Abstract
Granulins (GRN 1–7) are short (~6 kDa), cysteine-rich proteins that are generated upon the proteolytic processing of progranulin (PGRN). These peptides, along with their precursor, have been implicated in multiple pathophysiological roles, especially in neurodegenerative diseases. Previously we showed that GRN-3 and GRN-5 are fully disordered in the reduced form implicating redox sensitive attributes to the proteins. Redox-based modulations are often carried out by metalloproteins in mitigating oxidative stress and maintaining metal-homeostasis within cells. To probe whether GRNs play a role in metal sequestration, we tested the metal binding potential of the reduced form of GRNs −3 and −5 under neutral and acidic pH mimicking cytosolic and lysosomal conditions, respectively. We found, at neutral pH, both GRNs selectively bind Cu and no other divalent metal cations, with a greater specificity for Cu(I). Binding of Cu did not result in a disorder-to-order structural transition but partly triggered the multimerization of GRNs via uncoordinated cystines at both pH conditions. Overall, the results indicate that GRNs −3 and −5 have a surprisingly strong affinity for Cu in the pM range, comparable to other known copper sequestering proteins. The results also hint at a potential of GRNs to reduce Cu(II) to Cu(I), a process that has significance in mitigating Cu-induced ROS cytotoxicity in cells. Together, this report uncovers a metal-coordinating capability of GRNs for the first time, which could have profound significance in their structure and pathophysiological functions.
Keywords: Granulins, Progranulin, Metalloproteins, Cysteine-rich, Frontotemporal lobar degeneration, redox
Introduction
Progranulin (PGRN) is a 68 kDa, glycosylated protein that is ubiquitously expressed in many cells and has pleiotropic roles in physiology such as neuronal growth, differentiation and survival2–4, wound healing and repair5–6, and immunomodulation7. The protein is also known to play roles in tumor growth and metastasis due to its growth-promoting and angiogenic characteristics8. PGRN is also linked to neurodegenerative disorders with the haploinsufficiency of the protein being implicated in frontotemporal dementia (FTD)9 while a complete loss of PGRN leads to neuronal ceroid lipofuscinosis, a lysosomal storage disease10. PGRN consists of seven and a half tandem-repeats of cysteine-rich modules called granulins (GRNs 1–7) and a partial para-GRN module. PGRN is secreted extracellularly and proteolytically cleaved by enzymes such as matrix metalloproteinase-12 (MMP-12)11, neutrophil elastase, and proteinase 312 releasing the individual GRNs. In physiology, a variety of roles have been attributed to GRNs such as innate immune response13, neurotrophic roles4, while other studies ascribe pro-inflammatory functions, opposing that of the precursor14. Although the extracellular processing of PGRN and GRN formation is known14–15, recent studies have also revealed an intracellular localization of these modules. PGRN has been shown to be transported into the lysosome where it is processed by the cysteine peptidase cathepsin-L16–17 producing GRNs that are stable within the lysosomal environment18.
Individual GRNs (1–7) are 6 kDa proteins with a characteristic feature of unusually high cysteine content (~20%)1, and have a consensus sequence of X2–3CX5–6CX5CCX8CCX6CCXDXXHCCPX4CX5–6CX, with the twelve conserved cysteines forming six putative disulfide bonds19–20. Studies into the structural properties of human GRNs have revealed that GRN-2, 4 and 5 adopt partially folded structure with a defined interdigitated disulfide bonding pattern, while the structures of other GRNs (1,3,6 and 7) are dominated by loops20. Previously, we have investigated the structure-function relationship of two GRNs; GRN-321–24 and GRN-524. Both GRNs show characteristics of classical intrinsically disordered proteins (IDPs) having no secondary structure especially in the fully reduced form22, 24. Abrogation of disulfide bonds in the reduced state renders the protein to be disordered and thermally unstable22. On the other hand, the disulfide-bonded oxidized GRN-3 showed high thermal stability but with a structure dominated by disordered loops22. Interestingly, both reduced and oxidized GRN-3 showed cellular activity implicating a possible redox-based regulation as a mode of functioning for GRNs21, 23.
Recently, the potential functional implications of disorder promoting sequences interspersed with order-promoting cysteines have begun to emerge. In eukaryotes, widespread presence of proteins, which are either conditionally disordered or containing intrinsically disordered regions are observed25. Some of these proteins display redox sensitivity by undergoing a disorder-to-order transition in response to cellular stimuli such as oxidative stress and are implicated to play a role in plethora of biological processes. The ability of some of the cysteine-rich proteins to cycle between oxidized and reduced states has recently been identified as a mechanism of mitigating potential redox-induced stress and maintaining cellular homeostasis.26–27 The redox-sensitive regions are also often associated with metal binding; while proteins form disulfide bonds under oxidizing conditions (extracellular), metal binding is commonly observed under reducing conditions25. It is also noteworthy that the presence of redox-sensitive proteins increases with increasing organism complexity25. Furthermore in eukaryotes, there exists a linear correlation between proteins containing disordered sequences and degree of cysteines in them28. Among the proteins containing more than 20% cysteines, GRNs show similarities to metallothioneins (MTs), which are ~60 amino acid long proteins consisting of 20 cysteines (33%)29. MTs function as cellular reservoirs and scavengers of metal ions29–32 and as heavy metal detoxifiers33–34. GRNs have also been hypothesized to bind metals19, 35, and were identified as a member of metalloproteins isolated from rat liver36. Motivated by these and the observation that the cystine-rich GRNs are present both extracellularly and intracellularly within lysosomes, we surmise that GRNs sequester metals to function as redox-sensitive regulators of cellular toxicity. Here we tested our hypothesis that GRNs bind metal cations in reducing environments, in vitro. Results show that out of the seven metal ions tested, GRNs −3 and −5 specifically bind to Cu(I) in part via cysteine thiols. These results raise the question of whether copper binding is a key function of GRNs in their repertoire of biological function that has remained unnoticed thus far. The results presented here support this hypothesis and further elevates the relevance in their physiological functions.
Materials and Methods
Recombinant expression, purification, and generation of apoGRNs:
GRN-3 and GRN-5 were recombinantly expressed and purified as previously described22, 24. Briefly, GRN-3 was expressed in SHuffle™ cells (New England Biolabs), while GRN-5 was expressed in Origami 2 DE3 (Invitrogen) as fusion constructs with a thioredoxin-A and hexa-histidine tag (TrxA-Hisx6-GRN). The constructs were purified using Ni-NTA affinity chromatography and eluate was treated with 5 mM ethylenediaminetetraacetic acid (EDTA) to chelate residual metal-ions. The buffers used in purification protocol were prepared using deionized water which was passed through a chelex resin column (BioRad) to exclude metal contaminants and obtain metal-free eluate. The glassware used in purification and buffer preparation were rinsed with chelex-treated water to wash residual contaminants. After exhaustive dialysis, fusion construct was cleaved with restriction grade thrombin (Bovine, BioPharm Laboratories) at 3U per 1 mg of the protein to remove both trxA and the His-tag. The reaction was incubated at room temperature (~25°C) for 22–24 hours. Using a semi-preparative Jupiter® 5 μm 10×250 mm C18 reverse phase HPLC column (Phenomenex) protein was then purified by applying a gradient elution of 60 – 80 % acetonitrile containing 0.1% TFA on a UltiMate 3000 system (Thermo) as previously described22. The lyophilized apo-proteins were resuspended in either 20 mM HEPES-pH 7.0 or 20-mM ammonium formate-pH 4.5 prepared using chelex treated deionized water, as described before. Protein concentration was determined by measuring absorbance at 280 nm and using molar extinction coefficients of 6250 M−1cm−1 for GRN-3 and 7740 M−1cm−1 for GRN-5. Reduced forms of the proteins were used in experiments and were generated by incubating the proteins with a 20 stoichiometric excess of Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) for 2 hours at RT.
Metal-stock preparation for assays:
Stock solutions of metals were prepared from their respective salts such as calcium chloride (CaCl2), cobalt chloride (CoCl2), cadmium chloride (CdCl2), copper(II) acetate (Cu(II)Ac), manganous chloride (MnCl2), magnesium chloride (MgCl2), nickel sulfate (NiSO4), zinc chloride (ZnCl2), at a concentration of 2 mM, in either 20 mM HEPES-pH 7.0- or 20-mM ammonium formate-pH 4.5 in the presence of 12 mM glycine to avoid precipitation37–38. These buffers were prepared using deionized, Chelex treated water, as described before and were stored at 4°C in dark. Cu(I) stocks were prepared by dissolving required amount of Tetrakis(acetonitrile)copper(I) tetrafluoroborate in degassed, Chelex-treated, deionized water in either 20 mM HEPES-pH 7.0 or 20-mM ammonium formate-pH 4.5 in presence of 12 mM glycine and 2% acetonitrile. The prepared buffers were then purged with nitrogen for 1 hour at RT and used immediately for experiments to avoid oxidation of Cu(I).
MALDI-ToF mass spectrometry and Alkylation assay:
For determination of binding between metal-cations and GRNs, 1 mM metal-cations were incubated with the reduced, metal-free samples of 20 μM GRN-3 (MW 6367.7 Da) or GRN-5 (MW 6017.7 Da) in 20 mM HEPES buffer at pH 7.0 in the presence of 500 μM tris(2-carboxyethyl)phosphine hydrochloride Characterization of the protein-metal complexes was performed on a Bruker Datonics Microflex LT/SH ToF-MS system. For analysis of metal-protein reactions, 95.5 ng of GRN-3 and 90.2 ng of GRN-5 (15 pmol) were spotted separately onto a Bruker MSP 96 MicroScout Target with a 1:1:1 ratio of sample:sinapinic acid matrix(saturated with acetonitrile and water): acetone. Instrument calibration was performed using Bruker Protein Calibration Standard I (Bruker Daltonics). Alkylation reactions were carried out by incubating respective metal-GRN samples prepared as described above, with 1 mM iodoacetamide for 2h at room temperature. The samples were then prepared for analysis using MALDI-ToF-MS by spotting onto a Bruker MSP 96 MicroScout Target with a 1:1:1 ratio of sample:sinapinic acid matrix (saturated with acetonitrile and water): acetone. The MALDI-ToF data were processed and analyzed using the Bruker flexAnalysis software (Bruker Daltonics); the peak assignment and baseline subtraction were performed using the respective tools within the software.
Fluorescence spectroscopy and determination of apparent dissociation constant ):
Intrinsic tryptophan fluorescence assays were performed on a Cary Eclipse spectrometer (Agilent Inc.) by exciting the samples at 285 nm and monitoring emission from 320 to 400 nm. 20 μM GRN (3 or 5) were titrated with increasing metal concentrations (10–240 μM) at pH 7.0 and pH 4.5 and spectra were recorded. Each spectrum represents an average of four repeat scan. The collected spectrum for each titration was normalized by integrating the area under the curve and the normalized intensities were plotted against concentration of Cu. The normalized curves were fitted to the one site binding equation:
where r0 and rs are fluorescence intensities in the absence and saturated levels of the ligand, Cu, respectively, while Lt and Pt are the respective total ligand (Cu) and GRN concentrations and Kd represents the apparent dissociation constant . The data normalization and curve fitting were performed on Origin 8.5 graphing software. For obtaining the apparent dissociation constant with the incorporated dissociation constant of the glycine-Cu complex, following equation was considered; where K1 and K2 are affinity constants for GRN-metal and glycine-metal binding, respectively. Using a value of 2.6 μM for the dissociation constant of glycine-Cu complex37–38 and the values obtained for the respective GRN-Cu complexes from intrinsic fluorescence assays, obtaining the final values in the picomolar range.
Luminescence Spectroscopy:
Cu(I) Luminescence assay was performed on a Cary Eclipse spectrometer (Agilent Inc.) by exciting the samples at 325 nm and monitoring emission from 530 to 635 nm. GRNs (3 or 5) were incubated at a concentration of 20 μM at either pH 4.5 or pH 7.0 in presence of 0.5 mM TCEP. GRNs were titrated with increasing concentrations of Cu(I) or Cu(II); 10–240 μM and luminescence spectra were recorded. Each spectrum represents an average of four scans. The spectra were then blank corrected and luminescence at 625 nm plotted as a function of increasing Cu.
Circular dichroism (CD) spectroscopy:
Far UV-CD spectra were collected on Jasco J-815 instrument using a 0.1mm path length quartz cuvette (Precision cell). GRN samples incubated with respective buffers (20 mM HEPES pH 7.0 or 20 mM ammonium formate) with or without Cu as prepared previously for MALDI-MS characterization were suitably diluted from 20 μM to a concentration of 7 μM to avoid detector saturation at lower wavelengths. The samples were scanned from wavelengths 260 nm to 195 nm in the continuous scanning mode at a speed of 50nm/min. Each spectrum represents an average of three repeat scans.
Results
GRNs show redox sensitivity across their sequence similar to known metal binding proteins.
The unique chemistry of cysteine residues marked by their high reactivity and ability to form covalent bonds allows them to modulate the structure-function relation within proteins based on cellular cues. Such redox-responsive character has become associated with proteins that maintain cellular homeostasis, especially in response to oxidative stress or metallotoxicity39. To evaluate the potential redox sensitive character of GRNs −3 and −5, we utilized the IUPred2A computational platform, which predicts the disorder propensities as well as redox sensitivity of a protein40. As expected, analysis on MT-2, a well-known metal binding protein showed significant redox sensitivity across the entire stretch of the sequence (Fig 1 a). Analysis on GRNs revealed appreciable differences in the disorder propensities between the reduced and the oxidized forms (Fig 1b and c) for GRNs −3 and −5, that are deemed redox sensitive regions within the protein (Fig 1; purple area). The disorder propensities of the free-thiol forms for MT-2 (Fig 1a), GRN-3 (Fig 1b) and GRN-5 (Fig 1c) have values between 0.75 and 1.0 indicating significant structural disorder. The redox sensitivity arises from both disorder within the sequence and due to a high number of cysteines interspersed within them in the form of recognizable motifs25 Such an arrangement of cysteines within the sequence is highlighted in MTs, where two characteristic conserved motifs are utilized for metal coordination; xCCx and CxC (Fig 1a)42. The xCCx motif is also present in GRNs (Fig 1) raising the question if GRNs could bind metal ions. These in-silico assessments align well with the previous findings on the disordered structure of GRNs21–22, 24 and MTs43–44 in the reduced form.
Figure 1: Redox sensitivity of cysteine-rich proteins predicted by IUPred2A.
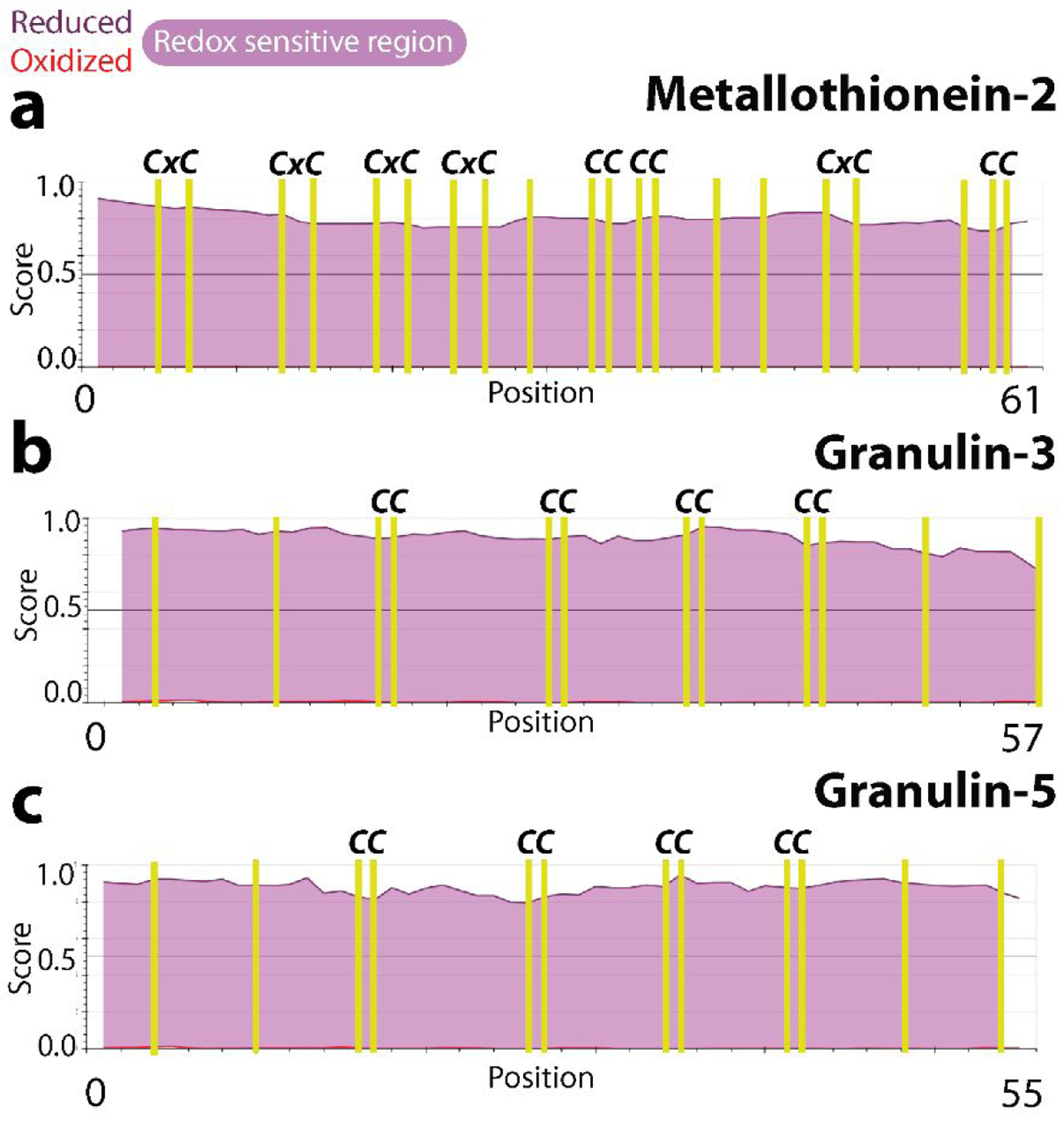
The disorder propensities of the redox forms of MT-2 (a), GRN-3 (b), and GRN-5 (c) are plotted with the disorder scores of the oxidized and reduced forms denoted by red and purple lines, respectively. Higher the score higher the disorder is. The positions of the cysteine residues are indicated by yellow vertical lines while the pink color between the red and purple lines denote the redox sensitive regions.
GRNs-3 and −5 bind Cu but no other divalent metal cations.
In order to see whether GRNs bind divalent metal cations, Cu along with Ca, Cd, Co, Mg, Mn, Ni and Zn were used. These metals were chosen for their relevance in pathophysiology; Cu, Zn, Co, Mn are cofactors for myriad enzymes, Ca and Mg are important in ionic balance and cellular signaling in addition to other important roles45 and Ni was chosen for testing the isoelectronic character and its involvement in toxicity45 while Cd was also chosen because of its toxicity45. These metal-ions were individually incubated in 50-fold molar excess with the reduced, Chelex-treated, metal-free samples of 20 μM GRN-3 (MW 6367.7 Da) or GRN-5 (MW 6017.7 Da) in 20 mM HEPES buffer at pH 7.0 in the presence of 500 μM tris(2-carboxyethyl)phosphine hydrochloride (see Methods). From these protein samples, aliquots of 15 pmols of GRN-3 and GRN-5 were then analyzed using MALDI-ToF MS (Fig 2, Fig S1; pH 7.0). At physiological pH of 7.0, both GRNs showed selective coordination up to 12 Cu for GRN-3 and −5 (Fig 2a and b). GRN-3 showed a higher co-ordination than GRN-5; for GRN-3, seven and eight Cu ions bound species were predominantly observed, while species with up to 12-Cu ions bound was also evident. GRN-5 showed 1-Cu bound species to be the most common one, with higher number of Cu-ions bound species, with up to 11-Cu bound forms, in an ascending order of abundance. GRN-3 showed more abundant Cu-bound forms of the protein with only 10% remaining in the unbound, oxidized form (6357.8 Da) (Fig 2a), while GRN-5 bound Cu, the unbound, oxidized form (6000.7 Da) was ~ 50% suggesting a diminished binding (Fig 2b). In contrast, no binding was observed for any of the other six divalent ions (Fig 2e, pH 7.0) with the exception of Zn and Cd; GRN-3 bound to one Zn was ~20% abundant while multiple Zn-bound protein constituted ~15% (Fig 2e). GRN-5 did not show the presence of Zn-bound species. Similarly, both GRNs showed weak coordination to Cd (Fig S1a, pH 7.0) where up to four Cd ions were bound to GRN-3 while GRN-5 bound up to two Cd ions although apo protein was predominant in both proteins (~ 80%) (Fig S1a). GRNs have recently been shown to be generated within the lysosomes also,17–18 while a central role of this organelle as a regulator of metal homeostasis has also come to the light46. To mimic the binding of GRNs to metalions within a lysosomal environment, 20 μM GRN-3 or 5 was incubated with 50-fold molar excess of metal ions at pH 4.5, buffered with 20 mM ammonium formate. In the lysosome-like acidic environment, Cu binding capability for GRN-3 and 5 were assessed by MALDI-ToF (Fig 2c and d). GRN-3 showed up to seven Cu bound to the protein but with significantly decreased intensities as compared to those at pH 7.0 indicating diminished binding capability at the lower pH (Fig 2c). GRN-5 also showed significantly diminished binding with only one and three Cu bound to the protein (Fig 2d). The relative abundance of the Cu-bound protein was not more than ~10% as compared to the apo form of the respective proteins, which showed ~90% abundance based on the spectral intensities. Unlike other metal ions, both GRNs displayed a weak but discernible coordination of Cd in the acidic environment, with up to two Cd ions being bound, which corresponded to ~10% abundance (Fig S1a, pH 4.5). A mass difference is observed for the non-coordinated species amongst the two pH conditions (pH 7.0 – pH 4.5) of both GRNs; 7.4 Da for GRN-3 (Fig 2a and c) and 4.1 Da for GRN-5 (Fig 2b and d) which can explained by the higher percentage of cysteines being present in the deprotonated thiolate form at the neutral pH47. Both proteins also showed inability to binding other metal cations at pH 4.5 (Fig 2e; pH 4.5). The attenuated metal binding at low pH is not surprising and could likely be due to chelating functional groups such as thiols being predominantly present in a protonated form47.
Figure 2: Metal binding analysis of GRNs by MALDI-ToF.
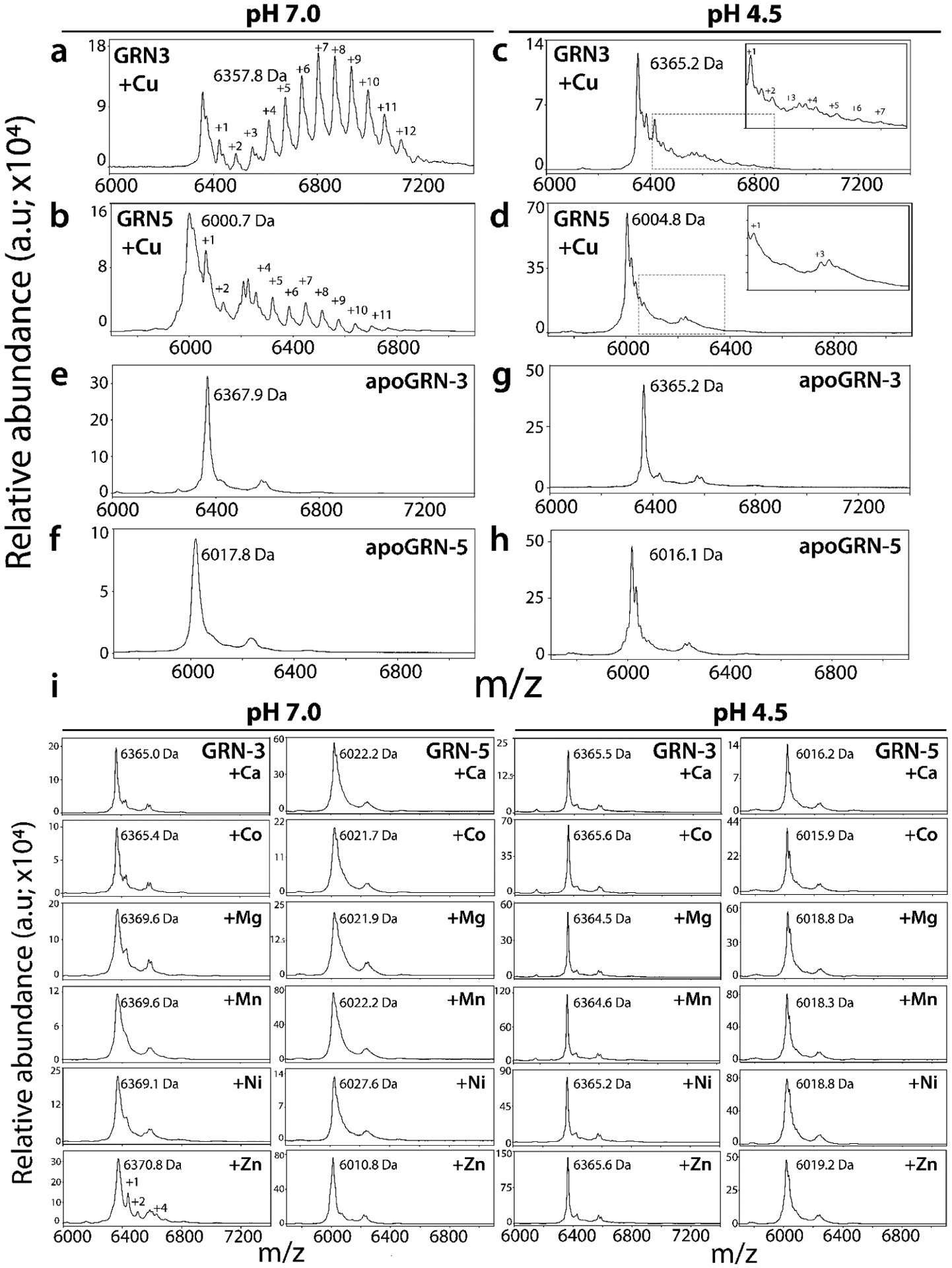
GRN-3 or GRN-5 (20 μM) were incubated separately with a 1mM (50-fold molar excess) of respective metal cations in 20 mM HEPES, 12 mM glycine at pH 7.0 or in 20 mM ammonium formate, 12 mM glycine at pH 4.5 individually, at 4°C overnight. For MALDI-ToF-MS analysis, aliquots containing 6 ×103 pmol/L of GRN-3 and GRN-5 from the respective reactions loaded on to the plate. Cu binding to GRN-3 (a) or GRN-5 (b) at neutral pH, and to GRN-3 (c) or GRN-5 (d) at pH 4.5. Apo GRN-3(e, g) or apo GRN-5 (f, h), prepared in similar buffers at the respective pH but without the metal cations. i) Spectra of Ca, Co, Mg, Mn, Ni and Zn binding to GRN-3 or GRN-5 at pH 7.0 (left panel) and at 4.5 (right panes).
Based on our hypothesis, free thiols in cystines are involved in metal coordination. Therefore, to determine how many cystines are involved in coordination, alkylation of free thiols in the Cu-bound holoproteins was conducted with iodoacetamide (IAA) for both GRNs. IAA reacts with free thiols to forms acetamide adduct (58 Da) with sulfur48. The Cu-bound GRN-3 at pH 7.0 displayed a gaussian distribution centered around ~two alkylated species (6490.8 Da) suggesting that most of the cystines were coordinated to Cu at pH 7.0 (Fig 3a). Heterogeneous population of species involving metal-bound and alkylated, only alkylated, or only metal-bound forms, should appear as clustered signals at m/z values of ~6700–7000 Da. Clearly, the gaussian curve is unsymmetrical towards the trailing edge suggesting this is indeed the case (Fig 3a). GRN-5 at the same pH showed no free thiols available for alkylation by IAA with a major peak of 6009.0 Da corresponding to the non-alkylated, oxidized form (Fig 3b). Similar to GRN-3, a well-defined non-alkylated signal is followed by trailing peaks indicating the presence of multiple alkylated, metal bound forms with a descending level of abundance (Fig 3b). Furthermore, despite attenuated binding of Cu, both GRNs under acidic conditions also show decreased alkylation suggesting that the interaction with the metal-ion leads to the unavailability of free thiols within GRN-3 (Fig 3c) and GRN-5 (Fig 3d), possibly due to induced oxidation (further elaborated in Discussion). The spectra of GRN-3 showed a defined signal at 6464.4 Da which corresponds to the monomeric protein without any alkylation (Fig 3c). Similar spectrum is observed for GRN-5, with a signal at 6014.9 Da (Fig 3d), again indicating the lack of thiols available for alkylation. The observation also suggests potential oxidation of cysteines induced by Cu at low pH. The alkylation of GRN-3 incubated with other metal ions at pH 7.0 indicated most of the thiols were free and available for labeling by IAA, except for Cd, Mn, and Zn. The sample with Mn showed a decrease of two free thiols, while the protein with Zn displayed a heterogenous mixture of multiply-alkylated forms; with four, seven and eleven free thiol species (6569.9 Da, 6738.1 Da, 7013.7 Da; Fig 3e, GRN-3 at pH 7.0) being the predominant ones. In the presence of Zn ion, a similar spectrum containing multiple alkylated species is observed for GRN-5, with four and eleven free thiols forms (6240.9 Da, 6660.1 Da; Fig 3e, GRN-5 at pH 7.0). For samples incubated with Cd (Fig S1b, pH 7.0), GRN-3 displays species with up to ten (6935.9 Da) to twelve (7051.8 Da) free thiols, while a signal for monomeric, non-alkylated form (6354.9 Da) is also visible (Fig S1b, pH 7.0, GRN-3). On the other hand, GRN-5 shows peaks corresponding to eleven (6651.5 Da) to twelve (6705.5 Da) free-thiol species at neutral pH (Fig S1b, pH 7.0, GRN-5) reflecting a weak affinity for the metal (Fig S1a, pH 7.0, GRN-5). These observations along with the detectable albeit weak binding of GRNs to Cd and Zn (Fig 2e, pH 7.0; GRN-3 and Fig S1b, pH 7.0; GRN-3 and GRN-5) further indicates potentially weak interactions of these metal-ions with GRNs at neutral pH. In acidic pH, GRNs show decreased availability of thiols for alkylation as indicated by the spectra of GRN-3 and 5 control samples without any metal-ions (Fig 3e, GRN-3 and GRN-5 at pH 4.5). The lower pH increases the concentration of the protonated thiols and hence, decreases alkylation by IAA, an observation that is in line with a decrease in rate of thiol alkylation by 2-vinylpyridine in an acidic pH49. Compared to the control reactions, no other metal-ion incubated samples showed any significant deviation in terms of alkylation patterns, except for Zn incubated samples, where both proteins showed an increase of two-three thiols available for alkylation (Fig 3e, Fig S1b, GRN-3 and GRN-5 at pH 4.5). Alkylation of thiols using IAA is widely performed, especially due to low levels of unwanted, side reactions that accompany this chemistry48, 50. Despite this, our results indicate the presence of alkylation of residues other than cysteine, as observed in Fig 3e, GRN-3 at pH 7.0, where a discernible signal from a species with 13-alkylated functional groups is present at a mass of 7121.3 Da. This points towards alkylation of either an acidic residue or histidine, along with cysteine thiols48. Another side reaction observed is the modification of N-terminal methionine residues of GRNs on reaction with the iodine containing alkylating agent resulting in the loss of a 47 Da fragment from these proteins51. This loss is evident in the IAA reactions of GRN-5 (Fig 3e, GRN-5 at pH 7.0 and pH 4.5) where a signal is observed at 5970.7 Da, preceding the monomeric, non-alkylated form of the protein (GRN-5 MW; 6017.7 Da).
Figure 3: Determination of free thiols by IAA alkylation of GRNs.
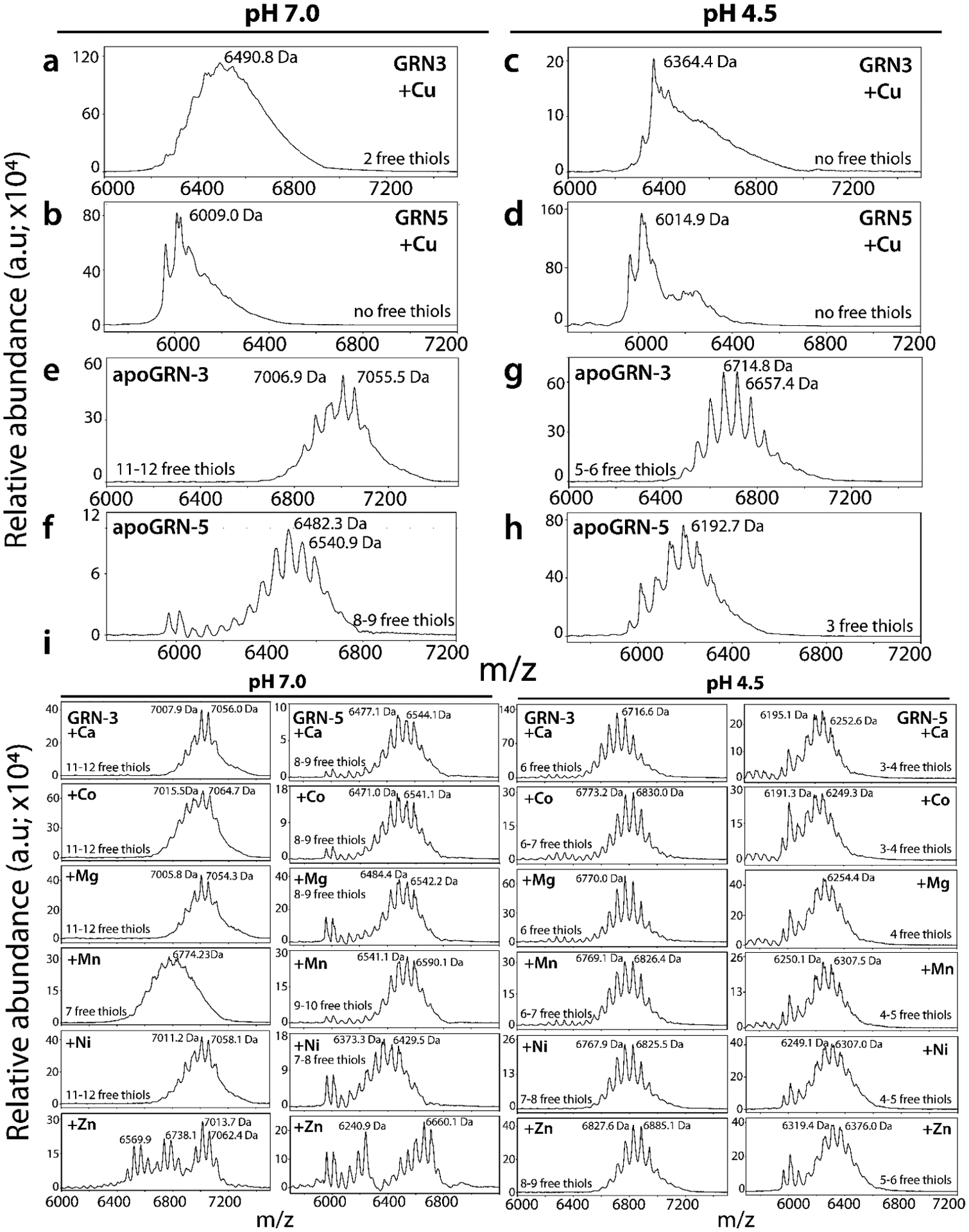
Holoproteins from Fig 2 were used for alkylation reactions using 1 mM iodoacetamide (IAA) and aliquots of samples were analyzed using MALDI-ToF-MS. The spectra of alkylated samples of Cu incubated GRN-3 (a) and GRN-5 (b) in a pH 7.0 buffer and GRN-3 (c) and GRN-5 (d) in pH 4.5. e-f) MALDI-ToF-MS spectra of apoGRNs at the two pH conditions tested. i) The spectra of alkylation reactions of GRN-3 and −5 incubated with other metal-ions (Ca, Co, Mg, Mn, Ni and Zn) at neutral and acidic pH.
Copper binding induces multimerization of GRNs.
As mentioned earlier, the interaction of GRNs with Cu decreased the availability of free thiols for alkylation suggesting their potential oxidation (Fig 3a–d). To determine whether the thiols are involved in intra or inter-molecular disulfide bonding following their interaction with Cu and other metal-ions, we subjected the samples from Fig 2 to polyacrylamide gel electrophoresis (PAGE). At neutral pH, GRN-3 (MW; 6367.7 kDa) is observed to undergo multimerization in the presence of Cu (Fig 4a, +Cu). Band corresponding to the dimeric form (MW; 12.6 kDa) is prominently visible while faint bands corresponding to monomeric and trimeric species (MW; 19.1 kDa) are also observed (Fig 4a). On the other hand, only a band corresponding to the monomeric form is seen in the presence of other metal cations, similar to apoGRN-3 (Fig 4a). Prior characterization of GRN-5 (MW; 6017.7 Da) in our lab revealed the monomeric protein displays a decreased electrophoretic mobility in PAGE, a commonly observed property among disordered proteins24, 52. In line with our previous observation, apoGRN-5 is observed as a band at ~30 kDa at neutral pH (Fig 4b, apoGRN-5). In the presence of Cu, a diffuse, ‘smear’ pattern is visible with no distinct band, indicating significant multimerization of the protein (Fig 4b, +Cu). All other metal cations, with the exception of Zn and Ni, showed only a single band similar to aopGRN-5 (Fig 4b). In the presence of Zn and Ni, bands are observed at ~17 and 20 kDa, respectively (Fig 4b). It is unclear as to why such an odd migration is observed for these metals. Nevertheless, multimerization is not observed with other metal cations in a manner that is observed with Cu. At pH 4.5, all metal-incubated GRN-3 samples show the presence of a distinct trimer at 18 kDa (Fig4c). In the presence of Cu, the degree of multimerization is prominent with a heterogenous mixture of multimeric species forming a ‘smear’ pattern, where the monomeric, dimeric and trimeric bands are still discernible (Fig4c, +Cu). Other metal-ions failed to induce the multimerization of GRN-3, similar to the observation at pH 7.0 (Fig 4c). At pH 4.5, GRN-5 shows an electrophoretic pattern similar to one observed at neutral pH (Fig 4b and d). The presence of Cu induces multimerization of GRN-5 but it is not as pronounced as that observed at pH 7.0 (Fig 4b and d, +Cu). A distinct band is observed at ~30 kDa, similar to apoGRN-5 and a characteristic smear of multimerized protein extending above 50 kDa. Sample with Zn shows higher electrophoretic mobility in the acidic regime as well, with the protein band present at ~17 kDa. At both the pH conditions tested, Mg and Mn are observed to induce a visible smearing of GRN-5 towards the lower molecular weight region, indicating higher electrophoretic mobility of the species formed (Fig 4b and d, +Mg, +Mn). These results indicate that interaction with Cu (most likely Cu(II)), but not necessarily binding, seems to induce the oxidation of cysteines within GRNs with consequent intermolecular disulfide crosslinks leading to multimerization of the protein.
Figure 4: Binding and metal-induced structural changes in GRNs.
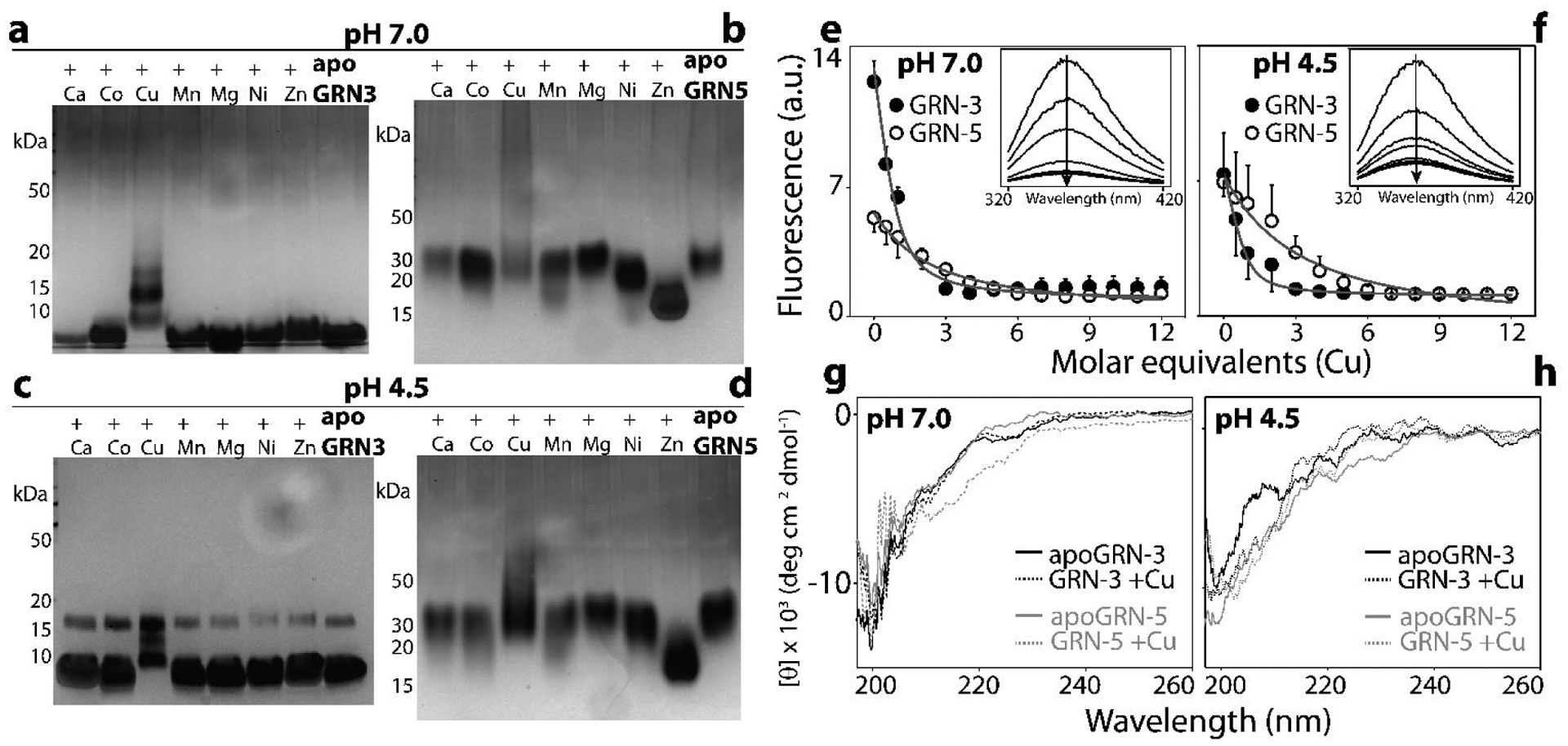
Metal-incubated samples of GRNs were subjected to PAGE in non-reducing conditions. (a-d) and spectroscopic analysis (e-h). Silver stained gels of Apo and holoGRN-3 (a) and apo and holoGRN-5 (b) at neutral pH; apo and holo GRN-3 (c) and GRN-5 (d) at pH 4.5. Intrinsic tryptophan fluorescence of GRNs (20 μM) measured as a function of increasing Cu (10–240 μM) concentration at pH 7.0 (e) and pH 4.5 (f) to evaluate tertiary structural changes. (inset): raw spectral scans shown as a function of increasing Cu concentrations (arrow). g and h) Far-UV circular dichroism (CD) spectra of apo and holoGRNs incubated with Cu(II at neutral pH (g) and at pH 4.5 (h).
To see whether metal binding induces changes to the conformational environment surrounding the lone tryptophan residue in the proteins, the reduced apo forms of 20 μM GRN-3 or GRN-5 were titrated with increasing molar concentrations (10–240 μM) of Cu and the intrinsic tryptophan fluorescence was measured. This informs about the changes in the environment around the tryptophan residue as a function of metal ion concentration, and therefore allows the calculation of apparent binding affinity for the metals21, 38, 53–54. At neutral pH, GRN-3 shows a higher intrinsic fluorescence than GRN-5 in absence of Cu (Fig 4e). Such a variation in fluorescence intensities suggests an inherent difference in the tryptophan environment amongst the two proteins54. Increasing Cu concentrations leads to the decrease in tryptophan fluorescence intensity in both GRNs (Fig 4e). This observation indicates an increased solvent exposure upon metal binding22. The data was fitted to a one-site binding equation (detailed in Methods) to obtain an apparent dissociation constant () for GRNs and Cu; 4.0 ± 1.5 μM for GRN-3 and 35.4 ± 8.2 μM for GRN-5. This however, does not account for the binding of Cu by glycine used in the buffer37–38. Upon correcting the binding constant for this nonspecific competitive binding of glycine (see Methods), 10.4 pM and 92.0 pM for GRN-3 and GRN-5, respectively are obtained. At acidic pH, both GRNs have equivalent fluorescence intensities in absence of Cu and show a similar trend of attenuation of fluorescence intensity with increasing Cu concentration. The apparent corrected dissociation constants for this interaction are 6.9 pM for GRN-3 and 130.5 pM for GRN-5. These apparent dissociation constants are comparable to those of known Cu-coordinating proteins, such as MT-255. It has to be borne in mind these values only reflect indirect affinities for Cu; at pH 4.5, binding of Cu is observed to be attenuated (Fig 2c and d), and yet quenching of fluorescence is pronounced. This anomaly indicates that the dissociation constants inform us about the structural changes around tryptophan, which may arise not only due to direct metal binding but also due to consequential multimerization.
Intrinsically disordered proteins have been known to obtain structural order upon metal coordination or binding to ligands52. To probe whether metal binding induces secondary structural changes within GRNs, far-UV circular dichroism (CD) was used samples from Fig 2, which were suitably diluted to 7 μM GRN concentrations. In agreement with previous biophysical studies on GRN-3 and GRN-5, at pH 7.0, both apo proteins display spectra characteristic of random coil with a minimum observed at 200 nm (Fig 4g). Cu-binding fails to induce any secondary structural elements within GRN-3 as evidenced by a spectrum similar to its apo form. At neutral pH, GRN-5 in presence of Cu shows a weak minimum at ~215 nm, indicative of some β-sheet content within the protein. Such induction of secondary structure within unstructured proteins by metal coordination is well known, although the intensity of the ellipticity observed here suggests GRN-5 remains mostly unstructured. At low pH also, apo forms of GRN-3 and GRN-5 display random coil spectra, with a minimum at 199nm, indicating these proteins remain unstructured in the acidic environment (Fig 4h). As would be expected, the Cu-incubated samples show overlapping signals with the apo-proteins (Fig 4h), since no coordination is detected with Cu at this pH (Fig 2 c and d). These results show that interaction with Cu brings about tertiary and quaternary structural changes within GRNs, as evidenced by the fluorescence quenching (Fig 4e and f) and multimerization (Fig 4a–d), respectively, but fails to induce significant secondary structural changes. These observations are similar to those observed for MTs, where these proteins do not attain any secondary structure on metal coordination but become conformationally constrained42, 44, 56.
GRNs show selective binding to Cu(I).
Since the reducing agent used in our study, TCEP has been shown to be able to reduce Cu(II) to Cu(I)57, so we questioned whether GRNs selectively bind Cu(II) or Cu(I). In order to address this, we performed luminescence spectroscopy where the complexation of diamagnetic Cu(I), and not Cu(II), leads to the emergence of a luminescence band in the wavelength range of 620–640 nm58–60. The luminescence band is also indictive of the environment of the complexed Cu(I); solvent exposed Cu(I)-complex leads to a quenching of the luminescence by the solvent58–59. In the luminescence assay, 20 μM GRN-3 or −5 sample was titrated with increasing concentrations of Cu(II) solution containing TCEP in the buffer. An emission band was observed at 625 nm that showed linear increase with the increase in Cu concentration followed by a sharp decrease for both GRNs (Fig 5a). Luminescence for GRN-3 in presence of Cu increased for up to 5–7 molar equivalents of Cu added and started to be quenched after (solid grey line), while for GRN-5, the luminescence increased up to 3 molar equivalents of Cu (dashed grey line) before being quenched (Fig 5a). To further confirm Cu(I) binding, the specific Cu(I) compound, tetrakis(acetonitrile)copper(I) tetrafluoroborate was used in the same buffer conditions which showed identical increases in the luminescence intensities (Fig 5b); increasing luminescence up to seven molar equivalents of Cu(I) for GRN-3 (solid grey line) and four molar equivalents for GRN-5 (dashed grey line), followed by quenching. Thus, the inflection point in the increase and decrease of the intensities provide insights into the binding stoichiometry. In acidic conditions, titration of Cu(II) on GRNs in TCEP containing buffer showed an inflection point that shifted to lower Cu equivalents; four molar equivalents for GRN-3 and two molar equivalents for GRN-5 (Fig 5c). Similarly, a shifted inflection point was observed on Cu(I) titration, where the luminescence signal peaked at two molar equivalents of Cu(I) and was followed by its quenching (Fig 5d). These results suggest a diminished coordination capacity of GRNs in an acidic environment. Together, based on these data, GRN-3 (grey lines) and GRN-5 (dashed grey lines) show binding stoichiometries of ~ 7 and 3 Cu(I) ions (Fig 5) at neural pH, and ~2–3 Cu(I) ions in an acidic environment, which seems to agree with the data obtained from mass spectrometry (Fig 2). To further investigate whether this selective Cu(I) can promote multimerization of GRNs, the samples from luminescence assays with Cu(I) were subjected to PAGE under denaturing, non-reducing conditions (Fig 5e). GRN-3, under both pH conditions tested undergoes multimerization, with multimers up to the tetrameric form distinctly visible GRN-3 (Fig 5e, pH 7.0 and 4.5, GRN-3). Similarly, for GRN-5 multimerization is evident from the characteristic smearing pattern which extends up to 50 kDa and higher(Fig 5e, pH 4.5 and 7.0, GRN-5). These observations are identical to those observed with Cu(II) salts in the presence of TCEP (Fig 4) and imply that Cu(I) coordination by GRNs induces their multimerization by free thiols present in the protein after metal coordination and/or by coupled redox chemistry involving disulfide bond formation induced by the reduction of Cu(II).
Figure 5: Determination of the oxidation state of Cu bound to GRNs.
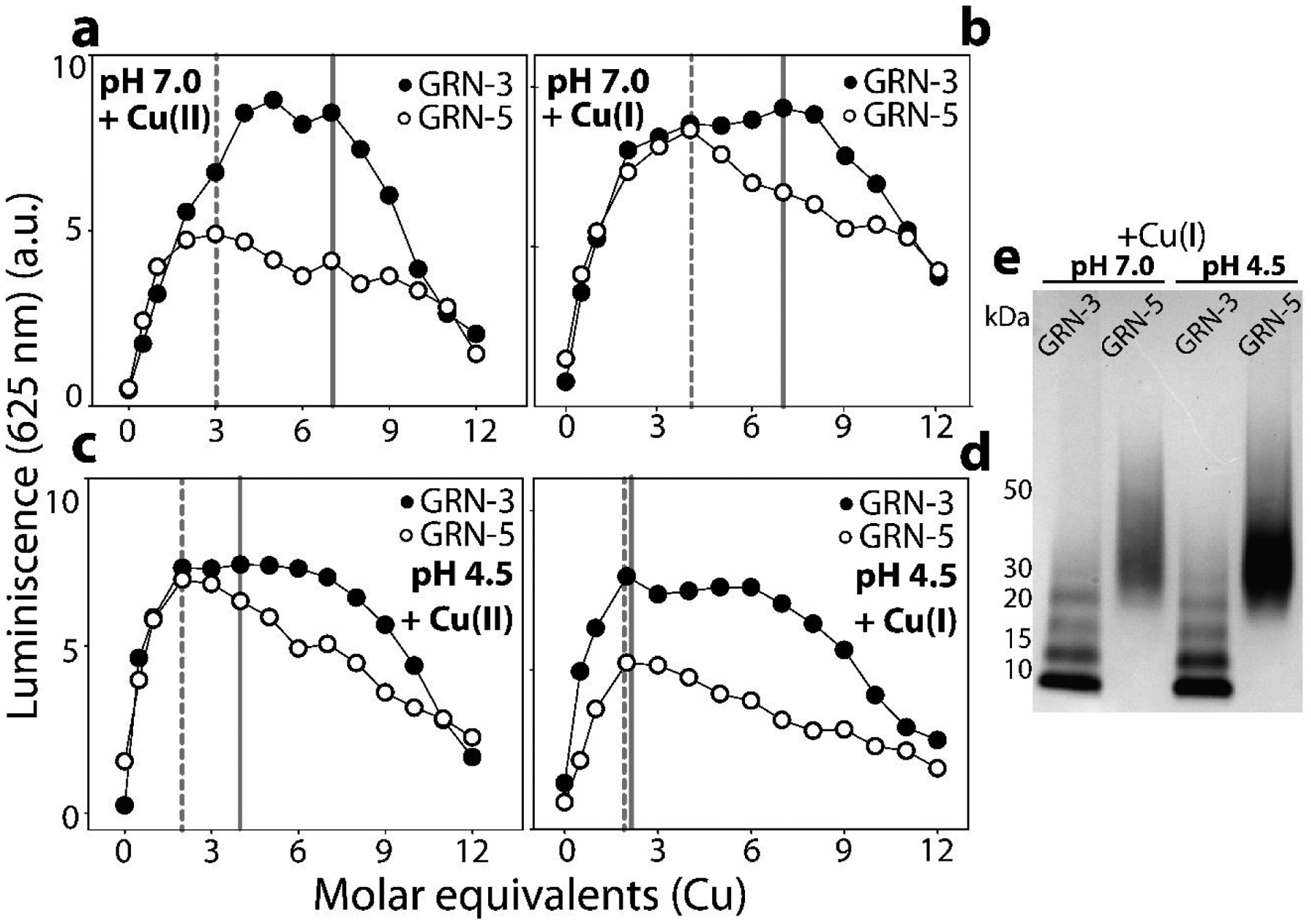
GRN-3 or 5 (20 μM) were incubated separately in 20 mM HEPES, 12 mM glycine at pH 7.0 or pH 4.5 in presence of 0.5 mM TCEP. The apo-proteins were titrated separately with increasing concentrations of either a) Cu(II) or b) Cu(I) at neutral pH and similarly with c) Cu(II) and d) Cu(I) at acidic pH and luminescence was recorded. e) Samples of GRNs incubated with Cu(I) at neutral and acidic pH were subjected to PAGE in non-reducing conditions.
Discussion
The potential of GRNs to coordinate metal ions has been hypothesized ever since their initial characterization35, 61, and a recent study by Fang and colleagues strengthened the rationale for such an investigation36. Our report brings forth a property of GRNs that has remained largely unexplored particularly at the protein level. Characteristics such as the high number of conserved cysteines and structural disorder in fully reducing conditions21 make GRNs similar to MTs, which are known metal sequesters of Cu, Zn and Cd ions, among others42. Further motivated by the IUPred2A predictions that show GRNs to be similar to MTs in redox sensitivity, we set out to test whether GRNs show affinity for divalent cations and reveal potential promiscuity or specificity of such metal coordination36. The results indicate that both GRNs −3 and −5 have affinity towards Cu with GRN-3 showing greater affinity than GRN-5 at neural pH. However, both GRNs, at acidic and neutral pH, failed to show a significant binding to any of the other metal cations, highlighting a selectivity towards Cu.
We also observe that binding of copper fails to induce any secondary structural changes in either GRN-3 or GRN-5, a characteristic similar to that of MTs56. Such conformational flexibility has become an identifiable property of IDPs that are often associated with pleiotropic roles52, 62–63. These observations align well with the multiple functional roles that GRNs are known to play and implicate copper-binding to be another function these peptides may have in their repertoire. Furthermore, interaction with Cu, but not other metals, induces the formation of multimeric GRN-3 or −5, under both pH conditions tested. Alkylation experiments using IAA suggest that the interaction with copper leaves no or few free cysteines within both GRNs. A majority of cysteines are either coordinated to copper or involved in intra- or inter-molecular disulfide bond formation, or both. This brings to light two possible scenarios that may contribute to the mechanism of GRN functions: i) A fraction of Cu(II) oxidizes thiols in some cysteines to form disulfide bonds leading to multimerization and the protein binds preferentially to Cu(I) (generated from the reduction of Cu(II) by thiols and TCEP) or ii) Cu(I) coordinates to cysteines from two GRN monomers forming a bridge ligand leading to multimerization. The former scenario is supported by the fact that decreasing the pH to 4.5 decreases the extent of multimerization of GRNs due to the protonation of thiols. The results here show the coordination of Cu at neutral pH, and its abrogation in an acidic pH, provides cues into their potential cellular roles; GRNs could mitigate cu-induced cytotoxicity by binding Cu(I) in the reducing, cytoplasmic environment and releasing them within the acidic, lysosomal milieu where GRNs are known to localize17–18. Secondly, the dissociation constants of Cu(I) with GRNs measured indirectly by intrinsic tryptophan fluorescence as a function of metal concentrations reveal that the affinities both GRNs for Cu(I) are in the picomolar range. This is comparable to the affinity of MTs for Cu(I), which is in the pM-fM range64. Mammalian cells mainly employ MTs and superoxide dismutases (SOD) to protect themselves from ROS activation by excess Cu. The comparable affinity range of copper for GRNs, MTs and SODs as well as the potential reduction of Cu(II) to Cu(I) by the cysteines, suggest that GRNs may play a protective role during Cu(II) induced oxidative stress to the cells similar to MTs and SODs by either promoting disulfide bonds or binding to free cystines or both. However, unlike MTs and SODs, GRNs seem to exhibit selectivity towards copper and only a negligible binding of Cd (supplementary data) based on the results obtained here. What precise physiological significance copper binding holds is unclear at this point but one can speculate that a network of copper binding proteins might be employed, driven by affinity gradients55. Intracellular GRNs may participate in mitigating ROS toxicity by behaving as additional metal scavengers which may be needed during acute Cu influx. Such a mechanism can bypass the ROS generation induced by free Cu and its associated Fenton-like chemistry. To our knowledge, this study is the first to uncover the Cu-binding ability of GRNs, adding it to the known functional repertoire of these proteins. Although intriguing, these results warrant further investigations into the interactions of GRNs with metal-cations to obtain a clear picture of what roles GRNs play in metal homeostasis in norm and pathology.
Supplementary Material
Acknowledgements
The authors would like to thank the following agencies for financial support: National Institute of Aging (1R56AG062292–01) and the National Science Foundation (NSF CBET 1802793) to VR. The authors also thank the National Center for Research Resources (5P20RR01647–11) and the National Institute of General Medical Sciences (8 P20 GM103476–11) from the National Institutes of Health for funding through INBRE for the use of their core facilities.
References
- 1.Palfree RG; Bennett HP; Bateman A, The Evolution of the Secreted Regulatory Protein Progranulin. PloS one 2015, 10 (8), e0133749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gass J; Lee WC; Cook C; Finch N; Stetler C; Jansen-West K; Lewis J; Link CD; Rademakers R; Nykjær A; Petrucelli L, Progranulin regulates neuronal outgrowth independent of Sortilin. Molecular Neurodegeneration 2012, 7 (1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X; Joselin AP; Wang L; Kar A; Ray P; Bateman A; Goate AM; Wu JY, Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK-3β. Protein Cell 2010, 1 (6), 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Damme P; Van Hoecke A; Lambrechts D; Vanacker P; Bogaert E; van Swieten J; Carmeliet P; Van Den Bosch L; Robberecht W, Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. The Journal of cell biology 2008, 181 (1), 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Z; Ong CH; Halper J; Bateman A, Progranulin is a mediator of the wound response. Nature medicine 2003, 9 (2), 225–9. [DOI] [PubMed] [Google Scholar]
- 6.Zhao YP; Tian QY; Frenkel S; Liu CJ, The promotion of bone healing by progranulin, a downstream molecule of BMP-2, through interacting with TNF/TNFR signaling. Biomaterials 2013, 34 (27), 6412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang W; Lu Y; Tian QY; Zhang Y; Guo FJ; Liu GY; Syed NM; Lai Y; Lin EA; Kong L; Su J; Yin F; Ding AH; Zanin-Zhorov A; Dustin ML; Tao J; Craft J; Yin Z; Feng JQ; Abramson SB; Yu XP; Liu CJ, The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science (New York, N.Y.) 2011, 332 (6028), 478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y; Bateman A, The Glycoprotein Growth Factor Progranulin Promotes Carcinogenesis and has Potential Value in Anti-cancer Therapy. Journal of Carcinogenesis & Mutagenesis 2012, 01. [Google Scholar]
- 9.Le Ber I; van der Zee J; Hannequin D; Gijselinck I; Campion D; Puel M; Laquerriere A; De Pooter T; Camuzat A; Van den Broeck M; Dubois B; Sellal F; Lacomblez L; Vercelletto M; Thomas-Anterion C; Michel BF; Golfier V; Didic M; Salachas F; Duyckaerts C; Cruts M; Verpillat P; Van Broeckhoven C; Brice A, Progranulin null mutations in both sporadic and familial frontotemporal dementia. Human mutation 2007, 28 (9), 846–55. [DOI] [PubMed] [Google Scholar]
- 10.Ward ME; Chen R; Huang HY; Ludwig C; Telpoukhovskaia M; Taubes A; Boudin H; Minami SS; Reichert M; Albrecht P; Gelfand JM; Cruz-Herranz A; Cordano C; Alavi MV; Leslie S; Seeley WW; Miller BL; Bigio E; Mesulam MM; Bogyo MS; Mackenzie IR; Staropoli JF; Cotman SL; Huang EJ; Gan L; Green AJ, Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Science translational medicine 2017, 9 (385). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh H-S; Choi N; Tarassishin L; Lee SC, Regulation of progranulin expression in human microglia and proteolysis of progranulin by matrix metalloproteinase-12 (MMP-12). PloS one 2012, 7 (4), e35115–e35115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessenbrock K; Frohlich L; Sixt M; Lammermann T; Pfister H; Bateman A; Belaaouaj A; Ring J; Ollert M; Fassler R; Jenne DE, Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. The Journal of clinical investigation 2008, 118 (7), 2438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park B; Buti L; Lee S; Matsuwaki T; Spooner E; Brinkmann MM; Nishihara M; Ploegh HL, Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity 2011, 34 (4), 505–13. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J; Nathan C; Jin W; Sim D; Ashcroft GS; Wahl SM; Lacomis L; Erdjument-Bromage H; Tempst P; Wright CD; Ding A, Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 2002, 111 (6), 867–78. [DOI] [PubMed] [Google Scholar]
- 15.Cenik B; Sephton CF; Kutluk Cenik B; Herz J; Yu G, Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. The Journal of biological chemistry 2012, 287 (39), 32298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CW; Stankowski JN; Chew J; Cook CN; Lam Y-W; Almeida S; Carlomagno Y; Lau K-F; Prudencio M; Gao F-B; Bogyo M; Dickson DW; Petrucelli L, The lysosomal protein cathepsin L is a progranulin protease. Molecular Neurodegeneration 2017, 12 (1), 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X; Paushter DH; Feng T; Sun L; Reinheckel T; Hu F, Lysosomal processing of progranulin. Molecular Neurodegeneration 2017, 12 (1), 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holler CJ; Taylor G; Deng Q; Kukar T, Intracellular Proteolysis of Progranulin Generates Stable, Lysosomal Granulins that Are Haploinsufficient in Patients with Frontotemporal Dementia Caused by GRN Mutations. eNeuro 2017, 4 (4), ENEURO.0100–17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrabal R; Chen Z; James S; Bennett HP; Ni F, The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nature structural biology 1996, 3 (9), 747–52. [DOI] [PubMed] [Google Scholar]
- 20.Tolkatchev D; Malik S; Vinogradova A; Wang P; Chen Z; Xu P; Bennett HP; Bateman A; Ni F, Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein science : a publication of the Protein Society 2008, 17 (4), 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghag G; Wolf LM; Reed RG; Van Der Munnik NP; Mundoma C; Moss MA; Rangachari V, Fully reduced granulin-B is intrinsically disordered and displays concentration-dependent dynamics. Protein engineering, design & selection : PEDS 2016, 29 (5), 177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghag G; Holler CJ; Taylor G; Kukar TL; Uversky VN; Rangachari V, Disulfide bonds and disorder in granulin-3: An unusual handshake between structural stability and plasticity. Protein science : a publication of the Protein Society 2017, 26 (9), 1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhopatkar AA; Ghag G; Wolf LM; Dean DN; Moss MA; Rangachari V, Cysteine-rich granulin-3 rapidly promotes amyloid-beta fibrils in both redox states. The Biochemical journal 2019, 476 (5), 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhopatkar AA; Uversky VN; Rangachari V, Granulins modulate liquid-liquid phase separation and aggregation of the prion-like C-terminal domain of the neurodegeneration-associated protein TDP-43. The Journal of biological chemistry 2020, 295 (8), 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erdős G; Mészáros B; Reichmann D; Dosztányi Z, Large-Scale Analysis of Redox-Sensitive Conditionally Disordered Protein Regions Reveals Their Widespread Nature and Key Roles in High-Level Eukaryotic Processes. PROTEOMICS 2019, 19 (6), 1800070. [DOI] [PubMed] [Google Scholar]
- 26.van der Reest J; Lilla S; Zheng L; Zanivan S; Gottlieb E, Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nature Communications 2018, 9 (1), 1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klomsiri C; Karplus PA; Poole LB, Cysteine-based redox switches in enzymes. Antioxid Redox Signal 2011, 14 (6), 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhopatkar AA; Uversky VN; Rangachari V, Disorder and cysteines in proteins: A design for orchestration of conformational see-saw and modulatory functions. In Progress in Molecular Biology and Translational Science, Academic Press: 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaegi JHR; Schaeffer A, Biochemistry of metallothionein. Biochemistry 1988, 27 (23), 8509–8515. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Menendez S; Garcia M; Fernandez B; Alvarez L; Fernandez-Vega-Cueto A; Coca-Prados M; Pereiro R; Gonzalez-Iglesias H, The Zinc-Metallothionein Redox System Reduces Oxidative Stress in Retinal Pigment Epithelial Cells. Nutrients 2018, 10 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suhy DA; Simon KD; Linzer DIH; O’Halloran TV, Metallothionein Is Part of a Zinc-scavenging Mechanism for Cell Survival under Conditions of Extreme Zinc Deprivation. Journal of Biological Chemistry 1999, 274 (14), 9183–9192. [DOI] [PubMed] [Google Scholar]
- 32.Sabolic I; Breljak D; Skarica M; Herak-Kramberger CM, Role of metallothionein in cadmium traffic and toxicity in kidneys and other mammalian organs. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine 2010, 23 (5), 897–926. [DOI] [PubMed] [Google Scholar]
- 33.Palmiter RD, The elusive function of metallothioneins. Proceedings of the National Academy of Sciences 1998, 95 (15), 8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L; Ma L; Bai Q; Zhu X; Zhang J; Wei Q; Li D; Gao C; Li J; Zhang Z; Liu C; He Z; Zeng X; Zhang A; Qu W; Zhuang Z; Chen W; Xiao Y, Heavy metal-induced metallothionein expression is regulated by specific protein phosphatase 2A complexes. The Journal of biological chemistry 2014, 289 (32), 22413–22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo F; Lai Y; Tian Q; Lin EA; Kong L; Liu C, Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis & Rheumatism 2010, 62 (7), 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang C; Zhang L; Zhang X; Lu H, Selective enrichment of metal-binding proteins based on magnetic core/shell microspheres functionalized with metal cations. Analyst 2015, 140 (12), 4197–4205. [DOI] [PubMed] [Google Scholar]
- 37.Thompsett AR; Abdelraheim SR; Daniels M; Brown DR, High affinity binding between copper and full-length prion protein identified by two different techniques. The Journal of biological chemistry 2005, 280 (52), 42750–8. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad MF; Singh D; Taiyab A; Ramakrishna T; Raman B; Rao CM, Selective Cu2+ Binding, Redox Silencing, and Cytoprotective Effects of the Small Heat Shock Proteins αA- and αB-Crystallin. Journal of Molecular Biology 2008, 382 (3), 812–824. [DOI] [PubMed] [Google Scholar]
- 39.Giles NM; Watts AB; Giles GI; Fry FH; Littlechild JA; Jacob C, Metal and Redox Modulation of Cysteine Protein Function. Chemistry & Biology 2003, 10 (8), 677–693. [DOI] [PubMed] [Google Scholar]
- 40.Mészáros B; Erdős G; Dosztányi Z, IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Research 2018, 46 (W1), W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semple F; Dorin JR, β-Defensins: Multifunctional Modulators of Infection, Inflammation and More? Journal of Innate Immunity 2012, 4 (4), 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vašák M; Meloni G, Chemistry and biology of mammalian metallothioneins. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry 2011, 16 (7), 1067–78. [DOI] [PubMed] [Google Scholar]
- 43.Romero-Isart N; Vašák M, Advances in the structure and chemistry of metallothioneins. Journal of Inorganic Biochemistry 2002, 88 (3), 388–396. [DOI] [PubMed] [Google Scholar]
- 44.Kägi JH; Vasák M; Lerch K; Gilg DE; Hunziker P; Bernhard WR; Good M, Structure of mammalian metallothionein. Environ Health Perspect 1984, 54, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maret W, The Metals in the Biological Periodic System of the Elements: Concepts and Conjectures. Int J Mol Sci 2016, 17 (1), 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaby-Haas CE; Merchant SS, Lysosome-related organelles as mediators of metal homeostasis. The Journal of biological chemistry 2014, 289 (41), 28129–28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole LB, The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 2015, 80, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suttapitugsakul S; Xiao H; Smeekens J; Wu R, Evaluation and optimization of reduction and alkylation methods to maximize peptide identification with MS-based proteomics. Molecular bioSystems 2017, 13 (12), 2574–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindorff-Larsen K; Winther JR, Thiol Alkylation below Neutral pH. Analytical Biochemistry 2000, 286 (2), 308–310. [DOI] [PubMed] [Google Scholar]
- 50.Sechi S; Chait BT, Modification of Cysteine Residues by Alkylation. A Tool in Peptide Mapping and Protein Identification. Analytical Chemistry 1998, 70 (24), 5150–5158. [DOI] [PubMed] [Google Scholar]
- 51.Müller T; Winter D, Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-containing Reagents. Molecular & cellular proteomics : MCP 2017, 16 (7), 1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tompa P, Fersht A, Structure and Function of Intrinsically Disordered Proteins. New York: Chapman and Hall/CRC, 2010. [Google Scholar]
- 53.Boteva R; Zlateva T; Dorovska-Taran V; Visser AJWG; Tsanev R; Salvato B, Dissociation Equilibrium of Human Recombinant Interferon γ. Biochemistry 1996, 35 (47), 14825–14830. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y; Barkley MD, Toward Understanding Tryptophan Fluorescence in Proteins. Biochemistry 1998, 37 (28), 9976–9982. [DOI] [PubMed] [Google Scholar]
- 55.Banci L; Bertini I; Ciofi-Baffoni S; Kozyreva T; Zovo K; Palumaa P, Affinity gradients drive copper to cellular destinations. Nature 2010, 465 (7298), 645–648. [DOI] [PubMed] [Google Scholar]
- 56.Irvine GW; Stillman MJ, Residue Modification and Mass Spectrometry for the Investigation of Structural and Metalation Properties of Metallothionein and Cysteine-Rich Proteins. Int J Mol Sci 2017, 18 (5), 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krȩżel A; Latajka R; Bujacz GD; Bal W, Coordination Properties of Tris(2-carboxyethyl)phosphine, a Newly Introduced Thiol Reductant, and Its Oxide. Inorganic Chemistry 2003, 42 (6), 1994–2003. [DOI] [PubMed] [Google Scholar]
- 58.Tarasava K; Loebus J; Freisinger E, Localization and Spectroscopic Analysis of the Cu(I) Binding Site in Wheat Metallothionein Ec-1. Int J Mol Sci 2016, 17 (3), 371–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dolderer B; Echner H; Beck A; Hartmann H-J; Weser U; Luchinat C; Del Bianco C, Coordination of three and four Cu(I) to the α- and β-domain of vertebrate Zn-metallothionein-1, respectively, induces significant structural changes. The FEBS Journal 2007, 274 (9), 2349–2362. [DOI] [PubMed] [Google Scholar]
- 60.Pountney DL; Schauwecker I; Zarn J; Vasák M, Formation of mammalian Cu8-metallothionein in vitro: evidence for the existence of two Cu(I)4-thiolate clusters. Biochemistry 1994, 33 (32), 9699–705. [DOI] [PubMed] [Google Scholar]
- 61.Hrabal R; Chen Z; James S; Bennett HP; Ni F, The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nature structural biology 1996, 3 (9), 747–752. [DOI] [PubMed] [Google Scholar]
- 62.Macossay-Castillo M; Marvelli G; Guharoy M; Jain A; Kihara D; Tompa P; Wodak SJ, The Balancing Act of Intrinsically Disordered Proteins: Enabling Functional Diversity while Minimizing Promiscuity. J Mol Biol 2019, 431 (8), 1650–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chapple CE; Robisson B; Spinelli L; Guien C; Becker E; Brun C, Extreme multifunctional proteins identified from a human protein interaction network. Nature Communications 2015, 6 (1), 7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheller JS; Irvine GW; Stillman MJ, Unravelling the mechanistic details of metal binding to mammalian metallothioneins from stoichiometric, kinetic, and binding affinity data. Dalton transactions (Cambridge, England : 2003) 2018, 47 (11), 3613–3637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


