Abstract
The role of non-SMC condensin I complex subunit G (NCAPG) in breast cancer remains unclear. The present study used online databases, reverse transcription-quantitative PCR, flow cytometry and western blotting to determine the expression levels, prognosis and potential molecular mechanisms underlying the role of NCAPG in breast cancer. The association between NCAPG expression and several different clinicopathological parameters in patients with breast cancer was determined, and the results revealed that NCAPG expression was negatively associated with estrogen receptor and progesterone receptor positive status, but was positively associated with HER2 positive status, Nottingham Prognostic Index score and Scarff-Bloom-Richardson grade status. Furthermore, upregulated expression levels of NCAPG resulted in a poor prognosis in patients with breast cancer. A total of 27 microRNAs (miRNAs/miRs) were predicted to target NCAPG, among which four miRNAs (miR-101-3p, miR-195-5p, miR-214-3p and miR-944) were predicted to most likely regulate NCAPG expression in breast cancer. A total of 261 co-expressed genes of NCAPG were identified, including cell division cyclin 25 homolog C (CDC25C), and pathway enrichment analysis indicated that these co-expressed genes were significantly enriched in the p53 signaling pathway. CDC25C expression was downregulated in breast cancer and was associated with a poor prognosis. These findings suggested that upregulated NCAPG expression may be a prognostic biomarker of breast cancer.
Keywords: microRNAs, non-SMC condensin I complex subunit G, cell division cyclin 25 homolog C, p53 signaling pathway, breast cancer
Introduction
Cancer has been one of the leading causes of disease-associated death worldwide since 2010, and the incidence and mortality rates associated with various types of cancer are increasing (1). Breast cancer is the most commonly diagnosed type of cancer and the leading cause of cancer-associated death in women, with ~17,000,000 new cases occurring annually in the world (2), and its incidence and mortality rates are expected to increase significantly in the next 5–10 years. In developing countries, the incidence rate will increase by 55% and the mortality rate will increase by 58% within 20 years (2). There are several causes of breast cancer, among which gene mutations are closely associated with the development and/or progression of breast cancer (3,4). The diagnosis, treatment and prognosis of patients with breast cancer has improved; however, the median survival of the majority of patients with metastatic cancer remains low (24 months) (5). Additionally, a majority of patients with breast cancer will experience a relapse following treatment. Therefore, identifying effective prognostic biomarkers may assist in predicting the prognosis of patients with cancer and the curative effects of several therapeutics, consequently providing a suitable treatment plan and ultimately improving the clinical efficacy and survival outcomes.
MicroRNAs (miRNAs/miRs) are endogenous non-coding RNAs 18–23 nucleotides in length that can regulate the expression of non-coding and coding RNAs (6,7). In human cells, a single miRNA can directly regulate a large number of RNAs (8). Thus, dysregulated miRNA expression can result in the aberrant expression levels of coding RNAs in cancer cells (5). In addition, dysregulation of miRNA expression may result in the acquisition of malignant properties, leading to cancer progression, metastasis and treatment resistance (9–12). Ke et al (13) revealed that miR-148b serves an inhibitory role in hepatocellular carcinoma (HCC) and that the miR-148b-CSF1 axis induces tumor-associated macrophage infiltration, thus promoting HCC. Han et al (14) demonstrated that miR-338-5p negatively regulates the inhibitor of DNA binding-1, altering 5-FU chemoresistance and suppressing the metastasis of esophageal squamous cell carcinoma. Additionally, it has been revealed that miR-1231 expression is downregulated in pancreatic cancer and serves a role in the TNM stage of pancreatic cancer, suggesting that miR-1231 exhibits an inhibitory role on the metastasis and development of pancreatic cancer (15).
Non-SMC condensin I complex subunit G (NCAPG) is a subunit of the condensed protein complex, which is responsible for the condensation and stabilization of chromosomes during mitosis and meiosis (16). According to Gene Ontology (GO) analysis, the pathways associated with NCAPG are involved in cell cycle progression, mitosis and concentration of cell cycle chromosomes in the prophase and metaphase (17). An increasing number of studies has revealed increased NCAPG expression in prostate cancer, glioma and lung cancer, indicating the notable involvement of NCAPG in various biological functions (18–20). For example, Arai et al (19) indicated that NCAPG was regulated by miR-99a-3p and that its overexpression was involved in the pathogenesis of castration-resistant prostate cancer. Liang et al (20) revealed that CENPE, KIF14 and NCAPG were direct targets of miR-137 or miR-6500-3p, and that knockdown of CENPE, KIF14 or NCAPG combined with temozolomide treatment resulted in a combined suppressive effect on pediatric high-grade glioma cell proliferation. Studies have revealed that upregulated NCAPG expression is associated with a poor prognosis in patients with prostate cancer (21). Ke et al (13) demonstrated that small homologous RNA-mediated knockdown of NCAPG significantly impaired cell viability, caused aberrant mitotic division, fragmented the mitochondrial network and increased cell death. Notably, upregulated NCAPG expression is significantly associated with a poor overall and disease-free survival in patients with HCC (22). However, to the best of our knowledge, previous studies on the role of NCAPG in breast cancer and the potential mechanisms by which NCAPG affects breast cancer are insufficient (23), and the association between the expression levels of NCAPG and prognosis in breast cancer has not been determined.
In the present study, NCAPG expression was assessed in breast cancer. Subsequently, the prognostic effect of NCAPG in patients with breast cancer based on their clinicopathological characteristics was determined. Additionally, the underlying mechanisms by which NCAPG regulated breast cancer development and progression were determined.
Materials and methods
Human protein atlas database analysis
Expression levels of NCAPG protein in different human normal and cancer tissues were determined using the Human Protein Atlas database (https://www.proteinatlas.org/) (24).
UALCAN database analysis
The UALCAN database (ualcan.path.uab.edu/) is an interactive network resource that provides a convenient method to obtain open cancer transcriptome data from The Cancer Genome Atlas (TCGA) (25). In the present study, the gene expression levels and the correlations between two genes were assessed using the UALCAN database. The UALCAN database performed the statistical analysis using a log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Breast cancer gene expression miner
Breast cancer gene expression miner (Bc-GenExMiner; bcgenex.centregauducheau.fr/) is an online platform that can analyze gene expression, prognosis and the associations in breast cancer (26,27). Based on the different clinicopathological characteristics of patients with breast cancer, Bc-GenExMiner was used to detect the expression levels of NCAPG in breast cancer. In addition, Bc-GenExMiner calculated the correlation between NCAPG and cell division cyclin 25 homolog C (CDC25C) expression. P<0.05 was considered to indicate a statistically significant difference.
Kaplan-Meier Plotter database analysis
Kaplan-Meier Plotter (kmplot.com/) is based on data obtained from Gene Expression Omnibus, which includes data on gene expression and survival information of patients with cancer (28). Two probes (218662_s_at and 218663_at) were used to analyze the association between NCAPG expression and the overall survival, relapse-free survival, distant metastasis-free survival and post-progression survival rates in patients with breast cancer. Briefly, NCAPG was selected in the database, and the Kaplan-Meier survival curves were plotted. The cases were classified into low and high expression groups based on the median NCAPG expression. Subsequently, the cohorts were compared using Kaplan-Meier survival plots, and the online tool calculated the hazard ratio (HR), 95% CI and log-rank P-values. Additionally, the Kaplan Meier-Plotter database was used to evaluate the prognostic values of the predicted miRNAs in breast cancer. A log-rank P<0.05 was considered to indicate a statistically significant difference.
PrognoScan database analysis
The PrognoScan database (dna00.bio.kyutech.ac.jp/PrognoScan/) collates clinically annotated publicly available cancer microarray datasets for bladder, brain, breast, blood, esophageal, colorectal and head and neck cancer (29). In the present study, PrognoScan was used to evaluate the biological association between the expression levels and prognosis of NCAPG in breast cancer. The datasets were as follows: GSE19615 (30), GSE12276 (31), GSE6532-GPL570 (32), GSE9195 (33), GSE12093 (34), GSE11121 (35), GSE1378 (36), GSE1379 (37), GSE2034 (38), GSE1456-GPL96 (39), GSE7378 (40), E-TABM-158 (41), GSE3494-GPL96 (42), GSE4922-GPL96 (43), GSE2990 (33) and GSE7390 (44). A log-rank P<0.05 was considered to indicate a statistically significant difference.
starBase database
The starBase database (starbase.sysu.edu.cn/) is an open-source platform that systematically defines the RNA-RNA and protein-RNA interactions from crosslinking-immunoprecipitation and high-throughput sequencing (45). In the present study, the upstream miRNAs of NCAPG were predicted using starBase, and the negative correlations between each miRNA expression and NCAPG expression were identified using starBase. R<-0.1 and P<0.05 were used as the thresholds for identification of potential miRNAs targeted by NCAPG for further investigation.
OncomiR database analysis
OncomiR (oncomir.org/) is an online resource for exploring dysregulated miRNA expression in several types of cancer. OncomiR acquires RNA-sequencing (seq), miRNA-seq and clinical data from TCGA, then performs a systematic statistical analysis to identify the dysregulated miRNAs associated with the development and progression of several types of cancer (46). OncomiR was used to explore the expression levels of miRNAs in breast cancer. A log-rank P<0.05 was considered to indicate a statistically significant difference.
Gene expression profiling interactive analysis (GEPIA) database
GEPIA (gepia.cancer-pku.cn/) is a database that provides key interactive and customizable functions, including correlation analysis, differential expression analysis, patient survival analysis, dimensionality reduction and similar gene detection analysis (47). In the present study, GEPIA was used to determine the co-expressed genes of NCAPG in breast cancer.
cBio cancer genomics portal (cBioPortal) database analysis
cBioPortal (cbioportal.org/) is an open online access resource for interactive analysis of multidimensional datasets for cancer genomics, acquiring data from >5,000 tumor samples from 172 cancer studies (48). cBioPortal was used to obtain the genes that were co-expressed with NCAPG in breast cancer, and the correlated genes with a Pearson's r>0.3 were selected for further analysis.
Enrichr database analysis
Enrichr (amp.pharm.mssm.edu/Enrichr/) is a comprehensive application, including new gene-set libraries, an optional approach to rank the enriched terms and a JavaScript library. Data-driven documents offer multiple interactive visualization approaches to show the enrichment results. The top 10 enriched GO items and pathways were displayed using Enrichr.
Human tissue samples and cell culture
A total of 24 pairs of breast cancer and adjacent normal tissues (>1 cm from the tumor) were collected from the Tongji Medical College of Huazhong Science and Technology University (Wuhan, China) between May 2019 and June 2019. Additionally, 113 paired cancer samples and adjacent tissue data were obtained from TCGA database. The 24 pairs of fresh breast cancer samples were acquired from excision surgery and immediately preserved in RNAlater Stabilization Solution (Qiagen GmbH) overnight at 4°C, and subsequently stored at −80°C until total RNA was extracted. All patients were female, with a median age of 57 years (range, 35–67 years). None of the patients had previously received pre-operative chemotherapy or radiotherapy. All procedures involving human participants in the present study were performed according to the ethical standards of the Ethics Committee of Tongji Medical College of Huazhong Science and Technology University, and written informed consent was provided by each participant. The human breast cancer MCF-7 cell line was obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. MCF-7 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2.
Cell transfection
Small interfering (si)RNAs targeting NCAPG and a scrambled control siRNA used as the negative control (NC) were purchased from Guangzhou RiboBio Co., Ltd. NCAPG siRNAs and the NC (50 nM) were transfected using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) into cells, according to the manufacturer's protocol. Each well was transfected with 2 µg siRNAs. After 48 h of transfection at 37°C, cells were collected for subsequent experiments. Expression levels of NCAPG following transfection were detected using reverse transcription-quantitative (RT-qPCR). The sequences of siRNAs and NC used are listed in Table SI.
RT-qPCR
Total RNA from the human samples or the cell lines was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse-transcribed according to the manufacturer's protocol using HiScript®II RT SuperMix (Vazyme Biotech Co., Ltd.). qPCR was performed in a Roche LightCycle480 II Real-Time PCR Detection System using SYBR Premix Ex Taq (cat. no. RR420A; Takara Biotechnology Co., Ltd.). The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 10 min, followed by 45 cycles at 92°C for 15 sec and 72°C for 5 min. All the measurements were performed in triplicate. The sequences of the primers used in the present study are listed in Table SI. Quantitative mRNA data were normalized and presented as a ratio to GAPDH, calculated using the 2−ΔΔCq method (49).
Flow cytometry
For cell cycle distribution, transfected cells were stained with propidium iodide (BD Biosciences) and fixed in ice-cold 75% ethanol overnight at 4°C. After fixing, the cells were washed and resuspended twice in PBS, and were then incubated with propidium iodide according to the manufacturer's protocol (BD Biosciences) and RNase for 30 min at room temperature. The cells were then analyzed using a FACSCalibur flow cytometer (BD Biosciences) with a laser beam at 488 nm. The data were analyzed using FlowJo v7.6 (FlowJo LLC).
Western blotting
Cells were lysed using RIPA lysis buffer (Thermo Fisher Scientific, Inc.) containing protease inhibitors. Total protein concentration was measured using a Bradford assay. Subsequently, 20 µg protein/lane was separated via 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked at room temperature in TBS-Tween (0.1% Tween-20) with 5% skimmed milk for 1 h and then incubated overnight at 4°C with rabbit anti-CDC25C (1:200; cat. no. ab32444; Abcam) and mouse anti-GAPDH (1:1,000; cat. no. ab8245; Abcam) primary antibodies. Subsequently, the membranes were washed with Tris-HCl solution + 0.1% Tween-20 three times for 10 min and were incubated with horseradish peroxidase-conjugated anti-rabbit (1:3,000; cat. no. ab150077; Abcam) and anti-mouse (1:3,000; cat. no. ab150113; Abcam) secondary antibodies for 1 h at room temperature. Signals were visualized using an enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) and imaged using a GelDocXR+ (Bio-Rad Laboratories, Inc.). GAPDH was used as the loading control.
Statistical analysis
Data are presented as the mean ± standard error of the mean. Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, Inc.). Unpaired Student's t-test was used to analyze the differences between two groups and paired Student's t-test was used to compare NCAPG expression between paired cancer and normal tissues. Multiple comparisons were analyzed using a one-way ANOVA with Tukey's post-hoc test. The χ2 test was used to analyze the association between NCAPG expression and clinicopathological features. The correlations between CDC25C and NCAPG expression were analyzed using Spearman's and Pearson's correlation analyses. Kaplan-Meier survival analysis and univariate/multivariate Cox proportional hazards analyses with log-rank method were used to compare the survival between the high and low NCAPG expression groups. The enrichment pathways of the co-expressed genes of NCAPG were analyzed using Fisher's exact test to obtain the Cox P-value. P<0.05 was considered to indicate a statistically significant difference.
Results
NCAPG expression is upregulated in breast cancer tissues
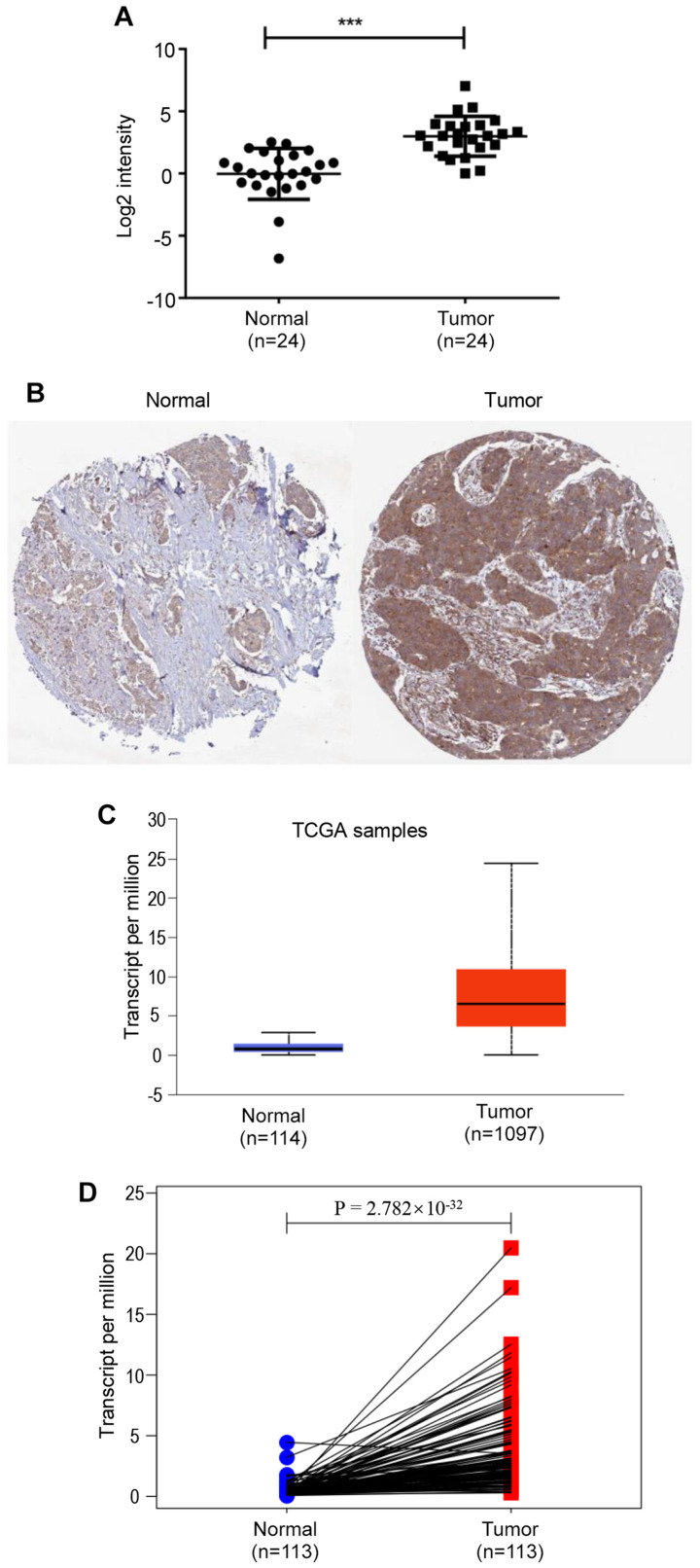
In the present study, the expression levels of NCAPG in 24 pairs of breast cancer and adjacent normal breast tissues were determined using RT-qPCR. NCAPG expression was significantly upregulated in breast cancer tissues compared with in normal breast tissues (Fig. 1A). Subsequently, NCAPG expression in breast cancer was analyzed using the UALCAN database based on data obtained from TCGA, revealing that NCAPG mRNA expression was significantly higher in breast cancer tissues compared with in normal tissues (Fig. 1C). Furthermore, the results from 113 paired cancer samples and adjacent tissues from TCGA database demonstrated that NCAPG mRNA expression was significantly upregulated in tumor tissues compared with in para-cancerous tissues (Fig. 1D). Additionally, the protein expression levels of NCAPG in breast cancer tissues were higher compared with in normal breast tissues (Fig. 1B), analyzed using the GEPIA database. Subsequently, the association between NCAPG expression and clinicopathological characteristics of patients with breast cancer was analyzed using TCGA breast cancer data. The results revealed that NCAPG expression was significantly associated with estrogen receptor (ER) status (P<0.0001), progesterone receptor (PR) status (P<0.0001), HER2 status (P=0.0034), T stage (P<0.001) and TNM pathological stage (50) (P=0.0002) (Table I). The current results indicated that NCAPG expression was significantly upregulated in breast cancer and was associated with the progression of breast cancer.
Figure 1.
NCAPG expression in breast cancer. (A) NCAPG expression in clinical breast cancer tissues compared with in matched adjacent normal tissues (n=24). (B) Protein expression levels of NCAPG in breast cancer and normal breast tissues were analyzed using immunohistochemical staining from the Human Protein Atlas database (https://www.proteinatlas.org/ENSG00000109805-NCAPG/pathology/breast+cancer#img). (C) NCAPG expression in breast cancer tissues compared with in normal tissues was analyzed using the UALCAN database. (D) NCAPG expression in 113 paired breast cancer samples and adjacent tissues from TCGA. ***P<0.001. NCAPG, non-SMC condensin I complex subunit G; TCGA, The Cancer Genome Atlas.
Table I.
Association between NCAPG expression and clinicopathological characteristics of patients with breast cancer in The Cancer Genome Atlas.
| NCAPG expression | |||
|---|---|---|---|
| Features | Cases, n | High/low, n | P-value |
| Age at diagnosis, years | |||
| ≤51 | 349 | 184/165 | 0.2035 |
| >51 | 686 | 333/353 | |
| Estrogen receptor status | |||
| Positive | 765 | 311/454 | <0.0001 |
| Negative | 221 | 180/41 | |
| Progesterone receptor status | |||
| Positive | 663 | 255/408 | <0.0001 |
| Negative | 320 | 235/85 | |
| HER2 status | |||
| Positive | 173 | 104/69 | 0.0034 |
| Negative | 721 | 344/377 | |
| Pathological stage | |||
| I | 171 | 63/108 | 0.0002 |
| II/III/IV | 841 | 443/398 | |
| T stage | |||
| 1 | 268 | 102/166 | <0.0001 |
| 2/3/4 | 764 | 413/351 | |
| N stage | |||
| N0/N1 | 823 | 413/410 | 0.9643 |
| N2/N3 | 192 | 96/96 | |
| M stage | |||
| M0 | 861 | 452/409 | 0.6740 |
| M1 | 21 | 12/9 | |
T, tumor; N, node; M, metastasis; NCAPG, non-SMC condensin I complex subunit G.
Association between NCAPG expression and clinicopathological characteristics of patients with breast cancer
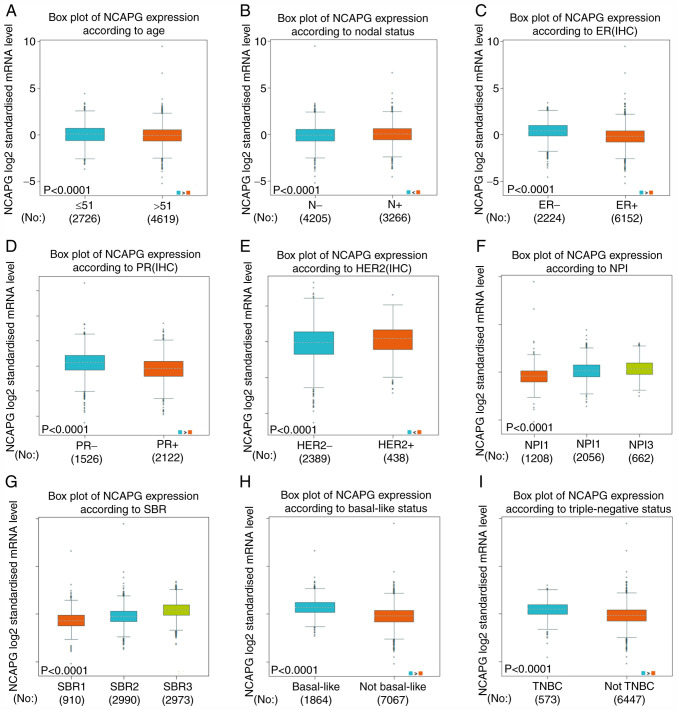
NCAPG expression and its association with the different clinicopathological parameters of patients with breast cancer were analyzed using the Bc-GenExMiner database. The average expression levels of NCAPG were significantly upregulated in patients <51 years old (Fig. 2A), nodal-positive (Fig. 2B), ER− (Fig. 2C), PR− (Fig. 2D) and HER2+ positive (Fig. 2E).
Figure 2.
Differences in NCAPG expression in patients with breast cancer based on different clinicopathological features from The Cancer Genome Atlas. NCAPG expression according to (A) age, (B) N status, (C) ER status, (D) PR status, (E) HER2 status, (F) NPI score, (G) SBR grade, (H) basal-like status and (I) triple-negative status. ER, estrogen receptor; PR, progesterone receptor; NPI, Nottingham Prognostic Index; SBR, Scarff-Bloom-Richardson; NCAPG, non-SMC condensin I complex subunit G; IHC, immunohistochemistry; TNBC, triple-negative breast cancer; N, nodal.
There was a positive association between the expression levels of NCAPG and the Nottingham Prognostic Index (NPI) score (51) and Scarff-Bloom-Richardson (SBR) grade (52), as shown in Fig. 2F and G. Additionally, the average NCAPG expression was significantly increased in basal-like (Fig. 2H) and triple negative breast cancer cases compared with in non-basal-like and non-triple-negative breast cancer cases, respectively (Fig. 2H and I).
The aforementioned results suggested that patients with higher expression levels of NCAPG were more likely to develop clinically advanced or more aggressive breast cancer types.
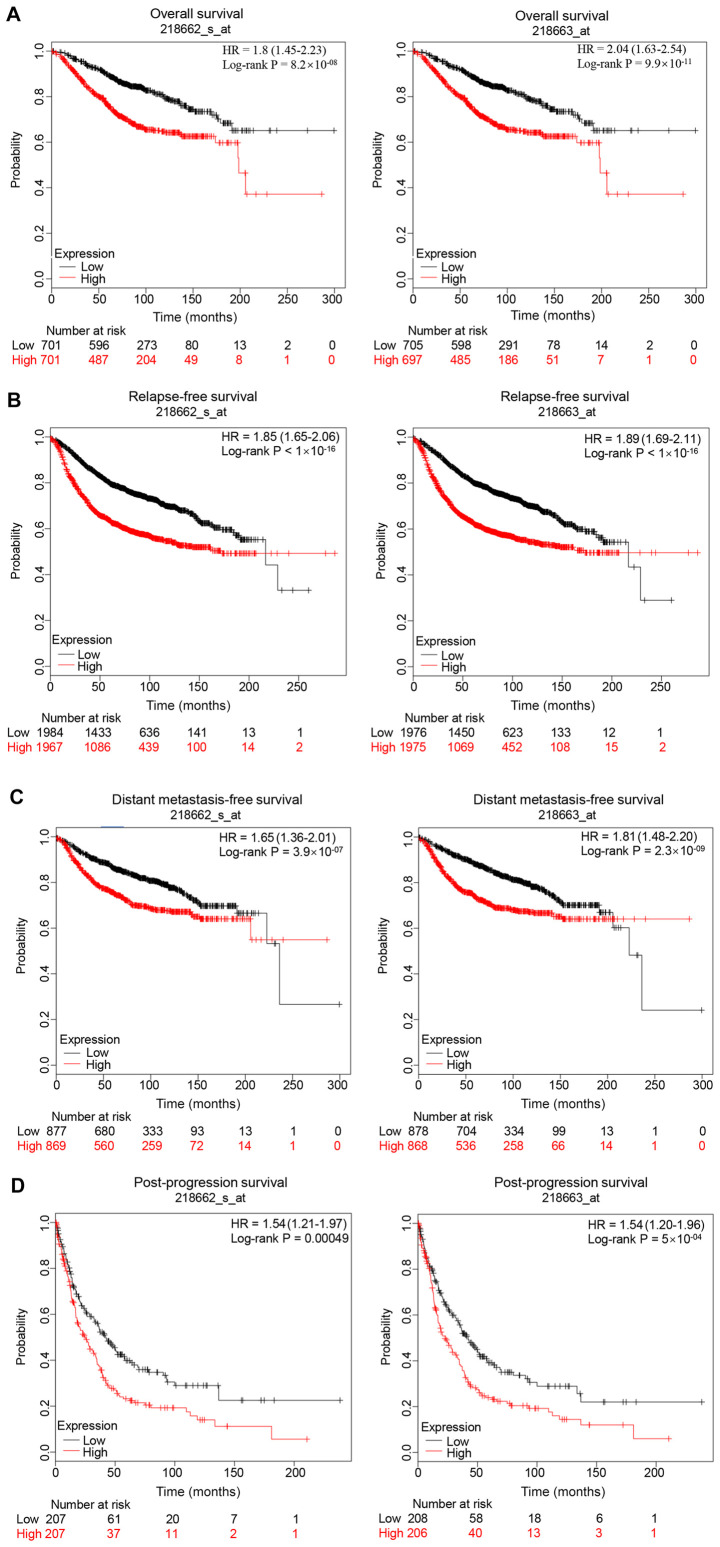
Patients with breast cancer with high expression levels of NCAPG have a poor prognosis
The prognostic value of NCAPG in breast cancer was evaluated. The Kaplan Meier-Plotter database contained two probes (218662_s_at and 218663_at) of NCAPG expression. As shown in Fig. 3, high expression levels of NCAPG resulted in a less favorable prognosis in patients with breast cancer compared with low expression levels, including less favorable overall survival, relapse-free survival, distant metastasis-free survival and post-progression survival rates in both probes. In addition, the association between NCAPG expression in breast cancer tissues and prognosis was determined using the PrognoScan database. The results revealed that the expression levels of NCAPG were negatively associated with overall survival (GSE1456-GPL96), disease-specific survival (GSE1456-GPL96), relapse-free survival (GSE12276, GSE6532-GPL570 and GSE1456-GPL96), disease-free survival (GSE7378) and distant metastasis-free survival (GSE6532-GPL570, GSE12093, GSE11121 and GSE2034) (Table II).
Figure 3.
Prognostic value of NCAPG expression (Affymetrix ID, 218662_s_at and 218663_at) in patients with breast cancer. (A) Overall survival, (B) relapse-free survival, (C) distant metastases-free survival and (D) post-progression survival curves of patients with breast cancer based on NCAPG expression. NCAPG, non-SMC condensin I complex subunit G; HR, hazard ratio.
Table II.
Association between NCAPG expression and prognosis in patients with breast cancer using the PrognoScan database.
| Dataset | Endpoint | Patient number | Probe ID | Cox P-value | Hazard ratio (95% CI) |
|---|---|---|---|---|---|
| GSE19615 | Distant metastasis-free survival | 115 | 218663_at | 0.3759 | 1.41 (0.66–3.05) |
| GSE19615 | Distant metastasis-free survival | 115 | 218662_s_at | 0.5339 | 1.32 (0.55–3.18) |
| GSE12276 | Relapse-free survival | 204 | 218663_at | 0.0001 | 1.53 (1.24–1.90) |
| GSE12276 | Relapse-free survival | 204 | 218662_s_at | 6.88×10−05 | 1.51 (1.23–1.85) |
| GSE6532-GPL570 | Distant metastasis-free survival | 87 | 218662_s_at | 0.1155 | 1.37 (0.93–2.03) |
| GSE6532-GPL570 | Distant metastasis-free survival | 87 | 218663_at | 0.0361 | 1.47 (1.03–2.11) |
| GSE6532-GPL570 | Relapse-free survival | 87 | 218662_s_at | 0.1155 | 1.37 (0.93–2.03) |
| GSE6532-GPL570 | Relapse-free survival | 87 | 218663_at | 0.0361 | 1.47 (1.03–2.11) |
| GSE9195 | Distant metastasis-free survival | 77 | 218663_at | 0.4349 | 1.30 (0.67–2.50) |
| GSE9195 | Distant metastasis-free survival | 77 | 218662_s_at | 0.1658 | 1.67 (0.81–3.43) |
| GSE9195 | Relapse-free survival | 77 | 218662_s_at | 0.2247 | 1.49 (0.78–2.83) |
| GSE9195 | Relapse-free survival | 77 | 218663_at | 0.4998 | 1.22 (0.68–2.21) |
| GSE12093 | Distant metastasis-free survival | 136 | 218662_s_at | 0.0062 | 3.38 (1.41–8.09) |
| GSE12093 | Distant metastasis-free survival | 136 | 218663_at | 0.0123 | 2.52 (1.22–5.19) |
| GSE11121 | Distant metastasis-free survival | 200 | 218662_s_at | 4.52×10−05 | 3.01 (1.77–5.12) |
| GSE11121 | Distant metastasis-free survival | 200 | 218663_at | 0.0099 | 1.78 (1.15–2.75) |
| GSE1378 | Relapse-free survival | 60 | 21899 | 0.7551 | 1.07 (0.71–1.59) |
| GSE1379 | Relapse-free survival | 60 | 21899 | 0.7284 | 1.08 (0.69–1.71) |
| GSE2034 | Distant metastasis-free survival | 286 | 218662_s_at | 0.0330 | 1.43 (1.03–1.98) |
| GSE2034 | Distant metastasis-free survival | 286 | 218663_at | 0.0025 | 1.67 (1.20–2.34) |
| GSE1456-GPL96 | Overall survival | 159 | 218662_s_at | 0.0008 | 2.91 (1.56–5.43) |
| GSE1456-GPL96 | Disease-specific survival | 159 | 218663_at | 0.0006 | 2.67 (1.52–4.71) |
| GSE1456-GPL96 | Overall survival | 159 | 218663_at | 0.0020 | 2.06 (1.30–3.27) |
| GSE1456-GPL96 | Relapse-free survival | 159 | 218662_s_at | 0.0003 | 3.16 (1.70–5.88) |
| GSE1456-GPL96 | Disease-specific survival | 159 | 218662_s_at | 0.0002 | 3.99 (1.90–8.34) |
| GSE1456-GPL96 | Relapse-free survival | 159 | 218663_at | 0.0005 | 2.26 (1.42–3.57) |
| GSE7378 | Disease-free survival | 54 | 218663_at | 0.0335 | 1.92 (1.05–3.52) |
| GSE7378 | Disease-free survival | 54 | 218662_s_at | 0.0709 | 1.74 (0.95–3.18) |
| E-TABM-158 | Overall survival | 117 | 218662_s_at | 0.2037 | 0.80 (0.56–1.13) |
| E-TABM-158 | Distant metastasis-free survival | 117 | 218662_s_at | 0.6812 | 1.09 (0.72–1.66) |
| E-TABM-158 | Relapse-free survival | 117 | 218663_at | 0.3616 | 0.84 (0.58–1.22) |
| E-TABM-158 | Disease-specific survival | 117 | 218663_at | 0.0900 | 0.67 (0.42–1.06) |
| E-TABM-158 | Distant metastasis-free survival | 117 | 218663_at | 0.7572 | 1.07 (0.70–1.63) |
| E-TABM-158 | Overall survival | 117 | 218663_at | 0.3616 | 0.84 (0.58–1.22) |
| E-TABM-158 | Disease-specific survival | 117 | 218662_s_at | 0.0352 | 0.63 (0.42–0.97) |
| E-TABM-158 | Relapse-free survival | 117 | 218662_s_at | 0.2038 | 0.80 (0.56–1.13) |
| GSE3494-GPL96 | Disease-specific survival | 236 | 218662_s_at | 0.0003 | 2.45 (1.51–3.96) |
| GSE3494-GPL96 | Disease-specific survival | 236 | 218663_at | 0.0016 | 1.80 (1.25–2.61) |
| GSE4922-GPL96 | Disease-free survival | 249 | 218663_at | 0.0007 | 1.61 (1.22–2.13) |
| GSE4922-GPL96 | Disease-free survival | 249 | 218662_s_at | 4.93×10−05 | 2.24 (1.52–3.32) |
| GSE2990 | Distant metastasis-free survival | 125 | 218662_s_at | 0.0760 | 1.42 (0.96–2.10) |
| GSE2990 | Relapse-free survival | 125 | 218662_s_at | 0.1234 | 1.27 (0.94–1.72) |
| GSE2990 | Relapse-free survival | 62 | 218663_at | 0.1058 | 1.65 (0.90–3.03) |
| GSE2990 | Distant metastasis-free survival | 54 | 218662_s_at | 0.0024 | 2.45 (1.37–4.37) |
| GSE2990 | Distant metastasis-free survival | 125 | 218663_at | 0.0797 | 1.51 (0.95–2.39) |
| GSE2990 | Relapse-free survival | 125 | 218663_at | 0.1058 | 1.37 (0.94–2.00) |
| GSE2990 | Distant metastasis-free survival | 54 | 218663_at | 0.1420 | 1.74 (0.83–3.67) |
| GSE2990 | Relapse-free survival | 62 | 218662_s_at | 0.0037 | 2.07 (1.27–3.38) |
| GSE7390 | Distant metastasis-free survival | 198 | 218663_at | 0.1751 | 1.15 (0.94–1.42) |
| GSE7390 | Overall survival | 198 | 218663_at | 0.0655 | 1.24 (0.99–1.55) |
| GSE7390 | Relapse-free survival | 198 | 218662_s_at | 0.2174 | 1.15 (0.92–1.44) |
| GSE7390 | Relapse-free survival | 198 | 218663_at | 0.1340 | 1.14 (0.96–1.34) |
| GSE7390 | Distant metastasis-free survival | 198 | 218662_s_at | 0.2798 | 1.16 (0.88–1.54) |
| GSE7390 | Overall survival | 198 | 218662_s_at | 0.2032 | 1.21 (0.90–1.63) |
Identification of miRNAs that potentially regulate NCAPG
The starBase database was used to predict the upstream miRNAs of mRNAs. The miRNAs that potentially regulated NCAPG were predicted using starBase. A total of 27 miRNAs (hsa-miR-488-3p, hsa-miR-181b-5p, hsa-miR-181a-5p, hsa-miR-15a-5p, hsa-miR-494-3p, hsa-miR-543, hsa-miR-495-3p, hsa-miR-422a, hsa-miR-497-5p, hsa-miR-27a-3p, hsa-miR-23a-3p, hsa-miR-181c-5p, hsa-miR-181d-5p, hsa-miR-128-3p, hsa-miR-124-3p, hsa-miR-124-5p, hsa-miR-15b-5p, hsa-miR-378a-3p, hsa-miR-340-5p, hsa-miR-340-3p, hsa-miR-590-5p, hsa-miR-27b-3p, hsa-miR-374b-5p, hsa-miR-424-5p, hsa-miR-506-3p, hsa-miR-374a-5p and hsa-miR-23b-3p) were identified to potentially regulate NCAPG expression. It is well known that certain miRNAs negatively regulate mRNA expression (53–55). Thus, the correlations (using Pearson's correlation analysis) between the expression levels of the predicted miRNAs and NCAPG were explored using TCGA. The results revealed that 12 miRNAs were inversely correlated with NCAPG expression (Table III). Subsequently, the prognostic values of the 12 miRNAs in patients with breast cancer were assessed using Kaplan-Meier Plotter. Low expression levels of miR-101-3p, miR-195-5p, miR-214-3p, miR-944 and miR-5691 were significantly associated with a poor prognosis in patients with breast cancer, compared with high expression levels (Fig. 4). The prognostic analyses of the other miRNAs are shown in Fig. S1. Overall, the present results and previous studies revealed that the expression levels of these 4 miRNAs (miR-101-3p, miR-195-5p, miR-214-3p and miR-944) were downregulated in breast cancer, resulting in a poor prognosis, and were also negatively correlated with NCAPG expression.
Table III.
Pearson correlation between predicted miRNAs and non-SMC condensin I complex subunit G expression.
| Predicted miRNA | R | P-value |
|---|---|---|
| miR-101-3p | −0.264 | 8.14×10−19 |
| miR-195-5p | −0.210 | 2.94×10−12 |
| miR-214-3p | −0.104 | 5.96×10−04 |
| miR-369-3p | −0.108 | 3.67×10−04 |
| miR-381-3p | −0.165 | 4.55×10−08 |
| miR-410-3p | −0.201 | 2.17×10−11 |
| miR-432-5p | −0.167 | 3.43×10−08 |
| miR-494-3p | −0.128 | 2.52×10−05 |
| miR-543 | −0.102 | 8.04×10−04 |
| miR-655-3p | −0.174 | 8.54×10−09 |
| miR-944 | −0.140 | 3.42×10−06 |
| miR-5691 | −0.234 | 6.31×10−15 |
miRNA/miR, microRNA.
Figure 4.
Prognostic values of five potential miRNAs in breast cancer analyzed using Kaplan-Meier plotter. (A) miR-101, (B) miR-195, (C) miR-214, (D) miR-944 and (E) miR-5691. miRNA/miR, microRNA; HR, hazard ratio.
GO functional annotation and pathway enrichment analysis of co-expressed genes of NCAPG
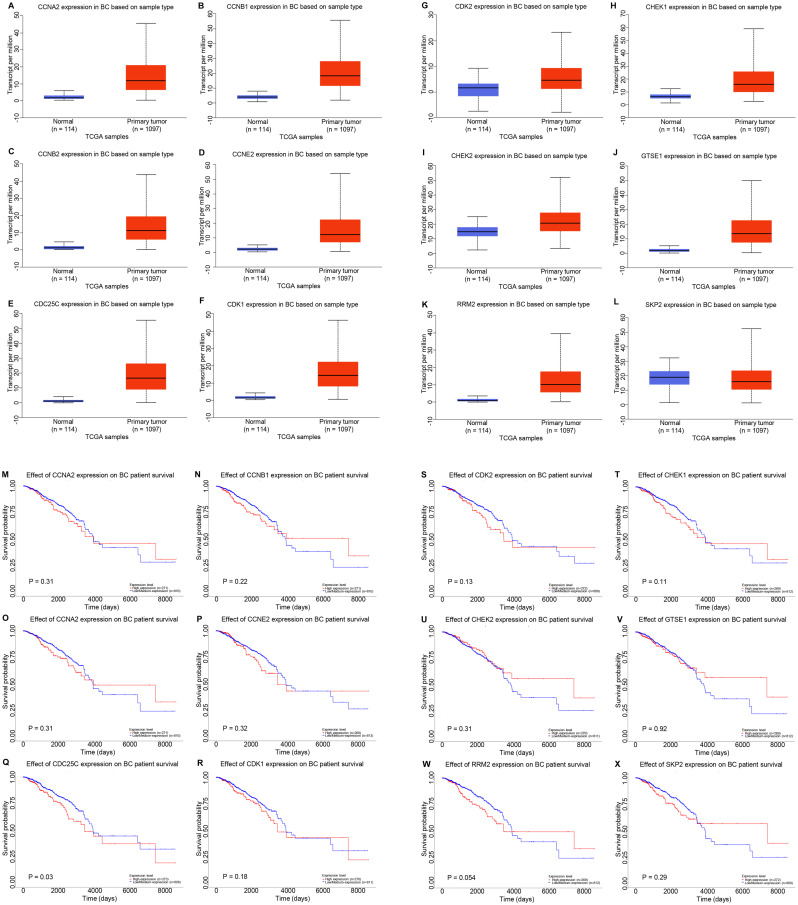
GEPIA, UALCAN and cBioPortal were used to analyze the co-expressed genes of NCAPG. A total of 261 co-expressed genes of NCAPG were identified in these three databases (Fig. 5A). To determine the functions of the co-expressed genes, GO functional annotation and pathway enrichment analysis was performed using Enrichr. The functional annotation was stratified into molecular function, cellular component and biological process. Additionally, the cell signaling pathways from the National Cancer Institute (NCI)-Nature, KEGG and Panther were analyzed for pathway enrichment using the Enrichr database. As shown in Fig. 5B, D and F, the top 10 enriched GO terms in each category included ‘DNA metabolic process’, ‘mitotic cell cycle phase transition’ and ‘mitotic sister chromatid segregation’ in the biological process category; ‘spindle’, ‘microtubule cytoskeleton’ and ‘nuclear chromosome part’ in the cellular component category; and ‘DNA-dependent ATPase activity’, ‘microtubule binding’ and ‘tubulin binding’ in the molecular function category. Fig. 5C, E and G presents the top 10 enriched signaling pathways (such as ‘cell cycle’, ‘DNA replication’ and ‘PLK1 signaling pathway’) in the NCI-Nature, KEGG and Panther databases, respectively. The p53 signaling pathway was the intersection of the pathways that were significantly enriched in the three databases (Table SII). Thus, the corresponding genes of the p53 signaling pathway were analyzed in KEGG, Panther and NCI-Nature pathways (Fig. 5H), revealing a total of 12 genes [cyclin (CCN)A2, CCNB1, CCNB2, CCNE2, CDC25C, CDK1, CDK2, checkpoint kinase (CHEK)1, CHEK2, G2 and S phase expressed 1 (GTSE1), ribonucleotide reductase regulatory subunit M2 (RRM2) and S-phase kinase-associated protein 2 (SKP2)] enriched in the p53 signaling pathway.
Figure 5.
Pathway enrichment and GO functional annotation analysis of the co-expressed genes of NCAPG. (A) Venn diagram of the co-expressed genes of NCAPG from GEPIA, UALCAN and cBioPortal databases. GO functional annotation according to (E) biological process, (F) cellular component and (G) molecular function of the 261 co-expressed genes of NCAPG. Pathway enrichment analysis of the 261 co-expressed genes of NCAPG according to (B) KEGG, (C) Panther and (D) NCI-Nature. (H) Venn diagram of the genes enriched in the p53 signaling pathway from Panther KEGG and NCI-Nature databases. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NCAPG, non-SMC condensin I complex subunit G; GEPIA, Gene Expression Profiling Interactive Analysis; cBioPortal, cBio Cancer Genomics Portal; NCI, National Cancer Institute.
Upregulated CDC25C expression is associated with dysregulation of the p53 signaling pathway
In order to determine the effects of p53 signaling on NCAPG-mediated progression of breast cancer, the UALCAN database was used to assess the expression levels of the 12 genes enriched in the p53 signaling pathway (Fig. 6A-L). The results revealed that SKP2 expression (Fig. 6L) was downregulated in breast cancer, while CCNA2 (Fig. 6A), CCNB1 (Fig. 6B), CCNB2 (Fig. 6C), CCNE2 (Fig. 6D), CDC25C (Fig. 6E), CDK1 (Fig. 6F), CDK2 (Fig. 6G), CHEK1 (Fig. 6H), CHEK2 (Fig. 6I), GTSE1 (Fig. 6J) and RRM2 (Fig. 6K) expression was upregulated in breast cancer compared with in normal breast tissue samples. The prognostic value of these 12 genes was assessed using Kaplan-Meier Plotter (Fig. 6M-X). High CDC25C expression resulted in a significantly worse prognosis in patients with breast cancer compared with low CDC25C expression (Fig. 6Q). For the other 11 genes, there were no significant associations based on expression and survival. Therefore, it was hypothesized that CDC25C expression was closely associated with NCAPG expression in breast cancer. GEPIA, UALCAN, bc-GenExMiner and cBioPortal were used to further detect the positive correlation between CDC25C and NCAPG expression in breast cancer (Fig. S2).
Figure 6.
Potential co-expressed genes of NCAPG in breast cancer. Expression levels of (A) CCNA2, (B) CCNB1, (C) CCNB2, (D) CCNE2, (E) CDC25C, (F) CDK1, (G) CDK2, (H) CHEK1, (I) CHEK2, (J) GTSE1, (K) RRM2 and (L) SKP2 in breast cancer analyzed using UALCAN. Prognostic roles of (M) CCNA2, (N) CCNB1, (O) CCNB2, (P) CCNE2, (Q) CDC25C, (R) CDK1, (S) CDK2, (T) CHEK1, (U) CHEK2, (V) GTSE1, (W) RRM2 and (X) SKP2 in breast cancer analyzed using Kaplan-Meier Plotter. NCAPG, non-SMC condensin I complex subunit G; BC, breast cancer; CCN, cyclin; CDC25C, cell division cyclin 25 homolog C; CHEK, checkpoint kinase; GTSE1, G2 and S phase expressed 1; RRM2, ribonucleotide reductase regulatory subunit M2; SKP2, S-phase kinase-associated protein 2; TCGA, The Cancer Genome Atlas.
To preliminarily assess the association between CDC25C and NCAPG in breast cancer, the expression levels of CDC25C and NCAPG were determined following knockdown of NCAPG using siRNA-NCAPG in the common breast cancer MCF-7 cell line. The knockdown effects of siRNA-NCAPG are presented in Fig. 7A. Additionally, a significant decrease in CDC25C mRNA and protein expression was observed following NCAPG-knockdown (Fig. 7B and C). Previous studies have demonstrated that CDC25C is a cell cycle regulatory protein and serves an important role in the cell cycle (56,57). Thus, the effects of NCAPG-knockdown on cell cycle progression in MCF-7 cells were determined. Cell cycle analysis revealed that NCAPG-knockdown resulted in an increased proportion of cells in the S phase (31.95% with siRNA-NCAPG1) and G1 phase (76.78% with siRNA-NCAPG2) compared with the control group (S phase, 12.95% and G1 phase, 67.45%), suggesting that downregulation of NCAPG resulted in S/M and G1/M arrest of MCF-7 cells (Fig. 7D-F). Overall, the current results suggested that NCAPG may be associated with p53 signaling through regulation of CDC25C, in turn promoting cell cycle progression in breast cancer.
Figure 7.
NCAPG-knockdown decreases CDC25C expression and results in cell cycle arrest at the G2/M phase. (A) Effect of siRNA-mediated knockdown of NCAPG in the MCF-7 cell line. CDC25C (B) mRNA and (C) protein expression following knockdown of NCAPG in MCF-7 cells. Effects of (D) siRNA-NC, (E) siRNA-NCAPG1 and (F) siRNA-NCAPG2 on cell cycle distribution in MCF-7 cells. *P<0.05. NCAPG, non-SMC condensin I complex subunit G; CDC25C, cell division cyclin 25 homolog C; siRNA, small interfering RNA; NC, negative control.
Discussion
NCAPG is a subunit of the condensin complex and is hypothesized to serve important roles in the mitosis and meiosis responsible for the condensation and stabilization of chromosomes (16). Several studies have revealed that higher expression levels of NCAPG serve a role in cell proliferation, angiogenesis and tumor therapy resistance in cancer (22,58,59). Wang et al (22) revealed that transient inhibition of NCAPG using specific siRNAs resulted in a significant decrease in cell proliferation and migration in vitro in hepatocellular carcinoma. Ai et al (59) reported that miR-181c induced hepatocellular carcinoma suppression via the regulation of NCAPG expression. However, at present, the specific mechanism by which NCAPG exerts its effects in cancer remains unclear.
To the best of our knowledge, the role and mechanism of NCAPG in the development and progression of breast cancer has not been extensively studied. NCAPG was upregulated in breast cancer clinical samples compared with in matched adjacent normal clinical samples. Additionally, NCAPG expression in breast cancer was upregulated based on analysis of the UALCAN database. The average NCAPG expression was negatively associated with PR and ER status, and positively associated with HER2 status, NPI score, SBR grade, basal-like status and triple-negative status in breast cancer. Thus, it was hypothesized that upregulated NCAPG expression was associated with the progression of breast cancer. Kaplan-Meier Plotter analysis results revealed that high NCAPG expression was associated with a less favorable progression in patients with breast cancer. Additionally, the prognostic role of NCAPG analyzed by PrognoScan suggested that low NCAPG expression was associated with a favorable prognosis in patients with breast cancer.
Subsequently, the potential mechanisms by which NCAPG exerted its effects in breast cancer were studied. miRNAs can regulate genes post-transcriptionally (60). Among the miRNAs predicted to potentially regulate NCAPG, 4 miRNAs (miR-101-3p, miR-195-5p, miR-214-3p and miR-944) were previously reported to function as tumor suppressor genes in breast cancer (Table IV) (4,61–65), combined with the prognosis analysis, text mining, expression analysis and correlation analysis.
Table IV.
Text mining the roles of potential miRNAs in breast cancer.
| First author, year | PubMed ID | miRNAs | Direct targets | Function | Overall effect | (Refs.) |
|---|---|---|---|---|---|---|
| Zhang et al, 2018 | 30583076 | miR-101-3p | Med19 | Inhibit tumor growth and metastasis | Tumor suppressor | (61) |
| Wang et al, 2020 | 32226515 | miR-101-3p | E2F8 | Inhibit tumor growth and metastasis | Tumor suppressor | (62) |
| Yang et al, 2019 | 30621700 | miR-195-5p | CCNE1 | Inhibit proliferation, migration, invasion | Tumor suppressor | (63) |
| Luo et al, 2014 | 24402230 | miR-195-5p | CCNE1 | Inhibit cell proliferation, reduced cell colony formation, suppressed cell migration | Tumor suppressor | (4) |
| Han et al, 2019 | 31539134 | miR-214-3p | Survivin | Inhibit proliferation | Tumor suppressor | (64) |
| Flores-pérez et al, 2016 | 27377268 | miR-944 | SIAH1, PTP4A1 | Inhibit migration and invasion | Tumor suppressor | (65) |
miRNA/miR, microRNA.
A total of 261 co-expressed genes of NCAPG were identified by co-expression analysis, and pathway enrichment analysis indicated that these co-expressed genes were significantly enriched in the p53 signaling pathway. Numerous studies have demonstrated that the activation of the p53 signaling pathway is associated with the development of various types of cancer, including breast cancer (66–68). Furthermore, the association between NCAPG and p53 has been previously reported. Liu et al (69) revealed that silencing NCAPG in SMMC-7721 and BEL-7404 cells resulted in decreased cell proliferation and increased apoptosis, and was associated with increased mRNA expression levels of p53, p27 and Bad. Additionally, 6 crucial proteins (NDC80, ESR1, ZWINT, NCAPG, ENO3 and CENPF) in HCC were primarily enriched in cell cycle regulation and p53 signaling pathway (70). A total of 12 genes (CCNA2, CCNB1, CCNB2, CCNE2, CDC25C, CDK1, CDK2, CHEK1, CHEK2, GTSE1, RRM2 and SKP2) were enriched in the p53 signaling pathway. TCGA breast cancer data, UALCAN and Kaplan-Meier Plotter were used to further determine the roles of these 12 genes, including their expression and prognostic value in breast cancer. Among these genes, CDC25C expression was upregulated in breast cancer, and high CDC25C expression resulted in a significantly worse prognosis compared with low CDC25C expression. CDC25C is a cell cycle regulatory protein that serves a role in cell cycle progression (4,63). In the present study, CDC25C expression was significantly decreased following knockdown of NCAPG. Additionally, downregulation of NCAPG resulted in G2/M arrest in MCF-7 breast cancer cells. It was preliminarily confirmed that NCAPG inhibited cell cycle progression in breast cancer by regulating CDC25C. Additionally, the present results indicated that NCAPG was downregulated by four targeted miRNAs, possibly by elevating CDC25C expression to promote the development of breast cancer.
Although the present study revealed that NCAPG expression was associated with the clinical prognosis of breast cancer, due to the limitations of online databases, only a single factor analysis was performed as opposed to a multifactor comparison of NCAPG expression. The grouping mode of the bc-GenExMiner database requires further optimization. Additionally, the association between NCAPG, CDC25C, the four identified miRNAs (mir-101-3p, mir-195-5p, mir-214-3p and mir-944) and the p53 signaling pathway in breast cancer samples and the potential therapeutic application of NCAPG needs to be further clarified.
In conclusion, the present study revealed that NCAPG expression was upregulated in breast cancer, and this upregulated expression was associated with a less favorable prognosis. In addition, NCAPG expression was targeted by four miRNAs and was associated with the dysregulation of the p53 signaling pathway, via increased expression levels of CDC25C. The current findings suggested that NCAPG may serve a key role in the development of breast cancer and may become a new therapeutic target for breast cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant on. 81802676) and the Wuhan Youth Cadre Project (grant nos. 2017zqnlxr01 and 2017zqnlxr02).
Funding
The present study was supported by the National Natural Science Foundation of China (grant on. 81802676) and the Wuhan Youth Cadre Project (grant nos. 2017zqnlxr01 and 2017zqnlxr02).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets (GSE19615, GSE12276, GSE6532-GPL570, GSE9195, GSE12093, GSE11121, GSE1378, GSE1379, GSE2034, GSE1456-GPL96, GSE7378, E-TABM-158, GSE3494-GPL96, GSE4922-GPL96, GSE2990 and GSE7390) generated and/or analyzed during the current study are available in The Cancer Genome Atlas (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Authors' contributions
MD conceived the study. MD and TX designed the study. XC and HL performed the experiments. MD analyzed the data and wrote the manuscript. XL and WX made substantial contributions to the conception of the study, secured funding and supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures involving human participants in the present study were performed according to the ethical standards of the Ethics Committee of Tongji Medical College of Huazhong Science and Technology University (Wuhan, China; approval no. S340), and written informed consent was provided by each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics for hispanics/latinos, 2012. CA Cancer J Clin. 2012;62:283–298. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495:1594–1600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Luo Q, Wei C, Li X, Li J, Chen L, Huang Y, Song H, Li D, Fang L. MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncol Rep. 2014;31:1096–1102. doi: 10.3892/or.2014.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaney ML, Sprunck-Harrild K, Ruddy KJ, Ligibel J, Barry WT, Baker E, Meyer M, Emmons KM, Partridge AH. Study protocol for young & strong: A cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health. 2015;15:37. doi: 10.1186/s12889-015-1346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 10.Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Ke M, Zhang Z, Cong L, Zhao S, Li Y, Wang X, Lv Y, Zhu Y, Dong J. MicroRNA-148b-colony-stimulating factor-1 signaling-induced tumor-associated macrophage infiltration promotes hepatocellular carcinoma metastasis. Biomed Pharmacother. 2019;120:109523. doi: 10.1016/j.biopha.2019.109523. [DOI] [PubMed] [Google Scholar]
- 14.Han L, Cui D, Li B, Xu WW, Lam AKY, Chan KT, Zhu Y, Lee NPY, Law SYK, Guan XY, et al. MicroRNA-338-5p reverses chemoresistance and inhibits invasion of esophageal squamous cell carcinoma cells by targeting Id-1. Cancer Sci. 2019;110:3677–3688. doi: 10.1111/cas.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang S, Wang J, Chen S, Tian R, Zeng H, Wang L, Xia M, Zhu H, Zuo C. Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer Med. 2019;8:7728–7740. doi: 10.1002/cam4.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlein A, Takasuga A, Setoguchi K, Pfuhl R, Flisikowski K, Fries R, Klopp N, Fürbass R, Weikard R, Kühn C. Dissection of genetic factors modulating fetal growth in cattle indicates a substantial role of the non-SMC condensin I complex, subunit G (NCAPG) gene. Genetics. 2009;183:951–964. doi: 10.1534/genetics.109.106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Thomas PD. GO functional similarity clustering depends on similarity measure, clustering method, and annotation completeness. BMC Bioinformatics. 2019;20:155. doi: 10.1186/s12859-019-2752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Xuan Y, Gao B, Sun X, Miao S, Lu T, Wang Y, Jiao W. Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res. 2018;10:3383–3392. doi: 10.2147/CMAR.S173941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arai T, Okato A, Yamada Y, Sugawara S, Kurozumi A, Kojima S, Yamazaki K, Naya Y, Ichikawa T, Seki N. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med. 2018;7:1988–2002. doi: 10.1002/cam4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai CF, Chao ME, Liu DJ, Chu SS, Chen W, Liu YR, et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget. 2016;7:19723–19737. doi: 10.18632/oncotarget.7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409–420. doi: 10.1038/bjc.2017.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Gao B, Tan PY, Handoko YA, Sekar K, Deivasigamani A, Seshachalam VP, OuYang HY, Shi M, Xie C, et al. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. FASEB J. 2019;33:8759–8770. doi: 10.1096/fj.201802213RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Ren L, Chen H, Pan J, Zhang Z, Kuang X, Chen X, Bao W, Lin C, Zhou Z, et al. NCAPG confers trastuzumab resistance via activating SRC/STAT3 signaling pathway in HER2-positive breast cancer. Cell Death Dis. 2020;11:547. doi: 10.1038/s41419-020-02753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontén F, Schwenk JM, Asplund A, Edqvist PH. The human protein atlas as a proteomic resource for biomarker discovery. J Intern Med. 2011;270:428–446. doi: 10.1111/j.1365-2796.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 25.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 27.Jézéquel P, Frénel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, Desmedt C, Sotiriou C, Szallasi Z, Iglehart JD, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 33.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Sieuwerts AM, McGreevy M, Casey G, Cufer T, Paradiso A, Harbeck N, Span PN, Hicks DG, Crowe J, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116:303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Böhm D, von Törne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kölbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 36.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, Gillet C, Ellis P, Ryder K, Reid JF, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 39.Hall P, Ploner A, Bjöhle J, Huang F, Lin CY, Liu ET, Miller LD, Nordgren H, Pawitan Y, Shaw P, et al. Hormone-replacement therapy influences gene expression profiles and is associated with breast-cancer prognosis: A cohort study. BMC Med. 2006;4:16. doi: 10.1186/1741-7015-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Yau C, Gray JW, Chew K, Dairkee SH, Moore DH, Eppenberger U, Eppenberger-Castori S, Benz CC. Enhanced NF kappa B and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer. 2007;7:59. doi: 10.1186/1471-2407-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 44.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 45.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database Issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong NW, Chen Y, Chen S, Wang X. OncomiR: An online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics. 2018;34:713–715. doi: 10.1093/bioinformatics/btx627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Martinez A, Krop IE, Hillman DW, Polley MY, Parker JS, Huebner L, Hoadley KA, Shepherd J, Tolaney S, Henry NL, et al. Survival, pathologic response, and genomics in CALGB 40601 (alliance), a neoadjuvant phase III trial of paclitaxel-trastuzumab with or without lapatinib in HER2-positive breast cancer. J Clin Oncol. 2020;38:4184–4193. doi: 10.1200/JCO.20.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsharawy KA, Mohammed OJ, Aleskandarany MA, Hyder A, El-Gammal HL, Abou-Dobara MI, Green AR, Dalton LW, Rakha EA. The nucleolar-related protein Dyskerin pseudouridine synthase 1 (DKC1) predicts poor prognosis in breast cancer. Br J Cancer. 2020;123:1543–1552. doi: 10.1038/s41416-020-01045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuba MG, Lester SC, Bowman T, Stokes SM, Taneja KL, Garber JE, Dillon DA. Histopathologic features of breast cancer in Li-Fraumeni syndrome. Mod Pathol. 2020 Jul 7; doi: 10.1038/s41379-020-0610-4. (Online ahead of print) [DOI] [PubMed] [Google Scholar]
- 53.Lou W, Chen J, Ding B, Chen D, Zheng H, Jiang D, Xu L, Bao C, Cao G, Fan W. Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation. J Transl Med. 2018;16:266. doi: 10.1186/s12967-018-1639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lou W, Liu J, Ding B, Xu L, Fan W. Identification of chemoresistance-associated miRNAs in breast cancer. Cancer Manag Res. 2018;10:4747–4757. doi: 10.2147/CMAR.S172722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lou W, Liu J, Gao Y, Zhong G, Ding B, Xu L, Fan W. MicroRNA regulation of liver cancer stem cells. Am J Cancer Res. 2018;8:1126–1141. [PMC free article] [PubMed] [Google Scholar]
- 56.Liao WL, Lin JY, Shieh JC, Yeh HF, Hsieh YH, Cheng YC, Lee HJ, Shen CY, Cheng CW. Induction of G2/M phase arrest by diosgenin via activation of Chk1 kinase and Cdc25C regulatory pathways to promote apoptosis in human breast cancer cells. Int J Mol Sci. 2019;21:172. doi: 10.3390/ijms21010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Z, Zhang X, Yu P, Chen X, Lu P, Li M, Liu X, Li Z, Wei F, Wang K, et al. Cinobufagin induces cell cycle arrest at the G2/M phase and promotes apoptosis in malignant melanoma cells. Front Oncol. 2019;9:853. doi: 10.3389/fonc.2019.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Su R, Shan C, Gao C, Wu P. Non-SMC condensin I complex, subunit G (NCAPG) is a novel mitotic gene required for hepatocellular cancer cell proliferation and migration. Oncol Res. 2018;26:269–276. doi: 10.3727/096504017X15075967560980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ai J, Gong C, Wu J, Gao J, Liu W, Liao W, Wu L. MicroRNA-181c suppresses growth and metastasis of hepatocellular carcinoma by modulating NCAPG. Cancer Manag Res. 2019;11:3455–3467. doi: 10.2147/CMAR.S197716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertoli G, Cava C, Castiglioni I. The potential of miRNAs for diagnosis, treatment and monitoring of breast cancer. Scand J Clin Lab Invest Suppl. 2016;245:S34–S39. doi: 10.1080/00365513.2016.1208444. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Gao D, Fang K, Guo Z, Li L. Med19 is targeted by miR-101-3p/miR-422a and promotes breast cancer progression by regulating the EGFR/MEK/ERK signaling pathway. Cancer Lett. 2018;444:105–115. doi: 10.1016/j.canlet.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, Wang L, Tang L, Luo J, Ji H, Zhang W, Zhou J, Li Q, Miao L. Long noncoding RNA SNHG6 promotes proliferation and angiogenesis of cholangiocarcinoma cells through sponging miR-101-3p and activation of E2F8. J Cancer. 2020;11:3002–3012. doi: 10.7150/jca.40592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Han LC, Wang H, Niu FL, Yan JY, Cai HF. Effect miR-214-3p on proliferation and apoptosis of breast cancer cells by targeting survivin protein. Eur Rev Med Pharmacol Sci. 2019;23:7469–7474. doi: 10.26355/eurrev_201909_18856. [DOI] [PubMed] [Google Scholar]
- 65.Flores-Pérez A, Marchat LA, Rodríguez-Cuevas S, Bautista VP, Fuentes-Mera L, Romero-Zamora D, Maciel-Dominguez A, de la Cruz OH, Fonseca-Sánchez M, Ruíz-García E, et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer. 2016;16:379. doi: 10.1186/s12885-016-2470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao J, Xia R, Chen J, Gao J, Luo X, Ke C, Ren C, Li J, Mi Y. Inhibition of esophageal-carcinoma cell proliferation by genistein via suppression of JAK1/2-STAT3 and AKT/MDM2/p53 signaling pathways. Aging (Albany NY) 2020;12:6240–6259. doi: 10.18632/aging.103019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang W, Hou L, Wei J, Du Y, Zhao Y, Deng X, Lin X. Hsa-miR-217 inhibits the proliferation, migration, and invasion in non-small cell lung cancer cells via targeting SIRT1 and P53/KAI1 signaling. Balkan Med J. 2020;37:208–214. doi: 10.4274/balkanmedj.galenos.2020.2019.9.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng X, Zhang J, Fang T, Wang X, Wang S, Ma Z, Xu Y, Han C, Sun M, Xu L, et al. The long non-coding RNA PIK3CD-AS2 promotes lung adenocarcinoma progression via YBX1-mediated suppression of p53 pathway. Oncogenesis. 2020;9:34. doi: 10.1038/s41389-020-0217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu K, Li Y, Yu B, Wang F, Mi T, Zhao Y. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can J Physiol Pharmacol. 2018;96:1246–1254. doi: 10.1139/cjpp-2018-0195. [DOI] [PubMed] [Google Scholar]
- 70.Liu ZK, Zhang RY, Yong YL, Zhang ZY, Li C, Chen ZN, Bian H. Identification of crucial genes based on expression profiles of hepatocellular carcinomas by bioinformatics analysis. PeerJ. 2019;7:e7436. doi: 10.7717/peerj.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets (GSE19615, GSE12276, GSE6532-GPL570, GSE9195, GSE12093, GSE11121, GSE1378, GSE1379, GSE2034, GSE1456-GPL96, GSE7378, E-TABM-158, GSE3494-GPL96, GSE4922-GPL96, GSE2990 and GSE7390) generated and/or analyzed during the current study are available in The Cancer Genome Atlas (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).