Abstract
Aptamers can serve as efficient bioreceptors for the development of biosensing detection platforms. Aptamers are short DNA or RNA oligonucleotides that fold into specific structures, which enable them to selectively bind to target analytes. The method used to identify aptamers is Systematic Evolution of Ligands through Exponential Enrichment (SELEX). Target properties can have an impact on aptamer efficiencies. Therefore, characteristics of water-borne microbial targets must be carefully considered during SELEX for optimal aptamer development. Several aptamers have been described for key water-borne pathogens. Here, we provide an exhaustive overview of these aptamers and discuss important microbial aspects to consider when developing such aptamers.
Keywords: aptamer, SELEX, water-borne pathogens, viable but non-culturable, coliforms, aptasensors
Introduction
Access to water that is safe for use and consumption is a basic human right. As a result, most countries have strict guidelines, regulations and standards for managing water sources and water distribution systems to supply high quality water free from chemical and microbial contaminants. In most cases, microbial contaminants must be removed from the water before distribution. These microbes include pathogens that cause gastroenteritis, such as Cryptosporidium, Giardia, Norovirus, Rotavirus, Campylobacter, and E. coli (WHO, 2017). Other water-borne diseases are caused by pathogens growing inside water distribution systems or within engineered water systems, such as cooling tower, fountains, spas and humidifiers (Wang H. et al., 2017). The latter include Legionella pneumophila, Pseudomonas spp. and non-tuberculosis mycobacteria. In recent years, several studies have shown that a high proportion of water associated deaths and illnesses are due to the aforementioned three environmental water-borne pathogens (Gargano et al., 2017; Greco et al., 2020). In fact, L. pneumophila, the causative agent of Legionnaires disease, has become the number one cause of water-borne outbreaks in recent years (McClung et al., 2017). The presence of coliforms is not indicative of the presence of several key water-based pathogens that are of significance to public health (Payment and Locas, 2011). Consequently, specific detection methods are needed to ensure safe water from the source to the point of use.
Monitoring and surveillance of specific water-borne microbes require robust detection methods. Challenges in select current detection methods for waterborne pathogens have been reviewed excellently in detail elsewhere (Ramírez-Castillo et al., 2015; Wang H. et al., 2017). In general, traditional microbial detection methods rely heavily on culture methods, which is fraught with several limitations. Culture methods are extremely time consuming and often require extensive material, specialized labor, and time. Culture recovery rates are also adversely affected by many factors such as the presence of competing microbes, the presence of viable but non-culturable (VBNC) cells, methods used for concentration of the sample or enrichment of the target microbe and sample type (bulk water or biofilm) (Wang H. et al., 2017). Drawbacks with culture techniques has led to a shift toward the use of molecular methods, including PCR, quantitative PCR (qPCR), high throughput sequencing, and immunoassays such as ELISA, immunochromatography and immuno-lateral flow assays. The most widely used molecular method is qPCR (Ramírez-Castillo et al., 2015; Wang H. et al., 2017). The advantage of qPCR, over conventional culture techniques, is more rapid turn-around times, high sensitivities and specificities, lower limits of detection, as well as an ability to detect VBNC cells. However, by detecting live, VBNC and dead cells qPCR leads to an overestimation of microbial burden. Additionally, qPCR involves multiple sample processing steps which requires specialized labor. qPCR is also inhibited by several compounds routinely found in water samples resulting in possible false negatives (Gentry-Shields et al., 2013).
Biosensors can mitigate some of the problems associated with traditional detection methods (Ahmed et al., 2014). They are analytical devices used to quantify or detect a specific analyte (Turner, 2013). Qualities of biosensors includes high specificity, high sensitivity, multiplexing capability, cost-effectiveness, portability and ease of use (Ahmed et al., 2014; Kumar et al., 2018; Cesewski and Johnson, 2020; McConnell et al., 2020). A biosensor set-up typically consists of three elements. A biorecognition element, which upon interaction with a target, produces a physico-chemical signal that is converted by a transducing element into a signal captured by a detection element (Turner, 2013). Biosensors are categorized based on either their transducing element (mechanical, optical, electrochemical) or the nature of the biorecognition element (affinity, catalytic) (Ahmed et al., 2014).
A versatile and stable biorecognition element is a critical component of any biosensing platform (Ahmed et al., 2014; Kumar et al., 2018). Antibodies are the most used bioreceptors in biosensor development and research, but aptamers are an increasingly widespread popular alternative (Song et al., 2008; Morales and Halpern, 2018). Aptamers are single stranded DNA or RNA oligonucleotides that fold into specific complex structures and interact with their targets via shape complementarity, hydrogen bonding, electrostatic interactions and stacking interactions (McKeague et al., 2015). Besides having high affinities and selectivity, they can bind to a wide range of targets from small non-immunogenic compounds to whole cells (McKeague et al., 2015). Aptamers can be generated in vitro in conditions one can preferentially select making them stable and versatile for a variety of applications (Song et al., 2008). They are cost-effective to synthesize with minimal batch to batch variation (Strehlitz et al., 2012; McConnell et al., 2020). Their easily modifiable nature facilitates functionalization on sensing surfaces (Song et al., 2008; McConnell et al., 2020). Their inherent small size also promotes high packing densities during functionalization (Song et al., 2008; Crivianu-Gaita and Thompson, 2016). In this minireview, we will briefly provide examples of aptamers with potential for detection of water-borne pathogens and discuss microbial determinants for the development of optimal aptamers and thus improved aptamer-coupled biosensors. Examples of aptamers is provided in Table 1 and a complete list of aptasensing platforms is provided in Supplementary Table 1.
TABLE 1.
Aptamers developed against water-borne bacteria.
| Aptamer name | Target | Culture conditiona | OD/Growth stagea,b | Counter-Selex Strainsc | Type of sensors | LOD | References |
| Norovirus | |||||||
| AG3 | MuNoV | NA | NA | Feline calicivirus (FCV) | Electrochemical | 180 virus particles | Giamberardino et al., 2013 |
| NA | Optical (colorimetric) | 200 virus/ml | Weerathunge et al., 2019 | ||||
| Aptamer 25/SMV-25 | SMV | NA | NA | HuNoV-negative human stool suspension, bead-antibody complex | NA | NA | Escudero-Abarca et al., 2014 |
| Non-toxic norovirus GII capsid recombinant | NA | Optical (Chemiluminescence) | 80 ng/ml | Kim B. et al., 2018 | |||
| Aptamer 21/SMV-21 | SMV | NA | NA | HuNoV-negative human stool suspension, bead-antibody complex | NA | NA | Escudero-Abarca et al., 2014 |
| Norovirus Group II (recombinant VLP) | NA | Electrochemical | 100 pM | Chand and Neethirajan, 2017 | |||
| C. parvum | |||||||
| R4-6 | Oocysts | NA | NA | Giardia duodenalis cysts | Electrochemical | 100 oocysts | Iqbal et al., 2015 |
| NA | Electrochemical | 50 oocysts | Iqbal et al., 2019 | ||||
| Min_Crypto2 | Oocysts | NA | NA | NA | Optical fluorescence | 5 oocysts | Hassan et al., 2021 |
| Acinetobacter | |||||||
| Aci49 | Whole-cell-A. baumannii (ATCC 19606) | BHI broth, 37°C, overnight | 0.4/E | Acinetobacter lwoffii, Acinetobacter calcoaceticus, and 11 species | Optical (colorimetric) | 103 CFU/ml | Rasoulinejad and Gargari, 2016 |
| NA | Optical (fluorescence) | 3 CFU/ml | Li et al., 2020 | ||||
| NA | Optical (fluorescent) | 10 CFU/ml | Yang et al., 2020 | ||||
| AB aptamer | Whole-cell A. baumannii | NR | NR | NR | Optical (colorimetric) | 450 CFU/rxn | Wu et al., 2018 |
| NA | Optical (fluorescence) | 105 CFU/ml | Su et al., 2020 | ||||
| NA | Optical (fluorescence) | 100 CFU/ml | Su et al., 2020 | ||||
| NA | Optical (fluorescence) | 300 CFU/ml | Bahari et al., 2021 | ||||
| Aeromonas | |||||||
| Apt1 | Whole-cell (A. hydrophila) | LB,37°C, 18 h | NR | NR | Optical (Fluorescence) | 1.5 CFU/ml | Zhu et al., 2019 |
| Campylobacter | |||||||
| Aptamer C2 and Aptamer C3 | Surface protein (C. jejuni) | NR | NR | NR | Optical (fluorescence) | 2.5 CFU/ml | Bruno et al., 2009 |
| NA | Optical (colorimetric) | 5–10 CFU/ml | Bruno and Sivils, 2017 | ||||
| ONS-23 | Whole-cell (C. jejuni A9a) | BBL brucella broth, 42°C, 48 h, microaerophillic conditions | PE* | 20 strains (enteric, non-enteric, lactic acid) | NA | NA | Dwivedi et al., 2010 |
| NA | Optical (colorimetric) | 10 CFU/ml | Dehghani et al., 2018 | ||||
| NA | Optical (colorimetric) | 7.2 × 105 CFU/ml | Kim Y. J. et al., 2018 | ||||
| CJA1 | Whole-cell (C. jejuni) | NR | Optical (colorimetric) | 10 CFU/ml | Chen et al., 2020 | ||
| Cyanobacteria | |||||||
| ATX8 | Anatoxin-a (ATX) | NA | NA | ATX free beads | Electrochemical | 0.5 nM | Elshafey et al., 2015 |
| MC-LR aptamer/AN6 | Microcystin-LR | NA | NA | Blank sepharose beads | Electrochemical | 10 pM | Ng et al., 2012 |
| NA | Optical (fluorescence) | 0.002 ng/ml | Lv et al., 2017 | ||||
| E. coli | |||||||
| L9F | O111-LPS (E. coli O111:K58) | 35°C, TSB, overnight | NR | NR | NA | NA | Bruno et al., 2008 |
| NA | Electrochemical | 112 CFU/ml | Luo et al., 2012 | ||||
| Eco4R/ECAII | Outer membrane protein (OMP)—E. coli 8739 | 37°C, blood agar, overnight | NR | NR | NA | NA | Bruno et al., 2010 |
| NA | Electrochemical | NR | Queirós et al., 2013 | ||||
| Eco4F | OMP-E. coli 8739 | 37°C, blood agar, overnight | NR | NR | NA | NA | Bruno et al., 2010 |
| NA | Optical (colorimetric/fluorescence) | 300 CFU/ml | Bruno, 2014 | ||||
| Eco3R/ECAI | OMP-E. coli 8739 | 37°C, blood agar, overnight | NR | NR | NA | NA | Bruno et al., 2010 |
| NA | Electrochemical | NR | Queirós et al., 2013 | ||||
| NA | Optical (colorimetric/fluorescence) | 300 CFU/ml | Bruno, 2014 | ||||
| NA | Optical (Evanescent wave fiber optics) | 0.1nM | Queirós et al., 2014 | ||||
| E1 | Whole cell (E. coli fecal isolate) | NB, 37°C | 0.45/E | E. coli (non-fecal isolate), other fecal isolates | NA | NA | Kim et al., 2013 |
| E2 | Whole cell (E. coli fecal isolate) | NB, 37°C | 0.45/E | E. coli (non-fecal isolate), other fecal isolates | NA | NA | Kim et al., 2013 |
| NA | Optical (fluorescence) | 3 CFU/ml | Jin et al., 2017 | ||||
| NA | Electrochemical | 100 CFU/ml | Wu et al., 2017 | ||||
| E10 | Whole cell (E. coli fecal isolate) | NB, 37°C | 0.45/E | E. coli (non-fecal isolate), other fecal isolates | NA | NA | Kim et al., 2013 |
| E1 + E2 + E10 (pooled) | NA | Electrochemical | 371 CFU/ml | Kim et al., 2014 | |||
| AptB12 | Whole cell (E. coli ETEC K88) | LB | E | ETEC K99, S. enteritidis, S. aureus, | Optical (fluorescence) | 1.1 × 103 CFU/ml | Peng et al., 2014 |
| RNAaptamer | NR | LB, 37°C, 2–3 h | NR | NA | Electrochemical | NR | So et al., 2008 |
| NA | Immunomagnetic separation and RT-PCR | 10 CFU/ml | Lee et al., 2009 | ||||
| NA | Electrochemical | 6–26 CFU/ml | Zelada-Guilleìn et al., 2010 | ||||
| Aptamer I-1 | O-antigen LPS (E. coli O157:H7) | Brucella broth,37°C,24 h (+0.04% formaldehyde) | NR | E. coli K12 | NA | NA | Lee et al., 2012 |
| NA | Electrochemical | 4 CFU/ml | Burrs et al., 2016 | ||||
| Ec3 (31) | Whole cell (E. coli DH5α) | LB | 0.4 | B. subtilis | Electrochemical | 2 × 104 CFU/ml | Dua et al., 2016 |
| P12-31 | Whole cell (E. coli O6) | 37°C, LB | 0.3 | NR | NA | NA | Marton et al., 2016 |
| AM-6 | Whole cell (E. coli O157:H7) | LB | 0.6 | E. coli strains O42, K12, Top10, DH5α, S. flexneri, S. Typhi | NA | NA | Amraee et al., 2017 |
| S1 | Whole cell (E. coli O157:H7) | BHI, 37°C | E | S. aureus, S. Typhyimurium, L. monocytogens | Mechanical (Quartz Crystal Microbalance-QCM) | 1.46 × 103 CFU/ml | Yu et al., 2018 |
| Apt-5 | whole cell (E. coli O157:H7) | LB, 37°C | NR | E. coli ETEC and 3 other species | NA | NA | Zou et al., 2018 |
| a-aptamer/E-17F72* | O157:H7 LPS | LB, 37°C | NR | NR | NA | NA | Bruno et al., 2009 |
| c-aptamer/E-18R72* | O157:H7 LPS | LB, 37°C | NR | NR | NA | NA | Bruno et al., 2009 |
| a-aptamer, c-aptamer | NA | Optical (colorimetric) | 10 CFU/ml | Wu et al., 2015 | |||
| a-aptamer, c-aptamer | NA | Optical (colorimetric) | 25 CFU/ml | Díaz-Amaya et al., 2019b | |||
| a-aptamer, c-aptamer | NA | Optical (surface enhanced raman spectroscopy-SERS) | 100 CFU/ml | Díaz-Amaya et al., 2019a | |||
| c-aptamer | NA | Optical (fluorescence) | 100 CFU/ml | Hao et al., 2019 | |||
| NA | Optical (fluorescence) | 80 CFU/ml | Jiang et al., 2020 | ||||
| Helicobacter pylori | |||||||
| Hp-Ag aptamer | Recombinant Hp surface antigen | NR | NR | BSA | NA | NA | Gu et al., 2018 |
| Hp4 | Recombinant Hp surface antigen | Blood agar, 37°C, 3 days | NR | BSA | NA | NA | Yan et al., 2019 |
| Legionella | |||||||
| R10C5, R10C1 | Whole cell (Lp 120292) | CYE agar plate, 37°C, 3 days followed by AYE media,37°C, 24 h | 2.5/PE | Pseudomonas putida KT2440, Pseudomonas fluorescens LMG1794 | NA | NA | Saad et al., 2020 |
| NTM | |||||||
| BM2/N31 | ManLAM, M. bovis (BCG) | L-J medium | E | NR | Optical (ELONA) | 104 CFU/ml | Sun et al., 2016 |
| Electrochemical | NR | Sodia et al., 2020 | |||||
| Pseudomonas aeruginosa | |||||||
| F23 | Whole cell (P. aeruginosa clinical isolate) | Mueller-Hinton (MH) media, 37°C, 24 h | NR | S. maltophilia, A. baumannii | Optical (fluorescence) | NR | Wang et al., 2011 |
| NA | Optical (fluorescence) | 100 CFU/ml | Gao et al., 2018 | ||||
| NA | Optical (Long range Surface Plasomon Resonance-LSPR) | 1 CFU/ml | Hu et al., 2018 | ||||
| NA | Optical (Fluorescence) | 1 CFU/ml | Zhong et al., 2018 | ||||
| NA | Electrochemical and Optical (colorimetric) | 60 CFU/ml | Das et al., 2019 | ||||
| NA | Electrochemical | 33 CFU/ml | Roushani et al., 2019 | ||||
| NA | Mechanical (piezoelectric quartz crystal) | 9 CFU/ml | Shi et al., 2019 | ||||
| St17Lp21, St21Lp17, St08Lp17 | Biofilm-derived whole cells (PA 692/ATCC 14502) | LB broth, 37°C, 16 h followed by 22°C, 42 h to make biofilm. | E | NR | NA | NA | Soundy and Day, 2017 |
| F23 + St08Lp17 (pool) | NA | Optical (Fluorescence) | 1 CFU/ml | Zhong et al., 2020 | |||
| Salmonella | |||||||
| Aptamer 33 | OMP (S. tyhpimurium PT10) | BHI, 37°C, 2–3 h | E. coli OMP and LPS, Salmonella LPS | Magnetic bead based pull down assay and qPCR | 10–100 CFU/ml | Joshi et al., 2009 | |
| NA | Optical (Fluorescence) | 5 CFU/ml | Duan et al., 2012 | ||||
| NA | Electrochemical | 3 CFU/ml | Ma et al., 2014 | ||||
| NA | Electrochemical | 55 CFU/ml | Hasan et al., 2018 | ||||
| NA | Optical (LSPR) | 30 CFU/ml | Yoo et al., 2015 | ||||
| NA | Optical (LSPR) | 104 CFU/ml | Oh et al., 2017 | ||||
| ST2P | Whole cell (S. typhimurium ATCC 50761) | BBL-BHI, 37°C, overnight | 0.3/E | L. monocytogenes, E. coli, S. aureus, S. pneumoniae, V. parahemolyticus, C. sakazakii | Optical (fluorescence) | 25 CFU/ml | Duan et al., 2013b |
| NA | Optical (Colorimetric, SERS) | 10 CFU/ml | Duan et al., 2016 | ||||
| NA | Optical (Fluorescence) | 25 CFU/ml | Duan et al., 2014 | ||||
| S8-7 | Whole cell (S. typhimurium S913) | TSB-amp, 37C, overnight | NR | L. monocytogenes Scott A E. coli O157: H7, B. cereus, E. faecalis | NA | NA | Dwivedi et al., 2013 |
| C4 | Whole cell (S. typhimurium) | BHI, 35°C, overnight | NR | E. coli, S. enteritidis, S. aureus | NA | NA | Moon et al., 2013 |
| Apt22 | Whole cell (S. paratyphi A) | NB,37C | 2.1/E | S. Enteritidis, S. Typhimurium, S. Cholerasuis, S. Arizonae | Optical (chemiluminescence) | 1000 CFU/ml | Yang et al., 2013 |
| S25 | Whole cell (S. enteriditis-multiple) | TSB, overnight | NR | Salmonella serovars-multiple | Hyeon et al., 2012 | ||
| SAL26 | Whole cell (S. typhimurium ATCC14028) | TSB,37°C, overnight culture followed by TSB,37°C, 3 h then fixing with methanol | E | 4 Salmonella enterica serovars and 9 bacterial species. | Optical (Colorimetric) | 100 CFU/ml | Lavu et al., 2016 |
| SAL1 | Whole cell (S. paratyphi-A ATCC 9150) | LB broth, 37°C | E | S. Typhimurium, S. flexneri, E. coli O157:H7, Yersinia enterocolitica | Optical (fluorescence) | 10 CFU/ml | Rm et al., 2020 |
| B5 | Whole cell (S. typhimurium ATCC14028) | BHI broth, 37°C | PE | S. aureus, L. monocytogenes, E. coli O157:H7 | Mechanical (QCM) | 1,000 CFU/ml | Wang L. et al., 2017 |
| Shigella | |||||||
| Aptamer S 1 | Whole cell (Shigella dysenteriae) | LB | E | S. aureus, S. typhimurium, E. coli, L. monocytogenes, V. parahaemolyticus | Optical (Fluorescence) | 50 CFU/ml | Duan et al., 2013a |
| NA | Electrochemical | 1 CFU/ml | Zarei et al., 2018 | ||||
| Sp1 | Whole cell (Shigella sonnei ATCC 51334) | LB, 37°C, overnight | NR | S. dysenteriae, S. flexneri, S. boydii, S. typhimurium, E. coli | Optical (fluorescence) | 30 CFU/ml | Gong et al., 2015 |
| NA | Optical (SERS) | 10 CFU/ml | Wu et al., 2020 | ||||
| Sp20 | Whole cell (Shigella sonnei ATCC 51334) | LB, 37°C, overnight | NR | S. dysenteriae, S. flexneri, S. boydii, S. typhimurium and E. coli | Optical (Fluorescence) | 30 CFU/ml | Gong et al., 2015 |
| S. flexneri aptamer1 | Whole cell (Shigella flexneri) | NR | NR | Optical (fluorescence) | 100 CFU/ml | Zhu et al., 2015 | |
| SS-3, SS-4 | Whole cell (Shigella sonnei) | NB, 37°C | NR | E. coli | Optical (Fluorescence) | 1,000 CFU/ml | Song et al., 2017 |
| S. flexneri aptamer‘ | Whole cell (Shigella flexneri ATCC 12022) | NB, 37°C, 12 h | NR | NR | Optical (colorimetric) | 80 CFU/ml | Feng et al., 2019 |
| Vibrio cholerae | |||||||
| CT916 | Cholerae toxin | NA | NA | Ethanolamine-blocked magnetic beads | Optical (colorimetric) | 2.1 ng/ml | Frohnmeyer et al., 2018 |
| NA | Optical (colorimetric) | 1–100 ng/ml | Frohnmeyer et al., 2019 | ||||
| Whole cell (V. cholerae O1 -Inaba, ATCC 39315 and Ogawa) | LB broth, 37°C | 0.4/E | E. coli O157:H7, S. a dysenteriae, S. enteritidis, S. Typhimurium, Yersinia spp., S. flexneri | Optical (colorimetric) | 104 CFU/ml | Mojarad and Gargaria, 2020 | |
| Yersinia | |||||||
| N30yc5, N71yc2 | Recombinant Yop51 | NR | NR | NR | NA | NA | Bell et al., 1998 |
| M1, M5, M7 | Whole cell (Yersinia entercolitica) | Specific media (NaCl, beef extract, peptone, pH 7.2-7.4), 26°C | 0.3 (L), 0.6 (E), 0.9 (PE) | B. cereus, S. dysenteriae, L. monocytogenes, S. typhimurium, S. aureus, and E. coli | NA | NA | Shoaib et al., 2020 |
NR, Not Reported, NA, Not applicable. *Extrapolated from culture conditions. aMicrobial culture conditions and growth conditions are listed for aptamer development only. The Microbial culture and growth conditions used for aptasensors development are listed in Supplementary Table S1. bState: L, lag phase; E, exponential; PE, post-exponential. cIf number of strains used for counter selection is higher than to 5, they are listed in Supplementary Table S1.
Aptamer Development
Aptamers are typically identified by SELEX (Systematic Evolution of Ligands through Exponential Enrichment). SELEX is an iterative process where repeated exposure of a target to a large pool of random oligonucleotides results in the gradual enrichment of specific sequences that bind with the highest affinity to the target. Since the technique’s inception in 1990, many variations of the original SELEX method have been published (Darmostuk et al., 2015). These experimental variations differ based on desired aptamer properties and details have been reviewed elsewhere (Wang et al., 2019). Of note, cell-SELEX can be used to select aptamers against whole cells in solution, to ensure cell surface target epitopes are in their native state (Kaur, 2018). This method is particularly useful for developing aptamers to detect water-borne pathogens. Cell-SELEX may include counter-selection steps to remove sequences binding to non-target microbes thus minimizing cross-reactivity and improving the specificity of the resulting aptamers (see Table 1 for examples).
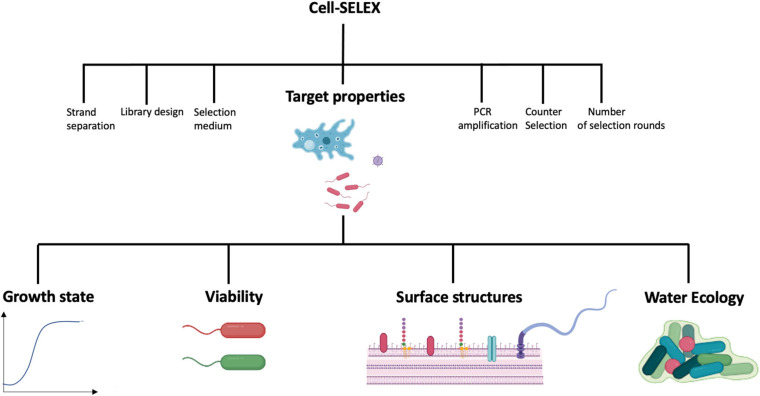
Several aptamer-coupled biosensing systems or aptasensors have been described for the detection of water-borne pathogens or toxins accumulating in water (Table 1 and Supplementary Table S1) with the majority targeting bacterial pathogens. Nevertheless, none have been officially adopted for routine detection of water-borne pathogens. The development of successful aptamer-coupled biosensors to detect water-borne pathogens requires a multi-pronged approach. Besides intricate knowledge of the sensing system, its transducer, the physico-chemical phenomenon that mediate signal responses, and a deep understanding of aptamer chemistries, careful consideration of the physiology and ecology of the target microorganism is required. This is because physio-ecological factors affect microbial morphologies and surface structures and thus the presence of aptamer targets (Figure 1). Although several works discuss transducing systems and aptamer design and chemistries in detail, relatively fewer studies consider the physio-ecological context of water-borne microbes for sensing platforms. Since most aptamers and aptasensing systems described in the literature detects water-borne bacterial pathogens, properties of bacteria are discussed in more detail to illustrate the importance of considering the target’s microbial characteristics for aptamer and aptasensor development.
FIGURE 1.
Factors affecting Cell-SELEX and thus the efficiency of aptamers targeting water-borne microbial pathogens. Most factors have been reviewed elsewhere, except the properties of the target, which are the topics of this review. Images were created in Biorender (https://biorender.com).
Aptamers Targeting Microbes in Specific States and Growth Conditions
Protozoan microbes have varying life cycles which can alternate between metabolically active feeding states, i.e., trophozoites, or inactive, dormant states such as oocysts or cysts (Aguilar-Díaz et al., 2011; Jain et al., 2019). Both oocysts and cysts are infectious forms that persist for long periods of time in environmental waters and resist a wide range of stressors (Omarova et al., 2018). The Cryptosporidium parvum oocyst-specific aptamer R4-6 was thus developed using cell-SELEX (Table 1; Iqbal et al., 2015). A counter selection step against Giardia duodenalis, another protozoan commonly found in water samples (Ongerth, 2013; WHO, 2017) was included to enhance aptamer specificity. This aptamer was first used in multiple assay formats on electrochemical biosensing platforms to detect oocysts of C. parvum down to 50 oocysts in river and lake water samples (Iqbal et al., 2015; Iqbal et al., 2019). Recently, a fluorescence plate assay coupled with magnetic beads labeled with a truncated version of the aptamer R4-6, named Min_Crypto2 achieved a detection limit of 5 oocysts (Hassan et al., 2021). The low LOD of this system is promising for oocyst detection in water given that the infectious dose of C. parvum is between 10 and 30 oocysts (Jain et al., 2019). Aptamer Min_Crypto2 was selective for Cryptosporiudum species, despite differences in size amongst species, but did not bind to Giardia oocysts. These features combined with its robust performance in water samples highlights its potential for oocyst detection in water.
Bacteria suspended in water are in a different metabolic state than bacteria growing in laboratory media. For example, L. pneumophila adopts a specific regulatory program when suspended in water due to starvation (Li et al., 2015). Consequently, the aptamers R10C5 and R10C1 were created by cell-SELEX using L. pneumophila suspended in water for 24 h, to allow the bacterium to adopt the associated metabolic state (Table 1). Counter selection was performed on two Pseudomonas spp. strains, prevalent in environmental waters (Paranjape et al., 2020). Both aptamers have excellent specificity for L. pneumophila (Saad et al., 2020).
Water borne bacteria can also be biofilm-associated. These bacteria can gain adaptive traits which make it harder to eliminate or disinfect them. To that end, biofilm-derived Pseudomonas aeruginosa cells were used to select aptamers through Cell-SELEX, without counter selection (Soundy and Day, 2017). The resulting aptamers were specific for 4 out of 5 clinical Pseudomonas aeruginosa isolates, minimally labeled non-Pseudomonas bacteria, and bound to both biofilm derived and planktonic Pseudomonas cells. The authors created chimeras and generated aptamers St17Lp21, St21Lp17. The chimeric aptamers showed improved binding and enhanced specificity for Pseudomonas bacteria as compared to the parent non-chimeric aptamers but were still unable to differentiate between biofilm and planktonic cells. This is not surprising since the biofilm-derived cells were washed and vortexed to release cells and remove alginate and exopolysaccharides. Mechanical stress induced by vortexing can destroy larger surface structures such as fimbriae and flagella. The lack of counter-selection coupled with the vigorous washing steps may have exposed cell surface structures not unique to the biofilm-derived Pseudomonas. Using counter selection could have eliminated sequences that bind to surface structures such as LPS or ubiquitous OMPS that are common in both planktonic and biofilm-derived Pseudomonas.
Aptamers against Yersinia enterocolitica were generated using Cell-SELEX with bacteria grown at 26°C (Shoaib et al., 2020). After counter selecting with several bacterial pathogens, the three aptamers M1, M5, and M7 were isolated (Table 1). Y. enterocolitica grown at 37°C showed reduced binding by the aptamers compared to bacteria grown at 25°C. Presumably this aptamer is specific for a cell surface component mostly expressed at low temperature. This study illustrates another characteristic of bacteria, which are temperature dependent surface structure and morphological changes. In the case of Y. enterocolitica specifically, the bacterium inhibits flagellum synthesis at 37°C (Kapatral et al., 1996). Components of the LPS are also temperature regulated (Białas et al., 2012).
Aptamers Targeting Viable Cells
The ability to differentiate between dead and viable cells has important implications when assessing the risk or hazard of a microbe. For example, it would be costly and inefficient to administer shutdowns or disinfection protocols for the presence of dead pathogens in a system. The detection of viable cells is also important to determine the efficacy of water disinfection protocols. Some aptamers are able to differentiate between live and dead cells. Aptamer 33, specific for Salmonella enterica serovar Typhimurium, does not bind heat-killed cells (Table 1; Joshi et al., 2009). This aptamer might therefore be useful for monitoring the efficiency of heat-based disinfection. This aptamer is described in more detail below. Another example is aptamer ONS-23 created against whole cell C. jejuni (Table 1; Dwivedi et al., 2010). This aptamer was developed, using cell-SELEX, against a chicken isolate showing characteristic C. jejuni morphology (Thomas et al., 2002). Given that C. jejuni is found on raw poultry as well as in the gastrointestinal tract and feces of animals (Mughini-Gras et al., 2016), 20 bacterial species were used for counter selection, including food-borne pathogens, enteric bacteria, non-enteric bacteria and lactic acid bacteria. ONS-23 is therefore highly specific to C. jejuni strains showing minimal binding to non-C. jejuni strains (Dwivedi et al., 2010). Furthermore, ONS-23 does not bind non-viable C. jejuni (Kim Y. J. et al., 2018) indicating that it is specific for a surface structure only present on live C. jejuni cells (Kim Y. J. et al., 2018). Though this aptamer was not tested for water application, its selective properties for viable C. jejuni makes it promising for monitoring disinfection processes.
Aptamers Targeting Source-Or Application-Specific Isolates
Isolates that are representative of the sample source of the downstream application should be used during aptamer development to ensure usefulness of the aptasensor. Aptamers E1, E2, and E10 were generated against a non-pathogenic E. coli strain of fecal origin (Crooks strain) using cell-SELEX (Table 1; Kim et al., 2013). For counter selection a combination of fecal coliform species and two Gram positives were used. The resulting aptamers were better at binding E. coli isolates of fecal origin than others and showed low binding to other species including laboratory strains of E. coli (Kim et al., 2013; Jin et al., 2017; Wu et al., 2017). A detection system using aptamer E2 was able to detect the Crooks strain in spiked tap water, pond water and milk, making it promising for E. coli detection in water (Jin et al., 2017).
Aptamers Targeting Specific Surface Structures
Surface structures can be differentially expressed in response to growth states and environment (Justice et al., 2004; Van Der Woude and Bäumler, 2004; Liu et al., 2012; Fonseca and Swanson, 2014; Li et al., 2015). If the aptamer surface target is not differentially regulated then aptamers may bind cells in several conditions, including exponential and post-exponential phase. Examples of these are the ST2P aptamer against whole cell S. enterica Typhimurium (Duan et al., 2013b, 2014, 2016) and the E. coli E2 aptamer (Kim et al., 2013; Jin et al., 2017; Wu et al., 2017). Instead of whole cells, surface structures related to virulence can also be used as aptamer targets. The pathotype EHEC (E. coli enterohemorrhagic) contains the infamous O157:H7 serotype which is strongly linked to deadly outbreaks from contaminated drinking water (Solomon et al., 2002; Ali, 2004; Saxena et al., 2015). For detecting this serotype, the specific variant of LPS can be exploited. E. coli aptamers a-aptamer and c-aptamer were created against LPS of E. coli O157:H7 (Table 1; Bruno et al., 2009). These aptamers were used in several aptasensing platforms to detect whole E. coli O157:H7 cells with great specificity, showing minimal signals with other serotypes (Wu et al., 2015; Díaz-Amaya et al., 2019a,b; Hao et al., 2019; Jiang et al., 2020). The aptamers could bind to heat-killed and formalin killed E. coli (Hao et al., 2019; Jiang et al., 2020). This is likely due to the fact that these treatments do not negatively affect the LPS (Gao et al., 2006; Chafin et al., 2013). This approach allowed for very specific aptamers to be developed; however, since the target persists after killing of cells, the aptamers are of limited use for monitoring the efficacy of disinfection programs in water. This illustrates the need for designing aptamers relevant to the downstream application.
Outer membrane proteins (OMP) of Typhimurium were used to create Aptamer 33. Counter selection was done with purified LPS of the Salmonella isolate as well as OMPs and LPS from E. coli. Aptamer 33 showed pan-serovar specificity, binding to seven different serovars of S. enterica in one study and four different S. enterica serovars in another study (Joshi et al., 2009; Hasan et al., 2018). The aptamer was used in a fluorescence aptasensor to detect whole Typhimurium in water samples from different sources highlighting its potential for detection in water (Duan et al., 2012). The aptamer does not bind to heat-killed Typhimurium which is to be expected as most OMPs are heat labile (Oh et al., 2017). The authors also observed that the aptamers could not bind S. enterica serovars Tennessee and Muenchen. This suggests that the aptamer may not have broad serovar specificity.
Discussion
Aptamer-coupled biosensors are promising systems for the detection of pathogens in water samples but are limited in real-world applications. There are a few things to consider to improve aptamers practicality in aptasensing technology (Figure 1). Many studies do not explicitly report the growth states and conditions used during cell-SELEX or during subsequent testing of the aptamers (Table 1 and Supplementary Table 1). For example, OD600 values are meaningless without details about the growth conditions, including medium, temperature and aeration. We suggest that instead of reporting OD600, the growth phase should be determined and reported, as done by Zou et al. (2018), as this would offer insight into an aptamer’s potential for specific applications. Regardless, it is important to keep the end goal in mind while developing aptamers. For example, monitoring efficiency of disinfection program will require discerning viable cells from dead cells. Aptamer ONS23 and Aptamer 33 are able to distinguish between live and dead cells (Joshi et al., 2009; Dwivedi et al., 2010; Oh et al., 2017; Kim Y. J. et al., 2018). A cell-SELEX strategy for such an application could use dead cells for counter selection. Another factor to consider is the physio-morphological state of microbes. This ensures that the microbial target possesses traits and characteristics that are representative of what’s typically found in the environment that will be sampled. For example, biofilm-derived cells might be used (Soundy and Day, 2017), but care must be taken not to remove the biofilm-specific target when preparing the target for cell-SELEX. Alternatively, if the end goal is to detect pathogens in water, then bacteria suspended in water may be used as the target (Saad et al., 2020). Lastly, it is not trivial to select appropriate strains for counter selection. This will impact aptamer affinities for targets in source environments. A possible approach is to use a cocktail of strains for the target species and a cocktail of species typically found in the same environment for counter-selection (Dwivedi et al., 2010; Kim et al., 2013). In conclusion, it is necessary to better elucidate the microbial target and the limitation of its cognate aptamer to help push microbial aptasensing platforms to market. As such a collaborative effort is needed between academics and stakeholders (governments, industry, engineers) to develop both transducer and aptamer technologies for specific microbial contaminants in the context of source water, taking into account the particularities of the microbe and its physiological state.
Author Contributions
MS reviewed the literature and compiled the information reported here, and wrote the first draft of the manuscript. MS and SPF edited the manuscript. Both authors approved submission of the manuscript.
Conflict of Interest
The authors, together with Maryam Tabrizian (McGill University, Department of Biomedical Engineering), are the inventors of aptamers R10C1 and R10C5, subject of patent applications filed in United States, patent application number US 16/850,355; and in Canada – patent application number pending at the time of revised manuscript submission. At the time of submission of the manuscript, the applications were under review. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Work in our laboratory was supported by an NSERC Strategic Partnership Grant No. STPGP 521532 to SPF. MS was supported by a CRIPA scholarship supported by the Fonds de recherche du Queìbec—Nature et technologies no. RS-170946. MS was also supported by a FRQNT doctoral scholarship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.643797/full#supplementary-material
References
- Aguilar-Díaz H., Carrero J. C., Argüello-García R., Laclette J. P., Morales-Montor J. (2011). Cyst and encystment in protozoan parasites: optimal targets for new life-cycle interrupting strategies? Trends Parasitol. 27 450–458. 10.1016/j.pt.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Ahmed A., Rushworth J. V., Hirst N. A., Millner P. A. (2014). Biosensors for whole-cell bacterial detection. Clin. Microbio. Rev. 27 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S. H. (2004). A socio-ecological autopsy of the E. coli O157: H7 outbreak in Walkerton, Ontario, Canada. Soc. Sci. Med. 58 2601–2612. 10.1016/j.socscimed.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Amraee M., Oloomi M., Yavari A., Bouzari S. (2017). DNA aptamer identification and characterization for E. coli O157 detection using cell based SELEX method. Anal. Biochem. 536 6–44. [DOI] [PubMed] [Google Scholar]
- Bahari D., Babamiri B., Salimi A., Salimizand H. (2021). Ratiometric fluorescence resonance energy transfer aptasensor for highly sensitive and selective detection of Acinetobacter baumannii bacteria in urine sample using carbon dots as optical nanoprobes. Talanta 221:121619. 10.1016/j.talanta.2020.121619 [DOI] [PubMed] [Google Scholar]
- Bell S. D., Denu J. M., Dixon J. E., Ellington A. D. (1998). RNA molecules that bind to and inhibit the active site of a tyrosine phosphatase. J. Biol. Chem. 273 14309–14314. 10.1074/jbc.273.23.14309 [DOI] [PubMed] [Google Scholar]
- Białas N., Kasperkiewicz K., Radziejewska-Lebrecht J., Skurnik M. (2012). Bacterial Cell Surface Structures in Yersinia enterocolitica. Archiv. Immunol. Ther. Exper. 60 199–209. 10.1007/s00005-012-0168-z [DOI] [PubMed] [Google Scholar]
- Bruno J. G. (2014). Application of DNA aptamers and quantum dots to lateral flow test strips for detection of foodborne pathogens with improved sensitivity versus colloidal gold. Pathogens 3 341–355. 10.3390/pathogens3020341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno J. G., Carrillo M. P., Phillips T. (2008). In vitro antibacterial effects of antilipopolysaccharide DNA aptamer-C1qrs complexes. Folia Microbiol. 53 295–302. 10.1007/s12223-008-0046-6 [DOI] [PubMed] [Google Scholar]
- Bruno J. G., Carrillo M. P., Phillips T., Andrews C. J. (2010). A novel screening method for competitive FRET-aptamers applied to E. coli assay development. J. Fluoresc. 20 1211–1223. 10.1007/s10895-010-0670-9 [DOI] [PubMed] [Google Scholar]
- Bruno J. G., Phillips T., Carrillo M. P., Crowell R. (2009). Plastic-adherent DNA aptamer-magnetic bead and quantum dot sandwich assay for Campylobacter detection. J. Fluoresc. 19 427–435. 10.1007/s10895-008-0429-8 [DOI] [PubMed] [Google Scholar]
- Bruno J. G., Sivils J. C. (2017). Further characterization and independent validation of a DNA aptamer-quantum dot-based magnetic sandwich assay for Campylobacter, Folia. Microbiology 62 485–490. 10.1007/s12223-017-0520-0 [DOI] [PubMed] [Google Scholar]
- Burrs S. L., Bhargava M., Sidhu R., Kiernan-Lewis J., Gomes C., Claussen J. C., et al. (2016). A paper based graphene-nanocauliflower hybrid composite for point of care biosensing. Biosens. Bioelectron. 85 479–487. 10.1016/j.bios.2016.05.037 [DOI] [PubMed] [Google Scholar]
- Cesewski E., Johnson B. N. (2020). Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020:112214. 10.1016/j.bios.2020.112214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin D., Theiss A., Roberts E., Borlee G., Otter M., Baird G. S. (2013). Rapid two-temperature formalin fixation. PLoS One 8:e54138. 10.1371/journal.pone.0054138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand R., Neethirajan S. (2017). Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens. Bioelectron. 98 47–53. 10.1016/j.bios.2017.06.026 [DOI] [PubMed] [Google Scholar]
- Chen W., Teng J., Yao L., Xu J., Liu G. (2020). Selection of specific DNA aptamers for hetero-sandwich-based colorimetric determination of Campylobacter jejuni in food. J. Agric. Food. Chem. 68 8455–8461. 10.1021/acs.jafc.0c02865 [DOI] [PubMed] [Google Scholar]
- Crivianu-Gaita V., Thompson M. (2016). Aptamers, antibody scFv, and antibody Fab’fragments: an overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 85 32–45. 10.1016/j.bios.2016.04.091 [DOI] [PubMed] [Google Scholar]
- Darmostuk M., Rimpelova S., Gbelcova H., Ruml T. (2015). Current approaches in SELEX: an update to aptamer selection technology. Biotechnol. Adv. 33 1141–1161. 10.1016/j.biotechadv.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Das R., Dhiman A., Kapil A., Bansal V., Sharma T. K. (2019). Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 411 1229–1238. 10.1007/s00216-018-1555-z [DOI] [PubMed] [Google Scholar]
- Dehghani Z., Hosseini M., Mohammadnejad J., Bakhshi B., Rezayan A. H. (2018). Colorimetric aptasensor for Campylobacter jejuni cells by exploiting the peroxidase like activity of Au@Pd nanoparticles. Mikrochim. Acta 185:448. [DOI] [PubMed] [Google Scholar]
- Díaz-Amaya S., Lin L. K., Deering A. J., Stanciu L. A. (2019a). Aptamer-based SERS biosensor for whole cell analytical detection of E. coli O157:H7. Anal. Chim. Acta 1081 146–156. 10.1016/j.aca.2019.07.028 [DOI] [PubMed] [Google Scholar]
- Díaz-Amaya S., Zhao M., Lin L. K., Ostos C., Allebach J. P., Chiu G. T., et al. (2019b). Inkjet printed nanopatterned aptamer-based sensors for improved optical detection of foodborne pathogens. Small 15:e1805342. [DOI] [PubMed] [Google Scholar]
- Dua P., Ren S., Lee S. W., Kim J. K., Shin H. S., Jeong O. C., et al. (2016). Cell-SELEX based identification of an RNA Aptamer for Escherichia coli and its use in various detection formats. Mol. Cells 39 807–813. 10.14348/molcells.2016.0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan N., Chang B., Zhang B., Wang H. Z., Wu S. (2016). Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor. Intern. J. Food Microbiol. 218 38–43. 10.1016/j.ijfoodmicro.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Duan N., Ding X., Wu S., Xia Y., Ma X., Wang Z., et al. (2013a). In vitro selection of a DNA aptamer targeted against Shigella dysenteriae. J. Microbiol. Methods 94 170–174. 10.1016/j.mimet.2013.06.016 [DOI] [PubMed] [Google Scholar]
- Duan N., Wu S., Chen X., Huang Y., Xia Y., Ma X., et al. (2013b). Selection and characterization of aptamers against Salmonella typhimurium using whole-bacterium systemic evolution of ligands by exponential enrichment (SELEX). J. Agric. Food Chem. 61 3229–3234. 10.1021/jf400767d [DOI] [PubMed] [Google Scholar]
- Duan N., Wu S., Ma X., Xia Y., Wang Z. (2014). A universal fluorescent aptasensor based on AccuBlue dye for the detection of pathogenic bacteria. Anal. Biochem. 454 1–6. 10.1016/j.ab.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Duan N., Wu S., Zhu C., Ma X., Wang Z., Yu Y., et al. (2012). Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Analyt. Chim. Acta 723 1–6. 10.1016/j.aca.2012.02.011 [DOI] [PubMed] [Google Scholar]
- Dwivedi H. P., Smiley R. D., Jaykus L. A. (2010). Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 87 2323–2334. 10.1007/s00253-010-2728-7 [DOI] [PubMed] [Google Scholar]
- Dwivedi H. P., Smiley R. D., Jaykus L. A. (2013). Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting. Appl. Microbiol. Biotechnol. 97 3677–3686. 10.1007/s00253-013-4766-4 [DOI] [PubMed] [Google Scholar]
- Elshafey R., Siaj M., Zourob M. M. (2015). DNA aptamers selection and characterization for development of label-free impedimetric aptasensor for neurotoxin anatoxin-a. Biosens. Bioelectron. 68 295–302. 10.1016/j.bios.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Escudero-Abarca B. I., Suh S. H., Moore M. D., Dwivedi H. P., Jaykus L. A. (2014). Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PLoS One 9:e106805. 10.1371/journal.pone.0106805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Shen Q., Wu J., Dai Z., Wang Y. (2019). Naked-eyes detection of Shigella flexneri in food samples based on a novel gold nanoparticle-based colorimetric aptasensor. Food Control 98 333–341. 10.1016/j.foodcont.2018.11.048 [DOI] [Google Scholar]
- Fonseca M. V., Swanson M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell. Infect. Microbiol. 4:12. 10.3389/fcimb.2014.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer E., Frisch F., Falke S., Betzel C., Fischer M. (2018). Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA. J. Biotechnol. 269 35–42. 10.1016/j.jbiotec.2018.01.012 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer E., Tuschel N., Sitz T., Hermann C., Dahl G. T., Schulz F., et al. (2019). Aptamer lateral flow assays for rapid and sensitive detection of cholera toxin. Analyst 144 1840–1849. 10.1039/c8an01616j [DOI] [PubMed] [Google Scholar]
- Gao B., Wang Y., Tsan M.-F. (2006). The heat sensitivity of cytokine-inducing effect of lipopolysaccharide. J. Leukocyte Biol. 80 359–366. 10.1189/jlb.1205738 [DOI] [PubMed] [Google Scholar]
- Gao R., Zhong Z., Gao X., Jia L. (2018). Graphene oxide quantum dots assisted construction of fluorescent aptasensor for rapid detection of Pseudomonas aeruginosa in food samples. J. Agric. Food. Chem. 66 10898–10905. 10.1021/acs.jafc.8b02164 [DOI] [PubMed] [Google Scholar]
- Gargano J. W., Adam E. A., Collier S. A., Fullerton K. E., Feinman S. J., Beach M. J. (2017). Mortality from selected diseases that can be transmitted by water – United States, 2003–2009. J. Water Health 15 438–450. 10.2166/wh.2017.301 [DOI] [PubMed] [Google Scholar]
- Gentry-Shields J., Wang A., Cory R. M., Stewart J. R. (2013). Determination of specific types and relative levels of QPCR inhibitors in environmental water samples using excitation-emission matrix spectroscopy and PARAFAC. Water Res. 47 3467–3476. 10.1016/j.watres.2013.03.049 [DOI] [PubMed] [Google Scholar]
- Giamberardino A., Labib M., Hassan E. M., Tetro J. A., Springthorpe S., Sattar S. A., et al. (2013). Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS One 8:e79087. 10.1371/journal.pone.0079087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Duan N., Wu S., Huang Y., Chen X., Wang Z. (2015). Selection, identification, and application of dual DNA aptamers against Shigella sonnei. Analyt. Methods 7 3625–3631. 10.1039/c5ay00214a [DOI] [Google Scholar]
- Greco S. L., Drudge C., Fernandes R., Kim J., Copes R. (2020). Estimates of healthcare utilisation and deaths from waterborne pathogen exposure in Ontario, Canada. Epidemiol. Infect. 148:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Yan W., Liu S., Ren W., Lyu M., Wang S. (2018). Trypsin enhances aptamer screening: a novel method for targeting proteins. Anal. Biochem. 561–562 89–95. 10.1016/j.ab.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Hao X., Yeh P., Qin Y., Jiang Y., Qiu Z., Li S., et al. (2019). Aptamer surface functionalization of microfluidic devices using dendrimers as multi-handled templates and its application in sensitive detections of foodborne pathogenic bacteria. Anal. Chim. Acta 1056 96–107. 10.1016/j.aca.2019.01.035 [DOI] [PubMed] [Google Scholar]
- Hasan M. R., Pulingam T., Appaturi J. N., Zifruddin A. N., Teh S. J., Lim T. W., et al. (2018). Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 554 34–43. 10.1016/j.ab.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Hassan E. M., Dixon B. R., Sattar S. A., Stalker A., Örmeci B., DeRosa M. C. (2021). Highly sensitive magnetic-microparticle-based aptasensor for Cryptosporidium parvum oocyst detection in river water and wastewater: effect of truncation on aptamer affinity. Talanta 222:121618. 10.1016/j.talanta.2020.121618 [DOI] [PubMed] [Google Scholar]
- Hu J., Fu K., Bohn P. W. (2018). Whole-cell Pseudomonas aeruginosa localized surface plasmon resonance aptasensor. Anal. Chem. 90 2326–2332. 10.1021/acs.analchem.7b04800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon J. Y., Chon J. W., Choi I. S., Park C., Kim D. E., Seo K. H. (2012). Development of RNA aptamers for detection of Salmonella Enteritidis. J. Microbiol. Methods 89 79–82. 10.1016/j.mimet.2012.01.014 [DOI] [PubMed] [Google Scholar]
- Iqbal A., Labib M., Muharemagic D., Sattar S., Dixon B. R., Berezovski M. V. (2015). Detection of Cryptosporidium parvum oocysts on fresh produce using DNA aptamers. PLoS One 10:e0137455. 10.1371/journal.pone.0137455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A., Liu J., Dixon B., Zargar B., Sattar S. A. (2019). Development and application of DNA-aptamer-coupled magnetic beads and aptasensors for the detection of Cryptosporidium parvum oocysts in drinking and recreational water resources. Can. J. Microbiol. 65 851–857. 10.1139/cjm-2019-0153 [DOI] [PubMed] [Google Scholar]
- Jain S., Costa Melo T. G., Dolabella S. S., Liu J. (2019). Current and emerging tools for detecting protozoan cysts and oocysts in water. TrAC Trends Analyt. Chem. 121:115695. 10.1016/j.trac.2019.115695 [DOI] [Google Scholar]
- Jiang Y., Qiu Z., Le T., Zou S., Cao X. (2020). Developing a dual-RCA microfluidic platform for sensitive E. coli O157:H7 whole-cell detections. Anal. Chim. Acta 1127 79–88. 10.1016/j.aca.2020.06.046 [DOI] [PubMed] [Google Scholar]
- Jin B., Wang S., Lin M., Jin Y., Zhang S., Cui X., et al. (2017). Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens. Bioelectron. 90 525–533. 10.1016/j.bios.2016.10.029 [DOI] [PubMed] [Google Scholar]
- Joshi R., Janagama H., Dwivedi H. P., Senthil Kumar T. M., Jaykus L. A., Schefers J., et al. (2009). Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes. 23 20–28. 10.1016/j.mcp.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Justice S. S., Hung C., Theriot J. A., Fletcher D. A., Anderson G. G., Footer M. J., et al. (2004). Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 101 1333–1338. 10.1073/pnas.0308125100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V., Olson J. W., Pepe J. C., Miller V. L., Minnich S. A. (1996). Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19 1061–1071. 10.1046/j.1365-2958.1996.452978.x [DOI] [PubMed] [Google Scholar]
- Kaur H. (2018). Recent developments in cell-SELEX technology for aptamer selection. Biochim. Biophys. Acta Gen. Subj. 1862 2323–2329. 10.1016/j.bbagen.2018.07.029 [DOI] [PubMed] [Google Scholar]
- Kim B., Chung K. W., Lee J. H. (2018). Non-stop aptasensor capable of rapidly monitoring norovirus in a sample. J. Pharm. Biomed. Anal. 152 315–321. 10.1016/j.jpba.2018.02.022 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Kim H. S., Chon J. W., Kim D. H., Hyeon J. Y., Seo K. H. (2018). New colorimetric aptasensor for rapid on-site detection of Campylobacter jejuni and Campylobacter coli in chicken carcass samples. Anal. Chim. Acta 1029 78–85. 10.1016/j.aca.2018.04.059 [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Chung J., Song M. Y., Jurng J., Kim B. C. (2014). Aptamer cocktails: enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 54 195–198. 10.1016/j.bios.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Song M. Y., Jurng J., Kim B. C. (2013). Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Analyt. Biochem. 436 22–28. 10.1016/j.ab.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Kumar N., Hu Y., Singh S., Mizaikoff B. (2018). Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst 143 359–373. 10.1039/c7an00983f [DOI] [PubMed] [Google Scholar]
- Lavu P. S., Mondal B., Ramlal S., Murali H. S., Batra H. V. (2016). Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica Serovar typhimurium. ACS Comb. Sci. 18 292–301. 10.1021/acscombsci.5b00123 [DOI] [PubMed] [Google Scholar]
- Lee H.-J., Kim B. C., Kim K. W., Kim Y. K., Kim J., Oh M. K. (2009). A sensitive method to detect Escherichia coli based on immunomagnetic separation and real-time PCR amplification of aptamers. Biosens. Bioelectron. 24 3550–3555. 10.1016/j.bios.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Han S. R., Maeng J. S., Cho Y. J., Lee S. W. (2012). In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochem. Biophys. Res. Commun. 417 414–420. 10.1016/j.bbrc.2011.11.130 [DOI] [PubMed] [Google Scholar]
- Li J., Yang S., Zuo C., Dai L., Guo Y., Xie G. (2020). Applying CRISPR-Cas12a as a signal amplifier to construct biosensors for Non-DNA targets in ultralow concentrations. ACS Sens. 5 970–977. 10.1021/acssensors.9b02305 [DOI] [PubMed] [Google Scholar]
- Li L., Mendis N., Trigui H., Faucher S. P. (2015). Transcriptomic changes of Legionella pneumophila in water. BMC Genom. 16:637. 10.1186/s12864-015-1869-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Hu B., Zhou Z., Guo D., Guo X., Ding P., et al. (2012). A novel non-homologous recombination-mediated mechanism for Escherichia coli unilateral flagellar phase variation. Nucleic Acids Res. 40 4530–4538. 10.1093/nar/gks040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Lei Y., Yan L., Yu T., Li Q., Zhang D., et al. (2012). A rapid and sensitive aptamer-based electrochemical biosensor for direct detection of Escherichia coli O111. Electroanalysis 24 1186–1191. 10.1002/elan.201100700 [DOI] [Google Scholar]
- Lv J., Zhao S., Wu S., Wang Z. (2017). Upconversion nanoparticles grafted molybdenum disulfide nanosheets platform for microcystin-LR sensing. Biosens. Bioelectron. 90 203–209. 10.1016/j.bios.2016.09.110 [DOI] [PubMed] [Google Scholar]
- Ma X., Jiang Y., Jia F., Yu Y., Chen J., Wang Z. (2014). An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 98 94–98. 10.1016/j.mimet.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Marton S., Cleto F., Krieger M. A., Cardoso J. (2016). Isolation of an Aptamer that binds specifically to E. coli. PLoS One 11:e0153637. 10.1371/journal.pone.0153637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung R. P., Roth D. M., Vigar M., Roberts V. A., Kahler A. M., Cooley L. A., et al. (2017). Waterborne disease outbreaks associated with environmental and undetermined exposures to water—United States, 2013–2014. Morbid. Mortal. Weekly Rep. 66:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell E. M., Nguyen J., Li Y. (2020). Aptamer-based biosensors for environmental monitoring. Front. Chem. 8:434. 10.3389/fchem.2020.00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeague M., McConnell E. M., Cruz-Toledo J., Bernard E. D., Pach A., Mastronardi E., et al. (2015). Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 81 150–161. 10.1007/s00239-015-9708-6 [DOI] [PubMed] [Google Scholar]
- Mojarad A. E., Gargaria S. L. M. (2020). Aptamer-nanobody based ELASA for detection of Vibrio cholerae O1. Iran J. Microbiol. 12 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Kim G., Lee S., Park S. (2013). Identification of Salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiol. Methods 95 162–166. 10.1016/j.mimet.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Morales M. A., Halpern J. M. (2018). Guide to selecting a biorecognition element for biosensors. Bioconjug. Chem. 29 3231–3239. 10.1021/acs.bioconjchem.8b00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini-Gras L., Penny C., Ragimbeau C., Schets F. M., Blaak H., Duim B., et al. (2016). Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 101 36–45. 10.1016/j.watres.2016.05.069 [DOI] [PubMed] [Google Scholar]
- Ng A., Chinnappan R., Eissa S., Liu H., Tlili C., Zourob M. (2012). Selection, characterization, and biosensing application of high affinity congener-specific microcystin-targeting aptamers. Environ. Sci. Technol. 46 10697–10703. 10.1021/es301686k [DOI] [PubMed] [Google Scholar]
- Oh S. Y., Heo N. S., Shukla S., Cho H. J., Vilian A. E., Kim J., et al. (2017). Development of gold nanoparticle-aptamer-based LSPR sensing chips for the rapid detection of Salmonella typhimurium in pork meat. Sci. Rep. 7 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omarova A., Tussupova K., Berndtsson R., Kalishev M., Sharapatova K. (2018). Protozoan parasites in drinking water: a system approach for improved water, sanitation and hygiene in developing countries. Intern. J. Environ. Res. Public Health 15:495. 10.3390/ijerph15030495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongerth J. E. (2013). The concentration of Cryptosporidium and Giardia in water–the role and importance of recovery efficiency. Water Res. 47 2479–2488. 10.1016/j.watres.2013.02.015 [DOI] [PubMed] [Google Scholar]
- Paranjape K., Bédard E., Whyte L. G., Ronholm J., Prévost M., Faucher S. P. (2020). Presence of Legionella spp. in cooling towers: the role of microbial diversity, Pseudomonas, and continuous chlorine application. Water Res. 169:115252. 10.1016/j.watres.2019.115252 [DOI] [PubMed] [Google Scholar]
- Payment P., Locas A. (2011). Pathogens in water: value and limits of correlation with microbial indicators. Groundwater 49 4–11. 10.1111/j.1745-6584.2010.00710.x [DOI] [PubMed] [Google Scholar]
- Peng Z., Ling M., Ning Y., Deng L. (2014). Rapid fluorescent detection of Escherichia coli K88 based on DNA aptamer library as direct and specific reporter combined with immuno-magnetic separation. J. Fluoresc. 24 1159–1168. 10.1007/s10895-014-1396-x [DOI] [PubMed] [Google Scholar]
- Queirós R. B., de-los-Santos-Álvarez N., Noronha J. P., Sales M. G. F. (2013). A label-free DNA aptamer-based impedance biosensor for the detection of E. coli outer membrane proteins. Sens. Actuat. B Chem. 181 766–772. 10.1016/j.snb.2013.01.062 [DOI] [Google Scholar]
- Queirós R. B., Gouveia C., Fernandes J. R. A., Jorge P. A. S. (2014). Evanescent wave DNA-aptamer biosensor based on long period gratings for the specific recognition of E. coli outer membrane proteins. Biosens. Bioelectron. 62 227–233. 10.1016/j.bios.2014.06.062 [DOI] [PubMed] [Google Scholar]
- Ramírez-Castillo F. Y., Loera-Muro A., Jacques M., Garneau P., Avelar-González F. J., Harel J., et al. (2015). Waterborne pathogens: detection methods and challenges. Pathogens 4 307–334. 10.3390/pathogens4020307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasoulinejad S., Gargari S. L. M. (2016). Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol. 231 46–54. 10.1016/j.jbiotec.2016.05.024 [DOI] [PubMed] [Google Scholar]
- Rm R., Maroli N., Achuth J., Ponmalai K., Kadirvelu K. (2020). Highly adaptable and sensitive FRET-based aptamer assay for the detection of Salmonella paratyphi A. Spectrochim. Acta A Mol. Biomol. Spectrosc. 243:118662. 10.1016/j.saa.2020.118662 [DOI] [PubMed] [Google Scholar]
- Roushani M., Sarabaegi M., Pourahmad F. (2019). Impedimetric aptasensor for Pseudomonas aeruginosa by using a glassy carbon electrode modified with silver nanoparticles. Mikrochim. Acta 186:725. [DOI] [PubMed] [Google Scholar]
- Saad M., Chinerman D., Tabrizian M., Faucher S. P. (2020). Identification of two aptamers binding to Legionella pneumophila with high affinity and specificity. Sci. Rep. 10:9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena T., Kaushik P., Mohan M. K. (2015). Prevalence of E. coli O157: H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagn. Microbiol. Infect. Dis. 82 249–264. 10.1016/j.diagmicrobio.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Shi X., Zhang J., He F. (2019). A new aptamer/polyadenylated DNA interdigitated gold electrode piezoelectric sensor for rapid detection of Pseudomonas aeruginosa. Biosens. Bioelectron. 132 224–229. 10.1016/j.bios.2019.02.053 [DOI] [PubMed] [Google Scholar]
- Shoaib M., Shehzad A., Mukama O., Raza H., Niazi S., Khan I. M., et al. (2020). Selection of potential aptamers for specific growth stage detection of Yersinia enterocolitica. RSC Adv. 10 24743–24752. 10.1039/d0ra00683a [DOI] [PMC free article] [PubMed] [Google Scholar]
- So H. M., Park D. W., Jeon E. K., Kim Y. H., Kim B. S., Lee C. K., et al. (2008). Detection and titer estimation of Escherichia coli using aptamer-functionalized single-walled carbon-nanotube field-effect transistors. Small 4 197–201. 10.1002/smll.200700664 [DOI] [PubMed] [Google Scholar]
- Sodia T., Poch D., Honda J. R., Bonham A. J. (2020). Detection of mycobacterial mannose-capped Lipoarabinomannan via Electrochemical Biosensor. FASEB J. 34:1. 10.1096/fasebj.2020.34.s1.07310 [DOI] [Google Scholar]
- Solomon E. B., Yaron S., Matthews K. R. (2002). Transmission of Escherichia coli O157: H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68 397–400. 10.1128/aem.68.1.397-400.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. S., Sekhon S. S., Shin W. R., Kim H. C., Min J., Ahn J. Y., et al. (2017). Detecting and discriminating Shigella sonnei using an aptamer-based fluorescent biosensor platform. Molecules 22:825. 10.3390/molecules22050825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wang L., Li J., Fan C., Zhao J. (2008). Aptamer-based biosensors. TrAC Trends Analyt. Chem. 27 108–117. [Google Scholar]
- Soundy J., Day D. (2017). Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS One 12:e0185385. 10.1371/journal.pone.0185385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlitz B., Reinemann C., Linkorn S., Stoltenburg R. (2012). Aptamers for pharmaceuticals and their application in environmental analytics. Bioanalyt. Rev. 4 1–30. 10.1007/s12566-011-0026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C. H., Tsai M. H., Lin C. Y., Ma Y. D., Wang C. H., Chung Y. D., et al. (2020). Dual aptamer assay for detection of Acinetobacter baumannii on an electromagnetically-driven microfluidic platform. Biosens. Bioelectron. 159:112148. 10.1016/j.bios.2020.112148 [DOI] [PubMed] [Google Scholar]
- Sun X., Pan Q., Yuan C., Wang Q., Tang X. L., Ding K., et al. (2016). A Single ssDNA aptamer binding to mannose-capped Lipoarabinomannan of Bacillus calmette-guérin enhances immunoprotective effect against Tuberculosis. J. Am. Chem. Soc. 138 11680–11689. 10.1021/jacs.6b05357 [DOI] [PubMed] [Google Scholar]
- Thomas C., Hill D., Mabey M. (2002). Culturability, injury and morphological dynamics of thermophilic Campylobacter spp. within a laboratory-based aquatic model system. J. Appl. Microbiol. 92 433–442. 10.1046/j.1365-2672.2002.01550.x [DOI] [PubMed] [Google Scholar]
- Turner A. P. (2013). Biosensors: sense and sensibility. Chem. Soc. Rev. 42 3184–3196. [DOI] [PubMed] [Google Scholar]
- Van Der Woude M. W., Bäumler A. J. (2004). Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17 581–611. 10.1128/cmr.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Bédard E., Prévost M., Camper A. K., Hill V. R., Pruden A. (2017). Methodological approaches for monitoring opportunistic pathogens in premise plumbing: a review. Water Res. 117 68–86. 10.1016/j.watres.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang R., Chen F., Jiang T., Wang H., Slavik M., et al. (2017). QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 221 776–782. 10.1016/j.foodchem.2016.11.104 [DOI] [PubMed] [Google Scholar]
- Wang K. Y., Zeng Y. L., Yang X. Y., Li W. B., Lan X. P. (2011). Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 30 273–278. 10.1007/s10096-010-1074-0 [DOI] [PubMed] [Google Scholar]
- Wang T., Chen C., Larcher L. M., Barrero R. A., Veedu R. N. (2019). Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 37 28–50. 10.1016/j.biotechadv.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Weerathunge P., Ramanathan R., Torok V. A., Hodgson K., Xu Y., Goodacre R., et al. (2019). Ultrasensitive colorimetric detection of murine norovirus using nanozyme aptasensor. Anal. Chem. 91 3270–3276. 10.1021/acs.analchem.8b03300 [DOI] [PubMed] [Google Scholar]
- WHO, (2017). Guidelines for Drinking Water Quality. Geneva: WHO. [Google Scholar]
- Wu G., Dai Z., Tang X., Lin Z., Lo P. K., Meyyappan M., et al. (2017). Graphene field-effect transistors for the sensitive and selective detection of Escherichia coli using pyrene-tagged DNA Aptamer. Adv. Healthc. Mater. 6:736. [DOI] [PubMed] [Google Scholar]
- Wu J. H., Wang C. H., Ma Y. D., Lee G. B. (2018). A nitrocellulose membrane-based integrated microfluidic system for bacterial detection utilizing magnetic-composite membrane microdevices and bacteria-specific aptamers. Lab. Chip 18 1633–1640. 10.1039/c8lc00251g [DOI] [PubMed] [Google Scholar]
- Wu S., Duan N., He C., Yu Q., Dai S., Wang Z. (2020). Surface-enhanced Raman spectroscopic-based aptasensor for Shigella sonnei using a dual-functional metal complex-ligated gold nanoparticles dimer. Colloids Surf. B Biointerf. 190:110940. 10.1016/j.colsurfb.2020.110940 [DOI] [PubMed] [Google Scholar]
- Wu W., Zhao S., Mao Y., Fang Z., Lu X., Zeng L. (2015). A sensitive lateral flow biosensor for Escherichia coli O157:H7 detection based on aptamer mediated strand displacement amplification. Anal. Chim. Acta 861 62–68. 10.1016/j.aca.2014.12.041 [DOI] [PubMed] [Google Scholar]
- Yan W., Gu L., Ren W., Ma X., Qin M., Lyu M., et al. (2019). Recognition of Helicobacter pylori by protein-targeting aptamers. Helicobacter 24:e12577. 10.1111/hel.12577 [DOI] [PubMed] [Google Scholar]
- Yang M., Peng Z., Ning Y., Chen Y., Zhou Q., Deng L. (2013). Highly specific and cost-efficient detection of Salmonella Paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 13 6865–6881. 10.3390/s130506865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Guo Y., Fan J., Yang Y., Zuo C., Bai S., et al. (2020). A fluorometric assay for rapid enrichment and determination of bacteria by using zirconium-metal organic frameworks as both capture surface and signal amplification tag. Mikrochim. Acta 187:188. [DOI] [PubMed] [Google Scholar]
- Yoo S. M., Kim D. K., Lee S. Y. (2015). Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 132 112–117. 10.1016/j.talanta.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Yu X., Chen F., Wang R., Li Y. (2018). Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. J. Biotechnol. 266 39–49. 10.1016/j.jbiotec.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Zarei S. S., Soleimanian-Zad S., Ensafi A. A. (2018). An impedimetric aptasensor for Shigella dysenteriae using a gold nanoparticle-modified glassy carbon electrode. Mikrochim. Acta 185:538. [DOI] [PubMed] [Google Scholar]
- Zelada-Guilleìn G. A., Bhosale S. V., Riu J., Rius F. X. (2010). Real-time potentiometric detection of bacteria in complex samples. Analyt. Chem. 82 9254–9260. 10.1021/ac101739b [DOI] [PubMed] [Google Scholar]
- Zhong Z., Gao R., Chen Q., Jia L. (2020). Dual-aptamers labeled polydopamine-polyethyleneimine copolymer dots assisted engineering a fluorescence biosensor for sensitive detection of Pseudomonas aeruginosa in food samples. Spectrochim. Acta A Mol. Biomol. Spectrosc. 224:117417. 10.1016/j.saa.2019.117417 [DOI] [PubMed] [Google Scholar]
- Zhong Z., Gao X., Gao R., Jia L. (2018). Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles. Mikrochim. Acta 185:377. [DOI] [PubMed] [Google Scholar]
- Zhu Q. Y., Zhang F. R. T., Du Y., Zhang X. X., Lu J. Y., Yao Q. F., et al. (2019). Graphene-based steganographically aptasensing system for information computing, encryption and hiding, fluorescence sensing and in vivo imaging of fish pathogens. ACS Appl. Mater. Interf. 11 8904–8914. 10.1021/acsami.8b22592 [DOI] [PubMed] [Google Scholar]
- Zhu W., Li Z., Liu X., Yan X., Deng L. (2015). Determination of Shigella flexneri by a novel fluorescent aptasensor. Analyt. Lett. 48 2870–2881. [Google Scholar]
- Zou Y., Duan N., Wu S., Shen M., Wang Z. (2018). Selection, Identification, and binding mechanism studies of an ssDNA Aptamer targeted to different stages of E. coli O157:H7. J. Agric. Food Chem. 66 5677–5682. 10.1021/acs.jafc.8b01006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.