Summary
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptor-type transcription factors with three subtypes (α, δ, and γ) that regulate cell differentiation and metabolism. Co-crystals of human PPARα-ligand-binding domain (LBD)-PPARα ligand for X-ray crystallography have been difficult to obtain. Recombinant human PPARα-LBD proteins contain intrinsic fatty acids (iFAs of Escherichia coli origin) and may be unstable without ligands during crystallization. To circumvent these limitations, we have successfully applied various crystallization techniques, including co-crystallization, cross-seeding, soaking, delipidation, and coactivator peptide supplementation.
For complete details on the use and execution of this protocol, please refer to Kamata et al. (2020).
Subject areas: Protein biochemistry, Protein expression and purification, Structural biology, X-ray Crystallography
Graphical Abstract
Highlights
-
•
Protocols for recombinant PPARα ligand-binding domain protein purification
-
•
Techniques to prepare PPARα-ligand co-crystals for high-resolution X-ray crystallography
-
•
Strategy to obtain PPARα co-crystals with low-affinity PPARα ligands
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptor-type transcriptional factors with three subtypes (α, δ, and γ) that regulate cell differentiation and metabolism. Co-crystals of human PPARα-ligand-binding domain (LBD)-PPARα ligand for X-ray crystallography have been difficult to obtain. Recombinant human PPARα-LBD proteins contain intrinsic fatty acids (iFAs of Escherichia coli origin) and may be unstable without ligands during crystallization. To circumvent these limitations, we have successfully applied various crystallization techniques, including co-crystallization, cross-seeding, soaking, delipidation, and coactivator peptide supplementation.
Before you begin
Preparation of recombinant hPPARα-LBD proteins
Protein expression and affinity column chromatography
Timing: 5 days
-
1.Transform Rosetta (DE3) pLysS competent E. coli cells with a pET28a vector containing cDNA encoding amino-terminal His-tagged human PPARα-LBD (amino acids 200–468) at the Nde I-Bam HI locus of the multi-cloning site.
-
a.Mix 5 μL of competent cells and 4.5 ng (in 0.3 μL) of vector in a 1.5-mL plastic tube.
-
b.Incubate on ice for 5 min.
-
c.Incubate at 42°C for 30 s.
-
d.Incubate on ice for 2 min.
-
e.Add 80 μL of LB broth.
-
f.Incubate at 37°C for 1 h.
-
a.
-
2.
Spread cells on an LB agar plate containing 15 μg/mL kanamycin and 34 μg/mL chloramphenicol.
-
3.
Invert the plate and incubate at 37°C. Approximately 100 colonies should appear in 12–16 h.
-
4.
Pick a single colony (1–2 mm in diameter) and transfer it into 100 mL of LB broth containing 15 μg/mL kanamycin in a 500-mL baffled flask.
-
5.
Incubate the culture overnight (12–16 h) at 30°C with vigorous shaking (150 cycles/min in a rotary shaker) until optical density at 600 nm (OD600) reaches approximately 1.1.
-
6.
Transfer 50 mL of the overnight culture into 1 liter of TB broth containing 15 μg/mL kanamycin in a 2-liter baffled flask (OD600 is 0.15 at start).
-
7.
Incubate the culture for 1.5 h at 30°C with vigorous shaking (120 cycles/min). OD600 reaches approximately 0.18.
-
8.
Incubate the culture at 15°C with vigorous shaking until OD600 reaches 0.27 (OD600 reaches 0.24 after 1 h and 0.27 after 2 h, approximately).
-
9.
Add 5 mL of 100 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (final 0.5 mM).
-
10.
Incubate the culture for 48 h at 15°C with vigorous shaking until OD600 reaches approximately 1.9.
-
11.
Chill the culture on ice.
-
12.
Centrifuge at 2,900 × g for 10 min at 4°C.
-
13.
Remove the supernatant and resuspend the pellet in 30 mL of ice-cold Buffer A (see materials and equipment).
-
14.
Transfer resuspended pellet to a 50-mL plastic (Falcon) tube.
-
15.
Centrifuge at 2,900 × g for 10 min at 4°C.
-
16.
Remove the supernatant and resuspend the pellet in 40 mL of ice-cold Buffer A plus cOmplete EDTA-free protease inhibitor.
-
17.
Sonicate the cells for 2 min (repeat for five sonications in total, with 8 min intervals between them) on ice at an output of 8 (in 1–10 range) and a 30% duty (cycles of 0.3 s pulse on/0.7 s pulse off) using a microtip-equipped 100-W output sonicator.
-
18.
Centrifuge at 12,000 × g for 20 min at 4°C.
-
19.
Transfer 40 mL of the supernatant to a 50-mL tube and add 1.2 mL of 5% (v/v) polyethyleneimine (final 0.15% [v/v]; adjusted to pH 8.0 by 6 M HCl), all on ice.
-
20.
Vortex for 10 s.
-
21.
Centrifuge at 12,000 × g for 20 min at 4°C.
-
22.
Transfer 35 mL of the supernatant to a 50-mL tube, and add 20 g of ammonium sulfate (80% saturation).
-
23.
Mix gently for 30 min at 4°C using a rotator.
-
24.
Centrifuge at 12,000 × g for 20 min at 4°C.
-
25.
Remove the supernatant, and add 10 mL of ice-cold Buffer B to the pellet (see materials and equipment).
-
26.
Resuspend the pellet by up-and-down pipetting with a 10-mL plastic pipette.
-
27.
Filter the resuspended sample through a 0.22 μm PVDF filter.
-
28.
Add 20 mL of ice-cold Buffer B, and mix by inversion.
-
29.
Pre-equilibrate a cobalt-based immobilized metal affinity column (HisTALON Superflow Cartridges, 1 mL) with 10 mL of Buffer B at 1 mL/min at 4°C using a GE Healthcare AKTA prime liquid chromatography system or similar.
-
30.
Inject 30 mL of the sample onto the affinity column at 1 mL/min at 4°C.
-
31.
Wash the column with 70 mL of Buffer B at 1 mL/min at 4°C.
-
32.
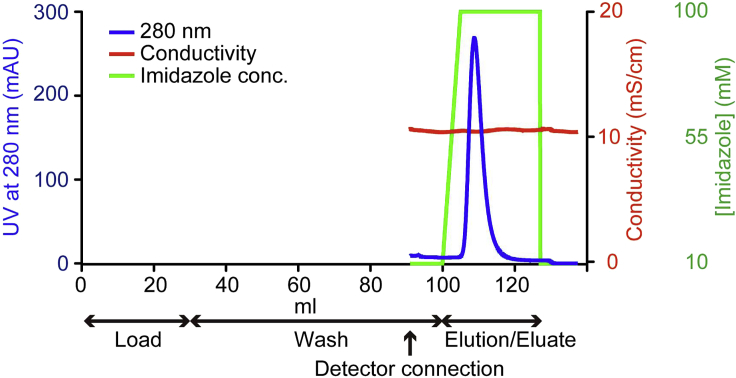
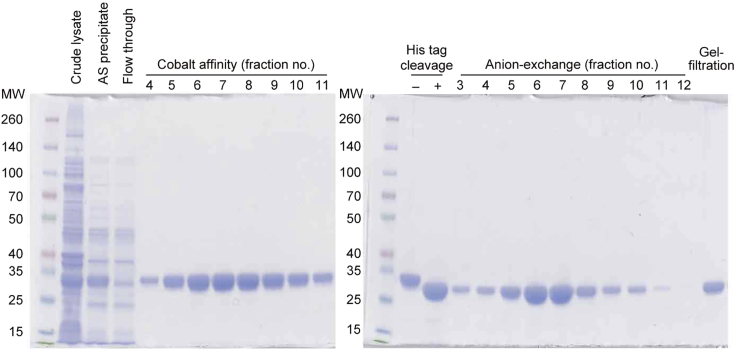
Elute His-tagged hPPARα-LBD proteins with 2.5 mL of a liner gradient of 10–100 mM imidazole/Buffer A followed by 20 mL of 100 mM imidazole/Buffer A at 4°C. Collect the target protein eluted by 100 mM imidazole/Buffer A solution (Figure 1).
Note: At this stage, approximately 8 mg (6 mL elutes) of hPPARα-LBD proteins are obtained.
CRITICAL: Before the following purification steps, the FPLC system should be washed with 0.5 M NaOH as part of routine maintenance.
Pause point: The eluted sample can be stored for a few days at 4°C. Even 10 mg/mL elutes do not precipitate in this step.
Optional: Delipidation to remove iFA(s) of E. coli origin
Timing: 3 days
-
33.
Divide 6 mL eluted samples into two 3 mL volumes in separate 50 mL tubes.
-
34.
Add 27 mL ethanol to each 3 mL sample in each 50 mL tube.
-
35.
Mix well and store for 2 h at room temperature (20°C–25°C).
-
36.
Centrifuge at 15,000 × g for 20 min at 4°C.
-
37.
Remove the supernatants, and add 5 mL ethanol to each white pellet.
-
38.
Repeat steps 36 and 37.
-
39.
Centrifuge at 15,000 × g for 20 min at 4°C.
-
40.
Remove the supernatants and add 10 mL of 6 M guanidine-HCl in Buffer A to each pellet (samples are processed in duplicate from here).
-
41.
Leave overnight (12–16 h) at 4°C (no need to agitate).
-
42.
Aliquot 5 mL of the sample to a new 50 mL tube.
-
43.
Add 45 mL of ice-cold Buffer A and vortex mix.
-
44.
Apply to an AMICON ULTRA-15 centrifugal filter.
-
45.
Centrifuge at 5,000 × g at 4°C to obtain 2.5–3.0 mL concentrates. Protein concentrations over 2 mg/mL tend to precipitate.
Note: The yield of delipidized hPPARα-LBD proteins here is approximately 4 mg from 8 mg hPPARα-LBD proteins isolated from 1 liter TB culture.
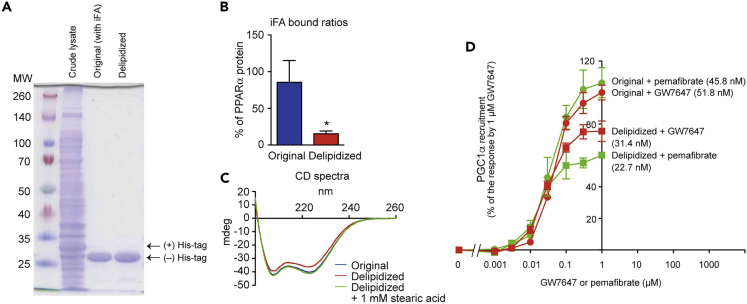
Note: Delipidation does not significantly affect hPPARα-LBD structure and functionality (Figure 2).
Figure 1.
The representative affinity column chromatography profile for hPPARα-LBD purification
Figure 2.
Impacts of delipidation on the hPPARα-LBD structure and functionality
(A) SDS-PAGE gel of original and delipidized hPPARα-LBD.
(B) iFA(s)-bound ratios revealed by fatty acid quantification. Data are represented as means ± SEM (n = 3). The difference was significant in ∗p < 0.05 in t test.
(C) Circular dichroism (CD) spectra. Stearic acid supplementation restores the CD shift by delipidation.
(D) PPARα coactivator recruitment (PPARα activation) assay. PPARα agonists (pemafibrate and GW7647) activate both original and delipidized PPARα-LBD in a similar concentration-dependent manner. Data are represented as mean ± SEM (n = 3 [pemafibrate] or 4 [GW7647]). Figure reprinted with permission from Kamata et al. (2020).
CRITICAL: The sample is prone to precipitation (even at concentrations lower than 0.1 mg/mL) after dilution with Buffer A (step 43 above), and the sample should filter-centrifuged as soon as possible after this. When the sample precipitates, increase the volume of 6 M guanidine-HCl solution to dissolve the aggregates.
Preparation of recombinant hPPARα-LBD proteins
His-tag removal and anion-exchange chromatography
Timing: 2 days
-
46.
Add 100 units of thrombin protease to 3 mL of sample (either lipidated [4 mg/3 mL] or not [8 mg/3 mL]) in a 50-mL tube.
-
47.
Dialyze sample against 500 mL Buffer A overnight (12–16 h) at 4°C using a Slide-A-Lyzer G2 Dialysis Cassette (20-kDa cutoff, 1–3-mL sample volume).
-
48.
Monitor the His-tag removal by SDS-PAGE with Coomassie Brilliant Blue staining.
-
49.
Dialyze against 500 mL Buffer C (Refer to materials and equipment) for 3 h at 4°C.
-
50.
Pre-equilibrate a HiTrap Q HP anion-exchange column with 5 mL of Buffer C at 0.5 mL/min at 4°C.
-
51.
Inject 5 mL of sample onto the anion-exchange column at 0.5 mL/min at 4°C.
-
52.
Wash the column with 30 mL Buffer C at 0.5 mL/min at 4°C.
-
53.
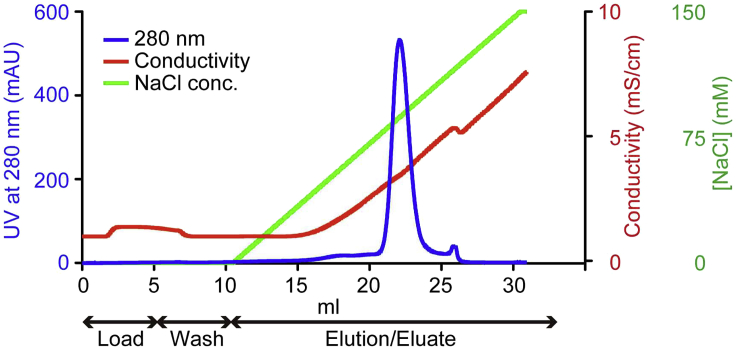
Elute with 20 mL of a linear gradient of 0–150 mM NaCl (from Buffer C to Buffer A). The target protein starts to elute around 2.9 mS/cm (Figure 3).
Figure 3.
The ion exchange column chromatography profile for hPPARα-LBD purification
Preparation of recombinant hPPARα-LBD proteins
Gel-filtration chromatography
Timing: 4 h
-
54.
Pre-equilibrate a HiLoad 16/600 Superdex 75 pg gel-filtration column with 130 mL Buffer A at 1 mL/min at 4°C.
-
55.
Inject 2 mL sample onto the gel-filtration column at 1 mL/min at 4°C.
-
56.
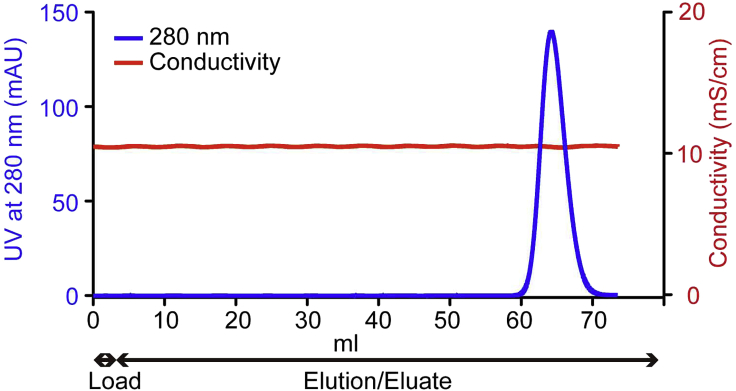
Elute with 70 mL Buffer A at 1 mL/min at 4°C. The target protein usually elutes in the last 10 mL (Figure 4).
-
57.
Apply the hPPARα-LBD-containing elutes to an AMICON ULTRA-4 centrifugal filter.
-
58.
Centrifuge at 7,500 × g at 4°C to obtain 0.3 mL concentrates (up to 30 mg/mL).
-
59.
Quantify protein concentrations and adjust to 20 mg/mL with Buffer A.
-
60.
Store at 4°C.
Pause point: The eluted sample can be stored for a few days at 4°C.
Note: Coomassie Brilliant Blue staining of SDS-PAGE gels shows only a single band of hPPARα-LBD after the first affinity column chromatography (Figure 5); however, we routinely apply anion-exchange and gel-filtration column chromatography to ensure highly purified protein is obtained. The three-step column chromatography is not always necessary but could be essential in some cases (as the case may be).
Figure 4.
The gel-filtration column chromatography profile for hPPARα-LBD purification
Figure 5.
SDS-PAGE gel of hPPARα-LBD preparations during three-step column chromatography
MW, molecular weight marker; AS, ammonium sulfate. Figure reprinted with permission from Kamata et al. (2020).
Note: The final yield of non-delipidized and delipidized hPPARα-LBD proteins here are approximately 4 mg and 2 mg, respectively, starting from 8 mg hPPARα-LBD proteins isolated from 1 liter TB culture.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli Rosetta (DE3) pLysS | Novagen | Cat# 70956 |
| Chemicals, peptides, and recombinant proteins | ||
| LB medium | Nacalai Tesque | Cat# 20068-75 |
| TB medium | Becton Dickinson | Cat# 243820 |
| Kanamycin | Fujifilm-Wako | Cat# 117-00341 CAS# 133-92-6 |
| Chloramphenicol | Fujifilm-Wako | Cat# 032-19451 CAS# 56-75-7 |
| Isopropyl β-d-galactopyranoside | Fujifilm-Wako | Cat# 099-05013 CAS: 367-93-1 |
| Tris | Nacalai Tesque | Cat# 35434-21 CAS# 77-86-1 |
| NaCl | Nacalai Tesque | Cat# 31320-34 CAS# 7647-14-5 |
| Tris 2-carboxyethylphosphine (TCEP)-HCl | Nacalai Tesque | Cat# 07277-16 CAS# 51805-45-9 |
| Glycerol | Nacalai Tesque | Cat# 17018-83 CAS# 56-81-5 |
| cOmplete EDTA-free protease inhibitor | Sigma-Aldrich | Cat# 05056489001 |
| Polyethyleneimine (MW 70,000) | Fujifilm-Wako | Cat# 167-11951 CAS# 9002-98-6 |
| Ammonium sulfate | Nacalai Tesque | Cat# 02619-15 CAS# 7783-20-2 |
| Imidazole | Fujifilm-Wako | Cat# 095-00015 CAS# 288-32-4 |
| Thrombin | Nacalai Tesque | Cat# 33842-44 CAS# 9002-04-4 |
| Guanidine hydrochloride | Fujifilm-Wako | Cat# 077-02435 CAS# 50-01-1 |
| Ethanol | Fujifilm-Wako | Cat# 057-04456 CAS# 64-17-5 |
| Bis-Tris | Hampton Research | Cat# HR2-783 CAS# 6976-37-0 |
| HEPES | Hampton Research | Cat# HR2-729 CAS# 7365-45-9 |
| Polyethylene glycol (PEG) 3350 | Hampton Research | Cat# HR2-144 CAS# 25322-68-3 |
| SRC1 peptide (LTERHKILHRLLQEG) | GenScript | N/A |
| DMSO | Nacalai Tesque | Cat# 13445-45 CAS# 67-68-5 |
| GW7647 | Cayman Chemical | Cat# 10008613 CAS# 265129-71-3 |
| Bezafibrate | Cayman Chemical | Cat# 10009145 CAS# 41859-67-0 |
| Tetradecylthioacetic acid (TTA) | Fujifilm-Wako | Cat# 209-18141 CAS# 2921-20-2 |
| Wy14643 | Cayman Chemical | Cat# 70730 CAS# 50892-23-4 |
| GW9662 | Cayman Chemical | Cat#70785 CAS# 22978-25-2 |
| Ciprofibrate | Fujifilm-Wako | Cat# 033-21191 CAS# 52214-84-3 |
| Fenofibric acid | Combi-Blocks | Cat# OR-1173 CAS# 42017-89-0 |
| 5,8,11,14-Eicosatetraynoic acid (ETYA) | Cayman Chemical | Cat# 90120 CAS# 1191-85-1 |
| Eicosapentaenoic acid (EPA) | Cayman Chemical | Cat# 90110 CAS# 10417-94-4 |
| Clofibric acid | LKT Labs | Cat# C4556 CAS# 882-09-7 |
| Gemfibrozil | Combi-Blocks | Cat# OR-0524 CAS# 25812-30-0 |
| Arachidonic acid | Cayman Chemical | Cat#10006607 CAS# 6610-25-9 |
| Pemafibrate | ChemScene | Cat# CS-6084 CAS# 848259-27-8 |
| Saroglitazar | ChemScene | Cat# CS-6149 CAS# 495399-09-2 |
| Palmitic acid | Sigma-Aldrich | Cat# P0500-10G CAS# 57-10-3 |
| Stearic acid | Sigma-Aldrich | Cat# S4751-1G CAS# 57-11-4 |
| Oleic acid | Nacalai Tesque | Cat# 25630-51 CAS# 112-80-1 |
| Deposited data | ||
| 34 novel human PPARα-LBD co-crystal structures reported in Kamata et al. (2020) | Protein Data Bank (PDB) | Codes: 6KAX, 6KAY, 6KAZ, 6KB0, 6KB1, 6KB2, 6KB3, 6KB4, 6KB5, 6KB6, 6KB7, 6KB8, 6KB9, 6KBA, 6KYP, 6L36, 6L37, 6L38, 6LX4, 6LX5, 6LX6, 6LX7, 6LX8, 6LX9, 6LXA, 6LXB, 6LXC, 7BPY, 7BPZ, 7BQ0, 7BQ1, 7BQ2, 7BQ3, 7BQ4 |
| Recombinant DNA | ||
| pET28a encoding human PPARα residues 200–468 | (Oyama et al., 2009) | N/A |
| Other | ||
| Bioshaker BR-43FL | TAITEC | Cat# 0053027-000 |
| Ultrasonic disrupter UD-201 with Micro Tip TP-040 | Tomy | Cat# UD-201 |
| AKTAprime plus | Cytiva (GE Healthcare) | N/A |
| HisTALON superflow cartridges (1 mL) | Clontech | Cat# 635650 |
| HiTrap Q HP (1 mL) | Cytiva (GE Healthcare) | Cat# 17115301 |
| HiLoad 16/600 Superdex 75 pg | Cytiva (GE Healthcare) | Cat# 28989333 |
| Slide-A-Lyzer G2 dialysis cassette | Thermo Fisher | Cat# 87735 |
| AMICON ULTRA-4, 4-mL; 3 kDa cutoff | Merck Millipore | Cat# UFC800324 |
| AMICON ULTRA-15, 15-mL; 3 kDa cutoff | Merck Millipore | Cat# UFC900324 |
| EasyXtal 15-Well Tool X-Seal (20) | QIAGEN | Cat# 132008 |
| Dual-thickness MicroMounts (75 μm) | MiTeGen | Cat# M2-L18SP-75 |
| 18 mm mounted CryoLoop - 20 μm | Hampton Research | Cat# HR4-972 |
| Bio-Rad protein assay | Bio-Rad | Cat# 5000006JA |
| Human hair (on the crown of the head) | Young adult males | N/A |
Materials and equipment
Buffers A–C
| Reagent (stock) | Amount (final concentration) |
||
|---|---|---|---|
| Buffer A | Buffer B | Buffer C | |
| Tris-HCl [pH 8.0] (1 M) | 20 mL (20 mM) | 10 mL (20 mM) | 20 mL (20 mM) |
| NaCl (5 M) | 30 mL (150 mM) | 15 mL (150 mM) | 0 |
| TCEP (100 mM) | 10 mL (1 mM) | 5 mL (1 mM) | 10 mL (1 mM) |
| Glycerol (50% [v/v]) | 200 mL (10% [v/v]) | 100 mL (10% [v/v]) | 200 mL (10% [v/v]) |
| Imidazole (2 M) | 0 | 2.5 mL (10 mM) | 0 |
| ddH2O | 740 mL | 367.5 mL | 770 mL |
| Total | 1,000 mL | 500 mL | 1,000 mL |
Crystallization Buffer
| Reagent (stock) | Final concentration | Amount |
|---|---|---|
| Bis-Tris-HCl [pH 6.5], HEPES-NaOH [pH 7.0 or 7.5], or Tris-HCl [pH 8.0 or 8.5] (1 M) | 100 mM | 1 mL |
| PEG 3350 (50%) | 25% | 5 mL |
| ddH2O | N/A | 4 mL |
| Total | 10 mL |
Soaking Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES-NaOH [pH 7.5] (1 M) | 100 mM | 1 mL |
| PEG 3350 (50%) | 20% | 4 mL |
| ddH2O | N/A | 5 mL |
| Total | 10 mL |
Step-by-step method details
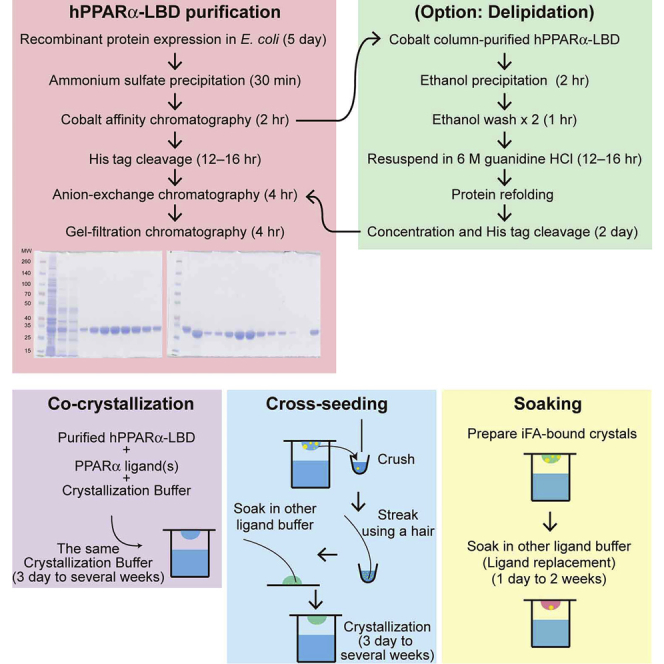
Case 1: Co-crystallization
Timing: 3 days to several weeks
Note: Co-crystallization is the most popular crystallization method. This method was used to prepare crystals deposited in PDB with codes: 6KBA (with Wy14643); 6LX4 (with fenofibric acid); 6LX5 (with ciprofibrate); 6KYP (with GW9662 + clofibric acid); 6L36 (with GW9662 + fenofibric acid); 6L37 (with GW9662 + ciprofibrate); and 6L38 (with GW9662 + gemfibrozil).
-
1.
Dilute ligand stock solution with Buffer A.
Note: A 2-mM ligand solution should be used in the first trial.
-
2.
Mix 25 μL each of hPPARα-LBD solution (20 mg/mL in Buffer A, either delipidized or not) and the ligand solution in a 1.5-mL plastic tube on ice.
Note: Concentrations of hPPARα-LBD and the ligand are estimated to be 320 μM (determined by Bradford method-based Bio-Rad Protein Assay using bovine serum albumin as a standard) and 1 mM, respectively.
-
3.
Dispense 200 μL of Crystallization Buffer (see materials and equipment) to each well of an EasyXtal 15-Well Tool X-Seal plate at 4°C.
-
4.
Mix 1 μL of Crystallization Buffer and 1 μL hPPARα-LBD-ligand mixed solution on the backside of the screw cap at 4°C.
-
5.
Fasten the screw cap.
-
6.
Incubate the plate at 4°C (see troubleshooting problem 1).
CRITICAL: Steps 3–6 above should be performed in the cold room (4°C). Crystals can be damaged quickly at room temperature (20°C–25°C).
Note: When crystals (such as Wy14643-bound PPARα-LBD crystals) are used for seeds in cross-seeding, tiny crystals obtained within 1–2 weeks are fine.
Optional: To prepare PPARα-LBD/ligand/coactivator (peptide) crystals, use 0.25 μL ligand solution and 0.25 μL peptide solution (4 mM in water) instead of the 0.5 μL ligand solution. This method was used to prepare crystals deposited in PDB with codes: 7BPY (with clofibric acid + SRC1); 7BQ0 (with fenofibric acid + SRC1); 7BQ1 (with iFA + SRC1); 7BQ3 (with GW7647 + SRC1); and 7BQ4 (with eicosapentaenoic acid [EPA] + SRC1).
Note: To obtain co-crystals with PPARα ligands whose binding affinities are lower than iFAs, the use of delipidized hPPARα-LBD was necessary. Clofibric acid-SRC1 (peptide)-bound delipidized hPPARα-LBD crystals that gave high (2.09 Å) resolution in X-ray crystallography were obtained after a 2-week incubation at 4°C in 0.1 M Tris-HCl (pH 8.5) and 30% (w/v) PEG 3350 Crystallization Buffer (Figure 6).
Figure 6.
Clofibric acid-SRC1 (peptide)-bound delipidized hPPARα-LBD crystals (PDB: 7BPY)
Clofibric acid is a relatively low-affinity PPARα ligand (the EC50 value for clofibric acid in coactivator recruitment assay is 574 μM, which is slightly higher than iFAs (428 μM for stearic acids and 471 μM for palmitic acid) (Kamata et al., 2020)). Clofibric acid-bound hPPARα-LBD co-crystals were only obtained in the presence of SRC1 or GW9662 (PDB: 6KYP) using delipidized proteins. These crystals were used for soaking in bezafibrate (another low-affinity PPARα ligand)-rich buffer to obtain bezafibrate-SRC1-bound crystals (PDB: 7BPZ). Bar, 200 μm.
Case 2: Cross-seeding
Timing: 3 days to several weeks
Note: Cross-seeding is generally applied when new protein-ligand co-crystals have not been obtained. Micro-seeds derived from previously obtained crystals of closely related complexes can be used for cross-seeding (Benvenuti and Mangani, 2007; Hassell et al., 2007; McPherson, 1999). This method was used to prepare crystals deposited in PDB with codes: 6KAX (with iFA); 6KB3 and 6KB8 (with GW7647); 6KB4 and 6KB9 (with pemafibrate); 6KB5 (with 5, 8, 11, 14-eicosatetraynoic acid [ETYA]); 6KB6 (with tetradecylthioacetic acid [TTA]); 6KB7 (with Wy14643); 6LX6 (with palmitic acid); 6LX7 (with stearic acid); 6LX8 (with oleic acid); 6LX9 (with arachidonic acid); 6LXA (with EPA); and 6LXC (with saroglitazar).
-
7.
Dilute ligand stock solution with Buffer A.
-
8.
Mix 25 μL each of hPPARα-LBD solution (20 mg/mL in Buffer A, either delipidized or not) and ligand solution in a 1.5-mL plastic tube on ice.
-
9.
Dispense 200 μL Crystallization Buffer to each well of an EasyXtal 15-Well Tool X-Seal plate at 4°C.
-
10.
Mix 1 μL of Crystallization Buffer and 1 μL PPAR-ligand mixed solution on the backside of the screw cap at 4°C.
-
11.
Fasten the screw cap.
-
12.
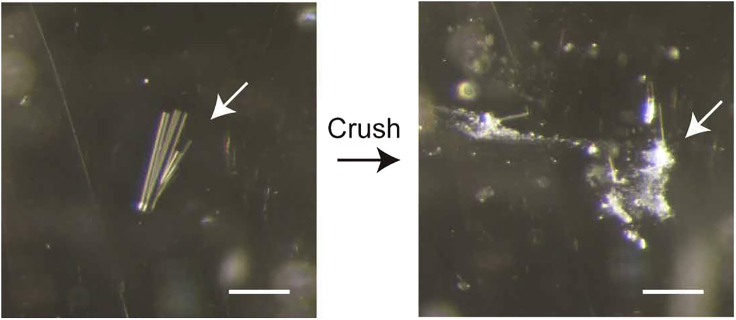
Crush the seed crystals (Wy14643 or iFA-bound crystals) in 10 μL of Crystallization Buffer (crystal nuclei solution) using a needle tip (Dual-Thickness MicroMounts) at 4°C (Figure 7).
Note: Crushing the crystals may produce too many crystal nuclei. Only small numbers of nuclei (obtained by serial dilutions with Crystallization Buffer) are needed. As a rough estimate, the nuclei obtained from the crush of a 10 x 10 x 50 μm crystal could provide 40–80 diffraction quality crystals.
-
13.
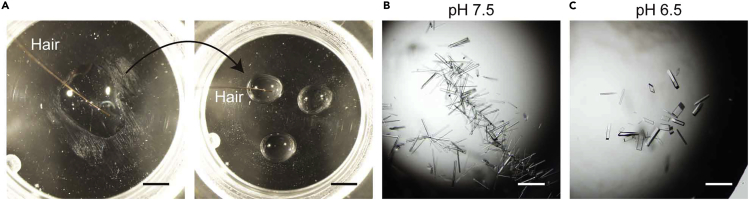
Transfer the crystal nuclei via a single streak with a human hair to 2 μL mixture of hPPARα-LBD/ligand in Crystallization Buffer (as used in step 10) at 4°C (Figure 8A).
Note: We use our own (young adult males') hairs and have not compared those with commercially available hairs/whiskers of the other animal species such as horses and cats.
-
14.
Fasten the screw cap.
-
15.
Incubate the plate at 4°C (see troubleshooting problem 1).
CRITICAL: Steps 9–15 above should be performed in the cold room (4°C). Crystals can be damaged quickly at room temperature (20°C–25°C).
Note: When only small crystals are obtained, repeat the procedure or try other conditions (different buffers) (Figures 8B and C) or lower protein concentrations (McPherson and Gavira, 2014) to obtain larger crystals.
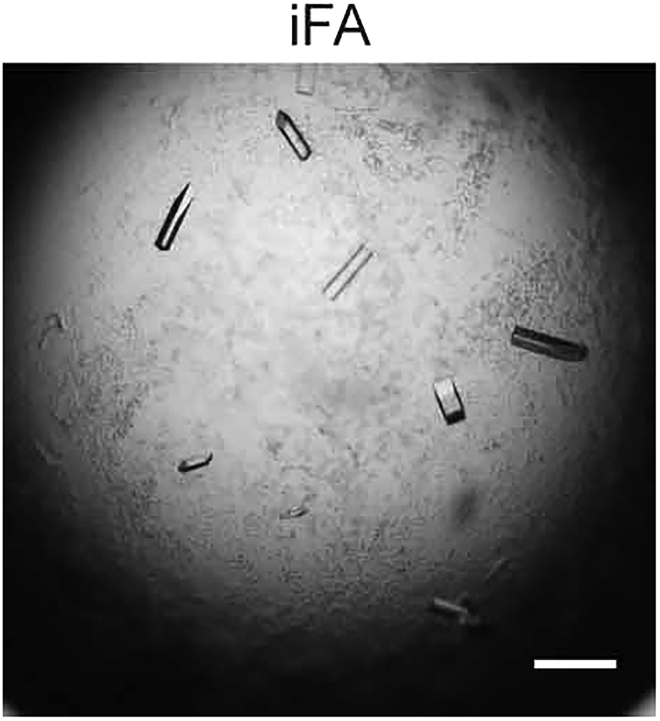
Note: The iFA-bound hPPARα-LBD crystals that gave very high (1.23 Å) resolution in X-ray crystallography were obtained after a 2-week incubation at 4°C in 0.1 M HEPES-NaOH (pH 7.0) and 25% (w/v) PEG 3350 Crystallization Buffer (Figure 9).
Figure 7.
Crush of crystals to prepare crystal nuclei
Scale bars, 200 μm
Figure 8.
Crystal nuclei transfer to another hPPARα-LBD/other ligand solution using a human hair
(A) A single streak with a human hair is used for crystal nuclei transfer.
(B) In some (bad) cases (such as iFA-bound hPPARα-LBD crystals in 0.1 M HEPES (pH 7.5)/25% PEG 3350 buffer), many small crystals form along the lines streaked by the hair.
(C) In some (good) cases (such as iFA-bound hPPARα-LBD crystals in 0.1 M Bis-Tris (pH 6.5)/25% PEG 3350 buffer), small numbers of large diffraction quality crystals form independent of the streaked lines. Scale bars, 2 mm (A) or 200 μm (B and C).
Figure 9.
The intrinsic fatty acid (iFA)-bound hPPARα-LBD crystals that gave the highest (1.23 Å) resolution among our 34 crystals
Reported in Kamata et al. (2020) (PDB: 6KAX). Scale bar, 200 μm.
Optional: To prepare hPPARα-LBD/ligand/coactivator (peptide) crystals, use 0.25 μL ligand solution and 0.25 μL peptide solution (4 mM in water) instead of the 0.5 μL ligand solution.
Case 3: Soaking
Timing: 1 day to 2 weeks
Note: The resolution of iFA-bound hPPARα-LBD crystals was the highest in our experience (Figure 6). Therefore, ligand replacement by soaking in iFA-bound crystal solution may produce high-resolution crystals with ligands that have higher affinities than iFAs (such as palmitic and stearic acid). This method was used to prepare crystals deposited in PDB with codes: 6KAY (with GW7647); 6KAZ (with pemafibrate); 6KB0 (with ETYA); 6KB1 (with TTA); 6KB2 (with Wy14643); and 6LXB (with saroglitazar).
Note: Obtaining crystals with ligands that are only slightly soluble may be difficult (Hassell et al., 2007).
-
16.
Prepare iFA-bound crystals by cross-seeding.
-
17.
Dilute ligand stock solution with Soaking Buffer (see materials and equipment). Try 1 mM initially.
-
18.
Dispense 200 μL Soaking Buffer to each well of an EasyXtal 15-Well Tool X-Seal plate at 4°C.
-
19.
Place 5 μL ligand solution (in Soaking Buffer) on the backside of the screw cap at 4°C.
-
20.
Pick up the iFA-bound crystals with a CryoLoop, wash the crystals with Soaking Buffer, and soak in 5 μL ligand solution at 4°C (Figure 10).
Note: The wash step may remove precipitates around crystal surfaces. Some crystals may be cracked in this transfer step, but such fragile crystals do not finally provide high-resolution profiles.
-
21.
Fasten the screw cap.
-
22.
Incubate the plate at 4°C (see troubleshooting problem 2).
Optional: The iFA-SRC1-bound and clofibric acid-SRC1-bound crystals (PDB: 7BQ1 and 7BPY) were used for soaking to obtain other SRC1-bound crystals; the former was used to prepare co-crystals with pemafibrate + SRC1 (PDB: 7BQ2) and the latter was used to prepare co-crystals with bezafibrate + SRC1 (PDB: 7BPZ).
CRITICAL: Steps 17–22 should be performed in the cold room (4°C). Crystals can be damaged quickly at room temperature (20°C–25°C).
Note: Ligand concentration and incubation time should be optimized for each ligand.
Figure 10.
Transfer of crystals with a CryoLoop for soaking in other ligand solutions
Scale bar, 5 mm.
Cryoprotection of crystals for X-ray crystallography
Timing: 1 h
-
23.
Mix 400 μL of Crystallization Buffer and 100 μL of 100% (v/v) glycerol (Cryoprotection Buffer) in a 1.5 mL plastic tube.
-
24.
Make 5 μL drops of Cryoprotection Buffer on the backside of the screw cap of an EasyXtal 15-Well Tool X-Seal plate at 4°C.
-
25.
Pick up the crystals with a CryoLoop, and soak into Cryoprotection Buffer at 4°C.
-
26.
Freeze under liquid nitrogen until X-ray diffraction data collection.
Note: Lowering PEG 3350 concentration (from 25% to 20%) by 100% glycerol dilution could damage crystals or even dissolve them in some cases but not in our case on hPPARα-LBD crystals.
CRITICAL: The use of liquid nitrogen in a walk-in cold room demands great attention. Only small volumes of liquid nitrogen should be brought into a large well-ventilated room by two or more researchers; oxygen concentrations in this room should be monitored during these procedures. Crystals can be damaged quickly at room temperature (20°C–25°C).
Expected outcomes
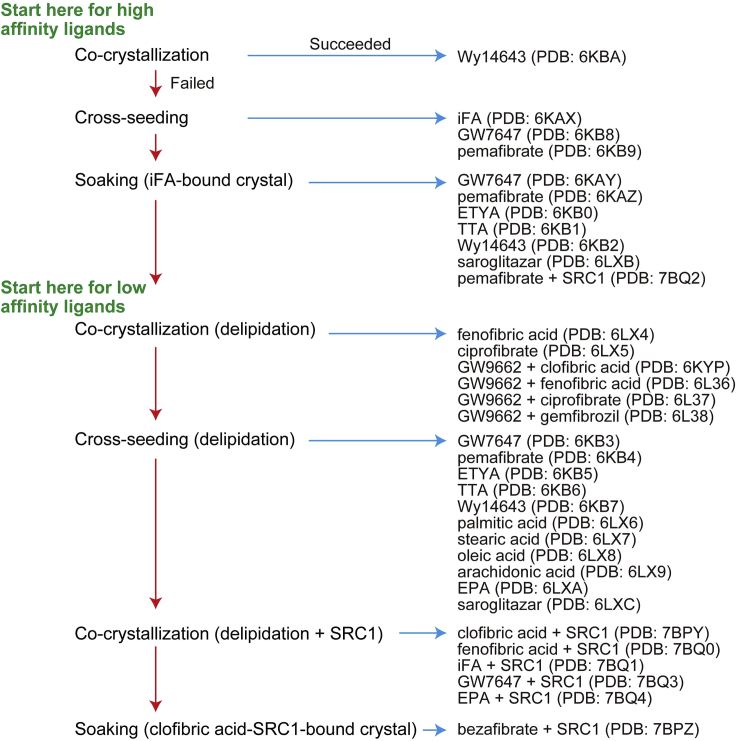
We utilized several crystallization methods for hPPARα, depending on PPARα ligands (Figure 11). Co-crystallization with Wy14643 is first recommended to check your techniques and obtain crystal nuclei for cross-seeding. Co-crystals with high-affinity ligands (such as GW7647 and pemafibrate) are obtained using cross-seeding. Co-crystals with iFAs (mixtures of abundant fatty acids of E. coli origin such as palmitic acid, stearic acid, and oleic acid [or vaccenic acid]) can also be obtained using cross-seeding and can be used for soaking with medium affinity ligands (such as ETYA, TTA, and saroglitazar).
Figure 11.
Strategy to produce new 34 hPPARα-LBD-ligand co-crystals
In Kamata et al. (2020).
With lower affinity (than iFAs) ligands, which include most of clinical fibrates (fenofibric acid, ciprofibrate, clofibric acid, and gemfibrozil), delipidation of hPPARα-LBD is necessary. Cross-seeding with delipidized hPPARα-LBD could help retain fatty acids such as palmitic acid, oleic acid, arachidonic acid, and EPA. Adding a SRC1 coactivator peptide could also provide crystals with lower affinity ligand during co-crystallization. Co-crystals with bezafibrate are only produced in the presence of SRC1 and by soaking with clofibric acid-SRC1-bound co-crystals.
Limitations
The 34 hPPARα-LBD structures we obtained (Kamata et al., 2020) represent a similar activated form with an Activation Function-2 (AF-2) helix 12. Structural analyses of antagonist-bound inactivated forms are awaited. We may use similar crystallization procedures using antagonists and corepressor peptides, or need different techniques. Currently, only one antagonist-bound inactivated form has been reported by (Xu et al., 2002).
Troubleshooting
Problem 1
Crystals do not form, or only many small (but not large well-formed) crystals occur.
Potential solution
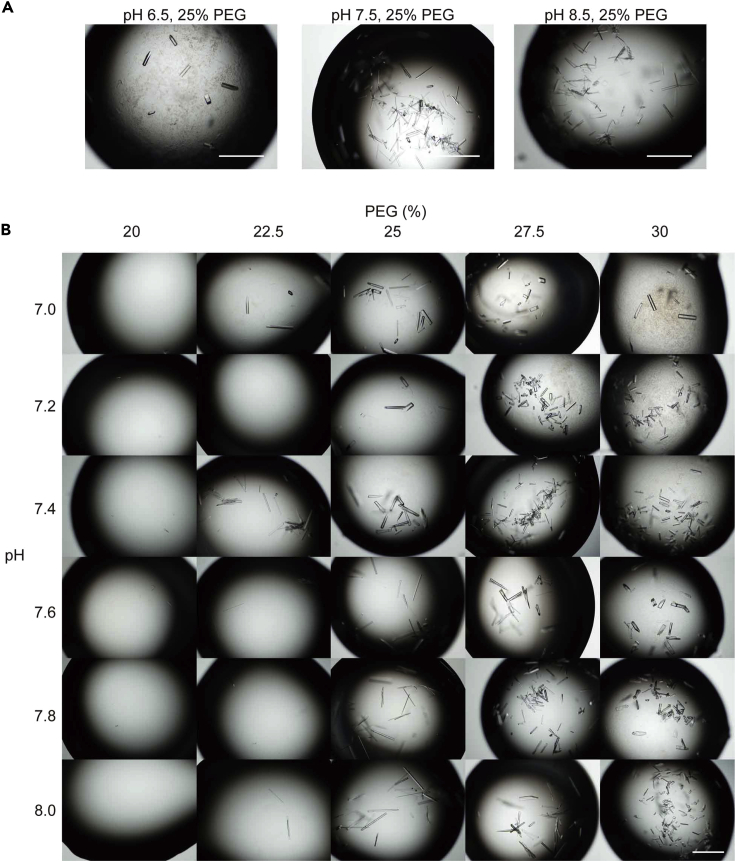
When crystals do not form, try a basic (approximately pH 8.5) Crystallization Buffer or decrease the ligand concentrations (Figure 12). When only many small crystals form, try an acidic (approximately pH 6.5) Crystallization Buffer or dilute the crystal nuclei solution. In our case, 25% PEG 3350 concentration was most effective; 20%–25% PEG 3350 produced fewer numbers of crystals and 27.5%–30% PEG 3350 only produced those with atypical appearances.
Figure 12.
Optimization of Crystallization Buffer in producing iFA-bound co-crystals
Varied concentrations of PEG 3350 and pH are tested. Scale bars, 500 μm.
Problem 2
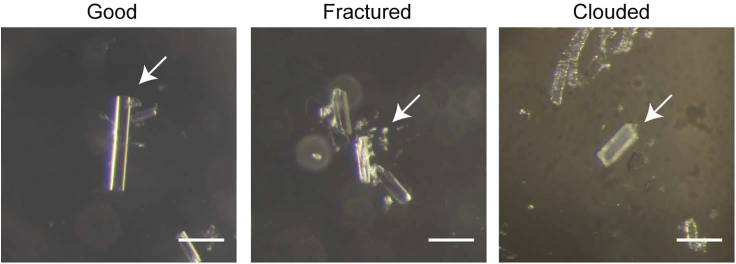
In soaking, the original ligand in hPPARα-LBD is not replaced with intended ligands; crystals break or become clouded; or crystal resolution decreases.
Potential solution
When ligands are not replaced, increasing ligand concentrations and incubation time may improve the situation (though we were not so successful with those methods). When crystals become fractured or clouded (Figure 13), or the resolution of crystals decreases, limit the incubation time with the ligand or reduce the concentration of ligand solvent as possible. Limiting the incubation time in particular often provides good results.
Figure 13.
Examples of good and bad (fractured or clouded) crystals for X-ray crystallography
Scale bars, 200 μm.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Isao Ishii (isao-ishii@umin.ac.jp).
Materials availability
This study did not generate unique reagents.
Data and code availability
The data supporting the findings of this study are available in Kamata et al. (2020). PDB ID codes for the 34 hPPARα-ligand structures reported in this paper are: 6KAX (iFA: intrinsic fatty acid); 6KAZ, 6KB9, and 6KB4 (pemafibrate); 6KAY, 6KB8, and 6KB3 (GW7647); 6KB0, 6KB5 (ETYA); 6KB1, 6KB6 (TTA); 6KXB, 6LXC (saroglitazar); 6KB2, 6KBA, and 6KB7 (Wy14643); 6LX4 (fenofibric acid); 6LX5 (ciprofibrate); 6LX6 (palmitic acid); 6LX7 (stearic acid); 6LX8 (oleic acid); 6LX9 (arachidonic acid); 6LXA (EPA); 6KYP (GW9662 + clofibric acid); 6L36 (GW9662 + fenofibric acid); 6L37 (GW9662 + ciprofibrate); 6L38 (GW9662 + gemfibrozil); 7BPY (clofibric acid + SRC1); 7BPZ (bezafibrate + SRC1); 7BQ0 (fenofibric acid + SRC1); 7BQ1 (intrinsic fatty acid + SRC1); 7BQ2 (pemafibrate + SRC1); 7BQ3 (GW7647 + SRC1); and 7BQ4 (EPA + SRC1).
Acknowledgments
S.K. and I.I. acknowledge funding from Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Sciences (JSPS) (grant numbers 19K16359 and 16H05107), Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED (grant number JP19am0101071; support number 1407), and research grants from Showa Pharmaceutical University. T.O. acknowledges funding from a Grant-in-Aid for Scientific Research from JSPS (grant number 18K06081) and Adaptable and Seamless Technology Transfer Program through Target-driven R&D (A-STEP) from Japan Science and Technology Agency (grant number JPMJTM19AT). This work was performed under the approval of the Photon Factory Program Advisory Committee (proposal number 2018G658).
Author contributions
S.K., T.O., and I.I. conceived the study and wrote the paper.
Declaration of interests
The authors declare no competing interests.
References
- Benvenuti M., Mangani S. Crystallization of soluble proteins in vapor diffusion for x-ray crystallography. Nat. Protoc. 2007;2:1633–1651. doi: 10.1038/nprot.2007.198. [DOI] [PubMed] [Google Scholar]
- Hassell A.M., An G., Bledsoe R.K., Bynum J.M., Carter H.L., 3rd, Deng S.J., Gampe R.T., Grisard T.E., Madauss K.P., Nolte R.T. Crystallization of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2007;63:72–79. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata S., Oyama T., Saito K., Honda A., Yamamoto Y., Suda K., Ishikawa R., Itoh T., Watanabe Y., Shibata T. PPARalpha ligand-binding domain structures with endogenous fatty acids and fibrates. iScience. 2020;23:101727. doi: 10.1016/j.isci.2020.101727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. Crystallization of Biological Macromolecules. Cold Spring Harbor Laboratory Press; 1999. Strategies and special approaches in growing crystals; pp. 271–330. [Google Scholar]
- McPherson A., Gavira J.A. Introduction to protein crystallization. Acta Crystallogr. F Struct. Biol. Commun. 2014;F70:2–20. doi: 10.1107/S2053230X13033141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T., Toyota K., Waku T., Hirakawa Y., Nagasawa N., Kasuga J.I., Hashimoto Y., Miyachi H., Morikawa K. Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2009;65:786–795. doi: 10.1107/S0907444909015935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.E., Stanley T.B., Montana V.G., Lambert M.H., Shearer B.G., Cobb J.E., McKee D.D., Galardi C.M., Plunket K.D., Nolte R.T. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available in Kamata et al. (2020). PDB ID codes for the 34 hPPARα-ligand structures reported in this paper are: 6KAX (iFA: intrinsic fatty acid); 6KAZ, 6KB9, and 6KB4 (pemafibrate); 6KAY, 6KB8, and 6KB3 (GW7647); 6KB0, 6KB5 (ETYA); 6KB1, 6KB6 (TTA); 6KXB, 6LXC (saroglitazar); 6KB2, 6KBA, and 6KB7 (Wy14643); 6LX4 (fenofibric acid); 6LX5 (ciprofibrate); 6LX6 (palmitic acid); 6LX7 (stearic acid); 6LX8 (oleic acid); 6LX9 (arachidonic acid); 6LXA (EPA); 6KYP (GW9662 + clofibric acid); 6L36 (GW9662 + fenofibric acid); 6L37 (GW9662 + ciprofibrate); 6L38 (GW9662 + gemfibrozil); 7BPY (clofibric acid + SRC1); 7BPZ (bezafibrate + SRC1); 7BQ0 (fenofibric acid + SRC1); 7BQ1 (intrinsic fatty acid + SRC1); 7BQ2 (pemafibrate + SRC1); 7BQ3 (GW7647 + SRC1); and 7BQ4 (EPA + SRC1).