Abstract
Background
Fibroblast growth factor receptor 2 (FGFR2) rearrangement is expected to be a novel therapeutic target in advanced/recurrent biliary tract cancer (BTC). However, efficient detection and the exact frequency of FGFR2 rearrangements among patients with advanced/recurrent BTC have not been determined, and the clinical characteristics of FGFR2 rearrangement-positive patients have not been fully elucidated. We aimed to determine the frequency of FGFR2 rearrangement-positive patients among those with advanced/recurrent BTC and elucidate their clinicopathological characteristics.
Methods
Paraffin-embedded tumor samples from formalin-fixed surgical or biopsy specimens of patients with advanced/recurrent BTC were analyzed for positivity of FGFR2 rearrangement by fluorescent in situ hybridization (FISH). RNA sequencing was performed on samples from all FISH-positive and part of FISH-negative patients.
Results
A total of 445 patients were enrolled. FISH was performed on 423 patients (272 patients with intrahepatic cholangiocarcinoma (ICC), 83 patients with perihilar cholangiocarcinoma (PCC), and 68 patients with other BTC). Twenty-one patients with ICC and four patients with PCC were diagnosed as FGFR2-FISH positive. Twenty-three of the 25 FISH-positive patients (20 ICC and 3 PCC) were recognized as FGFR2 rearrangement positive by targeted RNA sequencing. Younger age (≤ 65 years; p = 0.018) and HCV Ab- and/or HBs Ag-positivity (p = 0.037) were significantly associated with the presence of FGFR2 rearrangement (logistic regression).
Conclusions
FGFR2 rearrangement was identified in ICC and PCC patients, and was associated with younger age and history of hepatitis viral infection.
Electronic supplementary material
The online version of this article (10.1007/s00535-020-01735-2) contains supplementary material, which is available to authorized users.
Keywords: Advanced/recurrent biliary tract cancer, FGFR2 rearrangement, Fluorescent in situ hybridization, RNA sequencing
Introduction
Patients with biliary tract cancer (BTC) have a poor prognosis, with a 5 year survival rate of 22.5% [1]. BTC consists of intrahepatic cholangiocarcinoma (ICC), perihilar cholangiocarcinoma (PCC), gallbladder carcinoma (GBC), distal cholangiocarcinoma (DCC), and ampullary carcinoma (AC), and the biological characteristics and prognosis vary depending on the primary site [2].
The international standard first-line chemotherapy for advanced/recurrent BTC is gemcitabine plus cisplatin therapy; however, the median overall survival is only 11.7 months in the ABC-02 study, which established this therapy as its standard [3].
Genomic analyses of BTC have led to the development of molecular target therapy [4]. Especially, isocitrate dehydrogenase (IDH) mutations [5, 6] and fibroblast growth factor receptor 2 (FGFR2) fusion genes [7–12] in ICC have been identified as important driver alterations and are promising therapeutic targets. Actually, the US Food and Drug Administration (FDA) granted accelerated approval to pemigatinib for cholangiocarcinoma with an FGFR2 rearrangement or fusion in April 2020 based on the favorable results of a clinical trial [13].
Fibroblast growth factor/fibroblast growth factor receptor (FGF/FGFR) signaling plays a role in the development of normal organs and blood vessels, as well as in skeleton formation. FGFR rearrangements (fusions/truncations) autonomously activate the FGF signaling pathway and are involved in breast cancer, lung cancer, gastric cancer, and hematological tumors [10]. FGFR2 rearrangements are considered to be one of the important driver genes in ICC, with 9–14% of ICC cases reported as positive for FGFR2 rearrangements [11–14].
Several clinical trials targeting FGFR2 rearrangements have already been conducted [8, 9, 13], but there is only limited information of its positivity rate and the related clinical features in advanced/recurrent cases. In addition, although the positive rate of FGFR2 rearrangements has been reported in ICC, data are not available for the other BTCs. Thus, to verify whether, in addition to ICC, other BTCs are also associated with FGFR2 rearrangements, investigations should be extended to all advanced/recurrent BTCs, including those outside of ICC.
Various molecular diagnostic methods can be applied for the detection of the oncogenic fusion genes such as FGFR2 and ALK, i.e., multiplex RT-PCR, fluorescent in situ hybridization (FISH) and target-panel DNA sequencing or RNA sequencing from frozen or formalin-fixed paraffin-embedded (FFPE) specimens. However, more than 40 genes have been identified as fusion partner with FGFR2, but comprehensive identification of fusion genes takes time and often becomes a problem in medical practice. Among those candidates, we adopted FISH assay for screening FGFR2 rearrangements in this prospective study, because the majority of our cases were expected not to have archival surgical tissue and it is frequently difficult to obtain enough volume of tumor tissue for many tests including next-generation sequencing (NGS). FISH is also a highly preferable method for oncogenic gene rearrangement detection in the point of short turnaround time, low cost, and requiring smaller amounts of tissue (than NGS) [15]. We also performed post hoc RNA sequencing and validated the FISH results in this study.
Methods
Study design and patients
This was a prospective observational multicenter study conducted in Japan. The subjects were patients with advanced/recurrent BTC (ICC, PCC, DCC, GBC, and AC), histologically confirmed as adenocarcinoma or adenosquamous carcinoma, who were scheduled for or had received systemic chemotherapy. FISH analysis was performed for all BTCs between March 2014 and February 2016 (the first period). Since, in this period, positive patients were only found among those with ICC and PCC, only patients with these two BTCs were enrolled in the subsequent study period between October 2016 and November 2018.
This study was approved by the institutional review boards from all participating institutions. Written consent was obtained directly from patients, and the study was conducted in accordance with the Declaration of Helsinki, the “Ethical Guidelines for Epidemiology Research,” and the “Ethical Guidelines for Medical and Health Research Involving Human Subjects.” This trial was registered in UMIN Clinical Trials Registry (UMIN-CTR) under the registration number UMIN000014767.
Sample preparation
For efficient and prospective diagnosis of the status of FGFR2 rearrangement, we performed the break-apart FISH assays using paraffin-embedded tumor samples from formalin-fixed surgical or biopsy specimens as described below. After patients were enrolled, one hematoxylin and eosin (HE) stained, and five unstained sections, of 4 µm thickness, were prepared from the FFPE samples. The HE-stained section was used for marking tumors, while FISH analysis and targeted RNA sequencing were performed using one and two unstained sections, respectively.
To set the cutoff value for the FISH analysis, we used previously reported FGFR2 fusion-positive and -negative cases from surgically resected specimens [16]. Three FGFR2 fusion-positive cases and nine negative cases validated by RNA sequencing were used in the assay. In fusion-positive cases, two showed YGR FISH signal (80% and 91%) and one showed YR FISH signal (88%). Fusion-negative cases showed background FISH signals at 6.4% (mean + 2SD). From these results, we initially defined the cutoff value of ≥ 7% for the positive cells in the FISH analysis. We also performed RNA sequencing, targeting 1385 genes including FGFR2, for all 197 cases evaluated by FISH in the first period except 16 cases with low RNA-quantity specimens, and on 18 FISH-positive cases in the second period.
We collected clinical information of the patients who got FISH analyses. The following items were considered at enrollment: age, sex, family history, occupational history of working in the printing industry, disease status, degree of histological differentiation, macroscopic type, smoking history, drinking history, HCV antibody, HBs antigen, cholelithiasis, past history of primary sclerosing cholangitis (PSC), and presence/absence of pancreaticobiliary maljunction. In the second period, the survival of FGFR2 rearrangement-positive patients and the use of FGFR inhibitors were also investigated.
The primary end point was the frequency of FGFR2 rearrangement-positive patients among those with BTC. The secondary end point was the correlation between the presence of FGFR2 rearrangement and the clinical characteristics of the patients.
FISH analysis
To identify FGFR2 rearrangements, break-apart FISH assays were performed on FFPE tumors using a probe set, which hybridizes with the neighboring 5’-telomeric (RP11-78A18, labeled with Spectrum Green) and 3’-centromeric (RP11-7P17, labeled with Spectrum Red) sequences of the FGFR2 gene (Chromosome Science Labo Inc., Sapporo, Japan). One-hundred non-overlapping tumor cells with at least one 5’ and one 3’ signal, whether fused or separated, were examined and a detailed signal pattern was recorded at a clinical laboratory testing company with a turnaround time between 7 and 10 days (LSI medience, Tokyo, Japan). A fused 5’/3’ signal may appear yellow due to co-localization of green (5’ probe) and red (3’ probe) signals. A split signal was defined by 5’ and 3’ probes observed at a distance > 1 time the signal size, and signals separated by less than this distance were regarded as fused signals. The rearrangement-positive cells were defined as having any split signal (YGR FISH type) or isolated green signal (YG type), while any isolated red signal (YR type) was treated as rearrangement-negative, because this type denotes 5’ probe deletion of FGFR2 (Supple Fig. 1). The rate of rearrangement-positive cells was calculated for each case. These scoring criteria were developed and validated internally by using genotyped positive and negative controls from surgically resected specimens.
Targeted RNA sequencing
Total RNA was isolated from one or two FFPE tumor sections with 4 µm thickness using an miRNeasy FFPE kit (Qiagen, Hilden, Germany). The quantity of the RNA was determined with a NanoDrop instrument (ThermoFisher, Waltham, MA, USA). A targeted RNA sequencing library was prepared from 25–200 ng total RNA using a TruSight RNA PanCancer library kit (Illumina, San Diego, USA), which covers 1385 cancer-related genes including FGFR2. The library was subjected to paired-end sequencing of 151-bp fragments on a MiSeq DNA sequencer (Illumina). We obtained at least 50 million reads per sample, and the paired-end reads were mapped and aligned to known RNA sequences in the RefSeq, Ensembl, and LincRNA databases with the BWA-MEM program. After selecting the best hits with the proper spacing and orientation, gene expression values were calculated as reads per kilobase of exon per million mapped reads (RPKM). FGFR2 gene expression is indicated as the ratio (fold) of RPKM between each sample and the median value of FISH-negative cases (RPKM = 162.9). Aberrant paired reads that mapped to different transcription units of FGFR2 were identified as FGFR2 rearrangements. Junction reads revealed in-frame gene fusions between FGFR2 and other genes. In six cases (case nos. 2, 3, 4, 5, 24 and 25), another targeted RNA sequencing method of Anchored Multiplex PCR assay to detect FGFR fusions (AMP-FGFR), FusionPlex FGFR panel (Archer DX, Boulder, USA) was performed. Using 50 ng total RNA, sequencing libraries of targeting FGFR genes (FGFR1, 2 and 3) were sequenced and analyzed according to the manufacturer’s protocols.
Statistical analysis
The primary objective was to estimate the rate of FGFR2 rearrangement-positive patients. With an expected positive FGFR2 rearrangement rate of 20–40%, a total of 100 patients were planned for achieving a target width of less than 10% for the two-sided 95% confidence interval. The correlations between the presence of FGFR2 rearrangement and the clinical characteristics of the patients were analyzed using the Fisher’s exact test and logistic regression. All tests were two-sided, and a p value < 0.05 was considered statistically significant. Younger age was defined under the median age of 65 years, and excessive alcohol abuse was defined as consumption of 60 g or more of pure ethanol per day according to the WHO criteria. The FGFR2 rearrangement-positive patients enrolled in the second period were included in the survival analysis. The starting date for the overall survival analysis was the date of first-line treatment initiation, and the ending date was the date on which an event was censored, or death occurred. Survival was analyzed using the Kaplan–Meier method. The software used for analyses was SPSS 22.0 (IBM Corp. SPSS Inc., Armonk, NY, USA).
Results
Study flow
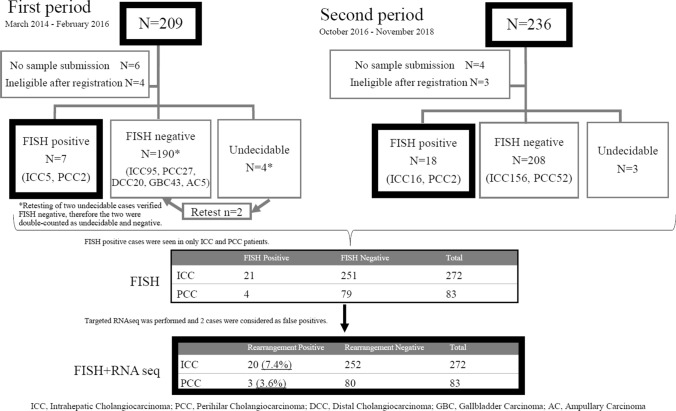
A total of 445 Japanese patients were enrolled from 20 institutions for this prospective study, and the analysis diagram is summarized in Fig. 1. There were seven patients in whom the FISH assay was undecidable (two cases were successfully reported in retest) due to no signal (potentially insufficient sample amount) in three cases, self-luminous primarily due to inappropriate marking (ink containing a fluorescence) during formation of the paraffin block in three cases, and peeling off of the paraffin section due to unsuitable slide glass lacking silane coating in one case. Results of the FISH assay were successfully reported on 423 patients (Suppl Fig. 1, 2). Table 1 shows the clinical characteristics of the patients. Regarding the primary sites, 64% and 20% of patients were ICC and PCC, respectively. The mean time between enrollment and FISH report delivery was 13 days (7–31 days). There are two periods of patient enrollment in this study, where 209 patients were in the first period and 236 patients were in the second period. Four patients in the first period and three in the second period failed to be analyzed by the FISH assay, and two patients in the first period were retested using a different tumor tissue with negative results obtained. The FISH assay was performed a total of 432 times, yielding 425 results (98.3%). The success rate of the FISH assay in the biopsy specimen and surgically resected specimens was 97.5% (273/280) and 100% (152/152), respectively.
Fig. 1 .
Study flow and summary of FGFR2 rearrangement detection
Table 1 .
Clinical characteristics of the study patients (N = 423)
| Characteristic | N (%) |
|---|---|
| Sex | |
| Male | 280 (66) |
| Female | 143 (34) |
| Age | |
| > 65 | 191 (45) |
| ≤ 65 | 232 (55) |
| Tested tumor specimen | |
| Resection | 152 (36) |
| Biopsy | 271 (64) |
| Stage | |
| Metastasis | 221 (52) |
| Recurrence after resection | 108 (26) |
| Locally advanced | 94 (22) |
| Differentiation | |
| Poorly differentiated | 75 (18) |
| Moderate/well differentiated | 166 (39) |
| Unknown | 182 (43) |
| Primary site | |
| ICC | 272 (64) |
| ECC | |
| PCC | 83 (20) |
| DCC | 20 (5) |
| GBC | 43 (10) |
| AC | 5 (1) |
ICC intrahepatic cholangiocarcinoma, ECC extrahepatic cholangiocarcinoma, PCC perihilar cholangiocarcinoma, DCC distal cholangiocarcinoma, GBC gallbladder carcinoma, AC ampullary carcinoma
FISH analysis
Of the 423 patients, 25 were diagnosed as FGFR2 rearrangement positive (FGFR2-FISH positive) using the proper FISH pattern criteria (YGR or YG, except YR), and the ≥ 7% cutoff value (Supple Fig. 1). The number of FGFR2-FISH positives was 21/272 cases and 4/83 cases among patients with ICC and PCC, respectively (Fig. 1).
RNA sequencing
RNA sequencing targeting 1385 genes including FGFR2 was performed on the 25 patients determined to be FGFR2-FISH positive, and in-frame FGFR2 fusion transcripts were detected in 19 cases (Table 2). Fourteen kinds of FGFR2 fusion partner genes were identified, and FGFR2-BICC1 was frequently detected (4/19). However, no FGFR2 fusion transcript was detected in the remaining six FGFR2-FISH-positive cases. One of the cases that was FGFR2-FISH positive and fusion transcript negative showed an YGR (split) FISH pattern; meanwhile the other five showed a YG (loss of 3’ probe) FISH pattern, with relatively low FGFR2 gene expression ratio (1.50 – 2.59) observed, except for one case (case 6, 7.56). In other two cases showing the YG pattern in FISH assay, FGFR2 fusion transcripts were detected as FGFR2-CCDC6 and FGFR2-BICC1, respectively. Moreover, the FGFR2 gene expression was high (5.10 and 18.33) in these two cases. Discrimination of FGFR2 fusion transcript positive or negative was validated in six cases (cases 2, 3, 4, 5, 24 and 25) by another RNA sequencing method, AMP-FGFR, targeting FGFR1, FGFR2 and FGFR3 genes, and the results of the identified FGFR2 fusion transcripts or no FGFR2 fusion transcripts were coincident completely with the prior ones. In two fusion transcript-negative cases (case 24, 25), the FISH-positive cell rates (8%, 9%) were close to the ≥ 7% preset cutoff value, causing us to treat them as false positives after RNA sequencing. In the remaining 190 FGFR2-FISH-negative cases from the first period, RNA sequencing targeting 1385 genes was also performed on all cases, except 16 with low RNA quantity, to survey the false negative cases in the FISH analysis. Results show that all cases were confirmed to be negative for FGFR2 fusion transcripts. This implies no false negative cases in the FISH assay for FGFR2 rearrangement. Taken together, 23 patients were identified as FGFR2 rearrangement positive including 20 (7.4%) ICC cases and three (3.6%) PCC cases.
Table 2 .
Details of all FGFR2 rearrangement-positive cases
| Case | Sex | Age | Stage | Primary site | HBV/ HCVa |
History of smoking | Alcoholb | FGFR TKIc | OS (days) | FISH + cell rate (%) | FISH type | Targeted RNAseq | FGFR2 expression (fold)d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 57 | Recurrence | PCC | + | Y | N | na | na | 94 | YGR | FGFR2-POC1B | 1.7 |
| 2 | M | 73 | Metastatic | ICC | − | Y | N | na | na | 89 | YG | No fusion | 1.5 |
| 3 | F | 75 | Metastatic | ICC | − | N | N | na | na | 90 | YGR | FGFR2-PAWR | 8.0 |
| 4 | M | 46 | Locally advanced | ICC | − | Y | N | na | na | 87 | YGR | FGFR2-AHCYL1 | 4.0 |
| 5 | M | 53 | Metastatic | ICC | − | Y | Y | na | na | 78 | YGR | FGFR2-SHC1 | 10.9 |
| 6 | M | 60 | Metastatic | ICC | − | N | N | − | 1547 | 24 | YG | No fusion | 7.6 |
| 7 | M | 52 | Metastatic | ICC | + | Y | N | + | 1081 | 90 | YGR | FGFR2-BICC1 | 27.1 |
| 8 | M | 61 | Recurrence | PCC | − | N | N | + | 462 | 15 | YG | No fusion | 2.6 |
| 9 | M | 69 | Metastatic | ICC | + | Y | N | − | 420† | 91 | YGR | FGFR2-ZMYM4 | 2.3 |
| 10 | M | 65 | Recurrence | ICC | − | Y | N | + | 551 | 87 | YGR | FGFR2-SLMAP | 1.7 |
| 11 | M | 41 | Recurrence | ICC | − | N | N | + | 716 | 87 | YG | FGFR2-CCDC6 | 5.1 |
| 12 | M | 59 | Metastatic | ICC | + | Y | Y | + | 1166† | 69 | YGR | FGFR2-BICC1 | 5.9 |
| 13 | M | 66 | Metastatic | ICC | − | Y | N | + | 920† | 93 | YGR | FGFR2-BICC1 | 5.7 |
| 14 | F | 57 | Metastatic | ICC | + | Y | N | + | 662 | 94 | YGR | FGFR2-TRIM55 | 2.2 |
| 15 | F | 40 | Metastatic | ICC | − | Y | Y | + | 430 | 89 | YGR | FGFR2-POC1B | 3.2 |
| 16 | M | 59 | Metastatic | ICC | − | Y | N | + | 676† | 85 | YGR | FGFR2-SLMAP | 1.5 |
| 17 | M | 53 | Metastatic | ICC | − | N | Y | + | 1304 | 41 | YGR | FGFR2-SMARCC1 | 5.4 |
| 18 | M | 62 | Recurrence | ICC | + | Y | N | + | 453 | 89 | YGR | FGFR2-AFF3 | 4.5 |
| 19 | M | 43 | Recurrence | ICC | − | Y | Y | + | 804 | 91 | YGR | FGFR2-HOOK1 | 5.5 |
| 20 | M | 66 | Recurrence | ICC | − | Y | Y | + | 559 | 91 | YG | FGFR2-BICC1 | 18.3 |
| 21 | M | 62 | Metastatic | ICC | − | Y | Y | − | 231 | 93 | YGR | FGFR2-SHROOM3 | 15.4 |
| 22 | M | 65 | Recurrence | ICC | − | N | N | − | 300† | 85 | YGR | FGFR2-MYH9 | 6.8 |
| 23 | M | 66 | Locally advanced | PCC | − | Y | N | − | 364 | 94 | YG | No fusion | 2.0 |
| 24 | F | 64 | Metastatic | PCC | − | Y | N | na | na | 8 | YG | No fusion | 1.8 |
| 25 | F | 39 | Metastatic | ICC | + | N | N | na | na | 9 (88) | YGR (YR) | No fusion | 1.8 |
M male, F female, ICC intrahepatic cholangiocarcinoma, PCC perihilar cholangiocarcinoma, Y smoker or drinker, N non-smoker or non-drinker, OS overall survival, FGFR TKI FGFR tyrosine kinase inhibitor, RPKM reads per kilobase of exon per million mapped reads, na no available data
aHBV or HCV hepatitis virus infection positive ( + ) or negative ( − ) was analyzed with HBs antigen and HCV antibody
bDrinker is defined as drinking equal or more than 60 g of ethanol a day
cTreatment with FGFR-TKI was performed ( + ) or not ( − )
dFGFR2 gene expression is indicated as the ratio (fold) of RPKM between each sample and the median value of FISH-negative cases
† dead
Clinical features and prognosis
The clinical characteristics were evaluated in the 23 cases with FGFR2 rearrangements. The macroscopic type of FGFR2 rearrangement-positive ICC was invariably the mass forming type (Table 3). Univariate analysis unveiled associations between the presence of FGFR2 rearrangement and two factors, i.e., younger age (≤ 65 years; p = 0.0085), and HCV Ab- and/or HBs Ag-positivity (p = 0.02). Although a history of heavy drinking (ethanol ≥ 60 g/day) also tended to be associated with positivity for FGFR2 rearrangement, no statistically significant difference was achieved (p = 0.06). Furthermore, multivariate analysis of these three factors identified younger age (≤ 65 years) and HCV Ab- and/or HBs Ag-positivity as associated with FGFR2 rearrangement (Table 4). The median age of FGFR2 rearrangement positive/negative patients was 60 years old (range 40–75)/67 years old (range 25–91), respectively. Thus, 10.3% (17/165) of younger patients and 16.6% (6/36) of hepatitis virus-positive patients were FGFR2 rearrangement positive. Of the patients who were positive for both factors, 26% showed FGFR2 rearrangements.
Table 3 .
Univariate analysis of FGFR2-positive patients determined by FISH and RNAseq results (N = 355). The analysis was limited to ICC and PCC
| Positive (N = 23) |
Negative (N = 332) |
P value | |
|---|---|---|---|
| Sex | |||
| Male | 19 | 225 | 0.167 |
| Female | 4 | 107 | |
| Age | |||
| > 65 | 6 | 184 | 0.009 |
| ≤ 65 | 17 | 148 | |
| Tested tumor specimen | |||
| Resection | 7 | 118 | 0.822 |
| Biopsy | 16 | 214 | |
| Percutaneous biopsy | 13 | 164 | |
| EUS-FNA | 2 | 24 | |
| Endoscopic biopsy | 1 | 26 | |
| Stage | |||
| Metastasis | 13 | 168 | 0.124 |
| Recurrence | 8 | 77 | |
| Locally advanced | 2 | 87 | |
| Differentiation | |||
| Poorly | 1 | 61 | 0.178 |
| Moderate/well | 9 | 129 | |
| Unknown | 13 | 142 | |
| Primary site | |||
| ICC | 20 | 252 | 0.311 |
| PCC | 3 | 80 | |
| Macroscopic typea | |||
| Mass forming | 20 | 219 | 0.221 |
| Periductal infiltrating | 0 | 27 | |
| Intraductal growth | 0 | 1 | |
| Unknown | 0 | 5 | |
| Smoking history | |||
| Positive | 17 | 204 | 0.372 |
| Negative | 6 | 122 | |
| Unknown | 0 | 6 | |
|
Alcohol abuse (ethanol/day) | |||
| ≥ 60 g | 7 | 38 | 0.060 |
| < 60 g | 16 | 289 | |
| Unknown | 0 | 5 | |
| HCV Ab and/or HBs Ag | |||
| Either positive | 6 | 30 | 0.020 |
| Others | 17 | 302 | |
| Pancreaticobiliary maljunction | |||
| + | 0 | 1 | 1.0 |
| − | 23 | 331 | |
| PSC | |||
| Positive | 0 | 1 | 1.0 |
| Negative | 23 | 331 | |
| Printing industry | |||
| Positive | 0 | 3 | 1.0 |
| Negative | 23 | 317 | |
| Unknown | 0 | 12 | |
| Cholelithiasis | |||
| + | 1 | 21 | 1.0 |
| − | 22 | 311 | |
| Family history about malignant tumor | |||
| + | 11 | 172 | 0.830 |
| − | 12 | 160 | |
EUS-FNA endoscopic ultrasound-fine needle aspiration, PSC primary sclerosing cholangitis
aFor macroscopic type, only ICC was analyzed
Table 4 .
Multivariate analysis of FGFR2-positive patients (logistic regression analysis)
| Multivariate analyses | Odds ratio | 95% CI | P value |
|---|---|---|---|
| Age | |||
| ≤ 65 / > 65 | 3.20 | (1.22 − 8.41) | 0.0181 |
| HCV Ab and/or HBs Ag | |||
| + / − | 2.98 | (1.07 − 8.28) | 0.0365 |
| Alcohol abuse (ethanol/day) | |||
| ≥ 60 g/ < 60 g | 1.32 | (0.42− 1.81) | 0.0702 |
In the second period, we investigated the details and clinical responses of first-line chemotherapy regimens for cases positive for FGFR2 rearrangements. The gemcitabine and cisplatin (GC) therapy was performed in 17/18 patients, and gemcitabine + S-1 therapy was performed in the remaining patient. The response rate and disease control rate of the first-line treatment were 22% (4/18) and 66.7% (12/18), respectively. The overall survival (OS), from the initiation of first-line chemotherapy, of FGFR2 rearrangement-positive patients is shown in Fig. 2. The median OS was 38.8 months for the 18 FGFR2 rearrangement-positive patients, 13 of whom received molecular targeting therapy with FGFR inhibitors.
Fig. 2 .
Overall survival (OS) curve for FGFR2 rearrangement-positive cases in the second period, and progression-free survival (PFS) curve for first-line chemotherapy
Discussion
FGFR2 gene alterations were reported to be associated with early stages [17]; however, no study has selectively examined advanced cases. In this study, the frequency of FGFR2 rearrangement-positive cases was 7.4% among patients with advanced/recurrent ICC, which was lower than those of previous reports (13–14%) that analyzed surgically resected cases [12, 14]. In contrast, the current study and previous reports [13] that targeted advanced/recurrent patients indicated lower frequency (7.4–9%). Churi et al. suggested that the activation of the FGF/FGFR pathway associates with better prognosis in patients with ICC [18]. Hayashi et al. reported that FGFR2 fusion gene-positive ICC develops from peripheral bile ducts and is found in type 2, which was characterized by low mucin production, mass forming macroscopic type and better prognosis [19]. The different frequency of FGFR2 rearrangements between patients undergoing resection, and those with advanced cancer, could be due to the preferential occurrence of FGFR2 rearrangements in patients with peripheral and mass forming type, which progress slowly and are frequently treated by surgical resection. This study also revealed that 3.6% of patients with PCC had FGFR2 rearrangements. In clinical practice, ICC and PCC are often difficult to discriminate.
Additionally, in the current study, three PCC patients were found to be FGFR2 rearrangement positive, two of which had intrahepatic extension. However, these two cases were resected and the diagnosis of the primary region were made via detailed pathological assessment of resected specimen. The remaining one case was diagnosed by ERCP, enhanced CT and biopsy, and was determined to have a perihilar lesion without mass formation of intrahepatic extension. Hence, the diagnosis of the primary region as “perihilar bile duct” for these three cases is reliable. Our results indicate that PCC could also be screened for FGFR2 rearrangements as a potential target category of FGFR inhibitors.
According to the post hoc targeted RNA sequencing using FFPE specimens, the sensitivity and specificity of the FISH assay to detect FGFR2 rearrangements was 100% and 99.0%, respectively, indicating that FISH is a reliable diagnostic assay method. Meanwhile, a recent report indicated that multiplex NGS panel testing is possible even with FNA samples, although it did not survey any FGFR2 fusions [20]. NGS is a useful and convenient method. However, there are also associated concerns regarding sample preparation, including the amount of DNA required (sufficient for tumor cell count and percent tumor content), sample storage time, and probe design according to the partner gene. Additionally, NGS requires longer turnaround time than the FISH technique. Therefore, the complementary use of several techniques including NGS and FISH is desirable in clinical settings.
Besides FGFR2 in-frame fusions, recurrent C-terminal truncation events translocating FGFR2, without its 3’-UTR, to intergenic regions were also reported in ICC [21]. ICC patients with 3’-UTR truncated FGFR2 transcripts exhibited higher RNA expression compared to wild-type FGFR2 transcripts. Meanwhile, C-terminal truncation of FGFR2 showed transforming ability in gastric cancer [22]. As target RNA sequencing in this study analyzed only coding exon sequences, no structural information was available regarding the 3’-UTR of FGFR2. Nevertheless, our data revealed that 4/6 FISH-positive cases exhibiting loss of the 3’-probe (YG signal) had no FGFR2 fusion transcript, which may reflect the 3’-UTR loss of FGFR2. Further analysis will clarify whether clinical responses to FGFR-targeted therapy differ depending on rearrangement pattern on FISH.
Over 40 genes have been detected as FGFR2 fusion partners in ICC [17, 23]. Although break-apart FISH assays cannot identify the fusion partner gene, it can clarify the presence/absence of FGFR2 rearrangements (fusions/truncations) including those involved with unknown partners. All FGFR2 fusion-positive cases showed YGR type in this FISH analysis, and samples showing YG type included both fusion and truncation. Therefore, FISH alone analysis can distinguish fusion from fusion or truncation if the FISH type is YGR. YR FISH type was rare, and treated as FGFR2 rearrangement ‘negative’ in this study. Besides case 25, two case showed YR FISH type (11% and 23%) with low FGFR2 rearrangement-positive signal (YGR or YG: 5% and 0%, respectively). YR type denotes 5ʹ probe deletion of FGFR2 and might lose oncogenic driver activity. Then we think YR FISH type is incidental and happens rare.
Break-apart ALK FISH assay was approved by the FDA as a companion diagnostic for detecting ALK rearrangements in lung cancer patients who may benefit from treatment of ALK tyrosine kinase inhibitor therapy using ≥ 15% as a cutoff value [24]. In meta-analysis of ALK rearrangement-positive non-small lung cancer, higher percentage of ALK rearrangement-positive cells tend to respond better to the crizotinib therapy [25]. At present, our analysis cannot judge whether the percentage of FISH-positive cell rate influence the outcome because of the limited cases. In this study, we preset the cutoff value as ≥ 7% for a positive FISH result. This threshold was obtained from the assay background (6.4%, mean + 2SD) in the analysis of a limited number of genotyped FGFR2 fusion-positive and -negative controls from surgically resected specimens. However, this ≥ 7% cutoff might be too low for the small biopsy samples. In two fusion transcript-negative cases (case 24, 25), the FISH-positive cell rate (8%, 9%) was close to the ≥ 7% preset cutoff value, and were thus treated as false positives after RNA sequencing. Hence, further optimization of diagnostic thresholds should be necessary, such as setting a higher cutoff value, as FISH-positive cell rate in all cases was higher than 15%, except for the two fusion transcript-negative cases.
FGFR2 rearrangement was more frequently detected in younger and hepatitis virus-positive patients. A previous study analyzing somatic mutations related to ICC with cirrhotic liver reported higher frequencies of either IDH mutations or FGFR2 alterations [26]. Both that report and our study suggest a possible association with a background of continuous damage to hepatocytes, such as those related to virus infection [27, 28] and the presence of FGFR2 rearrangements.
In FGFR2 rearrangement-positive 18 patients, the median OS was 38.8 months, and they have good prognosis as survival in advanced/recurrent ICC/PCC. It may be due to the clinical feature of FGFR2 fusion/rearrangement ICC/PCC patients [18] and therapeutic intervention of FGFR inhibitors. Among them, 17 patients received combination therapy of GC as first-line chemotherapy, and 1 patient received combination therapy of gemcitabine and S-1. The median progression-free survival (PFS) of these 18 patients in first-line chemotherapy was 8.9 months (Fig. 2). Valle et al. reported that the median PFS of GC therapy was 8.0 months in the ABC-02 trial [3]. The result of PFS in this study was similar to the previous reports. Therefore, we consider that a crucial factor for good prognosis in these 18 patients was treatment with FGFR inhibitors, as was reported in 13 of the 18 patients (72%).
Certain limitations were noted in this study. First, since the number of positive patients was small, the prognostic significance of FGFR2 rearrangement positivity must be confirmed in larger studies. Second, since clinical trials of FGFR inhibitors, including our cases, are ongoing, the clinical utility of our FISH assay as a companion diagnostic test to predict the therapeutic efficacy of FGFR inhibitors requires further evaluation in the future.
In conclusion, this study demonstrated the feasibility of FGFR2-FISH assay using biopsy specimens of BTC, and showed that 7.4% of cases in advanced/recurrent ICC retained FGFR2 rearrangements, and 3.6% cases in advanced/recurrent PCC also carried the alterations. Hence, this accounts for the first report of positive PCC cases. Moreover, younger age and a history of hepatitis viral infection are associated with the presence of FGFR2 rearrangements. These findings have important implications for elucidating the pathophysiology of FGFR2 rearrangements, and will be useful in developing targeted therapy for ICC and PCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 (TIFF 4381 kb)Supplementary Figure S1. Representative FISH images from FGFR2 rearrangement-positive/negative cases. Break-apart FISH assay revealed the rearrangements with the 5ʹ-probe of FGFR2 shown in green and the 3ʹ-probe shown in red. The rearrangement positive cells were defined as having any split signal (YGR type) as in (a) or any isolated green signal (YG type) as in (b), while any isolated red signal (YR type) as in (c) was treated as rearrangement-negative. (d) Schematic diagram of FGFR2 FISH probes and FISH patterns. Thin arrow, green signal; bold arrow, red signal; arrowhead, yellow signal
Acknowledgements
We thank all the patients who participated in this study, their families, the investigators and the study site personnel. We are also grateful to the members of the Department of Hepatobiliary and Pancreatic Oncology (Ms. Hiroko Hosoi), Division of Cancer Genomics (Mr. Hiroshi Chikuta, Ms. Wakako Mukai and Dr. Arisa Hara), Clinical Research Support Office (Ms. Keiko Wakakuwa and Ms. Suga Yamagami), and Data Management Section (Masahisa Kamikura) of the National Cancer Center for their support in collecting data, preparing the original report, and overseeing the study management (Dr. Ken Kato, Clinical Research Support Office). We also thank Dr. Atsushi Ohtsu and Dr. Koichi Goto who provided advice on study design.
Funding
This research was supported in part by the Ministry of Health, Labour and Welfare (MHLW) Health Labour Sciences Research Grants: “Research on the establishment of treatment for biliary tract cancer” and “Clinical Trial on Development of New Drugs and Medical Devices”, MEXT KAKENHI (18K07956) and The National Cancer Center Research and Development Fund 23-A-15.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The original online version of this article was revised due to the update in supplementary figure 2.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/30/2020
A Correction to this paper has been published: 10.1007/s00535-020-01755-y
References
- 1.Cancer Statistics in Japan 2018, The Editorial Board of the Cancer Statistics in Japan
- 2.Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 4.Valle JW, Lamarca A, Goyal L, et al. New horizons for precision medicine in biliary tract cancers. Cancer Discov. 2017;7:943–962. doi: 10.1158/2159-8290.CD-17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012;43:1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Abou-Alfa GK, Macarulla Mercade T, Javle M, et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:796–807. doi: 10.1016/S1470-2045(20)30157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borad MJ, Gores GJ, Roberts LR. Fibroblast growth factor receptor 2 fusions as a target for treating cholangiocarcinoma. Curr Opin Gastroenterol. 2015;31:264–268. doi: 10.1097/MOG.0000000000000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Javle M, Lowery M, Shroff RT, et al. Phase II study of BGJ398 in patients with FGFR-altered advanced Cholangiocarcinoma. J Clin Oncol. 2018;36:276–282. doi: 10.1200/JCO.2017.75.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollebecque A, Borad M, Sahai V, et al. Interim results of fight-202, a phase II open-label multicenter study of INCB054828 in patients (pts) with previously treated advanced/metastatic or surgically unresectable cholangiocarcinoma (CCA) with/without fibroblast growth factor (FGF)/FGF receptor (FGFR) genetic alterations. Ann Oncol. 2018;29(8):258. doi: 10.1093/annonc/mdy282.139. [DOI] [Google Scholar]
- 10.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 11.Mahipal A, Tella SH, Kommalapati A, et al. FGFR2 genomic aberrations: Achilles heel in the management of advanced cholangiocarcinoma. Cancer Treat Rev. 2019;78:1–7. doi: 10.1016/j.ctrv.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–1434. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 13.Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham RP, Barr Fritcher EG, Pestova E, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45:1630–1638. doi: 10.1016/j.humpath.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Weiss GJ, Hoff BR, Whitehead RP, et al. Evaluation and comparison of two commercially available targeted next-generation sequencing platforms to assist oncology decision making. Onco Targets Ther. 2015;8:959–967. doi: 10.2147/OTT.S81995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 17.Jain A, Borad MJ, Kelley RK, et al. Cholangiocarcinoma with FGFR genetic aberrations: a unique clinical phenotype. JCO Precis Oncol. 2018;2:1–12. doi: 10.1200/PO.17.00080. [DOI] [PubMed] [Google Scholar]
- 18.Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS ONE. 2014;9:e115383. doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi A, Misumi K, Shibahara J, et al. Distinct clinicopathologic and genetic features of 2 histologic subtypes of intrahepatic cholangiocarcinoma. Am J Surg Pathol. 2016;40:1021–1030. doi: 10.1097/PAS.0000000000000670. [DOI] [PubMed] [Google Scholar]
- 20.Hirata K, Kuwatani M, Suda G, et al. A novel approach for the genetic analysis of biliary tract cancer specimens obtained through endoscopic ultrasound-guided fine needle aspiration using targeted amplicon sequencing. Clin Transl Gastroenterol. 2019;10:e00022. doi: 10.14309/ctg.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and epigenomic landscapes of etiologically distinct subtypes of Cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cha JY, Maddileti S, Mitin N, et al. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J Biol Chem. 2009;284:6227–6240. doi: 10.1074/jbc.M803998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res. 2018;24:4154–4161. doi: 10.1158/1078-0432.CCR-18-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Wang L, Tang G, et al. Fluorescence in situ hybridization (FISH) for detecting anaplastic lymphoma kinase (ALK) rearrangement in lung cancer: clinically relevant technical aspects. Int J Mol Sci. 2019;20:3939. doi: 10.3390/ijms20163939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soria JC, Ho SN, Varella-Garcia M, et al. Correlation of extent of ALK FISH positivity and crizotinib efficacy in three prospective studies of ALK-positive patients with non-small-cell lung cancer. Ann Oncol. 2018;29:1964–1971. doi: 10.1093/annonc/mdy242. [DOI] [PubMed] [Google Scholar]
- 26.Joseph NM, Tsokos CG, Umetsu SE, et al. Genomic profiling of combined hepatocellular-cholangiocarcinoma reveals similar genetics to hepatocellular carcinoma. J Pathol. 2019;248:164–178. doi: 10.1002/path.5243. [DOI] [PubMed] [Google Scholar]
- 27.Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? a meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012;57:69–76. doi: 10.1016/j.jhep.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Li J, Li P, et al. Hepatitis B virus infection increases the risk of cholangiocarcinoma: a meta-analysis and systematic review. J Gastroenterol Hepatol. 2012;27:1561–1568. doi: 10.1111/j.1440-1746.2012.07207.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (TIFF 4381 kb)Supplementary Figure S1. Representative FISH images from FGFR2 rearrangement-positive/negative cases. Break-apart FISH assay revealed the rearrangements with the 5ʹ-probe of FGFR2 shown in green and the 3ʹ-probe shown in red. The rearrangement positive cells were defined as having any split signal (YGR type) as in (a) or any isolated green signal (YG type) as in (b), while any isolated red signal (YR type) as in (c) was treated as rearrangement-negative. (d) Schematic diagram of FGFR2 FISH probes and FISH patterns. Thin arrow, green signal; bold arrow, red signal; arrowhead, yellow signal