Abstract
Electrochemical aptamer-based (E-AB) sensors are a versatile sensing platform that can achieve rapid and robust target detection in complex matrices. However, the limited sensitivity of these sensors has impeded their translation from proof-of-concept to commercial products. Surface-bound aptamers must be sufficiently spaced to bind targets and subsequently fold for signal transduction. We hypothesized that electrodes fabricated using conventional methods result in sensing surfaces where only a fraction of aptamers are appropriately spaced to actively respond to the target. As an alternative, we presented a novel aptamer immobilization approach that favors sufficient spacing between aptamers at the microscale to achieve optimal target binding, folding, and signal transduction. We first demonstrated that immobilizing aptamers in their target-bound, folded state on gold electrode surfaces yields an aptamer monolayer that supports greater sensitivity and higher signal-to-noise ratio than traditionally prepared E-AB sensors. We also showed that performing aptamer immobilization under low ionic strength conditions rather than conventional high ionic strength buffer greatly improves E-AB sensor performance. We successfully tested our approach with three different small-molecule-binding aptamers, demonstrating its generalizability. On the basis of these results, we believe our electrode fabrication approach will accelerate development of high-performance sensors with the sensitivity required for real-world analytical applications.
Keywords: aptamer, electrochemistry, sensing, target-assisted aptamer immobilization, small molecules, bundling effect
Graphical Abstract
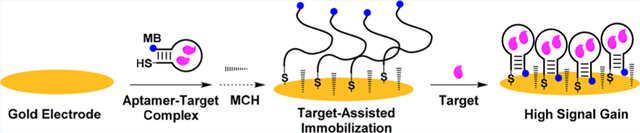
INTRODUCTION
Electrochemical biosensors that utilize bioreceptors, such as enzymes and antibodies, can achieve rapid, sensitive, and selective detection of targets via specific molecular recognition.1,2 For example, personal glucose meters utilize the enzyme glucose oxidase to quantify glucose concentrations directly in whole blood.2 However, the appeal of such sensors is diminished by the lack of enzymes available for detecting a broad range of analytes2 and the generally short shelf life and high cost of protein-based bioreceptors.3 Aptamers offer a promising alternative in this context; these are single-stranded DNA or RNA oligonucleotides that can be isolated from randomized libraries via an in vitro process to bind to virtually any target of interest.4,5 Aptamers can be isolated relatively quickly, have high chemical stability and long shelf-lives, and can be synthesized at low cost with minimal batch-to-batch variation.6 Electrochemical aptamer-based (E-AB) sensors have great potential for diagnostic, research, and therapeutic applications because they enable rapid detection of specific analytes directly in complex samples such as soil, foodstuffs, urine, and whole blood.7–12 E-AB sensors are fabricated by immobilizing aptamers that have been modified with a terminal thiol and a redox label (usually methylene blue) onto a gold electrode via thiol-gold chemistry. This step is typically followed by backfilling with alkanethiol diluents to mitigate the adsorption of oligonucleotide probes and interferents onto the electrode surface.13 The aptamers employed in E-AB sensors have structure-switching functionality, meaning that they are unfolded in their unbound state and undergo a conformational change when binding to the target. This structural change alters the distance between the redox label and the electrode surface, leading to a change in current that is proportional to the concentration of the analyte.
E-AB sensors have greatly evolved in the past decade, transitioning from macroscale sensors that can only perform in vitro detection to miniaturized devices that can detect analytes directly in vivo in real time.14–16 Despite these advances, many E-AB sensors reported to date are incapable of detecting analytes at relevant levels for specific applications in complex samples due to their poor sensitivity and low signal-to-noise ratios (SNRs).8,14,17 For instance, a previously reported cocaine-detecting E-AB sensor has a limit of detection (LOD) of 10 μM in blood,8 which is well outside the range of medically and forensically relevant blood concentrations of cocaine (0.1–1 μM).18 This can be primarily attributed to the biofouling that occurs when detection is performed in biological matrices and the low target affinity of structure-switching aptamers. The biofouling problem has been remedied through the development of new monolayer chemistries and membranes that mitigate protein and cell adsorption on the electrodes.19 For example, the Plaxco group has used biomimetic zwitterionic phospholipid-based thiols as backfillers20 and polysulfone membranes as a physical barrier16 to enable continuous detection of analytes directly in the circulating blood of live animals using E-AB sensors. However, the aptamer affinity issue has only been partially resolved. Although advances in aptamer selection protocols have facilitated the isolation of high-affinity aptamers,21,22 most aptamers still experience considerable reduction in binding affinity upon being engineered with structure-switching functionality via strategies, such as aptamer truncation23 or splitting,24 because of thermodynamic destabilization.
Previous reports have suggested that spacing of oligonucleotide probes on the electrode surface profoundly affects sensor performance.25,26 For example, the Fan group fabricated an E-AB sensor for cocaine by immobilizing a split cocaine-binding aptamer incorporated into a DNA tetrahedron construct tethered to the electrode surface.25 They hypothesized that the tetrahedron would provide the aptamer sufficient spacing to facilitate aptamer-target assembly, thereby augmenting sensor responsiveness. Indeed, they observed significant improvements in sensitivity compared to previous E-AB sensors for cocaine.8 In this work, we described a new method to improve the sensitivity and SNR of E-AB sensors based on the spatial distribution of aptamers on the electrode surface. Although the average spacing between aptamers can be tuned by adjusting the quantity of aptamer27 or buffer ionic strength28 employed during the immobilization step, these strategies do not allow control over the interoligonucleotide distance of probes at the microscopic level to favor optimal target binding and signal transduction. Here, we discovered two aspects of the electrode modification step that could optimize spacing of surface-bound aptamer probes. First, we determined that immobilizing the aptamer in its folded, target-bound state rather than its unfolded state—as is done conventionally9,13—improves both SNR and LOD, which is probably due to optimized spacing granted by the aptamer-target complex at the microscopic level. Next, we observed that the use of low ionic strength buffers during the aptamer immobilization process likewise greatly enhances E-AB sensing performance. This improvement in performance can likely be attributed to mitigation of the clustering of surface-bound aptamers, which commonly occurs when high ionic strength buffers are utilized in conventional protocols.26 These two changes to the immobilization process proved beneficial for E-AB sensing regardless of the aptamer sequence or structure, as we demonstrated in experiments with three different small-molecule-binding aptamers. We, therefore, believe that our enhanced immobilization protocols are generalizable, and will be highly valuable for the fabrication of E-AB sensors with greater sensitivity.
MATERIALS AND METHODS
Materials.
Potassium chloride, magnesium chloride, sodium chloride, monosodium hydrogen phosphate, sodium dihydrogen phosphate, Trizma preset crystals (Tris buffer, pH 7.4), tris(2-carboxyethyl) phosphine chloride, 6-mercapto-1-hexanol (MCH), cocaine hydrochloride, adenosine, sodium hydroxide, sulfuric acid (95–98%), and calf serum were purchased from Sigma-Aldrich. 3,4-Methylenedioxypyrovalerone hydrochloride (MDPV) was purchased from Cayman Chemicals. All solutions were prepared using Milli-Q (Millipore) water with resistivity of 18.2 MΩ × cm unless specified otherwise. Thiolated methylene blue-modified aptamers were synthesized by Biosearch Technologies and purified with HPLC. All other unmodified oligonucleotides were purchased from Integrated DNA Technologies with HPLC purification and dissolved in PCR-grade water. Oligonucleotides concentrations were measured using a NanoDrop 2000 spectrophotometer.
The names and sequences of DNA oligonucleotides used are as follows:
ADE-25:5′-CCTGGTGGAGTATTGCGGGGGAAGG-3′
COC-32:5′-AGACAAGGAAAATCCTTCAATGAAGTGGGTCT-3′
SC-34:5′-ACCTTAAGTGGGGTTCGGGTGGAGTTTATGGGGT-3′
ADE-25-MB: 5′-SH-C6-CCTGGTGGAGTATTGCGGGGGAAGG-MB-3′
COC-32-MB: 5′-SH-C6-AGACAAGGAAAATCCTTCAATGAAGTGGGTCT-MB-3′
SC-34-MB: 5′-SH-C6-ACCTTAAGTGGGGTTCGGGTGGAGTTTATGGGGT-MB-3′
(SH = thiol group, C6 = six carbon link, MB = methylene blue)
Buffers employed were as follows:
High-salt PBS: 1.6 mM NaH2PO4, 8.4 mM Na2HPO4, 1 M NaCl, 1 mM MgCl2, pH 7.2
Low-salt PBS: 1.9 mM NaH2PO4, 8.1 mM Na2HPO4, 1.9 mM NaCl, 0.5 mM MgCl2, pH 7.4
Low-salt Tris buffer: 10 mM Tris buffer, 20 mM NaCl, 0.5 mM MgCl2, pH 7.4
High-salt Tris buffer: 10 mM Tris buffer, 1 M NaCl, 1 mM MgCl2, pH 7.4
Fabrication of E-AB Sensors.
Gold disk electrodes (2 mm diameter) were purchased from CH Instruments and cleaned as previously reported.13 First, the electrode was polished on microcloth (Buehler) with 1-μm diamond suspension (BASi) and 0.05-μm gamma alumina suspension (Buehler). Each polishing step was followed by sonication in ethanol and distilled water for 5 min. Then, the electrode was electrochemically cleaned with a series of voltammetric scans performed in 0.5 M sodium hydroxide, 0.5 M sulfuric acid, and 0.1 M sulfuric acid solutions. To prepare the aptamers for electrode modification, the disulfide groups on the 5′-end of the thiolated, methylene blue-modified aptamers were reduced by incubation in 100 mM tris(2-carboxyethyl) phosphine chloride for 2 h in the dark at room temperature. The aptamers were then diluted to various concentrations (15–200 nM) in high-salt PBS, low-salt PBS with different pH values (pH = 6.0, 7.0, 7.4, or 8.0), or low-salt or high-salt Tris buffer with or without their respective target. The cleaned electrodes were rinsed with distilled water, dried with nitrogen, and immediately incubated in a solution of either aptamer or aptamer-target complexes for 13 h in the dark at room temperature. The electrodes were then backfilled with 1 mM 6-mercapto-1-hexanol solution containing the same concentration of respective target used in the aptamer immobilization step for 2 h at room temperature. Finally, the aptamer-modified electrodes were thoroughly washed with deionized water and then stored in 10 mM Tris (pH 7.4) at room temperature before performing electrochemical measurements.
Electrochemical Measurements.
All electrochemical measurements were carried out using a CHI760D electrochemical workstation (CH Instruments). We used a three-electrode system, including an Ag/AgCl reference electrode, platinum counter electrode, and aptamer-modified gold working electrode. The aptamer surface densities of the modified electrodes were measured via chronocoulometry using the method reported by Tarlov et al.29 Square wave voltammetry (SWV) was performed in the low-salt Tris buffer for the adenosine, cocaine, and MDPV E-AB sensors. Signal gain was calculated using the equation ((IT − I0)/I0)× 100%, where I0 and IT are the SWV peak currents in the absence and presence of target, respectively.
Aptamer Affinity Measurement via Isothermal Titration Calorimetry (ITC).
All ITC experiments were performed with a MicroCal ITC200 instrument (Malvern) at 23 °C. A summary of the experimental conditions employed for each experiment is shown in Table S1. In each experiment, the sample cell was loaded with an aptamer solution (final concentrations: 20 μM or 80 μM for COC-32, 20 μM for ADE-25, 20 μM for SC-34) and the syringe was loaded with the respective target (final concentrations: 800 or 4000 μM cocaine, 1200 or 2500 μM adenosine, 200, 300, 350, or 400 μM (−)-MDPV). Titrations involving COC-32 and SC-34 consisted of an initial purge injection of 0.4 μL, followed by 19 successive injections of 2 μL, with a spacing of either 120 or 180 s between each injection. Titrations involving ADE-25 consisted of an initial 0.4 μL purge injection followed by 38 successive 1 μL injections with a spacing of 120 s between each injection. For all experiments, if saturation was not reached after one series of injections, a second set of injections was performed in the same fashion after reloading the syringe with the target. The raw data were first corrected based on the dilution heat of each target and then analyzed with the MicroCal analysis kit integrated into Origin 7 software, with a single-site binding model for COC-32 and SC-34 or a two-site sequential binding model for ADE-25.
RESULTS AND DISCUSSION
Rationale for Enhancing E-AB Sensor Performance by Controlling Microscale Probe Spacing.
E-AB sensors utilize structure-switching aptamers, which undergo conformational changes when binding to a target. The termini of these aptamers are labeled with a thiol group for immobilization onto a gold electrode surface via thiol-gold bonding and an electroactive redox molecule, such as methylene blue for electrochemical reporting. Most E-AB sensors are fabricated using a previously reported protocol13 that entails aptamer immobilization in high ionic strength phosphate-buffered saline (pH 7.2) containing 1 M NaCl and 1 mM MgCl2 (high-salt PBS). High ionic strength buffers allow for high loading efficiencies of oligonucleotides on gold surfaces, because the negatively charged phosphate groups of the aptamers are shielded from each other by the relatively high concentration of cations. As a consequence, however, the aptamers are highly flexible with low persistence lengths,30 and immobilize onto the electrode surface as both individual probes with low interoligonucleotide proximities, as well as clusters of probes.31 Theoretically, limited spacing between aptamer probes imposes restrictions on target binding and folding that can thus affect signal transduction. The average spacing between aptamers on the electrode surface can be tuned at the macroscopic level by simply altering the quantity of aptamer used for immobilization.27 However, it is not possible to control local aptamer spacing at the microscale to maximize the number of active probes with optimal spacing for target binding and signaling and avoid the formation of dense clusters of inactive probes. Altering average surface densities alone also cannot overcome probe bundling, which occurs at high ionic strengths. To address this problem, we assessed two hypotheses to improve the robustness and sensitivity of E-AB sensors: (1) interoligonucleotide spacing can be optimized by immobilizing aptamer-target complexes rather than the aptamer alone and (2) bundling effects can be minimized by performing aptamer immobilization in low rather than high ionic strength conditions.
Effects of Target-Assisted Aptamer Immobilization on E-AB Sensor Performance.
To test our first hypothesis, we fabricated E-AB sensors from ADE-25, an engineered structure-switching adenosine-binding DNA aptamer32 derived from an ATP-binding aptamer reported by Huizenga and Szostak.33 ADE-25 specifically binds to adenosine with micromolar binding affinity (K1/2 = 27.6 ± 0.2 μM) (Figure S1A) in low-salt Tris buffer (10 mM Tris-HCl, 20 mM NaCl,0.5 mM MgCl2, pH 7.4) but has no affinity for phosphorylated nucleotide analogs such as ATP, ADP, and AMP.32 We first prepared E-AB sensors using thiolated, methylene blue-modified ADE-25 (ADE-25-MB) alone, and optimized aptamer surface coverage to tune average interoligonucleotide spacing by modifying electrodes with different concentrations of aptamer in high-salt PBS. Increases in aptamer concentration in the range of 15-75 nM resulted in monotonic increases in aptamer surface density (Figure S2A). We, then, evaluated the sensing performance of these E-AB sensors by performing detection in solutions containing various concentrations of adenosine (0–1000 μM), and found that the current increased with increasing target concentrations (Figure S2B). As expected, low signal gain was observed for sensors with very low or very high surface coverage because of insufficient probe loading or probe overcrowding, respectively. Signal gains at all target concentrations increased as surface density increased from 1.89 to 3.77 picomol/cm2, but a further increase to 11.1 picomol/cm2 resulted in decreased signal gains (Figure S2C). The impairment of aptamer functioning at higher surface densities is possibly due to lower interoligonucleotide spacing, which has been reported recently using high-resolution atomic force microscopy of oligonucleotide-modified gold surfaces.31 We predicted that by immobilizing the aptamer in its target-bound folded state, we could produce a self-assembled aptamer monolayer with optimal microscale spacing for target binding and signaling relative to unbound flexible aptamers, which are arbitrarily spaced on the surface in a nonoptimized fashion.
To test this, we modified an electrode using a solution of aptamer-target complexes at the optimal surface coverage determined above. Prior to aptamer immobilization, we first determined whether ADE-25 retains the ability to bind adenosine in high-salt PBS using isothermal titration calorimetry (ITC). The ITC results showed that ADE-25 binds adenosine with an affinity (K1/2) of 23.1 ± 0.8 μM under these conditions (Figure S1B). On the basis of this affinity measurement, we estimated that ~91% of the aptamer is bound to the target in the presence of 250 μM adenosine (Figure S3). For electrode fabrication, we first prepared a solution containing 25 nM freshly reduced ADE-25-MB in high-salt PBS with 250 μM adenosine and then submerged the electrodes in the aptamer-target complex solution (Figure 1A, right). Afterward, the electrodes were cleaned thoroughly with buffer to remove any residual adenosine. As a control, we prepared another set of gold electrodes modified with 25 nM ADE-25-MB alone using the traditional immobilization approach13 (Figure 1A, left). Both sets of electrodes had virtually the same average surface density (Figure S4). However, we observed that electrodes modified via target-assisted aptamer immobilization yielded higher SNR and had a lower LOD than electrodes modified with aptamer alone in both buffer (LOD = 1 vs 5 μM) and 50% serum (LOD = 5 vs 10 μM) (Figure 1B–C). Since the total amount of aptamer immobilized on both electrodes was essentially identical, the observed improvement in sensing performance can be most likely be attributed to optimized interoligonucleotide spacing.
Figure 1.
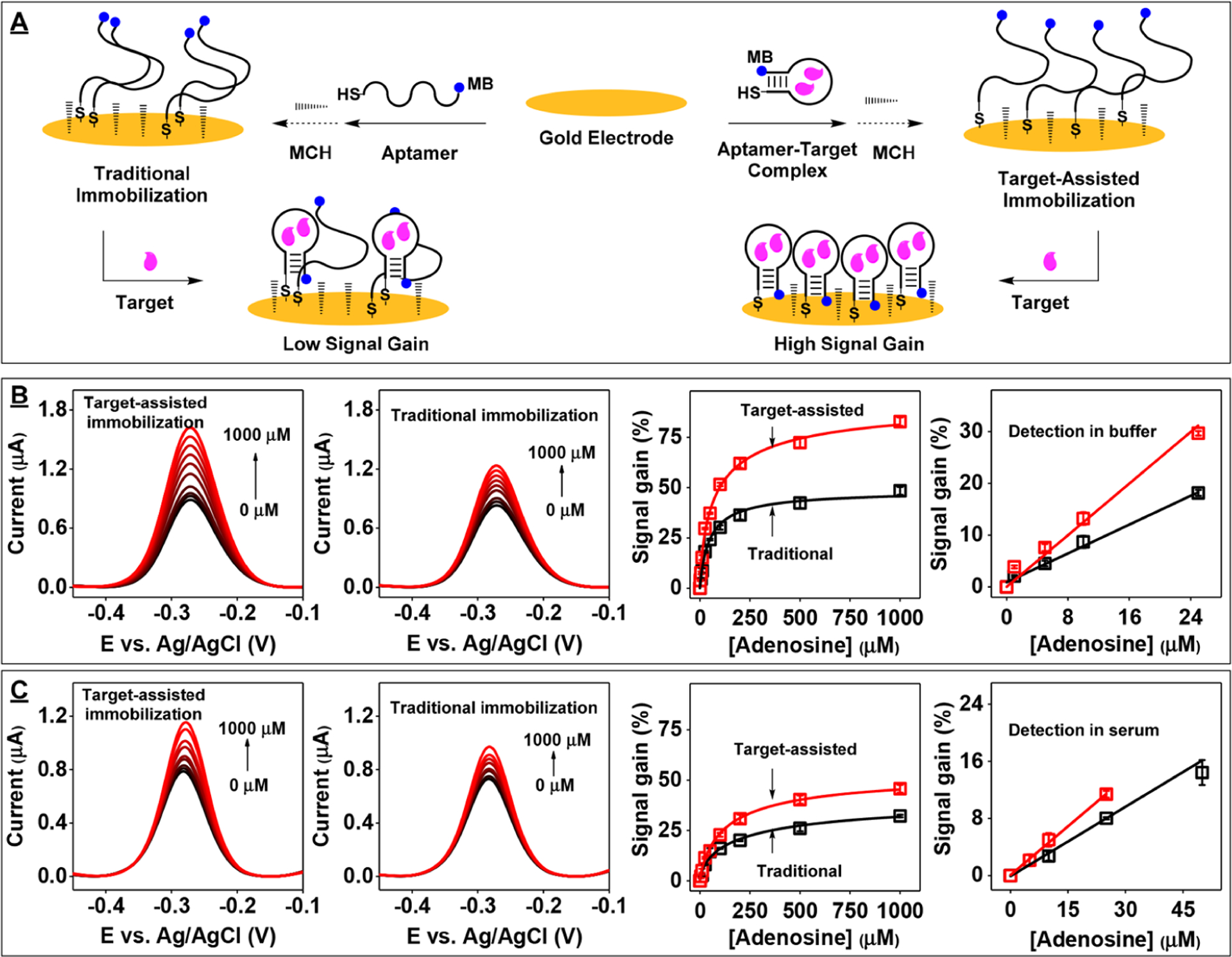
E-AB sensor performance using gold electrodes modified with ADE-25-MB either alone or bound to adenosine. (A) Modification of an electrode using either the traditional immobilization protocol (left) or our target-assisted immobilization strategy (right). Square-wave voltammetry (SWV) spectra of electrodes modified with the aptamer-target complex (left) or aptamer alone (middle) in (B) buffer or (C) 50% serum and corresponding calibration curves and linear ranges (right) collected using electrode prepared with target-assisted aptamer immobilization approach (red) or the traditional method (black). Error bars represent the standard deviation for three working electrodes from each measurement.
Confirmation of the Generalizability of Target-Assisted Aptamer Immobilization.
To demonstrate whether target-assisted aptamer immobilization can improve the performance of E-AB sensors regardless of the aptamer sequence and structure, we fabricated sensors with a well-studied three-way-junction-structured cocaine-binding aptamer34 with structure-switching functionality (COC-32).35 We prepared electrodes with the thiolated, methylene-blue labeled version of this aptamer (COC-32-MB) in the absence or presence of cocaine (250 μM) in high-salt PBS. Once again, although both sets of electrodes displayed similar aptamer surface densities (Figure S5A), the sensors prepared via target-assisted aptamer immobilization demonstrated greater signal gains at all tested concentrations of cocaine (0–1000 μM) (Figure S5B–D) and improved LODs relative to electrodes modified with aptamer alone (LOD = 1 vs 2 μM) (Figure S5E). This confirmed that target-assisted aptamer immobilization generally enhances the performance of E-AB sensors.
Relationship between the Concentration of Aptamer-Target Complex Used for Electrode Modification and E-AB Sensor Performance.
It has been reported that the cocaine-binding aptamer has weak affinity for its target at high salt concentrations.36 We confirmed via ITC that COC-32 binds cocaine with a KD of 70.4 ± 0.8 μM in the high-salt PBS commonly used for aptamer immobilization (Figure S6A). In these conditions, only ~78% of the aptamer is bound to the target in the presence of 250 μM cocaine (Figure S7), such that the resulting monolayer would be predicted to contain both active and inactive, nonoptimally spaced probes. We hypothesized that maximizing the quantity of target-bound aptamer during the aptamer immobilization process would result in the greatest quantity of immobilized active aptamers, thereby yielding the best attainable sensor performance. We therefore fabricated E-AB sensors by immobilizing COC-32-MB in the presence of 2 mM cocaine, where ~96% of the aptamer is in the target-bound state (Figure S7). Although electrodes prepared in the presence of 250 μM or 2 mM cocaine had the same aptamer surface densities (Figure S8), the latter sensors yielded higher SNRs at all target concentrations—especially in the range from 100–1000 μM cocaine, where we saw an 8% improvement in signal gain (Figure 2). This clearly demonstrates that more active aptamer probes were being immobilized on the electrode surface with immobilization solutions containing a higher proportion of aptamer-target complexes.
Figure 2.
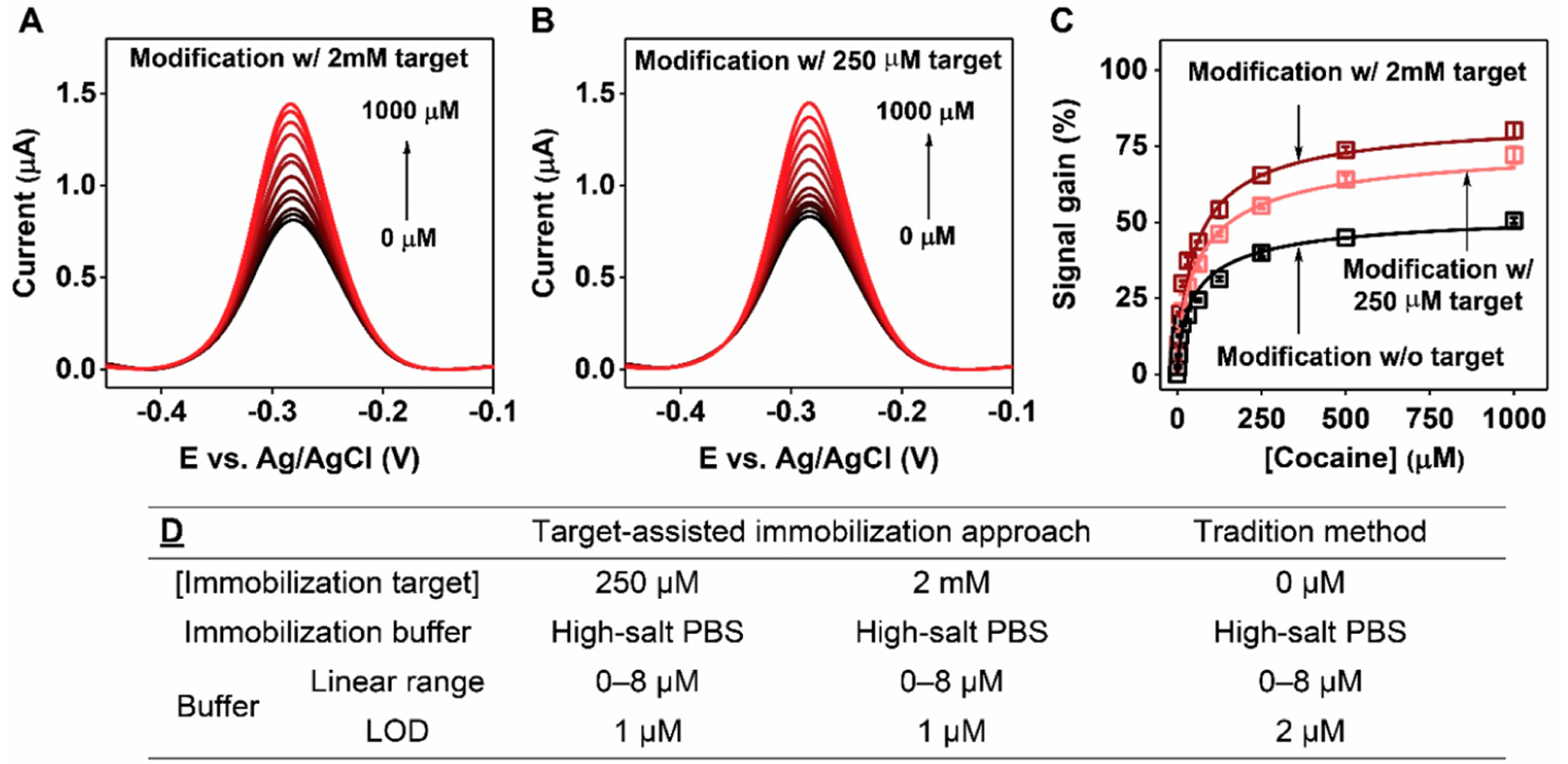
E-AB sensor performance using electrodes modified with COC-32-MB via target-assisted immobilization. SWV spectra collected at various concentrations of cocaine using electrodes modified in high-salt PBS with (A) 2 mM or (B) 250 μM cocaine. (C) Calibration curves derived from SWV spectra for electrodes modified with COC-32-MB plus 2 mM (brown) or 250 μM cocaine (pink) and electrodes modified with aptamer alone (black). (D) Linear ranges and LODs of electrodes fabricated via different methods. Error bars represent the standard deviation for three working electrodes from each measurement.
Dependency of E-AB Sensor Performance on the Ionic Strength of the Immobilization Buffer.
Having determined that target-assisted aptamer immobilization improves E-AB sensor performance, we next tested our hypothesis on the relationship between the ionic strength of the electrode modification buffer and E-AB sensor performance. It has been reported that local interoligonucleotide distances can be increased and probe bundling can be mitigated by decreasing the ionic strength of the buffer used for probe immobilization.31 To evaluate this, we modified electrodes with COC-32-MB alone in either high-salt PBS or low-salt PBS (10 mM PBS, 1.9 mM NaCl, 0.5 mM MgCl2, pH7.4). To ensure that the electrodes had similar surface densities, a greater concentration of aptamer was used for immobilization in the low ionic strength buffer. Although both electrodes had similar surface densities (Figure S9A), electrodes prepared in low-salt PBS had improved signal gain in the range of 50–1000 μM cocaine (Figure S9B–D). This indicates that the improvement in E-AB performance was most likely due to the mitigation of aptamer bundling and resulting increase in active probes on the electrode surface when lower ionic strength buffer was used. We then determined whether the addition of target-assisted immobilization would further improve sensor performance. Electrodes were modified with COC-32-MB in low-salt PBS containing 250 μM cocaine, as well as in high-salt PBS containing 2 mM cocaine as a control. In both cases, ~96% of the aptamer is bound to cocaine based on the target affinity of the aptamer (high-salt PBS KD = 70.4 ± 0.8 μM; low-salt PBS KD = 5.8 ± 0.3 μM) (Figure S6A and B) and the target concentration employed (Figures S7 and S10). Despite both sets of electrodes having similar surface coverages (Figure S11), we observed great improvements in SNR from electrodes immobilized with the aptamer in low ionic strength conditions, with approximately 10% improvement in signal gain in the concentration range of 50–1000 μM cocaine (Figure 3A–C). Given that the electrodes from both experiments demonstrated equivalent surface coverage and similar proportions of aptamer-target complexes for modification (~96%), the improvement in performance can be primarily attributed to the predominance of active aptamers on the electrode surface when the ionic strength of the immobilization buffer was reduced. These results confirm that buffer ionic strength has a significant effect on the performance of E-AB sensors.
Figure 3.
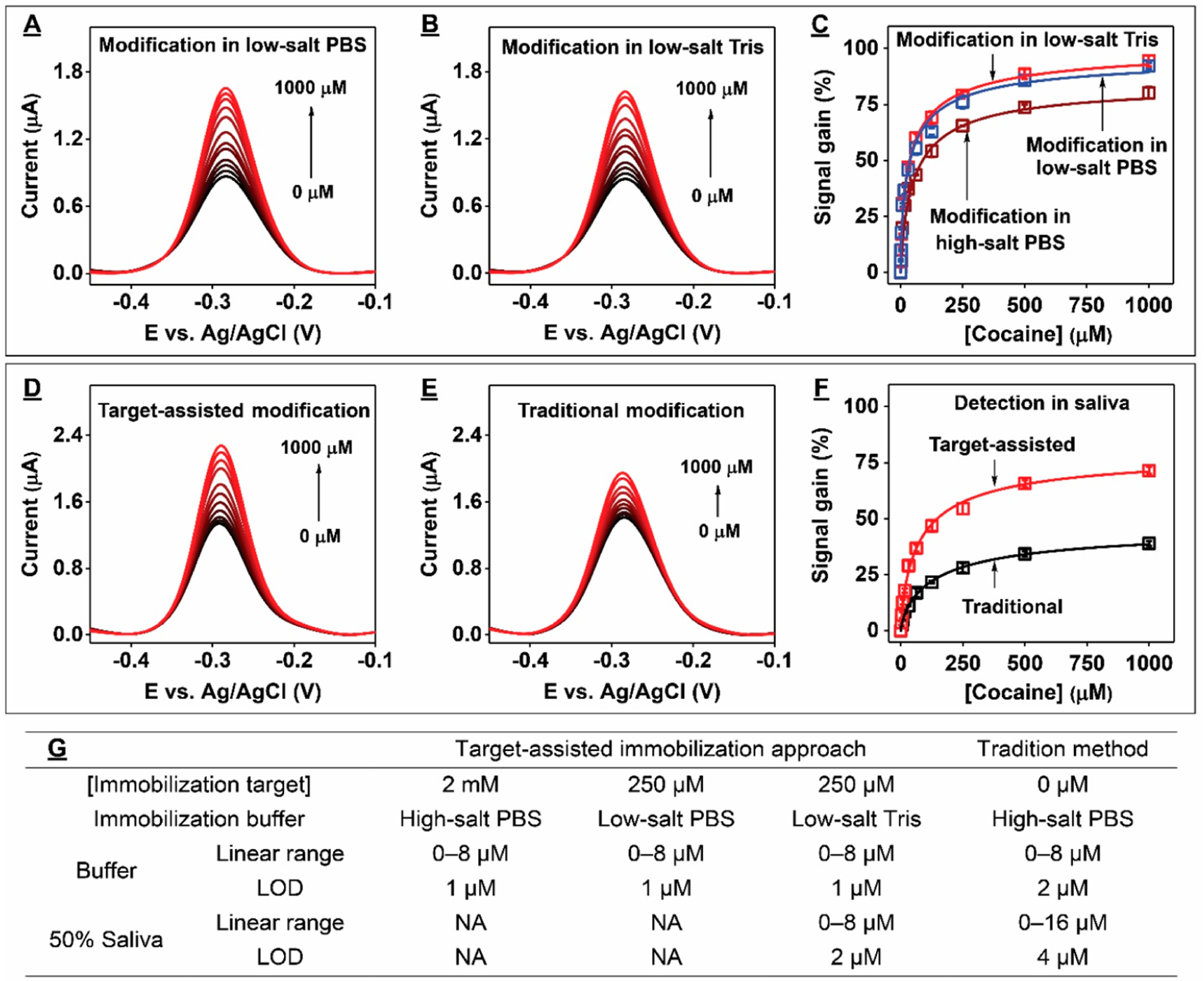
E-AB sensor performance using gold electrodes modified with target-bound COC-32-MB. SWV spectra at various concentrations of cocaine from electrodes modified with COC-32-MB plus cocaine in low-salt (A) PBS or (B) Tris. (C) Calibration curves derived from the SWV spectra shown in panel A (blue) and panel B (red) or from electrodes modified in cocaine-containing high-salt PBS (brown). Detection of cocaine in 50% saliva using electrodes modified with (D) aptamer-target complexes in low-salt Tris or (E) aptamer alone in high-salt PBS. (F) Calibration curves derived from the SWV spectra shown in panels D (red) and E (black). (G) Linear ranges and LODs of electrodes fabricated via different methods in buffer or 50% saliva. Error bars represent the standard deviation for three working electrodes from each measurement.
Effect of the Type of Buffering System Used for Aptamer Immobilization on E-AB Sensor Performance.
Changing the buffering system used for aptamer immobilization should not affect the performance of the resultant E-AB sensors if the ionic strength and pH remains the same. To confirm this, we modified electrodes with COC-32-MB in the presence of 250 μM cocaine in either low-salt Tris buffer or low-salt PBS with the same ionic strength and pH. As expected, at the same aptamer surface coverage (Figure S12A), both sensors exhibited similar performance (Figures S12B and C and 3C) due to the equivalent binding affinity of COC-32-MB for cocaine in both buffers (Figure S6B and C). Finally, we compared the performance of sensors prepared via target-assisted aptamer immobilization in low-salt Tris buffer versus conventionally prepared electrodes for the detection of cocaine in complex biosamples. We found that electrodes modified using our method demonstrated higher SNRs and greater sensitivity in 50% saliva relative to conventionally modified electrodes (LOD = 2 vs 4 μM) (Figures 3D–G and S13).
Target-Assisted Immobilization Is Effective Independent of Aptamer Sequence, Structure, and Affinity.
As a final demonstration of the generalizability of our method, we prepared electrodes using SC-34, a G-rich, high-affinity structure-switching DNA aptamer that binds the synthetic cathinone methylenedioxypyrovalerone (MDPV) with a KD of 300 nM in low-salt Tris buffer.22,37 We modified gold electrodes with the thiolated, methylene blue-modified version of SC-34 (SC-34-MB) in low-salt Tris buffer in the absence or presence of 5, 10, or 50 μM MDPV, respectively corresponding to 94%, 97%, 99% aptamer-target complex in solution (Figure S14). Increases in the concentration of MDPV during electrode modification resulted in slightly decreased aptamer surface coverage (Figure 4A). This is probably because the rigid aptamer-target complex occupies more space than the unbound aptamer, and thus slightly increasing the quantity of the complex results in lower surface coverage. Nevertheless, electrodes modified with aptamer-MDPV complexes consistently yielded higher signal gains at all tested target concentrations compared to electrodes modified with the aptamer alone (Figures 4B and S15). However, increasing the MDPV concentration beyond 5 μM during the modification step yielded no more than a 5% improvement in signal gain (at 100 μM MDPV), most likely because most aptamers (~94%) are already bound to the target at 5 μM MDPV. Using 50 μM MDPV for target-assisted aptamer immobilization, we also observed similar improvements in sensitivity for MDPV detection in 50% urine (LOD = 0.5 vs 1 μM) (Figures 4C and S16).
Figure 4.
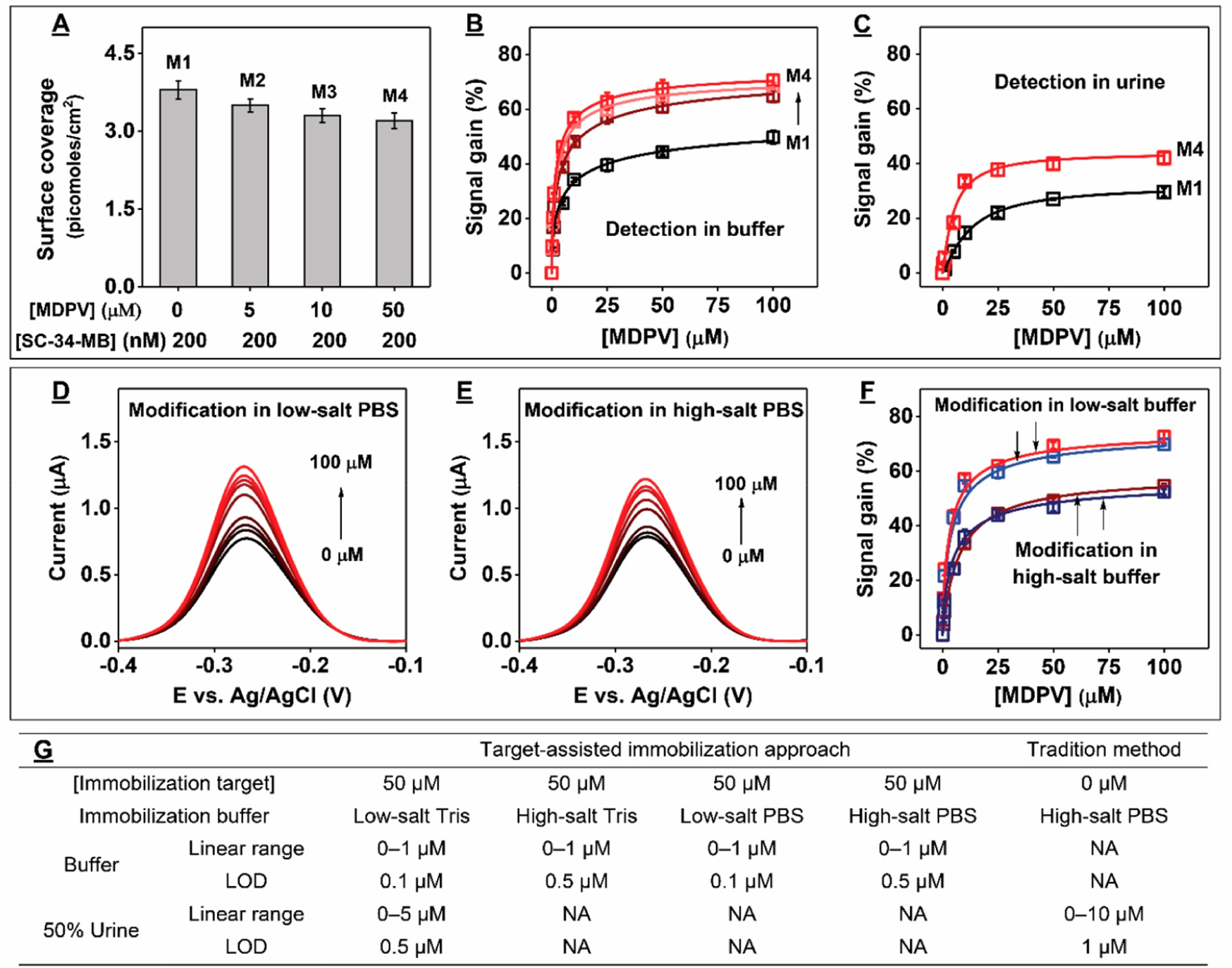
E-AB sensor performance using electrodes fabricated with SC-34-MB. (A) Effect of MDPV concentration on surface coverage during electrode modification. (B) Signal gain for the various electrodes prepared in panel A from various concentrations of MDPV in buffer. (C) Detection of MDPV in 50% urine using modified electrodes prepared in low-salt Tris with SC-34-MB alone (M1) or in the presence of 50 μM MDPV (M4). SWV spectra from various concentrations of MDPV using electrodes modified via target-assisted immobilization in either (D) low-salt or (E) high-salt PBS. (F) Calibration curves derived from the SWV spectra shown in D (red) and E (brown) or from electrodes prepared in low-salt (blue) or high-salt (navy) Tris buffer. (G) Linear ranges and LODs of electrodes fabricated via different methods in buffer or 50% urine. Error bars represent the standard deviation for three working electrodes from each measurement.
We then confirmed that the buffer system employed for aptamer immobilization has no effect on sensor performance in low-salt buffer; electrodes modified in either low-salt Tris buffer or PBS had nearly the same response at all tested target concentrations (Figure 4D and F). This is probably because the aptamer has the same Kd (~300 nM) in both buffers, as we confirmed by ITC (Figure S17A and B). Finally, to confirm that the ionic strength used during target-assisted aptamer immobilization effects E-AB sensing performance, we fabricated electrodes with SC-34-MB in either low-salt or high-salt PBS in the presence of 50 μM MDPV. In both buffers, ≥ 97% of the aptamer was bound to MDPV (Figures S14 and S18); based on ITC, the aptamer’s KD in high-salt PBS is 1.50 ± 0.04 μM (Figure S17C). Although both sets of electrodes had similar surface coverage (Figure S19A), electrodes modified in low-salt PBS were more sensitive than those modified in high-salt PBS (LOD = 0.1 μM vs 0.5 μM) (Figures 4E–G and S19B). Finally, we confirmed that immobilization buffer type also had no effect on E-AB sensor performance with high-salt condition. Specifically, we modified electrodes with SC-34-MB in high-salt Tris buffer in the absence and presence of 50 μM MDPV and challenged the resulting sensors with 0–100 μM MDPV. Although both sets of electrodes had similar surface coverages (Figure S20A), electrodes modified via target-assisted immobilization yielded higher signal gains (Figure S20B) and demonstrated lower limits of detection than traditionally modified electrodes (Figure S20C). These results further support the conclusion that target-assisted aptamer immobilization results in E-AB sensors with improved sensing performance. Notably, electrodes modified via target-assisted aptamer immobilization in high-salt Tris buffer or PBS exhibited similar performance (Figure 4F). ITC confirmed that the binding affinity of SC-34 was similar in high-salt PBS (Figure S17C, KD = 1.50 ± 0.05 μM) and high-salt Tris buffer (Figure S21, KD = 1.80 ± 0.04 μM), demonstrating that the use of different immobilization buffer systems does not affect sensor performance. These results further support the conclusion that sensor performance was primarily improved due to increased interoligonucleotide spacing and minimization of strand “bundling” in the low ionic strength buffer during modification.
Effect of Buffer pH Used for Aptamer Immobilization on E-AB Sensor Performance.
We finally tested the effect of altering the immobilization buffer pH on the performance of the resulting E-AB sensors. As a demonstration, we prepared gold electrodes with SC-34-MB in the absence or presence of 50 μM MDPV in low-salt PBS formulations with the same ionic strengths but different pH values (pH = 6.0, 7.0, 7.4, or8.0). Buffer pH did not significantly affect aptamer surface coverage, although electrodes fabricated via target-assisted immobilization consistently had slightly lower surface coverage (3.28 ± 0.07 pmol/cm2) than those modified via the traditional aptamer immobilization method (3.55 ± 0.06 pmol/cm2) (Figure S22). We first tested the performance of traditionally modified electrodes by challenging them with 0–100 μM MDPV. E-AB sensors fabricated at pH 8.0 (Figures 5A and S23A) or 7.4 (Figures 5B and S23B) produced nearly identical signal gains, while aptamer immobilization at pH 7.0 yielded sensors with only slightly inferior performance (Figures 5C and S23C). In contrast, sensors fabricated at pH 6.0 yielded noticeably lower signal gains at all target concentrations (Figures 5D and S23D). This may be because the phosphate groups of the aptamers are predominantly singly protonated at this pH, rather than being doubly protonated as occurs at pH ≥ 7.0. This would result in less electrostatic repulsion between aptamers, thus increasing the likelihood of bundling during immobilization. We then tested the performance of electrodes modified via target-assisted immobilization. As expected, regardless of immobilization buffer pH, these electrodes consistently yielded higher signal gains and had lower LOD compared to traditionally modified electrodes (Figure 5). Although we observed a similar pattern of pH effects on electrode performance, the magnitude of signal gain was consistently greater after target-assisted immobilization at every pH tested (Figure 5A and D).
Figure 5.
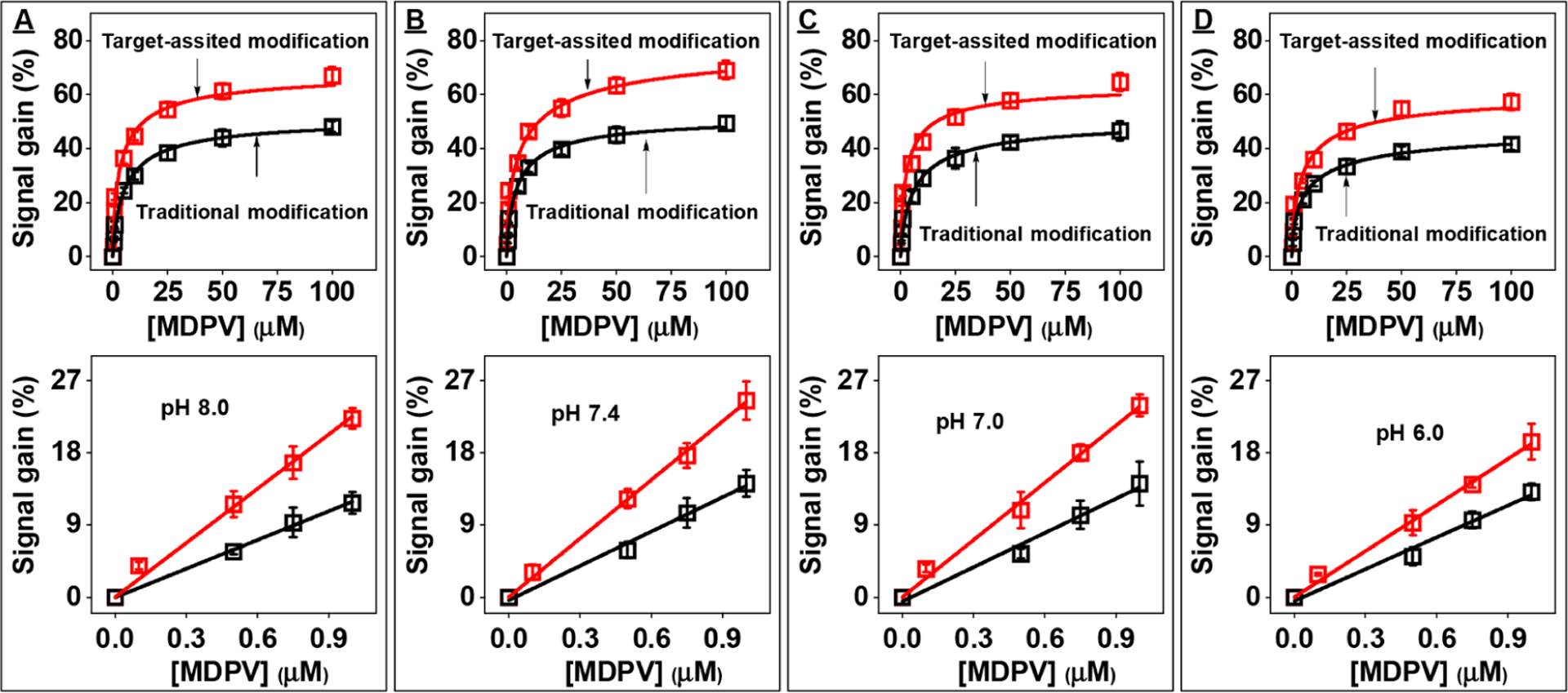
E-AB sensor performance using electrodes modified with SC-34-MB in the presence (target-assisted immobilization) or absence (traditional immobilization) of 50 μM MDPV in low-salt PBS at pH (A) 8.0, (B) 7.4, (C) 7.0, and (D) 6.0. The top panels show calibration curves for electrodes fabricated via target-assisted aptamer immobilization (red) or traditional modification method (black) produced by challenging with 0–100 μM MDPV, and bottom panels show their respective linear ranges. Error bars represent the standard deviation of measurements from three independently fabricated electrodes.
We finally used ITC to determine if buffer pH affected sensor performance by affecting the formation and stability of aptamer-target complexes during aptamer immobilization. Specifically, we measured the binding affinity of SC-34 to MDPV in low-salt PBS at pH 6.0, 7.0, 7.4, or 8.0. In all pH conditions, the binding affinity values did not greatly differ (Figure S24). This meant that in all buffers containing 50 μM MDPV, ≥97% of the aptamer was bound to the target, indicating that pH did not affect the extent of aptamer-target complexation. Our results therefore suggest that the use of relatively low-pH buffers for aptamer immobilization promotes aptamer bundling due to reduced electrostatic repulsion, which in turn negatively impacts sensor performance.
CONCLUSION
E-AB sensors are a promising class of analytical devices that have the potential for broad adoption in commercial applications due to their high selectivity, robustness, and ease of use. However, such devices are often hobbled by insufficient sensitivity and low SNRs, primarily because of the low target affinity of structure-switching aptamers and matrix effects. Several strategies have been devised to address these factors, but we have also determined here that an under-appreciated factor—the conditions employed in the aptamer immobilization step of the electrode fabrication process—can also profoundly affect sensor performance. We have analyzed the impact of two modifications to this process: immobilizing aptamers in a folded, target-bound state on the electrode rather than an unbound single-stranded state, and lowering buffer ionic strength to reduce aptamer bundling on the surface. Our results confirm that both target-assisted aptamer immobilization and the use of low ionic strength immobilization buffers generally improves the SNR and LOD of E-AB sensors relative to those fabricated using conventional immobilization methods. We also determined that the pH of the immobilization buffer can have a negative effect the performance of the resulting E-AB sensors if the pH is below 7. We attribute this to the protonation of the phosphate group of DNA, which reduces electrostatic repulsion and promotes aptamer bundling. Based on our findings, we believe the ideal buffer for aptamer immobilization should have relatively low ionic strength while also supporting aptamer-target binding. We suggest testing target-binding affinity at various ionic strengths, and then using the lowest ionic strength conditions at which >90% of aptamer bound to target can readily be achieved during the immobilization process. Although we do not currently have direct evidence for our proposed mechanisms by which E-AB sensing is altered, the use of surface analysis techniques, such as high-resolution atomic force microscopy, should allow for a more definitive understanding of these phenomenon.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health—National Institute on Drug Abuse R21DA04533401A1 and the National Science Foundation (1905143).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.0c20707
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.0c20707.
Experimental details for ITC; ITC characterization of the affinity of ADE-25 for adenosine, COC-32 for cocaine, SC-34 for MDPV in either PBS or Tris with various ionic strength and pH; performance of electrodes fabricated with ADE-25-MB, COC-32-MB, or SC-34-MB via different methods either in buffer or complex samples; simulated binding curve for ADE-25, COC-32, or SC-34 based on the affinity for their respective target in high-salt PBS; simulated binding curve for COC-32 or SC-34 based on the affinity for their respective target in either low-salt PBS or low-salt Tris; and aptamer surface coverage of electrodes fabricated with ADE-25-MB, COC-32-MB, or SC-34-MB via different methods (PDF)
REFERENCES
- (1).Privett BJ; Shin JH; Schoenfisch MH Electrochemical Sensors. Anal. Chem 2008, 80, 4499–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Labib M; Sargent EH; Kelley SO Electrochemical Methods for the Analysis of Clinically Relevant Biomolecules. Chem. Rev 2016, 116, 9001–9090. [DOI] [PubMed] [Google Scholar]
- (3).Zhou J; Rossi J Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discovery 2017, 16, 181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ellington AD; Szostak JW In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- (5).Tuerk C; Gold L Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- (6).Dunn MR; Jimenez RM; Chaput JC Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem 2017, 1, 0076. [Google Scholar]
- (7).Schoukroun-Barnes LR; Macazo FC; Gutierrez B; Lottermoser J; Liu J; White RJ Reagentless, Structure-Switching, Electrochemical Aptamer-Based Sensors. Annu. Rev. Anal. Chem 2016, 9, 163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Baker BR; Lai RY; Wood MS; Doctor EH; Heeger AJ; Plaxco KW An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. J. Am. Chem. Soc 2006, 128, 3138–3139. [DOI] [PubMed] [Google Scholar]
- (9).Xiao Y; Lubin AA; Heeger AJ; Plaxco KW Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew. Chem., Int. Ed 2005, 44, 5456–5459. [DOI] [PubMed] [Google Scholar]
- (10).Li H; Somerson J; Xia F; Plaxco KW Electrochemical DNA-Based Sensors for Molecular Quality Control: Continuous, Real-Time Melamine Detection in Flowing Whole Milk. Anal. Chem 2018, 90, 10641–10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xiao Y; Rowe AA; Plaxco KW Electrochemical Detection of Parts-Per-Billion Lead via an Electrode-Bound DNAzyme Assembly. J. Am. Chem. Soc 2007, 129, 262–263. [DOI] [PubMed] [Google Scholar]
- (12).Parolo C; Idili A; Ortega G; Csordas A; Hsu A; Arroyo-Currás N; Yang Q; Ferguson BS; Wang J; Plaxco KW Real-Time Monitoring of a Protein Biomarker. ACS Sens. 2020, 5, 1877–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xiao Y; Lai RY; Plaxco KW Preparation of Electrode-Immobilized, Redox-Modified Oligonucleotides for Electrochemical DNA and Aptamer-Based Sensing. Nat. Protoc 2007, 2, 2875–2880. [DOI] [PubMed] [Google Scholar]
- (14).Swensen JS; Xiao Y; Ferguson BS; Lubin AA; Lai RY; Heeger AJ; Plaxco KW; Soh HT Continuous, Real-Time Monitoring of Cocaine in Undiluted Blood Serum via a Microfluidic, Electrochemical Aptamer-Based Sensor. J. Am. Chem. Soc 2009, 131, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ferguson BS; Hoggarth DA; Maliniak D; Ploense K; White RJ; Woodward N; Hsieh K; Bonham AJ; Eisenstein M; Kippin TE; Plaxco KW; Soh HT Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Sci. Transl. Med 2013, 5, 213ra165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Arroyo-Currás N; Somerson J; Vieira PA; Ploense KL; Kippin TE; Plaxco KW Real-Time Measurement of Small Molecules Directly in Awake, Ambulatory Animals. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rowe AA; Miller EA; Plaxco KW Reagentless Measurement of Aminoglycoside Antibiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor. Anal. Chem 2010, 82, 7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jones AW; Holmgren A Concentrations of Cocaine and Benzoylecgonine in Femoral Blood from Cocaine-Related Deaths Compared with Venous Blood from Impaired Drivers. J. Anal. Toxicol 2014, 38, 46–51. [DOI] [PubMed] [Google Scholar]
- (19).Jiang C; Wang G; Hein R; Liu N; Luo X; Davis JJ Antifouling Strategies for Selective in Vitro and in Vivo Sensing. Chem. Rev 2020, 120, 3852–3889. [DOI] [PubMed] [Google Scholar]
- (20).Li H; Dauphin-Ducharme P; Arroyo-Currás N; Tran CH; Vieira PA; Li S; Shin C; Somerson J; Kippin TE; Plaxco KW A Biomimetic Phosphatidylcholine-Terminated Monolayer Greatly Improves the In Vivo Performance of Electrochemical Aptamer-Based Sensors. Angew. Chem., Int. Ed 2017, 56, 7492–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Nakatsuka N; Yang K-A; Abendroth JM; Cheung KM; Xu X; Yang H; Zhao C; Zhu B; Rim YS; Yang Y; Weiss PS; Stojanovic MN; Andrews AM Aptamer - Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science 2018, 362, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yang W; Yu H; Alkhamis O; Liu Y; Canoura J; Fu F; Xiao Y In Vitro Isolation of Class-Specific Oligonucleotide-Based Small-Molecule Receptors. Nucleic Acids Res. 2019, 47, No. e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Neves MAD; Reinstein O; Johnson PE Defining a Stem Length-Dependent Binding Mechanism for the Cocaine-Binding Aptamer. A Combined NMR and Calorimetry Study. Biochemistry 2010, 49, 8478–8487. [DOI] [PubMed] [Google Scholar]
- (24).Stojanovic MN; de Prada P; Landry DW Fluorescent Sensors Based on Aptamer Self-Assembly. J. Am. Chem. Soc 2000, 122, 11547–11548. [DOI] [PubMed] [Google Scholar]
- (25).Wen Y; Pei H; Wan Y; Su Y; Huang Q; Song S; Fan C DNA Nanostructure-Decorated Surfaces for Enhanced Aptamer-Target Binding and Electrochemical Cocaine Sensors. Anal. Chem 2011, 83, 7418–7423. [DOI] [PubMed] [Google Scholar]
- (26).Josephs EA; Ye T Nanoscale Spatial Distribution of Thiolated DNA on Model Nucleic Acid Sensor Surfaces. ACS Nano 2013, 7, 3653–3660. [DOI] [PubMed] [Google Scholar]
- (27).White RJ; Phares N; Lubin AA; Xiao Y; Plaxco KW Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir 2008, 24, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Petrovykh DY; Kimura-Suda H; Whitman LJ; Tarlov MJ Quantitative Analysis and Characterization of DNA Immobilized on Gold. J. Am. Chem. Soc 2003, 125, 5219–5226. [DOI] [PubMed] [Google Scholar]
- (29).Steel AB; Herne TM; Tarlov MJ Electrochemical Quantitation of DNA Immobilized on Gold. Anal. Chem 1998, 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- (30).Chen H; Meisburger SP; Pabit SA; Sutton JL; Webb WW; Pollack L Ionic Strength-Dependent Persistence Lengths of Single-Stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gu Q; Nanney W; Cao HH; Wang H; Ye T Single Molecule Profiling of Molecular Recognition at a Model Electrochemical Biosensor. J. Am. Chem. Soc 2018, 140, 14134–14143. [DOI] [PubMed] [Google Scholar]
- (32).Canoura J; Yu H; Alkhamis O; Roncancio D; Farhana R; Xiao Y Accelerating Post-SELEX Aptamer Engineering Using Exonuclease Digestion. J. Am. Chem. Soc 2020, DOI: 10.1021/jacs.0c09559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Huizenga DE; Szostak JW A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [DOI] [PubMed] [Google Scholar]
- (34).Stojanovic MN; de Prada P; Landry DW Aptamer-Based Folding Fluorescent Sensor for Cocaine. J. Am. Chem. Soc 2001, 123, 4928–4931. [DOI] [PubMed] [Google Scholar]
- (35).Wang Z; Yu H; Canoura J; Liu Y; Alkhamis O; Fu F; Xiao Y Introducing Structure-Switching Functionality into Small-Molecule-Binding Aptamers via Nuclease-Directed Truncation. Nucleic Acids Res. 2018, 46, No. e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Neves MAD; Slavkovic S; Churcher ZR; Johnson PE Salt-Mediated Two-Site Ligand Binding by the Cocaine-Binding Aptamer. Nucleic Acids Res. 2017, 45, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Liu Y; Yu H; Alkhamis O; Moliver J; Xiao Y Tuning Biosensor Cross-Reactivity Using Aptamer Mixtures. Anal. Chem 2020, 92, 5041–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


