Abstract
Inherited lung cancer risk, particularly in non-smokers, is poorly understood. Genomic and ancestry analysis of 1153 lung cancers from Latin America revealed striking associations between Native American ancestry and their somatic landscape, including tumor mutational burden (TMB), and specific driver mutations in EGFR, KRAS, and STK11. A local Native American ancestry risk score was more strongly correlated with EGFR mutation frequency compared to global ancestry correlation, suggesting that germline genetics (rather than environmental exposure) underlie these disparities.
Introduction:
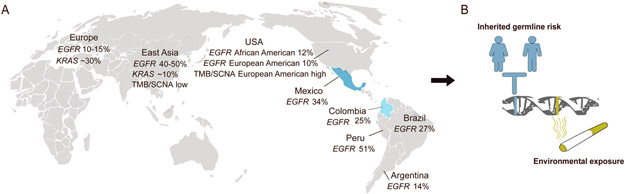
Lung cancer causes over 1.7 million deaths per year world-wide(1), and kills more people than any other malignancy in Latin America(2). Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer that is typically driven by genomic alterations of genes in the receptor tyrosine kinase (RTK)/RAS/RAF pathway(3) often allowing effective therapeutic targeting by RTK and other pathway inhibitors. It is well-known, but mysterious, that the frequency of somatic EGFR mutations is higher in LUADs from patients in East Asia (~45%) compared to LUADs from patients in Europe or patients of European (EUR) and/or African (AFR) descent in North America (~10%)(4-8). In Latin American countries, the frequency of somatic EGFR mutations in LUAD ranges from roughly 14% in Argentina, to 25-34% in Colombia, Brazil and Mexico, to 51% in Peru(9-11)(Fig. 1A). Moreover, recent genomic studies from East Asian (EAS) and African (AFR) populations have suggested different distributions of tumor mutation burden (TMB) and levels of somatic copy number alteration (SCNA)(12,13), compared to LUAD patients of European (EUR) ancestry.
Fig. 1: Genomic differences in LUAD across patient populations.
A) TMB, SCNA burden and the frequency of KRAS mutations are lower, while the frequency of EGFR mutations is higher, in lung cancers from East Asian patients, compared to lung cancers from patients of European and/or African origin. The somatic EGFR mutation rate in lung cancer varies among Latin American countries. Mexican and Colombian populations have varying degrees of admixed NAT ancestry, as indicated in blue. B) Both germline variations and environmental exposures such as smoking can predispose to somatic alterations driving the development of lung cancers, that may cause the genomic differences across populations.
Despite the differences in patterns of somatic mutation between LUAD from patients of different ethnicity, the landscape of ancestry effects on the lung cancer genomes for the Latin American populations has not been comprehensively described; and it remains unknown whether the differences are due to ancestry-specific germline variation, or rather to population-specific environmental exposures (Fig. 1B). This is of particular importance as Native American (NAT) ancestry -- which includes components of East Asian (EAS) ancestry derived through waves of migration(14) – is present to varying degrees in modern populations in Latin America, along with EUR and AFR ancestry(15).
Results:
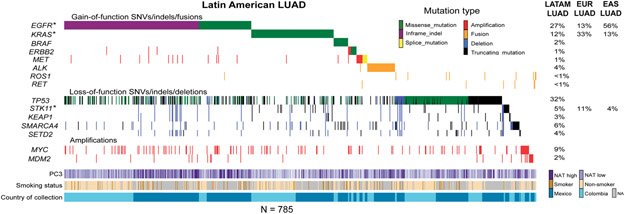
To explore the landscape of somatic cancer mutation in lung cancers from Latin America and to assess the influence of germline ancestry of genetically admixed patient populations on these somatic alterations, we performed genomic analysis of 601 lung cancer cases from Mexico and 552 from Colombia, including 499 self-reported non-smokers (Table S1). Next-generation sequencing targeting a panel of 547 cancer genes plus intronic regions of 60 cancer genes(16) was used to identify single nucleotide variants (SNVs), indels, SCNAs) and gene fusions. This gene panel covers all currently known lung cancer drivers(3), which are the focus of this work. Because we do not have matched germline samples, we applied a custom script to identify known, hotspot lung cancer driver mutations for the full 1153 samples to ensure the sensitivity for low coverage samples, as well as to avoid potential germline contamination (Methods). We found that 552 (48%) of all samples harbored oncogenic mutations in EGFR, KRAS, BRAF, ERBB2, or MET, or fusions in ALK, ROS1, or RET; 785 of 1153 samples harbored at least one detectable alteration in a broader set of known lung cancer driver genes also including TP53, STK11, KEAP1, SMARCA4, SETD2, MYC, and MDM2 (Fig. 2, Table S2-S3). The detected mutation frequencies of EGFR and KRAS were 30% and 10%, respectively, in the tested lung cancer samples from Mexican patients, and 23% and 13%, respectively, in the tested lung cancer samples from Colombian patients. SCNA analysis (Methods) identified 9% and 2% cases with high-level amplifications in MYC and MDM2, respectively. We did not observe novel amplification or deletion peaks in this Latin American lung cancer cohort as assessed by GISTIC analysis.
Fig 2: Somatic genome analysis of lung cancers from Mexico and Colombia.
Co-mutation plot displays alterations in known activators of the RTK/RAS/RAF pathway, tumor suppressor genes and significantly amplified genes. * indicates that oncogenic mutations in EGFR and KRAS as well as truncating mutations in STK11 are associated with NAT ancestry, but other somatic alterations are not; correlations with mutations are controlled for TMB. LATAM: Latin American. The mutation frequency for EUR LUAD is obtained from the TCGA dataset(51). The mutation frequency for EAS LUAD is obtained from Chen et al 2020(12).
Ancestry effects on somatic cancer genomes are understudied(17,18), and few genomic data sets have been developed from lung cancer patients with admixed ancestry. One potential source of samples for analysis of ancestry effects is discarded tissue or nucleic acids, left over after the clinical analysis of cancer samples. Here, we developed an analytical pipeline (https://github.com/jcarrotzhang/ancestry-from-panel) that offers the advantage of simultaneous measurement of global and local ancestry from sequencing tumor DNA only, without requiring matched germline samples (Fig. S1A-C). Briefly, we called the genotype of germline single nucleotide polymorphisms (SNP) using on-target and off-target reads from the sequencing panel, and measured global ancestry based on principal component analysis (PCA)(19) of the germline SNPs, in which principal components (PCs) 1, 2 and 3 captured prominently the axis of AFR, EUR and NAT ancestry, respectively (Fig. S2A). We then imputed missing SNPs using an external haplotype reference panel(15), and assigned local ancestry to each genomic region(20), based on the imputed variants. We validated our approach to ancestry analysis by performing whole-genome sequencing on a subset of 44 tumor samples, and SNP genotyping on 12 paired tumor and normal samples (Fig. S2B-C). We found a high accuracy of our off-target reads approach based on panel sequencing of tumor DNA, by comparing tumor and normal ancestry estimations (Pearson’s r>0.99), and by comparing panel sequencing to whole-genome sequencing (Pearson’s r>0.96). As panel testing of cancer genes has emerged as a practical diagnostic tool in clinical care, our off-target reads-based analytical method opens new avenues to explore the association of germline variants and somatic alterations by re-analyzing large, existing somatic sequencing datasets without matched germline sequencing data.
Having obtained data on both somatic alteration and genetic ancestry, our next step was to assess the correlation of these features, using multivariable regression controlling for self-reported smoking status and country of sample collection (Methods). As previous work focused on differences between populations, these associations with ancestry within a single admixed population provide more direct evidence of a putative genetic cause. First, we found a significant anti-correlation between TMB and PC3 representing the NAT ancestry (P=9x10−7, coef.=−0.02), in line with previous study of lung cancers from EAS patients(12); no correlation was found with the total SCNA burden, or with aneuploidy.
Evaluation of ancestry-mutation association, adjusting for sample-specific TMB, in each gene from Fig 2 showed that NAT ancestry was positively correlated with mutations in EGFR (FDR corrected p=9x10−5, coef.=0.005), and anti-correlated with mutations in KRAS (FDR corrected p=9x10−5, coef.=−0.007), and mutations in STK11 (FDR corrected p=7x10−4, coef.=−0.013), in line with previous studies focusing on EAS patients(12,21). Each feature (TMB, EGFR, KRAS, or STK11 mutation) was independently associated with NAT ancestry in a joint model (Table S4). In never smoking patients, the TMB and NAT ancestry association was stronger in EGFR-mutant (P=0.002, coef.=−0.031) than EGFR-wild type (P=0.038, coef.=−0.013). Moreover, the interaction of EGFR and NAT ancestry was significantly associated with TMB (P=0.04, coef.=−0.022) in a joint model (TMB ~ NAT ancestry + EGFR + EGFR*NAT ancestry), suggesting that the association of TMB and NAT ancestry is different in EGFR-mutant and EGFR-wild type samples. Furthermore, we demonstrated that NAT ancestry was predominantly associated with oncogenic, driver mutations in EGFR, but not with non-oncogenic, passenger mutations (Fig. S3). In addition, we did not observe SCNA of any lung cancer driver gene associated with ancestry (Methods). The observed correlations held in separate analyses of the Mexican and Colombian cohorts (Fig. S4A-B).
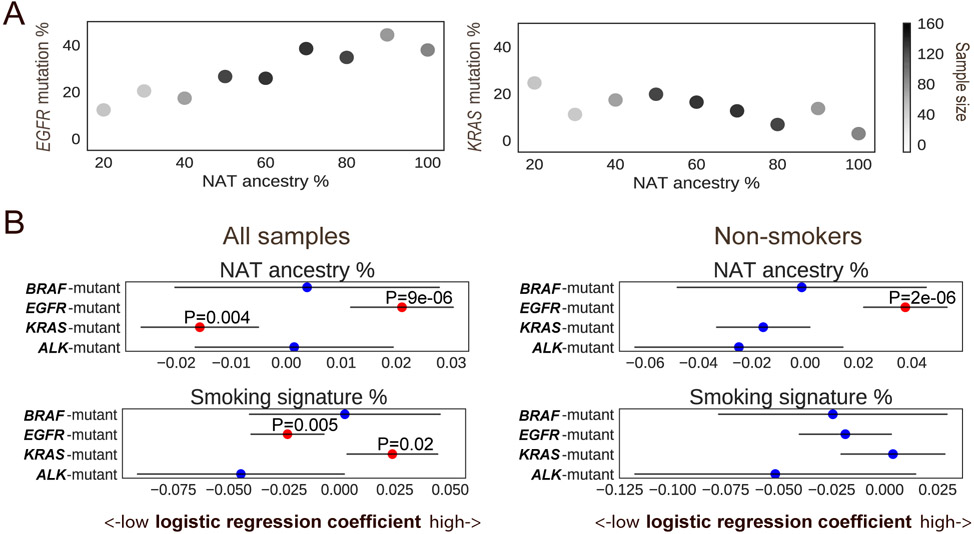
To better understand the relationship of ancestry and exposure-induced mutagenesis in the risk of developing LUADs through the activation of RTK/RAS/RAF pathway, we tested the ancestry associations with RTK/RAS/RAF pathway oncogene alterations adjusting for mutational signatures (Table S5). The positive correlation of NAT ancestry with EGFR mutation (OR=1.23 in every 10% increase of NAT ancestry, 95% CIs 1.12-1.35), and negative correlation of NAT ancestry with KRAS mutation (OR=0.85 in every 10% increase of NAT ancestry, 95% CIs 0.77-0.95) remained significant (Fig. 3A-B). The association of NAT ancestry with EGFR mutation was also observed in an analysis restricted to patients who reported themselves as never smokers (OR=1.46 in every 10% increase of NAT ancestry, 95% CIs 1.25-1.70, Fig. S5). This association was also observed in an analysis restricted to patients who reported themselves as smokers (OR=1.45 in every 10% increase of NAT ancestry, 95% CIs 1.08-1.94, Fig. 3B). When including smokers, KRAS mutation rate increased with the proportion of smoking signature (OR=1.27 in every 10% increase of smoking signature, 95% CIs 1.04-1.56, Fig. 3B). The ancestry effect on KRAS mutations in reported never smokers trended toward significance but was not significant (P=0.08) in this study, perhaps due to sample size (n=387). Moreover, the interaction of smoking signature and NAT ancestry did not modify the effect size of ancestry on KRAS (P=0.34, Methods). Gender and the APOBEC signature were not associated with mutations of any lung cancer oncogenes. Age of diagnosis was negatively associated with the risk of ALK-translocated cases (P=3x10−5, OR=0.97 in every 10-year increase of age, 95% CIs 0.96-0.99). Together, we conclude that NAT ancestry was associated with genomic differences in Latin American LUAD patients that are independent of smoking activity.
Fig. 3: Targetable LUAD driver genes associated with genetic ancestry.
A) The percentage of NAT germline ancestry is positively correlated with the percentage of somatic EGFR mutations, and negatively correlated with the percentage of somatic KRAS mutations. Color bar represents the number of samples in the NAT ancestry percentage range. B) Association of targetable LUAD driver genes with NAT ancestry, mutational signature and gender (n=705) (left), and the association in never smokers only (n=387) (right). Multivariable logistic regression P values are shown, with NAT ancestry percentage, gender, smoking and APOBEC signature as covariates. Red dots represent P value <0.05. Lines represent 95% confidence intervals.
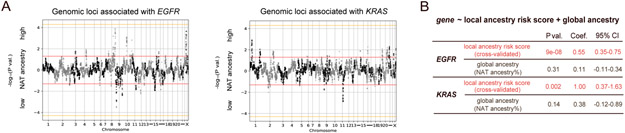
To assess whether the observed association with EGFR and KRAS mutations is due to NAT ancestry itself or to an environmental exposure/socioeconomic status related to the NAT ancestry, we next investigated the influence of local ancestry. Previous work has shown that associations between local ancestry and phenotype (while accounting for global ancestry) provide evidence of a genetically driven phenotype, as local ancestry is not expected to be causally associated with environmental exposure or socioeconomic status(22,23). We used RFMix(20) to map local ancestry, producing 5425 genomic regions with an assignment of AFR, EUR or NAT ancestral population for each parental chromosome (Table S6-S7). We performed a multivariable logistic regression of NAT ancestry for each genomic region correlating with the EGFR-mutant or KRAS-mutant samples, controlling for the global ancestry (Methods). We did not identify any region where correlation reached genome-wide significance of P<5x10−5 (Fig. 4A, S6).
Fig 4: Germline local ancestry in association with somatic EGFR and KRAS mutations.
A) Genome-wide association of local NAT ancestry with EGFR (left) and KRAS (right). “NAT ancestry high” indicates positive association, whereas “NAT ancestry low” indicates negative association. Red line indicates P=0.05. Orange line indicates genome-wide significance threshold (P<5x10−5). B) Association of local ancestry risk score with somatic EGFR or KRAS mutations, controlling for global ancestry (proportion of overall NAT ancestry).
We next evaluated whether local ancestries across multiple sub genome-wide significance threshold regions (5x10−5<P<0.05) were associated with the somatic mutation phenotype by constructing a polygenic ancestry score. This approach is conceptually similar to previous work leveraging local ancestry to quantifying phenotypic heritability(22), but we employ a risk score rather than variance partitioning as the former is more stable at low sample sizes. The local ancestry risk score was defined as the sum of NAT ancestry across each associated region weighted by the Z-score for the association of that region with the given mutation (Methods). To guard against overfitting, Z-scores and local ancestry risk scores were computed by cross-validation: splitting the dataset into ten subsets, obtaining Z-scores for the mutation-ancestry associations using nine subsets, and then calculating the local ancestry risk score on the held-out subset (Fig. S7). We then performed another logistic regression including both the cross-validated local ancestry risk score and global ancestry as covariates, and found that the local ancestry risk score was significantly associated with EGFR and KRAS mutations, respectively, whereas global ancestry was no longer significant in the joint model (Fig. 4B). In contrast, the local ancestry risk score was not associated with TMB and STK11 mutations in a joint model with global ancestry. Finally, although previous work suggested associations between cis alleles and EGFR mutations(24), we did not observe an association between the local ancestry of the EGFR locus and somatic mutations in EGFR (P=0.8). Moreover, when including the local ancestry of the EGFR locus as a covariate, the association of EGFR mutations and the local ancestry risk score remained significant (P=4x10−6). Our finding suggests that one or more genetic loci specific to NAT ancestry may modulate the evolution of lung cancer tumors to harbor EGFR or KRAS mutations in the Latin American populations.
Discussion:
In summary, the genomic landscape of LUADs is strikingly varied in Latin American patients with mixed ancestries. In our study of 1153 lung cancers, we demonstrated that NAT ancestry was correlated with somatic driver alterations, including EGFR and KRAS mutations that can be effectively targeted by small molecule inhibitors to prolong survival(4,25), and TMB and STK11 that are potential prognostic biomarkers in lung cancer patients(26,27). The ancestry and TMB association was independent of smoking-related mutational processes, and therefore, further investigation on the impact of ancestry-related TMB differences on the response to checkpoint inhibitors is needed(28). Of note, our TMB estimates may be susceptible to germline contaminations due to the lack of matched normal samples. If germline variants specific to the Mexican or Colombian population could not be sufficiently filtered (due to smaller germline reference panels), individuals with higher NAT ancestry would have more germline contamination and the anti-correlation between TMB and NAT ancestry may thus be even more significant than we have observed. Furthermore, due to the lack of matched germline samples and the use of panel sequencing data, we only tested known, hotspot mutations and protein truncating mutations in lung cancer driver genes. Future studies will be needed to comprehensively characterize lung cancer genomes from Latin American patients.
Disparities in access to genetic testing have been observed among Hispanic lung cancer patients in the United States(29). Our study shows that while controlling for global ancestry, local ancestry is associated with mutations in EGFR and KRAS, providing the first example, to our knowledge, of a germline influence, which may or may not act together with ancestry-specific environmental exposure, on targetable somatic events in lung cancer. These findings highlight the importance of providing somatic genetic testing for Latin American lung cancer patients with admixed ancestries. Given the limited sample size, we could not determine the precise risk loci for EGFR and KRAS mutation by local ancestry mapping. As low-dose CT scans have enabled lung cancer screening that can significantly reduce lung cancer mortality(30,31), we believe that the identification of germline allele(s) predisposing to the development of EGFR-mutant or KRAS-mutant LUAD from large, Latin American lung cancer sample sets may improve our understanding of the biological causes of EGFR and KRAS mutations and the evolutionary processes in lung cancers. Such findings could therefore shed light on prevention and early detection strategies for lung cancer in Latin America and beyond, particularly for non-smokers.
Methods:
Sample collection:
The protocol of this work was approved by the ethical and scientific committees of the Instituto Nacional de Cancerologia in Mexico City, Mexico, the Foundation for Clinical and Applied Cancer Research in Bogotá, Colombia, and the Dana-Farber Cancer Institute in Boston, Massachusetts, for detecting EGFR mutations and further genomic analysis. Biopsies were collected for histological diagnosis by the pathology departments of Instituto Nacional de Cancerologia and Foundation for Clinical and Applied Cancer Research.
Library preparation and sequencing:
Genomic DNA was extracted from fresh-frozen, blood and paraffin-embedded samples by a standard procedure using the Wizard Genomics DNA kit (Promega, Madison, USA) according to the manufacturer’s instructions. DNA was fragmented to 250 bp and size-selected DNA was ligated to sample-specific barcodes. A custom targeted hybrid capture sequencing platform (OncoPanel) was used to assay genomic alterations in tumor DNA(16). Each library was quantified by sequencing on an Illumina MiSeq nano flow cell (Illumina, San Diego, CA). Libraries were pooled in equal mass to a total of 500 ng for enrichment using the Agilent SureSelect hybrid capture kit (Agilent Technologies, Santa Clara, CA; cat. no. G9611A). Libraries were sequenced on an Illumina HiSeq2500 or HiSeq3000. Pooled samples were demultiplexed using the Picard tools (https://broadinstitute.github.io/picard/). Paired reads were aligned to the hg19 reference genome using BWA(32) with the following parameters “-q 5 -l 32 -k 2 -o 1”. Aligned reads were sorted and duplicate-marked using Picard. In each batch, we sequenced a control DNA sample as a “plate normal”. For a subset of 44 cases, the same libraries were sequenced on Illumina NovaSeq for low-coverage whole-genome sequencing at 1X coverage.
Mutation analysis:
Mutation detection for single nucleotide variants (SNVs) was performed using MuTect v1.1.4(33) in paired mode by pairing each sample to a control DNA sample profiled with the same OncoPanel. SomaticIndelDetector(34) was used for indel calling. Mutations were annotated by Variant Effect Predictor (VEP)(35) and Oncotator(36). Called variants with a frequency greater than or equal to 0.01% that were found in the Genome Aggregation Database (gnomAD)(37) containing 25,748 exomes, were excluded. TMB was calculated by dividing the total number of coding, non-silent mutations in an individual by the target size (3 MB). Mutational signatures were called using SignatureAnalyzer(38) with SNVs classified by 96 tri-nucleotide contexts. Prior studies have shown that mutational signature analysis can be inferred based on on-target reads from clinical panel sequencing(8,39). Smoking (COSMIC signature 4) and APOBEC signature (COSMIC signature 2) activities were inferred by the estimated number of mutations in a tri-nucleotide context associated with each signature.
A custom script (https://github.com/jcarrotzhang/ancestry-from-panel) was applied to inspect the sites of hotspot driver mutations in EGFR, KRAS, BRAF, ERBB2, MET, and TP53. For each mutation, we counted reads supporting the reference base and the altered base, after filtering out reads with base quality or mapping quality less than 20(40). A mutation was called if the total read count was greater than 5, the altered read count was greater than 2, and the mutant allele frequency was greater than 5%. Identified mutations with total coverage lower than 30X were manually inspected using IGV(41).
Copy number and rearrangements:
Read coverage was calculated at 1 MB bins across the genome and was corrected for GC content and mappability biases using ichorCNA version 0.1.0(42) using the plate normal as the matched control for each sample. GISTIC version 2.0.22(43) was applied to identify focal and arm-level SCNAs on ichorCNA generated copy number segments, with the high-level amplification defined as log2-transformed copy number ratio greater than 0.7. Rearrangement events were called by Breakmer(44) and filtered on discordant read counts and split read counts greater than 0. Total SCNA burden and the degree of aneuploidy was defined by the number of genes, or chromosomal arms affected by SCNAs, respectively (copy number ratio > 0.1 or copy number ratio > −0.1).
Ancestry analysis from genotyping array:
The Multi-Ethnic Genotyping Array (MEGA) was used for genotyping of paired fresh-frozen tumor tissue and blood samples. We used PLINK version 1.9(45) to filter out variants with missing rate greater than 2%, or failed Hardy-Weinberg equilibrium test (p<1x10−6). Markers with allele frequency less than 1% in the 1000 Genomes dataset were also excluded. The Mexican and Colombian samples were merged with samples from the 1000 Genomes phase 3(15), and PCA was performed on the merged data set using (GCTA) version 1.91.6(46).
Ancestry analysis from sequencing:
SAMtools(47) was used to genotype germline variants after filtering out reads with base quality or mapping quality less than 20. LASER version 2.04(48) was used to estimate overall ancestry based on 637,037 germline variants from all populations in HGDP(49). We obtained principal components from LASER results that place each sample into a reference PCA space using 939 HGDP samples as reference samples. For local ancestry identification, phasing and imputation were performed using Beagle version 4.1(50) based on SAMtools genotyped variants. We imputed missing variants using phased haplotypes from 1000 Genomes(15). Ancestry was assigned to each SNP using RFMix v2(20). For each parental population (NAT, AFR and EUR), 500 samples from 1000 Genomes were used as reference samples. Local ancestry regions spanning centromeres were filtered. RFMix outputted global ancestry estimates were used as the percentage of NAT ancestry.
Association analysis:
Multivariable logistic regression or linear regression was performed using a Python module (https://www.statsmodels.org). Because the Mexican population of lung cancer patients has a higher level of NAT ancestry than the Colombian population15, we accounted for the country of sample collection throughout our analyses. TMB was included as a covariate when associating PC3 to mutations. Total SCNA burden was included as a covariate when associating PC3 to SCNA of lung cancer genes. Gender, proportion of smoking and APOBEC signatures were included as covariates when associating the percentage of NAT ancestry with oncogenic mutations. To test whether smoking signature influence the relationship between ancestry and the KRAS mutations, the following model was performed:
Where gender and country of sample collection were considered as covariates, and NAT*smoking was included as an interaction effect. If the interaction term is not significant, that means that smoking signature activity does not modify the effect of ancestry.
To identify specific genomic region(s) associated with LUAD cases harboring certain somatic alterations, a logistic regression model was applied controlling for the percentage of NAT ancestry, TMB and country of sample collection, followed by genomic control . Ten-fold cross-validation was performed in the following steps (Fig. S7): the whole dataset was split into ten subsets. Z-scores for the mutation-ancestry associations for each genomic region were calculated using nine subsets, and a cross-validated local ancestry risk score (sum of the NAT ancestry across each associated region weighted by the Z-score of that region) was calculated for each sample on the held-out subset. These steps were repeated for ten times until a local ancestry risk score was generated for each sample:
Where n is the number of regions associated with the somatic feature (P<0.05), A is the ancestry of each associated region (NAT ancestry was coded as 1, and EUR or AFR ancestry was coded as 0), and Z is the z-score of that associated region.
Data and Code availability:
Raw sequencing data from cancer gene panel sequencing are being deposited at European Genome-phenome Archive (EGA) under the accession code EGAS00001004752. Ancestry identification method from panel sequencing and custom code used in the analyses are available at https://github.com/jcarrotzhang/ancestry-from-panel. All other data supporting the findings of this study are available upon reasonable request from the corresponding authors.
Supplementary Material
Significance:
The frequency of somatic EGFR and KRAS mutations in lung cancer varies by ethnicity but we do not understand why. Our study suggests that the variation in EGFR and KRAS mutation frequency is associated with genetic ancestry and suggests further studies to identify germline alleles that underpin this association.
Acknowledgements:
We would like to acknowledge the patients from Mexico and Colombia for their participation in this study. This study is supported by the V Foundation Translational Research Award (the Stuart Scott Memorial Fund) and by National Cancer Institute grants R35 CA197568, R01 CA116020 and R01 CA227237, as well as an anonymous gift to the Dana-Farber Cancer Institute. M.M. is an American Cancer Society Research Professor. J.C.-Z. holds the Banting fellowship. J.D.C. is funded by the LUNGevity Career Development award. We thank the Center for Cancer Genomics at Dana-Farber Cancer Institute and the Genomic Platform at Broad Institute for their sequencing and genotyping efforts, and we thank Aruna Ramachandran and Kar-Tong Tan for helpful discussions.
Footnotes
Conflicts of interest: M.M. is the scientific advisory board chair of OrigiMed; an inventor of patents licensed to LabCorp for EGFR mutation diagnosis and patent applications on EGFR inhibitors; and receives research funding from Bayer, Janssen, Novo Ventures, and Ono Pharmaceuticals.
References:
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Raez LE, Cardona AF, Santos ES, Catoe H, Rolfo C, Lopes G, et al. The burden of lung cancer in Latin-America and challenges in the access to genomic profiling, immunotherapy and targeted treatments. Lung Cancer. 2018;119:7–13. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Au JS-K, Thongprasert S, Srinivasan S, Tsai C-M, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5:2892–911. [PMC free article] [PubMed] [Google Scholar]
- 7.Steuer CE, Behera M, Berry L, Kim S, Rossi M, Sica G, et al. Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: Results from the Lung Cancer Mutation Consortium. Cancer. 2016;122:766–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell JD, Lathan C, Sholl L, Ducar M, Vega M, Sunkavalli A, et al. Comparison of Prevalence and Types of Mutations in Lung Cancers Among Black and White Populations. JAMA Oncol. 2017;3:801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta O, Cardona AF, Martín C, Más-López L, Corrales-Rodríguez L, Bramuglia G, et al. Updated Frequency of EGFR and KRAS Mutations in NonSmall-Cell Lung Cancer in Latin America: The Latin-American Consortium for the Investigation of Lung Cancer (CLICaP). J Thorac Oncol. 2015;10:838–43. [DOI] [PubMed] [Google Scholar]
- 10.Gimbrone NT, Sarcar B, Gordian ER, Rivera JI, Lopez C, Yoder SJ, et al. Somatic Mutations and Ancestry Markers in Hispanic Lung Cancer Patients. J Thorac Oncol. 2017;12:1851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leal LF, de Paula FE, De Marchi P, de Souza Viana L, Pinto GDJ, Carlos CD, et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep. 2019;9:3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52:177–86. [DOI] [PubMed] [Google Scholar]
- 13.Sinha S, Mitchell KA, Zingone A, Bowman E, Sinha N, Schäffer AA, et al. Higher prevalence of homologous recombination deficiency in tumors from African Americans versus European Americans. Nature Cancer. Nature Publishing Group; 2020;1:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, et al. Reconstructing Native American population history. Nature. 2012;488:370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium T 1000 GP, The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanna GJ, Lizotte P, Cavanaugh M, Kuo FC, Shivdasani P, Frieden A, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3:e98811. 10.1172/jci.insight.98811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell. 2018;34:549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrot-Zhang J, Chambwe N, Damrauer JS, Knijnenburg TA, Robertson AG, Yau C, et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell. 2020;37:639–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96:926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaitlen N, Pasaniuc B, Sankararaman S, Bhatia G, Zhang J, Gusev A, et al. Leveraging population admixture to characterize the heritability of complex traits. Nat Genet. 2014;46:1356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florez JC, Price AL, Campbell D, Riba L, Parra MV, Yu F, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52:1528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, He L, Ramírez J, Krishnaswamy S, Kanteti R, Wang Y-C, et al. Functional EGFR germline polymorphisms may confer risk for EGFR somatic mutations in non-small cell lung cancer, with a predominant effect on exon 19 microdeletions. Cancer Res. 2011;71:2423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DB, Frampton GM, Rioth MJ, Yusko E, Xu Y, Guo X, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res. 2016;4:959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res. 2018;24:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11:S71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch JA, Berse B, Rabb M, Mosquin P, Chew R, West SL, et al. Underutilization and disparities in access to EGFR testing among Medicare patients with lung cancer from 2010 - 2013. BMC Cancer. 2018;18:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovalchik SA, Tammemagi M, Berg CD, Caporaso NE, Riley TL, Korch M, et al. Targeting of low-dose CT screening according to the risk of lung-cancer death. N Engl J Med. 2013;369:245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duffy SW, Field JK. Mortality Reduction with Low-Dose CT Screening for Lung Cancer. N. Engl. J. Med 2020;382:572–3. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Touat M, Li YY, Boynton AN, Spurr LF, Iorgulescu JB, Bohrson CL, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580:517–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrot-Zhang J, Majewski J. LoLoPicker: detecting low allelic-fraction variants from low-quality cancer samples. Oncotarget. 2017;8:37032–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology. 2011;12:R41. 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Zhan X, Bragg-Gresham J, Kang HM, Stambolian D, Chew EY, et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet. 2014;46:409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012. 10.1126/science.aay5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data from cancer gene panel sequencing are being deposited at European Genome-phenome Archive (EGA) under the accession code EGAS00001004752. Ancestry identification method from panel sequencing and custom code used in the analyses are available at https://github.com/jcarrotzhang/ancestry-from-panel. All other data supporting the findings of this study are available upon reasonable request from the corresponding authors.