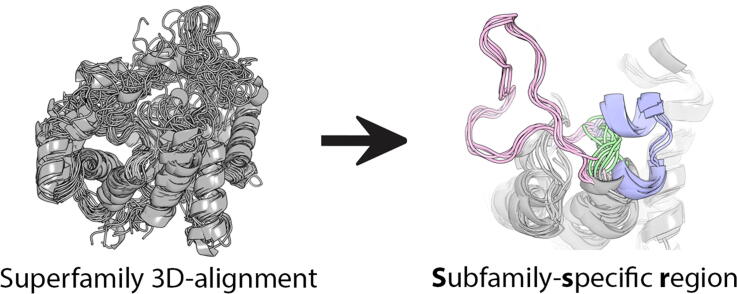
Graphical abstract
Keywords: Protein superfamilies, Structure-function relationship, 3D-structure analysis, Specificity-determining positions, Protein design, Drug discovery, Machine learning
Abbreviations: (H)DBSCAN, (Hierarchical) Density-Based Spatial Clustering of Applications with Noise; OPTICS, Ordering Points to Identify the Clustering Structure; RMSD, Root Mean Square Deviation; SSR, Subfamily-Specific Regions; SSP, Subfamily-Specific Position; SDR/SDP, Specificity-Determining Residue/Position
Abstract
Local 3D-structural differences in homologous proteins contribute to functional diversity observed in a superfamily, but so far received little attention as bioinformatic analysis was usually carried out at the level of amino acid sequences. We have developed Zebra3D – the first-of-its-kind bioinformatic software for systematic analysis of 3D-alignments of protein families using machine learning. The new tool identifies subfamily-specific regions (SSRs) – patterns of local 3D-structure (i.e. single residues, loops, or secondary structure fragments) that are spatially equivalent within families/subfamilies, but are different among them, and thus can be associated with functional diversity and function-related conformational plasticity. Bioinformatic analysis of protein superfamilies by Zebra3D can be used to study 3D-determinants of catalytic activity and specific accommodation of ligands, help to prepare focused libraries for directed evolution or assist development of chimeric enzymes with novel properties by exchange of equivalent regions between homologs, and to characterize plasticity in binding sites. A companion Mustguseal web-server is available to automatically construct a 3D-alignment of functionally diverse proteins, thus reducing the minimal input required to operate Zebra3D to a single PDB code. The Zebra3D + Mustguseal combined approach provides the opportunity to systematically explore the value of SSRs in superfamilies and to use this information for protein design and drug discovery. The software is available open-access at https://biokinet.belozersky.msu.ru/Zebra3D.
1. Introduction
Bioinformatic analysis of “specific positions” in multiple sequence alignments – also known as subfamily/family-specific positions (SSPs) or specificity-determining positions (SDPs) – that are conserved only within subfamilies/families, but different among them, is a widely used class of computational approaches to study functional diversity in proteins [1], [2], [3], [4], [5], [6]. Such SSPs/SDPs have both fundamental and practical value: they can help to understand how enzymes perform their natural functions, and can also be selected as hotspots for protein engineering experiments or as key residues involved in selective accommodation of ligand to assist drug discovery [7], [8]. Interest in the analysis of functionally important specific positions is a long-standing trend in computational biology: the concept was introduced in late 1990s [9], [10], the first systematic approach to identify such positions/residues in protein sequences was published in 2002 [11], followed by a variety of improvements to increase the accuracy of predictions and facilitate the ease-of-use in the daily routine [3], [5], [7], [12], [13]. In particular, the original Zebra/Zebra2 approach [14] to identify SSPs/SDPs in multiple sequence alignments was recognized as a tool [15], [16] to help studying structure–function relationship in protein superfamilies [17], [18], [19], and used to assist experimental design of improved enzymes [20] and ligand binding specificity [21].
Bioinformatic analysis of homologs implementing different properties within a shared superfamily fold has proven to be a useful tool to study the determinants of functional diversity as well as key catalytic and structural residues, but so far has usually operated at the level of amino acid sequences [3], [7], [8]. A 3D-structure is known to be a more content-rich representation of a protein, as even subtle differences in spatial arrangement of the backbone in homologs can significantly affect their function [22]. Yet, 3D-structure based methods in bioinformatics remain in the minority compared to sequence-based strategies. Probably, this is due to a modest amount of 3D-data that used to be available in the PDB databank [23]. Today, the number of non-redundant 3D-records of proteins representing functionally diverse superfamilies is approaching hundreds and thousands and continues to grow [24], requesting new bioinformatic solutions to address the emerging challenges and opportunities. So far, this demand has been met by only a handful of tools. Several strategies were proposed in an attempt to address the problem of protein structure flexibility [25], [26], [27] based on molecular modeling [28], [29] or comparative 3D-structure analysis [30], [31]. Most molecular visualization and manipulation packages implement tools to calculate root mean square deviation/fluctuation or perform principal component/normal mode analysis over the ensemble of full-size structures [32], [33]. These tools, in combination with visual expert inspection, can help to identify flexible parts in protein structures and to select regions that implement significantly different orientations among input 3D-entries. However, they were generally developed to study conformational plasticity in a given protein, as well as for global fold comparison, and are not well suited to study 3D-structural diversity among members of different families/subfamilies. Several methods are available for 3D-structure alignment of multiple proteins [34], [35], [36], [37], or that explicitly consider 3D-structural information while constructing a multiple sequence alignment [38], [39], [40]. However, the primary output of such tools is a sequence representation of such superimposition (e.g. a FASTA file), while its 3D-coordinate version in PDB format is commonly dismissed from further consideration. With the growing interest in 3D-structure analysis in the last few years, it became increasingly common to use 3D-record(s) of reference protein(s) to assist bioinformatic analysis, e.g. to map sequence alignment statistics onto such 3D-reference(s) to visualize results, or to improve accuracy of corresponding predictions by considering spatial features of a representative/query protein [14], [41], [42], [43]. These tools are interesting and practically useful, but in most cases consideration of 3D-data is focused on just one homolog or serves supplementary/illustrative purposes. Several algorithms have been developed to identify spatially equivalent 3D-motifs from similar constellations of amino acid main- or side-chains in 3D-structures of homologous or evolutionarily unrelated proteins [44], [45], [46], [47], [48]. Such 3D-motifs represent a three-dimensional analogue of conserved positions in a multiple sequence alignment, i.e. these can help to study key atoms/residues responsible for a function or property common to selected proteins, even in the absence of any detectable sequence or fold similarities, but can hardly explain functional diversity. 2StrucCompare web-server is available for analysis of 3D-structural differences in proteins, but can perform only pairwise comparisons of just two homologs with a particular focus on secondary structure elements, and thus is of limited productivity to study large superfamilies [49]. Finally, machine learning is emerging in structural bioinformatics as a powerful class of approaches to study the increasing abundance of 3D-data. Caretta is the most recent protein 3D-superimposition method [34]. In addition to the classic alignment function, it offers a feature extraction suite to be used in downstream steps for supervised or unsupervised machine learning. A related Geometricus software presents a novel approach to embed protein structures into fixed-length vectors, which can be used in machine learning algorithms aimed at predicting and understanding functional and physical properties, e.g. for fast structure similarity search, unsupervised clustering, and structure classification [50]. Nevertheless, despite recent progress in the development of advanced algorithms for studying proteins at the 3D-level, there is currently no tool aimed at a systematic analysis of specific 3D-structural differences among functionally diverse protein families.
Here we introduce Zebra3D – a novel bioinformatic software for systematic analysis of local structural differences in 3D-alignments of multiple proteins to determine subfamily-specific regions (SSRs) – patterns of local 3D-structure (i.e. single residues, loops, or secondary structure fragments) that are spatially equivalent within families/subfamilies, but are different among them. We further introduce the Zebra3D pipeline, describe practical means to facilitate its use by stacking with Mustguseal protein alignment web-engine, and discuss diverse case-studies to outline its application in daily laboratory routine.
2. Results
We have developed Zebra3D – a novel tool for bioinformatic analysis of 3D-alignments of homologous proteins to identify subfamily-specific regions in their structures, i.e. three-dimensional equivalents of SSPs/SDPs and plausible determinants of functional diversity in a superfamily. Zebra3D is a command-line Python3-based program that implements shared-memory parallel capabilities to scale efficiently on all CPU cores/threads hosted within a shared address space. The input to the program is a 3D-alignment of multiple protein structures. A companion Mustguseal web-server is available to handle automatic collection and superimposition of protein PDB entries within a superfamily of interest, thus reducing the minimal input required to operate the new tool to a single PDB code. Mustguseal web-server can also be used to run Zebra3D with default settings on-line. The Zebra3D classifies “columns” of a 3D-alignment into two categories: “common core” regions that are shared by homologs and “variable” regions featuring 3D-structural diversity. The latter are further subjected to a machine-learning cluster analysis technique in an attempt to classify the respective fragments of local structure into compact 3D-clusters, i.e. subfamilies (Fig. 1). “Variable” regions in which machine-learning identified at least two subfamilies are finally selected as SSRs and automatically prioritized according to their 3D-specificity S-scores and statistical significance Z-scores. The most visually prominent SSRs that are spatially consistent within clusters/subfamilies, but distant from each other, are ranked first to facilitate their expert analysis. This final statistical analysis step does not affect the number, length or content of SSRs, but only determines their ranking (i.e. the order of appearance in output files) and significance scores. The primary focus of Zebra3D is on 3D-specificity observed among members of different families/subfamilies and characterized by a significantly different organization of local 3D-structure (e.g. different length and/or organization/orientation of superimposed structural regions) in homologs. The currently implemented general-purpose statistical model attempts to discriminate SSRs, which feature 3D-specificity among protein families, from conformational variants, which are the result of average structural plasticity/flexibility not associated with a function. Top ranks and higher Z-scores are assigned to regions featuring 3D-structural diversity that is significantly different from the average level of random fluctuations in protein structures (see section “Algorithm”). For a particular purpose, user can manually implement a proprietary statistical model, e.g. to estimate significance of identified SSRs with respect to superfamily-specific or function-specific conformational plasticity. Overall, Zebra3D provides two types of useful results: a list of SSRs themselves and, for each such region, classification of proteins into subfamilies. Each region is evaluated independently; thus, the subfamily assignment may vary between finally selected SSRs. We further describe input and output in more detail, and discuss the potential of the new Zebra3D + Mustguseal combined approach on diverse case-studies.
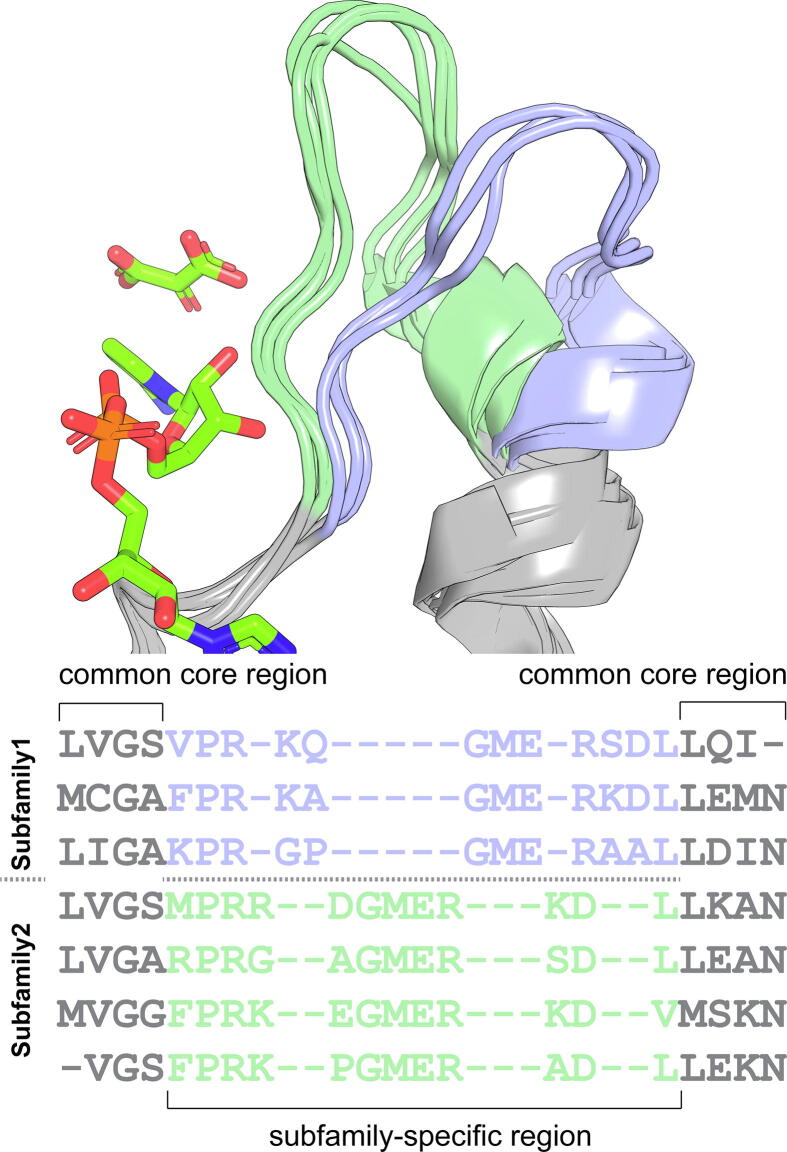
Fig. 1.
Scheme of a subfamily-specific region in 3D-structures of homologs. 3D-alignment (including selected crystallographic ligands) and its sequence representation are shown.
2.1. Input and output
The input to the program is a 3D-alignment of multiple protein structures provided as (1) a folder with superimposed PDB entries presented as separate files: if opened all at once in PyMol, the viewport should reveal a biologically meaningful 3D-superimposition; (2) a file with the corresponding sequence representation of 3D-alignment in FASTA format. SSR is a characteristic of consecutive “columns” in a 3D-alignment that is independent of spatial neighborhood. Thus, to simplify preparation of input data, the Zebra3D analysis is offered on a one-chain-at-a-time basis, i.e. separate chains of heteromeric proteins should be evaluated as independent tasks. The program itself does not impose limitations on the number of input PDB structures or their dimensions. The required input can be prepared by the user to meet the research objective and submitted to local installation of Zebra3D. Alternatively, a 3D-alignment of up to 128 diverse proteins can be automatically collected, constructed and then analyzed by Zebra3D with default settings fully on-line using the Mustguseal web-server [39]. The details are provided below.
The collection and subsequent 3D-alignment of diverse homologs can be handled fully on-line by the Mustguseal web-server [39]. To take advantage of that web-method, submit a PDB code of the query protein in the “Mode 4” (https://mustguseal.belozersky.msu.ru#mode=4). In this mode, only the first and second steps of the Mustguseal protocol will be carried out (i.e. 3D-similarity search and multiple 3D-alignment). By default, 3D-structure similarity matching of the query protein versus the PDB database is performed by the SSM algorithm with a general-purpose “70%–70%” thresholds [51] to collect evolutionarily distant proteins from functionally diverse families, yet similar enough for alignment, as recently discussed [52]. This step is followed by automatic selection of a non-redundant subset of no more than 16–128 PDB entries (the default value is 64) at the 95–40% pairwise sequence identity threshold [39], and finally concluded by a multiple 3D-alignment using parMATT [24]. The Mustguseal’s “Analysis” page is available on-line, to provide means for expert evaluation of the overall quality of the produced superimposition. That tool offers basic alignment statistics (e.g. alignment coverage and column conservation) and implements interactive sequence- and structure- manipulation tools. The automatically constructed 3D-alignment is further subjected to the Zebra3D bioinformatic analysis with default settings on-line. The user can download both the automatically prepared 3D-alignment and the default Zebra3D output from the Mustguseal web-server. Zebra3D results can be analyzed straightaway on a local computer. The superimposed 3D-entries and corresponding sequence representation of alignment in FASTA format can be used as input to a local installation of Zebra3D to further improve bioinformatic analysis by fine-tuning the parameters. The Mustguseal protocol [39], a step-by-step practical guide to its use and parameter selection [52], as well as a discussion of various case-studies [14], [41], [53], [54], [55] are available in our recent publications. An illustrated user guide for the input preparation is available on-line at https://biokinet.belozersky.msu.ru/Zebra3D-input.
Advanced users can manually prepare the input to Zebra3D by aligning a curated collection of protein 3D-entries with a tool of their choice. In particular, we have recently introduced parMATT [24] – the first parallel re-implementation of the highly successful MATT algorithm [35], [36], [37] to align large collections of protein 3D-structures by running on distributed-memory systems, i.e. computing clusters and supercomputers hosting memory-independent computing nodes. As previously noted, the primary focus of Zebra3D is on significantly different organization of local 3D-structure among members of different families/subfamilies (i.e. 3D-specificity). Thus, the use of redundancy filter when preparing a 3D-alignment for Zebra3D is generally recommended, and should be set at 40% pairwise sequence similarity or higher, as previously justified in the Mustguseal pipeline [39], [52]. The filter aims to reduce bias and computational complexity of Zebra3D analysis by excluding redundant proteins that belong to same subfamilies and therefore are likely to have low 3D-structural diversity. In principle, Zebra3D algorithm can also be used to assess conformational plasticity in 3D-structures, e.g. by analyzing molecular dynamics snapshots or PDB entries of the same or closely related proteins. In that case, the redundancy filter should not be used when preparing the input 3D-alignment.
Local installation of Zebra3D is straightforward and does not require significant investment of time from the user, as explained in the on-line manual available at https://biokinet.belozersky.msu.ru/Zebra3D-installation. The Zebra3D software logs all its activities to standard output stream. Users are advised to always check this log for warnings and errors. In particular, if input validation or further data analysis fail, the program will attempt to explain the problem in this log. If successful, plain text files and binary 3D-annonations with convenient visual representation of results will be created, describing location of each SSR in structures of homologs and predicted subfamilies (i.e. classification of protein fragments into clusters). Construction of 3D-annotations requires a local installation of the PyMol Molecular Graphics System. Illustrated guide to Zebra3D results is available on-line at https://biokinet.belozersky.msu.ru/Zebra3D-output.
2.2. Case-studies
To provide diverse examples of application, the Zebra3D + Mustguseal combined approach was used in ten case-studies of protein superfamilies to select SSRs previously shown to determine diverse properties in otherwise structurally similar ligand binding sites. All input alignments for the discussed case-studies were created automatically by the Mustguseal web-server with default settings, followed by on-line Zebra3D bioinformatic analysis with default settings. Proteins that represented families/superfamilies of interest were submitted as queries (Table 1). The automatic construction and analysis of each multiple 3D-alignment took at most 35 min. The results are summarized in Table1 and available on-line at https://biokinet.belozersky.msu.ru/Zebra3D-examples. The previously established functionally important subfamily-specific regions were ranked among the top-five hits in 11 cases out of 15. The ranking was improved even further by excluding SSRs formed by N-/C-terminal regions, which are generally known to be inaccurate in PDB records due to limitations of experimental methods and may contain very mobile residues not related to a function (Table 1). The estimated Z-scores/P-values in all cases indicated that the results were statistically significant at the widely used 0.05 threshold. The geometry-based subfamily classification of protein 3D-fragments in the selected SSRs was in agreement with functional annotation of the respective PDB records and provided clues as to interpretation of the results.
Table 1.
Functionally important SSRs selected by Zebra3D analysis of the case-studies.
| # | Query |
# of PDBs in aln. | # of SSRs | SSR |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PDB | Region | Rank | Z | P | Functional role | Interpretation of subfamily classification | |||
| 1 | H1N1 Neuraminidase | 3B7E:A | 29 | 19 (17) | 427–440 | 3 (1) | 6.00 | 9.6e−10 | Flexible loop-430 that drives formation of the cavity-430 | Neuraminidases from influenza strains/types with different pathogenicity |
| 136–156 | 10 (8) | 2.32 | 1.0e−02 | Flexible loop-150 that drives formation of the cavity-150 | Neuraminidases from influenza strains/types with different pathogenicity | |||||
| 2 | Ornithine Decarboxylase from Trypanosoma brucei | 1F3T:A | 31 | 23 (21) | 322–336 | 8 (6) | 2.37 | 8.8e−03 | The 310-helix shaping the binding cavity to discriminate substrate preference | β/α-barrel-fold basic amino acid decarboxylases with different substrate specificity |
| 3 | Human Aldose Reductase | 2ACQ:A | 62 | 14 (12) | 112–135 | 1 (1) | 10.11 | 2.5e−24 | Dynamic region involved in substrate binding | Aldo-keto reductases with different substrate specificity |
| 4 | Common ancestor of Haloalkane Dehalogenases and Renilla Luciferase | 6G75:A | 41 | 14 (13) | 146–176 | 3 (3) | 3.64 | 1.4e−04 | Region that includes L9-α4 loop-helix fragment involved in substrate binding | α/β-hydrolases with different catalytic activity |
| 222–239 | 5 (4) | 2.53 | 5.7e−03 | Region that includes L14 loop involved in substrate binding | α/β-hydrolases with different catalytic activity | |||||
| 5 | Polyester Hydrolase from Pseudomonas aestusnigri | 6SBN:A | 20 | 13 (11) | 127–134 | 5 (3) | 3.84 | 6.1e−05 | Substrate binding element of the active site | PET-hydrolases and closely related cutinases versus non-PET-hydrolase enzymes |
| 97–103 | 8 (6) | 1.66 | 4.8e−02 | Substrate binding element of the active site | PET-hydrolases and closely related cutinases versus non-PET-hydrolase enzymes | |||||
| 6 | Human Guanine Deaminase | 2UZ9:A | 23 | 22 (20) | 215–222 | 7 (5) | 3.32 | 4.5e−04 | Substrate-recognition element | Metal-dependent hydrolases with different substrate specificity |
| 7 | Human p38α MAP Kinase | 1R3C:A | 61 | 11 (10) | 169–185 | 3 (2) | 9.90 | 2.1e−23 | Kinase DFG motif and activation loop | PDB entries capturing different structural states of the activation loop |
| 8 | Human HSP90 | 1YET:A | 19 | 9 (7) | 107–136 | 3 (2) | 2.52 | 5.8e−03 | Flexible lid segment | PDB entries capturing different structural states of the lid segment |
| 9 | Malate Dehydrogenase from Sus scrofa | 5MDH:A | 60 | 21 (21) | 89–100 | 2 (2) | 3.83 | 6.5e−05 | Mobile region hosting residues involved in catalytic and binding functions | PDB entries capturing different structural states of the mobile region |
| 10 | Zinc Metallo-Beta-Lactamase from Bacillus cereus | 1BVT:A | 40 | 13 (12) | 30–39 | 3 (3) | 8.13 | 2.2e−16 | Functionally important L1 loop of the active site | Different classes/types of metallo-beta-lactamases |
| 181–184 | 4 (4) | 7.75 | 4.5e−15 | Part of functionally important L3 loop of the active site | Different classes/types of metallo-beta-lactamases | |||||
| 170–179 | 5 (5) | 6.42 | 6.8e−11 | Part of functionally important L3 loop of the active site | Different classes/types of metallo-beta-lactamases | |||||
“Query PDB” indicates PDB code and chain ID that was submitted as a query to the Mustguseal web-server to automatically collect and align a non-redundant set of 3D-structures of homologs. “# of PDBs in aln.” indicates the total number of finally selected 3D-entries in such alignment. “# of SSRs” indicates the total number of subfamily-specific regions identified by Zebra3D with default settings. “Region”, “Rank”, “Z”, and “P” indicate the first and last amino acid residues in SSR (according to numbering of query PDB record), its rank, statistical significance Z-score and P-value, respectively. The total number of SSRs and the ranking after excluding SSRs with N-/C-terminal regions (i.e. by running with the “exclude_ncterm = 5” parameter, see section “Algorithm”) is shown in parenthesis in the fields “# of SSRs” and “Rank”.
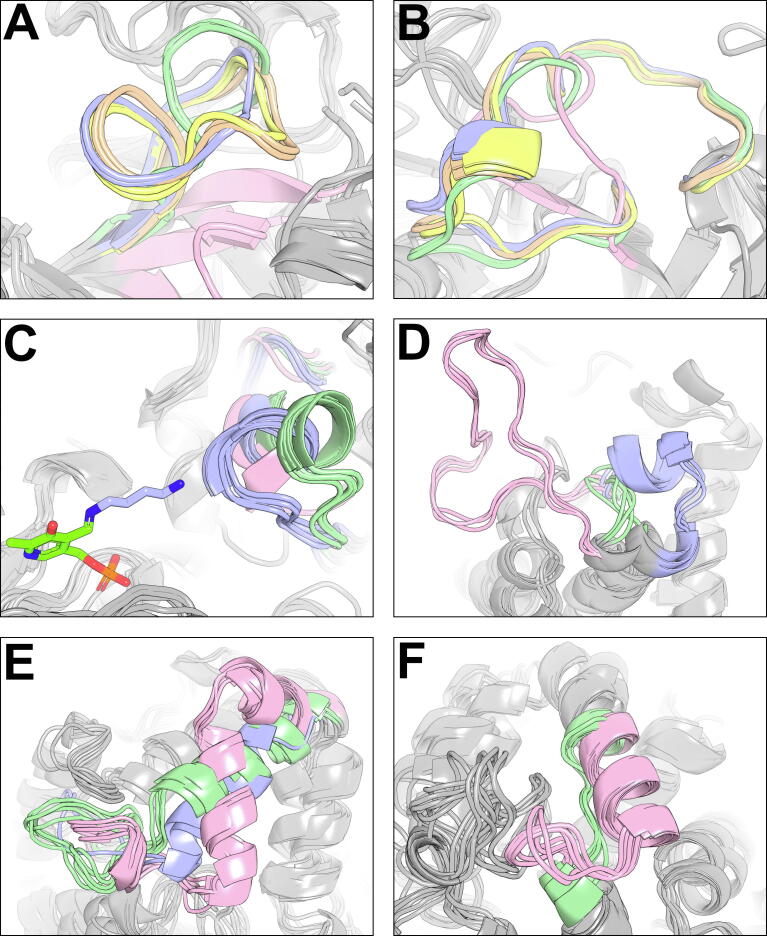
In the first case-study, Zebra3D was used to assess 3D-alignment of GH34 Neuraminidases, i.e. a family of pathogenicity enzymes from viruses. The SSR #3 (out of 19) was previously established as the flexible “loop-430” that drives formation of the “cavity-430” in active sites of prominent Influenza A virus Neuraminidases [56], [57], [58] (Fig. 2, A). The automatically proposed subfamily classification underlined the significant difference in size and configuration of the loop and the adjacent cavity among homologs. The N1-N9 Influenza A virus Neuraminidases were automatically classified into three subfamilies in agreement with their phylogenetic classification [59]. The N10 and N11 Neuraminidases from bats, which are devoid of Neuraminidase activity [60], as well as homologs from a less pathogenic Influenza B virus were assigned to another two subfamilies; in both cases, the “cavity-430” was either absent or significantly reduced, respectively. Previous studies suggested that the “cavity-430” may be one of intermediate binding sites for substrate on its way to the catalytic center, making it a promising drug target within active sites of N1-N9 Neuraminidases [61], [62]. The SSR #10 in the same 3D-alignment represented another prominent flexible “loop-150” driving the formation of the “cavity-150” in the active site of Neuraminidases [63], [64] (Fig. 2, B). In the second case-study, a 3D-alignment of Pyridoxal-Dependent β/α-Barrel-Fold Basic Amino Acid Decarboxylases were subjected to Zebra3D. The SSR #8 (out of 23) represented a previously established 310-helix that was located on one side of the substrate-binding cavity and implemented alternative orientations in subfamilies corresponding to enzymes with different substrate preference (Fig. 2, C). In particular, Ornithine Decarboxylases (colored blue in Fig. 2, C) bind a relatively short ligand (L-ornithine, showed as sticks colored blue in a covalent complex with the cofactor, colored chartreuse). Homologous Diaminopimelate Decarboxylases (colored green in Fig. 2, C) can accommodate ligands of a larger size due to a displacement of the 310-helix which frees up additional space at the binding site. The dimensions of the binding cavity between the PLP cofactor and the SSR #8 seem to serve as the key structural factor for discrimination of substrate preference in these homologs [65], [66]. In the third case-study, the SSR #1 (out of 14) was identified in the Aldo-Keto Reductase superfamily. It represented a region positioned at the top of the canonical (α/β)8-barrel structure that was significantly different between homologs with diverse substrate specificity (Fig. 2, D). Introduction of the corresponding flexible loop from Human Aldose Reductase (colored pink in Fig. 2, D) instead of the corresponding fragment in structure of hyperthermostable Alcohol Dehydrogenase D from Pyrococcus furiosus (which was equivalent to those colored green in Fig. 2, D) was one of the key steps in creation of a chimera that implemented substrate specificity of the donor enzyme and inherited thermostability of the parent enzyme [67]. In the fourth case-study, the identified SSR #3 and SSR #5 (out of 14) included L9-α4 loop-helix fragment (Fig. 2, E) and L14 loop (Fig. 2, F) in structure of Luciferase from Renilla reniformis that were spatially different in selected homologs from the α/β-hydrolases superfamily. A recently conducted multidisciplinary analysis revealed a crucial role of these dynamic regions in enzyme catalysis. They directly affected the opening/closing of the access tunnel that connected buried active site to the surrounding solvent, regulated the size of the active site cavity, and were involved in substrate/product binding [68]. Introduction of random insertions/deletions within these two regions in structure of thermostable ancestor of Haloalkane Dehalogenase and Renilla Luciferase (PDB code 6G75) by directed evolution led to 100-fold increase in luciferase activity. Further exchange of the L9-α4 region between the highly efficient modern luciferase and the poorly active ancestral enzyme produced a chimera characterized by a 7000-fold increase in catalytic efficiency [68]. In brief, another six case-studies of Polyester Hydrolase from Pseudomonas aestusnigri, Human Guanine Deaminase, p38α MAP Kinase and HSP90, Malate Dehydrogenase from Sus scrofa, and Zinc metallo-beta-lactamase from Bacillus cereus and their homologs by Zebra3D identified SSRs that were previously established as key substrate-recognition elements in the active sites of these enzymes (Table1) [69], [70], [71], [72], [73], [74], [75].
Fig. 2.
Illustrations of selected SSRs in the case-studies of (A) and (B) H1N1 Neuraminidase, (C) Ornithine Decarboxylase from Trypanosoma brucei, (D) Human Aldose Reductase, (E) and (F) Dehalogenase/Luciferase; see explanations in the text. Each figure represents one SSR in a 3D-alignment of homologs; protein fragments belonging to an SSR are colored according to the automatically proposed subfamily classification. For each subfamily, at most few representative configurations (i.e. protein structures) are shown, for clarity. The figures were prepared from the automatically generated 3D-annotation files using PyMol.
Finally, it can be noted that Zebra3D is not a competitor to sequence-based strategies to identify SSPs/SDPs. The output of Zebra3D and Zebra2 [14] was qualitatively different even when the same alignments from the case-studies were used as input. This was due to the fact that the two bioinformatic tools are focused on mutually exclusive parts of the alignment: sequence-based methods assess specificity in structurally similar regions (i.e. columns with a low content of gaps [11], [13], [14]) while 3D-specificity is a characteristic of regions that feature high structural diversity usually leading to a high content of gaps (Fig. 1).
3. Conclusions
Systematic bioinformatic analysis of protein superfamilies can provide mechanistic insights into the framework of a protein function and diversity, but so far has been limited to protein sequences. As a result, our understanding of how spatial arrangement of amino acid residues and regions in homologs affect their biological function remains incomplete and requires further attention. The Zebra3D + Mustguseal combined approach brings together an advanced bioinformatic engine for 3D-alignment of proteins, well-established algorithms for machine-learning 3D-cluster analysis, and a general-purpose statistical model that attempts to discriminate 3D-specificity among functionally diverse homologs from the average structural plasticity/flexibility not associated with a function. This combined approach offers an easy-to-use both on-line and standalone tool to study 3D-structural diversity in protein families systematically, thus taking advantage of the growing availability of 3D-data. The identified subfamily-specific regions are automatically prioritized according to spatial consistency of subfamilies and their dimensions, adjusted for the average structural plasticity/flexibility. The value of SSRs selected by the bioinformatic analysis can be assessed by an expert (similar to SSPs/SDPs, e.g. [54]), evaluated by directed evolution [68] or by exchange of equivalent regions between homologs with different properties [76], or can be further studied by molecular modelling to reveal the mechanisms of their involvement in a protein action [77], [78]. As we illustrated by the case-studies, Zebra3D can be used to identify 3D-determinants of protein functional diversity within a shared superfamily fold, assist protein engineering by loop re-design, and help to annotate sites/subsites characterized by an above average plasticity to facilitate accommodation of ligands. It is important to note that assessment of 3D-based specificity does not replace, but rather complements the previously developed sequence-based SSPs/SDPs-detection strategies. We believe that a combination of bioinformatic methods should be used to systematically study diverse protein superfamilies both at the sequence level and at the level of three-dimensional structure organization. The new Zebra3D software complements our family of bioinformatic methods [52] that is being built around the Mustguseal protein alignment web-engine [39] and already includes sequence-based Zebra2/pocketZebra web-tools [14], [79] to identify subfamily-specific determinants of functional diversity in protein sequences. We hope that a symbiosis of these tools will help to decipher the structure–function relationship, leading to the development of improved strategies for protein design and drug discovery [1], [4], [7], [8], [15], [16], [80], [81].
4. Algorithm
Zebra3D analysis is carried out at the 3D-level of protein backbone, i.e. only heavy backbone atoms (C, Cα, N, and O) are considered in each position and amino acid types and side-chain atoms are disregarded. Backbone atoms are defined by the exact XYZ coordinates derived from the input PDB files and further processed “as is”, in the same way for all regions and proteins, e.g. neither re-weighing of relative structural discrepancies nor correction for solvent-exposed loop regions is applied. The correspondence between positions (i.e. the alignment itself) is taken from the sequence representation (i.e. FASTA input file) as this information cannot be unambiguously recovered from a 3D-coordinate superimposition alone. The algorithm has three main steps: (1) selection of “common core” and “variable” regions in the 3D-alignment; (2) classification of “variable” regions to reveal the subfamily-specific pattern; (3) statistical analysis to evaluate significance of the discovered 3D-specificity. The algorithm details are further provided below.
In the first step, “common core” regions are selected as “columns” of 3D-alignment that contain low content of gaps in the sequence version, and low content of spatially misaligned residues in the 3D-superimposition. It is typical for a 3D-alignment software (e.g. MATT/parMATT [24], [37]) to assign spatially displaced residues into one column of sequence version of the output, making them appear as “aligned”; e.g. this can happen if the corresponding positions are located in alphabetically similar and/or gap-less regions. While this is usually beneficial when such sequence representation is considered for further analysis, it is not appropriate in the context of this study, as a shift in spatial arrangement of even identical amino acids/regions in homologs can significantly affect their function. Detection of misaligned residues in a 3D-aligment is not straightforward since protein-specific 3D-structural diversity/plasticity/fluctuation has to be taken into account, what presents a challenge. A commonly used solution would be to apply a hard cut-off, e.g. 5 Å [37]. Such hard cut-off approach generally led to poor performance of the Zebra3D algorithm in most case-studies (e.g. resulting in a significantly larger length of identified SSRs and lower ranking of the known functionally important regions). To improve the accuracy of Zebra3D, an alternative approach was developed that selects a threshold value specific to input alignment. By default, pairwise RMSD values are calculated between amino acid residues in columns of sequence version of the alignment with at most 5% of gaps. The largest value (i.e. “diameter”) is collected in each column. The diameter values are sorted in ascending order and plotted on the y-axis, and the corresponding column IDs are plotted on the x-axis. This plot is subjected to the commonly used “elbow method” heuristic to automatically detected its elbow/knee, similarly to what has been recently discussed [82]. Such bending point indicates the most significant change in ascending trend of RMSD metric. Assuming that the input superimposition of proteins contains both well-aligned regions with small diameters and mismatching (i.e. poorly/not-aligned) regions with large diameters (i.e. what would guarantee that such elbow exists), this automatically selected cut-off value is considered as a threshold to discriminate amino acid residues that appear to be spatially equivalent from those that are not. This threshold is further used to calculate the frequency of 3D-misaligned residues in each column, as follows. If the largest pairwise RMSD value in a column is above the cut-off, the amino acid residue with the largest sum of all pairwise RMSD values with other residues is considered as 3D-misaligned and dismissed from further consideration. This process iterates until all pairwise RMSD values between the remaining residues are below the threshold. Finally, columns that contain at most 5% of 3D-misaligned residues and at most 5% of gaps in the sequence representation are selected as the “common core”.
In the second step, “variable” regions located in-between the selected “common core” regions are subjected to machine learning to reveal whether they implement the 3D-specificity pattern, i.e. feature fragments of local structure that can be classified into spatially equivalent subfamilies/clusters. In each “variable” region, RMSD between every two protein fragments is calculated. If the corresponding segments have unequal lengths, the smaller one is matched with 103 randomly chosen subfragments of the same length within the larger one, and the respective values are averaged. The obtained distance matrix is further subjected to a machine learning cluster analysis technique. By default, the fully automated HDBSCAN algorithm from the “hdbscan” Python3 clustering library is used to produce “thicker” clusters and minimize the amount of outliers (i.e. unique/rare orientations), thus preserving as much data as possible for further expert analysis [83]. Two alternative methods can be switched on for a particular purpose. The OPTICS from the “scikit-learn” library is a fully automatic technique that tends to produce spatially more consistent (compact) but “thinner” clusters at the cost of data loss by throwing out a larger number of proteins as outliers [84]. Finally, the DBSCAN from the “scikit-learn” library is a curated technique dependent on the ‘eps’ parameter that can be manually calibrated to meet the particular research objective [85]. The minimal size of a cluster (i.e., minimal number of proteins in a subfamily), that is passed to either of the three algorithms, is automatically set to 10% of the total number of PDB entries in the input alignment, but not less than 2 proteins. Selection of default value for this parameter was based on the equivalent parameter in ideologically close sequence-based Zebra2 tool [14]. For a particular purpose, users can establish filtering rules to limit the length of “variable” regions (unlimited, by default), limit the number of outliers (unlimited, by default), or to specifically evaluate one manually defined region of a 3D-alignment. The user can also choose to exclude SSRs that contain N-/C-terminal regions of protein structures assigned to subfamilies (e.g. “exclude_ncterm=5” will dismiss SSRs containing the first five and the last five residues of any PDB entry included into the proposed subfamily classification). By default, N-/C-terminal regions are included in the analysis to preserve as much data as possible for further expert review.
The finally selected SSRs are ranked in descending order of the estimated specificity S-scores and statistical significance Z-scores (i.e. ranking by any of the two metrics has a one-to-one correspondence). The most visually prominent and statistically significant hits are shown first. The specificity score for each i-th SSR is calculated as
The silhouette-score for i-th SSR is a metric of how similar each structural fragment is to its own subfamily/cluster compared to other subfamilies/clusters [86], but does not explicitly take into account how far apart the subfamilies/clusters are from each other. The diameter of the i-th SSR is the largest average RMSD value between any two subfamilies/clusters, excluding outliers, and is implemented in the formula to consider spatial scale of the region. Since the two metrics are originally calculated on different scales (i.e. silhouette-scores take values within a range [−1; 1] and diameters are measured in angstroms), the raw values of and are standardized to and that each fits to the common range [0; 1], according to the formulas above. The respective standardization coefficients (i.e. and , and ) are selected over all SSRs “observed” in the input alignment and all “random” SSRs, as explained below. The finally calculated specificity scores take values in a range [0; 1], with larger values indicating more compact and spatially distant subfamilies/clusters.
The purpose of statistical analysis in predictive bioinformatics is to discriminate between significant and insignificant hits, given a specific context. In the case of Zebra3D, it is necessary to develop such a universal model that would be able to prioritize functionally significant 3D-variability observed between homologs and filter out functionally insignificant 3D-structural divergence which is due to random fluctuations in protein structures. This is a difficult task, as our understanding of the relationship between structure and function remains incomplete despite recent progress in the study of protein superfamilies and structural plasticity. There are at least are two main databases of protein conformational diversity: PDBFlex explores the intrinsic flexibility of protein structures by analyzing structural variations between different depositions and chains in asymmetric units of the same protein in PDB [87]; another useful resource is CoDNaS [31]. Based on the information contained in these bioinformatic resources and obtained from molecular modeling [28] it can be concluded that most parts of protein 3D-structures fluctuate at least to some degree. In majority of cases, there is no evidence to directly link this conformational plasticity to a function. Those cases where such evidence was obtained from experiments and modeling usually correspond to structural rearrangements with the largest amplitude, i.e. characterized by a difference in RMSD between conformations above the average [43], [87], [88]. Thus, for Zebra3D a general-purpose statistical model was developed based on the assumption that an average level of conformational flexibility in a region of a protein structure is unlikely to have a direct implication to a function. One hundred randomly selected sets were collected from the PDBFlex database, containing at least 20 PDB entries in each set, representing diverse examples of protein structural fluctuations. The finally collected sets contained 26–515 PDB entries per set, with the median value of 59 entries per set. Each set contained PDB structure snapshot of the same protein (e.g. 325 PDB entries corresponding to Human p38α MAP Kinase) and was subjected to 3D-alignment using parMATT [24]. Sets that featured obvious global structural rearrangements (e.g. domain movement) were dismissed. The remaining sets were submitted to Zebra3D to identify SSRs. The SSRs that included C/N-terminal regions or incomplete/missing protein fragments, or that were larger than 10% of the average protein length, or produced more than 40% outliers, were dismissed as undescriptive. In each set, one SSR with the median value of specificity score (i.e. the , , and standardization coefficients were acquired within the set) was selected and further considered as “random”, i.e. to be a result of random fluctuations in the protein structure. The median specificity was considered as a characteristic of “random” plasticity of each protein instead of the maximum value, since the largest and most noticeable fluctuations can actually correspond to functionally significant conformational rearrangements (e.g. movement of “activation loop” captured in different PDB entries of Human p38α MAP Kinase), as explained above. The selected regions were manually reviewed to the best of our ability to ensure that they do not correspond to previously established functionally important structural elements. The respective values of and scores of the “random” SSRs are stored in the Zebra3D source code to calculate standardization coefficients (i.e. and , and ) of specificity S-score during the “production” run (i.e. when the analysis of user data actually takes place). Assuming standard normal distribution of specificity metric, the respective σ and µ values of “random” SSRs are finally used to estimate statistical significance Z-scores and corresponding P-values of the “observed” SSRs. For a particular purpose, the user can provide proprietary statistical model for Zebra3D analysis by substituting the default and scores within the distribution package for manually selected values, as described in on-line tutorial available at https://biokinet.belozersky.msu.ru/Zebra3D-statistical-model.
CRediT authorship contribution statement
Daria Timonina: Conceptualization, Methodology, Software, Writing - original draft. Yana Sharapova: Validation, Investigation, Writing - original draft. Vytas Švedas: Conceptualization, Funding acquisition, Writing - original draft. Dmitry Suplatov: Supervision, Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Russian Foundation for Basic Research according to the research project [18-29-13060]. The use of HPC computing resources at the Lomonosov Moscow State University to accelerate the evaluation of Zebra3D during its development is acknowledged [89].
References
- 1.Sequeiros-Borja C.E., Surpeta B., Brezovsky J. Recent advances in user-friendly computational tools to engineer protein function. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang A., Troup M., Ho J.W. Scalability and validation of big data bioinformatics software. Comput Struct Biotechnol J. 2017;15:379–386. doi: 10.1016/j.csbj.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suplatov D., Kirilin E., Švedas V. Bioinformatic analysis of protein families to select function-related variable positions. In: Svendsen A., editor. Understanding Enzymes: Function, Design, Engineering, and Analysis. Pan Stanford Publishing; Singapore: 2016. pp. 351–385. [Google Scholar]
- 4.Pleiss J. Systematic analysis of large enzyme families: identification of specificity-and selectivity-determining hotspots. ChemCatChem. 2014;6(4):944–950. [Google Scholar]
- 5.De Juan D., Pazos F., Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14(4):249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 6.Kowarsch A., Fuchs A., Frishman D., Pagel P. Correlated mutations: a hallmark of phenotypic amino acid substitutions. PLoS Comput Biol. 2010;6(9) doi: 10.1371/journal.pcbi.1000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chagoyen M., García-Martín J.A., Pazos F. Practical analysis of specificity-determining residues in protein families. Brief Bioinform. 2016;17(2):255–261. doi: 10.1093/bib/bbv045. [DOI] [PubMed] [Google Scholar]
- 8.Suplatov D., Voevodin V., Švedas V. Robust enzyme design: Bioinformatic tools for improved protein stability. Biotechnol J. 2015;10(3):344–355. doi: 10.1002/biot.201400150. [DOI] [PubMed] [Google Scholar]
- 9.Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 10.Casari G., Sander C., Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995;2(2):171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- 11.Mirny L.A., Gelfand M.S. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002;321(1):7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 12.Karasev D., Sobolev B., Lagunin A., Filimonov D., Poroikov V. Prediction of Protein-Ligand Interaction Based on the Positional Similarity Scores Derived from Amino Acid Sequences. Int J Mol Sci. 2020;21(1):24. doi: 10.3390/ijms21010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalinina O.V., Mironov A.A., Gelfand M.S., Rakhmaninova A.B. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004;13(2):443–456. doi: 10.1110/ps.03191704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suplatov D., Sharapova Y., Geraseva E., Švedas V. Zebra2: advanced and easy-to-use web-server for bioinformatic analysis of subfamily-specific and conserved positions in diverse protein superfamilies. Nucleic Acids Res. 2020;48(W1):W65–W71. doi: 10.1093/nar/gkaa276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero-Rivera A., Garcia-Borràs M., Osuna S. Computational tools for the evaluation of laboratory-engineered biocatalysts. Chem Commun (Camb) 2017;53(2):284–297. doi: 10.1039/c6cc06055b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damborsky J., Brezovsky J. Computational tools for designing and engineering enzymes. Curr Opin Chem Biol. 2014;19:8–16. doi: 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Grahame D.S., Dupuis J.H., Bryksa B.C., Tanaka T., Yada R.Y. Comparative Bioinformatic and Structural Analyses of Pepsin and Renin. Enzyme Microb Technol. 2020;141 doi: 10.1016/j.enzmictec.2020.109632. [DOI] [PubMed] [Google Scholar]
- 18.Cao T.P., Choi J.M., Kim S.W., Lee S.H. The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MP T. J Microbiol. 2018;56(4):246–254. doi: 10.1007/s12275-018-7483-y. [DOI] [PubMed] [Google Scholar]
- 19.Popinako A., Antonov M., Tikhonov A., Tikhonova T., Popov V. Structural adaptations of octaheme nitrite reductases from haloalkaliphilic Thioalkalivibrio bacteria to alkaline pH and high salinity. PLoS ONE. 2017;12(5) doi: 10.1371/journal.pone.0177392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demming R.M., Hammer S.C., Nestl B.M., Gergel S., Fademrecht S., Pleiss J. Asymmetric enzymatic hydration of unactivated, aliphatic alkenes. Angew Chem. 2019;131(1):179–183. doi: 10.1002/anie.201810005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saroj Devi N., Shanmugam R., Ghorai J., Ramanan M., Anbarasan P., Doble M. Ligand-based modeling for the prediction of pharmacophore features for multi-targeted inhibition of the arachidonic acid cascade. Mol Inform. 2018;37(3):1700073. doi: 10.1002/minf.201700073. [DOI] [PubMed] [Google Scholar]
- 22.Li J., Koehl P. 3D representations of amino acids—applications to protein sequence comparison and classification. Comput Struct Biotechnol J. 2014;11(18):47–58. doi: 10.1016/j.csbj.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L. RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shegay M.V., Suplatov D.A., Popova N.N., Švedas V.K., Voevodin V.V. parMATT: parallel multiple alignment of protein 3D-structures with translations and twists for distributed-memory systems. Bioinformatics. 2019;35(21):4456–4458. doi: 10.1093/bioinformatics/btz224. [DOI] [PubMed] [Google Scholar]
- 25.Maria-Solano M.A., Serrano-Hervás E., Romero-Rivera A., Iglesias-Fernández J., Osuna S. Role of conformational dynamics in the evolution of novel enzyme function. Chem Commun. 2018;54(50):6622–6634. doi: 10.1039/c8cc02426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei G., Xi W., Nussinov R., Ma B. Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem Rev. 2016;116(11):6516–6551. doi: 10.1021/acs.chemrev.5b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monzon A.M., Zea D.J., Fornasari M.S., Saldaño T.E., Fernandez-Alberti S., Tosatto S.C. Conformational diversity analysis reveals three functional mechanisms in proteins. PLoS Comput Biol. 2017;13(2) doi: 10.1371/journal.pcbi.1005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suplatov D., Sharapova Y., Švedas V. EasyAmber: A comprehensive toolbox to automate the molecular dynamics simulation of proteins. J Bioinform Comput Biol. 2020;18(6):2040011. doi: 10.1142/S0219720020400119. [DOI] [PubMed] [Google Scholar]
- 29.Ganesan A., Coote M.L., Barakat K. Molecular dynamics-driven drug discovery: leaping forward with confidence. Drug Discovery Today. 2017;22(2):249–269. doi: 10.1016/j.drudis.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Monzon A.M., Zea D.J., Marino-Buslje C., Parisi G. Homology modeling in a dynamical world. Protein Sci. 2017;26(11):2195–2206. doi: 10.1002/pro.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monzon AM, Rohr CO, Fornasari MS, Parisi G (2016) CoDNaS 2.0: a comprehensive database of protein conformational diversity in the native state. Database, 2016. [DOI] [PMC free article] [PubMed]
- 32.Bakan A., Meireles L.M., Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Akdel M., Durairaj J., deRidder D., van Dijk A.D. Caretta-A Multiple Protein Structure Alignment and Feature Extraction Suite. Comput Struct Biotechnol J. 2020;18:981–992. doi: 10.1016/j.csbj.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpentier M., Chomilier J. Protein multiple alignments: sequence-based versus structure-based programs. Bioinformatics. 2019;35(20):3970–3980. doi: 10.1093/bioinformatics/btz236. [DOI] [PubMed] [Google Scholar]
- 36.Kalaimathy S., Sowdhamini R., Kanagarajadurai K. Critical assessment of structure-based sequence alignment methods at distant relationships. Brief Bioinform. 2011;12(2):163–175. doi: 10.1093/bib/bbq025. [DOI] [PubMed] [Google Scholar]
- 37.Menke M., Berger B., Cowen L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput Biol. 2008;4(1) doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozewicki J., Li S., Amada K.M., Standley D.M., Katoh K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 2019;47(W1):W5–W10. doi: 10.1093/nar/gkz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suplatov D.A., Kopylov K.E., Popova N.N., Voevodin V.V., Švedas V.K. Mustguseal: a server for multiple structure-guided sequence alignment of protein families. Bioinformatics. 2018;34(9):1583–1585. doi: 10.1093/bioinformatics/btx831. [DOI] [PubMed] [Google Scholar]
- 40.Pei J., Kim B.H., Grishin N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suplatov D., Timonina D., Sharapova Y., Švedas V. Yosshi: a web-server for disulfide engineering by bioinformatic analysis of diverse protein families. Nucleic Acids Res. 2019;47(W1):W308–W314. doi: 10.1093/nar/gkz385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumbalova L., Stourac J., Martinek T., Bednar D., Damborsky J. HotSpot Wizard 3.0: web server for automated design of mutations and smart libraries based on sequence input information. Nucleic Acids Res. 2018;46(W1):W356–W362. doi: 10.1093/nar/gky417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaillard T., Stote R.H., Dejaegere A. PSSweb: protein structural statistics web server. Nucleic Acids Res. 2016;44(W1):W401–W405. doi: 10.1093/nar/gkw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro V.S., Santana C.A., Fassio A.V., Cerqueira F.R., da Silveira C.H., Romanelli J.P. visGReMLIN: graph mining-based detection and visualization of conserved motifs at 3D protein-ligand interface at the atomic level. BMC Bioinf. 2020;21(2):1–12. doi: 10.1186/s12859-020-3347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He W., Liang Z., Teng M., Niu L. Lib ME—automatic extraction of 3D ligand-binding motifs for mechanistic analysis of protein–ligand recognition. FEBS Open Bio. 2016;6(12):1331–1340. doi: 10.1002/2211-5463.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilmeier J.P., Meng E.C., Polacco B.J., Babbitt P.C. 3D Motifs. In: Rigden D.J., editor. From Protein Structure to Function with Bioinformatics. Springer; Dordrecht: 2017. pp. 361–392. [Google Scholar]
- 47.Nadzirin N., Gardiner E.J., Willett P., Artymiuk P.J., Firdaus-Raih M. SPRITE and ASSAM: web servers for side chain 3D-motif searching in protein structures. Nucleic Acids Res. 2012;40(W1):W380–W386. doi: 10.1093/nar/gks401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanisenko V.A., Pintus S.S., Grigorovich D.A., Kolchanov N.A. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;40(suppl_1):D183–D187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drew E.D., Janes R.W. 2StrucCompare: a webserver for visualizing small but noteworthy differences between protein tertiary structures through interrogation of the secondary structure content. Nucleic Acids Res. 2019;47(W1):W477–W481. doi: 10.1093/nar/gkz456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durairaj J., Akdel M., de Ridder D., van Dijk A.D. Geometricus represents protein structures as shape-mers derived from moment invariants. Bioinformatics. 2020;36(Supplement_2):i718–i725. doi: 10.1093/bioinformatics/btaa839. [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 52.Suplatov D, Sharapova Y, Švedas V (2021) Mustguseal and Sister Web-methods: a Practical Guide to Bioinformatic Analysis of Protein Superfamilies. In: Multiple Sequence Alignment: Methods and Protocols, Methods in Molecular Biology, vol. 2231 (ed. Katoh K). Springer US, pp 179-200. [DOI] [PubMed]
- 53.Sharapova Y., Suplatov D., Švedas V. Catalytic and Lectin Domains in Neuraminidase A from Streptococcus pneumoniae are Capable of an Intermolecular Assembly: Implications for Biofilm Formation. FEBS J. 2021 doi: 10.1111/febs.15610. [DOI] [PubMed] [Google Scholar]
- 54.Fesko K., Suplatov D., Švedas V. Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in l-threonine aldolase from Aeromonas jandaei. FEBS Open Bio. 2018;8(6):1013–1028. doi: 10.1002/2211-5463.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharapova Y., Suplatov D., Švedas V. Neuraminidase a from Streptococcus pneumoniae has a modular organization of catalytic and lectin domains separated by a flexible linker. FEBS J. 2018;285(13):2428–2445. doi: 10.1111/febs.14486. [DOI] [PubMed] [Google Scholar]
- 56.Swaminathan K., Dyason J.C., Maggioni A., von Itzstein M., Downard K.M. Binding of a natural anthocyanin inhibitor to influenza neuraminidase by mass spectrometry. Anal Bioanal Chem. 2013;405(20):6563–6572. doi: 10.1007/s00216-013-7068-x. [DOI] [PubMed] [Google Scholar]
- 57.Landon M.R., Amaro R.E., Baron R., Ngan C.H., Ozonoff D., Andrew McCammon J. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des. 2008;71(2):106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amaro R.E., Minh D.D., Cheng L.S., Lindstrom W.M., Olson A.J., Lin J.H. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J Am Chem Soc. 2007;129(25):7764–7765. doi: 10.1021/ja0723535. [DOI] [PubMed] [Google Scholar]
- 59.Russell R.J., Haire L.F., Stevens D.J., Collins P.J., Lin Y.P., Blackburn G.M. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443(7107):45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y., Wu Y., Tefsen B., Shi Y., Gao G.F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014;22(4):183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diem-trang T.T., Le L.T., Truong T.N. Discover binding pathways using the sliding binding-box docking approach: application to binding pathways of oseltamivir to avian influenza H5N1 neuraminidase. J Comput Aided Mol Des. 2013;27(8):689–695. doi: 10.1007/s10822-013-9675-1. [DOI] [PubMed] [Google Scholar]
- 62.Le L., Lee E.H., Hardy D.J., Truong T.N., Schulten K. Molecular dynamics simulations suggest that electrostatic funnel directs binding of Tamiflu to influenza N1 neuraminidases. PLoS Comput Biol. 2010;6(9) doi: 10.1371/journal.pcbi.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y., Qin G., Gao F., Liu Y., Vavricka C.J., Qi J. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep. 2013;3:1551. doi: 10.1038/srep01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amaro R.E., Swift R.V., Votapka L., Li W.W., Walker R.C., Bush R.M. Mechanism of 150-cavity formation in influenza neuraminidase. Nat Commun. 2011;2(1):1–7. doi: 10.1038/ncomms1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng X., Lee J., Michael A.J., Tomchick D.R., Goldsmith E.J., Phillips M.A. Evolution of Substrate Specificity within a Diverse Family of β/α-Barrel-fold Basic Amino Acid Decarboxylases. J Biol Chem. 2010;285(33):25708–25719. doi: 10.1074/jbc.M110.121137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J., Michael A.J., Martynowski D., Goldsmith E.J., Phillips M.A. Phylogenetic diversity and the structural basis of substrate specificity in the β/α-barrel fold basic amino acid decarboxylases. J Biol Chem. 2007;282(37):27115–27125. doi: 10.1074/jbc.M704066200. [DOI] [PubMed] [Google Scholar]
- 67.Campbell E., Chuang S., Banta S. Modular exchange of substrate-binding loops alters both substrate and cofactor specificity in a member of the aldo-keto reductase superfamily. Protein Eng Des Sel. 2013;26(3):181–186. doi: 10.1093/protein/gzs095. [DOI] [PubMed] [Google Scholar]
- 68.Liskova V, Pluskal D, Vasina M, Emond S, Doerr M, Chaloupková R, Bednar D, Prokop Z, Hollfelder F, Bornscheuer U, Damborsky J (2020) Engineering Protein Dynamics of Ancestral Luciferase. ChemRxiv. Preprint. 10.26434/chemrxiv.12808295.v1.
- 69.Bollinger A., Thies S., Knieps-Grünhagen E., Gertzen C., Kobus S., Höppner A. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri–Structural and functional insights. Front Microbiol. 2020;11:114. doi: 10.3389/fmicb.2020.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy P.M., Bolduc J.M., Gallaher J.L., Stoddard B.L., Baker D. Alteration of enzyme specificity by computational loop remodeling and design. Proc Natl Acad Sci U S A. 2009;106(23):9215–9220. doi: 10.1073/pnas.0811070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 72.Amaral M., Kokh D.B., Bomke J., Wegener A., Buchstaller H.P., Eggenweiler H.M. Protein conformational flexibility modulates kinetics and thermodynamics of drug binding. Nat Commun. 2017;8(1):1–14. doi: 10.1038/s41467-017-02258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao M.L., Somero G.N., Dong Y.W. Comparing mutagenesis and simulations as tools for identifying functionally important sequence changes for protein thermal adaptation. Proc Natl Acad Sci U S A. 2019;116(2):679–688. doi: 10.1073/pnas.1817455116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palacios A.R., Mojica M.F., Giannini E., Taracila M.A., Bethel C.R., Alzari P.M. The reaction mechanism of metallo-β-lactamases is tuned by the conformation of an active-site mobile loop. Antimicrob Agents Chemother. 2019;63(1):e01754–18. doi: 10.1128/AAC.01754-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montagner C., Nigen M., Jacquin O., Willet N., Dumoulin M., Karsisiotis A.I. The role of active site flexible loops in catalysis and of zinc in conformational stability of Bacillus cereus 569/H/9 β-lactamase. J Biol Chem. 2016;291(31):16124–16137. doi: 10.1074/jbc.M116.719005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kundert K., Kortemme T. Computational design of structured loops for new protein functions. Biol Chem. 2019;400(3):275–288. doi: 10.1515/hsz-2018-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nussinov R., Tsai C.J., Shehu A., Jang H. Computational structural biology: Successes, future directions, and challenges. Molecules. 2019;24(3):637. doi: 10.3390/molecules24030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Childers M.C., Daggett V. Insights from molecular dynamics simulations for computational protein design. Mol Syst Des Eng. 2017;2(1):9–33. doi: 10.1039/C6ME00083E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suplatov D., Kirilin E., Arbatsky M., Takhaveev V., Švedas V. pocketZebra: a web-server for automated selection and classification of subfamily-specific binding sites by bioinformatic analysis of diverse protein families. Nucleic Acids Res. 2014;42(W1):W344–W349. doi: 10.1093/nar/gku448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ribeiro A.J., Tyzack J.D., Borkakoti N., Thornton J.M. Identifying pseudoenzymes using functional annotation. How loss of function correlates with mutations in the catalytic site. FEBS J. 2019;287(19):4128–4140. doi: 10.1111/febs.15142. [DOI] [PubMed] [Google Scholar]
- 81.Tawfik D.S., Gruic-Sovulj I. How evolution shapes enzyme selectivity–lessons from aminoacyl-tRNA synthetases and other amino acid utilizing enzymes. FEBS J. 2020;287(7):1284–1305. doi: 10.1111/febs.15199. [DOI] [PubMed] [Google Scholar]
- 82.Syakur MA, Khotimah BK, Rochman EMS, Satoto BD (2018) Integration k-means clustering method and elbow method for identification of the best customer profile cluster. In: IOP Conference Series: Materials Science and Engineering, vol. 336. IOP Publishing, p. 012017
- 83.McInnes L., Healy J., Astels S. HDBScan: Hierarchical density based clustering. J Open Source Softw. 2017;2(11):205. [Google Scholar]
- 84.Ankerst M., Breunig M.M., Kriegel H.P., Sander J. OPTICS: ordering points to identify the clustering structure. ACM Sigmod record. 1999;28(2):49–60. [Google Scholar]
- 85.Schubert E., Sander J., Ester M., Kriegel H.P., Xu X. DBSCAN revisited, revisited: why and how you should (still) use DBSCAN. ACM Trans Database Systems (TODS) 2017;42(3):1–21. [Google Scholar]
- 86.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 87.Hrabe T., Li Z., Sedova M., Rotkiewicz P., Jaroszewski L., Godzik A. PDBFlex: exploring flexibility in protein structures. Nucleic Acids Res. 2016;44(D1):D423–D428. doi: 10.1093/nar/gkv1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suplatov D., Kopylov K., Sharapova Y., Švedas V. Human p38α Mitogen-Activated Protein Kinase in the Asp168-Phe169-Gly170-in (DFG-in) state can bind allosteric inhibitor Doramapimod. J Biomol Struct Dyn. 2019;37(8):2049–2060. doi: 10.1080/07391102.2018.1475260. [DOI] [PubMed] [Google Scholar]
- 89.Voevodin V.V., Antonov A.S., Nikitenko D.A., Shvets P.A., Sobolev S.I., Sidorov I.Y. Supercomputer Lomonosov-2: large scale, deep monitoring and fine analytics for the user community. Supercomput Front Innov. 2019;6(2):4–11. [Google Scholar]