Abstract
T cell-based therapies have induced cancer remissions, though most tumors ultimately progress, reflecting inherent or acquired resistance including antigen escape. Better understanding of how T cells eliminate tumors will help decipher resistance mechanisms. We used a CRISPR/Cas9 screen and identified a necessary role for fas-fasL in antigen-specific T cell killing. We also found that fas-fasL mediated off-target ‘bystander’ killing of antigen-negative tumor cells. This localized bystander cytotoxicity enhanced clearance of antigen-heterogeneous tumors in vivo, a finding that has not been shown previously. Fas-mediated on-target and bystander killing were reproduced in chimeric antigen receptor (CAR-T) and bispecific antibody T cell models and were augmented by inhibiting regulators of fas signaling. Tumoral FAS expression alone predicted survival of CAR-T-treated patients in a large clinical trial (NCT02348216). These data suggest strategies to prevent immune escape by targeting both the antigen expression of most tumor cells and the geography of antigen-loss variants.
Introduction
T cell-based immunotherapies – including adoptive transfer of engineered T cells, bispecific antibodies, and checkpoint blockade – have revolutionized cancer treatment. However, even with the remarkably high response rates of CAR-T-treated patients, most either progress or relapse within 1 year (1–3). Microenvironmental factors contributing to T cell priming (4–6) and T cell-intrinsic factors (7,8) both influence anti-tumor immunity, but tumor cell-intrinsic factors have the most abundant clinical evidence for contributing to treatment potency and failures.
The clearest such mechanism is target antigen (Ag) modulation – expression downregulation, lineage switching, or emergence of splice variants – which is the most common cause of relapse following CAR-T therapy for B-cell acute lymphoblastic leukemia (B-ALL) (9). Similar mechanisms of Ag escape have also been noted in CAR-T therapy for non-Hodgkin lymphoma (NHL) (9), multiple myeloma (10), and glioblastoma multiforme (11). While downregulation of target Ag is a well-characterized occurrence, comparable rates of Ag+ relapses (12) that are unresponsive to CAR-T re-infusion (13–15) also indicate the presence of other immune evasion mechanisms in tumor cells such as suppression of apoptotic pathways.
The majority of preclinical and clinical data suggest that the tumoricidal effects of murine and human T cells are perforin- and granzyme-mediated (16–18). Consequently, expression of these and other effector molecules (e.g. IFNγ and TNFα) are the primary focus of clinical biomarker studies across T cell-based therapies (19–22). However, equal efficacy of adoptive cell therapy (23) and checkpoint blockade (24) in perforin-deficient murine models, as well as recent evidence of death receptor signaling gene signatures correlating with CAR-T efficacy (25), supports the notion that other mechanisms may have an underappreciated role in T cell cytotoxicity and warrant further investigation.
Additionally, even with optimal Ag-specific tumor clearance, emergence of pre-existing Ag-null variants is an expected mechanism of rapid relapse. Thus, the frequency of durable immunotherapy remissions despite tumoral Ag heterogeneity is surprising. A majority of B-ALL patients harbor >1% CD19− cells at diagnosis (26), though only ~20% of patients relapse with CD19− disease after CD19-directed therapy (9). Similarly, response rates of CAR-T therapy for NHL are comparable between subgroups with high and low/negative tumoral CD19 expression (27). Just as puzzling is that subclonal loss of β2m, effectively abolishing antigen presentation to CD8 T cells, in MSI-H colorectal tumors has no apparent effect on PD-1 blockade efficacy (28). These data are all highly suggestive of bystander cytotoxicity mediating clearance of Ag− cancer subclones. However, there are no published studies identifying tumor cell mechanisms of how this occurs in vivo despite its enormous therapeutic potential.
To dissect these mechanisms and address these important knowledge gaps, we performed a CRISPR/Cas9 screen using a targeted library of genes highly expressed by the A20 murine lymphoma cell line and identified the death receptor fas as crucial to T cell cytotoxicity. For the first time, we demonstrate that fas-mediated bystander killing enhances in vivo control of heterogenous tumors. We also demonstrate that pre-treatment tumoral FAS expression is an even better predictor of long-term CAR-T therapy outcomes than expression of the target molecule, which may suggest that homogeneity of Ag expression is not needed to achieve full efficacy of Ag-directed therapies. Finally, we present evidence and rationale for combination strategies that augment both on-target and bystander cytotoxic pathways in order to prevent both Ag+ and Ag− relapses after adoptive T cell therapies.
Results
Fas is a critical T cell effector molecule.
Using a reductionist 3-cell system comprising GFP-specific (JEDI) CD8 T cells (29,30), Ag+ A20-GFP cells, and Ag− A20-mCherry cells (Fig. 1A), we exerted strong iterative Ag-specific T cell selection pressure on the GFP+ population (Supplementary Fig. S1A) and sequenced surviving cells to measure changes in the frequency of targeted genes (Supplementary Fig. S1B). The 291 genes targeted in the screen (Supplementary Table S1) were manually curated for high expression in our cancer model and enriched for immune-related functional annotations. Significant deviations from the expected frequency of targeted genes in the GFP+ cells were internally controlled by the measured frequency in mCherry+ cells (Supplementary Fig. S1C). As expected, knockout clones of B2m and Tap1 (Supplementary Fig. S1D and E), genes crucial to Ag presentation, became highly enriched following selection (p<1e-14; Fig. 1B; Supplementary Fig. S1C). Enrichment of Ag+ cells that had lost MHC class I expression was confirmed at the protein level (Supplementary Fig. S1F and G), in addition to the expected relative loss of PD-L1-deleted cells (Supplementary Fig. S1F and H). Surprisingly, knockout clones of Fas became equally enriched (p<1e-14; Fig. 1B; Supplementary Fig. S1C and I), a finding unique in our model system compared to other recent T cell-based screens (31–34). For validation, we created polyclonally-derived stable Fas knockout lines of A20 lymphoma as well as 4T1 breast cancer (Fig. 1C) that had no measurable surface fas protein expression (Supplementary Fig. S2A) and were resistant to fas-induced cell death (Supplementary Fig. S2B). While WT GFP+ cells were depleted by JEDI, both Fas-negative lymphoma (Fig. 1D; quantified in Supplementary Fig. S2C) and breast cancer (Fig. 1E; quantified in Supplementary Fig. S2D) demonstrated resistance to killing under the same conditions, despite robust T cell proliferation, granzyme B production, and degranulation in both conditions (Fig. 1F). Similarly, superantigen-induced cytotoxicity by both CD8 and CD4 T cells – mediated by bacterial toxins that non-specifically bind MHC molecules to T cell receptors – was also highly dependent on Fas expression (Supplementary Fig. S2E), while irradiation-induced apoptosis was not (Supplementary Fig. S2F), demonstrating specificity to T cell-mediated killing.
Figure 1. A pooled CRISPR/Cas9 screen for T cell cytolytic mechanisms identifies Fas as crucial to on-target anti-cancer immunity.
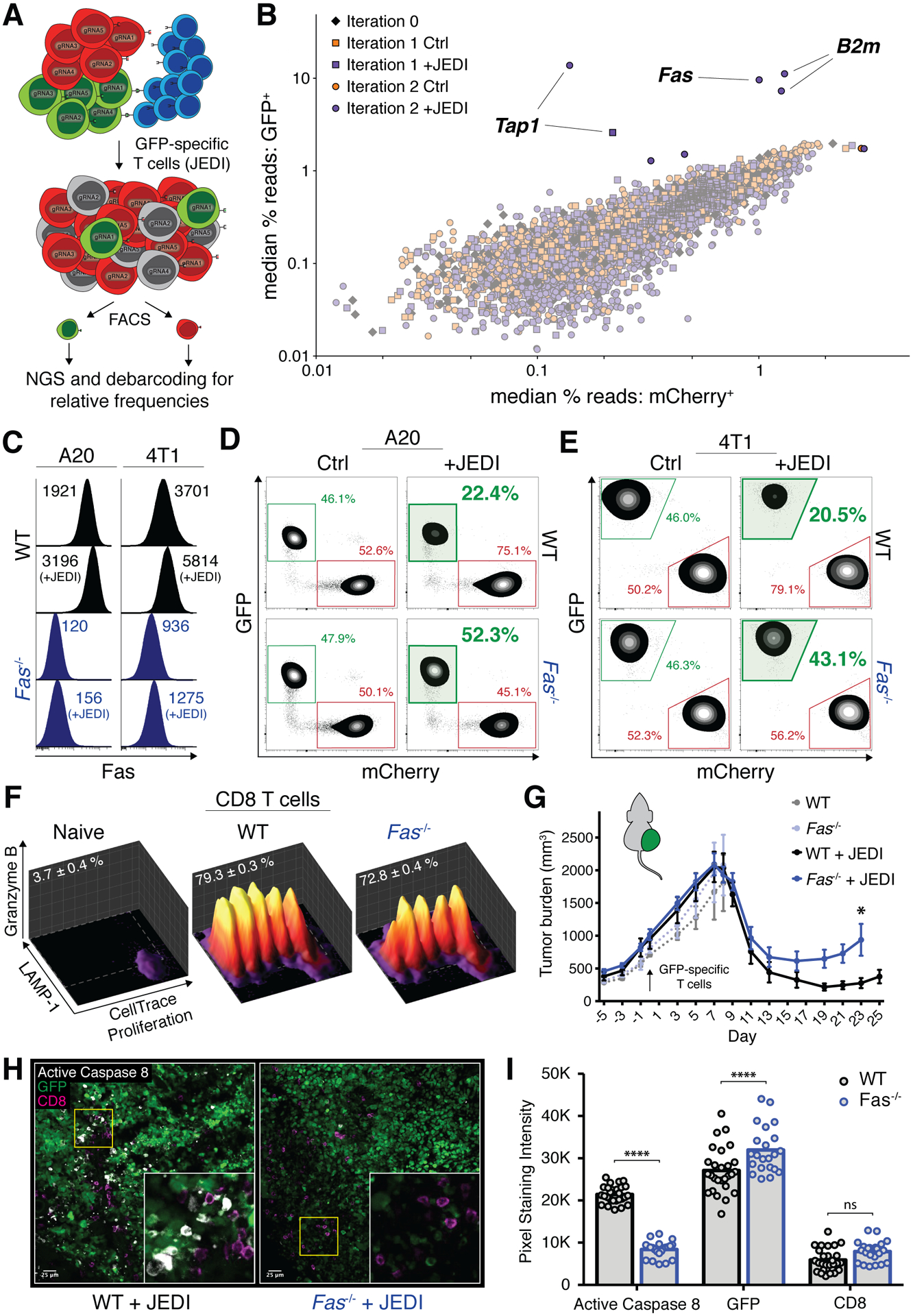
A, Schema depicting screening strategy; A20-GFP cells (Ag+; green), A20-mCherry cells (Ag−; red), GFP specific T cells (blue). B, Summary of 291-gene screen across 4 pooled libraries after 0, 1, or 2 iterations of selection with JEDI; each data point represents median read frequency of one small guide RNA in GFP-sorted vs. mCherry-sorted populations (n=5). C, Mean fluorescent intensity of surface fas expression in WT (black) and Fas-knockout (blue) lymphoma (left panel) and breast cancer (right panel) cell line models. D–E, Representative changes in percentage of surviving WT (top row) or Fas−/− (bottom row) GFP+ lymphoma (D) and breast cancer (E) relative to WT mCherry+ populations; co-culture conditions with JEDI T cells bolded and shaded in green. F, Representative proliferation, degranulation, and granzyme B expression of JEDI T cells, naïve or co-cultured with WT or Fas−/− GFP+ cancer; mean percent in gate ± SEM indicated (n=3). G, Tumor growth of WT (black) or Fas−/− (blue) GFP+ lymphoma in Rag1−/− mice adoptively transferred with tetramer-sorted GFP-specific CD8 T cells (arrow); error bars presented as mean ± SEM; two-way ANOVA with Sidak correction for multiple comparisons, *p<0.05 (n=4–12 mice/group; pooled from 2 independent experiments). Individual tumor curves in Supplemental Fig. S2G. H, Representative confocal IF of tumors cryopreserved from ‘+JEDI’ groups from G on day 8; area outlined in yellow magnified in inset. I, Quantification of fluorescent intensities from 22–27 fields of view as shown in H; pixel staining intensity units on a scale from 0 to 65,536 (16-bit); two-way ANOVA with Sidak correction for multiple comparisons, ****p<0.0001 (n=3–4 mice/group, pooled from 3 independent experiments).
Resistance to granzyme-secreting T cells was surprising given its recognition as a dominant pathway for killing transformed cells (35). Using an in vivo adoptive transfer model with tetramer-sorted GFP-specific CD8 T cells, we observed earlier recurrence of Fas−/− GFP+ tumors compared to the WT counterparts (Fig. 1G; individual curves in Supplementary Fig. S2G), validating our in vitro findings. Significantly decreased activation of the extrinsic apoptosis mediator caspase 8 in the dying Fas−/− tumor cells despite comparable T cell infiltration confirmed a role for fas-mediated cytotoxicity in situ (Fig. 1H and I). At the same time, higher staining intensity for GFP in Fas−/− tumors when challenged with T cells (Fig. 1I), but not in control mice (Supplementary Fig. S2H and I), confirmed increased survival of Fas−/− tumor cells. Taken together, these data validate fas as an important mediator of cytotoxicity in Ag-specific T cell-based immunotherapy, even in conditions with abundance of other mediators e.g. granzymes.
Fas mediates T cell bystander killing.
While our model system showed clear protection of Fas−/− Ag+ cells, it also demonstrated a significant, T cell-dependent increase in the GFP+/mCherry+ ratio (Supplementary Fig. S2C), indicating either increased proliferation of the Fas−/− GFP+ (Ag+) cells or increased clearance of the mCherry+ (Ag−) cells. Measurement of absolute cell counts confirmed the latter and demonstrated a surprising depletion of Ag− cells – in the setting of only partial protection for Fas−/− Ag+ cells – that was concealed by using the ratio as a surrogate for measuring cytotoxicity (Supplementary Fig. S3A). A close T cell dose titration revealed that conditions were possible in which the killing of mCherry+ cells could overtake the killing of GFP+ cells (Supplementary Fig. S3A), resulting in an increase of the GFP+/mCherry+ ratio. This phenomenon of Ag− tumor cell death – herein referred to as bystander killing – was observed even at very low effector:target cell ratios in the WT setting and only in the presence of Ag-specific (tetramer-sorted) T cells but not with pre-activated non-specific T cells (Fig. 2A). A role for MHC class II-mediated killing was ruled out by the lack of CD4 T cell contamination in our system (Supplementary Fig. S3B). Transwell assays confirmed that this bystander killing was contact-mediated and required co-localized Ag+ cells (Fig. 2B, purple highlight; quantified in Supplementary Fig. S3C) to induce bystander apoptosis in Ag− cells. Given our observation of fas-dependent Ag-specific cytotoxicity, we hypothesized that it may play a role in this bystander cytotoxicity as well. Co-culturing GFP(WT) with mCherry(Fas−/−) cells in the presence of JEDI T cells completely abrogated the depletion of the Ag− population observed with mCherry(WT) cells, confirming that this bystander effect was fas-mediated (Fig. 2C, left panel). Notably, mCherry(B2m−/−) cells were equally susceptible to bystander killing, ruling out Ag cross-reactivity or exogenous loading of MHC class I as possible explanations (Fig. 2C, left panel). Bystander killing was also nullified in the presence of GFP(B2m−/−) cells, demonstrating that local TCR-peptide-MHC activation by Ag+ cells is needed for T cells to exert contact-mediated cytotoxicity against Ag− cells (Fig. 2C, right panel). The lack of bystander apoptosis despite the presence of pre-activated JEDI in the chamber with Ag− cells confirmed that concurrent and local activation of the T cells was necessary (Fig. 2B, orange highlight; Supplementary Fig. S3C), consistent with the strict regulation of surface fasL activity (36) as well as ruling out distant delivery of active fasL via microvesicles (37) or exosomes (38).
Figure 2. Fas mediates geography-dependent off-target bystander killing of Ag− tumor by Ag-specific CD8 T cells.
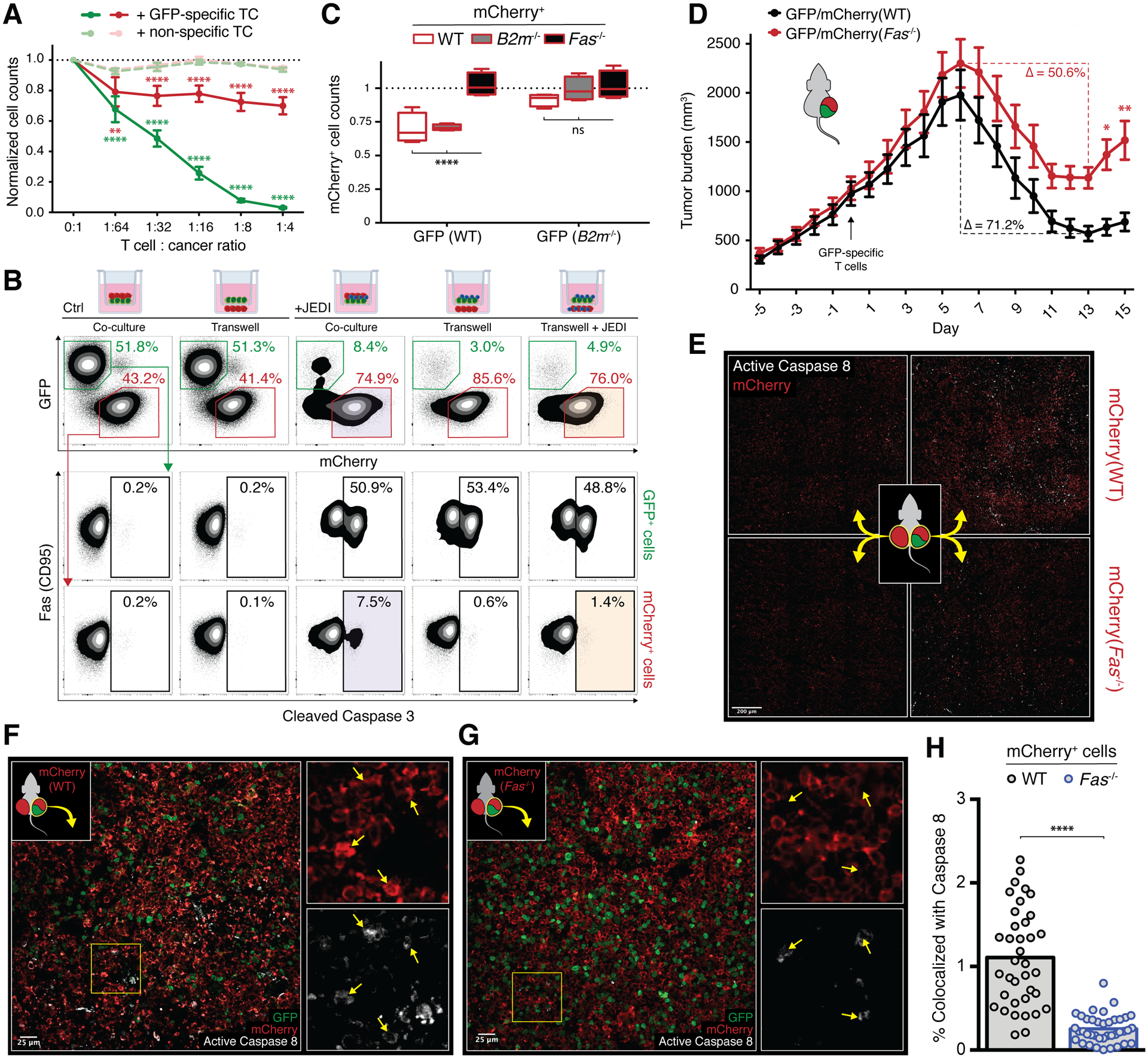
A, Cell counts of WT GFP+ (green) and WT mCherry+ (red) cells co-cultured with FACS purified tetramer+ (dark, GFP-specific) or tetramer− (light, non-specific) activated CD8 T cells at indicated ratios; counts relative to beads and normalized to 0:1 condition; error bars presented as mean ± SEM; two-way ANOVA with Sidak correction for multiple comparisons, **p<0.01, ****p<0.0001 (n=4). B, Representative cleaved caspase 3 staining of GFP+ cells (middle row) and mCherry+ cells (bottom row) after co-culture with activated JEDI T cells in transwell configurations (top row, from left to right: GFP+ and mCherry+ in top chamber; GFP+ in top chamber / mCherry+ in bottom chamber; GFP+, mCherry+, and JEDI in top chamber; GFP+ and JEDI in top chamber / mCherry+ in bottom chamber; GFP+ and JEDI in top chamber / mCherry+ and JEDI in bottom chamber). C, Normalized cell counts of WT (white), B2m−/− (gray), or Fas−/− (black) mCherry+ cells when co-cultured with WT (left panel) or B2m−/− (right panel) GFP+ cells in the presence of JEDI T cells; boxplots presented as minimum to maximum; two-way ANOVA with Sidak correction for multiple comparisons, ****p<0.0001 (n=4). D, Growth of mixed tumors with 50% WT GFP+ cells and 50% WT mCherry+ (black) or Fas−/− mCherry+ (red) cells in Rag1−/− mice adoptively transferred with tetramer-sorted GFP-specific CD8 T cells (arrow); percent change from maximum (day 6) to minimum (day 13) of each curve indicated; error bars presented as mean ± SEM; two-way ANOVA with Sidak correction for multiple comparisons, *p<0.05, **p<0.01 (n=9 mice/group; pooled from 2 independent experiments). Individual tumor curves in Supplemental Fig. S3D. E, Tiled confocal IF of tumors from mice bearing mixed primary GFP+/mCherry+ tumors (right column) and distant secondary mCherry+ tumors (left column) with either WT (top row) or Fas−/− (bottom row) mCherry+ cells; tumors cryopreserved on days 6–8 after adoptive transfer of tetramer-sorted GFP-specific CD8 T cells (Supplementary Fig. S3F). F–G, Higher power images of primary GFP+/mCherry+ tumors with either WT (F) or Fas−/− (G) mCherry+ cells from E (right column) with insets showing magnified single channel images of area outlined in yellow; arrows point to areas of active caspase 8 staining (bottom insets) and corresponding areas of mCherry staining (top insets). H, Quantification of percent area of mCherry staining that is colocalized with active caspase 8 staining from 37–39 fields of view as shown in F and G; unpaired two-tailed t test, ****p<0.0001 (n=3–5 mice/group, pooled from 3 independent experiments).
To assess off-target killing in vivo, we inoculated mixed tumors of GFP(WT) cells with either mCherry(WT) or mCherry(Fas−/−) cells in Rag1−/− mice lacking endogenous T cells, followed by adoptive transfer of GFP-specific T cells. Although tumors in both cohorts comprised equal numbers of Ag+ target cells, tumors with Fas−/− bystander cells demonstrated a blunted response to the therapy (Fig. 2D, 50.6% vs. 71.2% peak reduction in tumor volume; individual curves in Supplementary Fig. S3D) and quicker recurrence, confirming that Ag-specific T cells were capable of exerting fas-dependent bystander tumor clearance in vivo. Depletion studies ruled out a role for natural killer cells in this phenomenon (Supplementary Fig. S3E).
We next used a two-tumor model to determine whether this bystander effect was systemic or required the presence of local Ag+ cells like in our in vitro transwell experiments. In addition to the same mixed GFP+/mCherry+ tumors used in the prior model, which we now call the primary tumor (Fig. 2E, right flank on schema), we also inoculated a distant tumor on the opposite side (Fig. 2E, left flank on schema) comprising only mCherry+ cells of the corresponding WT or Fas−/−genotype. Lack of regression in these Ag− distant tumors, even while the primary tumors were rapidly shrinking in response to GFP-specific T cell transfer, demonstrated that bystander cytotoxicity was geographically restricted and dependent on co-localized Ag− and Ag+ tumor cells (Supplementary Fig. S3F). Imaging over a large area exhibited a corresponding lack of caspase 8-mediated tumor cell death in these distant tumors (Fig. 2E, left side; Supplementary Fig. S3G and H). Immunofluorescence of the primary tumors from the same mice, however, demonstrated widespread active caspase 8 activity in response to the transferred T cells (Fig. 2E, right side). Higher-power images of the mixed primary tumors confirmed colocalization of active caspase 8 with mCherry(WT) cells (Fig. 2F) while mCherry(Fas−/−) cells appeared completely protected despite being surrounded by Ag+ tumor undergoing on-target killing (Fig. 2G). Quantification of the staining revealed that more than four times as many mCherry(WT) tumor cells were undergoing apoptosis compared to mCherry(Fas−/−) cells at days 6–8 when the tumors had just begun regressing (Fig. 2H). This in situ evidence of mCherry+ bystander cell apoptosis correlates with and reasonably accounts for the fas-dependent differences in macroscopic tumor responses to T cell transfer (Fig. 2D).
Bystander killing can be potentiated.
These data introduce the idea that Ag-loss variants of tumor cells can be targeted based on their geographic proximity to other Ag+ tumor cells in vivo. We hypothesized that the efficacy of immunotherapies like checkpoint inhibitors may at least partially be due to unappreciated bystander effects. As expected, PD-1 blockade enhanced on-target apoptosis of GFP+ cells in our model (Fig. 3A, left panel), but surprisingly also nearly doubled bystander apoptosis of mCherry+ cells (Fig. 3A, right panel). This increased bystander cell death was perhaps partly due to the significant upregulation of fas protein levels on the cell surface (Fig. 3B, top panel, column 2). In contrast, neutralization of interferon gamma (IFNγ) blunted fas expression relative to the untreated condition (Fig. 3B, top panel, column 3) and decreased bystander apoptosis nearly 2-fold (Fig. 3A, right panel). These results suggest that levels of fas may be contributing to sensitivity to off-target killing, consistent with its role in other settings of fas-mediated apoptosis (39,40). We speculate that fasL expression on the T cells is also being modulated in these conditions and may be contributing to the effects as well, given the prominent roles of the PD-1 pathway and the cytokine IFNγ on CD8 T cell activation. All bystander apoptosis was nullified by treatment with a caspase 8 inhibitor or with mCherry(Fas−/−) cells, confirming that the observed killing was mediated by extrinsic apoptotic pathway machinery e.g. fas/fasL (Fig. 3A).
Figure 3. Cancer cells can be sensitized to on-target and bystander killing by induced upregulation of Fas or inhibition of downstream regulators.
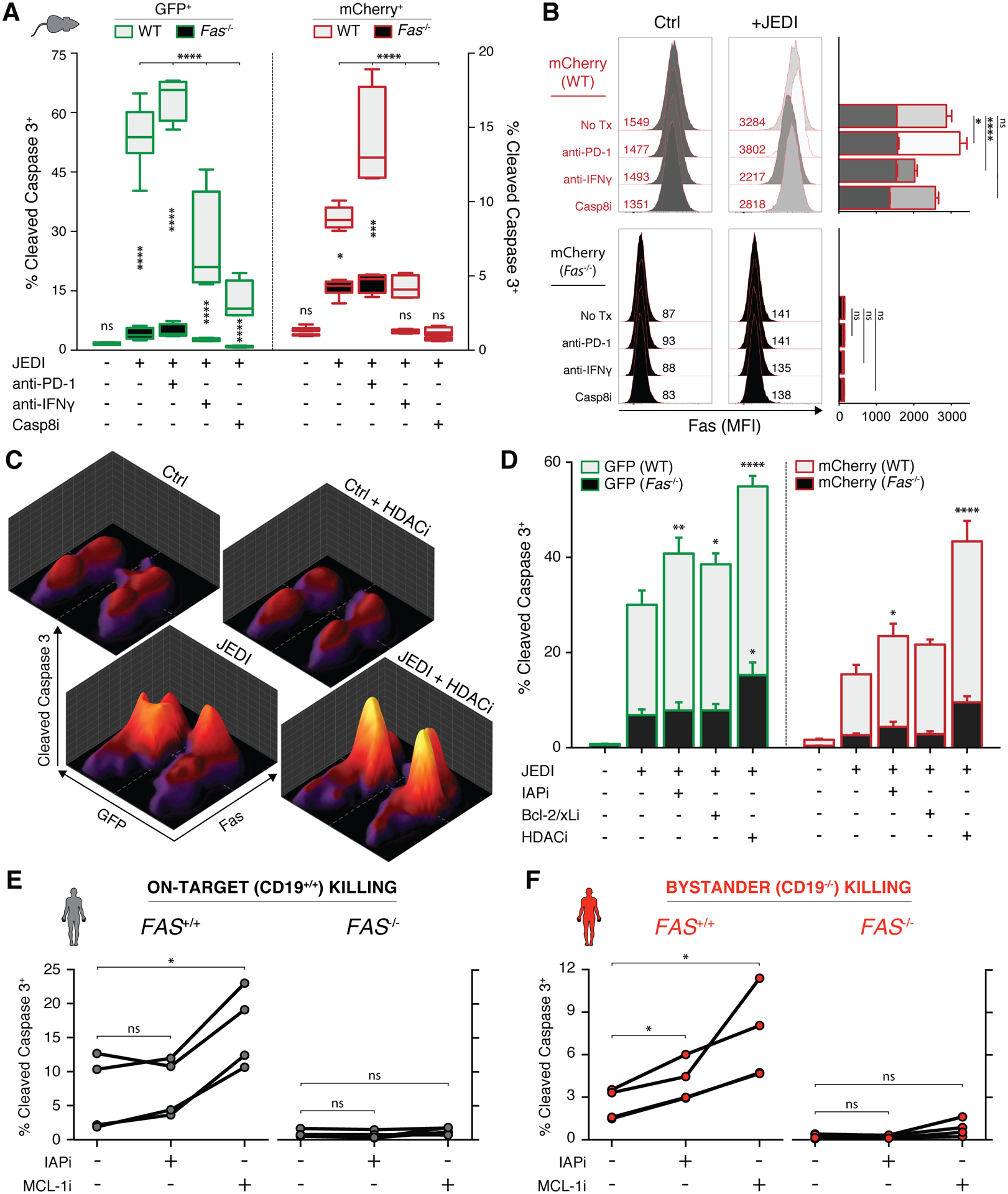
A, Percent of dying WT (gray) or Fas−/− (black) GFP+ (green, left panel) and mCherry+ (red, right panel) cells after 4 day co-culture with JEDI T cells, 100 μg/mL anti-PD-1, 100 μg/mL anti-IFNγ, and/or 25 μM caspase 8 inhibitor Z-IETD-FMK; boxplots presented as minimum to maximum; two-way ANOVA with Sidak correction for multiple comparisons, *p<0.05, ***p<0.001, ****p<0.0001 (n=4). B, Left panel: representative histograms and mean fluorescent intensities of surface fas expression of mCherry+ cells; right panel: quantification of replicates with error bars presented as mean ± SEM; two-way ANOVA with Sidak correction for multiple comparisons with indicated statistics for ‘+JEDI’ conditions only, *p<0.05, ****p<0.0001 (n=4). C, Representative cleaved caspase 3 expression (z-axis) simultaneously measured in 4 populations of target cancer cells (GFP+/− and fas+/−) when co-cultured with JEDI and/or 1 nM panobinostat (HDACi). D, Percent of dying WT (gray) or Fas−/− (black) GFP+ (green, left panel) and mCherry+ (red, right panel) cells after 2 day co-culture with JEDI T cells, 1 μM birinapant (IAPi), 100 nM ABT-737 (Bcl-2/xLi), and/or 1 nM panobinostat (HDACi); error bars presented as mean ± SEM; two-way ANOVA with Sidak correction for multiple comparisons with indicated statistics relative to second column of each panel, *p<0.05, **p<0.01, ****p<0.0001 (n=3). E–F, Percent of dying CD19+/+ (E) and CD19−/− (F) target Raji cells after co-culture with purified human CD8 T cells in the presence of 100 pM blinatumomab and treated with 100 nM birinapant (IAPi) or 10 nM S63845 (MCL-1i); matched data points from each healthy donor connected by lines; repeated measure one-way ANOVA with Holm-Sidak correction for multiple comparisons, *p<0.05 (n=4).
A critical difference between granzyme/perforin- and death receptor-mediated apoptosis is the well-characterized signaling of the latter and thus the potential to target downstream pathways (41). We therefore screened select families of proteins with known modulatory activity within the fas pathway and identified small molecule inhibitors that augmented fas-mediated cell death in our system (Supplementary Fig. S4A and B). We hypothesized that these drugs may sensitize cancer cells to on-target and bystander T cell killing. Using co-cultures of Ag+ or Ag− and WT or Fas−/− lymphoma cells, treatment with the histone deacetylase inhibitor (HDACi) panobinostat induced a significant increase in tumoral fas levels (Supplementary Fig. S4C). Concurrently, we saw increased caspase 3 cleavage in both Ag+ and Ag− populations when cultured with Ag-specific T cells, while Fas−/− populations in the same culture did not respond (Fig. 3C and D). Fas-dependent potentiation of on-target and bystander apoptosis was similarly observed after treatment with the inhibitor of apoptosis proteins antagonist (IAPi) birinapant and Bcl-2/xL inhibitor ABT-737 (Fig. 3D; Supplementary Fig. S4D). To ensure that various metrics support the same conclusions, we confirmed similar results with a different viability assay (Supplementary Fig. S4E). For example, 1nM panobinostat had negligible effect (<1%) on A20-GFP (columns 1 and 4) and A20-mCherry (columns 9 and 12) viability at baseline, but potentiated JEDI-induced on-target killing by 31% (columns 5 and 8) and bystander killing by 32% (columns 13 and 16). Similar effects which appeared greater-than-additive were seen with inhibitors of IAP and Bcl-2/xL (Supplementary Fig. S4E). Importantly, all compounds had little or no effect on the viability of the T cells in the same co-cultures (Supplementary Fig. S4F), suggesting lack of fas-mediated potentiation of T cell fratricide (42) with these combination therapies.
Bystander killing is critical to immunotherapies.
To translate these concepts to clinically relevant T cell therapies, we obtained healthy donor human T cells and measured cytotoxicity mediated by the CD3/CD19 bispecific T cell engager blinatumomab against on-target CD19+/+ and bystander CD19−/− Raji lymphoma cells (Supplementary Fig. S5A). As in the murine setting, we again observed potentiation of fas-dependent killing of both on-target (Fig. 3E) and bystander (Fig. 3F) cells using inhibitors to IAP and the Bcl-2 family member Mcl-1 without inducing any toxicity against the co-cultured T cells (Supplementary Fig. S5B and C).
To assess the relevance of these findings to CAR-T cells, which utilize activation signaling distinct from endogenous T cells that may result in different effector functionality, we tested a murine CD19-targeting CD3ζ-CD28 CAR construct (43) transduced into WT syngeneic CD8 T cells. Using co-cultures of CD19+ or CD19− and WT or Fas−/− lymphoma cells, we again observed resistance of Ag+ target cells lacking surface fas to CAR-T killing (Fig. 4A). Quantification of cell counts demonstrated that while fas-mediated cytotoxicity played a moderate role in CAR-T on-target killing (Fig. 4B, black/gray), it contributed to nearly all measured bystander killing (Fig. 4B, red/pink). To model in vivo therapy of Ag-loss escape tumors, we inoculated homogeneous (100% CD19+) or heterogeneous (95% CD19+/5% CD19−) tumors prior to CAR-T therapy. While untreated mice died similarly rapidly with both homogeneous and heterogenous tumors (median 24–25 days), CAR-T therapy prolonged survival of the homogeneous tumor cohort (median 42 days) and similarly prolonged survival of the heterogenous tumor cohort (median 41 days), suggesting some protective effect of anti-CD19 CAR-T cells against CD19− tumor (Fig. 4C, solid lines). Systemic fasL blockade severely impaired CAR-T efficacy, reaffirming the fas-fasL dependence of on-target CAR-T cytotoxicity (Fig. 4C, dashed lines). Most notably, however, with fasL blockade the heterogeneous tumor cohort had significantly worse survival than the homogeneous tumor cohort (Fig. 4C, highlighted dashed lines; median 30.5 vs. 34 days). These findings suggest that fas-fasL signaling is critical for CAR-T bystander cytotoxicity against Ag-loss variants implicated in disease relapse.
Figure 4. Fas-mediated on-target and bystander killing are critical for the efficacy of T cell immunotherapies.
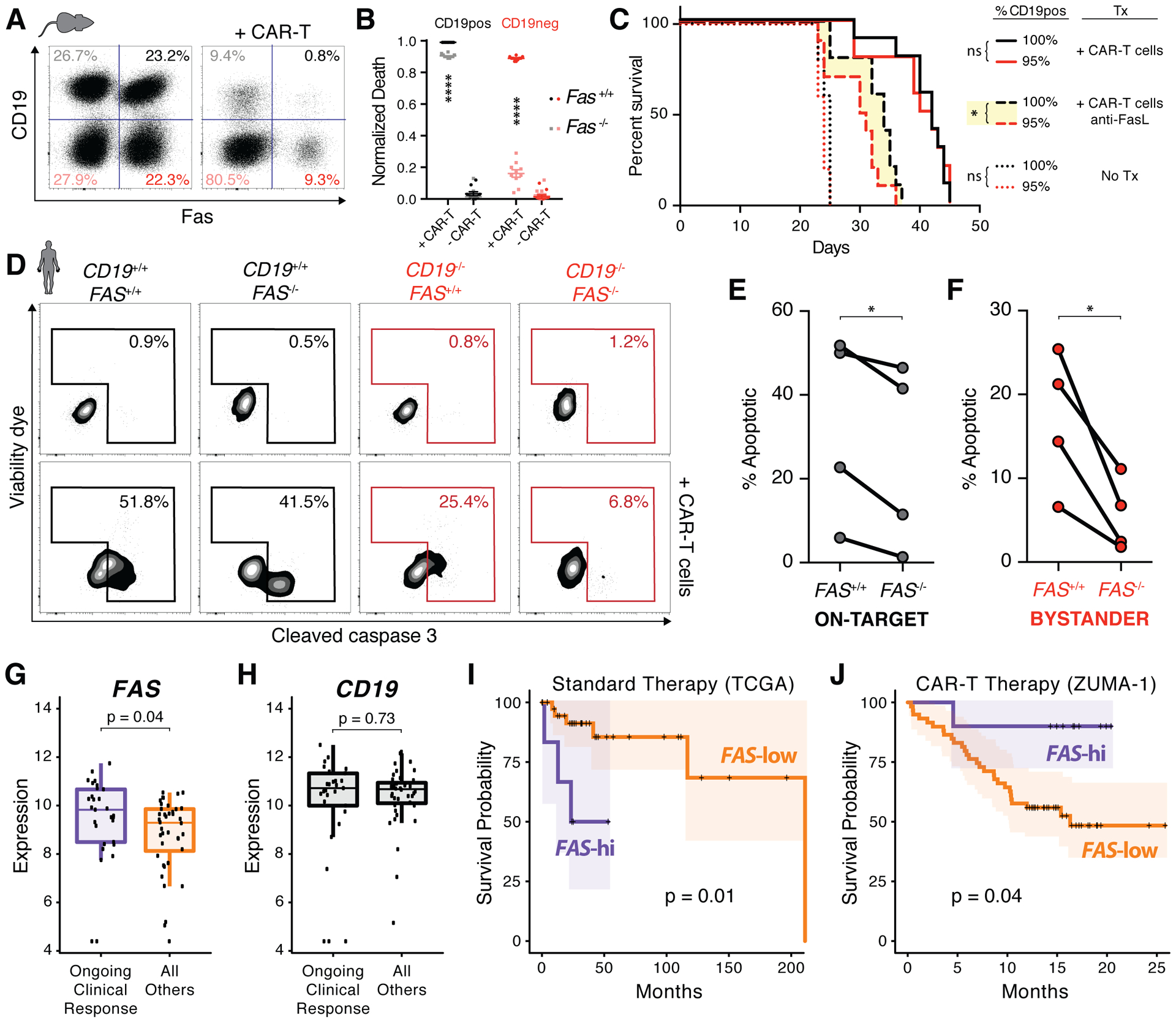
A, Representative changes in 4 populations of target cells (CD19+/− and fas+/−) after co-culture with balb/c CD8 T cells retrovirally transduced with a CD19-targeting CAR construct. B, Fraction of dying cells within each population from (A) based on normalized cell counts; two-way ANOVA with Sidak correction for multiple comparisons, ****p<0.0001 (n=10). C, Survival curves of mice bearing homogeneous (black) or heterogenous (red) tumor with no therapy (dotted), CAR-T therapy (solid), and CAR-T therapy with FasL antibody blockade (dashed); Gehan-Breslow-Wilcoxon test, *p<0.05 (n=5–10 mice/group). D, Percent of dying Ag+ on-target (black) and Ag− bystander (red) Raji cells when co-cultured with human CD19-targeting CAR-T cell product for 6 hours. E–F, Quantification of on-target (E) and bystander (F) apoptosis from D; matched data points from each healthy donor CAR-T cell product connected by lines; paired t-test, *p<0.05 (n=4). G–H, RNA expression (variance stabilizing transformation (VST) normalized count) of FAS (G) and CD19 (H) in pre-treatment tumor samples with ongoing clinical response (n=29) and all others (n=40) from ZUMA-1 CAR-T trial; Wilcoxon test. I–J, Kaplan-Meier survival curves with 95% confidence interval shading of TCGA DLBCL data (I; n=48, median follow-up 32 months) and ZUMA-1 data (J; n=69, median follow-up 16 months); top 14–15% stratified into FAS-hi group by maxstat package described in the methods; log-rank test.
To evaluate the clinical relevance of these findings, we tested anti-human CD19 CD3ζ-CD28 CAR-T cell products (44) made from multiple healthy donors. Consistent with the murine data, these CAR-T cells induced cytotoxicity against both CD19+/+ and CD19−/− Raji cells when co-cultured (Fig. 4D). Quantifying these effects reaffirmed the murine results: CAR-T on-target killing was moderately fas-dependent (Fig. 4E), but CAR-T bystander killing was far more sensitive to FAS expression (Fig. 4F). Similar to the bispecific antibody results, CAR-T bystander killing against Raji could also be potentiated by inhibition of Mcl-1 or IAP (Supplementary Fig. S5D). Importantly, we demonstrated that Mcl-1 inhibition can synergistically enhance both CAR-T-mediated and blinatumomab-mediated killing of primary malignant cells from two different CLL patients (Supplementary Fig. S5E), further strengthening the rationale of such combination strategies in the clinic.
To assess the impact of these concepts in patients, we analyzed pre-treatment tumoral RNA sequencing data in a subset of patients from the ZUMA-1 trial for refractory diffuse large B-cell lymphoma (DLBCL), which utilized the same CAR-T construct (3). We found that patients experiencing durable clinical responses had significantly elevated tumoral FAS expression compared to all others (Fig. 4G) despite the fact that the two cohorts had comparable tumor CD19 expression (Fig. 4H). Additionally, data from the TCGA DLBCL cohort undergoing standard therapies demonstrated that patients with high tumor FAS expression had significantly worse survival outcomes (Fig. 4I), consistent with pro-tumorigenic fas signaling in low membrane-fasL environments (45,46). By contrast, patients with high FAS expression receiving CAR-T therapy had the opposite correlation: a significantly prolonged survival relative to those with lower expression (Fig. 4J). These data provide evidence for the critical role of the fas pathway in the efficacy of T cell-based immunotherapies and perhaps even in the context of highly variable target Ag expression (Fig. 4H).
Discussion
Altogether, we present here that the fas-fasL pathway may be an undervalued effector mechanism of Ag-specific cytotoxicity and an underrecognized mechanism of bystander cytotoxicity in T cell-based immunotherapies. To our knowledge, this is the first report using in vivo models to implicate a role for the fas-fasL pathway in the clearance of Ag− tumor cells as well as clinical data to show the impact of tumoral FAS expression on survival after CAR-T therapy.
Prior in vitro work has suggested that T cells may exert non-canonical, indiscriminate, multi-directional killing (47–50) and that fasL-mediated cytotoxicity can be immune synapse independent (51). However, because not all in vitro observations translate (52), our in vivo data is critical evidence that tumoral factors can mediate bystander killing. Earlier findings of the importance of fas have been downplayed by acceptance that the perforin-granzyme pathway is the dominant mechanism for killing transformed cells (35). We believe that our in vivo data validate these decades-old in vitro observations, and the mechanisms proposed here have broad implications for any Ag-directed immunotherapy against heterogeneous tumors.
Target Ag loss is a primary mechanism of immune escape after CAR-T therapy, with 9–25% of patients in trials demonstrating Ag modulation (12). The primary strategy being pursued to treat Ag− relapses is targeting of alternative tumor Ag; a CAR-T product targeting CD22 has already shown promise for cancers resistant to CD19-targeting CARs (53). A primary limitation of this approach is that alternative, homogeneously expressed, highly tumor-specific surface Ag cannot be identified for most tumors. Therefore, Ag-agnostic approaches to preventing relapses are preferable to sequential Ag-based therapies.
Downplayed in the interpretation of CD19-CAR-T trial data is the heterogeneity of tumors and the lack of correlation between treatment efficacy and tumoral target expression (3,27). Our analysis of tumoral transcriptomic data from ZUMA-1 indicate that FAS expression predicts long-term outcomes of CAR-T treated patients, even though target Ag expression does not. Accordingly, our murine CAR-T model shows that Ag heterogenous and homogeneous tumors respond similarly only if fas signaling is intact. We therefore propose that fas-mediated bystander elimination of Ag-loss variants may already be occurring in CAR-T treated patients. This mechanism lays the groundwork for a novel therapeutic approach to improving Ag-directed therapies: targeting the proximity of tumor cells to each other rather than targeting new Ag. Future studies will benefit by quantifying the distribution and spatial relationship of tumoral fas and Ag expression with infiltrating Ag-specific T cells.
In vivo demonstration of tumorally-mediated bystander killing is novel. Previous studies closely measuring in vivo T cell killing with longitudinal imaging (54,55) did not observe bystander killing, possibly due to the use of fasL-insensitive tumor models (56). Other reports were able to demonstrate indirect clearance of Ag-loss variants by killing Ag+ cross-presenting tumor stroma (57,58), but these studies did not demonstrate any direct contact-mediated effects by T cells against tumor nor any potentiation of this bystander killing.
We show that surface fas expression is upregulated by IFNγ exposure, and together with recent studies demonstrating that T cell-secreted IFNγ affects tumor cells hundreds of microns away (59,60), our data suggest that this might augment subsequent fas/fasL-mediated bystander killing. Additionally, we demonstrate that known sensitizers of fas-mediated cell death, e.g. HDAC inhibitors (61) and SMAC mimetics (62), can enhance T cell-mediated killing, independent of Ag expression. We propose that investigating combinations of these clinical-stage small molecule fas signaling modulators with T cell therapies should be a focus of future research. These approaches may prevent – rather than treat – cancer relapse due to antigen escape by targeting both tumor cell antigens as well as tumor cell geography.
Methods
Ethical Compliance.
Protocols for the treatment of patients, as well as human sample collection and analysis, were approved by the Mount Sinai Institutional Review Board, and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. All experiments including human specimens were performed in compliance with the relevant ethical regulations.
Animals.
Balb/c and balb/c(Rag1−/−) were purchased from Jackson labs and housed at the animal facility of the Icahn School of Medicine at Mount Sinai. JEDI mice (29) were backcrossed onto the balb/c background for eight generations in our facility. All experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai.
Cell lines.
All cell lines (A20, 4T1, and Raji) were maintained at 37 °C with 5% CO2 in RPMI supplemented with 10% heat-inactivated FCS, penicillin/streptomycin and 50 μM beta-mercaptoethanol. Cell lines with stable expression of Cas9 were generated by transducing with a lentivirus encoding Cas9 (lentiCRISPRv2; Addgene plasmid #52961) and selecting for puromycin resistance. Single gene knockout lines were generated by cloning in the specific sgRNA (Supplementary Table 1; Fas 5’-GGCGTCCCAAAGCTTACCAG-3’) as previously described (63) prior to transduction and FACS purification. To control for clonal heterogeneity, 1e6 cells from the parent line were transduced with a multiplicity of infection (MOI) of 10. After 5 days of growth, 1e6 cells negative for surface fas expression were purified by FACS and propagated as the knockout line. Expression of Cas9 was confirmed by western blot (anti-Cas9, clone 7A9, Millipore). A20-Cas9 cells were then transduced with GFP or mCherry lentiviral vectors at 10 MOI, and stable GFP+ or mCherry+ cells (~10%) were purified by FACS 1 week post-transduction. Cells for library screening were maintained in puromycin until 1 day before transduction with libraries. A20(CD19+) and A20(CD19−) lines were generated by FACS purification. 4T1-GFP and 4T1-mCherry cell lines were a gift from the Brown lab (Mount Sinai). Raji cells were a gift from the Dominguez-Sola lab (Mount Sinai). Cell line authentication was not performed, but mycoplasma testing by PCR was performed annually. Cell lines were used for experiments within 2 weeks of thawing from frozen stocks.
Library design.
291 target genes for screening were manually curated and selected based on expression levels >1 RPKM in A20 lymphoma cells (accession ENCSR000CLV) and cross-referencing with the UniProt Knowledgebase (64) for the following annotations: secreted, membrane, immunity, inflammatory response, or cytokine. sgRNA sequences were obtained from previously described CRISPR libraries (65) and are listed in Supplementary Table 1. For increased sensitivity, targets genes were separated into 4 separate pools for independent screening.
Library preparation and lentiviral production.
The custom oligonucleotide libraries were reconstituted in water to a final concentration of 0.01 pmol/μl and PCR amplified using Q5 Hot Start Polymerase (New England Biolabs). Each PCR-amplified library was then purified using a PCR purification kit (Qiagen) following the manufacturer’s protocol. Subsequently, a restriction digest was performed using BbsI restriction enzyme (New England Biolabs) at 37°C overnight. The digested library was then purified by electrophoresis on a 2% agarose gel with 1× TBE running buffer and recovered using a gel extraction kit (Qiagen) following the manufacturer’s protocol. The library oligonucleotides were then cloned downstream of the human U6 promoter in a lentiviral vector that also contained a NGFR transgene downstream of the human PGK promoter. The vector backbone was digested with AgeI and EcoRI, treated with FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific), and purified on a 1% agarose gel and recovered using a gel extraction kit (Qiagen). Ligation was performed using Quick Ligase kit (New England Biolabs). To prevent loss of library diversity, colonies were collected from fifteen 10-cm plates after transformation of NEB 10-beta electrocompetent cells (New England Biolabs). The pool of plasmids was prepared for transfection using an endotoxin-free Maxi prep kit (Qiagen). Lentiviral vectors were produced as previously described (34). Briefly, 293T cells were seeded 24 hours before Ca3PO4 transfection with third-generation VSV-pseudotyped packaging plasmids and library transfer plasmids. Supernatants were then collected, passed through a 0.22-μm filter and purified by ultracentrifugation. Viral titer was estimated on 293T cells by limiting dilution.
Screening assay.
To ensure that a majority of transduced cells received only one vector and that there was proper library representation, 1e6 cells were transduced for each replicate at an MOI of 10 to ensure ~10% transduction efficiency. Transductions were done in a 12 well plate with a total volume of 500 μL in the presence of 5 μg/mL polybrene (Millipore). 5 separate transductions were done for each cell type (A20-Cas9-GFP and A20-Cas9-mCherry) and each library to generate replicates. 1 week after the transduction, cells that received and integrated vector were purified by flow sorting for NGFR+ cells. Purified cells were expanded in culture for 5 more days before cryopreservation and thawed 2 days prior to the screening assay.
20,000 A20-GFP-library+ cells, 20,000 A20-mCherry-library+ cells, and 40,000 preactivated JEDI T cells (pooled from 3 separate mice) were co-cultured per well in 96-well U-bottom plates. Each condition was done in 96 separate reactions (1 plate) and pooled together to ensure proper library representation. Each replicate consisted of 1 plate with (JEDI) and 1 plate without (Ctrl) T cells added into the culture. After 2 days, wells were pooled together and FACS purified for live NGFR+GFP+ cells and NGFR+mCherry+ cells. Purified cells were expanded in culture for 2 more days before cryopreservation for propagation into future iterations and preservation of a frozen pellet (2e6 cells) for DNA extraction.
Sequencing and screen analysis.
Library preparation was adapted from methods previously described (66). Briefly, genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen). The sgRNA sequence sites were amplified from genomic DNA as follows: in a 50-μl reaction, 25 μl Q5 High-Fidelity 2X Master Mix (NEB), 2.5 μl sense primer (5 μM), 2.5 μl antisense primer (5 μM), and 200 ng gDNA from the sample. Cycling parameters were 98°C for 30s; 28 cycles of 98°C for 10s, 61°C for 30s, 72°C for 25s; and 72°C for 2 min. The primers used to amplify the target sites contained sequences that hybridize directly to the Illumina flow cell. Barcodes were inserted immediately following the Illumina sequencing primer binding site to allow for multiplexing. PCR-amplified libraries were purified for a 250bp band on a 2% agarose gel using a gel extraction kit (Qiagen). Before sequencing, all purified library amplification products were analyzed on an Agilent 2100 Bioanalyzer. The prepared libraries were multiplexed and sequenced on the Illumina Hi-Seq2000.
Debarcoded raw read counts were normalized to the total number of mapped reads within each sample. For each condition, the median of 5 independent replicates within the GFP+ population was plotted against the paired mCherry+ population. A linear regression was performed on all 5,186 data points, and standardized residuals (Z-scores) of the observed percent reads in the GFP+ population for each target relative to the expected reads were calculated for determining significance of enrichment.
Preparation of JEDI T cells for use in vitro and in vivo.
Spleen and lymph nodes of JEDI mice were dissected, and single cell suspensions of leukocytes were obtained by mechanical disruption and filtering through a 70-mm cell strainer. Red blood cells were lysed using ACK buffer (Lonza), and CD8+ T cells were negatively selected using MagniSort Mouse CD8+ T Cell Enrichment Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. For in vitro experiments described in the text using JEDI, cells were activated for 2 days with 5 μg/mL plate-bound purified hamster anti-mouse CD3e (clone 145–2C11, BD Biosciences), 1 μg/mL purified hamster anti-mouse CD28 (clone 37.51, BD Biosciences), and 20 ng/mL recombinant mouse IL-2 (Gemini Bio-Products) in RPMI with 10% FBS, 100 U/ml penicillin/streptomycin/ampicillin, and 50 μM 2-mercaptoethanol. For in vitro experiments described in the text using GFP-specific T cells, cells were first FACS purified using H-2Kd-HYLSTQSAL tetramer reagent produced by the NIH Tetramer Core Facility prior to activation as described above. For all in vivo experiments, freshly isolated tetramer-sorted GFP-specific T cells were used without activation.
In vitro JEDI T cell killing assays.
3-cell co-culture cytotoxicity assays using A20 cancer cells were implemented with 2.5e4 A20-GFP + cells, 2.5e4 A20-mCherry+ cells, and 5e4 JEDI T cells (2:1 T:target cell ratio) unless otherwise noted and harvested for analysis by flow cytometry after 24 or 48 hours. For 5-cell co-culture experiments involving (WT), (Fas−/−), and/or (B2m−/−) target cell lines, a 2:1 T:target cell ratio was maintained unless otherwise noted. Assays using 4T1 cancer cells were similarly performed at 25:1 T:target cell ratio. When indicated, a homogeneous suspension of Precision Counting Beads (BioLegend) were uniformly added to each sample prior to analysis by flow cytometry. Normalized cell counts were calculated by dividing the number of event counts of a population of interest by the number of event counts of beads within the same sample.
Transwell assays.
3- and 4-cell co-culture cytotoxicity assays were performed as described above, except with the indicated populations seeded into a transwell insert separated by a permeable membrane with 1μM pores within a 24-well plate (ThermoFisher Scientific). After 48 hours, the cells from the top and bottom chambers were combined before analysis by flow cytometry as one sample.
In vitro superantigen-induced T cell killing assays.
CD4 and CD8 T cells were isolated from naïve balb/c mice using the MagniSort Mouse CD4+ or CD8+ T Cell Enrichment Kits (ThermoFisher Scientific) according to the manufacturer’s protocol. 5e4 purified T cells were co-cultured with 5e4 total A20 cells (a mixture of outlined genotypes) with 50 ng/mL staphylococcal enterotoxin B (Toxin Technology) for 48 hours before harvesting for analysis by flow cytometry.
Tumor induction and measurements.
3e6 total cells were injected in 100 μl HBSS subcutaneously on the flank. For two-tumor models, the same number of total cells was inoculated on the contralateral flank 10 days after the first tumor in order to delay lethality from the non-responding tumor during a critical time window of experimental observation of the responding tumor. Tumor size was determined via caliper measurements on the indicated days (length × width × height).
GFP-specific in vivo tumor models.
Balb/c(Rag1−/−) mice subcutaneously inoculated with homogeneous A20-GFP (WT or Fas−/−) tumors on the right flank were adoptively transferred with 5e3 GFP-specific T cells via tail vein injection on the indicated day. Balb/c(Rag1−/−) mice inoculated with mixed heterogeneous tumors, or two-tumor models, were adoptively transferred with 3e5 GFP-specific T cells via tail vein injection on the indicated day. Where indicated, depletion of natural killer cells was achieved by two intraperitoneal loading doses of 50 uL anti-asialo-GM1 (Wako) 6 and 2 days prior to T cell transfer, followed by 25 uL doses every 4 days. All tumors harvested for immunofluorescence were dissected on day 8 post-T cell transfer.
Immunofluorescence.
Whole tumors were dissected out, washed in PBS, and incubated in PLP buffer (0.05M phosphate buffer containing 0.1M L-lysine [pH 7.4], 2 mg/mL NaIO4, and 10 mg/mL paraformaldehyde) overnight at 4°C. Tissue was equilibrated sequentially in 10%, 20%, and 30% sucrose solutions for 2 hours each before embedding in OCT (ThermoFisher Scientific) and rapidly frozen on dry ice stored at −80°C. Slides with 10 um sections made with a cryostat were incubated in blocking buffer (2% FBS and 1% BSA in PBS) for 2 hours and then incubated with rat anti-GFP (FM264G, AlexaFluor 488, BioLegend), chicken anti-mCherry (ab205402, Abcam), rat anti-CD8 (53–6.7, AlexaFluor 647, BioLegend), rabbit anti-cleaved caspase 8 (polyclonal, Novus Biologicals), and/or rat anti-B220 (RA3–6B2, BV421, BioLegend) in 0.1X blocking buffer overnight. Slides were washed with PBS-Tween (0.1%) and incubated for 1 hour with donkey anti-rabbit (Poly4064, AlexaFluor 594 or 647, BioLegend) and/or goat anti-chicken (A32759, AlexaFluor 594, ThermoFisher Scientific) secondary antibodies where indicated. ProLong Gold antifade (ThermoFisher Scientific) was used as a mounting reagent, and 16-bit images were acquired on a Zeiss LSM780 confocal microscope. Images were analyzed using FIJI (67). For quantification, the mean intensity of the top 10% pixels of each channel were calculated on a standardized scale to remove background signal. For pixel colocalization analysis, Otsu and RenyiEntropy auto thresholding algorithms were applied before the Image Calculator function was used to perform logical operations on the binary images created from each channel. A total of at least 22–30 fields of view from 3–4 mice pooled from 3 independent experiments were included in the analysis.
Inhibition and potentiation of apoptosis.
The following concentrations of agents were added to co-cultures of T cells with target cells when indicated, except when specified differently: 100 ug/mL anti-PD-1 (RMP1–14, BioXcell), 100 ug/mL anti-IFNγ (XMG1.2, BioXcell), 25 uM Z-IETD-FMK (Selleck), 1 uM birinapant (APExBIO Technology), 100 nM ABT-737 (Selleck), 1 nM panobinostat (Selleck), and 10 nM S63845 (Selleck). Other agents used during screening include: venetoclax (Selleck), entinostat (Selleck), and fasentin (Sigma Aldrich). Fas-mediated apoptosis during compound screening was induced by agonistic antibodies to mouse (Jo2, BD Biosciences; 10 ng/mL) or human (CH11, Millipore; 100 ng/mL) CD95.
CD3/CD19 bispecific T cell engager assays.
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donor blood by density gradient centrifugation using Ficoll Paque Plus (GE Healthcare), followed by red blood cell lysis using ACK buffer (Lonza). CD8+ T cells were negatively selected using MojoSort Human CD8 T cell Isolation Kit (BioLegend) according to manufacturer’s protocol. CD8+ T cells were subsequently activated and expanded for 48 hours with 500U/mL IL-2 (R&D Systems) in RPMI with 10% FBS, 100 U/ml penicillin/streptomycin/ampicillin, and 50 μM 2-mercaptoethanol. 4e4 pre-activated CD8+ T cells were co-cultured with 1e4 each of CD19+/+FAS+/+, CD19−/−FAS+/+, CD19+/+FAS−/−, and CD19−/−FAS−/− Raji cells in the presence of 500U/mL IL-2 and 100 pM blinatumomab for 72 hours before analysis by flow cytometry. All Raji cells were labeled with CellTrace CFSE (ThermoFisher Scientific) and all CD19+/+ Raji cells were also labeled with CellTrace Violet (ThermoFisher Scientific) according to manufacturer’s protocol prior to seeding for gating purposes.
Generation of mouse CD19-targeting CAR-T cells.
Retroviral vector and CAR-T cells using a previously published anti-mouse CD19 CD3ζ-CD28 CAR construct (43) was generated as previously described (68) with the following adaptations. Briefly, leukocytes were isolated from spleens and lymph nodes of balb/c mice. Single cell suspensions were obtained by mechanical disruption and filtering through a 70 nm cell strainer. Red blood cells were lysed using ACK buffer (Lonza), and CD8+ T cells were negatively selected using MagniSort Mouse CD8+ T Cell Enrichment Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. Cells were activated in 6 well plates (1.5e6 cells per well) with 5 μg/mL plate-bound purified hamster anti-mouse CD3e (clone 145–2C11, BD Biosciences), 1 μg/mL purified hamster anti-mouse CD28 (clone 37.51, BD Biosciences), and 20 ng/mL recombinant mouse IL-2 (Gemini Bio-Products) in RPMI with 10% FBS, 100 U/ml penicillin/streptomycin/ampicillin, and 50 μM 2-mercaptoethanol. After one day, 1 mL/well of viral supernatant (thawed from stocks frozen at −80°C) was added to 6 well plates pre-coated with 15 μg/mL RetroNectin (Takara), and 2e6 cells in 1 mL complete media (supplemented with 80 ng/mL IL-2) were added on top. The plates were centrifuged at 2,000g at 30°C for 1 hour before returning to the incubator (1st spinoculation). The next day, 1 mL of media was carefully aspirated out and replaced with 1 mL new viral supernatant before centrifuging at 2,000g at 30°C for 1 hour again and returning to the incubator (2nd spinoculation). Cells were then expanded while maintaining at 1–2e6/mL in media supplemented with 20 ng/mL IL-2 for 4 more days before cryopreservation.
In vitro mouse CAR-T killing assay.
6e4 thawed mouse CAR-T cells were co-cultured with 1.5e4 each of [CD19+ or CD19−] and [Fas+ or Fas−] A20 cells for 72 hours before analysis by flow cytometry. A homogeneous suspension of Precision Counting Beads (BioLegend) were uniformly added to each sample prior to analysis by flow cytometry. Normalized cell counts were calculated by dividing the number of event counts of a population of interest by the number of event counts of beads within the same sample, and normalized death was calculated by calculating the percentage of reduction in counts compared to the control sample.
In vivo mouse CAR-T killing assay.
Balb/c mice were inoculated with homogeneous (100% A20 CD19+ cells) or heterogeneous (95% A20 CD19+/5% A20 CD19− cells) tumors immediately after 5 Gy total body irradiation (TBI). 2.5e6 CAR-T cells were adoptively transferred by tail vein injection 4 days post-TBI. For FasL blockade, 250 μg/mouse InVivoMab anti-mouse FasL (MFL3, BioXcell) was injected intraperitoneally every 3 days starting on the day of adoptive T cell transfer, except for the first loading dose of 500 μg/mouse. Mice were tracked for survival every day and closely monitored for any signs of systemic disease (e.g. leg paralysis), for which they were euthanized.
In vitro human CAR-T killing assay.
Human CAR-T cell products were manufactured as described previously (44) and cryopreserved. 1e6 thawed CAR-T cells were co-cultured with 5e4 each of CD19+/+FAS+/+, CD19−/−FAS+/+, CD19+/+FAS−/−, and CD19−/−FAS−/− Raji cells for 6 hours (unless otherwise indicated) before analysis by flow cytometry. All Raji cells were labeled with CellTrace CFSE (ThermoFisher Scientific) and all CD19+/+ Raji cells were also labeled with CellTrace Violet (ThermoFisher Scientific) according to manufacturer’s protocol prior to seeding for gating purposes.
In vitro killing assays using primary CLL cells.
Patient PBMCs were collected and cryopreserved. 2e5 thawed patient cells were co-cultured with 8e5 human CAR-T cells or 8e5 healthy donor CD8 T cells in the presence of 1 nM blinatumomab for 2 days before analysis by flow cytometry. Patient cells were labeled with CellTrace Violet (ThermoFisher Scientific) according to manufacturer’s protocol prior to seeding for gating purposes, and CD20+ cells were used for analysis of effects on malignant cells only.
Flow cytometry and cell sorting.
Viability staining was performed in HBSS using fixable viability stain 450 or 780 (BD Biosciences) at 1:1000 for 5 min at room temperature (RT). Surface staining of mouse leukocytes was performed in FACS blocking buffer (made in house) using monoclonal antibodies against B220 (RA3–6B2, BioLegend), CD8 (53–6.7, BioLegend), CD4 (RM4–5, BioLegend), CD19 (6D5, BioLegend), CD45.1 (A20, BioLegend), CD45.2 (104, BioLegend), CD95 (SA367H8, BioLegend), CD107a (1D4B, BioLegend), H-2Kd (SF1–1.1, BioLegend), H-2Kd-HYLSTQSAL tetramer (NIH tetramer core facility), PD-L1 (10 F.9G2, BioLegend), NGFR (C40–1457, BD Biosciences), and TCRb (H57–597, BioLegend). Surface staining of human leukocytes was performed in FACS blocking buffer (made in house) using monoclonal antibodies against CD3 (HIT3a, BD Biosciences), CD4 (RPA-T4, BD Biosciences), CD8 (RPA-T8, BD Biosciences), CD19 (HIB19, BioLegend), CD20 (2H7, BioLegend), and CD95 (DX2, BioLegend). All surface antibodies were used at a dilution of 1:400, and samples were incubated with antibodies for 15 min at RT in the dark. Hoechst 33258 (BD Biosciences), fixable viability stains (BD Biosciences), Annexin V (BioLegend), and/or 7-AAD (BioLegend) were added according to manufacturer’s protocols to analyze dead cells. For intracellular staining, surface-stained cells were fixed and permeabilized using commercial buffer sets (Invitrogen), then stained with anti-cleaved caspase 3 (C92–605.1, BD Biosciences), anti-Granzyme B (NGZB, ThermoFisher Scientific), or anti-IFNγ (XMG1.2, BioLegend) at 1:200 dilution for 30 min. Samples were acquired using a LSR-Fortessa (BD Biosciences) or Attune (ThermoFisher Scientific) and data analyzed with Cytobank. Cell sorting was performed on a FACSAria (BD Biosciences).
Analysis of RNA sequencing data.
TCGA gene expression (z-scores of RSEM RNA-seq V2) and survival data were downloaded from cBioPortal (69). For ZUMA-1 data, the paired-end reads were aligned to the Genome Reference Consortium Human Build 38 using STAR aligner (70). Gene counts for each sample were generated using the featureCounts function in the R/Bioconductor package “Rsubread” (71). The R/Bioconductor package “DESeq2” was used to apply the variance stabilizing transformation (VST) normalization on the count data (72). When multiple RNA-seq samples were available for one patient, the VST normalized expression counts were averaged. The VST normalized counts were used for comparing the expression distribution of FAS and CD19 genes among the patients with ongoing treatments vs. others using the Wilcoxon test. The boxplots were generated using the R package “ggpubr”. Expression thresholds for comparing survival outcomes were selected by implementing the “surv_cutpoint” function from the “maxstat” R package (73).
Statistical analyses.
Data analysis was performed using GraphPad Prism6. Unpaired two-tailed Student’s t test was used to compare two independent groups; one-way ANOVA with Sidak correction for multiple comparisons was used to compare multiple (>2) groups with one independent variable; two-way ANOVA with Sidak correction for multiple comparisons was used to compare multiple (>2) groups with two independent variables; one-way ANOVA with Holm-Sidak correction for multiple comparisons was used to compare multiple (>2) groups with matched data points. P values > 0.05 were considered statistically non-significant (ns).
Data and materials availability.
Human chimeric antigen receptor T cell products, clinical correlates data, and RNA sequencing data from ZUMA-1 used in this report are courtesy of Kite Pharma. Material requests should be directed to Kite Pharma. Results shown are in part based on data generated by the TCGA Research Network (www.cancer.gov/tcga). Otherwise, all other data supporting the findings presented in this study are available from the corresponding author upon reasonable request.
Supplementary Material
Significance.
This study demonstrates the first report of in vivo fas-dependent bystander killing of antigen-negative tumor by T cells, a phenomenon that may be contributing to the high response rates of antigen-directed immunotherapies despite tumoral heterogeneity. Small molecules that target the fas pathway may potentiate this mechanism to prevent cancer relapse.
Acknowledgments
We thank the flow cytometry core facility, microscopy core facility, and the CCMS animal facility at ISMMS. We also thank S. Hekmaty, G. Panda, and R. Sachidanandam for assistance with sequencing, J. Javier Bravo-Cordero for assistance with image analysis, and A. Kamphorst for critical review of the manuscript. Research reported in this article was supported by NIH P30CA196521; RU was supported by NIH 5T32GM007280 and 5T32AI007605; MM was supported by NIH R01CA154947 and R01CA190400; BDB was supported by the Cancer Research Institute (CRI), NIH R01AT011326, R01AI113221, and R33CA223947; BDB and MM were supported by NIH U19AI128949; JDB was supported by the Damon Runyon Cancer Research Foundation, Merck Investigator Studies Program, Cancer Research Institute (CRI), and NIH R37CA246239.
Footnotes
Conflicts of Interest: NSc, ABo, and JR are employees of Kite Pharma. All other authors declare no competing interests.
References
- 1.Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med 2018;378:449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in children and young adults with B-Cell lymphoblastic leukemia. N Engl J Med 2018;378:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-Cell therapy in refractory large B-Cell lymphoma. N Engl J Med 2017;377:2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerich L, Marron TU, Upadhyay R, Svensson-Arvelund J, Dhainaut M, Hussein S, et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat Med 2019;25:814–24. [DOI] [PubMed] [Google Scholar]
- 5.Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salmon H, Remark R, Gnjatic S, Merad M. Host tissue determinants of tumour immunity. Nat Rev Cancer 2019;19:215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med 2018;24:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall N, Hutchinson K, Marron TU, Aleynick M, Hammerich L, Upadhyay R, et al. Anti-tumor T-cell homeostatic activation is uncoupled from homeostatic inhibition by checkpoint blockade. Cancer Discov 2019;9:1520–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov 2018;8:1219–26. [DOI] [PubMed] [Google Scholar]
- 10.Ali SA, Shi V, Maric I, Wang M, Stroncek DF, Rose JJ, et al. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood 2016;128:1688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N Engl J Med 2016;375:2561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol 2019;16:372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 2015;385:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner RA, Finney O, Annesley C, Brakke H, Summers C, Leger K, et al. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood 2017;129:3322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle CJ, Hanafi L-A, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR–T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 2016;126:2123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kägi D, Ledermann B, Bürki K, Seiler P, Odermatt B, Olsen KJ, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 1994;369:31–7. [DOI] [PubMed] [Google Scholar]
- 17.Yasukawa M, Ohminami H, Arai J, Kasahara Y, Ishida Y, Fujita S. Granule exocytosis, and not the fas/fas ligand system, is the main pathway of cytotoxicity mediated by alloantigen-specific CD4(+) as well as CD8(+) cytotoxic T lymphocytes in humans. Blood 2000;95:2352–5. [PubMed] [Google Scholar]
- 18.Kochenderfer JN, Somerville RPT, Lu T, Shi V, Bot A, Rossi J, et al. Lymphoma remissions caused by anti-CD19 chimeric antigen receptor T Cells are associated with high serum interleukin-15 levels. J Clin Oncol 2017;35:1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daver N, Kantarjian H, Mackay S, Flynn B, Basu S, Garcia-Manero G, et al. Abstract LB-222: Polyfunctionality determined by single-cell proteomics of bone marrow-derived CD4 T cells from patients with acute myeloid leukemia identifies patients responding to anti-PD-1-based therapy and discovers profound T cell defect in mutant TP53 disease. Cancer Res 2019;79:LB-222. [Google Scholar]
- 20.Rossi J, Paczkowski P, Shen Y-W, Morse K, Flynn B, Kaiser A, et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 2018;132:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay S, Flynn B, Morse K, Paczkowski P, Bacchiocchi A, Fan R, et al. Single-cell cytokine profiling of tumor-infiltrating T cells to measure patient responses to anti-PD-1 therapy. J Clin Oncol 2017;35(7_suppl):49–49. [Google Scholar]
- 22.Zhou J, Bethune MT, Malkova N, Sutherland AM, Comin-Anduix B, Su Y, et al. A kinetic investigation of interacting, stimulated T cells identifies conditions for rapid functional enhancement, minimal phenotype differentiation, and improved adoptive cell transfer tumor eradication. PLoS One 2018;13:e0191634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seki N, Brooks AD, Carter CRD, Back TC, Parsoneault EM, Smyth MJ, et al. Tumor-specific CTL kill murine renal cancer cells using both perforin and fas ligand-mediated lysis in vitro, but cause tumor regression in vivo in the absence of perforin. J Immunol 2002;168:3484–92. [DOI] [PubMed] [Google Scholar]
- 24.Allard B, Pommey S, Smyth MJ, Stagg J. Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin Cancer Res 2013;19:5626–35. [DOI] [PubMed] [Google Scholar]
- 25.Singh N, Lee YG, Shestova O, Ravikumar P, Hayer KE, Hong SJ, et al. Impaired death receptor signaling in leukemia causes antigen-independent resistance by inducing CAR T-cell dysfunction. Cancer Discov 2020;10:552–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal J, Naqvi AS, Luo M, Wertheim G, Paessler M, Thomas-Tikhonenko A, et al. Heterogeneity of surface CD19 and CD22 expression in B lymphoblastic leukemia. Am J Hematol 2018;93:E352–5. [DOI] [PubMed] [Google Scholar]
- 27.Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med 2019;380:45–56. [DOI] [PubMed] [Google Scholar]
- 28.Middha S, Yaeger R, Shia J, Stadler ZK, King S, Guercio S, et al. Majority of B2M -mutant and -deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis Oncol 2019;3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agudo J, Ruzo A, Park ES, Sweeney R, Kana V, Wu M, et al. GFP-specific CD8 T cells enable targeted cell depletion and visualization of T-cell interactions. Nat Biotechnol 2015;33:1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agudo J, Park ES, Rose SA, Alibo E, Sweeney R, Dhainaut M, et al. Quiescent tissue stem cells evade immune surveillance. Immunity 2018;48:271–85.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature 2017;548:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547:413–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan D, Kobayashi A, Jiang P, De Andrade LF, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wroblewska A, Dhainaut M, Ben-Zvi B, Rose SA, Park ES, Amir E-AD, et al. Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell 2018;175:1141–55.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol 2018;18:527–35. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M, Itai T, Adachi M, Nagata S. Downregulation of fas ligand by shedding. Nat Med 1998;4:31–6. [DOI] [PubMed] [Google Scholar]
- 37.Monleón I, Martínez-Lorenzo MJ, Monteagudo L, Lasierra P, Taulés M, Iturralde M, et al. Differential secretion of fas ligand- or APO2 ligand/TNF-related apoptosis-inducing ligand-carrying microvesicles during activation-induced death of human T cells. J Immunol 2001;167:6736–44. [DOI] [PubMed] [Google Scholar]
- 38.Cai Z, Yang F, Yu L, Yu Z, Jiang L, Wang Q, et al. Activated T cell exosomespromote tumor invasion via fas signaling pathway. J Immunol 2012;188:5954–61. [DOI] [PubMed] [Google Scholar]
- 39.Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, Weissberg P. Cell surface trafficking of fas: a rapid mechanism of p53-mediated apoptosis. Science 1998;282:290–3. [DOI] [PubMed] [Google Scholar]
- 40.Maecker HL, Yun Z, Maecker HT, Giaccia AJ. Epigenetic changes in tumor fas levels determine immune escape and response to therapy. Cancer Cell 2002;2:139–48. [DOI] [PubMed] [Google Scholar]
- 41.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity 2009;30:180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su MWC, Pyarajan S, Chang JH, Yu CL, Jin YJ, Stierhof YD, et al. Fratricide of CD8+ cytotoxic T lymphocytes is dependent on cellular activation and perforin-mediated killing. Eur J Immunol 2004;34:2459–70. [DOI] [PubMed] [Google Scholar]
- 43.Kochenderfer JN, Yu Z, Frasheri D, Restifo NP, Rosenberg SA. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 2010;116:3875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kochenderfer JN, Feldman SA, Zhao Y, Xu H, Black MA, Morgan RA, et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother 2009;32:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Park SM, Tumanov AV., Hau A, Sawada K, Feig C, et al. CD95 promotes tumour growth. Nature 2010;465:492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, et al. Membrane-bound fas ligand only is essential for fas-induced apoptosis. Nature 2009;461:659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanzavecchia A Is the T-cell receptor involved in T-cell killing? Nature 1986;319:778–80. [DOI] [PubMed] [Google Scholar]
- 48.Fleischer B Lysis of bystander target cells after triggering of human cytotoxic T lymphocytes. Eur J Immunol 1986;16:1021–4. [DOI] [PubMed] [Google Scholar]
- 49.Smyth MJ. Fas ligand-mediated bystander lysis of syngeneic cells in response to an allogeneic stimulus. J Immunol 1997;158:5765–72. [PubMed] [Google Scholar]
- 50.Wiedemann A, Depoil D, Faroudi M, Valitutti S. Cytotoxic T lymphocytes kill multiple targets simultaneously via spatiotemporal uncoupling of lytic and stimulatory synapses. Proc Natl Acad Sci U S A 2006;103:10985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol 2004;5:524–30. [DOI] [PubMed] [Google Scholar]
- 52.Halle S, Halle O, Förster R. Mechanisms and dynamics of T cell-mediated cytotoxicity in vivo. Trends Immunol 2017;38:432–43. [DOI] [PubMed] [Google Scholar]
- 53.Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med 2018;24:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breart B, Lemaître F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest 2008;118:1390–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schietinger A, Arina A, Liu RB, Wells S, Huang J, Engels B, et al. Longitudinal confocal microscopy imaging of solid tumor destruction following adoptive T cell transfer. Oncoimmunology 2013;2:e26677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kataoka T, Ito M, Budd RC, Tschopp J, Nagai K. Expression level of c-FLIP versus fas determines susceptibility to fas ligand-induced cell death in murine thymoma EL-4 cells. Exp Cell Res 2002;273:256–64. [DOI] [PubMed] [Google Scholar]
- 57.Spiotto MT, Rowley DA, Schreiber H. Bystander elimination of antigen loss variants in established tumors. Nat Med 2004;10:294–8. [DOI] [PubMed] [Google Scholar]
- 58.Zhang B, Karrison T, Rowley DA, Schreiber H. IFN-γ- and TNF-dependent bystander eradication of antigen-loss variants in established mouse cancers. J Clin Invest 2008;118:1398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoekstra ME, Bornes L, Dijkgraaf FE, Philips D, Pardieck IN, Toebes M, et al. Long-distance modulation of bystander tumor cells by CD8+ T-cell-secreted IFN-γ. Nat Cancer 2020;1:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thibaut R, Bost P, Milo I, Cazaux M, Lemaître F, Garcia Z, et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat Cancer 2020;1:302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schimmer AD, Thomas MP, Hurren R, Gronda M, Pellecchia M, Pond GR, et al. Identification of small molecules that sensitize resistant tumor cells to tumor necrosis factor-family death receptors. Cancer Res 2006;66:2367–75. [DOI] [PubMed] [Google Scholar]
- 62.Dufva O, Koski J, Maliniemi P, Ianevski A, Klievink J, Leitner J, et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood 2020;135:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateman A UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 2012;9:840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee J, Sadelain M, Brentjens R. Retroviral transduction of murine primary T lymphocytes. Methods Mol Biol 2009;506:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019;47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lausen B, Schumacher M. Maximally selected rank statistics. Biometrics 1992;48:73–85. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


