Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the most common tumor subtype of pancreatic cancer, which exhibits poor patient prognosis due to the lack of effective biomarkers in the diagnosis and treatment. The present study aimed to identify the potential biomarkers of PDAC carcinogenesis and progression using three microarray datasets, GSE15471, GSE16515 and GSE28735, which were downloaded from the Gene Expression Omnibus database. The datasets were analyzed to screen out differentially expressed genes (DEGs) in PDAC tissues and adjacent normal tissues. A total of 143 DEGs were identified, including 132 upregulated genes and 11 downregulated genes. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes functional and signaling pathway enrichment analyses were performed on the DEGs, and the Search Tool for the Retrieval of Interacting Genes/Proteins database was used to construct a protein-protein interaction network. The main functions of DEGs include extracellular matrix degradation, and regulation of matrix metalloproteinase activity and the PI3K-Akt signaling pathway. The five hub genes were subsequently screened using Cytoscape software, and survival analysis demonstrated that abnormal expression levels of the hub genes was associated with poor disease-free survival and overall survival. Biological experiments were performed to confirm whether mesenchymal-to-epithelial transition (MET) factors promote the proliferation, migration and invasion of PDAC cells via the PI3K/AKT signaling pathway. In addition, six MET-targeted microRNAs (miRNAs) were identified, four of which had conserved binding sites with MET. Based on the signaling pathway enrichment analysis of these miRNAs, it is suggested that they can affect the progression of PDAC by targeting MET via the PI3K/AKT signaling pathway. In conclusion, the hub genes and miRNAs that were identified in the present study contribute to the molecular mechanisms of PDAC carcinogenesis and progression. They also provide candidate biomarkers for early diagnosis and treatment of patients with PDAC.
Keywords: bioinformatics analysis, pancreatic ductal adenocarcinoma, microarray, differentially expressed genes, microRNA
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common tumor subtype of pancreatic cancer accounting for ~85% of cases (1), which is observed in the digestive system. The global incidence rate of PDAC continues to increase annually, due in part to its extreme aggressiveness and early metastasis characteristics (2). The 5-year survival rate of patients with PDAC is ~5% (3). The outcomes of immunotherapy, surgery, chemotherapy and radiotherapy for PDAC remain unsatisfactory, and treatment side effects have resulted in patients experiencing a lower quality of life (4). Comprehensive genomic analysis of PDAC has demonstrated that PDAC occurrence and development is closely associated with multiple gene mutations and signal transduction pathways, which are involved in the mutant genes (5). In addition to the four common cancer mutant genes, KRAS, TP53, SMAD4 and CDKN2A, it also includes some genes that have a mutation rate of ~10%, including KDM6A, RBM10 and MLL3 (6). These mutant genes mainly influence PDAC progression in DNA damage repair, cell cycle regulation, chromatin regulation and TGF-β signaling pathway (7). Thus, understanding the underlying molecular mechanisms of PDAC is vital for improving current therapeutic options and developing novel therapeutic strategies.
Recently, notable progress has been made in microarray technology and bioinformatics methods. Unlike traditional experimental methods, where only either a single gene or several genes can be studied at once, large-scale research and analysis can be performed, and mutant genes can be screened out at the genomic level, which provides a better method for the identification of cancer biomarkers and the study of molecular mechanisms (8).
In the present study, three microarray datasets, GSE15471 (9), GSE16515 (10) and GSE28735 (11) were downloaded from the Gene Expression Omnibus (GEO) database to identify differentially expressed genes (DEGs) in PDAC tissues and adjacent normal tissues, and five hub genes were further screened from the DEGs using Cytoscape software. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional and signaling pathway enrichment analyses of the DEGs were performed, and a protein-protein interaction (PPI) network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database to assess PDAC development at the molecular level.
MicroRNAs (miRNAs/miRs) are non-coding single-stranded RNA molecules that are 20–24 nucleotides in length, which can be paired with the 3′-untranslated region of the target gene to regulate its expression (12). miRNAs are involved in several cellular biological processes, including cell proliferation, migration, apoptosis, invasion, angiogenesis and immune escape, and also participate in the occurrence and development of cancer through regulation of the expression of oncogenes and antioncogenes (13). Increasing evidence suggests that specific cancer-related miRNAs that are detected in body fluids, such as plasma, serum, urine and saliva can serve as biomarkers for cancer diagnosis and prognosis (14,15).
In the present study, six mesenchymal-to-epithelial transition (MET)-targeted miRNAs (miR-1-3p, miR-23b-3p, miR-31-5p, miR-34a-5p, miR-340-5p and miR-130a-3p) were screened using the DIANA, TargetScan, miRDB and miRTarBase databases, and the results demonstrated that miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p have conserved binding sites with MET. Thus, a miRNA-mRNA signaling pathway was assessed, which affects PDAC progression, thereby providing potential biomarkers for the early diagnosis and treatment of patients with PDAC.
Materials and methods
Microarray datasets
A total of three microarray datasets, GSE15471, GSE16515 and GSE28735, were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo), based on the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array and the Affymetrix Human Gene 1.0 ST Array. The GSE15471 dataset contained 36 PDAC tissues and 36 normal tissues, the GSE16515 dataset contained 36 PDAC tissues and 16 normal tissues, and the GSE28735 dataset contained 45 PDAC tissues and 36 normal tissues. The datasets were screened for the following criteria: i) Samples contained PDAC tissues and normal pancreatic tissues, ii) study type was restricted to expression profiling by array, iii) organism was restricted to homo sapiens and iv) original data were accessible.
GO and KEGG functional and signaling pathway enrichment analyses of the DEGs
GEO2R is an interactive web tool that allows users to compare two or more databases in a GEO series, and screen DEGs under different experimental conditions (8). Thus, the GEO2R database (http://www.ncbi.nlm.nih.gov/geo/geo2r) was used to screen the DEGs between PADC tissues and normal pancreatic tissues. The probe sets were converted into corresponding gene symbols based on the annotation information in the platform. Probe sets that did not have corresponding gene symbols or genes with more than one probe set were removed or averaged. |Log2 fold change| >1 and adjusted P<0.01 were considered to indicate a statistically significant difference.
The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov) was used to perform GO and KEGG functional and signaling pathway enrichment analyses of the DEGs. GO analysis (http://www.geneontology.org/) is used to predict the potential functions of target genes based on molecular function (MF), cell component (CC) and biological processes (BP) (16). KEGG (http://www.genome.jp/kegg/pathway.html) is a database that is associated with gene pathways and improves the understanding of metabolic pathways where genes are involved, and the associations between pathways by linking genomic information to a higher-level system (17).
PPI network construction
Analysis of the interactions between proteins can help uncover the molecular mechanisms of disease occurrence and development (18). In the present study, the STRING database (http://string-db.org) was used to construct the PPI network of the DEGs, and interaction with a combined score >0.4 was considered to indicate statistical significance. The Cytoscape software (version 3.6.1) (8) was subsequently used to visualize the PPI network.
Selection and analysis of hub genes
A total of five hub genes were selected from the DEGs using MCODE plug-in (version 1.5.1) within the Cytoscape software, and BiNGO plug-in (version 3.0.4) was subsequently used to perform biological process analyses of the hub genes. The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/detail.php) was used to assess the differences in expression of the five hub genes in PDAC tissues and normal pancreatic tissues. The Kaplan-Meier plotter database (http://kmplot.com) was used to perform disease-free survival and overall survival of the hub genes in PDAC. The Oncomine database (http://www.oncomine.com) is used to compute gene expression signatures, clusters, gene-set modules and automatically extract biological insights from data (19). The Oncomine database was used in the present study to analyze the DEGs between PDAC tissues and normal pancreatic tissues, and detect the expression levels of the hub gene MET in different types of tumors.
Cell culture and transfection
The PDAC cell line, PANC-1, was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of. Cells were maintained in DMEM (HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (cat. no. SV30010; Shanghai Canspec Scientific & Technology Co., Ltd.), at 37°C with 5% CO2. The medium was replaced every 2 days for 2 weeks.
Small interfering (si)RNAs were subsequently used to silence MET expression and assess the effect of MET on PDAC cells. Cells were seeded into 6-well plates at a density of 5×105 cells/ml and cultured until they reached 60–70% confluence. Cells were transfected at 37°C for 48 h with 5 µl MET siRNA (siRNA-MET) and 5 µl negative control (scrambled) siRNA (siRNA-NC) using 10 µl Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The following sequences were used: siRNA-MET sense, 5′-CACUAACUACAUUUAUGUUUU-3′ and antisense, 5′-AACAUAAAUGUAGUUAGUGGC-3′; siRNA-NC sense, 5′-AUAUAAUUCUACUUACUUGCU-3′, and antisense, 5′-CGAGUAAGUACAGUAAUGAUU-3′ (Shanghai GenePharma Co., Ltd.). Non-transfected cells were used as the control group. Transfection efficiency was assessed via reverse transcription-quantitative (RT-q)PCR analysis.
RT-qPCR
Total RNA was extracted from cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.), and NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.) was used to measure the concentration and purity. Total RNA (1 µg) was reverse transcribed into cDNA using TaqMan™ Reverse Transcription Reagents (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions, in a 20 µl volume of reaction mixture. qPCR was subsequently performed using the SYBR® Premix Ex Taq™ kit (cat. no. RR820A; Takara Bio, Inc.), according to the manufacturer's instructions. The following primer sequences were used for qPCR: MET forward, 5′-CCTGCGAAGTGAAGGGTCTCC-3′ and reverse, 5′-CTGGCAGCTTTGCACCTGTTT-3′; and GAPDH forward, 5′-CTTCAAGATCATCAGCAATGC-3′ and reverse, 5′-GTTGAAGTCAGAGGAGACCACC-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 5 min, followed by 39 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 60 sec, and a final extension step at 72°C for 5 min. Relative expression levels were calculated using the 2−ΔΔCq method (20) and normalized to the internal reference gene GAPDH. All experiments were performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
PANC-1 cells were seeded into 96-well plates at a density of 5,000 cells/well, with each sample containing three replicates. CCK-8 reagent (10%, Dojindo Molecular Technologies, Inc.) was added to each well following culturing for 0, 6, 12, 18, 24, 30, 36, 42 and 48 h. Following incubation with CCK-8 for 1 h, the results were analyzed at a wavelength of 450 nm, using a microplate reader (Bio-Rad Laboratories, Inc.). All experiments were performed in triplicate.
Wound healing assay
PANC-1 cells were seeded into 6-well plates (104/well) and cultured until they reached 90% confluence. Subsequently, the cells were serum-starved for 12 h, the cell monolayers were scratched using 200-µl sterile pipette tips, and the cells were continued to be cultured with serum-free DMEM. Wound healing was observed at 0, 24 and 48 h under an inverted light microscope (magnification, ×40; Olympus Corporation).
Transwell assay
Cell invasion was evaluated using Transwell chambers coated with Matrigel at 37°C for 48 h (BD Biosciences). A total of 2×105 PANC-1 cells were plated in the upper chambers of 24-well Transwell plates in serum-free medium and 200 µl cell suspension was subsequently added. DMEM (700 µl) supplemented with 20% FBS was plated in the lower chambers. Following incubation for 48 h at 37°C, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and subsequently stained with 0.1% crystal violet for 20 min at room temperature. The non-invading cells were removed using a wet cotton swab, whilst the stained cells were counted under an inverted light microscope (magnification, ×100; Olympus Corporation).
Western blotting
PANC-1 cells were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with protease and phosphatase inhibitors (Beyotime Institute of Biotechnology). Protein concentration was determined using the BCA protein assay kit (cat. no. P0012S; Beyotime Institute of Biotechnology) and 40 µg protein/lane was separated by 10% SDS-PAGE. The separated proteins were subsequently transferred onto PVDF membranes and blocked with 5% non-fat milk for 1 h at room temperature. The membranes were incubated with primary antibodies against rabbit anti-GAPDH (1:1,000; cat. no. ab9485; Abcam), PI3K (1:1,000; cat. no. 4249; Cell Signaling Technology, Inc.), phosphorylated (p)-PI3K (1:1,000; cat. no. 17366; Cell Signaling Technology, Inc.), Akt (1:1,000; cat. no. 4691; Cell Signaling Technology, Inc.) and p-Akt (1:1,000; cat. no. 4060; Cell Signaling Technology, Inc.) overnight at 4°C. Following the primary incubation, membranes were incubated with HRP-conjugated goat anti-rabbit (1:2,000; cat. no. ab150079; Abcam) secondary antibody at room temperature for 1 h. Protein bands were visualized using ECL reagents (Tanon, Science and Technology Co., Ltd.) and the intensity of each band was quantified using Image Lab software (version 4.1; Bio-Rad Laboratories, Inc.). All experiments were performed in triplicate.
miRNA-MET prediction
The DIANA (http://diana.imis.athena-innovation.gr), TargetScan (http://www.targetscan.org), miRDB (http://www.mirdb.org) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) databases were used to predict MET-targeted miRNAs. The results from the four databases were intersected to obtain six common miRNAs, and functional and signaling pathway enrichment analyses of the miRNAs were subsequently performed. The TargetScanHuman database (http://www.targetscan.org) was used to analyze and predict the MET-targeted miRNAs, as well as to obtain the sequences of fragments that contain binding sites.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software (IBM Corp.). All experiments were performed in triplicate and data are presented as the mean ± standard deviation. One-way ANOVA followed by Dunnett's multiple comparison test were used to compare differences among multiple groups, while differences between two groups were compared by Wilcoxon rank-sum test. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening DEGs in the microarray datasets
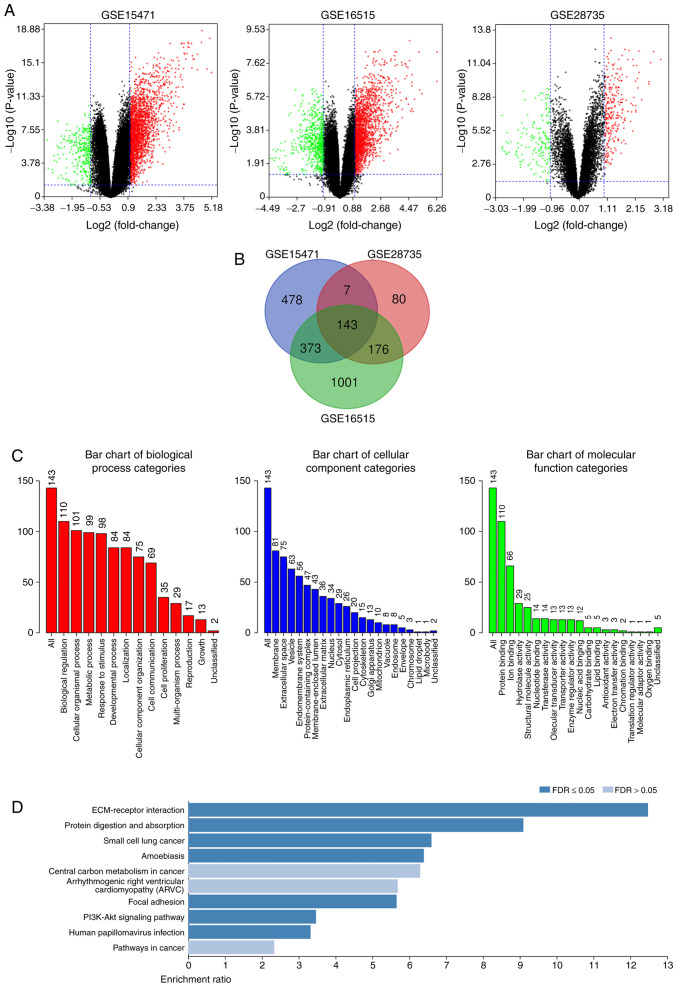
The present study analyzed the DEGs in PDAC tissues and normal pancreatic tissues. The DEGs were obtained following standardization of the microarray results, which demonstrated that the GSE15471 dataset contained 1,330 upregulated genes and 66 downregulated genes, the GSE16515 dataset contained 1,801 upregulated genes and 561 downregulated genes, and the GSE28735 dataset contained 267 upregulated genes and 156 downregulated genes (Fig. 1A). As presented in the Venn diagram, the intersection of the three microarray datasets contained 143 genes, composed of 132 upregulated genes and 11 downregulated genes (Fig. 1B).
Figure 1.
Identification of DEGs in PDAC, and functional and signaling pathway enrichment analyses. (A) Volcano plot of genome-wide gene expression profiles in pancreatic ductal adenocarcinoma tissues and adjacent normal tissues from the GSE15471, GSE16515 and GSE28735 datasets. Red plots represent upregulated genes with P<0.05 and log2 FC >1, black plots represent normally expressed genes and green plots represent downregulated genes with P<0.05 and log2 FC <-1. (B) The intersection of the three microarray datasets contained 143 DEGs. (C) Gene Ontology and (D) Kyoto Encyclopedia of Genes and Genomes functional and signaling pathway enrichment analyses of the DEGs were performed using the DAVID database. P<0.05 was set as the threshold. DEGs, differentially expressed genes.
Functional and signaling pathway enrichment analyses of the DEGs
The DAVID database was used to perform GO and KEGG functional and signaling pathway enrichment analyses of the DEGs. GO analysis demonstrated that the DEG BP are mainly focused on cell metabolism, intercellular communication and cell proliferation regulation. CC are mainly concentrated in the structural composition of extracellular matrix, cell membrane system, and the binding with collagen and integrin. MF is mainly focused on cell molecular transport, protein binding and enzyme activity regulation (Fig. 1C). KEGG analysis demonstrated that DEG signaling pathways mainly enriched extracellular matrix receptors, protein digestion and absorption and the PI3K-Akt signaling pathway (Fig. 1D).
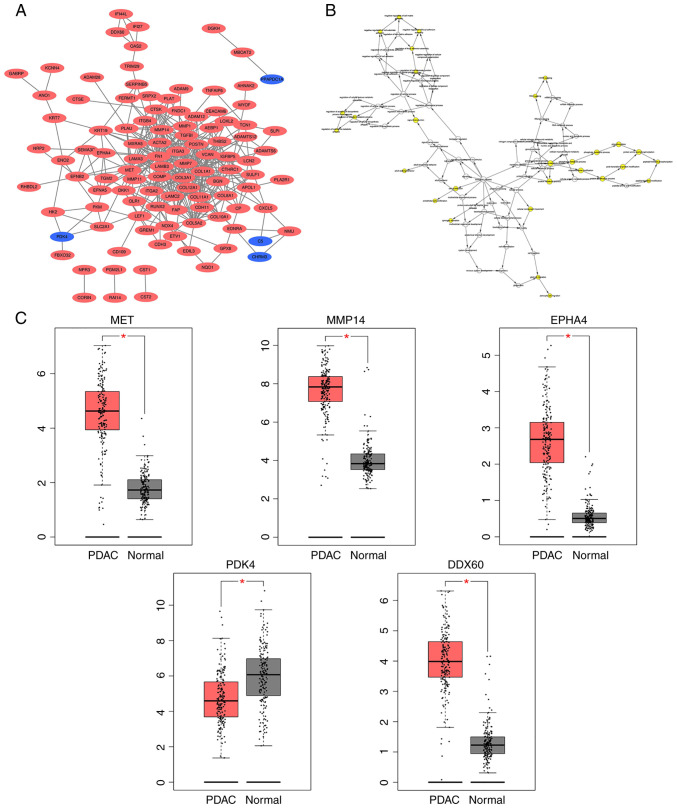
PPI network construction, and selection and analysis of hub genes
A DEG-based PPI network was constructed using the STRING database and visualized using Cytoscape software (Fig. 2A). MCODE was subsequently used to screen out five hub genes [MET, matrix metalloproteinase (MMP)14, EPHA4, pyruvate dehydrogenase kinase (PDK)4 and DDX60] from the DEGs. PDK4 expression was downregulated, while the expression levels of the other four hub genes were upregulated. BiNGO was subsequently used to assess the biological processes of the hub genes, revealing that their functions were mostly focused on cell proliferation and migration, phosphorylation metabolism process and matrix metalloproteinase activation (Fig. 2B). Analysis using the GEPIA database demonstrated that the differences in the expression levels of these five hub genes between PDAC tissues and normal pancreatic tissues were statistically significant (all P<0.05; Fig. 2C).
Figure 2.
PPI network of the DEGs, and the expression levels of the hub genes in PDAC tissues and normal tissues. (A) The Cytoscape software was used to construct a DEG-based PPI network. Upregulated genes are marked in red, while downregulated genes are marked in blue. (B) BiNGO was used to construct the biological processes of the hub genes. The color depth of the nodes refer to the corrected P-value of ontologies, while the size of the nodes refer to the number of genes that are involved in the ontologies. (C) The Gene Expression Profiling Interactive Analysis database was used to analyze the differences in the expression levels the hub genes between PDAC tissues and normal pancreatic tissues. *P<0.05. PPI, protein-protein interaction; DEGs, differentially expressed genes; PDAC, pancreatic ductal adenocarcinoma.
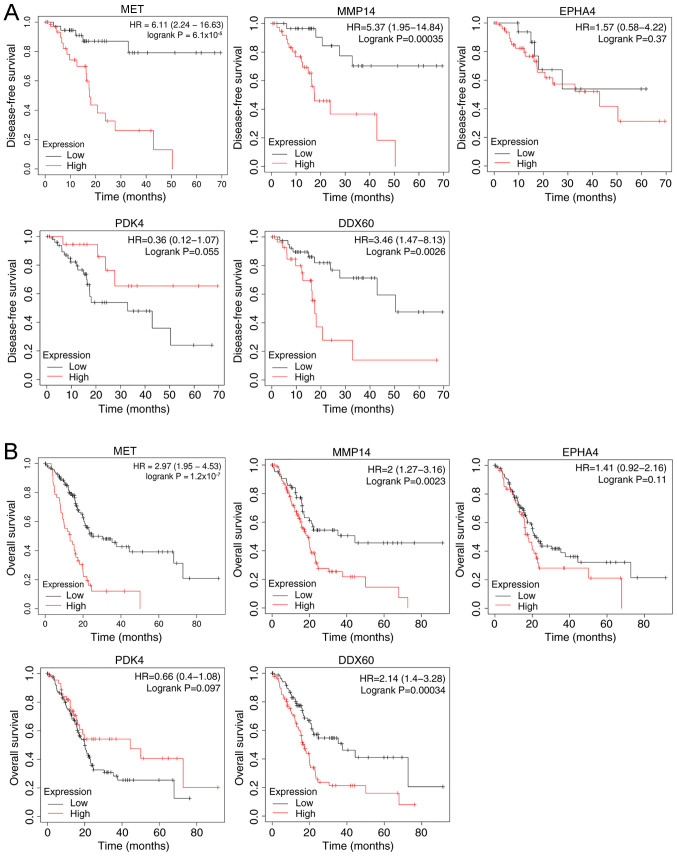
Kaplan-Meier survival analysis of the hub genes demonstrated that both high expression levels of MET, MMP14, EPHA4 and DDX60, and low PDK4 expression resulted in a shorter disease-free survival time (Fig. 3A) and overall survival time (Fig. 3B) of patients with PDAC.
Figure 3.
Kaplan-Meier survival analysis of the hub genes in pancreatic ductal adenocarcinoma. (A) Disease-free survival time. (B) Overall survival time. HR, hazard ratio.
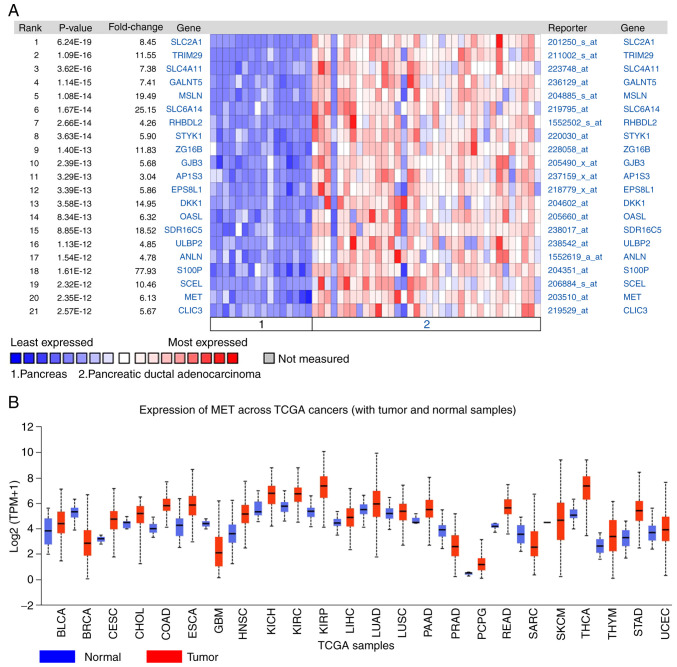
The DEGs between PDAC tissues and normal pancreatic tissues were analyzed using the Oncomine database. The results demonstrated that MET expression was 6.13 times higher in PDAC tissues compared with normal tissues (Fig. 4A). The expression of MET in different tumors was subsequently analyzed, and the results demonstrated that MET expression was upregulated in different types of cancer, including lung, cervical, gastric and colorectal cancer (Fig. 4B).
Figure 4.
MET expression in different types of cancer. (A) The differentially expressed genes were identified between PDAC tissues and normal pancreatic tissues using the Oncomine database. (B) MET expression in different types of cancer tissues and normal tissues.
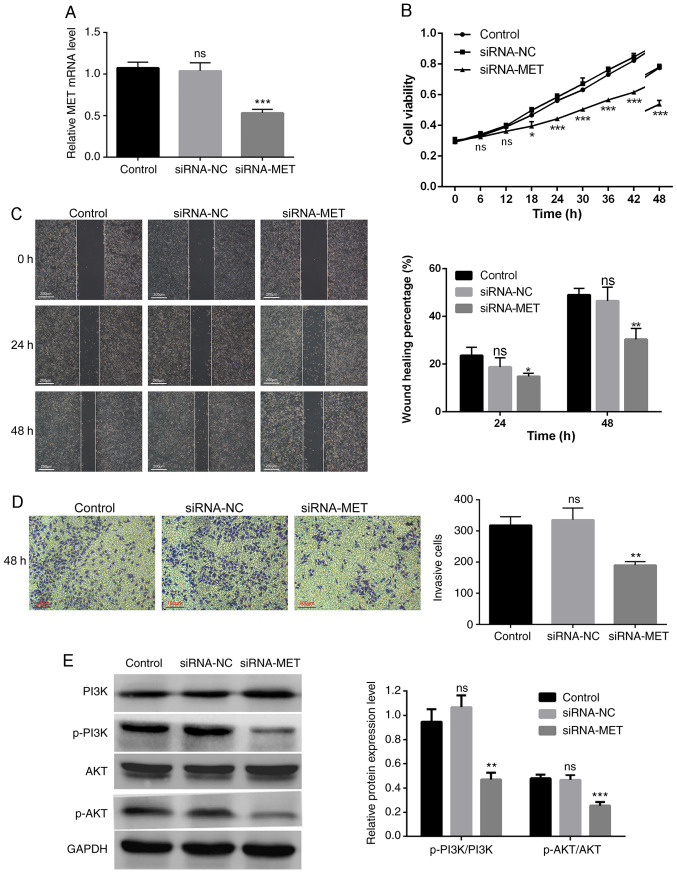
Silencing MET inhibits the proliferation, migration and invasion of PDAC cells
MET expression was knocked down in PANC-1 cells via siRNA transfection (Fig. 5A). The results of the CCK-8 assay confirmed that MET knockdown significantly inhibited the proliferation of PANC-1 cells (Fig. 5B). The results of the wound healing and Transwell assays demonstrated that high MET expression promoted the migration and invasion of PDAC cells. In addition, the wound healing rate of the siRNA-MET group significantly decreased compared with the control group (P<0.05), whereas the differences between the siRNA-NC group and the control group were not statistically significant (P>0.05; Fig. 5C). The number of invasive cells in the siRNA-MET group was significantly lower compared with the control group (P<0.01), and the differences between the siRNA-NC group and the control group were not statistically significant (P>0.05; Fig. 5D).
Figure 5.
Effect of MET knockdown on PANC-1 cells. (A) Transfection efficiency of MET knockdown was assessed via reverse transcription-quantitative PCR analysis. (B) The results of the Cell Counting Kit-8 assay demonstrated that MET knockdown significantly inhibited the proliferation of PANC-1 cells. (C) The results of the wound healing assay demonstrated that MET knockdown significantly inhibited cell migration. Scale bar, 200 µm. (D) The results of the Transwell assay demonstrated that MET knockdown significantly inhibited cell invasion. Scale bar, 100 µm. (E) Western blot analysis demonstrated that MET knockdown inhibited phosphorylation of the PI3K/AKT pathway. *P<0.05, **P<0.01, ***P<0.001 vs. control group. si, small interfering; NC, negative control; ns, no significance; p, phosphorylated.
MET promotes the progression of PDAC by activating the PI3K/AKT signaling pathway
To further study the molecular mechanism by which MET regulates the proliferation, migration and invasion of PDAC cells, the protein levels of the PI3K/AKT pathway-related molecules were assessed in PANC-1 cells transfected with siRNA-MET. The results demonstrated that MET knockdown significantly decreased the levels of phosphorylated PI3K and AKT, but not their total protein levels (Fig. 5E). Taken together, these results suggest that MET is an upstream signal factor that modulates the PI3K/AKT pathway in PDAC, and that MET promotes the proliferation, migration and invasion of PDAC cells through activation of the PI3K/AKT pathway.
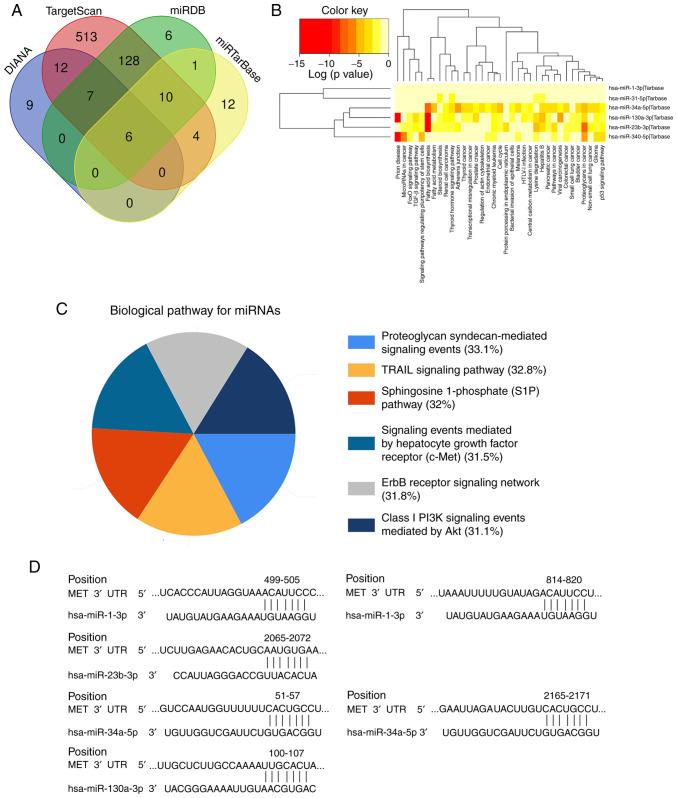
Prediction of the miRNA-MET signaling pathway
A total of six MET-targeted miRNAs (miR-1-3p, miR-23b-3p, miR-31-5p, miR-34a-5p, miR-340-5p and miR-130a-3p) were screened using the DIANA, TargetScan, miRDB, and miRTarBase databases (Fig. 6A). Functional and signaling pathway analyses of these six miRNAs demonstrated that their functions mainly focused on prion diseases, fatty acid synthesis and metabolism, and they are involved in the progression of thyroid, prostate, colorectal and pancreatic cancers. The signaling pathways mainly include the TRAIL-mediated external apoptosis signaling pathway, sphingosine-1-phosphate signaling pathway, c-MET/hepatocyte growth factor (HGF) signaling pathway, PI3K/Akt signaling pathway and ErbB receptor signaling pathway (Fig. 6B and C). A total of four miRNAs (miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p) were demonstrated to have conserved binding sites with MET, using the TargetScanHuman database (Fig. 6D). Thus, it is suggested that these four miRNAs can act as upstream regulators of the MET/PI3K/AKT pathway, and serve as potential biomarkers and therapeutic targets for patients with PDAC.
Figure 6.
Screening of miRNAs and signaling pathway prediction. (A) A total of six MET-targeted miRNAs were screened using online databases. (B and C) Functional and signaling pathway enrichment of the miRNAs were predicted using DIANA-miRPath v3.0. (D) The binding site sequence of MET mRNA and miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p. miRNA/miR, microRNA; UTR, untranslated region.
Discussion
Currently, there are still significant challenges facing the early diagnosis of PDAC. A lack of understanding of the molecular mechanism in PDAC progression and effective treatments for PDAC facilitates a poor prognosis in patients. Thus, the identification of signal factors that play a key regulatory role in the occurrence and development of PDAC is of great importance, and they may become pivotal early diagnostic biomarkers and targets in the treatment of PDAC.
Microarray technology can be used for studying gene transcription and epigenetic changes, and it is an incredibly effective method for identifying disease biomarkers (21). In the present study, microarray technology and bioinformatics methods were used to analyze three PDAC-related microarray datasets, which were downloaded from the GEO database, and 143 DEGs between PDAC tissues and normal tissue were screened out, including 132 upregulated genes and 11 downregulated genes. GO and KEGG functional and signaling pathway enrichment analyses were performed, and it was discovered that DEG's functions are mainly focused on the extracellular matrix (ECM), protein digestion and absorption, and the regulation of cellular metabolic processes. ECM is a network structure that is composed of collagen, glycoprotein and proteoglycan, and provides biomechanics for the regulation of cell behavior, while maintaining organs' morphology and integrity (22). ECM is in dynamic equilibrium under the influence of extracellular proteases and their inhibitors; however, when a tumor grows, the dynamic balance is broken, which triggers pathological ECM remodeling, and results in reduced adhesion of cells to the ECM (23). This is conducive to cancer cells invading nearby organs and blood vessels, thereby promoting cancer invasion and metastasis (24). This also explains the reason for PDAC's high invasiveness (23).
A total of five hub genes (MET, MMP14, EPHA4, PDK4 and DDX60) were screened out from the DEGs, PDK4 was downregulated and the other four genes were upregulated. MET is a potential therapeutic target for different types of cancer, and it can promote cell proliferation, migration and epithelial-to-mesenchymal transition (25). The binding of MET with its ligand, HGF, is a key driver in the development of cancer (26). MMP14 is a type of MMP, which is upregulated in different types of cancer, including colorectal, lung and nasopharyngeal cancer (27–29). MMP14 promotes angiogenesis, inflammation, invasion and metastasis of cancer cells, and severely affects the prognosis of patients (30). EPHA4 plays a significant role in neurodegenerative diseases, including Alzheimer's disease and amyotrophic lateral sclerosis (31). Previous studies have demonstrated that EPHA4 is upregulated in PDAC and promotes tumor growth (32–34). Its high expression is also associated with poor prognosis of patients with gastric cancer and breast cancer (35). PDK plays a vital role in the regulation of cell metabolism and mitochondrial function (36). PDK includes four isoenzymes (PDK1, 2, 3, and 4); PDK4 provides a tri-carbon substrate (pyruvate, lactic acid and alanine) for gluconeogenesis during fasting to maintain blood glucose levels (37). RIG-I-mediated type I interferon (IFN) and nuclease-mediated viral RNA degradation are of great importance for the innate antiviral immune response in vivo (38). DDX60 is an IFN-induced cytoplasmic helicase that participates in viral RNA degradation, performs a vital role in the innate immune response and is dependent on the RIG-I antiviral pathway (39). A previous study has demonstrated that DDX60 is highly expressed in oral cancer and is closely associated with tumor progression and poor prognosis (40). The results of the Oncomine and GEPIA analyses in the present study demonstrated that MET, MMP14, EPHA4 and DDX60 were highly expressed in PDAC, while PDK4 was expressed at low levels. The impact of hub genes on the survival time of patients with PDAC was subsequently analyzed and the results revealed that abnormal expression of these five hub genes was associated with poor disease-free survival time and overall survival time. Taken together, these results suggest that the five hub genes play a significant role in the progression of PDAC. Furthermore, through a series of biological experiments, it was confirmed that the hub gene, MET, can promote the proliferation, migration and invasion of PDAC cells via the PI3K/AKT signaling pathway.
miRNAs, as major regulators of several biological and pathological processes, including growth, development, metabolism, infection, immunity, cell death, messenger signaling, DNA repair and self-renewal, have exhibited carcinogenic or anticancer activity in different types of human tumors (41,42). Increasing evidence suggests that several miRNAs are crucial for cancer development as they regulate various biological processes in cancer cells, including proliferation, differentiation, apoptosis, cell cycle, migration and invasion, which has led to miRNAs becoming potential diagnostic biomarkers and therapeutic targets for different types of cancer (43). In the present study, six MET-targeted miRNAs were screened out using several online databases, and four (miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p) of them were demonstrated to have conserved binding sites with MET. Previous studies have demonstrated that miR-1-3p expression is downregulated in different types of cancer, including colon cancer, esophageal squamous cell carcinoma and bladder cancer, and that the overexpression of miR-1-3p can inhibit the growth of these tumors (44–46). Li et al (47) demonstrated that miR-1-3p can inhibit prostate cancer cell proliferation by regulating the cell cycle-related genes, E2F5 and PFTK1. miR-23b-3p is considered a tumor suppressor that affects the carcinogenesis and aggressiveness of liver cancer, and it can be used as a predictive biomarker and therapeutic target for liver cancer (48). Ge and Li (49) have demonstrated that long non-coding RNA SNHG17 can promote glioma cell proliferation, migration and invasion by regulating the miR-23b-3p/ZHX1 axis. Studies have reported that miR-34a-5p is involved in the occurrence and development of cancer through regulation of the Notch, PI3K/Akt, SIRT1/HIF-1α and Wnt/β-Catenin signaling pathways (50–52). As a tumor suppressor, miR-34a-5p is able to inhibit the growth of glioma and esophageal squamous cell carcinoma (53,54). miR-130a-3p is a key regulator of human cancer, and it can inhibit the proliferation, migration and invasion of various cancer cells, such as liver cancer, breast cancer and nasopharyngeal carcinoma (55–57). Notably, Dai et al (58) have also demonstrated that miR-130a-3p promotes the growth of gastric cancer. Several studies have reported that abnormal miRNA expression is closely associated with the occurrence and development of PDAC, and that miRNAs can be used as early detection biomarkers for potential treatment and prognostic monitoring targets (59,60). However, the miRNAs that play a regulatory role in the development of PDAC and their molecular mechanisms are rarely reported. The present study assessed four miRNAs (miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p) that can potentially be involved in PDAC development; however, further studies are required to determine their molecular mechanisms.
MET is a receptor for HGF, which is encoded by the proto-oncogene, c-Met. MET is a tyrosine kinase receptor that is mainly distributed in the membranes of epithelial-derived or endothelium-derived cells (61). MET is upregulated in different types of cancer, including breast cancer, colorectal cancer, glioma and non-small cell lung cancer (62–64). MET has the ability to help cells adapt to adverse environment, promote tumor angiogenesis, enhance tumor cell survival, and their metastatic and invasive abilities (65). This is consistent with the experimental results of the present study. When HGF ligand binds to a MET receptor, MET is activated and induces intracellular tyrosine residue phosphorylation, activating the downstream RAS/ERK/MAPK, PI3K/Akt, Wnt/β-catenin and STAT signaling pathways (64). These pathways can drive proliferation, migration, invasion, angiogenesis and epithelial-to-mesenchymal transition in a variety of tumors (66–68). According to the functional and signaling pathway enrichment analysis of DEGs and miRNAs in the present study, they were all involved in the PI3K/Akt signaling pathway. The PI3K/Akt signaling pathway plays a crucial regulatory role in different types of cancer, including PDAC (69). In the present study, western blot analysis demonstrated that MET activated the PI3K/AKT pathway in PDAC cells. Based on this study's experimental results and bioinformatics analysis, it can be speculated that under the modulation of miRNA, MET promotes PDAC progression through the PI3K/AKT signaling pathway. In addition, four alternative MET-targeted miRNAs, miR-1-3p, miR-23b-3p, miR-34a-5p and miR-130a-3p are provided in the present study.
Future studies will perform dual-luciferase reporter assays to verify the targeting association between these four miRNAs and MET. In addition, the expression levels of the miRNAs will be restrained in PANC-1 cells via transfection, and cell proliferation, migration and invasion will be assessed. Furthermore, prospective studies will perform western blot analysis to detect the expression levels of PI3K/AKT pathway-related proteins. Collectively, these experiments will help identify novel diagnostic biomarkers and therapeutic targets for patients with PDAC.
In conclusion, the present study aimed to identify DEGs and molecular mechanisms that affect the progression of PDAC to discover potential diagnostic biomarkers and therapeutic targets for patients with PDAC. A total of 143 DEGs were screened out, five hub genes and four miRNAs were further identified, and the cancer-promoting effect of MET in PDAC was proven. According to the functional and signaling pathway enrichment analysis of DEGs and miRNAs, it can be speculated that these four miRNAs affect PDAC progression by targeting MET via the PI3K/AKT signaling pathway. However, further biological experiments must be performed to confirm this speculation. Notably, the present study identified a novel mechanism that affects PDAC progression, and provided four potential miRNAs that can serve as promising biomarkers for the early diagnosis and treatment of patients with PDAC.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant no. LGF19H290004).
Funding
The present study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant no. LGF19H290004).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
LCY, XHJ, SSY, WW and ZGT contributed to experimental design, data acquisition, and drafting and editing the initial manuscript. LW, LLZ, FX, TJ and LY analyzed the data and assisted in the experiments. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21:7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, Zhan HX. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231–241. doi: 10.1002/ijc.30670. [DOI] [PubMed] [Google Scholar]
- 4.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 5.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyer SB, Chang DK, Bailey P, Biankin AV. Pancreatic cancer genomes: Implications for clinical management and therapeutic development. Clin Cancer Res. 2017;23:1638–1646. doi: 10.1158/1078-0432.CCR-16-2411. [DOI] [PubMed] [Google Scholar]
- 7.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 10.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian T, Wang J, Zhou X. A review: microRNA detection methods. Org Biomol Chem. 2015;13:2226–2238. doi: 10.1039/C4OB02104E. [DOI] [PubMed] [Google Scholar]
- 13.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S, Zhou L, Wei J, Chang H, Li K, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: Evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7:1692. doi: 10.1038/s41598-017-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabbani G, Baig MH, Ahmad K, Choi I. Protein-protein Interactions and their role in various diseases and their prediction techniques. Curr Protein Pept Sci. 2018;19:948–957. doi: 10.2174/1389203718666170828122927. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 22.Vogel V. Unraveling the mechanobiology of extracellular matrix. Annu Rev Physiol. 2018;80:353–387. doi: 10.1146/annurev-physiol-021317-121312. [DOI] [PubMed] [Google Scholar]
- 23.Tian C, Clauser KR, Öhlund D, Rickelt S, Huang Y, Gupta M, Mani DR, Carr SA, Tuveson DA, Hynes RO. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci USA. 2019;116:19609–19618. doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuzhalin AE, Lim SY, Kutikhin AG, Gordon-Weeks AN. Dynamic matrisome: ECM remodeling factors licensing cancer progression and metastasis. Biochim Biophys Acta Rev Cancer. 2018;1870:207–228. doi: 10.1016/j.bbcan.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Moosavi F, Giovannetti E, Saso L, Firuzi O. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit Rev Clin Lab Sci. 2019;56:533–566. doi: 10.1080/10408363.2019.1653821. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto K, Umitsu M, De Silva DM, Roy A, Bottaro DP. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017;108:296–307. doi: 10.1111/cas.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui G, Cai F, Ding Z, Gao L. MMP14 predicts a poor prognosis in patients with colorectal cancer. Hum Pathol. 2019;83:36–42. doi: 10.1016/j.humpath.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Stawowczyk M, Wellenstein MD, Lee SB, Yomtoubian S, Durrans A, Choi H, Narula N, Altorki NK, Gao D, Mittal V. Matrix metalloproteinase 14 promotes lung cancer by cleavage of heparin-binding EGF-like growth factor. Neoplasia. 2017;19:55–64. doi: 10.1016/j.neo.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan T, Lin Z, Jiang J, Lu S, Chen M, Que H, He X, Que G, Mao J, Xiao J, Zheng Q. MMP14 regulates cell migration and invasion through epithelial-mesenchymal transition in nasopharyngeal carcinoma. Am J Transl Res. 2015;7:950–958. [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez-Molina J, Gramolelli S, Liao Z, Carlson JW, Ojala PM, Lehti K. MMP14 in sarcoma: A regulator of tumor microenvironment communication in connective tissues. Cells. 2019;8:991. doi: 10.3390/cells8090991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas LM, Cerpa W, Muñoz FJ, Zanlungo S, Alvarez AR. Amyloid-β oligomers synaptotoxicity: The emerging role of EphA4/c-Abl signaling in Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1148–1159. doi: 10.1016/j.bbadis.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Huang H, Wang C, Kong Y, Zhang H. Involvement of ephrin receptor A4 in pancreatic cancer cell motility and invasion. Oncol Lett. 2014;7:2165–2169. doi: 10.3892/ol.2014.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iiizumi M, Hosokawa M, Takehara A, Chung S, Nakamura T, Katagiri T, Eguchi H, Ohigashi H, Ishikawa O, Nakamura Y, Nakagawa H. EphA4 receptor, overexpressed in pancreatic ductal adenocarcinoma, promotes cancer cell growth. Cancer Sci. 2006;97:1211–1216. doi: 10.1111/j.1349-7006.2006.00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giaginis C, Tsourouflis G, Zizi-Serbetzoglou A, Kouraklis G, Chatzopoulou E, Dimakopoulou K, Theocharis SE. Clinical significance of ephrin (eph)-A1, -A2, -a4, -a5 and -a7 receptors in pancreatic ductal adenocarcinoma. Pathol Oncol Res. 2010;16:267–276. doi: 10.1007/s12253-009-9221-6. [DOI] [PubMed] [Google Scholar]
- 35.Takano H, Nakamura T, Tsuchikawa T, Kushibiki T, Hontani K, Inoko K, Takahashi M, Sato S, Abe H, Takeuchi S, et al. Inhibition of Eph receptor A4 by 2,5-dimethylpyrrolyl benzoic acid suppresses human pancreatic cancer growing orthotopically in nude mice. Oncotarget. 2015;6:41063–41076. doi: 10.18632/oncotarget.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michelakis ED, Gurtu V, Webster L, Barnes G, Watson G, Howard L, Cupitt J, Paterson I, Thompson RB, Chow K, et al. Inhibition of pyruvate dehydrogenase kinase improves pulmonary arterial hypertension in genetically susceptible patients. Sci Transl Med. 2017;9:eaao4583. doi: 10.1126/scitranslmed.aao4583. [DOI] [PubMed] [Google Scholar]
- 37.Leem J, Lee IK. Mechanisms of vascular calcification: The pivotal role of pyruvate dehydrogenase kinase 4. Endocrinol Metab (Seoul) 2016;31:52–61. doi: 10.3803/EnM.2016.31.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan YK, Gack MU. RIG-I-like receptor regulation in virus infection and immunity. Curr Opin Virol. 2015;12:7–14. doi: 10.1016/j.coviro.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshiumi H, Miyashita M, Okamoto M, Morioka Y, Okabe M, Matsumoto M, Seya T. DDX60 is involved in RIG-I-dependent and independent antiviral responses, and its function is attenuated by virus-induced EGFR Activation. Cell Rep. 2015;11:1193–1207. doi: 10.1016/j.celrep.2015.04.047. [DOI] [PubMed] [Google Scholar]
- 40.Fu TY, Wu CN, Sie HC, Cheng JT, Lin YS, Liou HH, Tseng YK, Shu CW, Tsai KW, Yen LM, et al. Subsite-specific association of DEAD box RNA helicase DDX60 with the development and prognosis of oral squamous cell carcinoma. Oncotarget. 2016;7:85097–85108. doi: 10.18632/oncotarget.13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabekkodu SP, Shukla V, Varghese VK, D'Souza J, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc. 2018;93:1955–1986. doi: 10.1111/brv.12428. [DOI] [PubMed] [Google Scholar]
- 42.Romano G, Kwong LN. Diagnostic and therapeutic applications of miRNA-based strategies to cancer immunotherapy. Cancer Metastasis Rev. 2018;37:45–53. doi: 10.1007/s10555-017-9716-7. [DOI] [PubMed] [Google Scholar]
- 43.Liang B, Li Y, Wang T. A three miRNAs signature predicts survival in cervical cancer using bioinformatics analysis. Sci Rep. 2017;7:5624. doi: 10.1038/s41598-017-06032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JY, Huang JC, Chen G, Wei DM. Expression level and potential target pathways of miR-1-3p in colorectal carcinoma based on 645 cases from 9 microarray datasets. Mol Med Rep. 2018;17:5013–5020. doi: 10.3892/mmr.2018.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sang C, Chao C, Wang M, Zhang Y, Luo G, Zhang X. Identification and validation of hub microRNAs dysregulated in esophageal squamous cell carcinoma. Aging (Albany NY) 2020;12:9807–9824. doi: 10.18632/aging.103245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Shen F, Wang C, Lu W, Wei J, Shang A, Wang C. MiR-1-3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression. Tumour Biol. 2017;39:1010428317698383. doi: 10.1177/1010428317698383. [DOI] [PubMed] [Google Scholar]
- 47.Li SM, Wu HL, Yu X, Tang K, Wang SG, Ye ZQ, Hu J. The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J Exp Clin Cancer Res. 2018;37:219. doi: 10.1186/s13046-018-0895-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He RQ, Wu PR, Xiang XL, Yang X, Liang HW, Qiu XH, Yang LH, Peng ZG, Chen G. Downregulated miR-23b-3p expression acts as a predictor of hepatocellular carcinoma progression: A study based on public data and RT-qPCR verification. Int J Mol Med. 2018;41:2813–2831. doi: 10.3892/ijmm.2018.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge BH, Li GC. Long non-coding RNA SNHG17 promotes proliferation, migration and invasion of glioma cells by regulating the miR-23b-3p/ZHX1 axis. J Gene Med. 2020;22:e3247. doi: 10.1002/jgm.3247. [DOI] [PubMed] [Google Scholar]
- 50.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: A new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, Liang J, Zhang Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi: 10.1016/j.ebiom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci. 2018;11:225. doi: 10.3389/fnmol.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma S, Fu T, Zhao S, Gao M. MicroRNA-34a-5p suppresses tumorigenesis and progression of glioma and potentiates Temozolomide-induced cytotoxicity for glioma cells by targeting HMGA2. Eur J Pharmacol. 2019;852:42–50. doi: 10.1016/j.ejphar.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Zhao Y, Lu Q, Fei X, Lu C, Li C, Chen H. MiR-34a-5p inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in esophageal squamous cell carcinoma by targeting LEF1 and inactivation of the Hippo-YAP1/TAZ signaling pathway. J Cancer. 2020;11:3072–3081. doi: 10.7150/jca.39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G, Popovic B, Tao J, Jiang A. Overexpression of COX7RP promotes tumor growth and metastasis by inducing ROS production in hepatocellular carcinoma cells. Am J Cancer Res. 2020;10:1366–1383. [PMC free article] [PubMed] [Google Scholar]
- 56.Kong X, Zhang J, Li J, Shao J, Fang L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem Biophys Res Commun. 2018;501:486–493. doi: 10.1016/j.bbrc.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 57.Chen X, Yue B, Zhang C, Qi M, Qiu J, Wang Y, Chen J. MiR-130a-3p inhibits the viability, proliferation, invasion, and cell cycle, and promotes apoptosis of nasopharyngeal carcinoma cells by suppressing BACH2 expression. Biosci Rep. 2017;37:BSR20160576. doi: 10.1042/BSR20160576. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Dai X, Guo X, Liu J, Cheng A, Peng X, Zha L, Wang Z. Circular RNA circGRAMD1B inhibits gastric cancer progression by sponging miR-130a-3p and regulating PTEN and p21 expression. Aging (Albany NY) 2019;11:9689–9708. doi: 10.18632/aging.102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, Zöller M. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 60.Rawat M, Kadian K, Gupta Y, Kumar A, Chain PSG, Kovbasnjuk O, Kumar S, Parasher G. MicroRNA in pancreatic cancer: From biology to therapeutic potential. Genes (Basel) 2019;10:752. doi: 10.3390/genes10100752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouattour M, Raymond E, Qin S, Cheng AL, Stammberger U, Locatelli G, Faivre S. Recent developments of c-Met as a therapeutic target in hepatocellular carcinoma. Hepatology. 2018;67:1132–1149. doi: 10.1002/hep.29496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 63.Heist RS, Shim HS, Gingipally S, Mino-Kenudson M, Le L, Gainor JF, Zheng Z, Aryee M, Xia J, Jia P, et al. MET exon 14 skipping in non-small cell lung cancer. Oncologist. 2016;21:481–486. doi: 10.1634/theoncologist.2015-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raghav K, Bailey AM, Loree JM, Kopetz S, Holla V, Yap TA, Wang F, Chen K, Salgia R, Hong D. Untying the gordion knot of targeting MET in cancer. Cancer Treat Rev. 2018;66:95–103. doi: 10.1016/j.ctrv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 65.Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy. Nat Rev Cancer. 2018;18:341–358. doi: 10.1038/s41568-018-0002-y. [DOI] [PubMed] [Google Scholar]
- 66.Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in lung cancer: Will expectations finally be MET? J Thorac Oncol. 2017;12:15–26. doi: 10.1016/j.jtho.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deying W, Feng G, Shumei L, Hui Z, Ming L, Hongqing W. CAF-derived HGF promotes cell proliferation and drug resistance by up-regulating the c-Met/PI3K/Akt and GRP78 signalling in ovarian cancer cells. Biosci Rep. 2017;37:BSR20160470. doi: 10.1042/BSR20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer. 2013;32:253–265. doi: 10.5732/cjc.013.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.