stability of viral genome and mRNA. Interestingly, Subclade 1C variants were spread mainly in early phase of pandemic in USA, but not prevalent now. Thus, the low prevalence of Subclade 1C variants post early phase of pandemic might be need to be further investigated to understand about the transmission.
The fast increment of infection is giving more published sequences that may contribute to analyze some visibility and proof of populace structure, especially the chance of various presentations of SARS-CoV-2 into the commercial and comprehend future risks for biological reservoirs conveying. The virus spread among the travelers and by nearly contact in an exposure population but for compare the high spread in the same continent at the same time. The present study demonstrated the difference of SARS-CoV-2 clades and subclades in the prevalence and geographic spread in the global, as well as America, Europe, Asia, Africa and Oceania areas. This study provided the molecular epidemiology of SARS-CoV-2 post 6-month pandemic, particular genotyping, spread timeline and geographic grouping, which might be useful for preventing the transmission of SARS-CoV-2 and guiding the development of vaccines and therapeutics against COVID-19.
Credit author statement
Ngoc-Niem Bui: Software, investigation, formal analysis, Writing-original draft.
Yu-Tzu Lin: Validation, investigation, visualization, data curation.
Su-Hua Huang: investigation, visualization.
Cheng-Wen Lin: Conceptualization, Methodology, formal analysis, Resources, Writing-review and editing, supervision project, Funding acquisition.
Declaration of Competing Interest
All authors declare no competing financial interest.
Acknowledgments
This work was financially supported by China Medical University, Taiwan (CMU109-ASIA-07, CMU108-S-12, and CMU108-MF-42), and funded by grants from the Ministry of Science and Technology, Taiwan (MOST107-2923-B-039-001-MY3; MOST108-2320-B-039-039-MY3). The experiments and data analysis were performed in part using the Medical Research Core Facilities Center, Office of Research & Development at China Medical University, Taiwan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104800.
Appendix A. Supplementary data
Supplementary material
References
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/jvi.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N. Y). 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/msystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. 812-827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y.M., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lauring A.S., Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010 doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D., Toplak I., Ameneiros R.S., Pfeifer A., Thiel V., Drexler J.F., Müller M.A., Drosten C. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Pathirana P., Nguyen T., Nguyen H., Bhatti A., Nguyen D., Nguyen D.T., Nguyen N.D., Creighton D., Abdelrazek M. 2020. Genomic Mutations and Changes in Protein Secondary Structure and Solvent Accessibility of SARS-CoV-2 (COVID-19 Virus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J., Zhu W., Tonnu N., Singer O., Hunter T., Ryan A.L., Pao G.M. The D614G mutation in the SARS-CoV2 spike protein increases infectivity in an ACE2 receptor dependent manner. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.07.21.214932. 2020.07.21.214932. [DOI] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlikar A., Kalia K., Sinha S., Patnaik S., Sharma N., Vemuri S.G., Sharma G. Understanding genomic diversity, pan-genome, and evolution of SARS-CoV-2. PeerJ. 2020;8 doi: 10.7717/peerj.9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.-Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281-292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Qihui, Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Qisheng, Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F.K. The proteins of severe acute respiratory syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020 doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Yunru, Liu M., Mohammed A., Zhao C., Yang Yun, Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jin T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material