Abstract
Several attempts have been made over the past decade to explore the concept of prodrug strategies that exploit PSA as a molecular target for the release of anticancer drugs in prostate tumors using various prostate specific antigen (PSA)-cleavable peptide linkers, but the desired antitumor and antimetastatic efficacy has not yet been fully achieved. We set out to look for new PSA-cleavable peptide substrates that could be cleaved more rapidly and efficiently than the previously used peptides. To look for the most susceptible PSA-cleavable peptide substrates, we used the so-called spot technology. With the following general formula, we designed 25 different fluorogenic heptapeptides; Cellulose-P5-P4-P3-P2-P1-P1′-P2’ (Fluorophore). The increase of the fluorescence in the supernatant of the reaction mixture was monitored using a 96-well fluorometric plate reader with excitation of λex 485 nm and λem 535 nm. Three sequences showed a high fluorogenic liberation after incubation with PSA, i.e., Arg-Arg-Leu-His-Tyr-Ser-Leu (7), Arg-Arg-Leu-Asn-Tyr-Ser-Leu (8) and Arg-Ser-Ser-Tyr-Arg-Ser-Leu (23). Future incorporation of these optimized substrates in the PSA-cleavable prodrug formulations could further optimize the cleavage pattern and so the release characteristics of these prodrugs to rapidly and efficiently liberate the free cytotoxic agents inside the tumor tissues.
Keywords: Peptide substrate, Prodrug, Spot assay, Prostate-specific antigen (PSA), Prostate cancer
Highlights
-
•
Prostate-specific antigen (PSA) represents a molecular target for selectively releasing anticancer agents from prodrugs.
-
•
Optimal PSA-cleavable peptide substrates are not yet identified.
-
•
Spot technology is used to elucidate a new PSA-cleavable peptide substrates.
-
•
We describe new three peptide sequences with a maximal PSA cleavability.
-
•
These new peptide substrates could improve the antitumor efficacy of PSA-cleavable prodrugs.
1. Introduction
Prostate-specific antigen (PSA) is a 33 kDa single-chain glycoprotein [1] composed of 237 amino acid residues, four carbohydrate side chains, and multiple disulfide bonds [[2], [3], [4]]. The amino acid sequence of PSA is homologous to proteases of the kallikrein family and may be referred to as human glandular kallikrein-3 (hK-3) [5]. Whereas most of the other kallikreins have trypsin-like proteolytic activity, PSA is considered a chymotrypsin-like protease based on similarities with chymotrypsin in the specificity of the catalytic site and its preference for cleaving after hydrophobic residues in the P1 position [6].
PSA is aptly named; in that it is specifically and exclusively produced by normal and malignant prostate epithelial cells. Its blood levels used as a screening test for prostate cancer and is not produced in any significant amounts by other normal tissue in the human male [7,8]. The increased secretion of PSA by prostate carcinoma cells coupled with the disruption of normal tissue architecture in the prostate or distal sites by prostate cancer cells results in the leakage of increased amounts of PSA into circulation in a positive correlation with the stage of the disease [7,9].
Interestingly, these PSA molecules have been reported to be secreted in an active form only in the extracellular fluid surrounding prostate carcinoma cells. In the circulation, PSA loses its enzymatic activity as a result of binding to abundant serum proteases inhibitors α1-antichymotrypsin and α2-macroglobulin, which were reported to be at a 104– to 105-fold molar excess in the circulation of patients with prostate carcinoma [[9], [10], [11], [12]].
Many strategies have previously been investigated to inhibit the protease activity of PSA to control the prostate tumor progression and metastasis but as a result of heterogeneity and autocatalysis, no success was achieved [3,[13], [14], [15], [16]]. In our research, we had investigated the concept of prodrug strategies that exploit PSA as a molecular target for releasing anticancer drugs in prostate tumors. Consequently, and as a main goal of the present work, we set out to look for new optimized PSA-cleavable peptide substrates that could be cleaved more rapidly and efficiently.
2. Materials and methods
2.1. Materials
In collaboration with JPT Peptide Technologies GmbH (Berlin, Germany), we designed 25 different fluorogenic heptapeptides with the following general formula; Cellulose–P5-P4-P3-P2-P1-P1'-P2'–(Fluorophore) as seen in (Table 1). The rationale of this design considered that the major physiologic substrates for PSA appear to be the gel-forming proteins in freshly ejaculated semen, semenogelin I and semenogelin II which are synthesized and secreted by the seminal vesicles. Active PSA in the seminal fluid cleaves preferentially after tyrosyl and glutaminyl peptide bonds to generate several low-molecular-weight soluble protein fragments [6,17,18]. On this basis of the PSA cleavage map for semenogelin I and semenogelin II, so much information became available about the amino acid sequence proximal to the PSA proteolytic sites as described in (Table 1). With this knowledge, we used the so-called spot technology [[19], [20], [21], [22]], to guess or search for the most susceptible PSA-cleavable peptide substrates.
Table 1.
Identified peptide sequences from the natural PSA substrates Semenogelin I and II (SG-I and SG-II), which are split by PSA [44].
| P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | Cleavage site in Protein (P1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Lys | Gly | Gln | His | Tyr | Ser | Gly | Gln | Lys | Gly | SG-I 25 |
| Phe | Ser | Ile | Gln | Tyr | Thr | Tyr | His | Val | Asp | SG-I 44 |
| Lys | Ser | Gln | Gln | Tyr | Asp | Leu | Asn | Ala | Leu | SG-I 62; SG-II 62 |
| Ile | Ser | Ser | Gln | Tyr | Ser | Asn | Thr | Glu | Glu | SG-I 136 |
| Glu | Leu | Leu | Val | Tyr | Asn | Lys | Asn | Gln | His | SG-I 240 |
| Arg | Arg | Leu | His | Tyr | Gly | Glu | Asn | Gly | Val | SG-I 277, 337 |
| Gln | Ser | Ser | Ile | Tyr | Ser | Gln | Thr | Glu | Glu | SG-I 292 |
| Gln | Arg | Ser | Ile | Tyr | Ser | Gln | Thr | Glu | Glu | SG-I 352 |
| Lys | Gly | Gln | His | Tyr | Phe | Gly | Gln | Lys | Asp | SG-II 25 |
| Ser | Gln | Ser | Ser | Tyr | Val | Leu | Gln | Thr | Glu | SG-II 171 |
| Arg | Arg | Leu | Asn | Tyr | Gly | Gly | Lys | Ser | Thr | SG-II 457 |
| Lys | Gly | His | Tyr | Gln | Asn | Val | Val | Glu | Val | SG-I 198; SG-II 198 |
| Ser | Ser | Lys | Val | Gln | Thr | Ser | Leu | Cys | Pro | SG-I 212 |
| Lys | Ile | Ser | Tyr | Gln | Ser | Ser | Ser | Thr | Glu | SG-I 266, 326; SG-II 326, 386 |
| Ser | Ser | Lys | Leu | Gln | Thr | Ser | Leu | His | Pro | SG-II 212 |
| Lys | Ser | Gln | Asn | Gln | Val | Thr | Ile | His | Ser | SG-II 306 |
| Arg | Ile | Pro | Ser | Gln | Ala | Gln | Glu | Tyr | Gly | SG-II 372 |
| Lys | Met | Ser | Tyr | Gln | Ser | Ser | Ser | Thr | Glu | SG-II 446 |
| Phe | Ser | Ile | Gln | His | Thr | Tyr | His | Val | Asp | SG-II 44 |
| Lys | Ser | Lys | Gln | His | Leu | Gly | Gly | Ser | Gln | SG-II 76 |
| Leu | His | Pro | Ala | His | Gln | Asp | Arg | Leu | Gln | SG-II 219 |
| Glu | Arg | Gln | Leu | His | His | Gly | Glu | Lys | Ser | SG-II 276 |
| Gly | Gln | Ser | Thr | Asn | Arg | Glu | Gln | Asp | Leu | SG-I 389 |
| Glu | Arg | Arg | Leu | Asn | Ser | Gly | Glu | Lys | Asp | SG-II 396 |
| Gly | Val | Gln | Lys | Asp | Val | Ser | Gln | Ser | Ser | SG-I 285, 345 |
| Ser | Gln | Gln | Leu | Leu | His | Asn | Lys | Gln | Glu | SG-I 84 |
| Ile | Thr | Ile | Pro | Ser | Gln | Glu | Gln | Glu | His | SGI-I 311 |
Each heptapeptide is attached from its C-terminus with Alexa-Fluor-488 as a fluorescent label, whereas the N-terminus is attached to Cellulose disc (Each cellulose disc contains approximately 57 nmol peptide) which is accurately fixed to a well in a 96-well plate (Table 2).
Table 2.
The fluorogenic heptapeptides that were designed and examined as PSA substrates.
| Nr | Cellulose–P5-P4-P3-P2-P1-P1'-P2'–Alexa-Fluor-488 |
|---|---|
| 1 | Arg-Ala-Ser-Tyr-DGln-Ser-Leu |
| 2 | Gly-Gly-Gly-Gly-Gly-Ser-Leu |
| 3 | Arg-Ser-Ser-Tyr-Tyr-Ser-Leu |
| 4 | Lys-Gly-Gln-His-Tyr-Ser-Leu |
| 5 | Ile-Ser-Ser-Gln-Tyr-Ser-Leu |
| 6 | Gln-Ser-Ser-Ile-Tyr-Ser-Leu |
| 7 | Arg-Arg-Leu-His-Tyr-Ser-Leu |
| 8 | Arg-Arg-Leu-Asn-Tyr-Ser-Leu |
| 9 | Arg-Ser-Ser-Ser-Tyr-Ser-Leu |
| 10 | Arg-Ser-Ser-Tyr-Gln-Ser-Leu |
| 11 | Lys-Ile-Ser-Tyr-Gln-Ser-Leu |
| 12 | Lys-Met-Ser-Tyr-Gln-Ser-Leu |
| 13 | Ser-Ser-Lys-Tyr-Gln-Ser-Leu |
| 14 | Ser-Ser-Lys-Val-Gln-Ser-Leu |
| 15 | Lys-Ser-Gln-Asn-Gln-Ser-Leu |
| 16 | Arg-Ile-Pro-Ser-Gln-Ser-Leu |
| 17 | Arg-Ile-Arg-Ser-Gln-Ser-Leu |
| 18 | Arg-Ser-Ser-Tyr-His-Ser-Leu |
| 19 | Lys-Ser-Lys-Gln-His-Ser-Leu |
| 20 | Arg-Ser-Ser-Gln-His-Ser-Leu |
| 21 | Arg-Ser-Ser-Ala-His-Ser-Leu |
| 22 | Arg-Ser-Ser-Leu-His-Ser-Leu |
| 23 | Arg-Ser-Ser-Tyr-Arg-Ser-Leu |
| 24 | Ser-Ser-Ser-Arg-Arg-Ser-Leu |
| 25 | Gly-Gln-Ser-Thr-Asn-Ser-Leu |
Among the designed 25 fluorogenic substrates there were two PSA-uncleavable heptapeptides, i.e., Arg-Ala-Ser-Tyr-DGln-Ser-Leu (1) and Gly-Gly-Gly-Gly-Gly-Ser-Leu (2) that were included in the study as a negative control. All the other remaining 23 heptapeptides are expected to be PSA cleavable substrates including our previously used heptapeptide substrate i.e., Arg-Ser-Ser-Tyr-Tyr-Ser-Leu (3) which was incorporated to serve as a positive control. Each of the 25 heptapeptides is prepared as a triplicate (repeated three times) and placed consequently in the 96-well plate. Specifically, we decided to use the amino acids Ser in P1’ position and Leu in P2’ position in all prepared 25 heptapeptides (Table 2), depending on some emerging evidences that have been concluded in our previous work, as well as by other investigators. Additionally, these amino acids have been observed to occupy these positions in the natural PSA cleavable substrates and were also used in these positions by other investigators to design some PSA-cleavable substrates [[19], [20], [21], [22]].
The peptides were synthesized on the cellulose discs, starting with a linker system at the C-terminus. The peptides ended at the N-terminus with a free Cysteine residue, where the fluorophore (in this case the Alexa-Fluor 488) was coupled as final step. The evidence of purity for the prepared peptides was provided with their characteristic UV-trace showed that the target mass was found at the peak at 2.8 min as shown in the analysis curve [Fig. 1S].
Fig. 1.
Diagrammatic illustration for the principle of assay and monitoring the fluorescence increase in the reaction supernatant after PSA cleavage of the fluorogenic heptapeptides substrates.
Enzymatically active PSA was purchased from Calbiochem (Bad Soden, Germany). All other chemicals including methanol, Tris HCl buffer ingredients and solvents were of analytical grade and obtained from standard suppliers and were used without further purification. The buffers used were vacuum-filtered through a 0.2 μm membrane (Sartorius, Germany) and thoroughly degassed with ultrasound prior to use. A 96-well fluorometric plate reader with excitation of λex 485 nm and λem 535 nm was used to measure the fluorescence.
2.2. Enzymatic fluorogenic assay
The principle of assay depends on monitoring the increase of the fluorescence in the supernatant of the reaction mixture as a result of the fluorophore liberation after the cleavage of the fluorogenic heptapeptides substrates with PSA (Fig. 1).
Accordingly, each spot was rinsed with methanol for 5 min to solubilize the peptides. Subsequently, it was washed 4 times with 250 μL of 50 mM Tris buffer pH 7.5 with gentle agitation in each step. Afterwards, 250 μL of enzymatically active PSA solution (20 μg/ml) was added to each well. A 50 μL aliquot was transferred directly from each well to its corresponding well in a new 96-well plate containing 50 μL of 50 mM Tris buffer pH 7.5. After mixing, the fluorescence of the new plate was measured to be considered as a 0-time fluorescence level (blank) using a 96-well fluorometric plate reader with excitation of λex 485 nm and λem 535 nm. The reaction mixture was then sealed with a plastic seal and the reaction was allowed to proceed at room temperature without agitation. At 1-h intervals, an aliquot of 50 μL from the reaction supernatant was transferred to a new 96-well plate, diluted and mixed with 50 μL of 50 mM Tris buffer pH 7.5 [23], and its fluorescence was measured as described above. Analysis is performed at JPT using HPLC-MS at 220 nm, linear gradient, 5–95% acetonitrile in water +0.05% TFA.
2.3. Statistical analysis
The results were determined from the initial linear increase of fluorescence till 5 h and were expressed as the mean fluorescence change per hour (ΔF/h) after its analyses by GraphPad Prism version 5.
3. Results
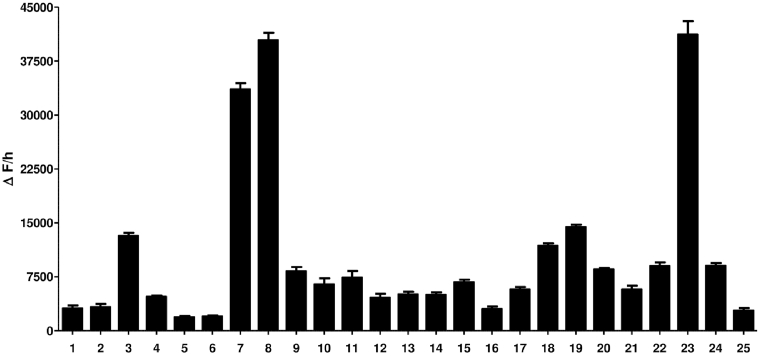
The obtained cleavage results revealed that among the examined 25 fluorogenic heptapeptide substrates, there were three sequences that showed the highest fluorogenic liberation after incubation with PSA (Fig. 2).
Fig. 2.
The mean fluorescence change per hour (ΔF/h) of each individual fluorogenic heptapeptide substrates afterPSA cleavage. Data are presented as the mean ± SEM (n = 3).
These heptapeptides are Arg-Arg-Leu-His-Tyr-Ser-Leu (7), Arg-Arg-Leu-Asn-Tyr-Ser-Leu (8) and Arg-Ser-Ser-Tyr-Arg-Ser-Leu (23). Of note that, this pronounced increase in the rate of fluorescence release in these three heptapeptides was, however, higher than that of our previously used substrate, i.e., Arg-Ser-Ser-Tyr-Tyr-Ser-Leu (3). Additionally, and in a clear conformation for the PSA cleavage liability, the two heptapeptides which were included as negative controls, i.e., Arg-Ala-Ser-Tyr-DGln-Ser-Leu (1) and Gly-Gly-Gly-Gly-Gly-Ser-Leu (2), did not show any significant increase in the florescence rate over the taken period. This can indicate that the three heptapeptides with the highest increase in the florescence rate could resemble the best substrates for PSA among the all examined heptapeptides.
4. Discussion
Based on unique biology of PSA (a serine protease), several studies have investigated prodrug strategies that exploit PSA as a molecular target for releasing anticancer drugs from enzymatically cleavable prodrug formulations inside the prostate tumors [12,[24], [25], [26], [27]]. PSA proteolysis of extracellular matrix proteins such as fibronectin and laminin can stimulate the detachment of tumour cells and promote tumour invasion and metastasis, ultimately contributing to the spread and progression of prostate cancer [28,29]. The inhibition and/or consumption of PSA at the tumour site can therefore be a useful method for curing prostate cancer.
PSA as a molecular target for releasing anticancer drugs is based mainly on the conjugation of the conventional anticancer drugs with a wide spectrum of low- and high-molecular-weight carriers including sugars, growth factors, vitamins, peptides, antibodies, polysaccharides, lectins, serum proteins, and synthetic polymers through a linker molecule that incorporates a pre-determined breaking point to ensure a tumor-specific release of the drug payload [30].
Denmeade research group introduced a relevant example of the low-molecular weight prodrug strategies that exploit PSA as a molecular target for the release of anticancer drugs from prodrug formulations when they produced an inactive doxorubicin prodrug that was synthesized by coupling the doxorubicin primary amine to the COOH-terminal carboxyl of a seven-amino acid peptide carrier [i.e., Mu-His-Ser-Ser-Lys-Leu-Gln-Leu (Mu = morpholinocarbonyl)] which was recorded to be precisely hydrolysable by PSA [1,31]. The resulting prodrug was shown to be efficiently activated by human prostate cancer cell lines producing PSA (such as PC-82 and LNCaP cells) to release the active cytotoxin L-leucyl-doxorubicin, whereas the prodrug had no cytotoxic effect on PSA-nonproducing TSU human prostate cancer cells in vitro [1].
In principle, in addition to the active cytotoxic agent, an enzymatically cleavable prodrug contains a peptide linker substrate that guarantees the subsequent release of the active compound within the tumour interstitium [30]. We and others have produced several prodrug formulations that integrate PSA-cleavable peptide substrates based on some available information from the proteolytic cleavage map for PSA's physiological substrate, human semenogelin I and II [24,26,32,33]. In these prodrugs, the most commonly used PSA-cleavable peptide linkers were the N-glutaryl-(4-hydroxyprolyl)AlaSer-cyclohexaglycyl-GlnSerLeu-COOH [27]. Mu-His-Ser-Ser-Lys-Leu-Gln-Leu [1], and our previously used heptapeptide substrate, i.e., Arg-Ser-Ser-Tyr-Tyr-Ser-Leu [34,35]. Despite the relatively promising results of these studies, further optimization of the PSA-cleavable peptide substrates remains important to achieve the maximum improved cleavage pattern and thus achieve the desired antitumor and antimetastatic efficacy of the corresponding prodrug in order to allow complete tumour remission. The lack of ideal optimization can be attributed to the fact that the main PSA-cleavage products of these prodrugs represent the drug monopeptides or dipeptides that still need further cleavage to release the active drug as a final product over time [24,33,35].
In the current study, as an option, we set out to look for new optimized PSA-cleavable peptide substrates that could be cleaved more quickly and efficiently than the peptides previously mentioned. The incorporation of these optimized substrates into prodrug formulations could further optimize the pattern of PSA cleavage and thus the release properties of these prodrugs to release the free cytotoxic agents within the tumour tissues quickly and efficiently and thus maximally improve their antitumor and antimetastatic efficacy.
Our study findings showed that after incubation with PSA, three sequences had high fluorogenic release, i.e., Arg-Arg-Leu-His-Tyr-Ser-Leu (7), Arg-Arg-Leu-Asn-Tyr-Ser-Leu (8) and Arg-Ser-Ser-Tyr-Arg-Ser-Leu (23).
Incubation studies with PSA showed that the albumin-bound form of the two drugs was quickly split, but at the P1–P1’ scissile bond, releasing the drug-dipeptides, i.e. H-Ser-Leu-PABC-doxorubicin and H-Ser-Leu-PABC-paclitaxel, in the same previous manner, which were further degraded to release the free drug as a final cleavage product within few hours in prostate tumor tissue homogenates as well as in PSA-positive LNCaP LN cell lysates [[33], [34], [35], [36]]. These data indicate that the N-glutaryl-(4-hydroxyprolyl) AlaSer-cyclohexaglycyl-GlnSerLeu-CO2HH [26], Mu-His-Ser-Ser-Lys-Leu-Gln-Leu [1], and our previously used heptapeptide substrate, i.e., Arg-Ser-Ser-Tyr-Tyr-Ser-Leu [[33], [34], [35]], were the most commonly used PSA-cleavable peptide linkers in previous studies to develop PSA-cleavable prodrugs. It is apparent that neither of these PSA-cleavable peptide binders has been sufficiently satisfactory to achieve the maximum optimized cleavage pattern of the corresponding prodrug, so the desired antitumor and antimetastatic efficacy has not yet been fully achieved.
In conclusion, there were at least three sequences of the various fluorogenic heptapeptide substrates tested in the current study that showed the highest fluorogenic release after PSA incubation, i.e., Arg-Arg-Leu-His-Tyr-Ser-Leu (7), Arg-Arg-Leu-Asn-Tyr-Ser-Leu (8) and Arg-Ser-Ser-Tyr-Arg-Ser-Leu-Leu (23). These three sequences may be considered, according to these findings, to be promising candidates for the future production of the corresponding PSA-cleavable prodrugs integrating these substrates.
Informed consent
This manuscript does not contain any studies with human subjects or animals performed by any of the authors.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This study had no funding from any resource.
Authors' contributions
Study concept and design: BEME and MHH; Data analysis and writing manuscript: BEME and MHH; All authors revised and approved the final version of the manuscript.
Statements
In this work we used the so-called Allexa-spot technology to look for new peptide substrates for the development of albumin binding anticancer pro-drugs that are cleaved by prostate-specific antigen (PSA) to improve the anti tumor efficacy.
Declaration of competing interest
The authors declare that they have no known competing financialinterestsor personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are deeply thankful to JPT Peptide Technologies GmbH (Berlin, Germany) for designing the fluorogenic heptapeptides.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.100966.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Denmeade S.R., Nagy A., Gao J., Lilja H., Schally A.V., Isaacs J.T. Enzymatic activation of a doxorubicin-peptide prodrug by prostate-specific antigen. Canc. Res. 1998;58:2537–2540. [PubMed] [Google Scholar]
- 2.Robert M., Gibbs B.F., Jacobson E., Gagnon C. Characterization of prostate-specific antigen proteolytic activity on its major physiological substrate, the sperm motility inhibitor precursor/semenogelin I. Biochemistry. 1997;36:3811–3819. doi: 10.1021/bi9626158. [DOI] [PubMed] [Google Scholar]
- 3.Hassan M.I., Kumar V., Singh T.P., Yadav S. Structural model of human PSA: a target for prostate cancer therapy. Chem. Biol. Drug Des. 2007;70:261–267. doi: 10.1111/j.1747-0285.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- 4.Yang C.F., Porter E.S., Boths J., Kanyi D., Hsieh M., Cooperman B.S. Design of synthetic hexapeptide substrates for prostate-specific antigen using single-position minilibraries. J. Pept. Res. 1999;54:444–448. doi: 10.1034/j.1399-3011.1999.00141.x. [DOI] [PubMed] [Google Scholar]
- 5.Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J. Clin. Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeBeau A.M., Singh P., Isaacs J.T., Denmeade S.R. Prostate-specific antigen is a "chymotrypsin-like" serine protease with unique P1 substrate specificity. Biochemistry. 2009;48:3490–3496. doi: 10.1021/bi9001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams S.A., Singh P., Isaacs J.T., Denmeade S.R. Does PSA play a role as a promoting agent during the initiation and/or progression of prostate cancer? Prostate. 2007;67:312–329. doi: 10.1002/pros.20531. [DOI] [PubMed] [Google Scholar]
- 8.Ideo H., Kondo J., Nomura T., Nonomura N., Inoue M., Amano J. Study of glycosylation of prostate-specific antigen secreted by cancer tissue-originated spheroids reveals new candidates for prostate cancer detection. Sci. Rep. 2020;10(1):2708. doi: 10.1038/s41598-020-59622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denmeade S.R., Sokoll L.J., Chan D.W., Khan S.R., Isaacs J.T. Concentration of enzymatically active prostate-specific antigen (PSA) in the extracellular fluid of primary human prostate cancers and human prostate cancer xenograft models. Prostate. 2001;48:1–6. doi: 10.1002/pros.1075. [DOI] [PubMed] [Google Scholar]
- 10.Pettersson K., Piironen T., Seppala M., Liukkonen L., Christensson A., Matikainen M.T., Suonpaa M., Lovgren T., Lilja H. Free and complexed prostate-specific antigen (PSA): in vitro stability, epitope map, and development of immunofluorometric assays for specific and sensitive detection of free PSA and PSA-alpha 1-antichymotrypsin complex. Clin. Chem. 1995;41:1480–1488. [PubMed] [Google Scholar]
- 11.Hakalahti, L., Vihko, P. , Henttu, P., Autio-Harmainen, H. , Soini, Y.,&Vihko, R. , Evaluation of PAP and PSA gene expression in prostatic hyperplasia and prostatic carcinoma using northern-blot analyses, in situ hybridization and immunohistochemical stainings with monoclonal and bispecific antibodies.Int. J. Canc., 55, 590-597. [DOI] [PubMed]
- 12.Diamandis E.P. New diagnostic applications and physiological functions of prostate specific antigen. Scand. J. Clin. Lab. Invest. Suppl. 1995;221:105–112. doi: 10.3109/00365519509090573. [DOI] [PubMed] [Google Scholar]
- 13.Diamandis E.P., Helle S.I., Yu H., Melegos D.N., Lundgren S., Lonning P.E. Prognostic value of plasma prostate specific antigen after megestrol acetate treatment in patients with metastatic breast carcinoma. Cancer. 1999;85:891–898. doi: 10.1002/(sici)1097-0142(19990215)85:4<891::aid-cncr17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 14.Baumgart Y., Otto A., Schafer A., Usbeck E., Cott C., Schott A., Tornack M., Wenzel A., Mossie A., Birkenmeier G. Characterization of novel monoclonal antibodies for prostate-specific antigen (PSA) with potency to recognize PSA bound to alpha 2-macroglobulin. Clin. Chem. 2005;51:84–92. doi: 10.1373/clinchem.2004.039636. [DOI] [PubMed] [Google Scholar]
- 15.Yang L., Zhang L.Y., Chen W.W., Kong F., Zhang P.J., Hu X.Y., Zhang J.Y., Cui F.A. Inhibition of the expression of prostate specific antigen by curcumin. Yao Xue Xue Bao. 2005;40:800–803. [PubMed] [Google Scholar]
- 16.Petit J.H., Chen M.H., Loffredo M., Sussman B., Renshaw A.A., D'Amico A.V. Prostate-specific antigen recurrence and mortality after conventional dose radiation therapy in select men with low-risk prostate cancer. Cancer. 2006;107:2180–2185. doi: 10.1002/cncr.22243. [DOI] [PubMed] [Google Scholar]
- 17.Lilja H., Laurell C.B. The predominant protein in human seminal coagulate. Scand. J. Clin. Lab. Invest. 1985;45:635–641. doi: 10.3109/00365518509155271. [DOI] [PubMed] [Google Scholar]
- 18.Lilja H., Abrahamsson P.A., Lundwall A. Semenogelin, the predominant protein in human semen. Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J. Biol. Chem. 1989;264:1894–1900. [PubMed] [Google Scholar]
- 19.Naus S., Reipschläger S., Wildeboer D., Lichtenthaler S.F., Mitterreiter S., Guan Z., Moss M.L., Bartsch J.W. Identification of candidate substrates for ectodomain shedding by the metalloprotease-disintegrin ADAM8. Biol. Chem. 2006;387(3):337–346. doi: 10.1515/BC.2006.045. [DOI] [PubMed] [Google Scholar]
- 20.Janssen, S., Jakobsen, C.M., Rosen, D.M., Ricklis RM., Reineke, U., Christensen, S.B., Lilja, H., &Denmeade, S.R. Screening a combinatorial peptide library to develop a human glandular kallikrein 2-activated prodrug as targeted therapy for prostate cancer. Mol. Canc. Therapeut., 3(11),1439-1450. [PubMed]
- 21.Kaup M., Dassler K., Reineke U., Weise C., Tauber R., Fuchs H. Processing of the human transferrin receptor at distinct positions within the stalk region by neutrophil elastase and cathepsin G. Biol. Chem. 2002;383(6):1011–1020. doi: 10.1515/BC.2002.108. [DOI] [PubMed] [Google Scholar]
- 22.Jones C.H., Dexter P., Evans A.K., Liu C., Hultgren S.J., Hruby D.E. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 2002;184(20):5762–5771. doi: 10.1128/JB.184.20.5762-5771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lea W.A., Simeonov A. Fluorescence polarization assays in small molecule screening. Expet Opin. Drug Discov. 2011 Jan;6(1):17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graeser R., Chung D.E., Esser N., Moor S., Schachtele C., Unger C., Kratz F. Synthesis and biological evaluation of an albumin-binding prodrug of doxorubicin that is cleaved by prostate-specific antigen (PSA) in a PSA-positive orthotopic prostate carcinoma model (LNCaP) Int. J. Canc. 2008;122:1145–1154. doi: 10.1002/ijc.23050. [DOI] [PubMed] [Google Scholar]
- 25.Denmeade S.R., Isaacs J.T. Enzymatic activation of prodrugs by prostate-specific antigen: targeted therapy for metastatic prostate cancer. Canc. J. Sci. Am. 1998;4(1):S15–S21. [PubMed] [Google Scholar]
- 26.DeFeo-Jones D., Garsky V.M., Wong B.K., Feng D.M., Bolyar T., Haskell K., Kiefer D.M., Leander K., McAvoy E., Lumma P., Wai J., Senderak E.T., Motzel S.L., Keenan K., Van Zwieten M., Lin J.H., Freidinger R., Huff J., Oliff A., Jones R.E. A peptide-doxorubicin 'prodrug' activated by prostate-specific antigen selectively kills prostate tumor cells positive for prostate-specific antigen in vivo. Nat. Med. 2000;6:1248–1252. doi: 10.1038/81351. [DOI] [PubMed] [Google Scholar]
- 27.Greene K.L., Albertsen P.C., Babaian R.J., Carter H.B., Gann P.H., Han M., Kuban D.A., Sartor A.O., Stanford J.L., Zietman A., Carroll P. Prostate specific antigen best practice statement: 2009 update. J. Urol. 2009;182:2232–2241. doi: 10.1016/j.juro.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 28.Caplan A., Kratz A. Prostate-specific antigen and the early diagnosis of prostate cancer. Am. J. Clin. Pathol. 2002;117:S104–S108. doi: 10.1309/C4UN-12LK-43HP-JXY3. [DOI] [PubMed] [Google Scholar]
- 29.Cheng C.W., Chan L.W., Ng C.F., Chan C.K., Tse M.K., Lai M.M. Breast metastasis from prostate cancer and interpretation of immunoreactivity to prostate-specific antigen. Int. J. Urol. 2006;13:463–465. doi: 10.1111/j.1442-2042.2006.01327.x. [DOI] [PubMed] [Google Scholar]
- 30.Webber M.M., Waghray A., Bello D. Prostate-specific antigen, a serine protease, facilitates human prostate cancer cell invasion. Clin. Canc. Res. 1995;1:1089–1094. [PubMed] [Google Scholar]
- 31.Kratz F., Muller I.A., Ryppa C., Warnecke A. Prodrug strategies in anticancer chemotherapy. ChemMedChem. 2008;3:20–53. doi: 10.1002/cmdc.200700159. [DOI] [PubMed] [Google Scholar]
- 32.Denmeade S.R., Lou W., Lovgren J., Malm J., Lilja H., Isaacs J.T. Specific and efficient peptide substrates for assaying the proteolytic activity of prostate-specific antigen. Canc. Res. 1997;57:4924–4930. [PMC free article] [PubMed] [Google Scholar]
- 33.Denmeade S.R., Jakobsen C.M., Janssen S., Khan S.R., Garrett E.S., Lilja H., Christensen S.B., Isaacs J.T. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. J. Natl. Cancer Inst. 2003;95:990–1000. doi: 10.1093/jnci/95.13.990. [DOI] [PubMed] [Google Scholar]
- 34.Elsadek B., Graeser R., Warnecke A., Unger C., Saleem T., El-Melegy N., Madkor H., Kratz F. Optimization of an albumin-binding prodrug of doxorubicin that is cleaved by prostate-specific antigen (PSA) ACS Med. Chem. Lett. 2010;1:234–238. doi: 10.1021/ml100060m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsadek B., Graeser R., Esser N., Schafer-Obodozie C., Ajaj K.A., Unger C., Warnecke A., Saleem T., El-Melegy N., Madkor H., Kratz F. Development of a novel prodrug of paclitaxel that is cleaved by prostate-specific antigen: an in vitro and in vivo evaluation study. Eur. J. Canc. 2010;46:3434–3444. doi: 10.1016/j.ejca.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 36.Elsadek B., Graeser R., Esser N., Schafer-Obodozie C., Tsurumi C., Abu Ajaj K., Warnecke A., Unger C., Saleem T., Kratz F. In vivo evaluation of a novel albumin-binding prodrug of doxorubicin in an orthotopic mouse model of prostate cancer (LNCaP) Prostate Cancer Prostatic Dis. 2011;14:14–21. doi: 10.1038/pcan.2010.43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.