Abstract
Objective
Islets of Langerhans contain heterogeneous populations of insulin-producing β-cells. Surface markers and respective antibodies for isolation, tracking, and analysis are urgently needed to study β-cell heterogeneity and explore the mechanisms to harness the regenerative potential of immature β-cells.
Methods
We performed single-cell mRNA profiling of early postnatal mouse islets and re-analyzed several single-cell mRNA sequencing datasets from mouse and human pancreas and islets. We used mouse primary islets, iPSC-derived endocrine cells, Min6 insulinoma, and human EndoC-βH1 β-cell lines and performed FAC sorting, Western blotting, and imaging to support and complement the findings from the data analyses.
Results
We found that all endocrine cell types expressed the cluster of differentiation 81 (CD81) during pancreas development, but the expression levels of this protein were gradually reduced in β-cells during postnatal maturation. Single-cell gene expression profiling and high-resolution imaging revealed an immature signature of β-cells expressing high levels of CD81 (CD81high) compared to a more mature population expressing no or low levels of this protein (CD81low/-). Analysis of β-cells from different diabetic mouse models and in vitro β-cell stress assays indicated an upregulation of CD81 expression levels in stressed and dedifferentiated β-cells. Similarly, CD81 was upregulated and marked stressed human β-cells in vitro.
Conclusions
We identified CD81 as a novel surface marker that labels immature, stressed, and dedifferentiated β-cells in the adult mouse and human islets. This novel surface marker will allow us to better study β-cell heterogeneity in healthy subjects and diabetes progression.
Keywords: β-Cells, CD81, Maturation, Immature, Heterogeneity, Dedifferentiation
Highlights
-
•
CD81 is differentially expressed in mouse adult pancreatic β-cells.
-
•
CD81 is downregulated during postnatal β-cell maturation.
-
•
CD81 is upregulated in in vivo and in vitro diabetic conditions.
-
•
The levels of CD81 are increased in stressed human β-cells.
1. Introduction
Diabetes mellitus is a complex metabolic disorder that results from the malfunction of the endocrine pancreas. In both type 1 (T1D) and type 2 diabetes (T2D), autoimmunity and glucolipotoxicity respectively lead to dedifferentiation, dysfunction, and ultimately loss of insulin-producing β-cells, key events in the progression [[1], [2], [3]]. These glucose-/nutrient-sensing and insulin-secreting cells are central for glucose homeostasis and exist as a heterogeneous population in health and disease [4,5]. In healthy islets, β-cells contain immature and mature subpopulations. Upon exposure to stress, β-cells upregulate a stress response and undergo dedifferentiation along a developmental trajectory, partially overlapping with the transcriptome of embryonic and neonatal β-cells [1,2]. Therefore, successful regenerative strategies can be achieved by stopping the stress and redifferentiation of dedifferentiated β-cells. Alternatively, triggering proliferation of immature and dedifferentiated β-cells, followed by a maturation step to recover β-cell identity, might be a therapeutic option. Along these lines, two separate studies in mice and humans have shown that upon non-canonical Wnt stimulation, the immature and more proliferative β-cell subpopulation can be directed to acquire a mature and functional phenotype [4,6].
During embryonic development, a common endocrine progenitor population gives rise to different endocrine hormone-producing cell types, including glucagon-secreting α-cells, insulin-producing β-cells, somatostatin-expressing δ-cells, pancreatic polypeptide-producing PP cells, and ghrelin-expressing ε-cells. These are the major hormone-producing cell types that cluster within the islets of Langerhans in the adult pancreas [7,8]. During development, β-cells are still immature and thus do not accurately secrete insulin in response to different glucose levels [9]. After birth, β-cells undergo a maturation process [10], which coincides with changes in the nutritional environment that result in a shift from amino acid-to glucose-stimulated insulin secretion (GSIS) [11,12]. Mature β-cells are defined by the expression of a set of maturation genes including Ucn3, Mafa, Glut2 (in rodents), and GLUT1 (in humans) [13,14]. The differential activities of mTORC1 and AMPK signaling play a crucial role in the β-cell maturation process [11,15]. Several recent studies have reported the existence of different β-cell subpopulations in the adult pancreas, including a minor subpopulation retaining an immature phenotype with increased proliferative potential [4,[16], [17], [18], [19], [20]]. It is essential to better characterize the different β-cell subpopulations and understand whether they can be targeted for β-cell regeneration by increasing cell proliferation. Uncovering markers and pathways distinguishing immature and mature β-cell subpopulations can offer molecular targets for redifferentiation of dedifferentiated β-cells and enforce stem cell-derived β-cell (SC-β) maturation to generate SC-β cells with improved functionality for in vitro studies and cell-replacement therapy.
Herein, we report cluster of differentiation 81 (CD81) as a novel surface marker that distinguishes two different β-cell subpopulations and identifies immature, stressed, and dedifferentiated β-cells in mice and humans. CD81 (Tspan-28, TAPA1) [21] belongs to the tetraspanin family of proteins, which contains 33 family members in humans. These proteins are involved in different cellular and physiological processes, such as controlling signaling pathways and regulating cell–cell fusion [22]. Tetraspanins are membrane proteins that comprise four transmembrane domains, a small and large extracellular loop and short carboxyl and amino cytoplasmic termini [23]. These proteins are organized in membrane regions rich in cholesterol, so-called tetraspanin-enriched microdomains (TEMs), in which they interact with partner proteins to exert their cellular functions by triggering intracellular signaling in response to extracellular stimuli [24]. Different members of tetraspanins can coexist in TEMs. For instance, the co-localization of CD9, CD63, CD81, and CD82 in the TEMs of HeLa cells has been shown [25]. Among these, CD81 is a 26 kDa protein that is expressed in several tissues, such as immune cells and hepatocytes [26]. CD81 functions as a docking receptor and mediates the infection of several human pathogens, such as hepatitis C virus (HCV) [27]. In immune cells, the interaction of CD81 with different protein partners regulates B-cell receptor signaling and T-cell activation [28].
The expression and function of CD81 in pancreatic endocrine cells have not been explored to date. In this study, we report CD81 as a novel marker that labels an immature population of β-cells. Importantly, CD81 levels were increased in β-cells from NOD and db/db diabetic mice as well as in in vitro stressed mouse and human β-cell lines, suggesting that this protein as a marker for stressed and dedifferentiated β-cells. Thus, the identification of CD81 as a surface marker will allow the specific targeting of the immature population or dedifferentiated β-cells for regenerative therapy.
2. Materials and methods
2.1. Mouse and cell lines
Mouse lines (males and females, age ranging from birth to 6 months old) were kept at the central facilities at Helmholtz Center Munich (HMGU) under SPF conditions in animal rooms with light cycles of 12/12 h, temperature of 20–24 °C, and humidity of 45–65%. The mice received sterile filtered water and a standard diet for rodents ad libitum. The experiments were conducted in agreement with German animal welfare legislation with the approved guidelines of the Society of Laboratory Animals (GV-SOLAS) and the Federation of Laboratory Animal Science Associations (FELASA). Post mortem examination of organs was not subject to regulatory authorization. We used the following mouse lines for this study: C57BL/6J, Flattop-Venus reporter [29], FltpiCre; mTmG [30,31], db/db leptin receptor mutant mice [32], and Nkx6-1-Venus fusion (Nkx6-1-VF) line [33]. The Min6 (clone K9) murine β-cell line and EndoC-βH1 human β-cell line were cultured in adherence as described previously [34,35]. In vitro differentiation of human iPSCs toward the pancreatic endocrine lineage was performed as described [36].
2.2. Islet isolation
Mouse pancreatic islets were isolated as previously described [37] using collagenase P (Roche) to digest the exocrine tissue and OptiPrep density gradient (Sigma) centrifugation to separate the islets from the rest of the digested tissue. Of note, the digestion time of postnatal pancreata was adjusted according to the dimensions of the organs. Islet culture medium was prepared with RPMI 1640 containing 1% penicillin/streptomycin and 10% fetal bovine serum (FBS).
2.3. Flow cytometry
Dispersed islets or cultured cells were sorted and analyzed using FACS-Aria III (BD Bioscience). TrypLE Express Enzyme (1X) (Thermo Fisher Scientific) was used to detach Min6 from the dish or dissociate islets into single cells. Staining was performed using FACS buffer (PBS, 2% FBS and 2 mM EDTA). Cells were incubated with rabbit monoclonal anti-PE-CD81 (#93765, Cell Signaling, 1:50) on ice for 30 min. Alternatively, cells were stained for 30 min on ice with primary antibody (rabbit monoclonal anti-CD81, #10037, Cell Signaling, 1:50) and subsequently for 20 min on ice and in the dark with donkey anti-rabbit IgG Alexa Fluor 647 (A31573, Invitrogen, 1:500). Post-processing analysis was performed using FlowJo software.
2.4. Western blotting
Min6 or sorted primary cells (sorted cells from isolated islets from 3 mice) were lysed in RIPA buffer (75 mM of NaCl, 6.37 mM of sodium deoxycholate, 0.005% NP40, 0.05% SDS, and 25 mM of Tris pH 8). Then 5–15 μg proteins per sample were loaded on SDS-PAGE gel and the samples were transferred to PVDF membranes (Bio-Rad), followed by overnight incubation with primary antibodies (Supplementary Table 1) at 4 °C. After washing, secondary antibodies (Supplementary Table 1) were added for 2 h at room temperature. Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) or SuperSignal West Pico PLUS Chemilumineszenz-Substrat (Thermo Fisher Scientific) were used and the signals were detected by enhanced chemiluminescence.
2.5. Quantitative PCR
RNA was extracted from cells using an miRNeasy Micro kit (Qiagen) and cDNA was generated via a SuperScript VILO cDNA Synthesis kit (Thermo Fisher Scientific). qPCR was performed using a mixture of 5–10 ng of RNA, water, TaqMan Probes (Supplementary Table 1), and TaqMan Advanced Master Mix (Life Technologies) using a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). The results were obtained with the ΔΔCt analysis method.
2.6. In vitro stress assay
Min6 cells were seeded (250,000/well) in 6-well plates as previously described [38,39]. After recovering for 24 h, control medium or medium containing glucose (additional 10 mM) and Palmitate (0.4 mM) were added to the cells for another 4 days. The medium was changed every 48 h. EndoC-βH1 cells were seeded (250,000/well) in 6-well plates as described [40] and after 24 h treated by control medium or media containing tunicamycin (5 μg/ml) or glucose (additional 30 mM) and palmitate (1 mM) for 4 days. The medium was changed every 48 h. Mouse adult islets were isolated and left to recover overnight. Then 60 islets per condition were treated with glucose (additional 25 mM) and palmitate (0.4 mM) for 2 days.
2.7. Immunostaining and microscopy
For cryo-sample preparation, pancreata were fixed in 4% paraformaldehyde (PFA), applied to sucrose gradients, embedded in Optimum Cutting Temperature (OCT) (Leica), and then cut in 20 μm sections. The samples were permeabilized (0.2% Triton X-100 and 0.1 M of glycin) for 30 min and incubated in blocking solution (10% fetal calf serum, 3% donkey serum, 0.1% BSA, and 0.1% Tween-20 in PBS) for 2 h at RT. They were then incubated with primary antibodies (Supplementary Table 1) overnight. After washing, the samples were incubated with secondary antibodies (Supplementary Table 1) for 2 h. Nuclei were stained with DAPI and the samples were embedded in ProLong Gold medium (Life Technologies). For paraffin-embedded samples, pancreata were fixed in formalin, then incubated in ethanol, xylol, and wax solutions and embedded in wax. The organs were then sectioned into 3 μm slides. Before staining, the sections went through a rehydration phase in xylol and ethanol and an antigen-retrieval process (citrate buffer). The rest of the protocol was performed as previously described. To stain Min6, EndoC-βH1, and primary islet cells, the samples were fixed in 4% PFA (10 min at 37 °C) or 100% methanol (15 min on ice) before staining. The cells fixed with PFA but not methanol were permeabilized for 10 min. The cells were then incubated for 1 h in blocking solution and then with primary antibodies (Supplementary Table 1) for 2–3 h at RT. After washing, the cells were stained with secondary antibodies (Supplementary Table 1) and DAPI followed by embedding in ProLong Gold (Life Technologies). All the images were obtained with a ZEISS LSM 880 confocal or Leica confocal SP5 microscopes. Images were analyzed using Fiji, ImageJ, or LAS X software.
2.8. Single-cell RNA sequencing
Libraries of single cells from isolated islets from postnatal day 16 (P16) FltpiCre; mTmG mice were produced using the Chromium Single-Cell 3′ library and gel bead kit v2 (PN #120237) from 10XGenomics. Briefly, we loaded 10,000 cells per channel (x3) of a 10x chip to produce gel bead-in-emulsions (GEMs). Then the samples underwent reverse transcription to barcode RNA followed by cleanup, cDNA amplification, enzymatic fragmentation, 5′ adaptor, and sample index attachment. The samples were sequenced using a HiSeq4000 (Illumina) with 150 bp paired-end sequencing of read2.
2.9. Analysis of scRNA-seq datasets
To analyze the scRNA-seq data from isolated islets from the P16 FltpiCre; mTmG mice, cell type annotation was performed using marker genes for endocrine cells after Louvain clustering. β-cell sub-clustering was performed using the Louvain clustering algorithm with a resolution of 0.5. Differential gene expression between β-cell subclusters was determined with scanpy rank_genes_groups function. For other analyses, processed, normalized, and annotated single-cell RNA sequencing data were downloaded from the following databases with the accession numbers GSE132188 [41], GSE128565 [2], GSE117770 [42], GSE114412 [43], and GSE84133 [44]. The scRNA-seq count data (10X Genomics Chromium protocol) from GSE117770 [41] were downloaded, filtered, normalized by library size (CPM normalization with target sum 10,000), and log+1 scaled. Cell-type annotation was performed using marker genes for endocrine cells after Louvain clustering. The scRNA-seq count data (inDrop protocol) from GSE84133 were provided in a pre-annotated format by the Hemberg lab (https://hemberg-lab.github.io/scRNA.seq.datasets/human/pancreas/retrieved on 08/28/2019). Consistent with other droplet-based datasets, the dataset was normalized by library size and log+1 scaled. In all of the droplet-based datasets, Cd81-positive (and CD81 resp.) cells were selected on the count matrix with a minimum count threshold of one. All the analyses were carried out in scanpy (version 1.4.5.2.dev6+gfa408dc7) in Python 3.7. Differential expression analysis was performed using t-tests as implemented in scanpy or limma (version 3.38.3) in R version 3.5.2. Pathway enrichment analysis was performed with Metascape [45].
2.10. Data availability
ScRNA-seq data from isolated islets from the P16 FltpiCre; mTmG mice were deposited in the Gene Expression Omnibus under accession number GSE161966. All of the codes to reproduce the results from this data as well as fully processed and annotated count matrices will also be publicly available upon acceptance of the manuscript.
3. Results
3.1. CD81 was heterogeneously expressed in mouse pancreatic β-cells
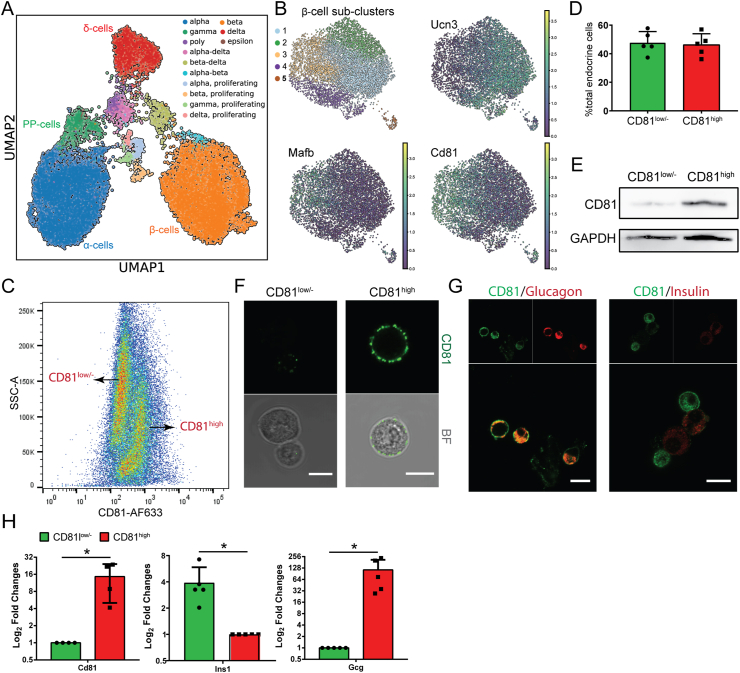
To dissect β-cell heterogeneity and identify novel markers for immature and mature β-cells, we performed high-throughput single-cell RNA sequencing (scRNA-seq) of mouse endocrine cells at postnatal (P) day 16, around which a major change in the expression of markers associated with β-cell maturation occurred [12,14]. Using droplet-based scRNA-seq, we transcriptionally profiled 18.716 single endocrine cells (after quality control). Unsupervised graph-based clustering [46] revealed four major cell clusters including α-, β-, δ-, and PP-cells (Figure 1A, Supplementary Fig. 1A, and Supplementary Table 2). A small fraction of each cluster expressed the cell cycle marker mKi67, indicating the presence of dividing cells at this early postnatal stage (Supplementary Fig. 1B). To explore postnatal β-cell heterogeneity, we performed Louvain clustering (resolution 0.5) of β-cells and identified five main cell clusters (Figure 1B). Among these, we found a differential expression of Cd81 in one of these clusters (cluster 2) compared to all of the others (Figure 1B and Supplementary Table 2). In contrast to a small fraction of Cd81-expressing β-cells, the majority of non-β-endocrine cells expressed this marker (Supplementary Fig. 1C). To validate these findings, we isolated adult mouse islets to analyze and sort by flow cytometry cells expressing CD81 using an anti-CD81 antibody. We found two endocrine subpopulations including a high-level CD81-expressing subpopulation (CD81high) and a subpopulation expressing no CD81 protein or very low levels (hereafter referred to as CD81low/-) (Figure 1C and D). The FACS results were confirmed by Western blotting (Figure 1E) and live cell imaging (Figure 1F) on the sorted populations. Immunostaining and qPCR analyses revealed that a main fraction of CD81low/- cells expressed insulin, whereas the majority of CD81high cells expressed glucagon (Figure 1G and H). Altogether, these findings demonstrate that CD81 has a heterogeneous pattern of expression in β-cells, thus enabling the separation of two distinct subpopulations.
Figure 1.
CD81 divided β-cells into two distinct subpopulations. (A) UMAP of 18.716 single cells from isolated islets from P16 mice. Different colors mark distinct endocrine cell types annotated based on the expression of signature hormone genes. (B) UMAPs representing only the β-cell cluster (7.275 single cells). On the top left, color-coded β-cell sub-clusters (1–5) identified by stochastic clustering (Louvain). The other 3 UMAPs indicate the mRNA expression levels (the lighter the color, the higher the expression) of Ucn3, Mafb, and Cd81 in β-cells. (C) Representative FACS plot of adult mouse isolated islets stained for CD81. (D) Quantification of CD81high and CD81low/- adult endocrine cells by FACS analysis. (E) Representative Western blotting image from sorted CD81high and CD81low/- endocrine cells. (F) Representative confocal pictures of single endocrine cells right after sorting. Scale bar 10 μm. (G) Immunostaining for insulin (Ins), glucagon (Gcg), and CD81 on dispersed islets. Scale bar 10 μm. (H) qPCR analysis of CD81-based sorted adult endocrine cells (∗P < 0.05; t-test).
3.2. CD81 expression levels was gradually reduced in β-cells after birth
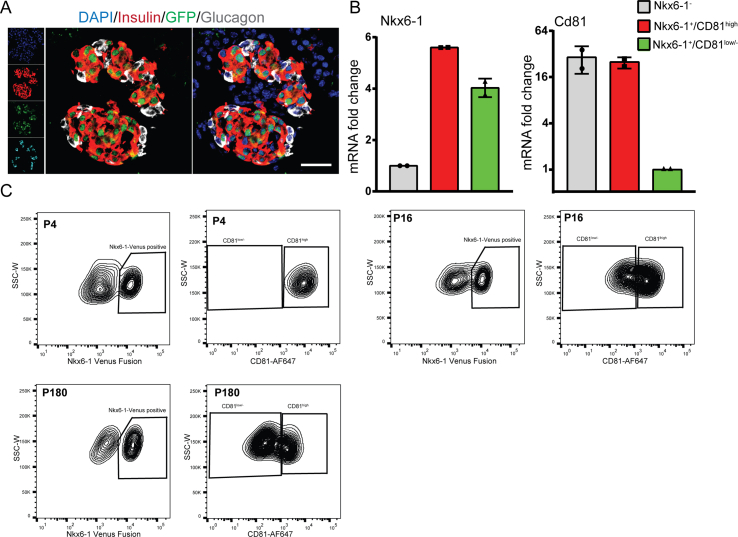
To assess the developmental changes in Cd81 expression in the endocrine lineage, we analyzed the expression of this gene in the scRNA-seq dataset derived from pancreas development at E12.5–15.5 [41] as well as P16 and adult islets [2]. We found that the majority of embryonic (E12.5–15.5) ductal, endocrine progenitors (EPs), Fev+ hormone−, and hormone+ endocrine cells expressed Cd81. However, at these stages, the expression levels of Cd81 were slightly higher in hormone+ endocrine cells than other clusters (Figure 2A). At these embryonic stages, a major fraction of all types of endocrine cells expressed Cd81 (Figure 2B). At P16, the majority of α-, δ-, and PP-cells expressed high levels of Cd81, whereas only a fraction of β-cells was Cd81high (Figure 2C). The portion of Cd81high β-cells was even further reduced in adult islets (Figure 2D). To further validate these findings, we used a novel knock-in Nkx6-1-Venus fusion reporter mouse line (hereafter referred to as Nkx6-1-VF) in which a bright fluorescence Venus protein was fused to the C-terminus of the endogenous β-cell-specific transcription factor (TF) Nkx6-1, allowing specific isolation of β-cells [33] (Supplementary Figs. 2A and B). Using this mouse model, we isolated β-cells at P4, P16, P21, and P180 and performed FACS analysis (Figure 2E and Supplementary Fig. 2C), which revealed a gradual reduction in the number of β-cells expressing high levels of CD81 (Figure 2F). The presence of Nkx6-1-VF+/CD81high and Nkx6-1-VF+/CD81low/- β-cells was also confirmed by immunofluorescence staining (IF) of single β-cells from isolated and dispersed islets (Figure 2G). Taken together, all endocrine cells are CD81high during pancreas development, whereas in early postnatal and adult islets, all endocrine non-β-cells and a small fraction of β-cells express high levels of CD81.
Figure 2.
CD81 expression levels gradually reduced in β-cells during development. (A–D) Violin plots indicating Cd81 expression levels in different pancreatic cell types at different developmental stages. (E) FACS counter plots representing P21 islets isolated from the Nkx6-1-VF mouse line and stained for CD81; the left plot displays the separation of two clusters based on the Venus fluorescence intensity; in the right plot, Nkx6-1-VF+ cells are separated based on CD81 expression levels. (F) Time course FACS analysis of CD81 expression levels in β-cells using the Nkx6-1-VF mouse line (N = 3 for P4 and N = 4 for the rest). (G) Representative image of dispersed β-cells isolated from Nkx6-1-VF islets stained for CD81 and Venus. Yellow asterisks mark CD81low/- β-cells, while white asterisks label CD81high β-cells. Scale bar 20 μm.
3.3. Increased Cd81 expression levels correlated with an immature β-cell profile
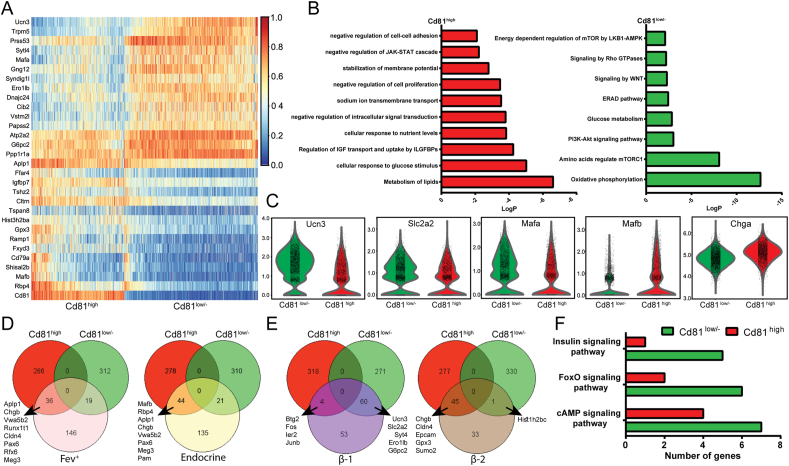
To provide a deeper insight into CD81-based β-cell heterogeneity, we performed differential gene expression analysis of P16 β-cells and identified 322 upregulated and 331 downregulated genes in Cd81high cells compared to Cd81low/- β-cells (Figure 3A and Supplementary Table 2). Gene ontology (GO)-term analysis revealed that Cd81low/- β-cells were enriched for terms related to oxidative phosphorylation, the PI3K pathway, and regulation of mTORC1 by amino acids or the LKB1-AMPK complex (Rheb, Rragd, and Fnip1 and Fnip2, Flcn, Prkab2, and many members of the Atp6v family) (Figure 3B and Supplementary Table 2). This suggested a more mature state of CD81low/- β-cells compared to CD81high cells. In support, we found increased levels of key β-cell maturation markers Ucn3, Glut2 (Slc2a2), Mafa, and Sytl4 in Cd81low/- cells. In contrast, immature β-cell markers Mafb [47], Rbp4 [48], and Chga were highly expressed in Cd81high β-cells (Figure 3A and C). We then compared the differentially expressed genes in Cd81low/- and Cd81high β-cells with the gene profile of embryonic Fev+/hormone− and hormone+ endocrine cells [41]. This analysis revealed a closer profile of Cd81high than Cd81low/- β-cells to the embryonic endocrine cells (Figure 3D and Supplementary Table 2). It has been shown that in healthy adult islets, β-cells cluster into more mature (β-1) and less mature (β-2) cells [2]. Comparing the differentially expressed genes in these clusters with those from postnatal Cd81low/- and Cd81high β-cells revealed a higher similarity of Cd81low/- with β-1 and Cd81high with β-2 cells (Figure 3E and Supplementary Table 3), highlighting the reduced mature phenotype of Cd81high β-cells. In support of this conclusion, targeted pathway enrichment analysis disclosed a higher expression of genes associated with cAMP [49,50], FoxO [51], and insulin-signaling pathways in Cd81low/- compared to Cd81high β-cells (Figure 3F and Supplementary Table 2).
Figure 3.
Cd81 expression levels correlated with the maturation state of β-cells. (A) Heat map representing the gene expression of the top 30 genes differentially regulated in P16 Cd81high and Cd81low/- β-cells. (B) Pathway enrichment analysis obtained from processing the differentially regulated genes of P16 Cd81high and Cd81low/- β-cells. (C) Violin plots of pivotal genes playing a crucial role during the maturation process of β-cells. Ucn3, Slc2a2 (Glut2), and Mafa are upregulated in Cd81low/- β-cells and Mafb and Cgha are upregulated in Cd81high β-cells. (D) Venn diagrams representing the comparison between differentially expressed genes of P16 Cd81high and Cd81low/- β-cells with highly upregulated genes from embryonic Fev+ (left) and endocrine cells (right) described in Bastidas-Ponce et al., 2019. (E) Venn diagrams representing the comparison between differentially expressed genes of P16 Cd81high and Cd81low/- β-cells with highly upregulated genes from β1 (left) and β2 (right) β-cells as described in Sachs et al., 2020. (F) Targeted analysis of specific signaling pathways differentially regulated between P16 Cd81high and Cd81low/- β-cells.
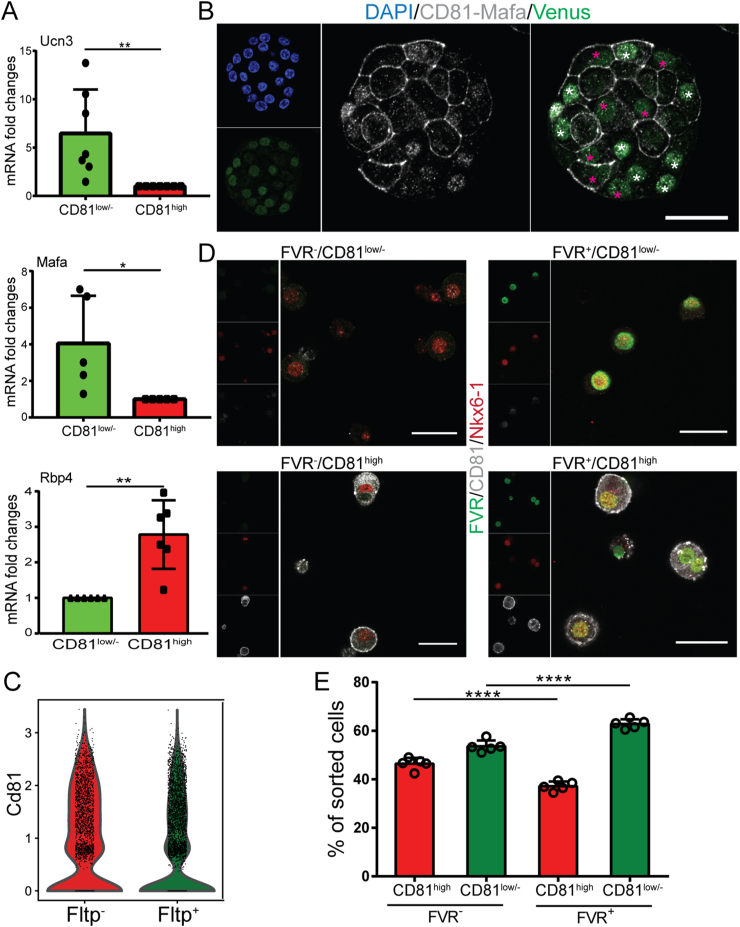
3.4. CD81low/- β-cells exhibited increased expression levels of Mafa and Fltp
To further validate the results from the scRNA-seq data, we first performed qPCR analysis on sorted β-cells from adult Nkx6-1-VF mice that unveiled higher expression levels of mature markers Ucn3 and Mafa in CD81low/- and increased transcriptome of immature marker Rbp4 [48] in CD81high β-cells (Figure 4A). IF staining also revealed a negative correlation between CD81 and Mafa expression in Nkx6-1-VF β-cells (Figure 4B). We previously identified the Wnt/planar cell polarity (PCP) effector protein Flattop (Fltp) as a marker for mature β-cells [4,29]. Hence, we explored the correlation between the expression levels of this marker and Cd81 using our P16 scRNA-seq dataset that was obtained from the FltpiCre; mTmG mouse line. In these animals, the expression of Cre under Fltp promoter activity resulted in excision and switching of the expression of the membrane Tomato (mT) to membrane GFP (mG). Therefore, upon Fltp expression, mT-expressing cells started to express mG, appeared yellow (mTmG), and were green afterward (expressing only mG). Based on this lineage-tracing system, we clustered P16 single β-cells and found increased Cd81 expression in Fltp− (expressing only mT) compared to Fltp+ (expressing mTmG and only mG) β-cells (Figure 4C). To support this data, we utilized the FltpZV knock-in/knock-out mouse line in which the open-reading frame of Fltp is substituted by histone 2B (H2B)-Venus reporter (FVR) [29]. We isolated islets from adult FltpZV/+ heterozygous mice and performed FACS analysis. Based on our sorting scheme, we found four endocrine cell groups including FVR−/CD81low/-, FVR−/CD81high, FVR+/CD81low/-, and FVR+/CD81high subpopulations (Figure 4D). As expected in adult islets, the majority of β-cells were CD81low/-. However, it was evident how the FVR− population was more enriched in CD81high compared to FVR+ (46.4% ± 2.442 vs 37.16% ± 1.936) (Figure 4E). Collectively, these experiments validated the scRNA-seq data and presented CD81 as a novel marker for immature β-cells.
Figure 4.
CD81 as immature β-cell markers. (A) qPCR analysis of maturation markers Ucn3 and Mafa and immature marker Rbp4 in sorted CD81high and CD81low/- β-cells isolated from Nkx6-1-VF mice (∗P < 0.05; ∗∗P < 0.01; t-test). (B) Immunostaining of dispersed islet cells from P21 Nkx6-1-VF mice. Pink asterisks indicate Mafa− β-cells and white asterisks show Mafa+ β-cells. CD81 and Mafa were imaged with the same confocal channel. Scale bar 20 μm. (C) Violin plots representing Cd81 expression levels in Fltp− and Fltp+ β-cells at P16. (D) Immunostaining of sorted islet cells from adult Fltp-Venus reporter (FVR) mice based on FVR and CD81 expression levels indicating the presence of four different subpopulations: FVR−/CD81low/-, FVR−/CD81high, FVR+/CD81low/-, and FVR+/CD81high. Scale bar 50 μm. (E) Quantification of FAC-sorted cells indicating the number of FVR+ and FVR− cells in CD81high (left) and CD81low/- (right) β-cell populations. (∗∗∗P < 0.001; t-test).
3.5. CD81 expression levels were increased in stressed and dedifferentiated β-cells
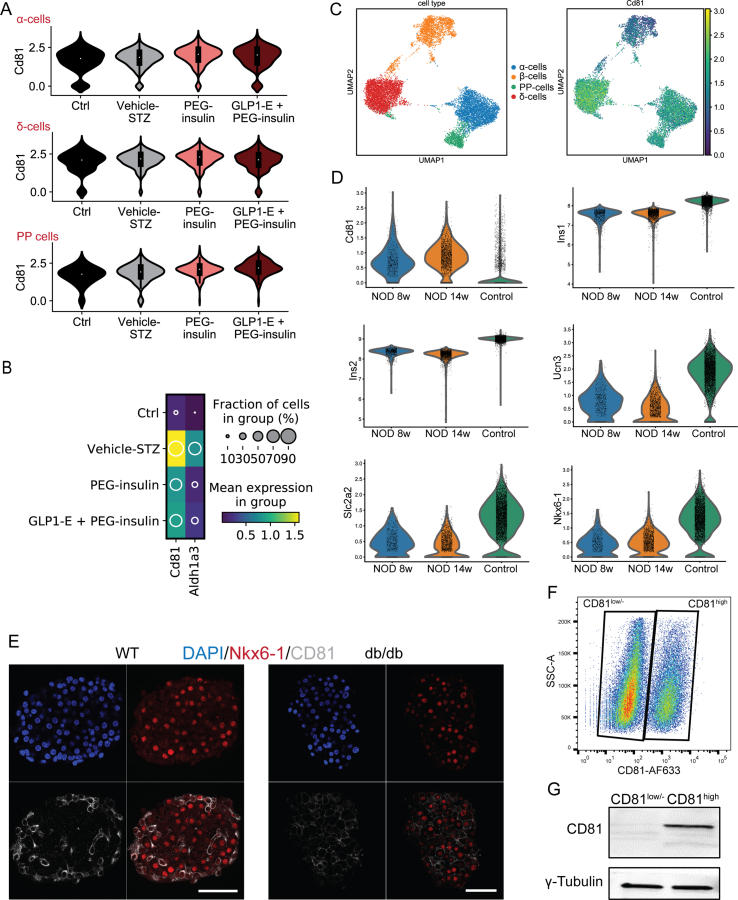
To check if Cd81 expression levels changed in diabetic conditions, we used our recently reported dataset from the multiple low-dose model of streptozotocin-induced (STZ) diabetic mice [2]. First, we plotted Cd81 expression in all endocrine cells from 4 major groups including WT and STZ treated with vehicle, long-acting pegylated insulin analog (PEG-Ins), and a combination of PEG-insulin and GLP-1-oestrogen conjugate (PEG-Ins/GLP-1-Oe) (Figure 5A and B). We found subtle changes in Cd81 transcripts in α-, δ-, and PP-cells between different groups (Figure 5A and Supplementary Fig. 3A). Strikingly, the levels of Cd81 were increased in β-cells from STZ mice but were reduced upon treatment with PEG-Ins and PEG-Ins/GLP-1-Oe (Figure 5A–C), suggesting CD81 as a marker for dedifferentiated β-cells. In support of this notion, we found a similar expression pattern of Cd81 and dedifferentiation marker Aldh1a3 in STZ β-cells (Supplementary Fig. 3B). When we compared the profile of β-cells from STZ mice to those from the P16 stage, they exhibited a higher overlap of differentially expressed genes to Cd81high β-cells. Conversely, STZ β-cells treated with PEG-Ins and PEG-Ins/GLP-1-Oe were transcriptionally more similar to P16 Cd81low/- cells (Figure 5D and E and Supplementary Table 3). Next, we explored the expression changes of Cd81 in the NOD mouse model, which mimics T1D. Although most β-cells undergo apoptosis in these animals, a subpopulation of β-cells resists immunological attacks due to the dedifferentiated phenotype [19,52]. We used a recent scRNA-seq dataset from NOD β-cells at 8 and 14 weeks to test our hypothesis of whether Cd81 was upregulated in a T1D disease model [42] and compared the Cd81 levels with β-cells of control healthy mice [2]. Importantly, we observed an increased level of Cd81 in β-cells but no other non-β endocrine cells from 8- to 14-week-old animals. (Supplementary Figs. 3C and D). To assess the changes in CD81 levels in a T2D model, we used islets from leptin receptor-deficient mice also known as db/db mice [53]. Immunostaining of pancreatic islets from these mice showed a striking increase in the number of CD81high β-cells compared to the WT control islets (Supplementary Fig. 3E). To validate these findings, we mimicked diabetic conditions in vitro by performing a stress assay of cultured Min6 mouse insulinoma cell line. We first showed that these cells also segregated into CD81high and CD81low/- subpopulations (Supplementary Figs. 3F and G). Treating Min6 cells with glucose/palmitate resulted in an increased number of CD81high cells (Figure 5F and G). The same result was also observed with primary isolated β-cells (Figure 5H and I). Altogether, these data demonstrated that exposure to stress increased the number of CD81high β-cells.
Figure 5.
CD81 levels increased in stressed β-cells. (A) The left UMAP represents the different endocrine populations in control and treated animals. The right UMAP indicates the expression levels of Cd81 in different endocrine cell types from different treatments. (B) The left UMAP shows β-cell clusters in control and treated animals. The right UMAP displays the expression levels of Cd81 in β-cells from different treatments. (C) Violin plots representing Cd81 expression levels in β-cells among the different treatments. (D) Venn diagrams showing the comparison between differentially expressed genes of P16 Cd81high and Cd81low/- β-cells with differentially expressed genes in β-cells from STZ, STZ treated with PEG-Ins, and STZ treated with PEG-Ins/GLP-1-Oe. The analyses indicated in A-D were performed from the database described in Sachs et al., 2020. (E) Violin plots of representative genes upregulated (Rbp4 and Aqp4) or downregulated (Ero1lb) in P16 Cd81high compared to Cd81low/- β-cells. Rbp4 and Aqp4 were differentially upregulated in STZ β-cells. Erol1b was differentially upregulated in β-cells from STZ treated with PEG-Ins and PEG-Ins/GLP-1-Oe. (F) Histogram representing FACS analysis of CD81 expression levels in control and glucose-palmitate (Glu.+Pal.)-treated Min6 cells. (G) Quantification of FACS analysis indicating increased CD81 levels in stressed Min6 cells compared to controls (∗P < 0.05; t-test). (H) Histogram showing FACS analysis of CD81 expression levels in control and Glu./Pal.-treated primary β-cells isolated from Nkx6-1-VF adult islets. (I) Quantification of FACS analysis revealing an increase in CD81 levels in stressed primary Nkx6-1-VF-derived β-cells (∗P < 0.05; t-test).
3.6. CD81 expression patterns in healthy and stressed human islets
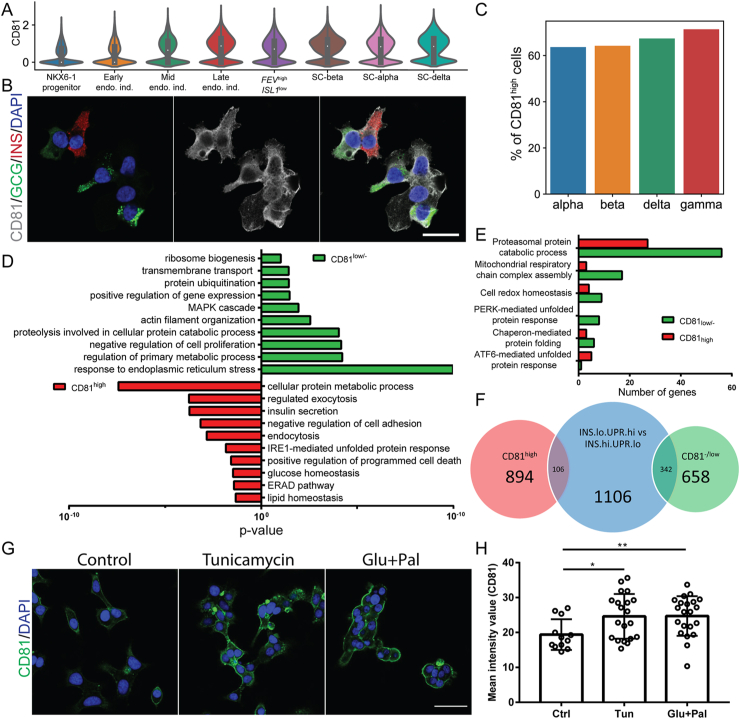
To translate our findings to humans, we first analyzed CD81 expression during human endocrinogenesis. Using the available scRNA-seq dataset from in vitro differentiated human endocrine cells [43], we found a gradual increase in CD81 transcripts during endocrinogenesis. Furthermore, a major fraction of stem cell-derived β–(SC–β), SC-α, and SC-δ cells expressed high levels of CD81 (Figure 6A). Immunostaining of in vitro differentiated endocrine cells also indicated the expression of CD81 protein in SC-β- and SC-α cells (Figure 6B). Next, we reanalyzed the scRNA-seq dataset of isolated islets from healthy adult human donors [44] and found that all of the endocrine cell types were segregated into CD81high and CD81low/- cells (Figure 6C). GO-term analysis of β-cells revealed terms associated with lipid and glucose homeostasis, programmed cell death, vesicle trafficking, secretion in CD81high, and terms related to transmembrane transport, cytoskeleton, and MAPK signaling in CD81low/- cells (Figure 6D and Supplementary Table 3). Both populations were enriched for terms associated with different modes of the ER stress response. While the CD81low/- subpopulation likely activated the PERK pathway in response to milder and physiological ER stress, the CD81high β-cell subpopulation possibly activated the ATF6-mediated unfolded protein response (together with IRE1 and ERAD) as a protective or compensatory response to ER stress (Figure 6D and E). Interestingly, a recent report showed the existence of β-cells with different status based on insulin secretion and unfolded protein response (UPR) in physiological conditions [54]. Pseudotime ordering of single human β-cells indicated that β-cells (not all at the same time) undergo a transition between low and high phases of insulin biosynthesis. The cells that produce high amounts of insulin undergo increased ER stress (INShigh/UPRlow). To cope with this, the cells downregulate insulin biosynthesis and upregulate ER stress coping mechanisms (UPR signaling) (INSlow/UPRhigh) [54]. When we compared the differentially expressed genes between CD81low/- and CD81high human β-cells with those between INSlow/UPRhigh and INShigh/UPRlow β-cells, we found a higher similarity between CD81low/- cells with INSlow/UPRhigh β-cells (Figure 6F and Supplementary Table 3). The data further highlighted the positive correlation between high levels of CD81 expression with increased ER stress levels of β-cells. To further support this concept, we performed an in vitro stress assay using cell line EndoC-βH1 and we found increased levels of CD81 protein upon exposure to different stress conditions including tunicamycin and glucose/palmitate (Figure 6G and H). Collectively, these data showed similar expression patterns of CD81 in healthy and stressed human β-cells as observed in mice.
Figure 6.
CD81 expression patterns in healthy and stressed human β-cells. (A) Violin plots indicating CD81 expression levels in different pancreatic cells during in vitro differentiation of endocrine cells from iPSCs. (B) Immunostaining of CD81 expression in stage (S) 6 of human endocrine cells derived from iPSCs. Scale bar 20 μm. (C) Bar plot showing the percentage of CD81high cells in different endocrine cell types using scRNA-seq of healthy adult human islets. (D) GO-terms enrichment analysis for differentially regulated genes of human CD81high and CD81low/- β-cells. (E) Targeted analysis of specific signaling pathways related to ER stress coping mechanisms differentially regulated between human CD81high and CD81low/- β-cells. (F) Venn diagram showing a comparison between differentially expressed genes of human CD81high and CD81low/- β-cells with β-cell clusters described in Xin et al., 2018. (G) Immunostaining for CD81 (green) in control and treated EndoC-βH1 cells. Scale bar 40 μm. (H) Quantification of the intensity levels of CD81 staining in EndoC-βH1 cells (2 independent experiments; ∗P < 0.05; ∗∗P < 0.01; t-test).
4. Discussion
The concept of β-cell heterogeneity was proposed and discovered several years ago [5,55]. This topic recently regained attention due to the development of single-cell sequencing technologies [48,[56], [57], [58]] and the identification of novel markers that have increased our understanding of β-cell heterogeneity in health and disease. The identification of the Wnt/PCP signaling pathway effector and reporter Fltp linked tissue architecture and polarity to β-cell maturation [4,59] and the identification of surface markers such as the tetraspanin CD9 and ST8SIA1 allow us to distinguish four subtypes of human β-cells [16]. An increasing number of studies have demonstrated functional heterogeneity regarding either metabolism or electrical activity [17,18,48,60]. In this context, identifying an immature β-cell subpopulation with increased mitotic potential suggests that they are ideal target cells for β-cell expansion strategies. Nevertheless, to the best of our knowledge, no surface marker that specifically labels immature β-cells has been reported to date. Therefore, the identification of CD81 in this study is of great importance because this protein not only marks immature β-cells but, due to its plasma membrane localization, can also be used to isolate and/or track this subpopulation. It should be noted that approximately 30% of adult β-cells still express CD81, while only less than 2% of β-cells effectively proliferate [61,62]. This observation could possibly indicate that while a subpopulation of β-cells remains proliferation competent, only a minor fraction is actively undergoing the cell cycle. This together with the observation of other markers differentially regulated within the Cd81high cells (Mafb, Cd79q, and Gpx3) highlights the extreme complexity of β-cell heterogeneity.
We found that the levels of CD81 increased during endocrinogenesis in mice and humans. This data proposes a possible important role of CD81 in endocrine lineage formation during development. A recent study identified a subset of CD81-positive beige adipocyte progenitor cells that are proliferative and contribute to maintenance cellular composition of adipose tissue [63]. This study illustrated that β-cells are not the only cell population exhibiting cellular heterogeneity based on CD81 expression levels. Importantly, CD81 was shown to play a role for de novo biogenesis of beige fat upon cold exposure. Mechanistically, this protein exerted its function by forming a complex with integrins to activate focal adhesion kinase (FAK) signaling. Integrin-mediating mechanosignaling has also been shown to dictate fate decisions of pancreatic progenitors [64]. The interaction between the extracellular matrix with integrin α5 activates a F-actin-YAP1-Notch signaling cascade that triggers differentiation of pancreatic progenitors toward the ductal lineage at the expense of endocrinogenesis. Future studies should address if CD81 mediates neighboring cell interactions and mechanosignaling during endocrine lineage decision. Furthermore, whether this protein plays a function in human endocrine lineage induction needs to be explored in more detail.
CD81 likely marks immature β-cells as its reduced expression levels are associated with increased gene regulatory networks involved with maintaining β-cell maturation [4,37]. A recent study showed that the maintenance of an immature β-cell subpopulation in adult healthy islets was connected to heterogeneity of PDX1/MAFA TFs, which is necessary for the normal physiological function of islets [65]. While the expression levels of CD81 were gradually restricted to a subpopulation of mouse β-cells, it was expressed in all non-β-endocrine cells in adult stages. This finding discloses that in the maturation process, β-cells but no other endocrine cells downregulate CD81 expression levels. If and how the expression levels of CD81 are associated with the maintenance of an immature state in β-cells is unknown. One possible scenario is that due to its surface localization, CD81 might be involved in triggering or repressing signaling cascades important for β-cell immaturity. Furthermore, the interaction of CD81 with other surface proteins may play a role. Along this line, we found that the two CD81 subpopulations differed in their surface molecule profiles (higher expression levels of Cldn3, Cldn4, Cldn6, Mslnl, Alcam, and Aqp4 in CD81high and higher expression levels of Ninj1, Tjp2, and Gabbr2 in CD81low/- β-cells; Supplementary Table 2). Moreover, CD81 might play a role in the interaction of β-cells with other endocrine and non-endocrine cell types, which might also influence the secretory capacity of β-cells. Generating a β-cell-specific CD81 knock-out mouse model will address whether CD81 function is essential for preserving β-cell immaturity. Notably, the expression pattern of CD81 differs in human endocrine cells, since both α- and β-cells are heterogeneous for CD81. This raises the question of why such differences exist and whether the islet architecture and neighboring cell-type interactions influence the expression of CD81 and determine an immature phenotype [5].
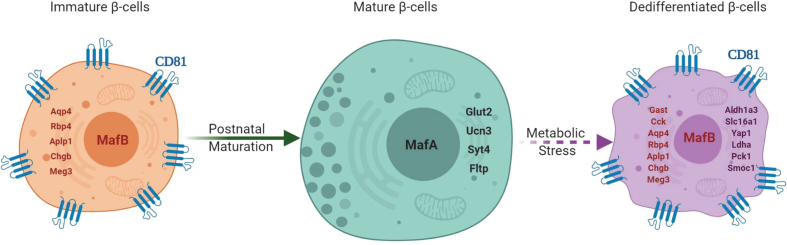
Healthy human β-cells have shown different levels of ER stress [54] that proposes an additional layer of β-cell heterogeneity, which correlates with CD81 expression levels. Moreover, the expression of CD81 was increased in different models of stressed β-cells in mice and humans. Although these findings clearly indicate a correlation between CD81 expression and β-cell stress levels, further investigations are required to dissect if and how CD81 might impact different routes of response to stress in β-cells. Because we identified CD81 as a marker for immature β-cells, the upregulation of this gene in stressed cells suggests an increase in their immaturity profile. Along this line, we recently reported that STZ treatment results in a significant shift in the β-cell profile toward a dedifferentiated state [2]. These adult dedifferentiated β-cells had a close transcriptional profile with immature β-cells. Similarly, dedifferentiated β-cells had a closer profile to P16 Cd81high than Cd81low/- cells, but the common shared upregulated genes between STZ and Cd81high β-cells were only a fraction of their total differentially expressed genes. Thus, these findings suggest that CD81 marks immature β-cells in healthy islets and labels dedifferentiated β-cells in metabolically stressed environments, such as during diabetes progression (Supplementary Fig. 4). Further studies are required to assess functional confirmation (GSIS, calcium activity, and mitochondria activity) of the transcriptomic data. Furthermore, as CD81 is expressed in immune cells [28], its identification on the surface of β-cells paves the way to several hypothesis regarding the pathogenesis of T1D. Provokingly, are CD81-expressing β-cells more or less visible to the primed immune system? Alternatively, is CD81 a possible entry point for viruses triggering the autoimmune response? Future studies should address these questions.
In summary, the identification of CD81 provides a valuable tool to target immature and diseased β-cells for regeneration. Because CD81 is a surface protein and is especially upregulated in dedifferentiated β-cells, it can offer a novel mediator for targeted delivery. Future studies should explore the possibility of conjugating antibodies against CD81 to small molecules, siRNA, and signaling factors capable of β-cell expansion or restoring their function. Along this line, a recent work reported the isolation of different endocrine cell types in mice and humans using a combination of CD24 and CD71 [66]. Based on this study, β-cells express low levels of CD24 and are positive for CD71. Although CD71 expression increases at the postnatal stage, it does not segregate β-cells into mature and immature populations. Thus, the combination of CD81 with these novel surface markers might provide a better strategy to target immature and dedifferentiated β-cells.
Author contributions
CS conceptualized and designed the study, performed the experiments, analyzed the data, and wrote the manuscript. M. Büttner analyzed the scRNA-seq data and helped draft the manuscript. ST reanalyzed the scRNA-seq data. PC, MTM, ABP, KS, IB, and AB performed the experiments. FJT supervised the scRNA-seq data analysis. M. Bakhti conceptualized and designed the study, interpreted the data, and wrote the manuscript. HL conceptualized the study, contributed to interpreting the results, provided financial support, and critically reviewed the manuscript. All of the authors approved the final version of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Mara Catani for her valuable comments on the manuscript. We thank Jessica Jaki for her technical support. This study was supported by the Helmholtz-Gemeinschaft (Helmholtz Portfolio Theme Metabolic Dysfunction and Common Disease) and Deutsches Zentrum für Diabetesforschung (DZD).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101188.
Contributor Information
Mostafa Bakhti, Email: mostafa.bakhti@helmholtz-muenchen.de.
Heiko Lickert, Email: heiko.lickert@helmholtz-muenchen.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig S1.
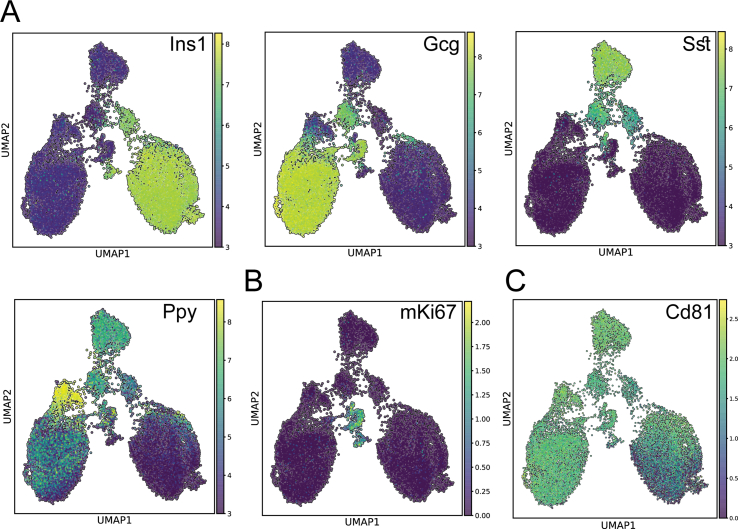
(A) UMAPs indicating the mRNA expression levels of the major islets’ hormones, respectively: insulin (Ins), glucagon (Gcg), somatostatin (Sst), and polypeptide y (Ppy). (B) UMAP depicting the expression levels of mKi67 as a marker for proliferating cells. (C) UMAP representing Cd81 expression levels in all of the endocrine lineages.
Fig S2.
(A) Immunofluorescence staining of cryosections from Nkx6-1-VF mouse pancreas depicting two islets of Langerhans. Insulin (red), glucagon (gray), Nkx6-1 Venus fusion (green), and DAPI (blue). Scale bar 50 μm. (B) Confirmation of the sorting strategy by RT-qPCR analysis of Nkx6-1 and Cd81 levels in sorted Nkx6-1 Venus fusion-negative and Nkx6-1 Venus fusion-positive CD81high and CD81low/- cells. (C) Representative FACS plots showing the separation of endocrine cells based on the Venus fluorescent signal (for Nkx6-1) and CD81 staining for three different ages (P4, P16, and P180).
Fig S3.
(A) Violin plots from the Sachs et al. dataset indicating no significant changes in mRNA expression levels of Cd81 in non-β-endocrine lineages (α-, δ-, and PP-cells) for the 4 different conditions. (B) Dot plot showing the expression levels of Cd81 and Aldh1a3 in the β-cell populations from different conditions from the Sachs et al. dataset. (C) UMAP depicting the 4 islet cell type populations in NOD mice at 8 and 14 weeks of age and the Cd81 expression levels for each cluster, both from Thompson et al. (D) Violin plots representing Cd81, Ins1, Ins2, Ucn3, Slc2a2, and Nkx6-1 expression levels in NOD β-cells at 8 and 14 weeks and control β-cells (from Sachs et al. 2020). (E) Whole mount immunofluorescent staining of an islet from adult WT and db/db mice showing an increased number of CD81high β-cells in the db/db mice. Scale bar 50 μm. (F) Representative FACS dot plot of Min6 K8 cells stained for CD81. (G) WB image of CD81-based sorted Min6 cells.
Fig S4.
A schematic image illustrating CD81 expression patterns during β-cell maturation and dedifferentiation. This schematic was designed using BioRender.com.
References
- 1.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs S., Bastidas-Ponce A., Tritschler S., Bakhti M., Böttcher A., Sánchez-Garrido M.A. Targeted pharmacological therapy restores β-cell function for diabetes remission. Nature Metabolism. 2020;2(2):192–209. doi: 10.1038/s42255-020-0201-1. [DOI] [PubMed] [Google Scholar]
- 3.Avrahami D., Wang Y.J., Schug J., Feleke E., Gao L., Liu C. Single-cell transcriptomics of human islet ontogeny defines the molecular basis of β-cell dedifferentiation in T2D. Molecular Metabolism. 2020;42:101057. doi: 10.1016/j.molmet.2020.101057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader E., Migliorini A., Gegg M., Moruzzi N., Gerdes J., Roscioni S. Identification of proliferative and mature β -cells in the islet of Langerhans. Nature. 2016;535(7612):430–434. doi: 10.1038/nature18624. [DOI] [PubMed] [Google Scholar]
- 5.Roscioni S.S., Migliorini A., Gegg M., Lickert H. Impact of islet architecture on β-cell heterogeneity, plasticity and function. Nature Reviews Endocrinology. 2016;12(12):695–709. doi: 10.1038/nrendo.2016.147. [DOI] [PubMed] [Google Scholar]
- 6.Yoshihara E., O'Connor C., Gasser E., Wei Z., Oh T.G., Tseng T.W. Immune-evasive human islet-like organoids ameliorate diabetes. Nature. 2020;86:606–611. doi: 10.1038/s41586-020-2631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastidas-Ponce A., Scheibner K., Lickert H., Bakhti M. Cellular and molecular mechanisms coordinating pancreas development. Development. 2017;144(16):2873–2888. doi: 10.1242/dev.140756. [DOI] [PubMed] [Google Scholar]
- 8.Bakhti M., Böttcher A., Lickert H. Modelling the endocrine pancreas in health and disease. Nature Reviews Endocrinology. 2019;15(3) doi: 10.1038/s41574-018-0132-z. [DOI] [PubMed] [Google Scholar]
- 9.Huang C., Walker E.M., Dadi P.K., Hu R., Xu Y., Zhang W. Synaptotagmin 4 regulates pancreatic β cell maturation by modulating the Ca2+ sensitivity of insulin secretion vesicles. Developmental Cell. 2018;45(3):347–361. doi: 10.1016/j.devcel.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salinno C., Cota P., Bastidas-ponce A., Tarquis-medina M., Lickert H., Bakhti M. β -cell maturation and identity in health and disease. International Journal of Molecular Sciences. 2019;20:5417. doi: 10.3390/ijms20215417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helman A., Cangelosi A.L., Davis J.C., Pham Q., Rothman A., Faust A.L. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metabolism. 2020;31(5):1004–1016. doi: 10.1016/j.cmet.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolovich-Rain M., Enk J., Vikesa J., Nielsen F., Saada A., Glaser B. Weaning triggers a maturation step of pancreatic β cells. Developmental Cell. 2015;32(5):535–545. doi: 10.1016/j.devcel.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura W., Takahashi S., Yasuda K. MafA is critical for maintenance of the mature beta cell phenotype in mice. Diabetologia. 2015;58(3):566–574. doi: 10.1007/s00125-014-3464-9. [DOI] [PubMed] [Google Scholar]
- 14.Blum B., Hrvatin S., Schuetz C., Bonal C., Rezania A., Melton D.a. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature Biotechnology. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaafar R., Tran S., Shah A.N., Sun G., Valdearcos M., Marchetti P. MTORC1-to-AMPK switching underlies βcell metabolic plasticity during maturation and diabetes. Journal of Clinical Investigation. 2019;129(10):4124–4137. doi: 10.1172/JCI127021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorrell C., Schug J., Canaday P.S., Russ H.A., Tarlow B.D., Grompe M.T. Human islets contain four distinct subtypes of [beta] cells. Nature Communications. 2016;7:1–9. doi: 10.1038/NCOMMS11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston N.R., Mitchell R.K., Haythorne E., Trauner D., Rutter G.A., Hodson Correspondence D.J. Beta cell hubs dictate pancreatic islet responses to glucose. Cell Metabolism. 2016;24:1–13. doi: 10.1016/j.cmet.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salem V., Silva L.D., Suba K., Georgiadou E., Neda Mousavy Gharavy S., Akhtar N. Leader β-cells coordinate Ca2+ dynamics across pancreatic islets in vivo. Nature Metabolism. 2019;1(6):615–629. doi: 10.1038/s42255-019-0075-2. [DOI] [PubMed] [Google Scholar]
- 19.Rui J., Deng S., Arazi A., Perdigoto A.L., Liu Z., Herold K.C. β cells that resist immunological attack develop during progression of autoimmune diabetes in NOD mice. Cell Metabolism. 2017;25(3):727–738. doi: 10.1016/j.cmet.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Meulen T., Mawla A.M., DiGruccio M.R., Adams M.W., Nies V., Dólleman S. Virgin beta cells persist throughout life at a neogenic niche within pancreatic islets. Cell Metabolism. 2017;25:911–926. doi: 10.1016/j.cmet.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren R., Takahashi S., Doss C., Levy R., Levy S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Molecular and Cellular Biology. 1990;10(8):4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charrin S., Jouannet S., Boucheix C., Rubinstein E. Tetraspanins at a glance. Journal of Cell Science. 2014;127(17):3641–3648. doi: 10.1242/jcs.154906. [DOI] [PubMed] [Google Scholar]
- 23.Hemler M.E. Tetraspanin functions and associated microdomains. Nature Reviews Molecular Cell Biology. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 24.Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends in Cell Biology. 2009;19(9):434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Nydegger S., Khurana S., Krementsov D.N., Foti M., Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. Journal of Cell Biology. 2006;173(5):795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y., Bhushan B., Mars W.M., Bowen W., Tao J., Orr A. Phosphorylated Ezrin (Thr567) regulates hippo pathway and yes-associated protein (yap) in liver. American Journal of Pathology. 2020;190(7):1427–1437. doi: 10.1016/j.ajpath.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruening J., Lasswitz L., Banse P., Kahl S., Marinach C., Vondran F.W. Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathogens. 2020;14(7) doi: 10.1016/j.ajpath.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy S. Function of the tetraspanin molecule CD81 in B and T cells. Immunologic Research. 2014;58(2–3):179–185. doi: 10.1007/s12026-014-8490-7. [DOI] [PubMed] [Google Scholar]
- 29.Gegg M., Böttcher A., Burtscher I., Hasenoeder S., Van Campenhout C., Aichler M. Flattop regulates basal body docking and positioning in mono- and multiciliated cells. ELife. 2014;3:1–24. doi: 10.7554/eLife.03842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange A., Gegg M., Burtscher I., Bengel D., Kremmer E., Lickert H. Fltp T2AiCre: a new knock-in mouse line for conditional gene targeting in distinct mono- and multiciliated tissues. Differentiation. 2012;83(2):S105–S113. doi: 10.1016/j.diff.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Muzumdar M.D., Tasic B., Miyamichi K., Li N., Luo L. A global double-fluorescent cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 32.Hummel K.P., Dickie M.M., Coleman D.L. Diabetes, a new mutation in the mouse. Science. 1966;153(3740):1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- 33.Burtscher I., Tarquis-medina M., Salinno C., Beckenbauer J., Bakhti M., Lickert H. Generation of a novel Nkx6-1 Venus fusion reporter mouse line. BioRxiv. 2021 doi: 10.1101/2021.01.29.428800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y. Establishment of a pancreatic b cell line that retains glucose inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 35.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhti M., Scheibner K., Tritschler S., Bastidas-Ponce A., Tarquis-Medina M., Theis F.J. Establishment of a high-resolution 3D modeling system for studying pancreatic epithelial cell biology in vitro. Molecular Metabolism. 2019;30(September):16–29. doi: 10.1016/j.molmet.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastidas-Ponce A., Roscioni S.S., Burtscher I., Bader E., Sterr M., Bakhti M. Foxa2 and Pdx1 cooperatively regulate postnatal maturation of pancreatic β-cells. Molecular Metabolism. 2017;6(6):524–534. doi: 10.1016/j.molmet.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki M., Minami K., Shibasaki T., Miki T., Miyazaki J., Seino S. Establishment of new clonal pancreatic β-cell lines (MIN6-K) useful for study of incretin/cyclic adenosine monophosphate signaling. Journal of Diabetes Investigation. 2010;1(4):137–142. doi: 10.1111/j.2040-1124.2010.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minami K., Yano H., Miki T., Nagashima K., Wang C.Z., Tanaka H. Insulin secretion and differential gene expression in glucose-responsive and -unresponsive MIN6 sublines. American Journal of Physiology - Endocrinology And Metabolism. 2000;279(4 42–4):E773–E781. doi: 10.1152/ajpendo.2000.279.4.e773. [DOI] [PubMed] [Google Scholar]
- 40.Tsonkova V.G., Sand F.W., Wolf X.A., Grunnet L.G., Kirstine Ringgaard A., Ingvorsen C. The EndoC-βH1 cell line is a valid model of human beta cells and applicable for screenings to identify novel drug target candidates. Molecular Metabolism. 2018;8(December 2017):144–157. doi: 10.1016/j.molmet.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bastidas-Ponce A., Tritschler S., Dony L., Scheibner K., Tarquis-Medina M., Salinno C. Comprehensive single-cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development. 2019;146(12):173849. doi: 10.1242/dev.173849. [DOI] [PubMed] [Google Scholar]
- 42.Thompson P.J., Shah A., Ntranos V., Van Gool F., Atkinson M., Bhushan A. Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metabolism. 2019;29(5):1045–1060. doi: 10.1016/j.cmet.2019.01.021. e10. [DOI] [PubMed] [Google Scholar]
- 43.Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H.-R., Harb G. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569(7756):368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Systems. 2016;3(4):346–360. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blondel V.D., Guillaume J.L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008(10) doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 47.Nishimura W., Kondo T., Salameh T., El Khattabi I., Dodge R., Bonner-Weir S. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Developmental Biology. 2006;293(2):526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camunas-Soler J., Dai X.Q., Hang Y., Bautista A., Lyon J., Suzuki K. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metabolism. 2020;31(5):1017–1031. doi: 10.1016/j.cmet.2020.04.005. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seino S., Shibasaki T., Minami K. Pancreatic β-cell signaling: toward better understanding of diabetes and its treatment. Proceedings of the Japan Academy Series B Physical and Biological Sciences. 2010;86(6):563–577. doi: 10.2183/pjab.86.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tengholm A. Cyclic AMP dynamics in the pancreatic β-cell. Upsala Journal of Medical Sciences. 2012;117(4):355–369. doi: 10.3109/03009734.2012.724732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glauser D.A., Schlegel W. The emerging role of FOXO transcription factors in pancreatic β cells. Journal of Endocrinology. 2007;193(2):195–207. doi: 10.1677/JOE-06-0191. [DOI] [PubMed] [Google Scholar]
- 52.Lee H., Lee Y.S., Harenda Q., Pietrzak S., Oktay H.Z., Schreiber S. Beta cell dedifferentiation induced by IRE1α deletion prevents type 1 diabetes. Cell Metabolism. 2020;31(4):822–836.e5. doi: 10.1016/j.cmet.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B., Chandrasekera P., Pippin J. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Current Diabetes Reviews. 2014;10(2):131–145. doi: 10.2174/1573399810666140508121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin Y., Gutierrez G.D., Okamoto H., Kim J., Lee A.H., Adler C. Pseudotime ordering of single human B-cells reveals states of insulin production and unfolded protein response. Diabetes. 2018;67(9):1783–1794. doi: 10.2337/db18-0365. [DOI] [PubMed] [Google Scholar]
- 55.Hellerstrom C., Petersson B., Hellman B. Some properties of the B cells in the islet of Langerhans studied with regard to the position of the cells. Acta Endocrinologica. 1960;34:449–456. doi: 10.1530/acta.0.xxxiv0449. [DOI] [PubMed] [Google Scholar]
- 56.Muraro M.J., Dharmadhikari G., Gr??n D., Groen N., Dielen T., Jansen E. A single-cell transcriptome atlas of the human pancreas. Cell Systems. 2016;3(4):385–394. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tritschler S., Theis F.J., Lickert H., Böttcher A. Systematic single-cell analysis provides new insights into heterogeneity and plasticity of the pancreas. Molecular Metabolism. 2017;6(9):974–990. doi: 10.1016/j.molmet.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y.J., Golson M.L., Schug J., Traum D., Liu C., Vivek K. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metabolism. 2016;24(4):616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghazizadeh Z., Kao D.I., Amin S., Cook B., Rao S., Zhou T. ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nature Communications. 2017;8(1):298. doi: 10.1038/s41467-017-00129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasteska D., Viloria K., Everett L., Hodson D.J. Informing β-cell regeneration strategies using studies of heterogeneity. Molecular Metabolism. 2019;27:S49–S59. doi: 10.1016/j.molmet.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulkarni R.N., Mizrachi E.B., Ocana A.G., Stewart A.F. Human β-cell proliferation and intracellular signaling: driving in the dark without a road map. Diabetes. 2012;61(9):2205–2213. doi: 10.2337/db12-0018. Epub 2012 Jun 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teta M., Long S.Y., Wartschow L.M., Rankin M.M., Kushner J.A. Very slow turnover of β-cells in aged adult mice. Diabetes. 2005;54(9):2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 63.Oguri Y., Shinoda K., Kim H., Alba D.L., Bolus W.R., Wang Q. CD81 controls beige fat progenitor cell growth and energy balance via FAK signaling. Cell. 2020;182(3):563–577. doi: 10.1016/j.cell.2020.06.021. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mamidi A., Prawiro C., Seymour P.A., Lichtenberg K.H. D.e., Jackson A., Serup P. Mechanosignaling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- 65.Nasteska D., Fine N.H.F., Ashford F.B., Cuozzo F., Viloria K., Smith G. PDX1LOW MAFALOW β-cells contribute to islet function and insulin release. Nature Communications. 2021;12(647) doi: 10.1038/s41467-020-20632-z. https://doi.org/10.1038/s41467-020-20632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berthault C., Staels W., Scharfmann R. Purification of pancreatic endocrine subsets reveals increased iron metabolism in beta-cells. Molecular Metabolism. 2020:101060. doi: 10.1016/j.molmet.2020.101060. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ScRNA-seq data from isolated islets from the P16 FltpiCre; mTmG mice were deposited in the Gene Expression Omnibus under accession number GSE161966. All of the codes to reproduce the results from this data as well as fully processed and annotated count matrices will also be publicly available upon acceptance of the manuscript.