Abstract
Pseudomonas putida is widely recognized as a spoiler of fresh foods under cold storage, and recently associated also with infections in clinical settings. The presence of antibiotic resistance genes (ARGs) could be acquired and transmitted by horizontal genetic transfer and further increase the risk associated with its persistence in food and the need to be deeper investigated. Thus, in this work we presented a genomic and phenotypic analysis of the psychrotrophic P. putida ITEM 17297 to provide new insight into AR mechanisms by this species until now widely studied only for its spoilage traits. ITEM 17297 displayed resistance to several classes of antibiotics and it also formed huge amounts of biofilm; this latter registered increases at 15 °C in comparison to the optimum growth condition (30 °C). After ITEM 17297 biofilms exposure to antibiotic concentrations higher than 10-fold their MIC values no eradication occurred; interestingly, biomasses of biofilm cultivated at 15 °C increased their amount in a dose-dependent manner. Genomic analyses revealed determinants (RND-systems, ABC-transporters, and MFS-efflux pumps) for multi-drugs resistance (β-lactams, macrolides, nalidixic acid, tetracycline, fusidic acid and bacitracin) and a novel ampC allele. Biofilm and motility related pathways were depicted underlying their contribution to AR. Based on these results, underestimated psychrotrophic pseudomonas, such as the herein studied ITEM 17297 strain, might assume relevance in relation to the risk associated with the transfer of antimicrobial resistance genes to humans through cold stored contaminated foods. P. putida biofilm and AR related molecular targets herein identified will provide a basis to clarify the interaction between AR and biofilm formation and to develop novel strategies to counteract the persistence of multidrug resistant P. putida in the food chain.
Keywords: Ready-to-eat foods, Antibiotic resistance, Genome sequencing, Lactamase, Cold storage, Biofilm eradication
Graphical abstract
Highlights
-
•
Multidrug resistant Pseudomonas putida ITEM 17297 was isolated from fresh vegetables.
-
•
Determinants for AR and biofilm formation were identified by genomic analysis.
-
•
Biofilm increased more than 10-fold antibiotic MIC value of planktonic cells.
-
•
Cold adapted biofilm increased its biomass under CHL, NA, and ERY pressure.
-
•
New insight into the risk for P. putida spread in the food chain were provided.
Abbreviations:
- RTE
ready-to-eat
- AR
antibiotic resistance
- ARGs
antibiotic resistance genes
- MDR
multi-drug-resistance
- NCBI
National Center for Biotechnology Information
- MAFFT
Multiple Alignment using Fast Fourier Transform
- MIC
minimal inhibitory concentration
- CV
crystal violet
- MBEC
minimum biofilm eradication concentration
- RND
resistance-nodulation-division
- ABC
ATP-binding cassette
- PBPs
penicillin-binding proteins
- EPS
exopolysaccharides
1. Introduction
Pseudomonas putida is a widespread Gram negative non-fermenting bacterium frequently occurring in a number of niches, especially in soil (Negi et al., 2011), wastewater and freshwater (Aresta et al., 2010; Igbinosa et al., 2019), as well as on fresh or minimally processed vegetables (Kaczmarek et al., 2019), fresh dairy and meat products (Gennari and Dragotto, 1992), indoor environments (Remold et al., 2015) and clinical settings (Molina et al., 2014; Peter et al., 2017).
Besides its biotechnological applications in bioremediation of polluted soils (Abatenh et al., 2017), and similarly to other Pseudomonas spp. with high metabolic versatility and psychrotrophic behaviour (Clarke, 1982; Quintieri et al., 2020), P. putida is also known to spoil fresh-cut vegetables (Baruzzi et al., 2015), dairy and meat products during cold storage (Odeyemi et al., 2020; Papadopoulou et al., 2020). To face this issue, new and sustainable approaches based on the application of antimicrobial peptides, neutral electrolyzed water and essential oils have been recently proposed (Baruzzi et al., 2015; Quintieri et al., 2015; György et al., 2020).
Although it is considered to be of low pathogenicity, this bacterium has been recently recognized as the causal agent of several hospital infections, above all in immunocompromised patients (Liu et al., 2014; Fernández et al., 2015; Chamon et al., 2020). In addition, the risk associated with this species is worsened by the spread of multi drug-resistant (MDR) P. putida strains, that are recently becoming a cause for concern (Peter et al., 2017; Raphael and Riley 2017; Tan et al., 2019); resistance factors that are located in mobile genetic elements can be also transmitted by P. putida to other bacterial species or even genera by horizontal gene transfer, giving this germ a role of “reservoir” for the spread of antibiotic resistance (Molina et al., 2014; Peter et al., 2017; Lu et al., 2020). Likewise to other microorganisms (Stewart and Costerton, 2001), multidrug resistance in P. putida was proved to be increased by the ability to form biofilm (Molina et al., 2014). Biofilm also promotes bacterial spread and colonization in the environment; for the switch from planktonic to sessile state, bacteria enable a sophisticated translocation machinery to reach a surface and promote attachment; motility also positively affects biofilm architecture and increased resistance to antibiotics (Wood et al., 2006; Lai et al., 2009).
Pseudomonas putida ITEM 17297 was isolated (Baruzzi et al., 2015) from refrigerated ready-to-eat curly endive (Cichorium endivia L. var. crispum Lam.); among the isolated strains, this one exhibited the highest spoilage activity during cold storage (Baruzzi et al., 2015). In several psychrotrophic pseudomonads isolated from spoiled foods, biofilm life style is favored at low temperatures and promotes the synthesis of spoilage-related enzymes (Quintieri et al., 2019a,b; Rossi et al., 2018). These traits together with the ability to produce pigments are also encountered in P. putida species. In addition to this, several studies provided information on plant resistome and its relationship with environmental bacteria: antibiotic resistance genes (ARGs)-bearing bacteria may attach to the leaf surfaces of vegetables, survive and colonize tissues or other parts of plants (Zhang et al., 2019). The transmission route of ARGs from soil to plants and vegetables highlights the risk of resistome migration to the food chain: considering that fresh vegetables are often eaten raw, this could represent a serious additional food safety concern (Chen et al., 2019; Quintieri et al., 2019a,b).
In light of these considerations, in this work P. putida ITEM 17297 was first genomically characterized aiming at revealing genetic resistance determinants for common antibiotics; then, susceptibility to the main antibiotics and the mutual influence with biofilm formation was studied under in vitro optimal growth conditions and at 15 °C.
2.1. Materials and methods
2.1.2. Culture conditions
Pseudomonas putida ITEM 17297, previously isolated from fresh vegetables (Baruzzi et al., 2015), was maintained at -80 °C as pure stock cultures in Nutrient Broth (NB; Oxoid S.p.A., Rodano, Milan, Italy) supplemented with glycerol 30% (vol/vol). The strain was routinely refreshed (30 °C, 24 h) by streaking onto Luria Bertani (LB; Sigma Aldrich, Milan, Italy) agar, and cultivated for 16 h at 30 °C and 150 strokes/min into 5 mL of LB broth to prepare the bacterial inocula for the subsequent experiments.
2.1.2. Genome sequencing and assembly
The DNA was extracted from a single colony of P. putida ITEM 17297 by using the Wizard® Genomic DNA Purification Kit (Promega). The integrity, purity and quantity of DNA were assessed as previously described by Fanelli et al. (2017) and submitted to IGA Technology Services (Udine, Italy) for whole-genome shotgun sequencing using the Illumina HiSeq2500 platform.
A preliminary evaluation of the quality of the raw data was performed with FastQC software. De novo assemblies were performed using SPAdes version 3.5.0 software within the Galaxy platform (Afgan et al., 2016). The overall contiguity of the assembly and genome statistics were determined with MIGA (Rodriguez et al., 2018).
2.1.3. Bioinformatic methods
Genes were predicted and annotated using the PROKKA pipeline implemented in the Galaxy platform (https://usegalaxy.org/; Galaxy Tool Version 1.0.0; Afgan et al., 2016). The predicted proteins were submitted to the PFAM annotator tool within the Galaxy platform in order to predict the pfam domains. Protein ID used in the manuscript indicated those obtained by NCBI (National Center for Biotechnology Information) Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016).
All the protein sequences used in this study were retrieved from GenBank (NCBI). The homology-based relationship of ITEM 17297 predicted proteins towards selected proteins was determined by the BLASTP algorithm on the NCBI site (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene models were manually determined, and clustering and orientation were subsequently deduced for the closely linked genes.
Phylogenetic analysis was performed as described in Quintieri et al. (2020) with modifications. The alignment of the concatenated dataset of 4 housekeeping genes (16S rRNA, rpoD, rpoB and gyrB), extracted from the genomic sequence of P. putida ITEM 17297 by BLASTn search and compared to type strains sequences retrieved from Mulet et al. (2010) and Gomila et al. (2015), was generated by MAFFT (Multiple Alignment using Fast Fourier Transform; Katoh et al., 2019). Phylogenetic tree was obtained by Maximum-Likelihood (ML) method by using the IQ-tree web server (Minh et al., 2020). Tree was graphically generated by iTOL version 5.5 (Letunic andBork, 2019).
Genome-based phylogeny was constructed by using The Phylogenetic Tree Building Service implemented in Patric platform (www.patric.org) considering the Cellvibrio japonicus Ueda 107 as outgroup according to Gomila et al. (2015) and ML method RAXML with progressive refinement (Stamatakis et al., 2014).
2.2. Phenotypic characterization
2.2.1. Antibiotic susceptibility testing
Antibiotic susceptibility testing was evaluated in triplicate by disk diffusion technique on Muller-Hinton agar II (MHA; Biolife Italiana, Milano, Italy), according to EUCAST guidelines version 6.0, which are essentially the same standards of the Clinical Laboratory Standard Institute (CLSI) guidelines (CLSI, 2018). In brief, bacterial suspensions of the target strain were manually adjusted to a turbidity equivalent to that of a 0.5 McFarland standard and manually spread on MHA plates. OxoidTM Antimicrobial Susceptibility Disks (Thermo Fisher Scientific) were impregnated with the following antibiotics: ß-lactams: ampicillin (10 μg), methicillin (10 μg), oxacillin (1 μg), penicillin G (10 μg); cephalosporins: ceftizoxime (30 μg); aminoglycosides: gentamicin (10 μg), tobramycin (10 μg), kanamycin (30 μg); quinolones: ciprofloxacin (5 μg), ofloxacin (5 μg), streptomycin (10 μg), nalidixic acid (30 μg); tetracycline (30 μg); glycopeptide antibiotics: vancomycin (30 μg); lincosamides: clindamycin (2 μg), lincomycin (2 μg); macrolides: erythromycin (15 μg); fusidanes: fusidic acid (10 μg); polypeptide antibiotics: nitrofurantoin (300 μg), bacitracin (130 μg) and nitrobenzenes: chloramphenicol (30 μg). Disks were placed on seeded MHA plates within 15 min. Blank disks were used as negative control. The plates were incubated overnight at 30 °C. The images of the plates were digitized and the calibrated diameter of inhibition halos around each antibiotic disk was measured using the UTHSCSA Image tool for Windows ver. 3.0.
2.2.2. Minimal inhibitory concentration (MIC) assay
This assay was performed in sterile 96 well plates (Corning®, NY, USA), using MH broth (MHB) added with increasing concentrations (1.95, 3.91, 7.81, 15.63, 31.25, 62.5, 125, 250, 500, 1.000 μg/mL) of the following antibiotics (at least one for each class): ampicillin, streptomycin, erythromycin, chloramphenicol and nalidixic acid. In particular, MHB was poured (200 μL) in each well with un-inoculated medium or inoculated medium (5–6 log CFU/mL) in presence or absence of antibiotics. Assays were performed in triplicate. Finally, 10 uL of an aqueous resazurin sodium salt (Merck Life Science S.r.l., Milano, Italy) solution (6.75 mg/mL) were added to each well. The plates were incubated at 30 °C for 24 h. The colour change was then visually assessed. Any colour change from purple to pink was recorded as positive of bacterial growth. The lowest concentration at which no colour change occurred was logged as the MIC value.
2.2.3. Biofilm formation and motility assay
Microtiter wells (Corning®, NY, USA) were inoculated with overnight LB-grown cultures diluted 1:100 in minimal MHB medium (Oxoid); the microtiter plates were then incubated at 30 °C and 15 °C for 72 h; biofilm biomass was measured as absorbance at 570 nm at 24, 48 and 72 h by following the crystal violet (CV)-based staining method (O’Toole, 2011). Swarming and swimming experiments were performed in polystyrene Petri dishes (50 mm diameter) containing 10 mL of MHB with 0.5 and 0.3% of agar, respectively. Then, plates were inoculated with 1 × 108 CFU/mL of bacterial broth culture (2.5 μL). The diameters of the swarming and swimming motility zones were measured after incubation (30 °C and 15 °C) for 24, 48, 72 h. Plates were also digitally acquired by the Image Scanner III (GE Healthcare, USA).
Twitching motility was assayed by subsurface agar method described by Köhler et al. (2000) with slight modifications. Briefly, 5 μL of each selected bacterial culture (1 × 108 CFU/mL) were stab inoculated through 1% agar plate with MHB. At 24, 48, 72 h of incubation at 15 °C and 30 °C, the hazy zone of twitching motility elaborated at the interstitial surface between the agar and the Petri dish was visualized by staining with a solution of 0.1% (wt/v) of CV. CV-binding biofilm solubilized with acetic acid (30%, v/v) was recorded spectrophotometrically at 570 nm.
2.2.4. Minimum biofilm eradication concentration (MBEC) assay
Measurements of the antimicrobial susceptibilities of the P. putida biofilms were assessed as described by Ceri et al. (1999) with some modifications. Briefly, the 24-h biofilms, formed at both temperatures (4 and 15 °C) in safe-lock Eppendorf™ tubes, were washed two times with 1 mL of sterile phosphate buffer solutions (PBS) to remove non-adherent cells. Then, concentrations of each antibiotic equal of MIC value or 2, 4, and 10-fold higher in MHB were added to test tubes and incubated at 30 °C for 24 h. Six replicates for each sample were carried out. MHB without antibiotics was also included as control. After the incubation, MHB supplemented or not with antibiotics was aspirated gently; 3 tubes were washed two times with sterile PBS solutions before being filled with fresh medium and biofilm was removed by ultrasonic disruption using Ultrasonic Cleaner (AGF Electronica Srl, Muggio, Milan, Italy) for 5 min; samples (190 μL) were transferred to 96 well plates and supplemented with 10 μL of resazurin solution prepared as described above. Plates were incubated at 30 °C for 24 h. The MBEC was defined as the lowest concentration of antibiotic preventing regrowth of bacteria from surviving biofilm. In addition, CV-staining was performed in conjunction with the MBEC assay to evaluate biofilm biomass by the remaining three replicates; then, samples were stained with a solution of 0.1% (wt/v) of CV following the method reported above.
2.3. Statistical analysis
Two-way ANOVA was carried out with SPSS 22.0 program (IBM, Armonk, NY, USA) in order to examine the effects of different MIC folds of each assayed antibiotic on biofilm amounts produced by P. putida ITEM 17297 at 15 and 30 °C. Post-hoc HSD Tukey’s test (P < 0.05) was performed to compare mean values. If homogeneity variance was not found (Levene’s test, P < 0.05) non-parametric Kruskal-Wallis H test followed by post-hoc multicomparison Dunn’s test was applied to provide a significant difference among mean ranks of MIC levels within each antibiotic treatment. The same statistical procedure was applied to compare biofilm amounts and motility shown by the strain during incubation at the two different temperatures.
3.1. Results and discussion
3.1.2. General features of P. putida ITEM 17297 genome
Pseudomonas putida ITEM 17297 genome was sequenced using a whole genome shotgun approach. Genome was assembled using Spades v5.0 software for a total of 118 contigs (>500 bp). The average GC content was 61.9% and the assembly length of 5.1 Mb (Table 1). The quality of the assembly was excellent with 94.6% of genome completeness and N50 of 389 kb. The whole genome shotgun project has been deposited at DDBI/ENA/GenBank under the accessions WJRT00000000. The version described in this paper is WJRT01000000.
Table 1.
Genome features of P. putida ITEM 17297.
| Features | ITEM 17297 |
|---|---|
| Genome size (bp) | 5,115,953 |
| GC (%) | 61.91 |
| Number of contigs | 118 |
| Completeness | 94.6% |
| Quality | 90.1 (excellent) |
| Conting N50 | 389,568 |
| Genes (total) | 4666 |
| Genes (coding) | 4536 |
| Coing density | 90.26% |
| rRNAs | 1, 1, 1 (5S, 16S, 23S) |
| tRNAs | 59 |
Phylogenetic analysis places P. putida ITEM 17297 in the same clade of all the P. putida included in the analysis (Supplementary Figures SF1 and SF2). The closest genetic relative is Pseudomonas sp. 02C_26 (Accession NZ_CP025262), also according to ANI calculation (Rodriguez and Konstantinidis, 2016) which showed a nucleotide identity of 97.35%.
3.1.2. Antibiotic susceptibility and resistance genes
In the last decades, food-related psychrotrophic Pseudomonas spp. have emerged for their resistance against multiple groups of antibiotics (Mah and O’Toole, 2001; Quintieri et al., 2019a; Meng et al., 2020). To date few pertinent studies have searched for similar abilities in non-P. aeruginosa species; this is particularly relevant from a clinical point of view, as most such species are important emerging pathogens and are being encountered in medical practice (health care facilities) with rising incidence (Docquier et al., 2003; Sperandio et al., 2012). Pseudomonas putida, widespread both in foods or environment, raises concerning questions about its persistence and the associated risk of antibiotic resistance spreading. Thus, the selected strain P. putida ITEM 17297, responsible for spoilage of vegetables under cold storage (Baruzzi et al., 2015), was analyzed for genetic AR determinants and tested for its susceptibility towards different classes of antibiotics.
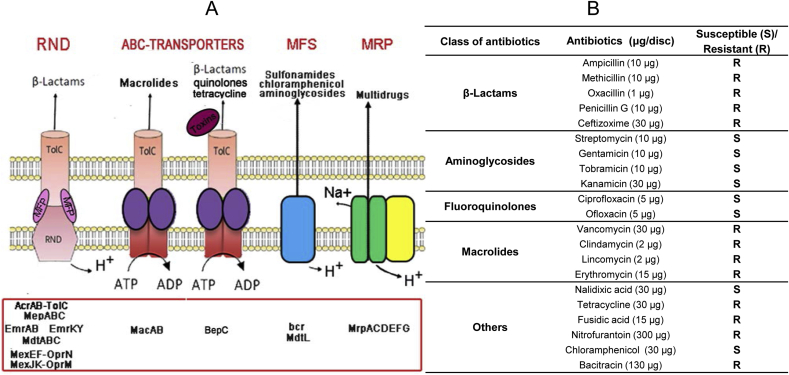
Several efflux pumps, peptidases, and other genetic determinants were found and summarized in Fig. 1, panel A and Supplementary Table SF1. As in other Gram negative bacteria (Nikaido 2011; Greene et al., 2018), many families of transporters in P. putida included tripartite forms traversing both the outer and the inner membrane and releasing drugs directly into the external medium, thus contributing to regulate the outer membrane permeability and reduce the diffusion of molecules across the cell envelope. This general mechanism of antibiotic resistance includes different systems with a broad substrate specificity, such as the several resistance-nodulation-division (RND) systems and the ATP-binding cassette (ABC)-transporter macAB; the MFS (major facilitator superfamily) efflux pump is instead located in the cytoplasmic membrane and transfers drugs into the periplasm (Fig. 1, panel A and Supplementary Table SF1). In addition to these, the MRP (Multiple resistance and pH) antiporter was also retrieved; the main physiological roles played by the MRP Na+/H+ antiporter are as regulators of the intracellular pH homeostasis and Na+ efflux in response to changes in the growth environment and its associated stresses (Ito et al., 2017). Furthermore, some homologues of the same family were implicated in multidrug tolerance (Ito et al., 2017). In this regard, ITEM 17297 resistance to beta-lactam class (penicillins and cephalosporins; Fig. 1, panel B) could be explained by the identification of acrAB-TolC, mepABC, emrAB, emrKY in the list of efflux pumps-related genes (Fig. 1, Supplementary Tables SF1 and SF2; Poole, 2001; Li and Nikaido, 2009; Thomson and Bonomo, 2005). In addition, several penicillin-binding proteins (PBPs), involved in peptidoglycan synthesis, responsible for the selective interaction with penicillin and exhibiting a beta-lactamase/transpeptidase like structural motif, were also listed (Supplementary Table SF1); PBP mechanism has been deeper described for resistant P. aeruginosa (Smith et al., 2013).
Fig. 1.
Panel A): genetic determinants of antibiotic resistance in P. putida ITEM 17297 and main classes of antibiotic developing resistance; Panel B): antibiotic susceptibility of P. putida ITEM 17297 (raw data are reported in Supplementary Table SF2).
In the genome of this strain we identified one ampC gene coding for a class C β-lactamase (GIB23_14635), which is responsible for the resistance to ß-lactam antibiotics (ampicillin, penicillin G, methicillin and oxacillin; Supplementary Table SF2). GIB23_14635 represents a novel variant of this class of enzyme. The multialignment of beta-lactamase retrieved from other P. putida strains (Fig. 2) shows that compared to the highest similar sequence of P. putida strain KT-27, isolated from Japanese soybean soil, they differ for a E195A substitution. The novel ampC allele was deposited under the GenBank accession number MW036223. MIC value for ampicillin was also determined and reported in Table 2.
Fig. 2.
Multiple sequence alignment of class C beta-lactamase in Pseudomonas spp. Conservation and consensus tracks were obtained by using Jalview 2.11.1.0 (Waterhouse et al., 2009) setting the color scheme used for alignments in ClustalX. Alignment conservation annotation is a quantitative numerical index reflecting the conservation of the physico-chemical properties for each column of the alignment. The conserved columns with a score of 11 are indicated by ‘∗‘. Columns with a score of 10 have mutations but all properties conserved are marked with a ‘+‘. Alignment consensus annotation reflects the percentage of the different residue per column. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Minimal inhibitory concentration (MIC) of the selected antibiotics (at least one for each class) in MHB inoculated with Pseudomonas putida ITEM 17297 and incubated for 24 h at 30 °C.
| Antibiotic | Minimal inhibitory Concentration (MIC) (μg/mL) |
|---|---|
| Ampicillin | 250 |
| Streptomycin | 30 |
| Erythromycin | 125 |
| Chloramphenicol | 15 |
| Nalidixic acid | 30 |
Antibiotic susceptibility tests also revealed resistance to macrolides (Fig. 1, panel B and Table SF2) previously correlated with the role of the efflux systems MacAB (Fitzpatrick et a l., 2017). By contrast the strain was sensitive to aminoglycosides and fluoroquinolones although MexEF-OprN and multidrug transporters (MdfA), responsible for resistance of Pseudomonas spp. to these antibiotic classes, were found (Edgar and Bibi, 1997; Poole, 2000) (Table SF1).
Our data strengthen the concern raised by the increasing of food-borne health threats (http://www.efsa.europa.eu/en/efsajournal/pub/4006.htm) that represent a reservoir of antimicrobial bacterial resistome potentially transmittable to the microbiome of consumers (Capita and Alonso-Calleja, 2013). Pseudomonas spp. strains contaminating pasteurised milk and dairy products were found resistant to penicillin G, sulfamethoxazole/trimethoprim and ceftazidime. Likewise, antibiotic resistant Pseudomonas spp. were also isolated from fresh vegetables and other foods, while co- or cross resistance between different antimicrobials was detected in goat and lamb slaughterhouse environments and meat products purchased from market (Estepa et al., 2015). Moreover, if on the one hand some Authors recently identified P. putida implicated in human bacteremia (Liu et al., 2014; Thomas et al., 2013; Chamon et al., 2020), on the other hand, other studies have indicated P. putida as a reservoir for ARGs (Marchiaro et al., 2014; Juan et al., 2010). Thus, based on these results and in the light of these considerations, the occurrence in food of psychrotrophic pseudomonads displaying a broad antibiotic resistance profile, such as the strain ITEM 17297, should concern more than their spoilage ability in the perspective for improving both the risk assessment and the control strategies.
3.1.3. Biofilm by P. putida ITEM 17297 and related genes: a contribution to AR
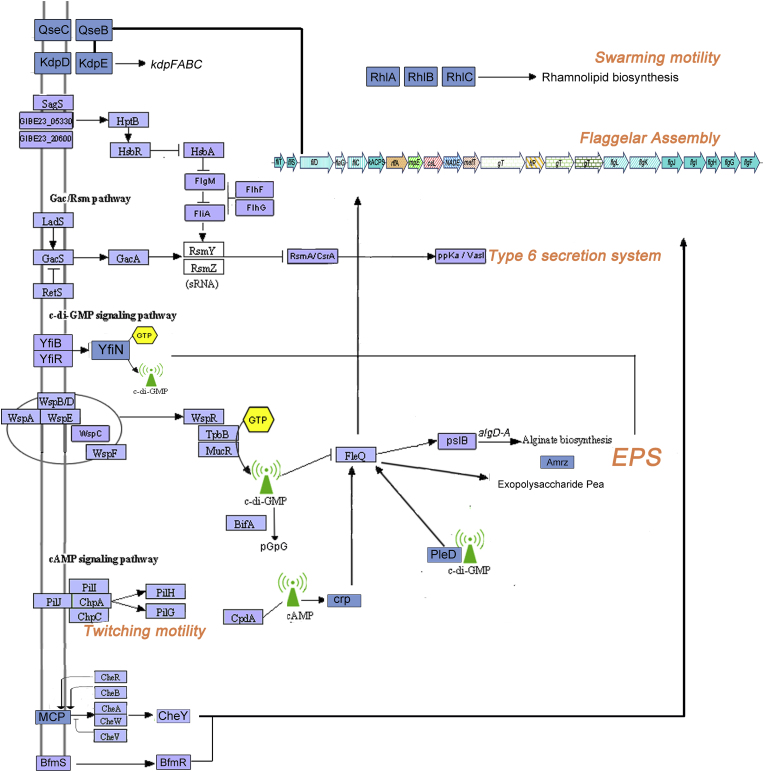
Biofilm growth confers several advantages to bacteria, including protective and adaptive mechanisms towards hostile environmental conditions such as osmotic stress, metal toxicity, and antibiotic exposure. Determinants of biofilm formation in ITEM 17297 genome were shown in Fig. 3 and Supplementary Table SF3. Biofilm-associated drug resistance and tolerance takes place by a) the failure to penetrate EPS matrix, b) the increase of antibiotic-degrading enzymes in the matrix, c) the occurrence of a metabolically inactive (viable-but-nonculturable or dormant cells) or slow-growing state, favored by the oxygen and other nutrients gradient occurring from the surface to the depth of the biofilm. This latter altered microenvironment (oxygen depletion, accumulation of acids, osmotic stress) within the biofilm can by itself directly antagonize the action of an antibiotic (Nadell et al., 2015; Bagge et al., 2004; Stewart et al., 2001). Evidence on the increased antibiotic resistance of cells in biofilm state rather than living as planktonic cells were also reported for clinical strains of P. putida (Molina et al., 2014). The genome of ITEM 17297 showed several two-component systems involved in adaptive response to external stimuli by signalling transduction in cellular compartments; among these we identified SagS and BarA/GacS, activating CsrA-regulated pathways by modulation of small RNA levels; as reported for P. aeruginosa, most likely, these signalling cascades intersect each other and promote motility, EPS production and biofilm formation (Petrova and Sauer, 2011). In addition to these, other two-component systems (e.g. Wsp and Che systems, CheY-like BfmR/S, QseCB) putatively activated biofilm-related pathways by the modulation of cyclic di-GMP and cAMP levels (Fig. 3).
Fig. 3.
Metabolic pathways involved in biofilm formation by P. putida ITEM 17297.
As in other P. putida strains, in ITEM 17297 the psl locus is poorly conserved. Indeed, we retrieved only homologues of the P. aeruginosa PAO1 pslB coding for a bifunctional isomerase/guanylyltransferase (GIB23_09065) and pslH, coding for a glycosyltransferase (GIB23_09060). The pea locus, responsible for the production of the exopolysaccharide A, is located in the operon GIB23_20405-GIB23_20455. This locus was shown to play a role in P. putida biofilm-related phenotypes (Nielsen et al., 2011). Our strain missed the bacterial cellulose bcs locus, the wss locus, responsible for cellulose production in P. fluorescens SBW25 (Spiers et al., 2003; Spiers and Rainey 2005) which impacts biofilm formation at the air-liquid interface, and the pel operon. As shown in Supplementary Fig. SF3, the alg operon (GIB23_11545-GIB23_11460) is instead conserved in content and organization with respect to the P. aeruginosa locus, while the flanking arn locus, which in the pathogenic species mediates resistance to polymyxins and other cationic antimicrobial peptides, is not present in the P. putida ITEM 12797 genome. Furthermore, additionally to P. aeruginosa PAO1 and P. putida KT2440, there are flanking genes upstream algD conserved in arrangement and predicted function with respect to Pseudomonas sp. 02C 26 (Supplementary Fig. SF3).
In this study we confirmed the biofilm production by ITEM 17297 both at low and optimal growth temperature of incubation; interestingly, biofilm amount formed at 15 °C was statistically higher by an average 1.3 times than that found at 30 °C (F(1, 12) = 321.517, p = 4.972 × 10-10) without a significant (P = 0.071) effect due to incubation time (Supplementary Fig. SF4, panel A).
This result was in accordance with those obtained for other psychrotrophic Pseudomonas spp. isolated from fresh foods and responsible for spoilage phenomena at temperatures ranged from 10 to 15 °C (Quintieri et al., 2020; Rossi et al., 2018). As concern motility, ITEM 17297 colony diameter markedly increased on low-agar medium than on semi-solid surface, suggesting that the strain preferred swimming rather than swarming; this behaviour was recorded at both assayed temperatures with significant time-dependent increases for swimming (τb = 0.820, p = 8.023 × 10-5; Supplementary Fig. SF4, panel B). Significant differences were also registered in twitching motility, a surface-associated movement due to the extension and retraction of type IV pili, which propels bacteria across a surface (Mattick, 2002); in particular, starting from 48 h of incubation, twitching motility by ITEM 17297 was displayed only at 15 °C (CV absorbance at 570 nm was 0.15 ± 0.10 and 0.30 ± 0.02 at 48 and 72 h, respectively; Supplementary Fig. SF5).
Pseudomonas putida motility has been extensively studied in the biotechnological important soil strain KT2440. Martínez-García et al. (2014) demonstrated that with specific knockout of genes related to flagellar export and assembly as well as regulatory elements, flagellar motility still provides a great environmental advantage in P. putida KT2440 despite the high metabolic costs. These results were recently confirmed by Navarrete et al. (2019) that characterized, by transcriptomic analysis, major components of flagellar-related proteins. In P. putida ITEM 17297 content and organization of the loci comprising flagella genes are conserved with respect to KT2440 (Supplementary Table SF4), although with relevant differences below described. Flagellar genes are in fact located in a large operon including those coding for proteins involved in the biosynthesis, assembly, export and motor and energizing enzymes. The master regulator of flagella biosynthesis FleQ (Arora et al., 1997) has been identified as GIB23_04455, while FlhF and FleN, described as necessary to attain proper flagellar location and numerically regulators of flagellar biogenesis, were GIB23_04600 and GIB23_04605, respectively. Outside the operon two extra copies of the motor genes motA (GIB23_17450) and motB (GIB23_17455), homologues to those found in the P. aeruginosa genome (Toutain et al., 2005), and the operon coding for the twitching motility proteins PilJLHGT are located.
The flagellin FliC, the subunit protein which polymerizes to form the filaments of bacterial flagella, has 61% identity with respect to P. putida KT2440 and it is of 283 aa length compared to the 687 aa of KT2440. As shown in Supplementary Fig. SF6, between the ketoacyl-ACP synthase gene and flgL a locus not present neither in P. putida KT2440 nor in P. aeruginosa PAO1 is included. This locus comprises genes coding for the transaminase RffA, one metallophosphoesterase, one carbamoyl-phosphate synthase subunit L, one NAD-dependent epimerase/dehydratase family protein, one methyltransferase domain-containing protein and 3 glycosyltransferases, interspaced by a gene coding for a hypothetical protein (InterPro domain IPR014985: WbqC-like protein family). This content was also retrieved by genomic comparative analysis in P. chlororaphis from a soybean field soil (Deng, 2015) and in the phytopathogen P. brassicacearum Wood3 (Gislason and de Kievit, 2020). In both strains these genes have a different positioning with respect to the flagellar genes, and in P. brassicacearum the ketoacyl-ACP synthase, one glycotransferase and flgL are distantly located. This locus was also partially present in P. alkylphenolica KL28 and P. plecoglossicida MR70, although their content and organization are not completely conserved (Supplementary Fig. SF6).
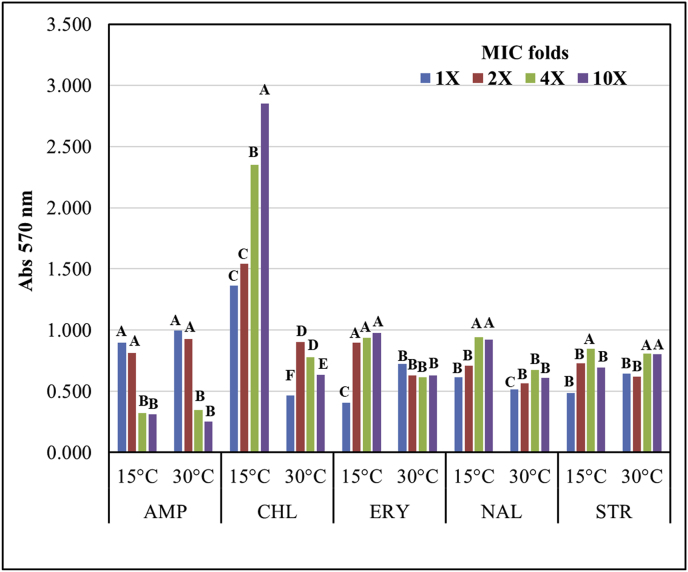
After evaluation of ITEM 17297 biofilm formation, biofilm phenotype was investigated in respect to antibiotic tolerance. Thus, in the light of results described above, MBEC of the selected antibiotics was determined against biofilm produced at both temperatures of incubation. Kruskal-Wallis test followed by post-hoc Dunn’s test within each antibiotic treatment showed significant differences in median biofilm amount produced at different MIC levels (Fig. 4, Kruskal-Wallis H test, χ2(2) = 21.553–22.573, p ≤ 0.004). In particular, at 15 °C the amount of biofilm linearly increased starting from 2X MIC of chloramphenicol, reaching the highest values at 10X MIC; by contrast, at 30 °C biofilm production was markedly less affected by chloramphenicol levels displaying a slight but significant (P < 0.05) decrease at high concentrations. At lower extent this behaviour was also observed for the strain assayed in presence of erythromycin and nalidixic acid. Slight but significant (P < 0.05) increases in biofilm were recorded at higher concentrations for streptomycin at both temperatures. Conversely, under ampicillin treatment the amount of biofilm was reduced as MIC levels increased regardless of temperature.
Fig. 4.
Biofilm amounts produced by P. putida ITEM 17297 grown at 15 and 30 °C in presence of different folds (2, 4, 10X) of minimal inhibitory concentration (MIC) of several antibiotics. Bars represent medians (N = 3). Bars with different letters are values statistically different within each antibiotic treatment (Kruskal-Wallis H test, χ2(2) = 21.553–22.573, p ≤ 0.004 followed by post-hoc Dunn’s test). AMP: ampicillin; CHL: chloramphenicol; ERY: erythromycin; NAL: nalidixic acid; STR: streptomycin.
Thus, although increasing concentrations of antibiotics were assayed, reaching the ten fold-MIC value, all the assayed antibiotics failed to eradicate biofilm, and biofilm-detached cells were viable when refreshed under optimal growth condition. These results were in accordance with those registered by Molina et al. (2014) for clinical isolates of P. putida; indeed, antibiotic concentrations required to eradicate clinical P. putida biofilm were from 3 to 40 fold higher than MIC values displayed for planktonic cells (Molina et al., 2014).
The ability of microbial biofilm to survive under high doses of antibiotics is reported for several bacteria, including P. aeruginosa (Ciofu and Tolker-Nielsen, 2019); in this species mechanisms of antibiotic tolerance were attributed to the over-production of Pel and Psl exopolysaccharide mediated by GacA-GacS, and the expression of specific genes as mexAB-oprM and mexEF-oprN, as well as ABC-multidrug transporters under the regulation of c-di-GMP levels. Likewise, Fernandez et al. (2012) reported that in P. putida KT2440 biofilm chloramphenicol resistance was attributed to the positive regulation of the efflux pump TtgABC biosynthesis, that herein was identified in GIB23_12100, GIB23_12105, GIB23_12110; ttgABC-overexpression was also correlated with biofilm biomass increases in mutant P. putida strains (Tettmann et al., 2014).
For the first time, P. putida biofilms growing at low temperature were investigated for their resistance to some antibiotics; interestingly, under selective pressure of 3 out of 5 antibiotics, biomass biofilm increased significantly in a dose dependent manner. Although no previous data have been reported for this behaviour in Pseudomonas spp., its AR might represent a beneficial strategy involved in adaptive response to other stressors such as temperature changes, nutrient and iron starvation, as suggested for other bacteria (Cruz-Loya et al., 2019). For example, in E. coli temperatures lower than the optimum clustered with resistance to antibiotics, which affect the early stages of protein synthesis or act as DNA gyrase inhibitors. This functional overlapping was attributed to the mainteinment of the optimal energy efficiency (Cruz-Loya et al., 2019). Recalcitrant biofilms can be also favored by adaptive response to oxidative stresses; upregulation of antioxidant systems, such as catalase and superoxide dismutase, in P. aeruginosa biofilms improved their antioxidant capacity and their tolerance to antibiotics (Ciofu and Tolker-Nielsen, 2019). Both these systems were also found upregulated in psychrotrophic pseudomonas with increased biofilm biomass under low temperature growth conditions (Quintieri et al., 2019b).
In this work we focused our study on one strain of P. putida which displayed the highest spoilage activity among previously analyzed P. putida strains. As reported by previous works, Pseudomonas spp. belonging to the same species and sharing spoilage activities (such as proteolytic, lipolytic ones and pigment release) exhibited also similar resistance to antibiotics Quintieri et al., 2019a; 2020); these characteristics could be attributed to the ability of these strains to form biofilm. Indeed, signalling networks at the basis of biofilm formation have been found correlated with spoilage traits as well as AR (Quintieri et al., 2020). Thus, in light of these considerations and of the results presented here, ITEM 17297 can be considered a model for analyzing the correlation among spoilage, AR and biofilm related pathways in P. putida.
4.1. Conclusions
Our data demonstrated the occurrence of multidrug resistance in P. putida ITEM 17297, a strain contaminating fresh vegetables and responsible for their decay under cold storage. A positive interaction between AR and biofilm formation was also found; no biofilm eradication was indeed obtained by increasing antibiotic concentrations up to 10-fold MIC value. Biofilm cultivated at 15 °C also increased its biomasses in a dose-dependent manner for most antibiotics.
In addition to favour P. putida persistence and related spoilage ability, these alarming adaptive mechanisms might worsen AR spread in the food chain by increasing the health risk for unaware consumers.
In the light of these evidences, underestimated psychrotrophic pseudomonas, such as the herein studied ITEM 17297 strain, assume relevance in relation to the risk related to their persistence in several human settings. In addition, the molecular mechanisms underlying tolerance to antibiotics are targets for therapeutic interventions for potentiating the anti-biofilm effect of antibiotics or novel sanitizers.
Funding
This research received no external funding.
CRediT authorship contribution statement
Francesca Fanelli: Conceptualization, Methodology, Investigation, Writing - original draft, Formal analysis, Data curation, Writing - review & editing, Supervision, Visualization. Leonardo Caputo: Investigation, Writing - original draft, Formal analysis, Data curation, Writing - review & editing, Supervision. Laura Quintieri: Conceptualization, Methodology, Investigation, Writing - original draft, Formal analysis, Data curation, Writing - review & editing, Supervision, Visualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
It is a pleasure to acknowledge Dr. Pasquale Del Vecchio for the bibliometric analysis support and Dr. Maria Morea for reviewing English throughout the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2021.02.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
MLSA phylogeny inferred by using the Maximum-Likelihood (ML) method. The tree is drawn to scale, rooted to P. aeruginosa PAO1, with branch lengths measured in the number of substitutions per site. Support values are represented by scaled circles at each node.
Genome-based phylogenetic tree inferred by using the Maximum Likelihood method RAxML with progressive refinement. Celvibrio japonicus Ueda 107 was used as an outgroup. The tree is drawn to scale. Support values are represented by scaled circles at each node.
Genomic organization of alginate operon in P. aeruginosa and P. putida strains. Gene clustering is represented by the arrows superposed on the black horizontal line. Intergenic spaces are not drawn in scale; argF: ornithine carbamoyltransferase; PA3538: ABC transporter ATP-binding protein; yaaA: peroxide stress protein; algD: GDP-mannose 6-dehydrogenase AlgD; alg8: glycosyltransferase Alg8; alg44: alginate biosynthesis protein Alg44; algK: alginate biosynthesis protein AlgK; algE; alginate production protein AlgE; algG: alginate-c5-mannuronan-epimerase; algX: alginate biosynthesis protein AlgX; algL: alginate lyase precursor; algI: alginate O-acetyltransferase AlgI; algJ: alginate O-acetyltransferase AlgJ; algF: alginate O-acetyltransferase AlgF; algA: Alginate biosynthesis protein AlgA; arnB: UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase; arnC: undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase; arnA: bifunctional UDP-glucuronic acid decarboxylase/UDP-4-amino-4-deoxy-L-arabinose formyltransferase; arnD: 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase ArnD; arnT: 4-amino-4-deoxy-L-arabinose lipid A transferase; arnE: 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol flippase subunit ArnE; arnF: 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol flippase subunit ArnF; PA3559: nucleotide sugar dehydrogenase; pho-H: PhoH-like protein; pdaC: peptidoglycan-N-acetylmuramic acid deacetylase PdaC; hlyD: HlyD family efflux transporter periplasmic adaptor subunit; tolC: TolC family protein; fts-X: FtsX-like permease family protein; yknY: ABC transporter ATP-binding protein YknY; hp; hypothetical protein; mex: multidrug transporter; ox: SDR family oxidoreductase; opmA: efflux transporter outer membrane subunit; hlyD: HlyD family efflux transporter periplasmic adaptor subunit; MFS: MFS transporter; lysR; LysR family transcriptional regulator; usp: universal stress protein; RND: efflux RND transporter periplasmic adaptor subunit
panel A) Biofilm biomass by P. putida ITEM 17297 produced up to 72 h at 15 and 30 °C. Different letters represent values statistically different according post-hoc HSD Tukey’s test, p ≤ 0.001; panel B) Swimming and swarming motility of P. putida ITEM 17297 in MHB performed at 15 °C and 30 °C for 24, 48 and 72 h. Bars represent the mean diameter of corresponding motility zones (N = 3). Different letters are statistically different values within each motility according to Kruskal-Wallis H test (χ2(2) = 15.277–22.295, p ≤ 0.009) followed by post-hoc pairwise Dunn test (P < 0.05).
Twitching motility of P. putida ITEM 17297 at 15 and 30 °C measured by crystal violet (CV) absorbance at 570 nm; Bars are average values (N = 3) and different superscript letters represent statistically different values according to (Kruskal-Wallis H test, (χ2(5) = 16.801, p ≤ 0.005) followed by post-hoc pairwise Dunn test (P < 0.05)
Genomic organization of fli locus in Pseudomonas spp.. Gene clustering is represented by the arrows superposed on the horizontal black line. Intergenic spaces are not drawn in scale. fliT: flagellar protein T; fliS: flagellar export chaperone protein S; fliD; flagellar filament capping protein D; flaG: flagellar biosynthesis protein FlaG; fliC: flagellin FliC; kACPS: ketoacyl-ACP synthase; rffA: transaminase RffA; mpE: metallophosphoEsterase; csL: carbamoyl-phosphate synthase subunit L; NADE: NAD-dependent Epimerase; metT: methyltransferase; gT: glycosyltransferase; hP: hypothetical Protein; flgL: flagellar hook-associated protein FlgL; flgK: flagellar hook-associated protein FlgK; flgJ: flagellar assembly peptidoglycan hydrolase FlgJ; flgI: flagellar basal body P-ring protein FlgI; flgH: flagellar basal body L-ring protein FlgH; flgG: flagellar basal-body rod protein FlgG; flgF: flagellar hook-basal body complex protein FlgF; ox: oxidoreductase; fgtA: flagellar glycosyltransferase FgtA.
References
- Abatenh E., Gizaw B., Tsegaye Z., Wassie M. The role of microorganisms in bioremediation-A review. Open J. Environ. Biol. 2017;2(1) doi: 10.17352/ojeb.000007. 030-046. [DOI] [Google Scholar]
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C., Grüning B., Guerler A., Hillman-Jackson J., Von Kuster G., Rasche E., Soranzo N., Turaga N., Taylor J., Nekrutenko A., Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aresta M., Acquaviva M.I., Baruzzi F., Noce R.L., Matarante A., Narracci M. Isolation and characterization of polyphenols-degrading bacteria from olive-mill wastewaters polluted soil. World J. Microbiol. Biotechnol. 2010;4:639–647. [Google Scholar]
- Arora S.K., Ritchings B.W., Almira E.C., Lory S., Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagge N., Hentzer M., Andersen J.B., Ciofu O., Givskov M., Hoiby N. Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2004;48(4):1168–1174. doi: 10.1128/aac.48.4.1168-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruzzi F., Pinto L., Quintieri L., Carito A., Calabrese N., Caputo L. Efficacy of lactoferricin B in controlling ready-to-eat vegetable spoilage caused by Pseudomonas spp. Int. J. Food Microbiol. 2015;215:179–186. doi: 10.1016/j.ijfoodmicro.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Capita R., Alonso-Calleja C. Antibiotic-resistant bacteria: a challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013;53(1):11–48. doi: 10.1080/10408398.2010.519837. [DOI] [PubMed] [Google Scholar]
- Chamon R.C., da Rocha J.A., Martins I.A., Pires L.L., de Almeida B.M., Leite N.S. KPC-2 producing Pseudomonas putida as an unexpected pathogen of catheter-associated bloodstream infection. J. Infect. Develop. Countries. 2020;14:411–414. doi: 10.3855/jidc.12145. 04. [DOI] [PubMed] [Google Scholar]
- Chen Q.L., Cui H.L., Su J.Q., Penuelas J., Zhu Y.G. Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 2019;24(6):530–541. doi: 10.1016/j.tplants.2019.02.010. [DOI] [PubMed] [Google Scholar]
- Ceri H., Olson M.E., Stremick C., Read R.R., Morck D., Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37(6):1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-How P. aeruginosa can escape antibiotics. Front. Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P.H. The metabolic versatility of pseudomonads. Antonie Leeuwenhoek. 1982;48(2):105–130. doi: 10.1007/BF00405197. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . CLSI standard M02. 13th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2018. Performance standards for antimicrobial disk susceptibility tests. [Google Scholar]
- Cruz-Loya M., Kang T.M., Lozano N.A., Watanabe R., Tekin E., Damoiseaux R., Savage V.M., Yeh P.J. Stressor interaction networks suggest antibiotic resistance co-opted from stress responses to temperature. ISME J. 2019;13(1):12–23. doi: 10.1038/s41396-018-0241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Wang X., Baird S.M., Lu S.E. Complete genome of Pseudomonas chlororaphis strain UFB2, a soil bacterium with antibacterial activity against bacterial canker pathogen of tomato. Standards Genom. Sci. 2015;10:117. doi: 10.1186/s40793-015-0106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier J.D., Riccio M.L., Mugnaioli C., Luzzaro F., Endimiani A. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 2003;47(5):1522–1528. doi: 10.1128/aac.47.5.1522-1528.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 1997;179(7):2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estepa V., Rojo-Bezares B., Torres C., Sáenz Y. Genetic lineages and antimicrobial resistance in Pseudomonas spp. isolates recovered from food samples. Foodborne Pathoghens. Dis. 2015;12(6):486–491. doi: 10.1089/fpd.2014.1928. [DOI] [PubMed] [Google Scholar]
- Fanelli F., Liuzzi V.C., Quintieri L., Mulè G., Baruzzi F., Logrieco A.F., Caputo L. Draft genome sequence of Pseudomonas fluorescens strain ITEM 17298, associated with cheese spoilage. Genome Announc. 2017;5(43) doi: 10.1128/genomeA.01141-17. e01141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández M., Porcel M., de la Torre J., Molina-Henares M.A., Daddaoua A., Llamas M.A. Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains. Front. Microbiol. 2015;6:871. doi: 10.3389/fmicb.2015.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M., Conde S., de la Torre J., Molina-Santiago C., Ramos J.L. Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob. Agents Chemother. 2012;56:1001–1009. doi: 10.1128/AAC.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick A.W., Llabrés S., Neuberger A., Blaza J.N., Bai X.C., Okada U. Structure of the MacAB–TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017;2(7):1–8. doi: 10.1038/nmicrobiol.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennari M., Dragotto F. A study of the incidence of different fluorescent Pseudomonas species and biovars in the microflora of fresh and spoiled meat and fish, raw milk, cheese, soil and water. J. Appl. Bacteriol. 1992;72(4):281–288. doi: 10.1111/j.1365-2672.1992.tb01836.x. [DOI] [PubMed] [Google Scholar]
- Gislason A.S., de Kievit T.R. Friend or foe? Exploring the fine line between Pseudomonas brassicacearum and phytopathogens. J. Med. Microbiol. 2020;69:3. doi: 10.1099/jmm.0.001145. [DOI] [PubMed] [Google Scholar]
- Gomila M., Peña A., Mulet M., Lalucat J., García-Valdés E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015;6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N.P., Kaplan E., Crow A., Koronakis V. Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front. Microbiol. 2018;9:950. doi: 10.3389/fmicb.2018.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy E., Laslo E., Kuzman I.H., Dezso Andras C. The effect of essential oils and their combinations on bacteria from the surface of fresh vegetables. Food Sci. Nutr. 2020;8(10):5601–6561. doi: 10.1002/fsn3.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa I.H., Nwodo U.U., Sosa A., Tom M., Okoh A.I. Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa, as reservoir of antibiotic resistant determinants. Int. J. Environ. Res. Publ. Health. 2019;9(7):2537–2549. doi: 10.3390/ijerph9072537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Morino M., Krulwich T.A. Mrp antiporters have important roles in diverse bacteria and archaea. Front. Microbiol. 2017;8:2325. doi: 10.3389/fmicb.2017.02325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan C., Zamorano L., Mena A., Alberti S., Perez J.L., Oliver A. Metallo-beta-lactamase-producing pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J. Antimicrob. Chemother. 2010;65(3):474–478. doi: 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- Kaczmarek M., Avery S.V., Singleton I. Microbes associated with fresh produce: sources, types and methods to reduce spoilage and contamination. Adv. Appl. Microbiol. 2019;107:29–82. doi: 10.1016/bs.aambs.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Curty L.K., Barja F., Van Delden C., Pechère J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000;182(21):5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S., Tremblay J., Déziel E. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009;11(1):126–136. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Z., Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69(12):1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu K., Yu X., Li B., Cao B. Identification and control of a Pseudomonas spp (P. fulva and P. putida) bloodstream infection outbreak in a teaching hospital in Beijing, China. Int. J. Infect. Dis. 2014;23:105–108. doi: 10.1016/j.ijid.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Lu J., Wang Y., Jin M., Yuan Z., Bond P., Guo J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020;169:115–229. doi: 10.1016/j.watres.2019.115229. [DOI] [PubMed] [Google Scholar]
- Mah T.F., O’Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Marchiaro P.M., Brambilla L., Moran-Barrio J., Revale S., Pasteran F., Vila A.J., Viale A.M., Limansky A.S. The complete nucleotide sequence of the carbapenem resistance-conferring conjugative plasmid pLD209 from a Pseudomonas putida clinical strain reveals a chimeric design formed by modules derived from both environmental and clinical bacteria. Antimicrob. Agents Chemother. 2014;58(3):1816–1821. doi: 10.1128/AAC.02494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., Nikel P.I., Chavarría M., de Lorenzo V. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ. Microbiol. 2014;16(1):291–303. doi: 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Meng L., Liu H., Lan T., Dong L., Hu H., Zhao S., Zhang Y., Zheng N., Wang J. Antibiotic resistance patterns of Pseudomonas spp. isolated from raw milk revealed by whole genome sequencing. Front. Microbiol. 2020;11:1005. doi: 10.3389/fmicb.2020.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L., Udaondo Z., Duque E., Fernández M., Molina-Santiago C., Roca A., Porcel M., de la Torre J., Segura A., Plesiat P., Jeannot K., Ramos J.L. Antibiotic resistance determinants in a Pseudomonas putida strain isolated from a hospital. PloS One. 2014;9(1) doi: 10.1371/journal.pone.0081604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet M., Lalucat J., mGarcía-Valdés E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- Nadell C.D., Drescher K., Wingreen N.S., Bassler B.L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015;9:1700–1709. doi: 10.1038/ismej.2014.246l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete B., Leal-Morales A., Serrano-Ron L., Sarrió M., Jiménez-Fernández A., Jiménez-Díaz L., López-Sánchez A., Govantes F. Transcriptional organization, regulation and functional analysis of flhF and fleN in Pseudomonas putida. PloS One. 2019;14(3) doi: 10.1371/journal.pone.0214166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi Y.K., Prabha D., Garg S.K., Kumar J. Genetic diversity among cold-tolerant fluorescent Pseudomonas isolates from Indian Himalayas and their characterization for biocontrol and plant growth-promoting activities. J. Plant Growth Regul. 2011;30(2):128–143. doi: 10.1007/s00344-010-9175-7. [DOI] [Google Scholar]
- Nielsen L., Li X., Halverson L.J. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ. Microbiol. 2011;13:1342–1356. doi: 10.1111/j.1462-2920.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Area Mol. Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odeyemi O.A., Alegbeleye O.O., Strateva M., Stratev D. Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 2020;19(2):311–331. doi: 10.1111/1541-4337.12526. [DOI] [PubMed] [Google Scholar]
- O’Toole G.A. Microtiter dish biofilm formation assay. JoVE. 2011;47 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou O.S., Iliopoulos V., Mallouchos A., Panagou E.Z., Chorianopoulos N., Tassou C.C., Nychas G.-J.E. Spoilage potential of Pseudomonas (P. fragi, P. putida) and LAB (Leuconostoc mesenteroides, Lactobacillus sakei) strains and their volatilome profile during storage of sterile pork meat using GC/MS and data analytics. Foods. 2020;9:633. doi: 10.3390/foods9050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter S., Oberhettinger P., Schuele L., Dinkelacker A., Vogel W., Dörfel D. Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa. BMC Genom. 2017;18(1):859. doi: 10.1186/s12864-017-4216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova O.E., Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193(23):6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated resistance to fluoroquinolones in Gram-negative bacteria. Antimicrob. Agents Chemother. 2000;44(9):2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001;3(2):255–264. [PubMed] [Google Scholar]
- Quintieri L., Caputo L., De Angelis M., Fanelli F. Genomic analysis of three cheese-borne Pseudomonas lactis with biofilm and spoilage-associated behavior. Microorganisms. 2020;8(8):1208. doi: 10.3390/microorganisms8081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintieri L., Carito A., Pinto L., Calabrese N., Baruzzi F., Caputo L. Application of lactoferricin B to control microbial spoilage in cold stored fresh foods. In: Mendèz-Vilas A., editor. Multidisciplinary Approach for Studying and Combating Microbial Pathogens. Microbiology Series. Brown Walker Press; 2015. pp. 58–62. [Google Scholar]
- Quintieri L., Fanelli F., Caputo L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: an underestimated risk and the control strategies. Foods. 2019;8(9):372. doi: 10.3390/foods8090372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintieri L., Zühlke D., Fanelli F., Caputo L., Liuzzi V.C., Logrieco A.F. Proteomic analysis of the food spoiler Pseudomonas fluorescens ITEM 17298 reveals the antibiofilm activity of the pepsin-digested bovine lactoferrin. Food Microbiol. 2019;82:177–193. doi: 10.1016/j.fm.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Raphael E., Riley L.W. Infections caused by antimicrobial drug-resistant saprophytic Gram-negative bacteria in the environment. Front. Med. 2017;4:183. doi: 10.3389/fmed.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remold S.K., Purdy-Gibson M.E., France M.T., Hundley T.C. Pseudomonas putida and Pseudomonas fluorescens species group recovery from human homes varies seasonally and by environment. PloS One. 2015;10(5) doi: 10.1371/journal.pone.0127704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R L.M., Gunturu S., Harvey W.T., Rosselló-Mora R., Tiedje J.M., Cole J.R., Konstantinidis K.T. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018;46(W1):W282–W288. doi: 10.1093/nar/gky467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R L.M., Konstantinidis K.T. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. Peer J. 2016;4 doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- Rossi C., Serio A., Chaves-López C., Anniballi F., Auricchio B., Goredo E., Cenci-Goga B.T., Lista F., Fillo S., Paparella A. Biofilm formation, pigment production and motility in Pseudomonas spp. isolated from the dairy industry. Food Contr. 2018;86:241–248. doi: 10.1016/j.foodcont.2017.11.018. [DOI] [Google Scholar]
- Smith J.D., Kumarasiri M., Zhang W., Hesek D., Lee M., Toth M., Vakulenko S., Fisher J.F., Mobashery S., Chen Y. Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in β-lactam resistance. Antimicrob. Agents Chemother. 2013;57(7):3137–3146. doi: 10.1128/AAC.00505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio D., Decoin V., Latour X., Mijouin L., Hillion M., Feuilloley M.G. Virulence of the Pseudomonas fluorescens clinical strain MFN1032 towards Dictyostelium discoideum and macrophages in relation with type III secretion system. BMC Microbiol. 2012;12(1):1–10. doi: 10.1186/1471-2180-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers A.J., Bohannon J., Gehrig S.M., Rainey P.B. Biofilm formation at the air–liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 2003;50:15–27. doi: 10.1046/j.1365-2958.2003.03670.x. [DOI] [PubMed] [Google Scholar]
- Spiers A.J., Rainey P.B. The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology. 2005;151:2829–2839. doi: 10.1099/mic.0.27984-0. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P.S., Costerton J.W. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Tan G., Xi Y., Yuan P., Sun Z., Yang D. Risk factors and antimicrobial resistance profiles of Pseudomonas putida infection in Central China, 2010–2017. Medicine. 2019;98(44) doi: 10.1097/MD.0000000000017812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettmann B., Dötsch A., Armant O., Fjell C.D., Overhage J. Knockout of extracytoplasmic function sigma factor ECF-10 affects stress resistance and biofilm formation in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2014;80(16):4911–4919. doi: 10.1128/AEM.01291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.S., Okamoto K., Bankowski M.J., Seto T.B. A lethal case of Pseudomonas putida bacteremia due to soft tissue infection. Infect. Dis. Clin. Pract. 2013;21(3):147–213. doi: 10.1097/IPC.0b013e318276956b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.M., Bonomo R.A. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 2005;8(5):518–524. doi: 10.1016/j.mib.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Toutain C.M., Zegans M.E., O’Toole G.A. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 2005;187(2):771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.K., Barrios A.F.G., Herzberg M., Lee J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006;72(2):361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Hu H.W., Chen Q.L., Singh B.K., Yan H., Chen D., He J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019;130:104912. doi: 10.1016/j.envint.2019.104912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MLSA phylogeny inferred by using the Maximum-Likelihood (ML) method. The tree is drawn to scale, rooted to P. aeruginosa PAO1, with branch lengths measured in the number of substitutions per site. Support values are represented by scaled circles at each node.
Genome-based phylogenetic tree inferred by using the Maximum Likelihood method RAxML with progressive refinement. Celvibrio japonicus Ueda 107 was used as an outgroup. The tree is drawn to scale. Support values are represented by scaled circles at each node.
Genomic organization of alginate operon in P. aeruginosa and P. putida strains. Gene clustering is represented by the arrows superposed on the black horizontal line. Intergenic spaces are not drawn in scale; argF: ornithine carbamoyltransferase; PA3538: ABC transporter ATP-binding protein; yaaA: peroxide stress protein; algD: GDP-mannose 6-dehydrogenase AlgD; alg8: glycosyltransferase Alg8; alg44: alginate biosynthesis protein Alg44; algK: alginate biosynthesis protein AlgK; algE; alginate production protein AlgE; algG: alginate-c5-mannuronan-epimerase; algX: alginate biosynthesis protein AlgX; algL: alginate lyase precursor; algI: alginate O-acetyltransferase AlgI; algJ: alginate O-acetyltransferase AlgJ; algF: alginate O-acetyltransferase AlgF; algA: Alginate biosynthesis protein AlgA; arnB: UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase; arnC: undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase; arnA: bifunctional UDP-glucuronic acid decarboxylase/UDP-4-amino-4-deoxy-L-arabinose formyltransferase; arnD: 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase ArnD; arnT: 4-amino-4-deoxy-L-arabinose lipid A transferase; arnE: 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol flippase subunit ArnE; arnF: 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol flippase subunit ArnF; PA3559: nucleotide sugar dehydrogenase; pho-H: PhoH-like protein; pdaC: peptidoglycan-N-acetylmuramic acid deacetylase PdaC; hlyD: HlyD family efflux transporter periplasmic adaptor subunit; tolC: TolC family protein; fts-X: FtsX-like permease family protein; yknY: ABC transporter ATP-binding protein YknY; hp; hypothetical protein; mex: multidrug transporter; ox: SDR family oxidoreductase; opmA: efflux transporter outer membrane subunit; hlyD: HlyD family efflux transporter periplasmic adaptor subunit; MFS: MFS transporter; lysR; LysR family transcriptional regulator; usp: universal stress protein; RND: efflux RND transporter periplasmic adaptor subunit
panel A) Biofilm biomass by P. putida ITEM 17297 produced up to 72 h at 15 and 30 °C. Different letters represent values statistically different according post-hoc HSD Tukey’s test, p ≤ 0.001; panel B) Swimming and swarming motility of P. putida ITEM 17297 in MHB performed at 15 °C and 30 °C for 24, 48 and 72 h. Bars represent the mean diameter of corresponding motility zones (N = 3). Different letters are statistically different values within each motility according to Kruskal-Wallis H test (χ2(2) = 15.277–22.295, p ≤ 0.009) followed by post-hoc pairwise Dunn test (P < 0.05).
Twitching motility of P. putida ITEM 17297 at 15 and 30 °C measured by crystal violet (CV) absorbance at 570 nm; Bars are average values (N = 3) and different superscript letters represent statistically different values according to (Kruskal-Wallis H test, (χ2(5) = 16.801, p ≤ 0.005) followed by post-hoc pairwise Dunn test (P < 0.05)
Genomic organization of fli locus in Pseudomonas spp.. Gene clustering is represented by the arrows superposed on the horizontal black line. Intergenic spaces are not drawn in scale. fliT: flagellar protein T; fliS: flagellar export chaperone protein S; fliD; flagellar filament capping protein D; flaG: flagellar biosynthesis protein FlaG; fliC: flagellin FliC; kACPS: ketoacyl-ACP synthase; rffA: transaminase RffA; mpE: metallophosphoEsterase; csL: carbamoyl-phosphate synthase subunit L; NADE: NAD-dependent Epimerase; metT: methyltransferase; gT: glycosyltransferase; hP: hypothetical Protein; flgL: flagellar hook-associated protein FlgL; flgK: flagellar hook-associated protein FlgK; flgJ: flagellar assembly peptidoglycan hydrolase FlgJ; flgI: flagellar basal body P-ring protein FlgI; flgH: flagellar basal body L-ring protein FlgH; flgG: flagellar basal-body rod protein FlgG; flgF: flagellar hook-basal body complex protein FlgF; ox: oxidoreductase; fgtA: flagellar glycosyltransferase FgtA.