Fig. 1. Comparison between IG/TR rearrangements detected by amplicon-based assays (‘DNAamp’) and RNAseq in 165 ALL patients.
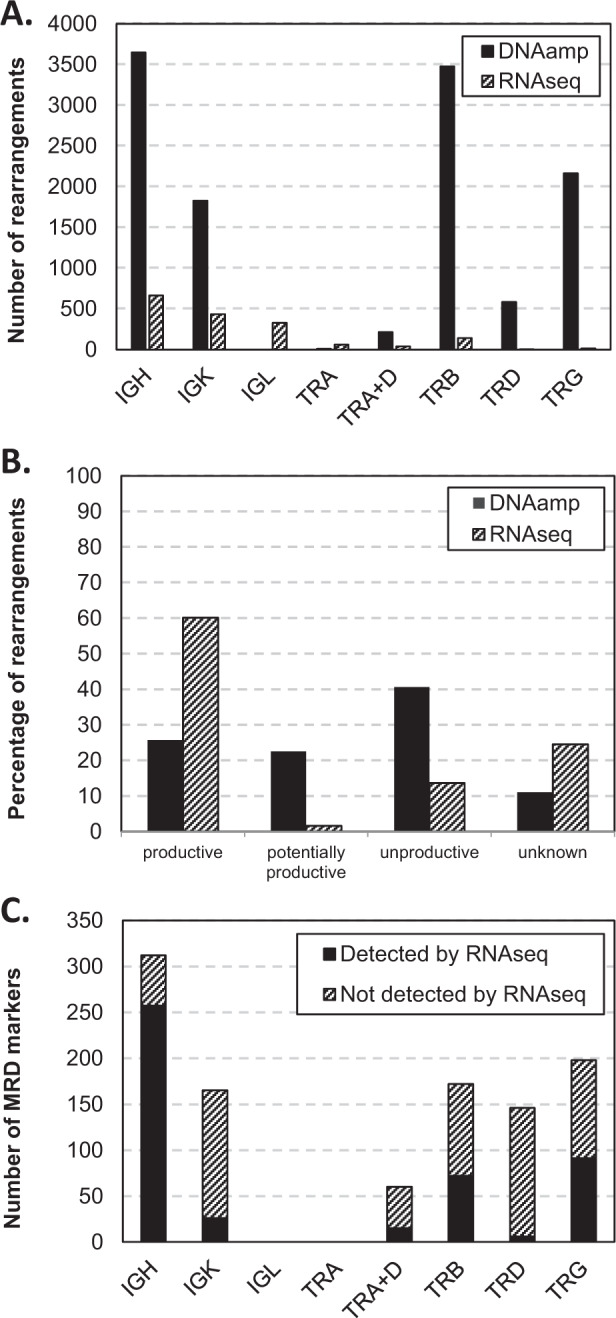
A Average number of rearrangements detected for the various IG/TR loci per case. B Frequencies of productive, potentially productive (incomplete sequences that have the potential to be productive when they would be complete), and unproductive rearrangements. Unknown: non-VJ and non-DJ classes, and “damaged” rearrangements from RNAseq. C Number of RNAseq re-identifiable MRD markers per target according to DNAamp benchmark.
