Abstract
Introduction
One of the remaining barriers to reaching WHO elimination targets of achieving global hepatitis C (HCV) cure is a lack of an established lower limit of detection (LLOD) to confirm cure post-treatment in near-patient technologies. Determining a LLOD at virologic failure aids in increasing testing feasibility through point-of-care assays in resource-limited settings.
Methods
We described the level of viremia in 69 patients experiencing virologic failure across 20 clinical trials (ENDURANCE-1, ENDURANCE-2, ENDURANCE-3, ENDURANCE-4, ENDURANCE 5–6, MAGELLAN-1, MAGELLAN-2, EXPEDITION-1, EXPEDITION-2, EXPEDITION-3, EXPEDITION-4, EXPEDITION-5, EXPEDITION-8, SURVEYOR-1, SURVEYOR-2, VOYAGE-1, VOYAGE-2, CERTAIN-1, CERTAIN-2 and APRI). These findings were categorized as on-treatment, post-treatment week (PTW) 4 or PTW12 failures.
Results
The mean HCV RNA level at baseline in the overall population of 5033 patients was 4,193,712 IU/ml ± 5,955,028 (6.2 log10 IU/ml ± 0.8) compared to 9,585,957 IU/ml ± 8,247,669 (6.8 log10 IU/ml ± 0.5) in 69 patients experiencing virologic failure by PTW12. The mean HCV RNA level at the time of virologic failure for all patients was 6,004,980 IU/ml ± 7,077,728 (6.4 log10 IU/ml ± 0.7). Twenty patients had on-treatment virologic failure with a mean HCV RNA level at the time of failure of 9,136,360 IU/ml ± 8,572,113 (6.7 log10 IU/ml ± 0.7), 36 patients had relapsed by PTW4 with a mean HCV RNA level at the time of relapse of 4,131,344 IU/ml ± 5,246,954 (6.3 log10 IU/ml ± 0.6), and 13 patients, who experienced relapse between PTW4 and PTW12, had a mean HCV RNA at relapse of 6,376,003 IU/ml ± 7,758,968 (6.3 log10 IU/ml ± 1.0).
Conclusions
At PTW12, 100% of virologic failures had an HCV RNA > 3.0 log10 IU/ml. The data are encouraging that with a LLOD of 3.0 log10 IU/ml, a point-of-care test could identify all treatment failures accurately; larger studies, including real-world data, are needed to confirm these findings.
Keywords: Direct-acting antivirals, HCV RNA, Hepatitis C virus, Virologic failure
Key Summary Points
| Why carry out this study? |
| Aid in establishing the lower limit of detection (LLoD) in order to confirm for HCV cure. |
| Determining LLOD at virologic failure aids in increasing testing feasibility through point-of-care assays in resource-limited settings. |
| What was learned from the study? |
| At post-treatment week 12, 100% of virologic failures had an HCV RNA > 3.0 log10 IU/ml. |
| The data are encouraging that a point-of-care test with a LLOD of 3.0 log10 IU/ml would likely identify all treatment failures accurately. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.13677418.
Introduction
Globally, there are approximately 71 million individuals infected with hepatitis C virus (HCV) [1]. The World Health Organization (WHO) initiated global elimination targets to help achieve HCV elimination by 2030 [1–3]. While the addition of direct-acting antiviral (DAA) treatment therapy has resulted in efficacy rates routinely exceeding 95% with favorable safety and tolerability profiles, a remaining barrier to HCV elimination is the limited access to medications and the limited ability of public health programs to confirm sustained virologic response (SVR), HCV cure, after treatment [4]. Currently, confirming SVR requires the use of nucleic acid amplification testing (NAAT) with complex procedures and reagents [5]. In many resource-limited settings, such technology is not readily available in close proximity to patients; consequently, many patients cannot obtain confirmation of HCV cure after treatment, resulting in an unknown infection status and whether the possibility of re-treatment is necessary [4, 6, 7]. Establishing a lower limit of detection (LLOD) in affordable, WHO pre-qualified, point-of-care assays to measure the presence of HCV viremia is needed for large-scale diagnosis and confirmation of cure, helping achieve WHO HCV elimination targets and reduce mortality [6, 8].
Typically, point-of-care assays do not include molecular amplification steps and are less sensitive than gold-standard, laboratory-based HCV NAAT [4]. Currently, for diagnostic point-of-care assays, a qualitative HCV RNA assay needs to have a LLOD of ≤ 1000 IU/ml (3.0 log10 IU/ml); however, there is limited evidence for the LLOD for assays testing for a cure [8]. Therefore, to establish LLOD in order to test for HCV cure via point-of-care assays, it is essential to characterize the level of viremia at the time of detecting HCV treatment failure to understand the sensitivity required to identify those who are not cured. Furthermore, it is important to identify the demographic and clinical correlates of those who have a low level of HCV viremia at the time of treatment failure, such that it is possible to make guidance for sub-populations that may not be appropriate for point-of-care testing.
The aim of this study is to describe population characteristics and the viral load at DAA failure to determine its implications for establishing LLOD at the time of SVR in simplified testing.
Methods
In this descriptive analysis, we constructed a cohort of patients with HCV virologic failure, selected from the overall number of patients completing phase II and III clinical trials; no new patients were enrolled. All patients provided written, informed consent to participate in the previous studies; each study included in this analysis was consistent with the ethical guidelines of the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice Guidelines. All previously conducted studies were approved at their respective sites by their independent ethics committee or institutional review board prior to enrollment. Inclusion criteria for this analysis were on-treatment HCV virologic failure or post-treatment HCV relapse. We defined on-treatment virologic failure as any one of the following: (1) confirmed increase in HCV RNA level of > 1 log10 IU/ml above the nadir during treatment, (2) confirmed HCV RNA level ≥ 100 IU/ml after the level had been < 15 IU/ml during treatment or (3) an HCV RNA level ≥ 15 IU/ml at the end of treatment (EoT) (with at least 6 weeks of treatment) [9–19]. Some studies required two consecutive results [10–12, 18]. Post-treatment relapse was defined as a confirmed HCV RNA level ≥ 15 IU/ml between the EoT and 12 weeks after the last dose of trial drug among patients who had both completed treatment and had an HCV RNA of < 15 IU/ml at the EoT [9, 13–19]. Plasma HCV RNA levels were determined by COBAS Ampliprep/Taqman® real-time reverse transcriptase polymerase-chain-reaction (RT-PCR) assay, v. 2.0 (Roche Molecular Diagnostics); two studies used COBAS TaqMan® RT-PCR assay v. 2.0 (Roche Molecular Diagnostics), where the lower limit of quantification (LLOQ) was 25 IU/ml [9, 10, 12–25]. In some studies, for patients receiving 12 weeks of glecaprevir/pibrentasvir (G/P) treatment, plasma HCV RNA levels were determined by using COBAS Ampliprep/Taqman® RT-PCR assay, v. 2.0, with a LLOD of 15 IU/ml, regardless of genotype (GT); in those studies, for patients receiving 8 weeks of G/P treatment, the COBAS TaqMan® RT-PCR assay v. 2.0 was used, which had a LLOQ of 25 IU/ml, regardless of GT. The LLOD was 5.6, 12, 3.7 and 20.4 IU/ml for HCV GT2 and GT4, 5 and 6, respectively [26, 27]. Patients were excluded from the study analysis if they did not achieve SVR12 for reasons other than virologic failure, such as treatment discontinuation or loss to follow-up.
Study Population
Patients were included from 20 phase II and phase III clinical trials including ENDURANCE-1 (NCT02604017), ENDURANCE-2 (NCT02640482), ENDURANCE-3 (NCT02640157), ENDURANCE-4 (NCT02636595), ENDURANCE 5–6 (NCT02966795), MAGELLAN-1 (NCT02446717), MAGELLAN-2 (NCT02692703), EXPEDITION-1 (NCT02642432), EXPEDITION-2 (NCT02738138), EXPEDITION-3 (NCT03219216), EXPEDITION-4 (NCT02651194), EXPEDITION-5 (NCT03069365), EXPEDITION-8 (NCT03089944), SURVEYOR-1 (NCT02243280), SURVEYOR-2 (NCT02243293), VOYAGE-1 (NCT03222583), VOYAGE-2 (NCT03235349), CERTAIN-1 (NCT02707952), CERTAIN-2 (NCT02723084) and APRI (NCT03212521). All patients had chronic HCV; HCV RNA > 1000 IU/ml was required at baseline for screening [9, 21, 23, 28]. In certain studies, an HCV RNA ≥ 1000 IU/ml was required, and in one study, an HCV RNA > 10,000 IU/ml was required at screening [10, 11, 13–15, 18, 19, 22, 24–27, 29, 30]. As per study protocols, patients were excluded from clinical trial participation if they had hepatitis B (HBV) co-infection or decompensated cirrhosis of the liver.
Statistical Analyses
The distribution of log10 HCV RNA level (IU/ml) at time of virologic failure was summarized by a histogram. Data were descriptive and summarized by means and standard deviations as well as medians and ranges (minimum and maximum), frequencies and percentages.
Results
Overall Patient Population
Across 20 clinical trials, 5033 patients with chronic HCV infection were enrolled. HCV DAA therapy was administered for 8 weeks in 50% (n = 2539) of patients, 12 weeks in 47% (n = 2360) of patients and for 16 weeks in 3% (n = 134) of patients; 96% of all patients received G/P. Fifty-five percent (n = 2774) of patients were male, 62% (n = 3123) were white, and 31% (n = 1543) were Asian. The majority of patients were GT1 (47%, n = 2388), treatment-naïve (76%, n = 3843) and non-cirrhotic (80%, n = 4042). Most patients were human immunodeficiency virus (HIV) negative (95%, n = 3455). In the overall population, 26% (n = 1321) of patients were from the USA, 10% (n = 512) and 9% (n = 430) of patients were from China and Japan, respectively; 5% (n = 253) were from Korea. The remainder of patients were from 32 countries across North America, South America, Asia, Africa, Australia and Europe. Further baseline characteristics of the overall population are described in Table 1.
Table 1.
Baseline demographics for the overall population
| Characteristic | Overall N = 5033 n (%) |
|---|---|
| Male | 2774 (55) |
| Race | |
| White | 3123 (62) |
| Black or African American | 291 (6) |
| Asian | 1543 (31) |
| Age ≥ 65 years | 888 (18) |
| BMI (kg/m2) (median, range) | 25.4 (14.2–65.7) |
| GT | |
| 1 | 2388 (47) |
| 2 | 1054 (21) |
| 3 | 1140 (23) |
| 4–6 | 451 (9) |
| Treatment-naïve | 3843 (76) |
| IFN or SOF-based treatment experience | 1014 (20) |
| DAA-based treatment experience | 175 (4) |
| Country (≥ 5% of patient population) | |
| USA | 1321 (26) |
| China | 512 (10) |
| Japan | 430 (8.5) |
| Korea, Republic of | 253 (5) |
| Fibrosis score | |
| F0–F1 | 2972 (62) |
| F2 | 343 (7) |
| F3 | 497 (10) |
| F4 | 979 (20) |
| Baseline HCV RNA level (IU/ml) (mean, SD) | 4,193,712 ± 5,955,028 |
| Baseline HCV RNA level (IU/ml) (median, range) | 1,930,000 (5.6–56,600,000) |
| < 1,000,000 | 1809 (36) |
| ≥ 1,000,000—< 2,000,000 | 751 (15) |
| ≥ 2,000,000 | 2473 (49) |
| Baseline HCV RNA level (log10 IU/ml) (mean, SD) | 6.2 ± 0.8 |
| Baseline HCV RNA (log10 IU/ml) (median, range) | 6.3 (0.8–7.8) |
| HIV co-infectiona | 188 (5) |
| Recent injection drug Useb | 66 (1) |
Patients were excluded from clinical trials if they had comorbid HBV or decompensated cirrhosis
BMI body mass index, DAA direct-acting antiviral, GT genotype, HBV hepatitis B, HIV human immunodeficiency virus, IFN interferon, SD standard deviation, SOF sofosbuvir
a1390 patients had missing HIV data
bWithin the last 12 months
HCV RNA Levels in the Overall Population at Baseline
The mean (± standard deviation) HCV RNA level at baseline in the overall population was 4,193,712 IU/ml ± 5,955,028 (6.2 log10 IU/ml ± 0.8). The median HCV RNA level at baseline was 1,930,000 IU/ml [range 5.6 IU/ml–56,600,000 IU/ml] (6.3 log10 IU/ml, 0.8–7.8). Baseline HCV RNA levels were ≥ 2,000,000 IU/ml in 49% of patients, ≥ 1,000,000 IU/ml – < 2,000,000 IU/ml in 15% of patients and < 1,000,000 IU/ml in 36% of patients.
Patients Experiencing Virologic Failure
Of the 5033 patients enrolled across 20 phase II and phase III clinical trials, a total of 69 patients experienced virologic failure. Of those, 66 patients received G/P therapy for 8, 12 or 16 weeks and 3 patients received a DAA other than G/P. Of the three patients who did not receive G/P treatment, one patient received treatment with sofosbuvir (SOF) + daclatasvir (DCV) for 12 weeks and two patients received treatment with SOF + ribavirin (RBV) for 12 weeks. Of the patients who received G/P and had virologic failure, 32% (n = 22) received 8 weeks of therapy, 48% (n = 33) received 12 weeks of therapy and 16% (n = 11) received therapy for 16 weeks. The majority of virologic failures were GT3 (n = 42, 61%) and GT1 (n = 17, 25%); 65% (n = 45) of patients were white and 33% (n = 23) were Asian. Ninety-eight percent (n = 47) of patients were negative for HIV; 21 patients had missing baseline HIV status. The majority of virologic failures were from the USA (n = 21, 30%), China (n = 13, 19%), New Zealand (n = 8, 12%) and Japan (n = 6, 9%). Further baseline characteristics of patients who experienced virologic failure are shown in Table 2.
Table 2.
Baseline demographics in patients with virologic failure
| Characteristic | Virologic failure at PTW12 N = 69 n (%) |
|---|---|
| Male | 51 (74) |
| Race | |
| White | 45 (65) |
| Black or African American | 1 (1) |
| Asian | 23 (33) |
| Age ≥ 65 years | 8 (12) |
| BMI (kg/m2) (median, range) | 25.1 (17.0–42.6) |
| GT | |
| 1 | 17 (25) |
| 2 | 7 (10) |
| 3 | 42 (61) |
| 4–6 | 3 (4) |
| Treatment-naïve | 33 (48) |
| IFN or SOF-based treatment experience | 23 (33) |
| DAA-based treatment experience | 13 (19) |
| Country (≥ 5% of patient population) | |
| USA | 21 (30) |
| China | 13 (19) |
| New Zealand | 8 (12) |
| Australia | 7 (10) |
| Japan | 6 (9) |
| Fibrosis score | |
| F0–F1 | 33 (49) |
| F2 | 9 (13) |
| F3 | 11 (16) |
| F4 | 14 (21) |
| Baseline HCV RNA level (IU/ml) (mean, SD) | 9,585,957 ± 8,247,669 |
| Baseline HCV RNA level (IU/ml) (median, range) | 8,140,000 (135,000–36,100,000) |
| < 1,000,000 | 5 (7) |
| ≥ 1,000,000— < 2,000,000 | 7 (10) |
| ≥ 2,000,000 | 57 (83) |
| HCV RNA level (IU/ml) (median, range) | |
| On-treatment virologic failurea | 7,425,000 (61,200–28,500,000) |
| Relapse by PTW12b | 2,130,000 (7,040–27,900,000) |
| Relapse by PTW4c | 1,780,000 (72,400–20,300,000) |
| Relapse between PTW4 and PTW12d | 3,990,000 (7040–27,900,000) |
| All virologic failure subjects | 3,350,000 (7040–28,500,000) |
| Baseline HCV RNA level (log10 IU/ml) (mean, SD) | 6.8 ± 0.5 |
| Baseline HCV RNA level (log10 IU/ml) (median, range) | 6.9 (5.1–7.6) |
| HCV RNA level (log10 IU/ml) (median, range) | |
| On-treatment virologic failurea | 6.9 (4.8–7.5) |
| Relapse by PTW12b | 6.3 (3.8–7.4) |
| Relapse by PTW4c | 6.3 (4.9–7.3) |
| Relapse between PTW4 and PTW12d | 6.6 (3.8–7.4) |
| All virologic failure subjects | 6.5 (3.8–7.5) |
| HIV co-infectione | 1 (2) |
| Recent injection drug usef | 1 (1) |
Patients were excluded from clinical trials if they had comorbid HBV or decompensated cirrhosis
BMI body mass index, DAA direct-acting antiviral, GT genotype, HBV hepatitis B virus, HIV human immunodeficiency virus, IFN interferon, PTW post-treatment week, SD standard deviation, SOF sofosbuvir
aN = 20
bN = 49
cN = 36
dN = 13
eTwenty-one patients had missing HIV data
fWithin the last 12 months
HCV RNA Levels in Patients Experiencing Virologic Failure
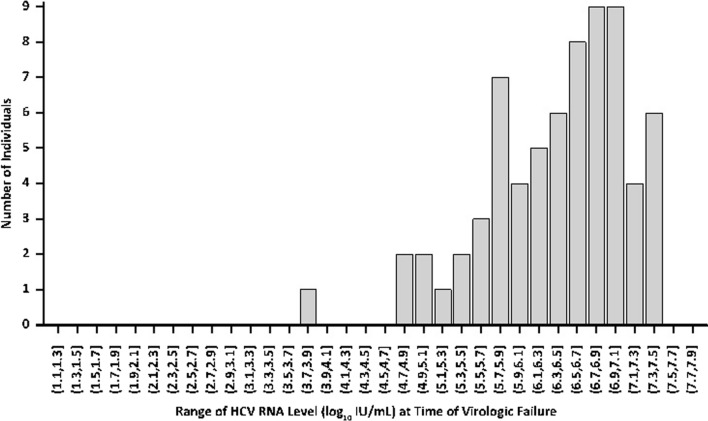
The mean baseline HCV RNA level for all patients experiencing virologic failure by post-treatment week (PTW) 12 in an intention-to-treat (ITT) population was 9,585,957 IU/ml ± 8,247,669 (6.8 log10 IU/ml ± 0.5). The mean HCV RNA level at the time of virologic failure for all patients was 6,004,980 IU/ml ± 7,077,728 (6.4 log10 IU/ml ± 0.7). The median baseline HCV RNA level was 8,140,000 IU/ml [135,000 IU/ml–36,100,000 IU/ml] (6.9 log10 IU/ml, 5.1–7.6). The median HCV RNA level at the time of virologic failure for all patients was 3,350,000 IU/ml [7,040 IU/ml–28,500,000 IU/ml] (6.5 log10 IU/ml, 3.8–7.5) (Fig. 1).
Fig. 1.
Histogram of HCV RNA level (log10 IU/ml) at time of virologic failure (ITT population with virologic failure at PTW12) in 69 patients across 20 phase II/III clinical trials. ITT intention to treat, PTW post-treatment week
Out of the 69 patients who experienced virologic failure, 20 patients had on-treatment virologic failure with a mean HCV RNA level at the time of virologic failure of 9,136,360 IU/ml ± 8,572,113 (6.7 log10 IU/ml ± 0.7) and a median HCV RNA level at the time of virologic failure of 7,425,000 IU/ml [61,200 IU/ml–28,500,000 IU/ml] (6.9 log10 IU/ml, 4.8–7.5). Forty-nine patients experienced relapse by PTW12 with a mean HCV RNA level of 4,726,866 IU/ml ± 6,010,599 (6.3 log10 IU/ml ± 0.7). The median HCV RNA level of patients experiencing relapse by PTW12 was 2,130,000 IU/ml [7040 IU/ml–27,900,000 IU/ml] (6.3 log10 IU/ml, 3.8–7.4). The 36 patients who had a relapse by PTW4 had a mean HCV RNA level at the time of relapse of 4,131,344 IU/ml ± 5,246,954 (6.3 log10 IU/ml ± 0.6) and a median HCV RNA level at the time of relapse of 1,780,000 IU/ml [72,400 IU/ml–20,300,000 IU/ml] (6.3 log10 IU/ml, 4.9–7.3). There were 13 patients who experienced relapse between PTW4 and PTW12; these patients had a mean HCV RNA at relapse of 6,376,003 IU/ml ± 7,758,968 (6.3 log10 IU/ml ± 1.0) and a median HCV RNA at relapse of 3,990,000 IU/ml [7040 IU/ml–27,900,000 IU/ml] (6.6 log10 IU/ml, 3.8–7.4).
Discussion
We analyzed data from > 5000 patients treated for HCV with DAA therapies to identify 69 patients with either on-treatment virologic failure or post-treatment HCV relapse. We then characterized the degree of HCV viremia at the time of virologic failure to help inform the LLOD for HCV cure.
Patients who experienced on-treatment virologic failure had the highest mean HCV RNA levels at the time that treatment failure was identified (6.7 log10 IU/ml). Patients who relapsed by PTW4, by PTW12 or between PTW4 and PTW12 had a similar mean HCV RNA (6.3 log10 IU/ml) as patients who relapsed between PTW4 and PTW12 having 6,376,003 IU/ml compared to 4,131,344 IU/ml and 4,726,866 IU/ml in patients who relapsed by PTW4 and PTW12, respectively. Understanding the HCV RNA level at various time points of virologic failure can aid in determining the LLOD at SVR to check for HCV cure. In all cases, 100% of individuals with a detectable viral load had an HCV RNA > 3.0 log10 IU/ml. Because the general limit of detection of near-patient technologies is approximately 3.0 log10 IU/ml, these data are reassuring that while point-of-care testing for HCV viremia may have a lower sensitivity compared the gold standard RNA testing, it may still be sensitive enough to accurately identify nearly all treatment failures, whether HCV viremia is tested while the patient is on treatment or after treatment is complete [8].
There are limitations to this study. Most notably, these data are from randomized controlled trials and may not reflect clinical treatment experience in real-world settings [31]. Treatment adherence was very high in these clinical trial cohorts, and treatment failure reflects virologic breakthrough despite treatment, as opposed to non-adherence to medication therapy; additionally, re-infection was not explored in the context of this analysis. In the majority of clinical trials assessed, patients were excluded at the time of screening if they had a baseline HCV RNA ≤ 1000 IU/ml [10, 13–15, 18, 22, 24, 29, 30]. In another study looking at HCV RNA levels at the time of virologic failure, baseline and PTW12 HCV RNA levels were highly correlated; in patients with low baseline HCV RNA levels experiencing virologic failure, low baseline HCV RNA levels may be associated with low PTW12 HCV RNA levels [32]. The potential association of low-level viremia at baseline and low-level HCV RNA at PTW12 raises the concern of potential misdiagnosis with a low-sensitivity point-of-care assay. However, with the high expected effectiveness rates of DAA therapy in patients with low baseline HCV RNA levels, the overall risk is likely minimal. The LLOD in the clinical trials varied at 3.7, 5.6, 12, 15 and 20.4 IU/ml depending on the HCV RNA assay used and patient GT; however, the variation in LLOD levels was not significant as the lower end of LLOD values was not explored for the purposes of this analysis. Additionally, all patients experiencing virologic failure had an HCV RNA level > 1000 IU/ml. Furthermore, while we characterized viral load for on-treatment failure as well as post-treatment relapse, these were distinct cohorts with one observation per failure. As there were no data available on individuals at multiple time points, we were unable to determine whether viral load changes from the on-treatment period to the post-treatment period in individual patients. The small number of patients who experienced on-treatment virologic failure or post-treatment relapse may limit both statistical power and generalizability of results to real-world populations.
Conclusion
We analyzed data from 20 clinical trials to characterize the level of HCV viremia at the time of treatment failure to inform the LLOD needed to monitor HCV cure after completion of treatment. The data are encouraging that a point-of-care test with a LLOD of 3.0 log10 IU/ml would likely identify all treatment failures accurately in a similar patient population. The reproducibility in other patient populations is yet to be determined, and a larger study combining data from multiple contexts, including real-world settings in underserved, global populations, is necessary to confirm these findings; however, these preliminary data are encouraging.
Acknowledgements
The authors would like to express their gratitude to the patients who participated in the previous studies (NCT02604017, NCT02640482, NCT02640157, NCT02636595, NCT02966795, NCT02446717, NCT02692703, NCT02642432, NCT02738138, NCT03219216, NCT02651194, NCT03069365, NCT03089944, NCT02243280, NCT02243293, NCT03222583, NCT03235349, NCT02707952, NCT02723084, NCT03212521), and their families, as well as the study investigators and coordinators of the studies included in this publication. Glecaprevir was identified by AbbVie and Enanta.
Funding
The design, study conduct, analysis, and financial support of the study was provided by AbbVie. AbbVie participated in the interpretation of data, review, and approval of the content. All authors had access to all relevant data and participated in writing, review, and approval of this manuscript. No honoraria or payments were made for authorship. Academic partners received no funding from AbbVie for this work. This work and the journal's Rapid Service and Open Access fees were supported by AbbVie.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Medical Writing and Editorial Assistance
Medical writing support was provided by Sneh Mody, PharmD, MBA, BCCCP of AbbVie, and funded by AbbVie.
Disclosures
Jake R Morgan, Alexandra Savinkina, Sonjelle Shilton and Benjamin Linas: nothing to disclose; Ana Gabriela Pires dos Santos, Zhenyi Xue: employees of AbbVie Inc. and may hold stock or options.
Compliance with Ethics Guidelines
All patients provided written, informed consent to participate in the previous studies; each study included in this analysis was consistent with the ethical guidelines of the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice Guidelines. All previously conducted studies were approved at their respective sites by their independent ethics committee or institutional review board prior to enrollment.
Data Availability
AbbVie is committed to responsible data sharing regarding the clinical trials we sponsor. This includes access to anonymized, individual and trial-level data (analysis data sets), as well as other information (e.g., protocols and Clinical Study Reports), as long as the trials are not part of an ongoing or planned regulatory submission. This includes requests for clinical trial data for unlicensed products and indications. These clinical trial data can be requested by any qualified researchers who engage in rigorous, independent scientific research and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). Data requests can be submitted at any time and the data will be accessible for 12 months, with possible extensions considered. For more information on the process, or to submit a request, visit the following link: https://www.abbvie.com/our-science/clinical-trials/clinical-trials-data-and-information-sharing/data-and-information-sharing-with-qualified-researchers.html.
References
- 1.Polaris Observatory HCVC. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol. 2017;2(3):161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Hepatitis Report 2017. Available at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. Accessed Jan 27, 2021.
- 3.Polaris Observatory. Centers for Disease Analysis. http://cdafound.org/polaris/. Accessed Jan 27, 2021.
- 4.Fourati S, Feld JJ, Chevaliez S, Luhmann N. Approaches for simplified HCV diagnostic algorithms. J Int AIDS Soc. 2018;21(Suppl 2):e25058. doi: 10.1002/jia2.25058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta E, Bajpai M, Choudhary A. Hepatitis C virus: screening, diagnosis, and interpretation of laboratory assays. Asian J Transfus Sci. 2014;8(1):19–25. doi: 10.4103/0973-6247.126683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freiman JM, Wang J, Easterbrook PJ, Horsburgh CR, Marinucci F, White LF, et al. Deriving the optimal limit of detection for an HCV point-of-care test for viraemic infection: analysis of a global dataset. J Hepatol. 2019;71(1):62–70. doi: 10.1016/j.jhep.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. Progress report on access to hepatitis C treatment: focus on overcoming barriers in low- and middle-income countries. 2018. Available from: http://www.who.int/hepatitis/publications/hep-c-access-report-2018/en/. Accessed on Jan 27, 2021.
- 8.European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatol [ahead of print]. 2020. 10.1016/j.jhep.2020.08.018. [DOI] [PubMed]
- 9.Zeuzem S, Foster GR, Wang S, Asatryan A, Gane E, Feld JJ, et al. Glecaprevir-Pibrentasvir for 8 or 12 Weeks in HCV Genotype 1 or 3 Infection. N Engl J Med. 2018;378(4):354–369. doi: 10.1056/NEJMoa1702417. [DOI] [PubMed] [Google Scholar]
- 10.Asselah T, Lee SS, Yao BB, Nguyen T, Wong F, Mahomed A, et al. Efficacy and safety of glecaprevir/pibrentasvir in patients with chronic hepatitis C virus genotype 5 or 6 infection (ENDURANCE-5,6): an open-label, multicentre, phase 3b trial. Lancet Gastroenterol Hepatol. 2019;4(1):45–51. doi: 10.1016/S2468-1253(18)30341-8. [DOI] [PubMed] [Google Scholar]
- 11.Poordad F, Felizarta F, Asatryan A, Sulkowski MS, Reindollar RW, Landis CS, et al. Glecaprevir and pibrentasvir for 12 weeks for hepatitis C virus genotype 1 infection and prior direct-acting antiviral treatment [supplement] Hepatology. 2017;66(2):389–397. doi: 10.1002/hep.29081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gane E, Poordad F, Wang S, Asatryan A, Kwo PY, Lalezari J, et al. High Efficacy of ABT-493 and ABT-530 Treatment in Patients With HCV Genotype 1 or 3 Infection and Compensated Cirrhosis. Gastroenterology. 2016;151(4):651–9 e1. [DOI] [PubMed]
- 13.Wei L, Wang G, Alami NN, Xie W, Heo J, Xie Q, et al. Glecaprevir-pibrentasvir to treat chronic hepatitis C virus infection in Asia: two multicentre, phase 3 studies- a randomised, double-blind study (VOYAGE-1) and an open-label, single-arm study (VOYAGE-2) Lancet Gastroenterol Hepatol. 2020;5(9):839–849. doi: 10.1016/S2468-1253(20)30086-8. [DOI] [PubMed] [Google Scholar]
- 14.Reau N, Kwo PY, Rhee S, Brown RS, Jr, Agarwal K, Angus P, et al. Glecaprevir/pibrentasvir treatment in liver or kidney transplant patients with hepatitis C virus infection. Hepatology. 2018;68(4):1298–1307. doi: 10.1002/hep.30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forns X, Lee SS, Valdes J, Lens S, Ghalib R, Aguilar H, et al. Glecaprevir plus pibrentasvir for chronic hepatitis C virus genotype 1, 2, 4, 5, or 6 infection in adults with compensated cirrhosis (EXPEDITION-1): a single-arm, open-label, multicentre phase 3 trial. Lancet Infect Dis. 2017;17(10):1062–1068. doi: 10.1016/S1473-3099(17)30496-6. [DOI] [PubMed] [Google Scholar]
- 16.Gane E, Lawitz E, Pugatch D, Papatheodoridis G, Brau N, Brown A, et al. Glecaprevir and pibrentasvir in patients with HCV and severe renal impairment. N Engl J Med. 2017;377(15):1448–1455. doi: 10.1056/NEJMoa1704053. [DOI] [PubMed] [Google Scholar]
- 17.Lawitz E, Flisiak R, Abunimeh M, Sise ME, Park JY, Kaskas M, et al. Efficacy and safety of glecaprevir/pibrentasvir in renally impaired patients with chronic HCV infection. Liver Int. 2020;40(5):1032–1041. doi: 10.1111/liv.14320. [DOI] [PubMed] [Google Scholar]
- 18.Brown RS, Jr, Buti M, Rodrigues L, Chulanov V, Chuang WL, Aguilar H, et al. Glecaprevir/pibrentasvir for 8 weeks in treatment-naive patients with chronic HCV genotypes 1–6 and compensated cirrhosis: the EXPEDITION-8 trial. J Hepatol. 2020;72(3):441–449. doi: 10.1016/j.jhep.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Peribanez-Gonzalez M, Cheinquer H, Rodrigues L, Lima MP, Alvares-da-Silva MR, Madruga J, et al. Efficacy and safety of glecaprevir/pibrentasvir in treatment-naive adults with chronic hepatitis C virus genotypes 1–6 in Brazil. Ann Hepatol. 2020;20:100257. doi: 10.1016/j.aohep.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Poordad F, Felizarta F, Asatryan A, Sulkowski MS, Reindollar RW, Landis CS, et al. Glecaprevir and pibrentasvir for 12 weeks for hepatitis C virus genotype 1 infection and prior direct-acting antiviral treatment. Hepatology. 2017;66(2):389–397. doi: 10.1002/hep.29081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poordad F, Pol S, Asatryan A, Buti M, Shaw D, Hezode C, et al. Glecaprevir/Pibrentasvir in patients with hepatitis C virus genotype 1 or 4 and past direct-acting antiviral treatment failure. Hepatology. 2018;67(4):1253–1260. doi: 10.1002/hep.29671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockstroh JK, Lacombe K, Viani RM, Orkin C, Wyles D, Luetkemeyer AF, et al. Efficacy and safety of glecaprevir/pibrentasvir in patients coinfected with hepatitis C virus and human immunodeficiency virus Type 1: the EXPEDITION-2 study. Clin Infect Dis. 2018;67(7):1010–1017. doi: 10.1093/cid/ciy220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyles D, Poordad F, Wang S, Alric L, Felizarta F, Kwo PY, et al. Glecaprevir/pibrentasvir for hepatitis C virus genotype 3 patients with cirrhosis and/or prior treatment experience: a partially randomized phase 3 clinical trial. Hepatology. 2018;67(2):514–523. doi: 10.1002/hep.29541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chayama K, Suzuki F, Karino Y, Kawakami Y, Sato K, Atarashi T, et al. Efficacy and safety of glecaprevir/pibrentasvir in Japanese patients with chronic genotype 1 hepatitis C virus infection with and without cirrhosis. J Gastroenterol. 2018;53(4):557–565. doi: 10.1007/s00535-017-1391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumada H, Watanabe T, Suzuki F, Ikeda K, Sato K, Toyoda H, et al. Efficacy and safety of glecaprevir/pibrentasvir in HCV-infected Japanese patients with prior DAA experience, severe renal impairment, or genotype 3 infection. J Gastroenterol. 2018;53(4):566–575. doi: 10.1007/s00535-017-1396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asselah T, Kowdley KV, Zadeikis N, Wang S, Hassanein T, Horsmans Y, et al. Efficacy of glecaprevir/pibrentasvir for 8 or 12 Weeks in patients with hepatitis C virus genotype 2, 4, 5, or 6 infection without cirrhosis [supplement] Clin Gastroenterol Hepatol. 2018;16(3):417–426. doi: 10.1016/j.cgh.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 27.Kwo PY, Poordad F, Asatryan A, Wang S, Wyles DL, Hassanein T, et al. Glecaprevir and pibrentasvir yield high response rates in patients with HCV genotype 1–6 without cirrhosis. J Hepatol. 2017;67(2):263–271. doi: 10.1016/j.jhep.2017.03.039. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda H, Chayama K, Suzuki F, Sato K, Atarashi T, Watanabe T, et al. Efficacy and safety of glecaprevir/pibrentasvir in Japanese patients with chronic genotype 2 hepatitis C virus infection. Hepatology. 2018;67(2):505–513. doi: 10.1002/hep.29510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gane E, Lawitz E, Pugatch D, Papatheodoridis G, Brau N, Brown A, et al. Glecaprevir and pibrentasvir in patients with HCV and severe renal impairment [supplement] N Engl J Med. 2017;377(15):1448–1455. doi: 10.1056/NEJMoa1704053. [DOI] [PubMed] [Google Scholar]
- 30.Fontana RJ, Lens S, McPherson S, Elkhashab M, Ankoma-Sey V, Bondin M, et al. Efficacy and safety of 8 weeks of glecaprevir/pibrentasvir in treatment-naive, HCV-infected patients with APRI </= 1 in a single-arm, open-label. Multicenter Study Adv Ther. 2019;36(12):3458–3470. doi: 10.1007/s12325-019-01123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan JR, Servidone M, Easterbrook P, Linas BP. Economic evaluation of HCV testing approaches in low and middle income countries. BMC Infect Dis. 2017;17(Suppl 1):697. doi: 10.1186/s12879-017-2779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrington PR, Komatsu TE, Sun H, Naeger LK. Hepatitis C virus RNA levels following virologic failure with direct-acting antivirals: implications for lower sensitivity diagnostic assays. Clin Infect Dis. 2020;70(2):327–330. doi: 10.1093/cid/ciz385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
AbbVie is committed to responsible data sharing regarding the clinical trials we sponsor. This includes access to anonymized, individual and trial-level data (analysis data sets), as well as other information (e.g., protocols and Clinical Study Reports), as long as the trials are not part of an ongoing or planned regulatory submission. This includes requests for clinical trial data for unlicensed products and indications. These clinical trial data can be requested by any qualified researchers who engage in rigorous, independent scientific research and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). Data requests can be submitted at any time and the data will be accessible for 12 months, with possible extensions considered. For more information on the process, or to submit a request, visit the following link: https://www.abbvie.com/our-science/clinical-trials/clinical-trials-data-and-information-sharing/data-and-information-sharing-with-qualified-researchers.html.