TO THE EDITOR:
von Willebrand factor (VWF) monomers dimerize through their C-terminal domain in the endoplasmic reticulum (ER). The unusual process of disulfide bond formation between N-terminal D′D3 assemblies (Figure 1) of neighboring dimers during tubule formation in the Golgi apparatus then forms the ultralong, tail-to-tail, head-to-head concatemers required for VWF activation in hemostasis.1-5 C-terminally truncated VWF fragments are secreted as mixtures of monomers and dimers; the monomers contain 2 free cysteines, Cys-1099 and Cys-1142, which were proposed to form the dimerizing interchain disulfide bonds.6 However, chemical determination of disulfide bonds in VWF is challenging7-9 and only the Cys-1142/Cys-1142′ disulfide has been confirmed.1 Surprisingly, a recent VWF D′D3 monomer crystal structure showed burial of Cys-1099 and Cys-1142, thus revealing how these residues are protected from disulfide bond formation in the ER, but little about disulfide bond formation in the Golgi apparatus.9 Gel-forming mucins contain D assemblies homologous to those of VWF and also form multimers in the Golgi apparatus10; however, in mucin MUC2, homologs of VWF Cys-1142 and Cys-1097 but not Cys-1099 formed dimerizing disulfides. Multiple explanations for the discrepancy were proposed but not resolved.11 Here, we present evidence suggesting that disulfide exchange between 3 Cys residues in VWF frees Cys-1097 to form a dimerizing disulfide bond.
Figure 1.
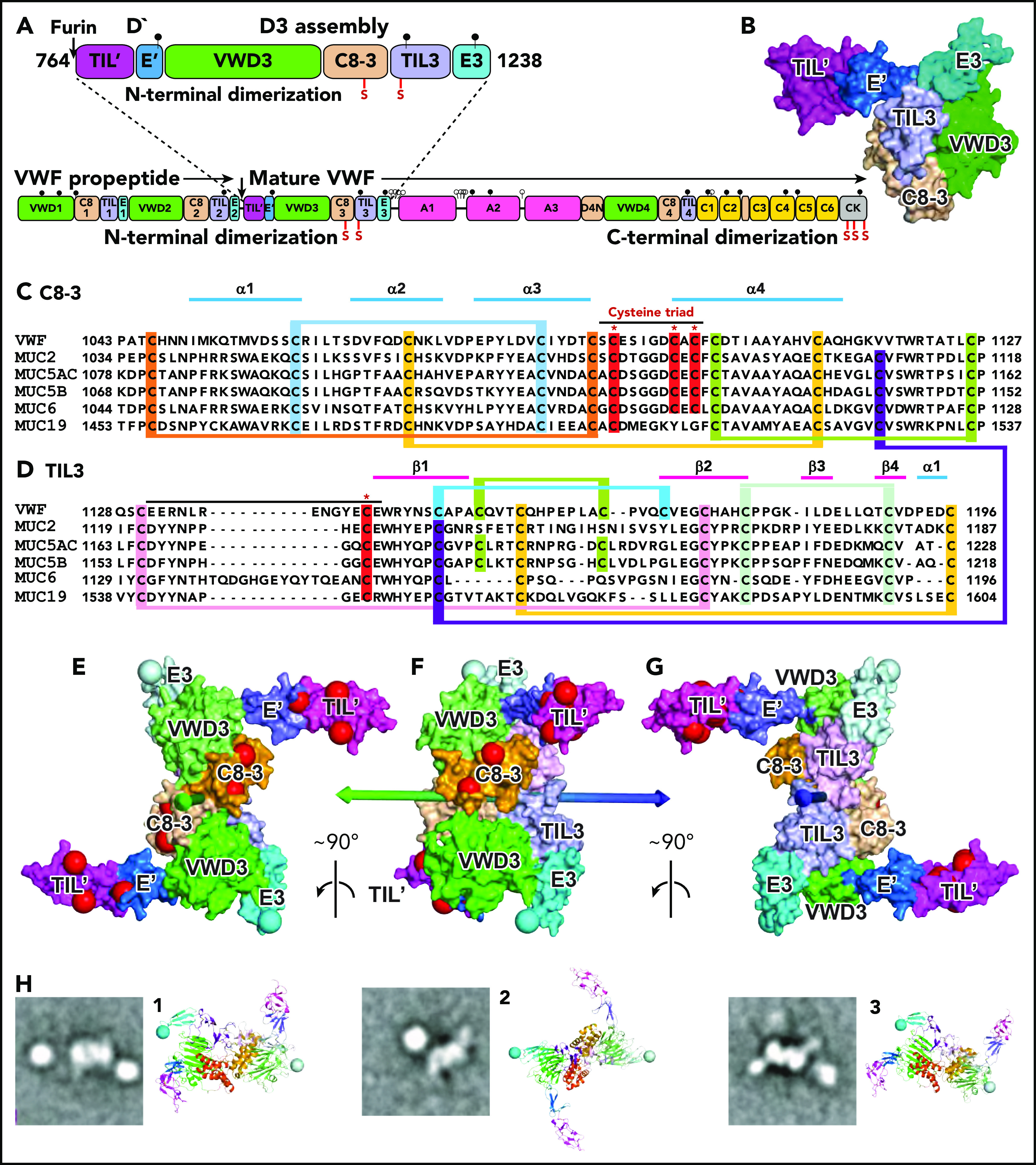
Modeling D′D3 dimerization. (A) Domain architecture and position of dimerizing cysteine residues (red vertical lines with S) in D′D3 assembly (top) and VWF monomer (bottom). (B) Solvent-accessible surface of D′D3.9 (C-D) Sequence alignments of D3 C8-3 and TIL3 modules in VWF and gel-forming mucins (see supplemental Figure 1 for VWD3 and E3). Cysteines implicated in disulfide exchange and interchain disulfides are noted with asterisks (*) and highlighted in red, whereas those that form intrachain disulfides are linked and highlighted in other colors. (E-G) The D′D3 dimer model is shown as a solvent-accessible surface colored by module with its twofold rotational axis as a double-ended arrow colored green on one end and blue on the other. D3 C termini in E3 are marked with large light-blue spheres. Residues mutated in VWF disease that cause loss of VWF binding (type 2M) and no other phenotypes (types 1, 3, 2A, and 2B) are marked with large red spheres. VWF tubules in Weibel-Palade bodies also have a dyad symmetry axis3,4 to which the D′D3 dimer axis must be parallel. VWF biology suggests that the dimer axis end colored green in panels E through G would orient toward the outside rather than the inside of tubules. This side links to A1 and the VWF C terminus and bears VWF disease mutations that specifically abolish factor VIII binding,9 which occurs during biosynthesis.19 A model of VWF tubules based on MUC2 filaments also supports this orientation.20 The dimer model was created with Modeler 9.621 as described in the text with each monomer templated with the segments of VWF and MUC2 (PDB ID 6N29 and 6RBF, respectively) shown in supplemental Figure 2. (H) Cross-correlation of the D′D3 dimer model to electron microscopy class averages of D′D3-A1 dimers12 was as described.12,22 The best-correlating D′D3 dimer orientation is shown enlarged as a ribbon diagram with coloring similar to that in panels E through G and with spheres at C termini of E3 to show where the mucin linker to A1 attaches. Owing to flexibility of this linker, A1 (a round globule) appears in different orientations or does not appear in class averages. CK, C-terminal cystine-knot domain. Modified from Springer.5
In the monomeric D′D3 crystal structure, Cys-1099 and Cys-1142 were mutated to alanine and modeled as Cys. Cys-1099, with disulfide-bonded Cys-1091 and Cys-1097, forms a triad of 3 Cys residues that locate close to one another in the C8-3 module (Figures 1C and 2). The free sulfhydryl (SH) group of Cys-1099 is shielded by the von Willebrand factor D 3 (VWD3) module, making Cys-1099 inaccessible for disulfide bond formation.9 Furthermore, movement of Cys-1099 is limited by its position in an α-helix with 2 disulfide bonds to other structural elements in C8-3.
Figure 2.
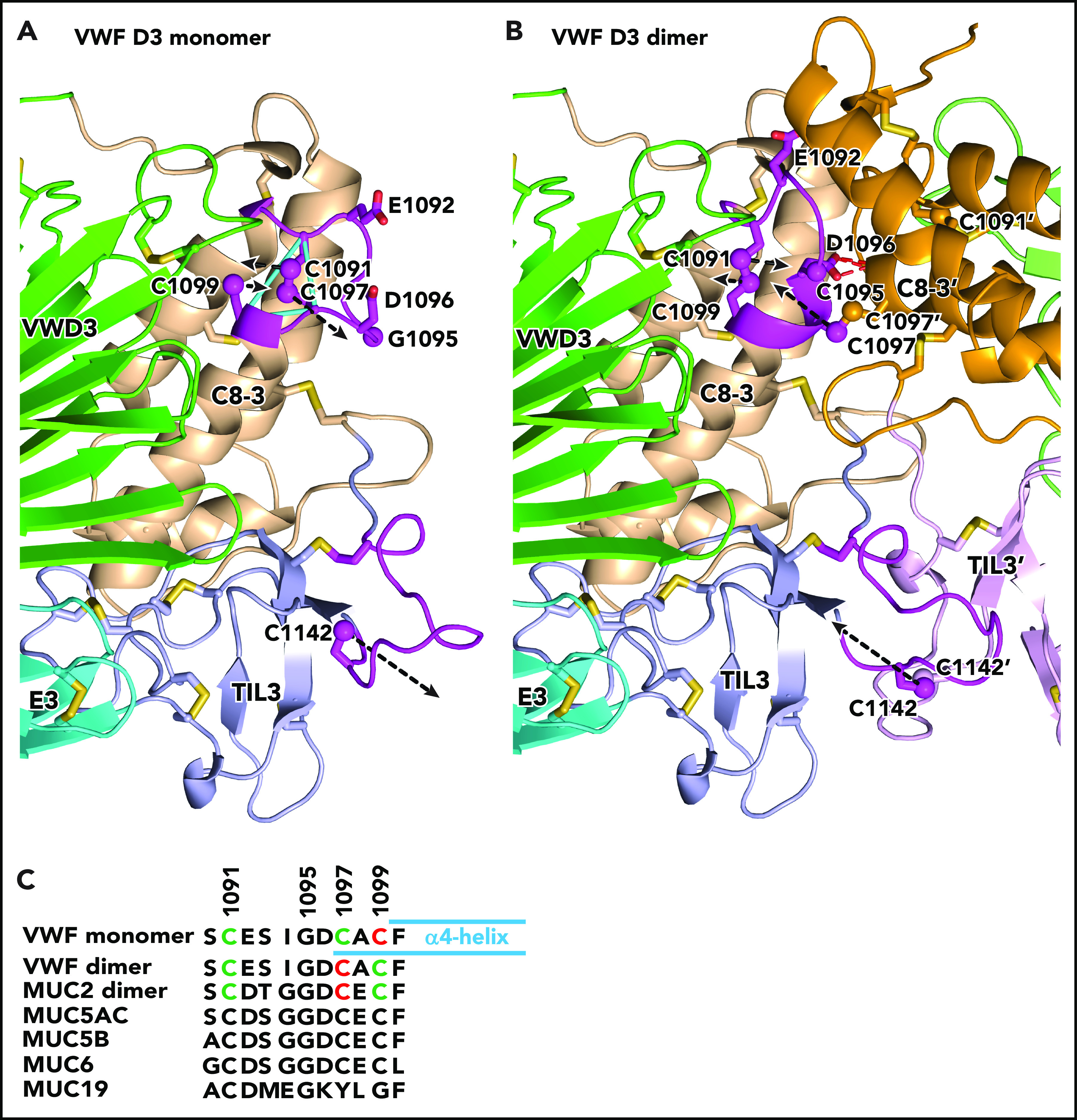
Conformational change in VWF C8-3 and TIL3 during dimerization. (A-B) Comparison of monomeric crystal structure model (A) and dimeric homology model (B) of D3. The monomer on the left is color-coded identically in panels A and B. Each module is labeled and has a distinct color for carbon atoms, except regions that change substantially in conformation upon dimerization are colored magenta. The other monomer on the right in the dimer in panel B lacks the color code for conformational change and its modules are shown in slightly different colors than in panel A. Its module and side-chain names are distinguished with an appended apostrophe. Side chains of cysteines and other selected residues are shown as sticks with yellow sulfurs and red oxygens. Additionally, sulfur atoms of cysteines in the triad in C8-3 and C1142 in TIL3 are shown as spheres. Dashed black arrows represent vectors for change in position of the sulfur atoms. Backbone Cα atoms of Cys triad residues are connected with cyan lines in panel A. The hydrogen bonds between the Asp-1096 side chain and main chain of the other monomer in the dimer are shown as red dashed lines for 1 of 2 reciprocal interactions in panel B. (C) Sequence alignment of the cysteine triad in human VWF and gel-like mucins. In VWF and MUC2, cysteines in intrachain disulfides are green and those as free cysteines in the VWF monomer or in interchain linkages in dimers are red. Cysteines in other mucins are black; however, 4 have cysteine triad sequences highly identical among VWF and 4 mucins and are predicted to undergo disulfide exchange between monomers and dimers, like VWF. MUC19 differs substantially in the same region, including having only 1 cysteine. It is predicted to be free in the monomer and disulfide linked in the dimer, much like Cys-1142 in VWF and its equivalent in all gel-like mucins.
In contrast, rearrangement of disulfide bonds within the cysteine triad is highly feasible, a possibility we were careful not to exclude with the previous conclusion “that structural rearrangements are required for D3 dimerization in the Golgi.”9(p1525) The Cys-1099 SH group is only 4.7 Å away from the disulfide-bonded sulfur atoms of Cys-1091 and Cys-1097, with no nearby atoms to hinder nucleophilic attack by Cys-1099 on the disulfide. Furthermore, the Cys-1091/Cys-1097 disulfide is exposed to solvent in a loop that contains a Gly residue that can confer flexibility and is invariant in VWF and mucins (Figure 2C). Thus, the most plausible mechanism for exposure of cysteine for dimerization is nucleophilic attack by the S− anion of C1099 on the disulfide, thereby freeing Cys-1097 for formation of a disulfide bond to another monomer. Given the high local concentrations of these residues within the triad loop, disulfide exchange among them is likely to be in rapid equilibrium. At neutral pH, the equilibrium favors free Cys-1099.6 At acidic pH, interactions among D assemblies at the ends of growing VWF tubules in the Golgi apparatus may favor free Cys-1097, the homolog of which in MUC2 forms the interchain disulfide.11 The triad loop also contains Glu-1092 and Asp-1096, which are always acidic or invariant, respectively, in VWF and gel-forming mucins (Figure 2C). These acidic residues might have a dual function in deprotonating the Cys SH group for nucleophilic attack and ionically repelling interactions between triad loops in different monomers in the ER to prevent premature D3 dimerization.
To test the feasibility of VWF dimerization through Cys-1097, we built a dimer model. The VWF D3 assembly aligns well by sequence with mucins (38% identity with MUC2) with no sequence insertions or deletions in the key C8-3 module among VWF and all 5, human, gel-forming mucins (Figure 1C-D; supplemental Figure 1, available on the Blood Web site). Monomeric VWF D′D39 was superimposed on each monomer of dimeric MUC2 D311 with a root-mean-square deviation of 1.3 Å over 330 residues per monomer. Using this dimeric template and homology modeling, we constructed a robust model of dimeric VWF D′D3 (Figures 1E-G and 2B; supplemental Figure 1).
The feasibility of the model is supported by the structure of the dimeric interface. The only significant structural differences between the monomer and dimer in this interface are in C8-3 module residues 1090 to 1099, which comprise the loop containing the cysteine triad, and in trypsin inhibitor-like 3 (TIL3) module residues 1131 to 1143, which contain Cys-1142 (black overlines in Figure 1C-D) (Figure 2). In between, residues 1122 to 1130 are in the interface but are in identical backbone conformations in dimeric MUC2 and monomeric VWF. Preservation of the geometry in monomeric VWF at dimer interfacial residues 1122 to 1130, with the ability to properly model interfacial disulfide formation in the dimer, supports our model of disulfide exchange to free Cys-1097 prior to 1097-1097′ disulfide formation.
The model suggests that D3 dimerization is accompanied by no change in backbone position of Cys-1099, which is well embedded in an α-helix. Only the side chain of Cys-1099 rotates to form a disulfide to Cys-1091, which moves in its loop. Residues 1096 to 1098 in the cysteine triad loop alter to α-helical conformation and add on to the end of the α4-helix bearing Cys-1099 (Figure 2C). In its new helical conformation, Cys-1097 disulfide bonds across the dimer interface to its mate in the other monomer. Furthermore, Asp-1096 in each monomer forms hydrogen bonds to backbone across the dimer interface (Figure 2B). To make the turn between the α4-helix and the loop bearing Cys-1091, Gly-1095 changes to the Lα conformation, which is only allowed for glycine, explaining the invariance of this residue in VWF and gel-forming mucins. Dimerization further requires the loop that buries Cys-1142 in VWF monomers to partially unfurl with an 11-Å movement of Cys-1142 to disulfide bond across the dimer interface.
In the loop bearing the cysteine triad, the 3 cysteines, Gly-1095, and Asp-1096 and their equivalents, are invariant in all sequenced animal species in VWF, MUC2, MUC5AC, MUC5B, and MUC6. Our results thus suggest that not only VWF, but also 4 of the 5 gel-forming mucins, have a free, buried Cys-1099 equivalent that is protected from disulfide formation in the ER and that undergoes disulfide exchange in the Golgi apparatus so that the more exposed Cys-1097 equivalent is free to form the dimerizing disulfide bond. In contrast, Muc19 contains only the Cys-1091 equivalent (Figure 2C), has Lys in place of VWF Asp-1096, and thus has a different conformation at the dimerization interface, which might involve dimerization through the equivalent of Cys-1091. We welcome confirmation of our model by determining whether single or double mutations in the cysteine triad in VWF or mucins compromise normal multimerization in the Golgi apparatus or lead to aberrant multimerization such as by exposing Cys-1097 or its equivalent in mucins in the ER.
As originally proposed by Sadler and colleagues1 and found in MUC2 dimers,11 monomers align parallel to one another in D′D3 dimers, with the twofold rotational symmetry (dyad) axis passing between the Cys-1097/Cys-1097′ and Cys-1142/Cys-1142′ pairs (Figure 1E-G). Electron microscopy class averages of dimers containing D′D3 linked to A1 through the flexible mucin segment12 cross-correlate well to the model, show horn-like D′ projections and A1 domain positions not far from the C terminus of D3, and provide further confirmation for the proposed dimerization mode (Figure 1H).
What triggers D3 dimerization? In the trans–Golgi apparatus, the D1D2 prodomain packs against D′D3 and these D1 to D3 units in turn pack further with one another and assemble into tubules with helical symmetry that characterize Weibel-Palade bodies.3-5 D1D2 is required for both tubule assembly3 and D3-D3 dimer formation.13-15 D1D2 was proposed to be an oxidoreductase with cysteine residues in a CGLC sequence that functioned, as in protein disulfide isomerases, to catalyze oxidation of the dimerizing disulfides in D3.16 D3 is highly homologous to D1 and D2 and has the same CGLC sequence.5 The D3 structure shows that the CGLC sequence is highly buried in the VWD module and that its cysteines disulfide-link to other VWD cysteines distal in sequence and could not be alternately reduced and disulfide-linked to one another as required in an oxidoreductase.9 Disruption of VWF multimer formation when an extra glycine is inserted into D1 or D2 CGLC sequences16 therefore should be reinterpreted as a disruption of domain conformation that might alter packing in tubules.
This reinterpretation supports the model by Sadler and colleagues in which tubule formation in the trans–Golgi apparatus facilitates VWF N-terminal dimerization by juxtaposing D3 domains.3 Between the ER and the trans–Golgi apparatus, the pH decreases from 7.2 to 5.8, and a pH of ∼5.8 is required for both VWF tubule formation in vitro and multimerization in vivo and in vitro.3,15,17 Histidine has a pKa of ∼6. Mutation of His residues that are conserved in VWF and mucins has shown that histidines in D2 and D1 regulate VWF multimerization10; furthermore, homology to D3 shows that these His residues are distributed around the periphery of the D1 and D2 assemblies, where contacts between neighboring D assemblies in tubules are expected to occur.3,5 Therefore, we propose that the pH-dependent contacts between D assemblies as they assemble on the growing ends of tubules in the trans–Golgi apparatus, including between juxtaposed D3 assemblies, trigger conformational changes that stabilize D3 dimer formation. Juxtaposition of D3 domains in tubules would stabilize a new conformation of the cysteine triad loop and disulfide exchange within it, freeing Cys-1097; and would also stabilize a new conformation of the loop containing Cys-1142. Thus, Cys-1097 and Cys-1142 would be juxtaposed with their mates in the neighboring D3 assembly.
How are the Cys-1097/Cys-1097′ and Cys-1142/Cys-1142′ disulfides oxidized? As we propose that disulfide exchange in the cysteine triad loop occurs directly, the redox function of a protein disulfide isomerase is not required; its oxidase function would suffice. Alternatively, a sulfhydryl oxidase, such as QSOX1, which localizes to the Golgi apparatus, might catalyze disulfide formation.18
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgment
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grant R01-HL148755.
Footnotes
For original data, please e-mail the corresponding author.
Authorship
Contribution: T.A.S. and X.D. contributed to conception, experimental design, and writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Springer, Center for Life Sciences, Boston Children’s Hospital/Harvard Medical School, Room 3103, 3 Blackfan Circle, Boston, MA 02115; e-mail: springer@crystal.harvard.edu.
REFERENCES
- 1.Dong Z, Thoma RS, Crimmins DL, McCourt DW, Tuley EA, Sadler JE. Disulfide bonds required to assemble functional von Willebrand factor multimers. J Biol Chem. 1994;269(9):6753-6758. [PubMed] [Google Scholar]
- 2.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395-424. [DOI] [PubMed] [Google Scholar]
- 3.Huang R-H, Wang Y, Roth R, et al. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci USA. 2008;105(2):482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berriman JA, Li S, Hewlett LJ, et al. Structural organization of Weibel-Palade bodies revealed by cryo-EM of vitrified endothelial cells. Proc Natl Acad Sci USA. 2009;106(41):17407-17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Springer TA. von Willebrand factor, Jedi knight of the bloodstream. Blood. 2014;124(9):1412-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purvis AR, Gross J, Dang LT, et al. Two Cys residues essential for von Willebrand factor multimer assembly in the Golgi. Proc Natl Acad Sci USA. 2007;104(40):15647-15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marti T, Rösselet SJ, Titani K, Walsh KA. Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry. 1987;26(25):8099-8109. [DOI] [PubMed] [Google Scholar]
- 8.Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. Sequence and structure relationships within von Willebrand factor. Blood. 2012;120(2):449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong X, Leksa NC, Chhabra ES, et al. The von Willebrand factor D’D3 assembly and structural principles for factor VIII binding and concatemer biogenesis. Blood. 2019;133(14):1523-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang LT, Purvis AR, Huang RH, Westfield LA, Sadler JE. Phylogenetic and functional analysis of histidine residues essential for pH-dependent multimerization of von Willebrand factor. J Biol Chem. 2011;286(29):25763-25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javitt G, Calvo MLG, Albert L, et al. Intestinal gel-forming mucins polymerize by disulfide-mediated dimerization of D3 domains. J Mol Biol. 2019;431(19):3740-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou YF, Eng ET, Nishida N, Lu C, Walz T, Springer TA. A pH-regulated dimeric bouquet in the structure of von Willebrand factor. EMBO J. 2011;30(19):4098-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verweij CL, Hart M, Pannekoek H. Expression of variant von Willebrand factor (vWF) cDNA in heterologous cells: requirement of the pro-polypeptide in vWF multimer formation. EMBO J. 1987;6(10):2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise RJ, Pittman DD, Handin RI, Kaufman RJ, Orkin SH. The propeptide of von Willebrand factor independently mediates the assembly of von Willebrand multimers. Cell. 1988;52(2):229-236. [DOI] [PubMed] [Google Scholar]
- 15.Mayadas TN, Wagner DD. In vitro multimerization of von Willebrand factor is triggered by low pH. Importance of the propolypeptide and free sulfhydryls. J Biol Chem. 1989;264(23):13497-13503. [PubMed] [Google Scholar]
- 16.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci USA. 1992;89(8):3531-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner DD, Mayadas T, Marder VJ. Initial glycosylation and acidic pH in the Golgi apparatus are required for multimerization of von Willebrand factor. J Cell Biol. 1986;102(4):1320-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horowitz B, Javitt G, Ilani T, et al. Quiescin sulfhydryl oxidase 1 (QSOX1) glycosite mutation perturbs secretion but not Golgi localization. Glycobiology. 2018;28(8):580-591. [DOI] [PubMed] [Google Scholar]
- 19.Turecek PL, Mitterer A, Matthiessen HP, et al. Development of a plasma- and albumin-free recombinant von Willebrand factor. Hamostaseologie. 2009;29(suppl 1):S32-S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javitt G, Khmelnitsky L, Albert L, et al. Assembly mechanism of mucin and von Willebrand factor polymers [published online ahead of print 2 October 2020]. Cell. doi:10.1016/j.cell.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23(3):318-326. [DOI] [PubMed] [Google Scholar]
- 22.Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18(9):984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


