Abstract
Maternal postpartum depression (PPD) is a significant public health concern due to the severe negative impact on maternal and child health and well-being. In this study, we aimed to identify genes associated with PPD. To do this, we investigated genome-wide gene expression profiles of pregnant women during their third trimester of pregnancy and tested the association of gene expression with perinatal depressive symptoms. A total of 137 women from a cohort from the University of North Carolina, USA were assessed. The main phenotypes analysed were Edinburgh Postnatal Depression Scale (EPDS) scores at 2 months postpartum and PPD (binary yes/no) based on an EPDS cutoff of 10. Illumina NextSeq500/550 transcriptomic sequencing from whole blood was analysed using the edgeR package. We identified 71 genes significantly associated with postpartum depression scores at 2 months, after correction for multiple testing at 5% FDR. These included several interesting candidates including TNFRSF17, previously reported to be significantly upregulated in women with PPD and MMP8, a matrix metalloproteinase gene, associated with depression in a genome-wide association study. Functional annotation of differentially expressed genes revealed an enrichment of immune response-related biological processes. Additional analysis of genes associated with changes in depressive symptoms from recruitment to 2 months postpartum identified 66 genes significant at an FDR of 5%. Of these genes, 33 genes were also associated with depressive symptoms at 2 months postpartum. Comparing the results with previous studies, we observed that 15.4% of genes associated with PPD in this study overlapped with 700 core maternal genes that showed significant gene expression changes across multiple brain regions (P = 7.9e-05) and 29–53% of the genes were also associated with estradiol changes in a pharmacological model of depression (P values range = 1.2e-4–2.1e-14). In conclusion, we identified novel genes and validated genes previously associated with oestrogen sensitivity in PPD. These results point towards the role of an altered immune transcriptomic landscape as a vulnerability factor for PPD.
Subject terms: Depression, Comparative genomics
Introduction
The perinatal period spanning across pregnancy and postpartum is a vulnerable time for increased mental health issues, which presents several challenges for women and families1. Postpartum depression (PPD) is a common mood disorder that occurs in 10–15% of pregnant women2. As per the Diagnostic and Statistical Manual (DSM-5, fifth edition), PPD is defined as a major depressive episode that occurs within 4 weeks post-delivery3, however many studies investigate depressive symptoms up to 1 year postpartum4,5.
Children who are born to mothers with PPD have poor health outcomes compared to the general population including shorter gestation periods, impaired cardiovascular functioning, increased gastrointestinal infections, reduced weight gain and lower respiratory tract infections6–8. PPD is also associated with a range of cascading long-term negative health outcomes for the mother and infant including an increased risk of psychiatric disorders and neurodevelopmental deficits such as behavioural problems and learning difficulties in the offspring9–11 and perturbed immune system in mothers and maternal suicide12. In addition, partners of women with PPD are also at risk of having higher rates of depression than expected13.
Despite the high prevalence and negative consequences of PPD, very little is currently known about its underlying biology. Several studies suggest that screening early on in pregnancy can reduce anxiety or depressive symptoms; however, there is very little research and intervention for primary prevention14,15. Potential causes for PPD include psychosocial factors such as a history of depression or a psychiatric disorder during pregnancy, inadequate social support, emotional isolation, financial strain, stressful life events and biological factors such as genetic risk and sensitivity to hormonal changes have been proposed16–18. Studies have reported the association between single nucleotide polymorphisms in genes including the serotonin transporter-linked promoter region polymorphism (5-HTTLPR)19, catechol-O-methyl transferase (COMT), monoamine oxidase (MAO)20 and brain-derived neurotrophic factor (BDNF)21. However, the pathogenesis of PPD still remains largely obscure.
One potential mechanism for biological changes associated with PPD are changing in the gene expression and/or DNA methylation of certain genes across pregnancy and postpartum. We and others have previously identified a panel of predictive gene expression markers whose transcriptome profiles in the third trimester of pregnancy could predict, with 88% accuracy (within sample), which women were at a higher risk of PPD; the panel was also replicated in an independent sample22. In the same study, we further investigated the functional relevance of genes associated with PPD and identified a role for the oestrogen signalling pathway in PPD. In a separate study, we confirmed that the predictive gene expression panel for PPD was also associated with transcriptomic and epigenetic sensitivity to sex-steroid manipulation and mapped onto depressive symptoms and serotonin transporter binding in the brain23. Additionally, we have also previously demonstrated that there is an altered peripheral blood transcriptomic landscape in a pharmacological model of sex-steroid induced depression24.
To the best of our knowledge, there is no large study that has investigated genome-wide gene expression profiles in PPD using the latest sequencing technologies and there is still a gap in our knowledge of the understanding of the biological mechanisms underlying PPD.
The aim of this study was to identify genes and pathways associated with PPD by investigating global changes in peripheral blood gene expression in association with depressive symptoms in a large cohort of pregnant women. We also aimed to evaluate and cross-validate earlier observations in independent clinical cohorts and test if patterns translate to (estradiol) sex hormone manipulation model.
Methods
Participants
Women who were pregnant and had a previous history of a clinical diagnosis of depression were recruited to this study as part of the Mood, Mother and Infant (MMI) study (R01HD073220, mmi.web.unc.edu), an ongoing longitudinal cohort study of mother–infant dyads who were extensively phenotyped during the first postpartum year25. The total cohort comprised of 222 mother–infant dyads enrolled in the MMI study. Of these, 164 women drawn from a longitudinal cohort of mother–infant dyads were followed from the third trimester through 12 months postpartum. From these, we selected women who agreed to participate in the genetic component of the study and provided blood sample (n = 159) and those with good RNA quality with RNA integrity numbers of over 8, amounting to a total of 137 women. These were the women included in this study. Participants underwent a baseline visit with a structured clinical interview in the third trimester of pregnancy and also postpartum via trained clinicians. The study was approved by the Human Research Ethics Committees of the University of North Carolina. All subjects gave written informed consent to participation. To reduce technical variation, all the patients were recruited from one site, and all had blood collected with the same protocol using the same tubes.
Clinical measurements
Maternal depressive symptoms during pregnancy and postnatal depressive symptoms were the main outcomes, measured using the Edinburgh Postnatal Depression Scale (EPDS), which has been validated in the antenatal and postnatal periods26,27. The EPDS has been recommended as a screening tool for perinatal depression among women worldwide.
Data on postpartum-depressive symptoms were collected 2 months after birth. The total number of postpartum-depressive symptoms was also tallied to obtain a total score (out of 30) and was then coded as a categorical variable (score <10 or score ≥10) to indicate scores suggestive of postpartum-depressive symptoms. The EPDS cut-points used to indicate probable depression were selected based on previously published studies28,29. For the study, the EPDS was used as the primary assessment measure and the PPD case definition defined as EPDS score of ≥10 postpartum.
Gene expression experiments
The total RNA was extracted from whole blood in PAXgene tubes using the PAXgene Blood miRNA Kit from Qiagen. RNA was quantified on the Nanodrop1000 and Qubit™ fluorimeter using the RNA HS Assay Kit followed by a quality check using the Agilent Bioanalyzer RNA 6000 Nano kit. mRNA libraries were prepared using the TruSeq® Stranded mRNA Library Prep Kit for NeoPrep™ following the manufacturer’s instructions. The Qubit Fluorometric quantitation was used to determine the 100 ng input for each sample. The libraries were sequenced as pools of up to 24 libraries. Post library construction each library was quantified using the Qubit™ dsDNA HS Assay Kit, followed by size determination using the Agilent High Sensitivity DNA Kit. Up to 24 unique indexed libraries were equimolar pooled and run on the NextSeq500/550 using NextSeq500/550 High Output v2 kit (75 cycles) following the manufacturer’s instructions at a final concentration of 1.5 pM. Sequencing runs were monitored in real time through Illumina BaseSpace.
RNASeq read filtering, alignment and normalisation were carried out using the RNA Aligner application hosted on BaseSpace. The average percentage of reads passing filter including abundant reads = 21,049,665, with >98% aligned to the human reference genome.
Statistical analysis
The QC steps were initially performed in R. Raw count data were available from 22,459 genes. The gene-level counts were imported into edgeR and pre-processing involved a filter threshold of >0.7 counts per million (CPM) to remove low expressed genes in at least 50% of the samples followed by trimmed mean of M-values (TMM) normalisation to scale for library size, allowing a total of 12,121 genes for further analysis.
The edgeR package30 was used to test for differential expression by fitting a model to the negative binomial distribution with the model including the CPM data against the phenotype of interest (PPD) and adjusting for maternal age, gestational age, current and past use of medication and psychological treatment, BMI and ethnicity. Multiple testing corrections were performed using the false discovery rate (FDR) at 5% threshold.
In silico functional annotation of the differentially expressed genes was performed using the Webgestalt interface31. Overrepresentation analysis of the biological processes was done as per the Gene Ontology Functional Database using an FDR for multiple testing correction.
Comparison of differentially expressed genes to previous studies was performed on a gene-level and merging was done using R. First, genes overlapping in the current dataset were merged with those assessed in the previously published study. Next, using R, the two results files from this study and previous study were merged to confirm whether genes differentially expressed in the current study were also significant in that study based on the P values of significance. Next, to test whether the overlap of significant genes was more than expected by chance, enrichment testing was performed using 1000 permutations (using random sets) and applying a two-sided Binominal test in R to give a P value of enrichment for the comparisons.
Results
Demographics
The study comprised 137 women from the MMI study cohort at the University of North Carolina, USA, including 15 women with PPD and 122 with no PPD. Details of the women included in the study are provided in Table 1. All gene expression profiles were measured during recruitment during the third trimester of pregnancy.
Table 1.
Demographics and clinical characteristics of the samples included in the study.
| Phenotype | Mean [SE]/N [%] |
|---|---|
| Overall sample | |
| Age (in years) | 31.39 [0.44] |
| Gestational age (in weeks) | 36.86 [0.12] |
| Ethnicity—non-Hispanic | 110 [86.6%] |
| BMI pre-pregnancy | 26.20 [0.67] |
| BMI baseline | 31.20 [0.62] |
| Pregnancy number | |
| 1 | 37 [29.1%] |
| 2 | 37 [29.1%] |
| 3 | 28 [22%] |
| >3 | 25 [19.8%] |
| Employment status | |
| Full-time working | 102 [80.3%] |
| Not working | 24 [18.9%] |
| Other | 1 [0.8%] |
| Highest level of education: high school graduate | 10 [7.9%] |
| Trade or Business college | 1 [0.8%] |
| College | 20 [15.7%] |
| College grad 4th yr | 38 [29.9%] |
| University Postgrad | 58 [45.7%] |
| EPDS baseline | 5.20 [0.38] |
| EPDS 2 months postpartum | 4.53 [0.33] |
| Current smoker | |
| Current psychotherapy | 23 [18.1%] |
| Current medication | 24 [18.9%] |
| Past psychotherapy | 77 [60.6%] |
| Past medication | 70 [55.1%] |
Gene expression associated with postpartum-depressive symptoms
First, we tested if gene expression profiles of women during the third trimester of pregnancy were significantly associated with depressive symptoms at 2 months postpartum. Through negative binomial regression models using the edgeR package in R30, the gene expression profiles were regressed against the quantitative EPDS scores, with maternal age, ethnicity, gestational age in weeks, current and past medications and psychological treatments and BMI included as covariates. In the MMI cohort, a total of 71 genes were significantly associated with depression scores at 2 months postpartum, after correction for multiple testing at 5% FDR (Fig. 1). Among the top genes were TNFRSR17, GYPA, JCHAIN, SMIM1, TRPM8, SPATA17, HEMGN, PRRT4 and ADAMS15 (Table 2).
Fig. 1. Differentially expressed genes in PPD.
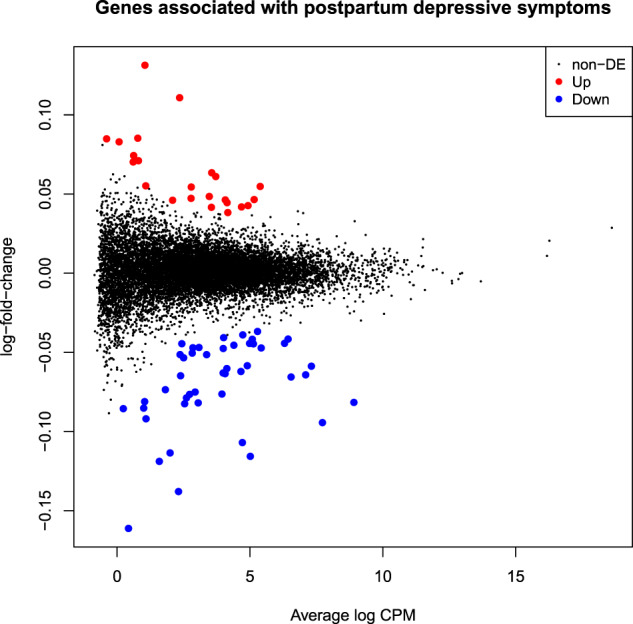
MA plot showing up and downregulated genes associated with depressive symptoms at 2 months postpartum.
Table 2.
List of differentially expressed genes at the third trimester of pregnancy associated with depressive symptoms at 2 months postpartum at 5% FDR.
| Symbol | GnRHa_estradiol | Probe | Chr | P value_EPDSpp | FDR_EPDSpp | logFC_diffEPDS | P value_diffEPDS | FDR_diffEPDS | logFC_PPD | P value_PPD | FDR_PPD | logFC_Dep | P value_Dep | FDR_Dep |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBZ | x | 3050 | 16 | 1.81E-13 | 7.73E-10 | 0.08198997 | 3.96E-11 | 8.47E-08 | 1.240623973 | 8.58E-19 | 2.75E-15 | −0.8915749 | 1.76E-05 | 0.02832105 |
| GYPA | 2993 | 4 | 3.65E-05 | 0.010910599 | 0.023796545 | 0.104618903 | 0.86184816 | 0.304788526 | 0.02543134 | 0.99980319 | −0.0668562 | 0.73764206 | 0.99997367 | |
| JCHAIN | 3512 | 4 | 3.02E-07 | 0.000204157 | 0.006836049 | 0.606968986 | 0.999245113 | 0.277591843 | 0.03477346 | 0.99980319 | −0.1611497 | 0.41273937 | 0.99997367 | |
| SMIM1 | 388588 | 1 | 1.08E-05 | 0.004463077 | −0.046924669 | 0.000526408 | 0.081415689 | 0.522340142 | 0.00011953 | 0.04456932 | −0.2791593 | 0.15879646 | 0.99997367 | |
| LOC153910 | 153910 | NA | 7.17E-08 | 7.08E-05 | 0.038517757 | 0.037839691 | 0.647066685 | 0.017750207 | 0.92289001 | 0.99980319 | −0.0583912 | 0.80144348 | 0.99997367 | |
| TRPM8 | 79054 | 2 | 2.95E-05 | 0.009184138 | 0.01408958 | 0.386065356 | 0.999245113 | 0.276389445 | 0.10047632 | 0.99980319 | −0.0771887 | 0.72789056 | 0.99997367 | |
| TNFRSF17 | 608 | 16 | 2.87E-05 | 0.009184138 | 0.023785396 | 0.126783514 | 0.901943031 | 0.377871402 | 0.01599402 | 0.95884504 | −0.2241632 | 0.28855385 | 0.99997367 | |
| SPATA17 | 128153 | 1 | 0.000145236 | 0.032868929 | −0.015977987 | 0.205361204 | 0.961442886 | 0.146092805 | 0.24794379 | 0.99980319 | 0.0772507 | 0.68551058 | 0.99997367 | |
| HLA-DQA2 | 3118 | 6 | 9.23E-06 | 0.00394781 | −0.03402622 | 0.004663188 | 0.273869178 | −0.23589098 | 0.07684565 | 0.99980319 | 0.39120209 | 0.05814488 | 0.99997367 | |
| NA | 54094 | NA | 6.80E-08 | 7.08E-05 | 0.107471759 | 3.55E-09 | 4.55E-06 | −0.077278025 | 0.65099901 | 0.99980319 | 0.11434726 | 0.6089463 | 0.99997367 | |
| HEMGN | 55363 | 9 | 1.16E-06 | 0.000678668 | 0.009033051 | 0.498373154 | 0.999245113 | 0.081613389 | 0.52344137 | 0.99980319 | 0.12426508 | 0.52030819 | 0.99997367 | |
| PRRT4 | 401399 | 7 | 7.30E-06 | 0.003231945 | −0.019808695 | 0.224306613 | 0.965527011 | −0.268941122 | 0.09621023 | 0.99980319 | 0.29661201 | 0.16607037 | 0.99997367 | |
| TMEM158 | x | 25907 | 3 | 0.000220881 | 0.044129491 | 0.004957364 | 0.718388328 | 0.999245113 | −0.08837544 | 0.50416126 | 0.99980319 | 0.26662194 | 0.17104819 | 0.99997367 |
| PF4V1 | 5197 | 4 | 0.000244208 | 0.046140633 | −0.012353244 | 0.364124795 | 0.999245113 | −0.091543585 | 0.49948912 | 0.99980319 | 0.16507481 | 0.40091434 | 0.99997367 | |
| DIAPH3 | 81624 | 13 | 0.000196028 | 0.039943103 | 0.000619347 | 0.970040483 | 0.999245113 | 0.109238181 | 0.4880814 | 0.99980319 | −0.0752023 | 0.72118901 | 0.99997367 | |
| CLRN1-AS1 | 116933 | 3 | 5.26E-05 | 0.015332746 | 0.030399958 | 0.033386246 | 0.621163618 | −0.379805572 | 0.01376195 | 0.92011568 | 0.42482939 | 0.04285816 | 0.99997367 | |
| GDF15 | 9518 | 19 | 0.000145948 | 0.032868929 | −0.034962226 | 0.038091465 | 0.647066685 | −0.427801989 | 0.01955796 | 0.99980319 | 0.44876145 | 0.05279787 | 0.99997367 | |
| ADAMTS15 | 170689 | 11 | 1.38E-07 | 0.000111272 | 0.066022647 | 1.68E-05 | 0.006532214 | 0.174319176 | 0.28210173 | 0.99980319 | −0.1148235 | 0.59692752 | 0.99997367 | |
| ELK2AP | 2003 | 14 | 3.58E-06 | 0.001767033 | −0.041376595 | 0.005329177 | 0.289875588 | 0.311367498 | 0.02115396 | 0.99980319 | −0.2259639 | 0.25708473 | 0.99997367 | |
| ZNF519 | x | 162655 | 18 | 0.000159288 | 0.034657239 | 0.002937227 | 0.845295248 | 0.999245113 | −0.202383477 | 0.20580551 | 0.99980319 | 0.23272153 | 0.2727202 | 0.99997367 |
| AZU1 | 566 | 19 | 6.52E-10 | 1.20E-06 | −0.034626939 | 0.018511889 | 0.497148793 | 0.203930581 | 0.13679875 | 0.99980319 | −0.0489381 | 0.8063294 | 0.99997367 | |
| PRTN3 | 5657 | 19 | 3.36E-06 | 0.001750825 | −0.037571179 | 0.025310749 | 0.573662105 | 0.006556881 | 0.96895741 | 0.99980319 | 0.14420354 | 0.49847204 | 0.99997367 | |
| C4BPA | x | 722 | 1 | 2.91E-10 | 6.22E-07 | −0.051502467 | 3.73E-06 | 0.002174605 | −0.640051256 | 6.58E-06 | 0.00384058 | 0.82369396 | 0.00010817 | 0.11571456 |
| FAM83A | 84985 | 8 | 6.56E-06 | 0.003005253 | 0.035483408 | 0.024659247 | 0.565101277 | 0.682420779 | 3.88E-06 | 0.00237108 | −0.4020439 | 0.04915913 | 0.99997367 | |
| CD177 | 57126 | NA | 3.00E-05 | 0.009184138 | −0.014578688 | 0.203567924 | 0.961442886 | 0.229194386 | 0.07083359 | 0.99980319 | −0.4054746 | 0.03687446 | 0.99997367 | |
| ELANE | 1991 | 19 | 1.15E-13 | 7.39E-10 | −0.055711655 | 0.000193525 | 0.042832372 | 0.120832578 | 0.37480411 | 0.99980319 | 0.09448629 | 0.63103503 | 0.99997367 | |
| SLC14A1 | 6563 | 18 | 1.84E-05 | 0.006388527 | 0.012590081 | 0.353435297 | 0.999245113 | −0.119878519 | 0.35197831 | 0.99980319 | 0.35220808 | 0.06875925 | 0.99997367 | |
| MFSD9 | 84804 | 2 | 0.000248219 | 0.046179506 | 0.033478011 | 0.019777123 | 0.512886733 | 0.338344196 | 0.01966212 | 0.99980319 | −0.2829192 | 0.15720327 | 0.99997367 | |
| ATOH8 | 84913 | 2 | 1.26E-07 | 0.000111272 | −0.026585915 | 0.054860764 | 0.72205758 | 0.488108301 | 0.00120052 | 0.23350081 | −0.6060181 | 0.0030407 | 0.99997367 | |
| FSTL5 | 56884 | 4 | 8.78E-05 | 0.023000406 | −0.033767859 | 0.050527822 | 0.708162007 | −0.473378851 | 0.00970811 | 0.8009674 | 0.4768658 | 0.03873353 | 0.99997367 | |
| LOC100132062 | 100132062 | NA | 2.74E-59 | 3.52E-55 | −0.09642385 | 1.86E-08 | 1.99E-05 | −2.079430726 | 1.58E-33 | 1.01E-29 | 2.03759936 | 5.40E-19 | 6.94E-15 | |
| ALK | 238 | 2 | 0.000259845 | 0.047184832 | −0.005844187 | 0.730382863 | 0.999245113 | −0.223973153 | 0.19563757 | 0.99980319 | 0.19379859 | 0.38496151 | 0.99997367 | |
| PTPRT | 11122 | 20 | 7.94E-05 | 0.021243225 | −0.009658525 | 0.540753469 | 0.999245113 | −0.117452457 | 0.4719397 | 0.99980319 | 0.10753096 | 0.61900473 | 0.99997367 | |
| DEFA1 | 1667 | 8 | 8.27E-09 | 9.65E-06 | −0.034105055 | 0.011155124 | 0.406813419 | 0.324107256 | 0.01418311 | 0.92830067 | −0.1781284 | 0.36501703 | 0.99997367 | |
| TPTE | 7179 | NA | 0.000260974 | 0.047184832 | −0.02190591 | 0.263909072 | 0.983409969 | −0.555091903 | 0.0068122 | 0.66302035 | 0.52221024 | 0.0339904 | 0.99997367 | |
| MXRA7 | 439921 | 17 | 1.39E-07 | 0.000111272 | 0.026352653 | 0.027553678 | 0.59646976 | 0.332170197 | 0.01493829 | 0.92830067 | −0.3610542 | 0.06986465 | 0.99997367 | |
| GPRC5C | x | 55890 | 17 | 8.18E-09 | 9.65E-06 | −0.074305012 | 1.07E-05 | 0.005302851 | 0.377528437 | 0.02896948 | 0.99980319 | −0.3655967 | 0.09786856 | 0.99997367 |
| COBL | 23242 | 7 | 1.32E-05 | 0.004967603 | −0.010669626 | 0.534510566 | 0.999245113 | −0.044677683 | 0.79962307 | 0.99980319 | 0.05461506 | 0.80848884 | 0.99997367 | |
| OLFM4 | 10562 | 13 | 1.18E-05 | 0.004714007 | −0.09056149 | 1.85E-12 | 1.09E-08 | −0.047752911 | 0.71452355 | 0.99980319 | 0.03402996 | 0.86046026 | 0.99997367 | |
| NELL1 | 4745 | 11 | 0.000236768 | 0.046051276 | 0.00942249 | 0.545557875 | 0.999245113 | 0.192429941 | 0.22244205 | 0.99980319 | −0.1997864 | 0.34726165 | 0.99997367 | |
| CEBPE | 1053 | 14 | 0.000223449 | 0.044129491 | −0.031019039 | 0.018250128 | 0.496108596 | 0.008593278 | 0.94802827 | 0.99980319 | −0.0084758 | 0.96512905 | 0.99997367 | |
| SEC14L3 | 266629 | 22 | 1.24E-09 | 1.99E-06 | 0.015828663 | 0.21812967 | 0.965527011 | 0.130489444 | 0.32437928 | 0.99980319 | 0.05236892 | 0.78723598 | 0.99997367 | |
| CTSG | 1511 | 14 | 4.15E-12 | 1.06E-08 | −0.047720951 | 0.002517574 | 0.198318861 | 0.256940153 | 0.08033028 | 0.99980319 | 0.00679864 | 0.97110331 | 0.99997367 | |
| LIPN | 643418 | 10 | 2.61E-05 | 0.008594769 | 0.015875193 | 0.215011061 | 0.965527011 | −0.104090347 | 0.43029098 | 0.99980319 | 0.1067839 | 0.57903016 | 0.99997367 | |
| NRN1 | 51299 | 6 | 0.000190372 | 0.03941628 | 0.025810875 | 0.090631657 | 0.832799778 | −0.036423626 | 0.80920195 | 0.99980319 | 0.08824533 | 0.6667064 | 0.99997367 | |
| GRIN2B | 2904 | 12 | 3.41E-06 | 0.001750825 | −0.048033232 | 0.016751319 | 0.476799743 | −0.452027194 | 0.03771923 | 0.99980319 | 0.41741015 | 0.10248191 | 0.99997367 | |
| MZB1 | 51237 | 5 | 2.17E-07 | 0.000154727 | 0.035070019 | 0.009931335 | 0.380562826 | 0.316097491 | 0.02091221 | 0.99980319 | −0.2712525 | 0.17173474 | 0.99997367 | |
| USP32P1 | 162632 | 17 | 1.23E-05 | 0.004803545 | −0.02265204 | 0.11552771 | 0.880715801 | 0.249454841 | 0.10004919 | 0.99980319 | −0.4216523 | 0.04835357 | 0.99997367 | |
| DEFA1B | 728358 | 8 | 2.73E-09 | 3.90E-06 | −0.082334647 | 2.83E-08 | 2.80E-05 | 0.535661022 | 5.17E-05 | 0.02287903 | −0.505308 | 0.00958881 | 0.99997367 | |
| XKR3 | 150165 | 22 | 3.51E-12 | 1.06E-08 | 0.020147807 | 0.157623182 | 0.94242466 | −0.090729408 | 0.55190992 | 0.99980319 | 0.12220984 | 0.55636885 | 0.99997367 | |
| SHISA6 | 388336 | 17 | 0.00017958 | 0.03812032 | −0.049507351 | 0.004773282 | 0.274774096 | −0.057699705 | 0.74644417 | 0.99980319 | 0.10304895 | 0.64837734 | 0.99997367 | |
| RAB3IL1 | 5866 | 11 | 9.19E-05 | 0.023607002 | −0.002823157 | 0.833233945 | 0.999245113 | −0.018794745 | 0.88737441 | 0.99980319 | 0.16526773 | 0.39493394 | 0.99997367 | |
| BPI | 671 | 20 | 0.000128913 | 0.031223752 | −0.047161513 | 0.000484145 | 0.076727959 | −0.373528161 | 0.00363043 | 0.45246397 | 0.34982249 | 0.06941875 | 0.99997367 | |
| OR8U8 | 504189 | NA | 7.64E-05 | 0.020856842 | 0.042692465 | 0.000391896 | 0.065334587 | 0.516211389 | 9.78E-05 | 0.03924713 | −0.3413738 | 0.08193914 | 0.99997367 | |
| TFF3 | 7033 | 21 | 0.000181144 | 0.03812032 | −0.027929543 | 0.05632791 | 0.723949792 | −0.539616026 | 0.00039369 | 0.10313927 | 0.5280528 | 0.01157969 | 0.99997367 | |
| CNBD1 | 168975 | 8 | 1.73E-05 | 0.006183318 | 0.01670876 | 0.332781984 | 0.999245113 | 0.022683504 | 0.9025405 | 0.99980319 | 0.03581888 | 0.87235199 | 0.99997367 | |
| PPARGC1A | x | 10891 | 4 | 7.58E-05 | 0.020856842 | −0.020666423 | 0.220000414 | 0.965527011 | −0.181135049 | 0.30375862 | 0.99980319 | 0.19450487 | 0.38919359 | 0.99997367 |
| CPA5 | x | 93979 | NA | 0.000105965 | 0.026159008 | −0.069099835 | 4.80E-06 | 0.002567843 | −0.119843608 | 0.44801674 | 0.99980319 | 0.19377932 | 0.35625076 | 0.99997367 |
| CLEC12A | x | 160364 | 12 | 0.000137021 | 0.031980631 | −0.046454778 | 3.37E-05 | 0.0109922 | 0.209864679 | 0.08989106 | 0.99980319 | −0.0452303 | 0.81056457 | 0.99997367 |
| DEFA4 | 1669 | 8 | 3.58E-07 | 0.000230067 | −0.065234631 | 3.52E-06 | 0.002148751 | 0.161740923 | 0.22345996 | 0.99980319 | −0.0154213 | 0.93716872 | 0.99997367 | |
| BCAM | 4059 | 19 | 2.22E-05 | 0.007486807 | 0.018351203 | 0.158160908 | 0.94242466 | −0.189520866 | 0.15008212 | 0.99980319 | 0.37806366 | 0.05108819 | 0.99997367 | |
| SHISA7 | 729956 | 19 | 4.27E-06 | 0.002031316 | 0.025612122 | 0.154200044 | 0.938547656 | 0.243275156 | 0.16087353 | 0.99980319 | −0.1664301 | 0.44848875 | 0.99997367 | |
| PLCB4 | x | 5332 | 20 | 0.000244416 | 0.046140633 | −0.019949231 | 0.273279271 | 0.989116508 | 0.141980609 | 0.45338217 | 0.99980319 | −0.0934137 | 0.68937109 | 0.99997367 |
| LTF | 4057 | 3 | 0.000136734 | 0.031980631 | −0.058823471 | 1.34E-05 | 0.005903106 | −0.464815718 | 0.00024729 | 0.07054463 | 0.38819801 | 0.0434471 | 0.99997367 | |
| RASAL2 | 9462 | 1 | 1.59E-05 | 0.005845119 | −0.096580109 | 2.55E-12 | 1.09E-08 | −0.496078584 | 0.00030139 | 0.08060174 | 0.63127969 | 0.00207093 | 0.99997367 | |
| TACSTD2 | 4070 | 1 | 0.000104739 | 0.026159008 | −0.001236843 | 0.932804732 | 0.999245113 | 0.212247984 | 0.18805412 | 0.99980319 | −0.2836035 | 0.18378057 | 0.99997367 | |
| PRSS33 | x | 260429 | 16 | 1.82E-06 | 0.00101726 | −0.017543158 | 0.204453261 | 0.961442886 | 0.669401492 | 7.80E-07 | 0.00077909 | −0.8128711 | 3.66E-05 | 0.05215136 |
| AOC1 | 26 | 7 | 2.14E-07 | 0.000154727 | −0.03220397 | 0.058722646 | 0.736808196 | −0.038105907 | 0.82650662 | 0.99980319 | −0.1190881 | 0.58675507 | 0.99997367 | |
| LINC01122 | 400955 | 2 | 0.000151692 | 0.033573551 | −0.010025233 | 0.562863428 | 0.999245113 | −0.365763605 | 0.04535562 | 0.99980319 | 0.38442049 | 0.09844397 | 0.99997367 | |
| TRPM3 | 80036 | 9 | 6.48E-05 | 0.018486308 | 0.029584207 | 0.051525843 | 0.709258838 | 0.126190993 | 0.41924413 | 0.99980319 | −0.0668866 | 0.75091325 | 0.99997367 | |
| TMTC1 | 83857 | 12 | 4.15E-07 | 0.000253841 | 0.008189727 | 0.481246148 | 0.999245113 | 0.067323981 | 0.6281319 | 0.99980319 | −0.3685774 | 0.06898419 | 0.99997367 |
EPDS Edinburgh Postnatal Depression Scale symptoms at 2 months postpartum, diffEPDS difference in EPDS symptoms from third trimester pregnancy to 2 months postpartum, PPD EPDS symptoms in the PPD group, Dep EPDS symptoms in the depression group, GnRHa_estradiol genes associated with estradiol in the GnRHa study, FDR P values after false discovery rate multiple testing.
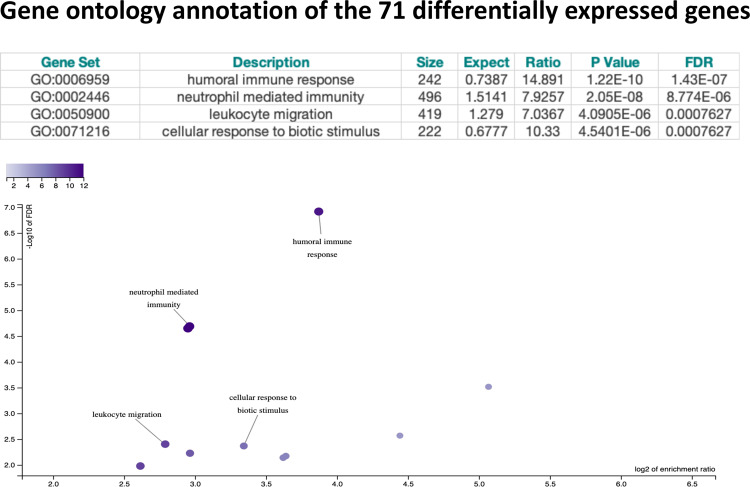
Using the Webgestalt interface, we found that the 71 genes were enriched for several immune-related categories as per the Gene Ontology Functional Database (Fig. 2) and the genes were overrepresented within the neutrophil degranulation (P = 1.1E-9, n = 13 genes), innate immunity (P = 1.8E-6, n = 14 genes) and fibrin clot formation (P = 4.5E-5, n = 3 genes) pathways using the Reactome Pathway database.
Fig. 2. Functional annotation.
Functional annotation of the differentially expressed genes associated with depressive symptoms at 2 months postpartum.
Gene expression associated with a change in postpartum-depressive symptoms
The gene expression profiles were regressed against the changes in EPDS scores, with maternal age, ethnicity, gestational age in weeks, current and past medications and psychological treatments and BMI included as covariates. When assessing gene expression profiles significantly associated with changes in depressive symptoms from recruitment to 2 months postpartum (2 months postpartum EPDS–EPDS at recruitment), a total of 66 genes were significant at an FDR of 5% in the MMI cohort.
Of these genes, 33 genes were also significantly associated (P < 0.05) with depressive symptoms at 2 months postpartum and of these 14 remained significant at 5% FDR. The genes included OLFM4, RASAL2, BHZ, DEFA1B, DEFA4, C4BPA, CPA5, GPRC5C, LTF, ADAMTS15, CLEC12A and ELANE (Table 3).
Table 3.
List of differentially expressed genes at the third trimester of pregnancy associated with changes in depressive symptoms (from the third trimester to 2 months postpartum) at 5% FDR.
| Symbol | GnRHa_estradiol | Probe | Chr | P value_EPDSv2 | FDR_EPDSv2 | logFC_diffEPDSv2 | P value_diffEPDSv2 | FDR_diffEPDSv2 | logFC_PPD | P value_PPD | FDR_PPD | logFC_Dep | P value_Dep | FDR_Dep |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC12A1 | x | 6557 | 15 | 0.738374001 | 0.999797241 | −0.143662302 | 3.04E-23 | 3.91E-19 | −1.514853079 | 3.68E-24 | 1.58E-20 | 1.52947173 | 1.01E-10 | 4.31E-07 |
| OLFM4 | 10562 | 13 | 1.18E-05 | 0.004714007 | −0.09056149 | 1.85E-12 | 1.09E-08 | −0.047752911 | 0.714523555 | 0.99980319 | 0.03402996 | 0.86046026 | 0.99997367 | |
| RASAL2 | 9462 | 1 | 1.59E-05 | 0.005845119 | −0.096580109 | 2.55E-12 | 1.09E-08 | −0.496078584 | 0.000301385 | 0.08060174 | 0.63127969 | 0.00207093 | 0.99997367 | |
| ZFP57 | x | 346171 | 6 | 0.688934236 | 0.999797241 | 0.113752939 | 9.57E-12 | 3.07E-08 | 0.629257704 | 0.000143791 | 0.04988752 | −0.6042326 | 0.0056805 | 0.99997367 |
| MUC6 | x | 4588 | 11 | 0.032615955 | 0.835444211 | 0.107037187 | 2.20E-11 | 5.65E-08 | 0.068004697 | 0.67455073 | 0.99980319 | −0.2445742 | 0.26286793 | 0.99997367 |
| HBZ | x | 3050 | 16 | 1.81E-13 | 7.73E-10 | 0.08198997 | 3.96E-11 | 8.47E-08 | 1.240623973 | 8.58E-19 | 2.75E-15 | −0.8915749 | 1.76E-05 | 0.02832105 |
| PNMA3 | 29944 | X | 0.588321779 | 0.999797241 | 0.084017297 | 1.38E-09 | 2.52E-06 | −0.048544382 | 0.737626973 | 0.99980319 | −0.0740358 | 0.71439951 | 0.99997367 | |
| RAP1GAP | 5909 | 1 | 0.000417001 | 0.066086904 | −0.084706388 | 1.69E-09 | 2.71E-06 | −0.700111217 | 1.09E-07 | 0.0001401 | 0.95024605 | 2.17E-06 | 0.00537669 | |
| IFI27 | 3429 | NA | 0.491717731 | 0.999797241 | 0.088816879 | 2.79E-09 | 3.98E-06 | 1.079393826 | 5.46E-16 | 1.40E-12 | −0.7622081 | 8.16E-05 | 0.10472842 | |
| NA | 54094 | NA | 6.80E-08 | 7.08E-05 | 0.107471759 | 3.55E-09 | 4.55E-06 | −0.077278025 | 0.650999011 | 0.99980319 | 0.11434726 | 0.6089463 | 0.99997367 | |
| ENC1 | 8507 | 5 | 0.005321833 | 0.336533844 | −0.072184102 | 1.08E-08 | 1.26E-05 | −0.282262065 | 0.032073728 | 0.99980319 | 0.22215453 | 0.25746315 | 0.99997367 | |
| LOC100132062 | 100132062 | NA | 2.74E-59 | 3.52E-55 | −0.09642385 | 1.86E-08 | 1.99E-05 | −2.079430726 | 1.58E-33 | 1.01E-29 | 2.03759936 | 5.40E-19 | 6.94E-15 | |
| DEFA1B | 728358 | 8 | 2.73E-09 | 3.90E-06 | −0.082334647 | 2.83E-08 | 2.80E-05 | 0.535661022 | 5.17E-05 | 0.02287903 | −0.505308 | 0.00958881 | 0.99997367 | |
| SLC35F1 | 222553 | 6 | 0.012780901 | 0.533803126 | −0.070659963 | 1.20E-07 | 0.000110253 | 0.899503922 | 2.61E-10 | 4.78E-07 | −0.7774074 | 0.00015966 | 0.14639261 | |
| FAM3B | 54097 | 21 | 0.000506343 | 0.073862832 | −0.081062951 | 4.36E-07 | 0.000357438 | −1.206842502 | 2.86E-11 | 6.13E-08 | 1.29211562 | 4.09E-08 | 0.00013115 | |
| A2M | 2 | 12 | 0.125486579 | 0.999797241 | 0.074797133 | 4.46E-07 | 0.000357438 | 0.066016866 | 0.674833048 | 0.99980319 | −0.1488668 | 0.48318013 | 0.99997367 | |
| RNF182 | 221687 | 6 | 0.983664265 | 0.999797241 | −0.08587623 | 5.84E-07 | 0.000441361 | −0.820514565 | 1.54E-08 | 2.47E-05 | 0.9490668 | 7.75E-06 | 0.01421399 | |
| CLDND2 | 125875 | 19 | 0.401412478 | 0.999797241 | 0.070517883 | 1.26E-06 | 0.000899767 | 0.016645892 | 0.912012943 | 0.99980319 | −0.1021826 | 0.6269187 | 0.99997367 | |
| COL5A3 | x | 50509 | 19 | 0.172251596 | 0.999797241 | 0.071123394 | 1.60E-06 | 0.001082012 | 0.059865205 | 0.690177553 | 0.99980319 | −0.2220476 | 0.27428873 | 0.99997367 |
| ANKRD36B | 57730 | 2 | 0.403733504 | 0.999797241 | 0.083729733 | 1.92E-06 | 0.001230403 | −0.148062332 | 0.425223241 | 0.99980319 | 0.19293391 | 0.40812337 | 0.99997367 | |
| DEFA4 | 1669 | 8 | 3.58E-07 | 0.000230067 | −0.065234631 | 3.52E-06 | 0.002148751 | 0.161740923 | 0.223459958 | 0.99980319 | −0.0154213 | 0.93716872 | 0.99997367 | |
| C4BPA | x | 722 | 1 | 2.91E-10 | 6.22E-07 | −0.051502467 | 3.73E-06 | 0.002174605 | −0.640051256 | 6.58E-06 | 0.00384058 | 0.82369396 | 0.00010817 | 0.11571456 |
| MICU1 | 10367 | 10 | 0.008452083 | 0.439583873 | 0.071183702 | 4.67E-06 | 0.002567843 | −0.393099561 | 0.01809445 | 0.99980319 | 0.4094252 | 0.05992712 | 0.99997367 | |
| CPA5 | x | 93979 | NA | 0.000105965 | 0.026159008 | −0.069099835 | 4.80E-06 | 0.002567843 | −0.119843608 | 0.448016742 | 0.99980319 | 0.19377932 | 0.35625076 | 0.99997367 |
| NT5C3A | 51251 | 7 | 0.002112895 | 0.191346638 | 0.064676607 | 5.26E-06 | 0.002701077 | 0.00076943 | 0.995962869 | 0.99980319 | 0.01731807 | 0.935702 | 0.99997367 | |
| GPRC5C | x | 55890 | 17 | 8.18E-09 | 9.65E-06 | −0.074305012 | 1.07E-05 | 0.005302851 | 0.377528437 | 0.028969482 | 0.99980319 | −0.3655967 | 0.09786856 | 0.99997367 |
| LINC00854 | 100874261 | 17 | 0.89885143 | 0.999797241 | 0.063317275 | 1.17E-05 | 0.005562922 | 0.107322993 | 0.468550742 | 0.99980319 | −0.1956506 | 0.34744068 | 0.99997367 | |
| VNN1 | 8876 | 6 | 0.046944791 | 0.982428275 | −0.049143107 | 1.27E-05 | 0.005837152 | −0.032198571 | 0.799444025 | 0.99980319 | 0.04643599 | 0.80871155 | 0.99997367 | |
| LTF | 4057 | 3 | 0.000136734 | 0.031980631 | −0.058823471 | 1.34E-05 | 0.005903106 | −0.464815718 | 0.000247294 | 0.07054463 | 0.38819801 | 0.0434471 | 0.99997367 | |
| FAM153B | 202134 | 5 | 0.011646873 | 0.515005387 | 0.076226915 | 1.38E-05 | 0.005903106 | −0.354084609 | 0.063289465 | 0.99980319 | 0.35484633 | 0.13436291 | 0.99997367 | |
| BRSK2 | 9024 | 11 | 0.184299576 | 0.999797241 | 0.076659523 | 1.52E-05 | 0.006217663 | −0.440448095 | 0.0242044 | 0.99980319 | 0.37176057 | 0.12460352 | 0.99997367 | |
| FAM53A | 152877 | 4 | 0.798983611 | 0.999797241 | 0.075963322 | 1.55E-05 | 0.006217663 | 0.2412146 | 0.188313624 | 0.99980319 | −0.268703 | 0.24084011 | 0.99997367 | |
| ADAMTS15 | 170689 | 11 | 1.38E-07 | 0.000111272 | 0.066022647 | 1.68E-05 | 0.006532214 | 0.174319176 | 0.282101731 | 0.99980319 | −0.1148235 | 0.59692752 | 0.99997367 | |
| COL6A3 | 1293 | 2 | 0.307133445 | 0.999797241 | 0.062579979 | 1.91E-05 | 0.007027999 | 0.146175885 | 0.347679407 | 0.99980319 | −0.0608776 | 0.77358719 | 0.99997367 | |
| LOC100507424 | 100507424 | NA | 0.123121497 | 0.999797241 | 0.054632332 | 1.92E-05 | 0.007027999 | −0.142127217 | 0.304077778 | 0.99980319 | 0.03929275 | 0.84720725 | 0.99997367 | |
| OBSCN | 84033 | 1 | 0.722086924 | 0.999797241 | 0.050750803 | 2.28E-05 | 0.008116835 | 0.071759756 | 0.571960158 | 0.99980319 | −0.1721324 | 0.37273278 | 0.99997367 | |
| HS2ST1 | 9653 | 1 | 0.013911554 | 0.553838011 | 0.064543649 | 2.43E-05 | 0.008422227 | 0.06095681 | 0.703847531 | 0.99980319 | −0.071729 | 0.73247663 | 0.99997367 | |
| NPR2 | x | 4882 | 9 | 0.313616353 | 0.999797241 | 0.07470813 | 2.91E-05 | 0.009837403 | 0.015369885 | 0.929490553 | 0.99980319 | −0.1003678 | 0.66059056 | 0.99997367 |
| CLEC12A | x | 160364 | 12 | 0.000137021 | 0.031980631 | −0.046454778 | 3.37E-05 | 0.0109922 | 0.209864679 | 0.089891059 | 0.99980319 | −0.0452303 | 0.81056457 | 0.99997367 |
| TMEM91 | 641649 | 19 | 0.404895127 | 0.999797241 | 0.058584845 | 3.43E-05 | 0.0109922 | −0.070196956 | 0.64309492 | 0.99980319 | −0.0135671 | 0.94490559 | 0.99997367 | |
| LINC00189 | 193629 | 21 | 0.010618077 | 0.493855975 | 0.056343712 | 3.87E-05 | 0.012116049 | −0.298187073 | 0.037427634 | 0.99980319 | 0.29577551 | 0.1414945 | 0.99997367 | |
| C4B_2 | 100293534 | NA | 0.007119309 | 0.388896016 | 0.055595437 | 3.96E-05 | 0.012116049 | 0.495547097 | 0.000181471 | 0.05762403 | −0.170722 | 0.37935001 | 0.99997367 | |
| PLCH2 | 9651 | 1 | 0.121882192 | 0.999797241 | 0.053982765 | 5.72E-05 | 0.017080892 | −0.123053219 | 0.378586968 | 0.99980319 | −0.0175891 | 0.92956679 | 0.99997367 | |
| KCNMA1 | 3778 | 10 | 0.004948314 | 0.321229175 | 0.055323213 | 6.44E-05 | 0.018566121 | 0.115752688 | 0.43053291 | 0.99980319 | −0.2240934 | 0.28222647 | 0.99997367 | |
| NPIPB5 | 100132247 | 16 | 0.363761914 | 0.999797241 | 0.053370079 | 6.51E-05 | 0.018566121 | 0.233122434 | 0.094712932 | 0.99980319 | −0.358436 | 0.07588194 | 0.99997367 | |
| LOC100132077 | 100132077 | NA | 0.003168783 | 0.252656315 | 0.055771593 | 8.95E-05 | 0.024969396 | −0.290148219 | 0.053014626 | 0.99980319 | 0.28559265 | 0.17196614 | 0.99997367 | |
| CLEC12B | 387837 | 12 | 0.000839057 | 0.105597818 | −0.045399984 | 0.000104155 | 0.028447571 | −0.125781635 | 0.339340239 | 0.99980319 | 0.28429311 | 0.14471212 | 0.99997367 | |
| DTX2P1-UPK3BP1-PMS2P11 | 441263 | NA | 0.697163808 | 0.999797241 | 0.054641303 | 0.00010951 | 0.029287154 | 0.019767196 | 0.891894338 | 0.99980319 | −0.0486238 | 0.80942813 | 0.99997367 | |
| PRKXP1 | 441733 | 15 | 0.989146813 | 0.999797241 | 0.047857927 | 0.000122714 | 0.03195716 | 0.264657472 | 0.053607631 | 0.99980319 | −0.3048762 | 0.12455578 | 0.99997367 | |
| MAML2 | 84441 | 11 | 0.054972619 | 0.999797241 | 0.047067694 | 0.000125186 | 0.03195716 | −0.202750932 | 0.115280757 | 0.99980319 | 0.17016595 | 0.3787149 | 0.99997367 | |
| DDR1 | x | 780 | 6 | 0.528160627 | 0.999797241 | 0.056339661 | 0.000127967 | 0.03195716 | 0.10239741 | 0.500742883 | 0.99980319 | −0.2130413 | 0.30183844 | 0.99997367 |
| CDC42BPG | 55561 | 11 | 0.740909675 | 0.999797241 | 0.055678667 | 0.000129452 | 0.03195716 | 0.144699948 | 0.344577413 | 0.99980319 | −0.2834911 | 0.17133707 | 0.99997367 | |
| KIF19 | 124602 | 17 | 0.674283974 | 0.999797241 | 0.064295989 | 0.000149316 | 0.036165474 | −0.31711123 | 0.086336356 | 0.99980319 | 0.36479985 | 0.11773515 | 0.99997367 | |
| UBAC2 | x | 337867 | 13 | 0.776875995 | 0.999797241 | 0.064645112 | 0.000163489 | 0.038864885 | −0.115184307 | 0.521453911 | 0.99980319 | 0.09990956 | 0.66283344 | 0.99997367 |
| FIBCD1 | 84929 | 9 | 0.01477133 | 0.571957401 | 0.054000943 | 0.00016668 | 0.038903107 | 0.72292732 | 2.77E-06 | 0.00197483 | −0.5574507 | 0.007439 | 0.99997367 | |
| KLRC3 | x | 3823 | 12 | 0.956279673 | 0.999797241 | 0.0509244 | 0.000178701 | 0.040964013 | 0.108077118 | 0.445135655 | 0.99980319 | −0.0246341 | 0.9013113 | 0.99997367 |
| RASA2 | x | 5922 | 3 | 0.033272719 | 0.844114418 | 0.047235684 | 0.000183238 | 0.041267076 | −0.267466884 | 0.043001433 | 0.99980319 | 0.26920447 | 0.16864055 | 0.99997367 |
| ELANE | 1991 | 19 | 1.15E-13 | 7.39E-10 | −0.055711655 | 0.000193525 | 0.042832372 | 0.120832578 | 0.374804112 | 0.99980319 | 0.09448629 | 0.63103503 | 0.99997367 | |
| PVALB | 5816 | 22 | 0.006261676 | 0.365368784 | −0.057766662 | 0.000215201 | 0.045767566 | 0.141207644 | 0.392691699 | 0.99980319 | 0.01047018 | 0.9611797 | 0.99997367 | |
| MS4A3 | x | 932 | 11 | 0.000798942 | 0.10346369 | −0.049997885 | 0.000218028 | 0.045767566 | −0.082803294 | 0.53179566 | 0.99980319 | 0.19154916 | 0.3263714 | 0.99997367 |
| SLC26A3 | 1811 | 7 | 0.672369503 | 0.999797241 | 0.054041842 | 0.0002224 | 0.045767566 | 0.284708866 | 0.063130844 | 0.99980319 | −0.1980125 | 0.34245232 | 0.99997367 | |
| ACHE | 43 | NA | 0.994744858 | 0.999797241 | −0.060368494 | 0.000222854 | 0.045767566 | −0.310915524 | 0.053342115 | 0.99980319 | 0.46295071 | 0.03073028 | 0.99997367 | |
| ISL2 | 64843 | 15 | 0.001486002 | 0.153837193 | −0.048382632 | 0.000224613 | 0.045767566 | −0.273752693 | 0.049356113 | 0.99980319 | 0.25044517 | 0.21361948 | 0.99997367 | |
| KRT73 | 319101 | 12 | 0.820103696 | 0.999797241 | 0.051206891 | 0.000229713 | 0.046075372 | 0.14722346 | 0.313700428 | 0.99980319 | −0.2500906 | 0.21348809 | 0.99997367 | |
| ARRDC4 | 91947 | 15 | 0.377622927 | 0.999797241 | −0.045100521 | 0.00023539 | 0.046487629 | −0.486360588 | 0.000256439 | 0.07156327 | 0.43111988 | 0.02688404 | 0.99997367 | |
| MYH7B | 57644 | 20 | 0.703269593 | 0.999797241 | 0.049995062 | 0.000256825 | 0.049952474 | −0.018903827 | 0.89554412 | 0.99980319 | −0.0695272 | 0.73062912 | 0.99997367 |
EPDS Edinburgh Postnatal Depression Scale symptoms at 2 months postpartum, diffEPDS difference in EPDS symptoms from the third trimester of pregnancy to 2 months postpartum, PPD EPDS symptoms in the PPD group, Dep EPDS symptoms in the depression group, GnRHa_estradiol genes associated with estradiol in the GnRHa study, FDR P values after False Discovery rate multiple testing.
Gene expression associated with postpartum depression onset
Next, we assessed the onset of PPD at 2 months postpartum. First, the women were split into three groups
-
i.
PPD-onset group—these were women with EPDS < 10 at recruitment and EPDS ≥ 10 at postpartum (anytime between 1 and 12 months postpartum) (N = 25)
-
ii.
Depressed group—women with ≥ 10 EPDS at recruitment and ≥10 EPDS at postpartum (anytime between 1 and 12 months postpartum) (N = 44)
-
iii.
Control/euthymic group—women with <10 EPDS at recruitment and postpartum (anytime between 1 and 12 months postpartum) (N = 68).
Separate regressions were performed to compare the PPD and depressed groups to the control groups, to identify PPD-onset specific genes. The gene expression profiles were regressed against the group status, with maternal age, ethnicity, gestational age in weeks, current and past medications and psychological treatments and BMI included as covariates. A total of 38 genes were significantly differentially expressed between the PPD and the control groups (Table 4), and of these, 8 genes overlapped with those differentially expressed between the depressed and control groups, indicating that 30 genes showed PPD-onset specific gene expression. Of the 30 genes, 16 genes were also significant in the difference in depressive symptoms (P < 0.05). The full analysis results are shown in Supplementary Table 1.
Table 4.
List of differentially expressed genes at the third trimester of pregnancy associated with changes in depressive symptoms (from the third trimester to 2 months postpartum) at 5% FDR.
| Symbol | GnRHa_estradiol | Probe | Chr | P value_EPDSv2 | FDR_EPDSv2 | logFC_diffEPDSv2 | P value_diffEPDSv2 | FDR_diffEPDSv2 | logFC_PPD | P value_PPD | FDR_PPD | logFC_Dep | P value_Dep | FDR_Dep |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOC100132062 | 100132062 | NA | 2.74E-59 | 3.52E-55 | −0.09642385 | 1.86E-08 | 1.99E-05 | −2.0794307 | 1.58E-33 | 1.01E-29 | 2.03759936 | 5.40E-19 | 6.94E-15 | |
| GSTA7P | 730152 | 6 | 0.147252834 | 0.999797241 | 0.072537172 | 0.000661439 | 0.094343313 | 2.272172 | 1.65E-36 | 2.12E-32 | −1.865593 | 1.46E-14 | 9.40E-11 | |
| SLC12A1 | x | 6557 | 15 | 0.738374001 | 0.999797241 | −0.143662302 | 3.04E-23 | 3.91E-19 | −1.5148531 | 3.68E-24 | 1.58E-20 | 1.52947173 | 1.01E-10 | 4.31E-07 |
| FAM3B | 54097 | 21 | 0.000506343 | 0.073862832 | −0.081062951 | 4.36E-07 | 0.000357438 | −1.2068425 | 2.86E-11 | 6.13E-08 | 1.29211562 | 4.09E-08 | 0.00013115 | |
| RAP1GAP | 5909 | 1 | 0.000417001 | 0.066086904 | −0.084706388 | 1.69E-09 | 2.71E-06 | −0.7001112 | 1.09E-07 | 0.0001401 | 0.95024605 | 2.17E-06 | 0.00537669 | |
| MYOM2 | 9172 | NA | 0.558974564 | 0.999797241 | 0.045488778 | 0.002563317 | 0.19942602 | 0.66107629 | 8.04E-07 | 0.00077909 | −0.9456007 | 2.51E-06 | 0.00537669 | |
| RNF182 | 221687 | 6 | 0.983664265 | 0.999797241 | −0.08587623 | 5.84E-07 | 0.000441361 | −0.8205146 | 1.54E-08 | 2.47E-05 | 0.9490668 | 7.75E-06 | 0.01421399 | |
| HBZ | 3050 | 16 | 1.81E-13 | 7.73E-10 | 0.08198997 | 3.96E-11 | 8.47E-08 | 1.24062397 | 8.58E-19 | 2.75E-15 | −0.8915749 | 1.76E-05 | 0.02832105 | |
| PRSS33 | x | 260429 | 16 | 1.82E-06 | 0.00101726 | −0.017543158 | 0.204453261 | 0.961442886 | 0.66940149 | 7.80E-07 | 0.00077909 | −0.8128711 | 3.66E-05 | 0.05215136 |
| IFI27 | 3429 | NA | 0.491717731 | 0.999797241 | 0.088816879 | 2.79E-09 | 3.98E-06 | 1.07939383 | 5.46E-16 | 1.40E-12 | −0.7622081 | 8.16E-05 | 0.10472842 | |
| PTGDR2 | 11251 | 11 | 0.07345098 | 0.999797241 | −0.009817193 | 0.48417276 | 0.999245113 | 0.68209765 | 9.10E-07 | 0.00077909 | −0.7728273 | 9.83E-05 | 0.11468724 | |
| C4BPA | x | 722 | 1 | 2.91E-10 | 6.22E-07 | −0.051502467 | 3.73E-06 | 0.002174605 | −0.6400513 | 6.58E-06 | 0.00384058 | 0.82369396 | 0.00010817 | 0.11571456 |
| MAPK8IP1P2 | 644172 | NA | 0.038012933 | 0.90051655 | 0.000303088 | 0.987530803 | 0.999245113 | −0.9800435 | 2.22E-06 | 0.001679 | 0.97855722 | 0.00012366 | 0.12211055 | |
| SLC35F1 | 222553 | 6 | 0.012780901 | 0.533803126 | −0.070659963 | 1.20E-07 | 0.000110253 | 0.89950392 | 2.61E-10 | 4.78E-07 | −0.7774074 | 0.00015966 | 0.14639261 | |
| CMBL | 134147 | 5 | 0.475930622 | 0.999797241 | −0.033792967 | 0.036959174 | 0.639413638 | −0.7375122 | 3.29E-06 | 0.00211048 | 0.74263329 | 0.00044747 | 0.36562804 | |
| SFRP2 | 6423 | 4 | 0.422133267 | 0.999797241 | −0.019762377 | 0.182110005 | 0.957043264 | −0.5742315 | 2.74E-05 | 0.01256455 | 0.70217911 | 0.00045572 | 0.36562804 | |
| GALNT15 | 117248 | 3 | 0.661378861 | 0.999797241 | 0.005687522 | 0.71076802 | 0.999245113 | 0.73127667 | 5.79E-07 | 0.00067543 | −0.678052 | 0.00076939 | 0.57668405 | |
| ALOX15 | 246 | 17 | 0.006381561 | 0.367354704 | −0.004218688 | 0.757440501 | 0.999245113 | 0.57396501 | 1.79E-05 | 0.00886108 | −0.6573506 | 0.00080862 | 0.57668405 | |
| RAVER2 | 55225 | 1 | 0.122015102 | 0.999797241 | 0.026473215 | 0.082808829 | 0.811263458 | 0.73076056 | 8.57E-07 | 0.00077909 | −0.6602258 | 0.00122902 | 0.83036233 | |
| HES6 | 55502 | 2 | 0.616984359 | 0.999797241 | −0.023906291 | 0.104221966 | 0.860937819 | 0.78046567 | 2.83E-08 | 4.04E-05 | −0.5834678 | 0.00300491 | 0.99997367 | |
| SPTBN4 | x | 57731 | 19 | 0.853952516 | 0.999797241 | 0.04814081 | 0.003906481 | 0.251997482 | 0.84111298 | 1.57E-06 | 0.00125925 | −0.5715084 | 0.01091626 | 0.99997367 |
| FIBCD1 | 84929 | 9 | 0.01477133 | 0.571957401 | 0.054000943 | 0.00016668 | 0.038903107 | 0.72292732 | 2.77E-06 | 0.00197483 | −0.5574507 | 0.007439 | 0.99997367 | |
| OLR1 | 4973 | 12 | 0.137268091 | 0.999797241 | −0.040708626 | 0.002826989 | 0.21222253 | −0.6437111 | 3.00E-06 | 0.00202536 | 0.62579802 | 0.00157598 | 0.99997367 | |
| FAM83A | 84985 | 8 | 6.56E-06 | 0.003005253 | 0.035483408 | 0.024659247 | 0.565101277 | 0.68242078 | 3.88E-06 | 0.00237108 | −0.4020439 | 0.04915913 | 0.99997367 | |
| RSAD2 | 91543 | 2 | 0.001262375 | 0.135042605 | −0.003101777 | 0.802973454 | 0.999245113 | 0.55847735 | 1.04E-05 | 0.00581173 | −0.423053 | 0.02966682 | 0.99997367 | |
| NA | 654341 | NA | 0.040041391 | 0.918606088 | 0.043642091 | 0.002347612 | 0.194240656 | −0.5985625 | 1.47E-05 | 0.00779562 | 0.62713957 | 0.00188074 | 0.99997367 | |
| DDX11L10 | 100287029 | 16 | 0.027398604 | 0.771306761 | 0.019363117 | 0.196566971 | 0.961362007 | −0.7114069 | 1.52E-05 | 0.00779562 | 0.60633309 | 0.00556542 | 0.99997367 | |
| NCAPG2 | 54892 | NA | 0.118956575 | 0.999797241 | 0.035486631 | 0.013137006 | 0.436890536 | 0.60702078 | 2.18E-05 | 0.01035033 | −0.4730099 | 0.01883019 | 0.99997367 | |
| DEFA1B | 728358 | 8 | 2.73E-09 | 3.90E-06 | −0.082334647 | 2.83E-08 | 2.80E-05 | 0.53566102 | 5.17E-05 | 0.02287903 | −0.505308 | 0.00958881 | 0.99997367 | |
| MMP8 | 4317 | 11 | 0.093862874 | 0.999797241 | −0.042863638 | 0.001119109 | 0.133018019 | −0.5140184 | 6.88E-05 | 0.02943342 | 0.50413614 | 0.00877009 | 0.99997367 | |
| NEBL | 10529 | 10 | 0.177260886 | 0.999797241 | −0.030514743 | 0.029510787 | 0.604610542 | 0.59512746 | 7.33E-05 | 0.03037117 | −0.6234225 | 0.00358599 | 0.99997367 | |
| OR8U8 | 504189 | NA | 7.64E-05 | 0.020856842 | 0.042692465 | 0.000391896 | 0.065334587 | 0.51621139 | 9.78E-05 | 0.03924713 | −0.3413738 | 0.08193914 | 0.99997367 | |
| CLC | 1178 | 19 | 0.04198233 | 0.934015898 | 0.001050171 | 0.933703936 | 0.999245113 | 0.48764167 | 0.00010431 | 0.0405769 | −0.5586975 | 0.00285235 | 0.99997367 | |
| SMIM1 | 388588 | 1 | 1.08E-05 | 0.004463077 | −0.046924669 | 0.000526408 | 0.081415689 | 0.52234014 | 0.00011953 | 0.04456932 | −0.2791593 | 0.15879646 | 0.99997367 | |
| ISG15 | 9636 | 1 | 0.335644265 | 0.999797241 | 0.016722871 | 0.173604797 | 0.957043264 | 0.49763474 | 0.00012152 | 0.04456932 | −0.3898466 | 0.04277624 | 0.99997367 | |
| KIAA1324L | 222223 | 7 | 0.940093172 | 0.999797241 | 0.011875341 | 0.433620456 | 0.999245113 | −0.6272798 | 0.0001271 | 0.04532091 | 0.603388 | 0.00521569 | 0.99997367 | |
| ZFP57 | x | 346171 | 6 | 0.688934236 | 0.999797241 | 0.113752939 | 9.57E-12 | 3.07E-08 | 0.6292577 | 0.00014379 | 0.04988752 | −0.6042326 | 0.0056805 | 0.99997367 |
| TMEM176B | x | 28959 | 7 | 0.003678342 | 0.266546287 | −9.93E-05 | 0.993610049 | 0.999245113 | 0.48636354 | 0.00014793 | 0.04997309 | −0.6027717 | 0.0021531 | 0.99997367 |
EPDS Edinburgh Postnatal Depression Scale symptoms at 2 months postpartum, diffEPDS difference in EPDS symptoms from the third trimester of pregnancy to 2 months postpartum, PPD EPDS symptoms in the PPD group, Dep EPDS symptoms in the depression group, GnRHa_estradiol genes associated with estradiol in the GnRHa study, FDR P values after false discovery rate multiple testing.
Overlap with previous postpartum depression onset candidate genes
We compared our results to our previously reported candidate genes whose third trimester of pregnancy gene expression profiles were significantly associated with PPD-onset22. Of the 116 genes associated with PPD onset in our previous study, 84 genes were present in this study. From these, three genes (TMEM189, GALNT10 and FBXL20) were also associated with PPD onset in this study and not with depression per se (enrichment P = 0.08, indicating no significant overlap than expected by chance).
Next, we compared genes associated with PPD in this study with our previous study investigating gene expression and DNA methylation changes in a pharmacological model of depression where women were treated with the gonadotrophin-releasing hormone agonist (GnRHa) to induce depressive symptoms24. In the GnRHa study, we have previously shown that gene expression changes post-GnRHa induction directly relate oestrogen-induced biological changes with depressive symptoms and associated markers of serotonin-signalling in the brain, suggesting that individual variations in molecular sensitivity to oestrogen are associated with susceptibility to hormone-induced mood changes23.
A total of 7552 evaluated genes in this study overlapped with the GnRHa study. Of the genes associated with depressive symptoms (Table 2), change in depressive symptoms at 2 months postpartum (Table 3) or PPD onset (Table 4) at FDR P < 0.05, 10 (of 34, 29% overlap, P value = 4.2e-06), 17 (of 32, 53% overlap, P value = 2.1e-14) and 6 (of 17, 35% overlap, P = 1.2e-4) genes respectively were significantly associated with estradiol changes among the GnRHa-treated women.
Since the GnRhHa model in particular provoked biphasic changes in estradiol, these findings provide further evidence of the role of oestrogen signalling in PPD.
Overlap with the previously reported list of core maternal genes
We investigated a list of 700 core maternal genes identified by Gammie and colleagues32 that showed significant gene expression changes across multiple brain regions. This list was synthesised via bioinformatics analysis using findings from four microarray studies of different maternal brain regions. Of the 700 maternal genes from Gammie et al.32, we had 455 in the current gene expression study that was detected above background and after quality control. Of these, 70/455 (15.4%) were significantly differentially expressed in one of the above analysis (Table 5) of association with depressive symptoms at 2 months postpartum or changes in depressive symptoms from recruitment to 2 months postpartum or differentially expressed between the PPD and the control groups. This is a significant enrichment than expected by chance (P = 7.9e-05).
Table 5.
Comparison of differentially expressed genes at the third trimester of pregnancy associated with PPD with previously reported maternal genes by Gammie et al. 2016.
| Symbol | Probe | Chr | P value_EPDSv2 | FDR_EPDSv2 | P value_diffEPDSv2 | FDR_diffEPDSv2 | P value_PPD | FDR_PPD | P value_Dep | FDR_Dep |
|---|---|---|---|---|---|---|---|---|---|---|
| ADCY5 | 111 | 3 | 0.014050053 | 0.554047576 | 0.002962235 | 0.216262025 | 0.91178575 | 0.99980319 | 0.98965523 | 0.99997367 |
| ASAP2 | 8853 | 2 | 0.23315749 | 0.999797241 | 0.832816451 | 0.999245113 | 0.4734246 | 0.99980319 | 0.73193125 | 0.99997367 |
| ATP10A | 57194 | 15 | 0.071056678 | 0.999797241 | 0.041084579 | 0.660254558 | 0.99541483 | 0.99980319 | 0.94005609 | 0.99997367 |
| BCL2L1 | 598 | 20 | 0.011365889 | 0.515005387 | 0.914054201 | 0.999245113 | 0.98761521 | 0.99980319 | 0.32804887 | 0.99997367 |
| CADPS | 8618 | 3 | 0.212269446 | 0.999797241 | 0.795011 | 0.999245113 | 0.77701682 | 0.99980319 | 0.81620633 | 0.99997367 |
| CCDC85B | 11007 | 11 | 0.025452704 | 0.739222531 | 0.363283011 | 0.999245113 | 0.05392936 | 0.99980319 | 0.26021851 | 0.99997367 |
| CD200 | 4345 | 3 | 0.093184684 | 0.999797241 | 0.232688233 | 0.969329781 | 0.00999878 | 0.80474158 | 0.02667391 | 0.99997367 |
| CNTNAP3 | 79937 | 9 | 0.016675728 | 0.616660406 | 0.024084768 | 0.559265472 | 0.71961007 | 0.99980319 | 0.3454232 | 0.99997367 |
| DAAM2 | 23500 | 6 | 0.618068822 | 0.999797241 | 0.465553179 | 0.999245113 | 0.33148853 | 0.99980319 | 0.43262365 | 0.99997367 |
| DDAH2 | 23564 | 6 | 0.121695981 | 0.999797241 | 0.633074639 | 0.999245113 | 0.10762088 | 0.99980319 | 0.34735339 | 0.99997367 |
| DLC1 | 10395 | 8 | 0.18544046 | 0.999797241 | 0.020026641 | 0.514369946 | 0.19652235 | 0.99980319 | 0.42960279 | 0.99997367 |
| DSCAM | 1826 | 21 | 0.002077118 | 0.191346638 | 0.675666126 | 0.999245113 | 0.62745455 | 0.99980319 | 0.97833471 | 0.99997367 |
| DZIP1 | 22873 | 13 | 0.268435651 | 0.999797241 | 0.567421782 | 0.999245113 | 0.70184475 | 0.99980319 | 0.95756208 | 0.99997367 |
| ENG | 2022 | 9 | 0.852264699 | 0.999797241 | 0.230231174 | 0.966439689 | 0.65484722 | 0.99980319 | 0.49902445 | 0.99997367 |
| FBN1 | 2200 | 15 | 0.030794244 | 0.826999396 | 0.9026211 | 0.999245113 | 0.88818414 | 0.99980319 | 0.89595002 | 0.99997367 |
| FLT1 | 2321 | 13 | 0.570727567 | 0.999797241 | 0.038153876 | 0.647066685 | 0.00242766 | 0.38734395 | 0.06800417 | 0.99997367 |
| FRAS1 | 80144 | 4 | 0.008458146 | 0.439583873 | 0.39781099 | 0.999245113 | 0.95921892 | 0.99980319 | 0.93647918 | 0.99997367 |
| FSTL5 | 56884 | 4 | 8.78E-05 | 0.023000406 | 0.050527822 | 0.708162007 | 0.00970811 | 0.8009674 | 0.03873353 | 0.99997367 |
| FXYD1 | 5348 | 19 | 0.018867223 | 0.646818417 | 0.762825669 | 0.999245113 | 0.92048491 | 0.99980319 | 0.88225019 | 0.99997367 |
| GCNT2 | 2651 | 6 | 0.212632395 | 0.999797241 | 0.024135941 | 0.559265472 | 0.91377176 | 0.99980319 | 0.72771845 | 0.99997367 |
| GLDC | 2731 | 9 | 0.018982052 | 0.646818417 | 0.039814013 | 0.653570948 | 0.2049579 | 0.99980319 | 0.36900048 | 0.99997367 |
| GPR27 | 2850 | NA | 0.703341699 | 0.999797241 | 0.008356958 | 0.348306075 | 0.26369055 | 0.99980319 | 0.56382651 | 0.99997367 |
| GPRIN3 | 285513 | 4 | 0.158689278 | 0.999797241 | 0.007280647 | 0.331404856 | 0.5722268 | 0.99980319 | 0.74894925 | 0.99997367 |
| HIF1A | 3091 | 14 | 0.083348331 | 0.999797241 | 0.805687086 | 0.999245113 | 0.00235364 | 0.38734395 | 0.05975394 | 0.99997367 |
| HIST1H1C | 3006 | 6 | 0.371095763 | 0.999797241 | 0.91982595 | 0.999245113 | 0.00481477 | 0.56509769 | 0.19892836 | 0.99997367 |
| HIST1H2BK | 85236 | 6 | 0.418176509 | 0.999797241 | 0.449859358 | 0.999245113 | 0.0580796 | 0.99980319 | 0.33328433 | 0.99997367 |
| HMGB3 | 3149 | X | 0.001054142 | 0.116964982 | 0.170650959 | 0.957043264 | 0.57580736 | 0.99980319 | 0.49913663 | 0.99997367 |
| HOMER1 | 9456 | 5 | 0.002422146 | 0.207713945 | 0.928286192 | 0.999245113 | 0.99781621 | 0.99980319 | 0.99212125 | 0.99997367 |
| HPCAL4 | 51440 | 1 | 0.962836118 | 0.999797241 | 0.404980957 | 0.999245113 | 0.00948858 | 0.79615389 | 0.07530006 | 0.99997367 |
| HSPB1 | 3315 | 7 | 0.774603671 | 0.999797241 | 0.008067963 | 0.341040786 | 0.02324166 | 0.99980319 | 0.24558202 | 0.99997367 |
| LRRC29 | 26231 | 16 | 0.399050799 | 0.999797241 | 0.022297823 | 0.540070111 | 0.33786985 | 0.99980319 | 0.62596094 | 0.99997367 |
| MAP3K15 | 389840 | X | 0.420717446 | 0.999797241 | 0.958895197 | 0.999245113 | 0.22671791 | 0.99980319 | 0.45515549 | 0.99997367 |
| MCF2L | 23263 | 13 | 0.398696592 | 0.999797241 | 0.001172201 | 0.13316409 | 0.25535639 | 0.99980319 | 0.20225951 | 0.99997367 |
| MERTK | 10461 | 2 | 0.101097195 | 0.999797241 | 0.044390268 | 0.678278078 | 0.63289989 | 0.99980319 | 0.40555607 | 0.99997367 |
| NEURL1B | 54492 | 5 | 0.036201122 | 0.879054327 | 0.11249574 | 0.877771504 | 0.94603704 | 0.99980319 | 0.9241486 | 0.99997367 |
| NEXN | 91624 | 1 | 0.388511732 | 0.999797241 | 0.021162427 | 0.524521154 | 0.3702592 | 0.99980319 | 0.6356162 | 0.99997367 |
| NR1D1 | 9572 | 17 | 0.244295838 | 0.999797241 | 0.032187846 | 0.615427276 | 0.48714116 | 0.99980319 | 0.99394571 | 0.99997367 |
| NR4A1 | 3164 | 12 | 0.54874689 | 0.999797241 | 0.811163817 | 0.999245113 | 0.03001189 | 0.99980319 | 0.08679762 | 0.99997367 |
| NSUN3 | 63899 | 3 | 0.586484515 | 0.999797241 | 0.459638237 | 0.999245113 | 0.08934916 | 0.99980319 | 0.08254255 | 0.99997367 |
| OLIG2 | 10215 | 21 | 0.063562425 | 0.999797241 | 0.026373718 | 0.585711441 | 0.00299288 | 0.42688463 | 0.0149956 | 0.99997367 |
| PDP1 | 54704 | 8 | 0.885184882 | 0.999797241 | 0.669359061 | 0.999245113 | 0.64335095 | 0.99980319 | 0.58963618 | 0.99997367 |
| PHLPP2 | 23035 | 16 | 0.755938138 | 0.999797241 | 0.596468698 | 0.999245113 | 0.03355505 | 0.99980319 | 0.09609877 | 0.99997367 |
| PLCB1 | 23236 | 20 | 0.348381116 | 0.999797241 | 0.020000685 | 0.514369946 | 0.50119888 | 0.99980319 | 0.79923428 | 0.99997367 |
| PLIN4 | 729359 | 19 | 0.408653941 | 0.999797241 | 0.033853456 | 0.62292633 | 0.48816799 | 0.99980319 | 0.35878095 | 0.99997367 |
| PLLP | 51090 | 16 | 0.746268876 | 0.999797241 | 0.006307858 | 0.305379969 | 0.75307827 | 0.99980319 | 0.66400787 | 0.99997367 |
| PPP1R12B | 4660 | 1 | 0.04338289 | 0.947889322 | 0.039379693 | 0.652530615 | 0.28069488 | 0.99980319 | 0.38859106 | 0.99997367 |
| PSD | 5662 | 10 | 0.923810949 | 0.999797241 | 0.042759347 | 0.672382742 | 0.6654271 | 0.99980319 | 0.45005221 | 0.99997367 |
| PSPH | 5723 | 7 | 0.686104071 | 0.999797241 | 0.873717886 | 0.999245113 | 0.42450925 | 0.99980319 | 0.92148676 | 0.99997367 |
| RAB30 | 27314 | 11 | 0.71805606 | 0.999797241 | 0.061149329 | 0.745952208 | 0.01561673 | 0.94562271 | 0.06022857 | 0.99997367 |
| RASA3 | 22821 | 13 | 0.772698055 | 0.999797241 | 0.011718405 | 0.413266918 | 0.68529277 | 0.99980319 | 0.88951742 | 0.99997367 |
| RASGEF1B | 153020 | 4 | 0.606014303 | 0.999797241 | 0.900764539 | 0.999245113 | 0.01995988 | 0.99980319 | 0.04431899 | 0.99997367 |
| RFTN1 | 23180 | 3 | 0.509223491 | 0.999797241 | 0.001629844 | 0.158502285 | 0.566442 | 0.99980319 | 0.97046749 | 0.99997367 |
| SBK1 | 388228 | 16 | 0.814900153 | 0.999797241 | 0.032447549 | 0.615427276 | 0.94079819 | 0.99980319 | 0.76509714 | 0.99997367 |
| SERINC2 | 347735 | 1 | 0.001346019 | 0.142800404 | 0.098238466 | 0.848244582 | 0.93519291 | 0.99980319 | 0.3510916 | 0.99997367 |
| SGSM1 | 129049 | 22 | 0.426624611 | 0.999797241 | 0.347460956 | 0.999245113 | 0.96549994 | 0.99980319 | 0.8219647 | 0.99997367 |
| SLC2A1 | 6513 | 1 | 0.197794608 | 0.999797241 | 0.73633701 | 0.999245113 | 0.00110525 | 0.21827851 | 0.0049442 | 0.99997367 |
| SLC38A5 | 92745 | X | 0.009995578 | 0.474076994 | 0.893153414 | 0.999245113 | 0.84453604 | 0.99980319 | 0.35205746 | 0.99997367 |
| SLC4A4 | 8671 | 4 | 0.960039114 | 0.999797241 | 0.458677999 | 0.999245113 | 0.02456949 | 0.99980319 | 0.04855063 | 0.99997367 |
| SLC4A8 | 9498 | 12 | 0.466064002 | 0.999797241 | 0.029660741 | 0.604610542 | 0.02353749 | 0.99980319 | 0.15735593 | 0.99997367 |
| SMOX | 54498 | 20 | 0.060547007 | 0.999797241 | 0.033388089 | 0.621163618 | 0.19395148 | 0.99980319 | 0.93632637 | 0.99997367 |
| SNRK | 54861 | 3 | 0.221630481 | 0.999797241 | 0.052251277 | 0.711293364 | 0.00027156 | 0.07416982 | 0.01404638 | 0.99997367 |
| SOCS3 | 9021 | 17 | 0.902530531 | 0.999797241 | 0.090285457 | 0.832799778 | 0.97152061 | 0.99980319 | 0.99901805 | 0.99997367 |
| SORCS2 | 57537 | 4 | 0.1797705 | 0.999797241 | 0.001092399 | 0.133018019 | 0.01452102 | 0.92830067 | 0.30719731 | 0.99997367 |
| TMEM165 | 55858 | 4 | 0.990113374 | 0.999797241 | 0.323611169 | 0.999245113 | 0.69745403 | 0.99980319 | 0.67185592 | 0.99997367 |
| TNFRSF12A | 51330 | 16 | 0.69170039 | 0.999797241 | 0.213123462 | 0.964674548 | 0.68818696 | 0.99980319 | 0.83076288 | 0.99997367 |
| TRIM9 | 114088 | 14 | 0.642086487 | 0.999797241 | 0.206442423 | 0.961442886 | 0.01238762 | 0.88646873 | 0.04950842 | 0.99997367 |
| ZER1 | 10444 | 9 | 0.008606227 | 0.443784651 | 0.288011553 | 0.994394748 | 0.4064001 | 0.99980319 | 0.27713802 | 0.99997367 |
| ZFP57 | 346171 | 6 | 0.688934236 | 0.999797241 | 9.57E-12 | 3.07E-08 | 0.00014379 | 0.04988752 | 0.0056805 | 0.99997367 |
| ZNF512B | 57473 | 20 | 0.188375011 | 0.999797241 | 0.005563967 | 0.293928586 | 0.93831436 | 0.99980319 | 0.56044337 | 0.99997367 |
| ZXDB | 158586 | X | 0.309677897 | 0.999797241 | 0.862922218 | 0.999245113 | 0.39484065 | 0.99980319 | 0.6766835 | 0.99997367 |
EPDS Edinburgh Postnatal Depression Scale symptoms at 2 months postpartum, diffEPDS difference in EPDS symptoms from the third trimester of pregnancy to 2 months postpartum, PPD EPDS symptoms in the PPD group, Dep EPDS symptoms in the depression group, GnRHa_estradiol genes associated with estradiol in the GnRHa study, FDR P values after false discovery rate multiple testing.
Among genes that showed significant maternal brain gene expression changes32 were interesting candidate genes including the ZFP57 gene, a maternal-zygotic effect gene involved in the stable maintenance of methylation imprints during development33 and the NR1D1 gene which has been highlighted as a key PPD gene and is linked to the steroid receptor.
Discussion
PPD is a common disorder that is frequently underdiagnosed and can lead to severe long-term negative health outcomes of the mother, infant and the family hence early identification of women at high risk for PPD is essential to allow timely preventive intervention and treatment before the symptoms of PPD begin to manifest.
The aim of this study was to evaluate genome-wide gene expression profiles of women from their third trimester of pregnancy to identify transcriptomic changes associated with PPD. To the best of our knowledge, this is the largest PPD gene expression study to date (n = 137), assessing early gene expression profiles during late pregnancy and their association with PPD.
A total of 71 genes were significantly associated with depression scores at 2 months postpartum, after correction for multiple testing. The PPD related genes were enriched for immune-related gene ontology biological processes including humoral immune response and neutrophil-mediated immunity and were overrepresented in the neutrophil degranulation and innate immune system pathways, suggesting a role of immune-pathway genes in PPD. These findings suggest that an altered immunological profile during pregnancy might play a role in PPD vulnerability. There are several theories linking depression with immune responses including the Pathogen Host Defense (PATHOS-D) hypothesis that suggests that depressive symptoms are integral components of immune-mediated host defence against pathogens, having an underlying evolutionary significance34. It remains to be established whether and at what extent the immune abnormalities observed are pre-existing even before pregnancy and how these change across pregnancy, possibly contributing to the development of PPD; larger longitudinal studies will uncover the exact mechanisms.
The top genes associated with 2 months postpartum-depressive scores included TNFRSF17, GYPA, JCHAIN, SMIM1, TRPM8, SPATA17, HEMGN, PRRT4 and ADAMS15. The TNFRSF17 gene is a member of the TNF-receptor superfamily, specifically leading to NF-kappaB and MAPK8/JNK activation, with a likely role in cell survival and proliferation. In another study35, during the third trimester of pregnancy, the levels of TNFRSF17 gene expression significantly upregulated in women with PPD as compared to euthymic pregnant women and depressed patients (depressed during pregnancy and postpartum). TNFRSF17 was also one of the immune response genes upregulated in patients with bipolar disorder36.
Next, we tested whether gene expression in the third trimester of pregnancy was associated with a difference in the third trimester of pregnancy to 2 months postpartum-depressive symptoms, as this was more likely to be of clinical relevance. We identified 66 genes whose gene expression profiles were associated with changes in depressive symptoms at a 5% FDR, these were substantially overlapped (50% at P < 0.05 and 21% at FDR 5%) with genes associated with only 2 months postpartum-depressive symptoms. The genes included OLFM4, RASAL2, BHZ, DEFA1B, DEFA4, C4BPA, CPA5, GPRC5C, LTF, ADAMTS15, CLEC12A and ELANE. The C4BPA has been reported in studies of individuals with psychotic experiences37 and schizophrenia38. The ELANE gene encodes for a protein that is found in neutrophils and plays a role in inflammation and in fighting infection.
When comparing the postpartum-onset group to euthymic controls (and using the depression group as a negative control), the study revealed 30 PPD-onset specific gene expression. Of the 30 genes, 16 genes were also significant in the difference in depressive symptoms (P < 0.05), 15 were also associated with depressive symptoms at 2 months postpartum and 8 of the genes were associated with both. Genes included MMP8, a matrix metalloproteinase–collagen degradation gene, previously reported to be associated with MDD in a genome-wide association study39. Other genes included SLC35F1, which was one of the significant genes for extraversion and neuroticism, and symptoms of anxiety, depression and psychological distress in a genome-wide association uncovering shared genetic effects among personality traits and mood states40.
Our previous research has investigated the transcriptomic landscape among women with PPD and identified that oestrogen signalling plays a key role in vulnerability to PPD. For instance, we and others had identified a panel of 116 genes significantly differentially expressed between women with PPD and euthymic controls22. These genes could predict with 88% accuracy which women went on to develop PPD within the sample and were enriched for those belonging to the oestrogen signalling pathway. In this study, we found three of these predictive PPD genes (TMEM189, GALNT10 and FBXL20) were also associated with PPD onset in the current study and not with depression per se. This overlap is consistent with chance alone (P = 0.80). The lack of enrichment in gene associations across the two studies could be the result of different samples and different technologies (microarray versus next-generation sequencing methods). Nevertheless, when comparing the gene expression profiles in PPD identified in this study with our previous pharmacological model of depression via GnRHa24, we found evidence that 29–53% of the genes were also associated with estradiol changes among the GnRHa-treated women (P values range = 1.2e-4–2.1e-14). Therefore, these results again emphasise the role of oestrogen signalling in PPD.
To further extrapolate and understand the function of the differentially expressed genes associated with PPD, the comparison was performed with the maternal genes identified by Gammie et al.32. This included a list of 700 core maternal genes that showed significant gene expression changes across multiple brain regions. Of the 455 genes present in the current gene expression study, 15.4% were significantly associated with PPD (enrichment P = 7.9e-05). Among genes that showed significant maternal brain gene expression changes32 was the ZFP57 gene, a maternal-zygotic effect gene that contributes towards the stable maintenance of methylation imprints during development33. This gene was significantly associated with depressive symptoms at 2 months postpartum in both discovery and replication cohorts, changes in depressive symptoms from late pregnancy to 2 months postpartum and was differentially expressed between the PPD and the control groups. Another interesting gene from the Gammie et al. study was the NR1D1 gene which was highlighted by the authors as a key PPD gene and is linked to the steroid receptor. NR1D1 gene expression is affected by estradiol, glucocorticoids and progesterone treatment, making this a key maternal brain gene, whose expression can be moderated by placenta-produced hormones and changes in these hormones. In this study, the NR1D1 gene was associated with depressive symptoms at 2 months postpartum.
This study has several limitations. Since this was a cohort-based study, some women (18%) were on medications and this likely influenced gene expression profiles, however, we did correct for medication status in the analysis. Given that only 11% of the women had PPD, to overcome the consequential issue of low power, we analysed both quantitative depression symptoms as well as PPD onset to better understand the influence of depressive symptoms on the transcriptome. Strengths of the study include that this is the largest gene expression study in PPD to date and we validate our results in other independent samples and show enrichment of the identified PPD genes in relevant studies.
In conclusion, this study interrogated the transcriptomic landscape during pregnancy and tested its association with PPD revealing novel genes and pathways associated with PPD and validating previous PPD candidates. Consistent with previous studies including our own research, this study strengthened the findings of altered immune genes in PPD and the role of oestrogen in the pathogenesis of PPD, suggesting avenues for treatment of PPD. These findings add to the existing knowledge of the molecular mechanisms and pathways associated with PPD, providing avenues for improved diagnosis and treatment.
Supplementary information
Acknowledgements
The authors would like to thank all the participants involved in this study. This project was funded by the Brain Behaviour Foundation NARSAD Young Investigator grant (grant number-22509) to Divya Mehta. N.R.W. acknowledges funding from the National Health and Medical Research Council (Grants: 1173790, 1078901, 1113400).
Author contributions
D.M. analysed and interpreted the data and wrote the first draft of the paper. K.G., B.P., S.M.B. and A.M.S. provided the UNC samples and supervised the project. S.W., L.W. and A.H. performed the experiments in the lab of N.R.W. D.M. and N.R.W. conceived the study and supervised the project. V.F. and E.B. provided the GnRHa data. All authors provided critical comments on the paper.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01270-5.
References
- 1.O’Hara MW, Wisner KL. Perinatal mental illness: definition, description and aetiology. Best. Pr. Res Clin. Obstet. Gynaecol. 2014;28:3–12. doi: 10.1016/j.bpobgyn.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sockol LE, Epperson CN, Barber JP. Preventing postpartum depression: a meta-analytic review. Clin. Psychol. Rev. 2013;33:1205–1217. doi: 10.1016/j.cpr.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th edn. (American Psychiatric Association, 2013.
- 4.Lapato DM, et al. DNA methylation associated with postpartum depressive symptoms overlaps findings from a genome-wide association meta-analysis of depression. Clin. Epigenet. 2019;11:169. doi: 10.1186/s13148-019-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putnam KT, et al. Clinical phenotypes of perinatal depression and time of symptom onset: analysis of data from an international consortium. Lancet Psychiatry. 2017;4:477–485. doi: 10.1016/S2215-0366(17)30136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ban L, Gibson JE, West J, Tata LJ. Association between perinatal depression in mothers and the risk of childhood infections in offspring: a population-based cohort study. BMC Public Health. 2010;10:799. doi: 10.1186/1471-2458-10-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gump BB, et al. Trajectories of maternal depressive symptoms over her child’s life span: relation to adrenocortical, cardiovascular, and emotional functioning in children. Dev. Psychopathol. 2009;21:207–225. doi: 10.1017/S0954579409000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman A, et al. Mothers’ mental health and infant growth: a case-control study from Rawalpindi, Pakistan. Child Care Health Dev. 2004;30:21–27. doi: 10.1111/j.1365-2214.2004.00382.x. [DOI] [PubMed] [Google Scholar]
- 9.Malm H, et al. Gestational exposure to selective serotonin reuptake inhibitors and offspring psychiatric disorders: a national register-based study. J. Am. Acad. Child Adolesc. Psychiatry. 2016;55:359–366. doi: 10.1016/j.jaac.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rai D, et al. Parental depression, maternal antidepressant use during pregnancy, and risk of autism spectrum disorders: population based case-control study. BMJ. 2013;346:f2059. doi: 10.1136/bmj.f2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein A, et al. Effects of perinatal mental disorders on the fetus and child. Lancet. 2014;384:1800–1819. doi: 10.1016/S0140-6736(14)61277-0. [DOI] [PubMed] [Google Scholar]
- 12.Sit D, et al. Suicidal ideation in depressed postpartum women: associations with childhood trauma, sleep disturbance and anxiety. J. Psychiatr. Res. 2015;66-67:95–104. doi: 10.1016/j.jpsychires.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulson JF, Bazemore SD. Prenatal and postpartum depression in fathers and its association with maternal depression: a meta-analysis. J. Am. Med. Assoc. 2010;303:1961–1969. doi: 10.1001/jama.2010.605. [DOI] [PubMed] [Google Scholar]
- 14.Austin MP. Antenatal screening and early intervention for “perinatal” distress, depression and anxiety: where to from here? Arch. Women’s Ment. Health. 2004;7:1–6. doi: 10.1007/s00737-003-0034-4. [DOI] [PubMed] [Google Scholar]
- 15.O’Connor E, et al. Primary care screening for and treatment of depression in pregnant and postpartum women: evidence report and systematic review for the US preventive services task force. J. Am. Med. Assoc. 2016;315:388–406. doi: 10.1001/jama.2015.18948. [DOI] [PubMed] [Google Scholar]
- 16.Guintivano J, Manuck T, Meltzer-Brody S. Predictors of postpartum depression: a comprehensive review of the last decade of evidence. Clin. Obstet. Gynecol. 2018;61:591–603. doi: 10.1097/GRF.0000000000000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guintivano J, et al. Adverse life events, psychiatric history, and biological predictors of postpartum depression in an ethnically diverse sample of postpartum women. Psychol. Med. 2018;48:1190–1200. doi: 10.1017/S0033291717002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leigh B, Milgrom J. Risk factors for antenatal depression, postnatal depression and parenting stress. BMC Psychiatry. 2008;8:24. doi: 10.1186/1471-244X-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta D, et al. The 5-HTTLPR polymorphism modulates the influence on environmental stressors on peripartum depression symptoms. J. Affect Disord. 2012;136:1192–1197. doi: 10.1016/j.jad.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Doornbos B, et al. The development of peripartum depressive symptoms is associated with gene polymorphisms of MAOA, 5-HTT and COMT. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2009;33:1250–1254. doi: 10.1016/j.pnpbp.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Figueira P, et al. An association study between the Val66Met polymorphism of the BDNF gene and postpartum depression. Arch. Women’s Ment. Health. 2010;13:285–289. doi: 10.1007/s00737-010-0146-6. [DOI] [PubMed] [Google Scholar]
- 22.Mehta D, et al. Early predictive biomarkers for postpartum depression point to a role for estrogen receptor signaling. Psychol. Med. 2014;44:2309–2322. doi: 10.1017/S0033291713003231. [DOI] [PubMed] [Google Scholar]
- 23.Mehta D, et al. Evidence for oestrogen sensitivity in perinatal depression: pharmacological sex hormone manipulation study. Br. J. Psychiatry. 2019;215:519–527. doi: 10.1192/bjp.2018.234. [DOI] [PubMed] [Google Scholar]
- 24.Mehta D, et al. Genome-wide gene expression in a pharmacological hormonal transition model and its relation to depressive symptoms. Acta Psychiatr. Scand. 2019;140:77–84. doi: 10.1111/acps.13038. [DOI] [PubMed] [Google Scholar]
- 25.Stuebe AM, et al. The mood, mother, and infant study: associations between maternal mood in pregnancy and breastfeeding outcome. Breastfeed. Med. 2019;14:551–559. doi: 10.1089/bfm.2019.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox, J. & Holden, J. Perinatal Mental Health: A Guide to the Edinburgh Postnatal Depression Scale (EPDS) (Royal College of Psychiatrists, 2003).
- 27.Pearson RM, et al. Maternal depression during pregnancy and the postnatal period: risks and possible mechanisms for offspring depression at age 18 years. JAMA Psychiatry. 2013;70:1312–1319. doi: 10.1001/jamapsychiatry.2013.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HH, Kim TH. Screening depression during and after pregnancy using the EPDS. Arch. Gynecol. Obstet. 2014;290:601–602. doi: 10.1007/s00404-014-3334-1. [DOI] [PubMed] [Google Scholar]
- 29.Meijer JL, et al. Predictive accuracy of Edinburgh postnatal depression scale assessment during pregnancy for the risk of developing postpartum depressive symptoms: a prospective cohort study. BJOG. 2014;121:1604–1610. doi: 10.1111/1471-0528.12759. [DOI] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gammie SC, et al. Genetic and neuroendocrine regulation of the postpartum brain. Front. Neuroendocrinol. 2016;42:1–17. doi: 10.1016/j.yfrne.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raison CL, Miller AH. The evolutionary significance of depression in Pathogen Host Defense (PATHOS-D) Mol. Psychiatry. 2013;18:15–37. doi: 10.1038/mp.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petralia MC, et al. Retrospective follow-up analysis of the transcriptomic patterns of cytokines, cytokine receptors and chemokines at preconception and during pregnancy, in women with post-partum depression. Exp. Ther. Med. 2019;18:2055–2062. doi: 10.3892/etm.2019.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan MM, et al. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol. Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- 37.Focking M, et al. Complement pathway changes at age 12 are associated with psychotic experiences at age 18 in a longitudinal population-based study: evidence for a role of stress. Mol. Psychiatry. 2019;26:524–533. doi: 10.1038/s41380-018-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, et al. An evaluation of association between common variants in C4BPB/C4BPA genes and schizophrenia. Neurosci. Lett. 2015;590:189–192. doi: 10.1016/j.neulet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Song GG, Kim JH, Lee YH. Genome-wide pathway analysis in major depressive disorder. J. Mol. Neurosci. 2013;51:428–436. doi: 10.1007/s12031-013-0047-z. [DOI] [PubMed] [Google Scholar]
- 40.Luciano M, et al. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:684–695. doi: 10.1002/ajmg.b.32072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.