Abstract
Eosinophilic esophagitis (EoE) is a chronic, food-triggered, immune-mediated disease of the oesophagus, clinically characterized by symptoms referred to oesophagal dysfunction, and histologically defined by an eosinophil productive inflammation of the oesophagal mucosa, among other cell types. The involvement of an adaptive Th2-type response to food antigens in EoE was known since 2000; several cytokines and chemokines promote food-specific responses, during which local production of IgE, but also IgG4 derived from plasma cells in lamina propria of oesophagal mucosa might play an important role. Evidence pointing towards a possible role for the innate immunity in EoE has arisen recently. Together, this evidence gives rise to a potential role that the innate immune system in general, and also the microbial pattern recognition receptors (PRRs) might play in EoE pathogenesis. Among PRRs, Toll-like receptors (TLRs) are type-I transmembrane receptors expressed both on epithelial and lamina propria cells with the capacity to distinguish between pathogen and commensal microbes. As TLRs in the different intestinal epithelia represent the primary mechanism of epithelial recognition of bacteria, this evidence underlines that oesophagal TLR-dependent signaling pathways in EoE support the potential implication of microbiota and the innate immune system in the pathogenesis of this disease. The oesophagal mucosa hosts a resident microbiota, although in a smaller population as compared with other districts of the gastrointestinal tract. Few studies have focused on the composition of the microbiota of the normal oesophagus alone. Still, additional information has come from studies investigating the oesophagal microbiota in disease and including healthy patients as controls. Our review aims to describe all the evidence on the oesophagal and intestinal microbiota in patients with EoE to identify the specific features of dysbiosis in this condition.
Keywords: microbiota, eosinophilic esophagitis, probiotics, dysbiosis, toll-like receptor
Microbiota in Eosinophilic Esophagitis: A Revision of Hygiene Hypothesis
Eosinophilic esophagitis (EoE) is a chronic immune-mediated disease of the oesophagus, characterized by oesophageal dysfunction and by a productive eosinophil inflammation of the oesophageal mucosa (1–3). The incidence of EoE has increased in recent years (4).
The involvement of an adaptive Th2-type response to food antigens in EoE is well demonstrated (5, 6); several cytokines and chemokines promote food-specific responses (7, 8), during which local production of IgE (9), but also IgG4 in lamina propria of oesophageal mucosa (10) may play an important role. Pro-fibrogenic factors released by inflammatory cells can provoke fibrous remodeling of the oesophageal mucosa (11, 12). Avoiding specific food triggers is, in some contexts, the first line of therapy for EoE (13, 14).
Rather than the specific adaptive immunity, the innate immune system recognizes and reacts to ecological insults and microbes without the need for an immunoglobulin-driven antigen-specific reaction. Proof pointing towards a possible role for the innate immunity in EoE has emerged. Oesophageal epithelial cells appear to be critical effectors provoking the inflammatory phenomena in EoE, not directly through eotaxin-3 release and other chemoattractants for eosinophils (15), but also by the recruitment of invariant natural killer T (iNKT) cells toward the oesophageal epithelium (16), which constitutes a crucial cytokine source. A pivotal role for mast cells (MCs) has also been recognized in the pathophysiology and symptoms of EoE, which reverse after effective dietary treatment (17). This evidence gives rise to a possible role that the innate immune system and raises some possible questions regarding the role of the microbial pattern recognition receptors (PRRs) in EoE pathogenesis.
Among PRRs, Toll-like receptors (TLRs) are type-I transmembrane receptors expressed both on epithelial and lamina propria cells with the ability to recognize microorganism and commensal organisms (18). In humans, there is an aggregate of 11 distinctive TLR, each having various specificities which, when activated, promote intracellular sign transduction pathways intervened by MAP kinases and NF-κB, at last setting off a supportive of inflammatory reaction. TLRs initiation is mindful, among different capacities, for setting off provocative reactions by going about as a connection among adaptive and innate immunity (19–21). Enactment and development of antigen-presenting cells and regulatory T cells (Tregs) rely in part upon TLR-intervened flagging, featuring their job on mucosal resistant homeostasis. A few investigations have assessed the association between hypersensitivity and TLR activation (19, 22, 23). TLR activity in oesophageal epithelial samples has been described (24).
Arias et al. shown that bacterial load and TLR1, TLR2, TLR4, and TLR9 were overexpressed on oesophageal biopsies with EoE compared to controls. Muc1 and Muc5B genes were downregulated while Muc4 was overexpressed. Upregulation of MyD88 and NFκB was discovered along with IL-1β, IL-6, IL-8, and IL-10 and PER-1, iNOS, and GRZA effectors. NG-K2D (KLRK1, IL-15, MICB) were likewise upregulated. In all cases, changes in EoE were neutralized after six food elimination diet (SFED) and mucosal healing.
As TLRs in the different intestinal epithelia represent the essential instrument of epithelial recognition of microbes (25), this proof underlines that oesophageal TLR-subordinate flagging pathways in EoE support the implication of microbiota and the innate immune system in the advancement of this condition.
Sterility in infant mice can prompt a move in the IgE-basophil axis, an unevenness in Th1/Th2 activation, just as inappropriate implication of Tregs. Human examinations to assess these components are not many (26, 27). These varieties may be suggestive of a connection between commensal microscopic organisms and hereditary demeanor to atopic illness. These bacterial ligands additionally could represent therapeutic targets in atopic disease.
Therefore, it appears potentially useful to understand the function of microbial flora in healthy human beings and the specific alterations in atopic diseases.
The White House Office of Science and Technology declared that the Fact Sheet for the National Microbiome Initiative (NMI) was intended to satisfy three explicit objectives: (1) to address central inquiries concerning the microbiome in different biological systems; (2) to create stage innovations for upgrading information sharing on microbiomes; and (3) to grow the microbiome workforce.
Since then, the interest in microbiome composition in different allergic conditions has grown (28–32).
The examination concerning how the microbiome can change, even in healthy individuals, is critical to improve comprehension of how the microbiome unthinkingly causes infection.
Oesophageal Microbiota in Health
Introduction Comments
Once thought composed of few microbes, the oesophageal mucosa showed a composition of around 300 bacteria species. New culture-independent techniques have allowed scientists to identify the microbial composition of the oesophagus.
The oesophageal mucosa hosts a resident microbiota, although in a smaller population as compared with other districts of the gut. The human microbiota of the digestive tract exhibits considerable qualitative and quantitative differences, with communities starting from 10 cells per g/mL of sampled material within the oesophagus and stomach to 1012 per g/mL of tested material in the large intestine (33, 34).
The Main Findings
It was initially not clear whether the oesophagus was characterized by a defined microbiota. The first studies on the oesophageal microbiota and based on cultivation methods demonstrated that the oesophagus did not merely contain a transient microbial population originating from the oral cavity by swallowing or from the stomach by gastroesophageal reflux (GER) (35–37). It was later observed that bacteria were associated with the oesophageal mucosal surface, confirming the presence of a resident microbiota at this site (38). Knowledge regarding the composition of the microbiota of healthy individuals has been expanded using investigations based on metagenomics approaches (39–41).
In general, the distal oesophageal microbiota was described as simply like that of the oropharynx, yet not indistinguishable (38, 42, 43).
Scarcely any examinations have concentrated on the organization of the microbiota of healthy oesophagus, (44–46) but extra data has originated from considers researching the oesophageal microbiota in disease compared to controls (47–56).
Influencers and Limitations
The accessible data on the microbiota are biased by various methodologies, contrasts in the tested parts of the oesophagus, and the heterogeneity of consideration/avoidance rules utilized in the different investigations don’t permit comparisons and make it hard to agree on the general microbiota synthesis of the healthy oesophagus.
The presence of Streptococcus spp. was described by all studies: along these lines, members from this genus, seem to be a dominant taxon in the microbiota of the healthy oesophagus. Other bacterial genera frequently identified in association with streptococci, albeit in lower extents, incorporate Fusobacterium, Veillonella and Prevotella. The nearness of different genera (e.g., Neisseria, Haemophilus, Gemella, Granulicatella, Actinomyces, Lactobacillus, Bacteroides, Porphyromonas, and Staphylococcus), was lower.
Outstandingly, the commonness of Fusobacterium, Streptococcus, Prevotella and Veillonella has reliably been accounted in several studies based on either culture-dependent or culture-independent methodologies and on various examples (biopsies, aspirates, brushes), subsequently giving a reliable sign of their commitment to the piece of the microbiota that colonizes the healthy oesophageal mucosa. The strength of streptococci and the continuous nearness of other taxa regular of the oropharyngeal microbiota have been identified with the piece of the microbial networks of the oropharyngeal cavity, where a high commonness of streptococci is discovered, along with Gemella, Fusobacterium, Veillonella, Rothia and Granulicatella, (57, 58) have bolstered the idea that the oesophageal microbiota is basically of oral inception.
In any case, not every single oral bacterium can colonize the oesophageal mucosa, while a few individuals from the oesophageal microbiota appear to be underrepresented in the oral cavity, highlighting an alternate microbiota variation in the two-body destinations (38, 45, 46).
Oesophageal Microbiome and Diet
There is no sufficient data on the effect of the diet on the oesophageal microbiota. A study of 47 patients showed that a diet higher in fiber was associated with a decrease of Proteobacteria and an increase of the Firmicutes phylum in the oesophagus (59). Additional studies correlating oesophageal microbiome and nutrition are needed. Studies that further define the stability of the oesophageal microbiome over time as well as other factors that determine inter-individual microbiome composition will aid in our understanding of the role of the diet.
Oesophageal Microbiota in EoE
The Main Findings
In 2015, Benitez et al. described the bacterial composition of the oral and oesophageal mucosa through 16S rRNA assessment of buccal swabs and oesophageal biopsies from 33 pediatric EoE subjects compared to 35 non-EoE healthy controls. They applied a longitudinal model before and after defined dietary changes.
In this study, Firmicutes were more abundant in oesophageal compared to oral samples, and oesophageal microbiota was more abundant of Proteobacteria in controls than EoE. The authors detected a significant difference between activated EoE and controls biopsies. The targeted dietary intervention did not produce substantial differences in either oesophageal or oral microbiota; the reintroduction of allergens led to enrichment in Campylobacter and Ganulicatella genera in the oesophagus (55).
Corynebacterium was enriched in the EoE samples, as was Neisseria genus, as previously described in different inflammatory conditions (60, 61). The Atopobium and Streptococcus genera were consistently enriched in non-EoE control samples.
The oesophageal microbiome of non-EoE control subjects showed a prevalence of Gram (+) bacteria of the Streptococcus genus, in agreement with previous findings in adult (62) and pediatric individuals (45) without oesophageal inflammation. Most of Benitez et al. study subjects were pediatric males with nearly 100% documented and concurrent proton-pump inhibitors (PPI) use, providing a further indication of the resilience of the Streptococcus-dominated microbiome in the healthy oesophagus in all genders and age groups.
They did not detect differences in the oral microbiome between inactive EoE, active EoE or non-EoE control samples, suggesting that in pediatric EoE, bacterial communities are stable and might not be altered by dietary modification. Data do not support the use of oral samples for EoE surveillance instead of biopsies.
In the same year, Harris et al. performed a prospective evaluation of secretions from EoE adult and children oesophageal biopsies, Gastro-oesophageal Reflux Disease (GERD) and healthy mucosa through Esophageal String Test (EST). Bacterial load was determined by quantitative PCR. Bacterial communities, determined by 16S rRNA and 454 pyrosequencing, were compared between disease and health. The bacterial amount was increased in both GERD and EoE and compared to healthy subjects. In EoE individuals, the amount was increased regardless of treatment status or level of mucosal eosinophilia. Haemophilus was significantly increased in untreated EoE individuals as compared with healthy subjects. Streptococcus was diminished in GERD subjects in PPI therapy as compared with healthy subjects. These data affirmed that diseases related to mucosal eosinophilia are characterized by a different microbiome from that found in the ordinary mucosa (56).
The EoE activity did not seem to directly affect a load of bacteria in EoE. However, eosinophils possess extracellular DNA traps and numerous anti-microbial properties with the release of defensins (63–65). The microbiota of untreated EoE subjects showed a shift from a mostly Gram (+) population to an increase in Gram (−) bacteria similar to what has been described in GERD (49). The implication of Gram (−) bacterial involvement in reflux esophagitis (66) is consistent with the observed increase in Haemophilus and Proteobacteria in EoE. These data suggest that treatment could affect the microbiota.
Norder Grusell et al. enrolled 17 subjects with GERD and 10 with EoE. All patients performed endoscopic brush sampling and biopsies from the upper and lower oesophagus and the oral cavity. Bacterial growth was identified to the species or genus level. The major part of bacterial groups or species was found in specimens from the lower oesophagus in EoE subjects compared to GERD subjects. Streptococcus viridans was the most common bacteria in both groups. GERD individuals had significantly inferior bacterial diversity in both oesophageal and oral samples. This discrepancy could depend on the protective mucosal biofilm by the acid content in GERD patients. The authors endorsed cultural method and speculated that bacteria identified by 16SrDNA/RNA techniques might not be alive, and there might be amplification bias following the processing steps. Nevertheless, they acknowledge that it is impossible to cultivate some bacterial species, and essential bacteria might, therefore, be overlooked (67).
Table 1 summarizes the main findings about microbiota in EoE.
Table 1.
Main findings about oesophageal Microbiota in EoE.
| Author, year | Study population | Method | Microbial differences |
|---|---|---|---|
| Benitez AJ. et al, 2015 (55) | Non-EoE pediatric controls and pediatric EoE subjects before and after defined dietary changes. | Bacterial composition through 16S rRNA gene sequencing of the oral and esophageal microenvironments using oral swabs and esophageal biopsies |
|
| Harris JK. et al., 2015 (56) | EoE adult and children, Gastro-oesophageal Reflux Disease and healthy mucosa. | Bacterial composition of secretions and biopsies through 16S rRNA gene amplification from samples obtained with Esophageal String Test (EST). Bacterial load was determined by quantitative PCR. |
|
| Norder Grussell E. et al., 2018 (67) | Subjects diagnosed with GERD and with EoE | Brush sampling and biopsies from the oral cavity, upper and lower esophagus. The samples were cultivated on agar plates, and bacterial growth was identified to the genus or species level and semi-quantified. |
|
| Hiremath G. et al., 2019 (68) | Non-EoE pediatric controls and pediatric EoE. | The salivary microbiome was determined through 16S rRNA gene sequencing. |
|
Helicobacter Pylori: A Controversial Role
Helicobacter pylori infection often occurs in early childhood and it seems to enhance immune-tolerance driving immune-mediated diseases in a susceptible host (69–71).
In this context, previous or current infection with Helicobacter pylori (exposure) has been reported to protect against EoE, perhaps owing to H. pylori-induced immunomodulation. In 2019 a meta-analysis evaluated 11 observational studies comprising data on 377,795 individuals worldwide. H. pylori exposure vs non-exposure was associated with a 37% reduction in odds of EoE (odds ratio, 0.63; 95% CI, 0.51–0.78) and a 38% reduction in odds of esophageal eosinophilia (odds ratio, 0.62; 95% CI, 0.52–0.76). Fewer prospective studies found a significant association between H. pylori exposure and EoE (p= .06) than retrospective studies. Effect estimates were not affected by study location, whether the studies were performed in pediatric or adult populations, time period, or prevalence of H. pylori in the study population (72). The role of H. pylori would therefore deserve more evidence.
Possible Alternatives to the Oesophageal Biopsies for Microbiota Evaluation
In 2012 Fillon et al. described the oesophageal microbiome in healthy children through the Enterotest™ (EST), a minimally invasive string technology. EST samples and mucosal biopsies were collected from healthy children (n=15) and their microbiome composition determined by 16S rRNA gene sequencing. Microbiota from oesophageal biopsies and ESTs produced nearly identical profiles of bacterial genera and were different from the bacterial contents of oral and nasal cavity samples. They concluded that the EST could be a useful device for the study of the oesophageal microbiome (45).
In 2019 Hiremath et al. collected saliva samples from 19 non-EoE controls and 26 children with EoE. The salivary microbiome was determined through 16S rRNA gene sequencing, and disease activity was assessed through the Eosinophilic Esophagitis Histologic Scoring System (EoEHSS) and the Eosinophilic Esophagitis Endoscopic Reference Score.
A trend toward lower microbial richness was recognized in EoE children. The salivary microbiome was similar between children with and without EoE.
Streptococcus tended to be more abundant in children with active EoE compared with non-EoE controls. Haemophilus was significantly plentiful in active EoE compared with inactive EoE and increased with the increasing Eosinophilic Esophagitis Histology Scoring System and EoEHSS. Besides, four broad salivary microbial communities correlated with the EoEHSS.
The authors concluded that the composition of the salivary microbiome community structure could be different in children with EoE. The disease activity positively correlates with the relative abundance of Haemophilus. Perturbations in the salivary microbiome may play a role in EoE pathobiology and could be a noninvasive marker of disease activity. The disease activity in the oesophagus does not seem to affect a load of bacteria in EoE directly (68).
Attempts at the Modulation of EoE with Probiotic
Recently, it was demonstrated that there are huge contrasts in gut microbial community structure, microbial abundance, and uniformity in patients with EoE.
Faecal microbiota was evaluated through 16SrRNA amplification from 12 EoE patients and 12 controls. Patients with EoE showed inferior gut microbiota alpha diversity. The authors observed at the phylum level an important increase in Bacteroidetes and a decrease in Firmicutes and a significant reduction in Clostridiales and Clostridia at the order and family level in patients with EoE.
The authors speculated that Clostridia based interventions could be tested as adjuncts to current therapeutic strategies in EoE (73).
The probiotic Lactococcus lactis NCC 2287 has previously been shown to decrease clinical scores in a food allergy model based on co-administration of cholera toxin and ovalbumin (74). Also, NCC 2287 is a potent inhibitor of the eosinophil survival cytokine IL-5 and an inducer of the immune-modulatory cytokine IL-10 and in Th2-skewed cultures of peripheral blood mononuclear cells (PBMC) (75). The probiotic Bifidobacterium lactis NCC 2818 is also known for its immunomodulatory properties in allergy (76).
Holvoet et al. in 2016 tested L. lactis NCC 2287 and B. lactis NCC 2818, for their capacity to decrease oesophageal inflammation in EoE murine model (77).
To test whether probiotics could decrease oesophageal eosinophilia in an EoE animal model, the strain NCC 2287 was added to the drinking water as prevention (day 0 to day 28; n = 8), as a treatment (day 28 to day 38; n = 10) or as continuous exposure (day 0 to day 38; n = 10). The maximum eosinophil count was significantly greater in the oesophagus of sensitized mice challenged with Af extract than in the oesophagus of non-sensitized mice. Interestingly, sensitized mice receiving NCC 2287 from day 28 to 38 had significantly less oesophageal eosinophilia than the non-supplemented sensitized group. However, there was no significant effect on oesophageal eosinophilia when the NCC 2287 strain was administered as a preventive measure or when it was given continuously, throughout the study.
This study demonstrates that the time frame of supplementation is fundamental: the beneficial effect of L. lactis NCC 2287 was only observed when it was administered as a treatment. Altogether, the data suggest that L. lactis NCC 2287 may be an exciting candidate for reducing EoE inflammation.
B. lactis NCC 2818 and L. lactis NCC 2287 both induce an increase of IL-10 in a Th2-skewed PBMC model, suggesting a possible immunoregulatory effect of these strains (76–78). However, in the same model, L. lactis NCC 2287 is more potent at reducing IL-5 levels and a stronger inducer of IFN-c than B. lactis NCC 2818 (36). It seems debatable the evaluation of the predictive value of these assays in human disease.
Probiotics seem to stabilize IL10 mRNA expression and to dysregulate microRNAs in human monocyte (78). These results suggest that other epigenetic mechanisms could explain the effect observed in the oesophagus and that the understanding of probiotic impact is at the beginning. Akei et al. have demonstrated that oesophageal eosinophilia is IL-5-dependent (79).
The balancing impact of NCC 2287 on IL-5 expression seen in Th2-slanted PBMCs (75) and an ovalbumin/cholera toxin-induced food hypersensitivity model (74) may somewhat clarify the abatement in oesophageal eosinophilia seen with NCC 2287 treatment in this study.
These results suggest that L. lactis NCC 2287 may lead to a steady decrease in gastrointestinal Th2 inflammation independently of the level of antigen sensitization in experimental allergy models.
Previous studies have highlighted the noticeable link between oesophageal and lung inflammation (45). Furthermore, 14–70% of EoE patients also presented with asthma (1, 46). The effect of L. lactis NCC 2287 on lung eosinophilia and oesophageal eosinophilia could be referred to a common consequence of the probiotic on eosinophil recruitment. Nevertheless, oral supplementation with B. lactis NCC 2818 had no effect on EoE, although it significantly reduced bronchoalveolar eosinophilia.
Their data suggest that a decrease in eosinophils in the lung is not concomitant with a significant decline in oesophageal eosinophilia. These outcomes are to be expected as various preclinical examinations have indicated that lung eosinophilia can diminish with probiotic supplementation (80, 81) or auxiliary to changes in the gut microbiota (82, 83).
Probiotics could vary in their ability to prevent or deal with the allergic response.
The proof in the animal model recommends that specific probiotics might be gainful in lessening oesophageal eosinophilic inflammation.
Instead, probiotics seem to permit a more comprehensive approach which may restore and maintain homeostasis in humans.
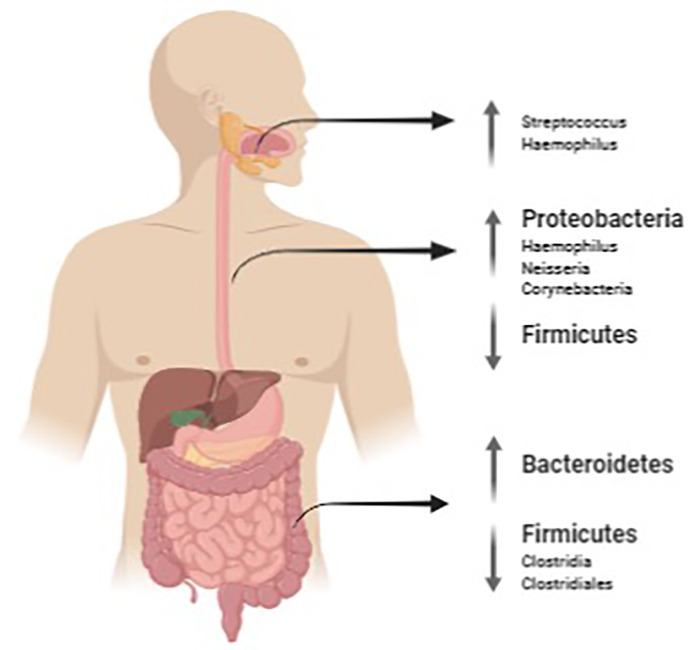
Figure 1 summarizes the main findings of microbiota found in EoE at oral, esophageal and intestinal level.
Figure 1.
Salivary, oesophageal and gut microbiota mutations in EoE. The image was created with Biorender.com.
Conclusion
In conclusion, the oesophageal mucosa hosts a resident microbiota and there is evidence that it may change in the presence of EoE with an increase of bacterial load (56, 67) and with Streptococcus as recurrent taxa (67, 68) and with Haemophilus as possible marker of disease activity in different studies (56, 68).
It is not yet clear what is the cause-effect link that regulates these changes.
It is therefore not yet possible to imagine possible rational interventions for modulating the esophageal microbiota.
It therefore appears essential to carry out immunological studies that clarify this phenomenon and that allow to hypothesize also possible alternative therapies for EoE.
Author Contributions
MM, AA, and AF conceived the review and research method of bibliographic sources. MM and RT performed the research, the analysis and the selection of the sources. MM and AA wrote the first draft of the manuscript. CR, FR, and PA performed the critical analysis of the sources and the final revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The Scientific Direction of Bambino Gesù Children's Hospital approved and contributed to the payment of the publication fee of this scientific contribution.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Lucendo AJ, Molina-Infante J, Arias A, von Arnim U, Bredenoord AJ, Bussmann C, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur Gastroenterol J (2017) 5(3):335–58. 10.1177/2050640616689525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Molina-Infante J, Bredenoord AJ, Cheng E, Dellon ES, Furuta GT, Gupta SK, et al. Proton pump inhibitor-responsive oesophageal eosinophilia: an entity challenging current diagnostic criteria for eosinophilic oesophagitis. Gut (2016) 65:521–31. 10.1136/gutjnl-2015-310991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lucendo AJ, Navarro M, Comas C, Pascual JM, Burgos E, Santamaría L, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol (2007) 31:598–606. 10.1097/01.pas.0000213392.49698.8c [DOI] [PubMed] [Google Scholar]
- 4. Arias Á, Pérez-Martínez I, Tenías JM, Lucendo AJ. Systematic review with meta-analysis: The incidence and prevalence of eosinophilic oesophagitis in children and adults in populationbased studies. Aliment Pharmacol Ther (2016) 43:3–15. 10.1111/apt.13441 [DOI] [PubMed] [Google Scholar]
- 5. Straumann A, Bauer M, Fischer B, Blaser K. Simon HU Idiopathic eosinophilic esophagitis is associated with a T (H)2-type allergic inflammatory response. J Allergy Clin Immunol (2001) 108:954–61. 10.1067/mai.2001.119917 [DOI] [PubMed] [Google Scholar]
- 6. Bullock JZ, Villanueva JM, Blanchard C, Filipovich AH, Putnam PE, Collins MH, et al. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr (2007) 45:22–31. 10.1097/MPG.0b013e318043c097 [DOI] [PubMed] [Google Scholar]
- 7. Lucendo AJ, Lucendo B. An update on the immunopathogenesis of eosinophilic esophagitis. Expert Rev Gastroenterol Hepatol (2010) 4:141–8. 10.1586/egh.10.9 [DOI] [PubMed] [Google Scholar]
- 8. Lucendo AJ. Cellular and molecular immunological mechanisms in eosinophilic esophagitis: an updated overview of their clinical implications. Expert Rev Gastroenterol Hepatol (2014) 8:669–85. 10.1586/17474124.2014.909727 [DOI] [PubMed] [Google Scholar]
- 9. Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut (2009) 51:12–20. 10.1136/gut.2009.178020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology (2014) 147:602–9. 10.1053/j.gastro.2014.05.036 [DOI] [PubMed] [Google Scholar]
- 11. Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol (2007) 119:206–12. 10.1016/j.jaci.2006.10.016 [DOI] [PubMed] [Google Scholar]
- 12. Lucendo AJ, Arias A, De Rezende LC, Yagüe-Compadre JL, Mota-Huertas T, González-Castillo S, et al. Subepithelial collagen deposition, profibrogenic cytokine gene expression, and changes after prolonged fluticasone propionate treatment in adult eosinophilic esophagitis: a prospective study. J Allergy Clin Immunol (2011) 128:1037–46. 10.1016/j.jaci.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 13. Arias A, González-Cervera J, Tenias JM, Lucendo AJ. Efficacy of dietary interventions for inducing histologic remission in patients with eosinophilic esophagitis: a systematic review and metaanalysis. Gastroenterology (2014) 146:1639–48. 10.1053/j.gastro.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 14. Molina-Infante J, Arias Á, Alcedo J, Garcia-Romero R, Casabona-Frances S, Prieto-Garcia A, et al. Step-up empiric elimination diet for pediatric and adult eosinophilic esophagitis: the 2-4-6 Study. J Allergy Clin Immunol (2017) 141(4):1365–72. 10.1016/j.jaci.2017.08.038 [DOI] [PubMed] [Google Scholar]
- 15. Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol (2007) 120:1292–300. 10.1016/j.jaci.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 16. Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, Blumberg RS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol (2014) 109:646–57. 10.1038/ajg.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arias Á, Lucendo AJ, Martínez-Fernández P, González-Castro AM, Fortea M, González-Cervera J, et al. Dietary treatment modulates mast cell phenotype, density, and activity in adult eosinophilic oesophagitis. Clin Exp Allergy (2016) 46:78–91. 10.1111/cea.12504 [DOI] [PubMed] [Google Scholar]
- 18. Jiménez-Dalmaroni MJ, Gerswhin ME, Adamopoulos IE. The critical role of toll-like receptors–from microbial recognition to autoimmunity: a comprehensive review. Autoimmun Rev (2016) 15:1–8. 10.1016/j.autrev.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Satoh T, Akira S. Toll-like receptor signaling and its inducible proteins. Microbiol Spectr (2016) 4(6). 10.1128/microbiolspec.MCHD-0040-2016 [DOI] [PubMed] [Google Scholar]
- 20. Kumagai Y, Akira S. Identification and functions of pattern-recognition receptors. J Allergy Clin Immunol (2016) 125:985–92. 10.1016/j.jaci.2010.01.058 [DOI] [PubMed] [Google Scholar]
- 21. van Egmond M, Vidarsson G, Bakema JE. Cross-talk between pathogen recognizing Toll-like receptors and immunoglobulin Fc receptors in immunity. Immunol Rev (2015) 268:311–27. 10.1111/imr.12333 [DOI] [PubMed] [Google Scholar]
- 22. Tunis MC, Marshall JS. Toll-like receptor 2 as a regulator of oral tolerance in the gastrointestinal tract. Mediators Inflammation (2014) 2014:606383. 10.1155/2014/606383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsybikov NN, Petrisheva I, Fefelova EV, Kuznik B, II, Magen E. Expression of TLR2 and TLR4 on peripheral blood monocytes during exacerbation of atopic dermatitis. Allergy Asthma Proc (2015) 36(6):e140–5. 10.2500/aap.2015.36.3901 [DOI] [PubMed] [Google Scholar]
- 24. Mulder DJ, Lobo D, Mak N, Justinich CJ. Expression of toll-like receptors 2 and 3 on esophageal epithelial cell lines and on eosinophils during esophagitis. Dig Dis Sci (2012) 57(3):630–42. 10.1007/s10620-011-1907-4 [DOI] [PubMed] [Google Scholar]
- 25. Burgueño JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol (2020) 17(5):263–78. 10.1038/s41575-019-0261-4 [DOI] [PubMed] [Google Scholar]
- 26. Woo JG, Assa’ad A, Heizer AB, Bernstein JA, Hershey GK. The -159 C–>T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol (2003) 112(2):438–44. 10.1067/mai.2003.1634 [DOI] [PubMed] [Google Scholar]
- 27. Liu J, Rädler D, Illi S, Klucker E, Turan E, von Mutius E, et al. TLR2 polymorphisms influence neonatal regulatory T cells depending on maternal atopy. Allergy (2011) 66(8):1020–9. 10.1111/j.1398-9995.2011.02573.x [DOI] [PubMed] [Google Scholar]
- 28. Bunyavanich S, Berin MC. Food allergy and the microbiome: Current understandings and future directions. J Allergy Clin Immunol (2019) 144(6):1468–77. 10.1016/j.jaci.2019.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanabe H, Sakurai K, Kato T, Kawasaki Y, Nakano T, Yamaide F, et al. Association of the maternal microbiome in Japanese pregnant women with the cumulative prevalence of dermatitis in early infancy: A pilot study from the Chiba study of Mother and Child Health birth cohort. World Allergy Organ J (2019) 12(10):100065. 10.1016/j.waojou.2019.100065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo J, Lv Q, Ariff A, Zhang X, Peacock CS, Song Y, et al. Western oropharyngeal and gut microbial profiles are associated with allergic conditions in Chinese immigrant children. World Allergy Organ J (2019) 12(8):100051. 10.1016/j.waojou.2019.100051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiu CY, Chan YL, Tsai MH, Wang CJ, Chiang MH, Chiu CC. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ J (2019) 12(3):100021. 10.1016/j.waojou.2019.100021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature (2016) 535(7610):94–103. 10.1038/nature18850 [DOI] [PubMed] [Google Scholar]
- 33. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep (2006) 7(7):688–93. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol (2015) 21(29):8787–803. 10.3748/wjg.v21.i29.8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau WF, Wong J, Lam KH, Ong GB. Oesophageal microbial flora in carcinoma of the oesophagus. Aust New Z J Surg (1981) 51(1):52–5. 10.1111/j.1445-2197.1981.tb05905.x [DOI] [PubMed] [Google Scholar]
- 36. Finlay IG, Wright PA, Menzies T, McArdle CS. Microbial flora in carcinoma of oesophagus. Thorax (1982) 37(3):181–4. 10.1136/thx.37.3.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mannell A, Plant M, Frolich J. The microflora of the oesophagus. Ann R Coll Surg Engl (1983) 65(3):152–4. [PMC free article] [PubMed] [Google Scholar]
- 38. Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci United States America (2004) 101(12):4250–5. 10.1073/pnas.0306398101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet (2011) 13(1):47–58. 10.1038/nrg3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgan XC, Huttenhower C. Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology (2014) 146(6):1437–48.e1. 10.1053/j.gastro.2014.01.049 [DOI] [PubMed] [Google Scholar]
- 41. Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol (2014) 22:267–74. 10.1016/j.tim.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 42. Baghdadi J, Chaudhary N, Pei Z, Yang L. Microbiome, innate immunity, and esophageal adenocarcinoma. Clin Lab Med (2014) 34:721–32. 10.1016/j.cll.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. May M, Abrams JA. Emerging Insights into the Esophageal Microbiome. Curr Treat Options Gastroenterol (2018) 16:72–85. 10.1007/s11938-018-0171-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gagliardi D, Makihara S, Corsi PR, Viana Ade T, Wiczer MV, Nakakubo S, et al. Microbial flora of the normal esophagus. Dis Esophagus (1998) 11:248–50. 10.1093/dote/11.4.248 [DOI] [PubMed] [Google Scholar]
- 45. Fillon SA, Harris JK, Wagner BD, et al. Novel device to sample the esophageal microbiome–the esophageal string test. PloS One (2012) 7:e42938. 10.1371/journal.pone.0042938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Norder Grusell E, Dahlén G, Ruth M, Ny L, Quiding-Järbrink M, Bergquist H, et al. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis Esophagus (2013) 26:84–90. 10.1111/j.1442-2050.2012.01328.x [DOI] [PubMed] [Google Scholar]
- 47. Pei Z, Yang L, Peek RM C.OMMAJ.R.X.X.X, Levine SM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett’s esophagus. World J Gastroenterol (2005) 11:7277–83. 10.3748/wjg.v11.i46.7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin Infect Dis (2007) 45:29–38. 10.1086/518578 [DOI] [PubMed] [Google Scholar]
- 49. Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology (2009) 137:588–97. 10.1053/j.gastro.2009.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu N, Ando T, Ishiguro K, Maeda O, Watanabe O, Funasaka K, et al. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett’s esophagus. BMC Infect Dis (2013) 13:130. 10.1186/1471-2334-13-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther (2013) 37:1084–92. 10.1111/apt.12317 [DOI] [PubMed] [Google Scholar]
- 52. Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ Microbiol (2014) 16:2905–14. 10.1111/1462-2920.12285 [DOI] [PubMed] [Google Scholar]
- 53. Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, et al. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer (2016) 16:52. 10.1186/s12885-016-2093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pajecki D, Zilberstein B, dos Santos MA, Ubriaco JA, Quintanilha AG, Cecconello I, et al. Megaesophagus microbiota: a qualitative and quantitative analysis. J Gastrointest Surg (2002) 6:723–9. 10.1016/s1091-255x(02)00028-8 [DOI] [PubMed] [Google Scholar]
- 55. Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome (2015) 3:23. 10.1186/s40168-015-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, et al. Esophageal microbiome in eosinophilic esophagitis. PloS One (2015) 10:e0128346. 10.1371/journal.pone.0128346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wade WG. The oral microbiome in health and disease. Pharmacol Res (2013) 69:137–43. 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 58. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol (2010) 192:5002–17. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nobel YR, Snider EJ, Compres G, Freedberg DE, Khiabanian H, Lightdale CJ, et al. Increasing Dietary Fiber Intake Is Associated with a Distinct Esophageal Microbiome. Clin Transl Gastroenterol (2018) 9:199. 10.1038/s41424-018-0067-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol (2012) 9:219–30. 10.1038/nrgastro.2012.14 [DOI] [PubMed] [Google Scholar]
- 61. Larsen JM, Musavian HS, Butt TM, Ingvorsen C, Thysen AH, Brix S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology (2015) 144:333–42. 10.1111/imm.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang L, Chaudhary N, Baghdadi J, Pei Z. Microbiome in reflux disorders and esophageal adenocarcinoma. Cancer J (2014) 20:207–10. 10.1097/PPO.0000000000000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol (1989) 142:4428–34. [PubMed] [Google Scholar]
- 64. Driss V, Legrand F, Hermann E, Loiseau S, Guerardel Y, Kremer L, et al. TLR2-dependent eosinophil interactions with mycobacteria: role of alpha-defensins. Blood (2009) 113:3235–44. 10.1182/blood-2008-07-166595 [DOI] [PubMed] [Google Scholar]
- 65. Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med (2008) 14:949–53. 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]
- 66. Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin Cancer Res (2012) 18:2138–44. 10.1158/1078-0432.CCR-11-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Norder Grusell E, Dahlén G, Ruth M, Bergquist H, Bove M. The cultivable bacterial flora of the esophagus in subjects with esophagitis. Scand J Gastroenterol (2018) 53:650–6. 10.1080/00365521.2018.1457712 [DOI] [PubMed] [Google Scholar]
- 68. Hiremath G, Shilts MH, Boone HH, Correa H, Acra S, Tovchigrechko A, et al. The Salivary Microbiome Is Altered in Children With Eosinophilic Esophagitis and Correlates With Disease Activity. Clin Transl Gastroenterol (2019) 10:e00039. 10.14309/ctg.0000000000000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Arnold IC, Hitzler I, Müller A. The immunomodulatory properties of Helicobacter pylori confer protection against allergic and chronic inflammatory disorders. Front Cell Infect Microbiol (2012) 2:10. 10.3389/fcimb.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kyburz A, Urban S, Altobelli A, Floess S, Huehn J, Cover TL, et al. Helicobacter pylori and its secreted immunomodulator VacA protect against anaphylaxis in experimental models of food allergy. Clin Exp Allergy J Br Soc Allergy Clin Immunol (2017) 47(10):1331–41. 10.1111/cea.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu YZ, Tan G, Wu F, Zhi FC. H. pylori attenuates TNBS-induced colitis via increasing mucosal Th2 cells in mice. Oncotarget (2017) 8(43):73810–6. 10.18632/oncotarget.17962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shah SC, Tepler A, Peek R. M. C.OMMAJ.R.X.X.X, Colombel JF, Hirano I, Narula N. Association Between Helicobacter pylori Exposure and Decreased Odds of Eosinophilic Esophagitis-A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc (2019) 17(11):2185–98.e3. 10.1016/j.cgh.2019.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kashyap PC, Johnson S, Geno DM, Lekatz HR, Lavey C, Alexander JA, et al. A decreased abundance of clostridia characterizes the gut microbiota in eosinophilic esophagitis. Physiol Rep (2019) 7:e14261. 10.14814/phy2.14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zuercher AW, Weiss M, Holvoet S, Moser M, Moussu H, van Overtvelt L, et al. Lactococcus lactis NCC 2287 alleviates food allergic manifestations in sensitized mice by reducing IL-13 expression specifically in the ileum. Clin Dev Immunol (2012) 2012:485750. 10.1155/2012/485750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Holvoet S, Zuercher AW, Julien-Javaux F, Perrot M, Mercenier A. Characterization of candidate anti-allergic probiotic strains in a model of th2-skewed human peripheral blood mononuclear cells. Int Arch Allergy Immunol (2013) 161:142–54. 10.1159/000343703 [DOI] [PubMed] [Google Scholar]
- 76. Singh A, Hacini-Rachinel F, Gosoniu ML, Bourdeau T, Holvoet S, Doucet-Ladeveze R, et al. Immune-modulatory effect of probiotic Bifidobacterium lactis NCC2818 in individuals suffering from seasonal allergic rhinitis to grass pollen: an exploratory, randomized, placebo-controlled clinical trial. Eur J Clin Nutr (2013) 67:161–7. 10.1038/ejcn.2012.197 [DOI] [PubMed] [Google Scholar]
- 77. Holvoet S, Doucet-Ladevèze R, Perrot M, Barretto C, Nutten S, Blanchard C. Beneficial effect of Lactococcus lactis NCC 2287 in a murine model of eosinophilic esophagitis. Allergy (2016) 71(12):1753–61. 10.1111/all.12951 [DOI] [PubMed] [Google Scholar]
- 78. Demont A, Hacini-Rachinel F, Doucet-Ladevèze R, Ngom-Bru C, Mercenier A, Prioult G, et al. Live and heat-treated probiotics differently modulate IL10 mRNA stabilization and microRNA expression. J Allergy Clin Immunol (2016) 137:1264–1267.e10. 10.1016/j.jaci.2015.08.033 [DOI] [PubMed] [Google Scholar]
- 79. Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology (2005) 129:985–94. 10.1053/j.gastro.2005.06.027 [DOI] [PubMed] [Google Scholar]
- 80. Pellaton C, Nutten S, Thierry AC, Boudousquié C, Barbier N, Blanchard C, et al. Intragastric and Intranasal Administration of Lactobacillus paracasei NCC2461 Modulates Allergic Airway Inflammation in Mice. Int J Inflam (2012) 2012:686739. 10.1155/2012/686739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Feleszko W, Jaworska J, Rha RD, Steinhausen S, Avagyan A, Jaudszus A, et al. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin Exp Allergy (2007) 37:498–505. 10.1111/j.1365-2222.2006.02629.x [DOI] [PubMed] [Google Scholar]
- 82. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med (2014) 20:159–66. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 83. Hogenkamp A, Thijssen S, van Vlies N, Garssen J. Supplementing pregnant mice with a specific mixture of nondigestible oligosaccharides reduces symptoms of allergic asthma in male offspring. J Nutr (2015) 145:640–6. 10.3945/jn.114.197707 [DOI] [PubMed] [Google Scholar]