Abstract
This study aimed to investigate the frequency and characteristics of respiratory co-infections in COVID-19 patients in the intensive care unit (ICU). In this retrospective observational study, pathogens responsible for potential co-infections were detected by the bacterial culture, real-time polymerase chain reaction (RT-PCR), or serological fungal antigen tests. Demographic and clinical characteristics, as well as microbial results, were analyzed. Bacterial culture identified 56 (58.3%) positive samples for respiratory pathogens, with the most common bacteria being Burkholderia cepacia (18, 18.8%). RT-PCR detected 38 (76.0%) and 58 (87.9%) positive results in the severe and critical groups, respectively. Most common pathogens detected were Stenotrophomonas maltophilia (28.0%) and Pseudomonas aeruginosa (28.0%) in the severe group and S. maltophilia (45.5%) in the critical group. P. aeruginosa was detected more during the early stage after ICU admission. Acinetobacter baumannii and Staphylococcus aureus were more frequently identified during late ICU admission. Fungal serum antigens were more frequently positive in the critical group than in the severe group, and the positive rate of fungal serum antigens frequency increased with prolonged ICU stay. A high frequency of respiratory co-infections presented in ICU COVID-19 patients. Careful examinations and necessary tests should be performed to exclude these co-infections.
Keywords: COVID-19, ICU, Critically ill patient, Co-infection, Respiratory pathogen
Coronavirus disease 2019 (COVID-19) is a worldwide pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In addition to SARS-CoV-2, bacteria and fungi are reported to cause co-infections in critically ill patients with COVID-19, which increases its morbidity and mortality [1,2]. Within the first few days after SARS-CoV-2 infection, critically ill patients with COVID-19 often develop respiratory tract distortion or pulmonary dysbiosis, which can further progress into a secondary bacterial or fungal infection few weeks later [[3], [4], [5]]. A study from Cambridge, UK, reported that a high percentage (9/14) of patients with COVID-19 in the intensive care unit (ICU) had confirmed secondary ventilator-associated pneumonia (VAP) [6]. The rapid and accurate identification of bacteria or fungi that present as pathogenic or resident microorganisms during the course of COVID-19 should be an important step during the management of COVID-19 patients [[7], [8], [9]]. However, the exact frequency of co-infections, the pathogens responsible for the co-infections, and the progress of the co-infections in critically ill patients with COVID-19 remain largely unknown.
In the present study, we analyzed the frequency and characteristics of respiratory co-infections among critically ill COVID-19 patients in the ICU.
1. Materials and methods
1.1. Study design, participant selection, and data collection
This retrospective observational study included patients with confirmed COVID-19 who were admitted to the ICU at the Beijing Ditan Hospital, Capital Medical University, Beijing, China, from January 30, 2020 to April 13, 2020. The study was approved by the Ethics Committee of Beijing Ditan Hospital (KT2020-036-02).
The diagnosis of COVID-19 and the severity assessment were defined in accordance with the National Clinical Guidance for COVID-19 Diagnosis and Treatment of China (seventh edition) by the National Health Commission [10]. Severe patients were those patients with one of the following criteria: 1) no requirement for supplemental oxygen, 2) oxygen saturation at rest ≤93%, 3) respiratory rate more than 30 breaths/min, 4) respiratory distress. Critical patients were those with acute respiratory failure (PaO2/FiO2 < 300, 30 > breaths/min), shock, organ failure required mechanical ventilation or intensive care management considered as critical. The demographics, gender, age, clinical data, and microbiology results were obtained from the electronic medical records.
1.2. Laboratory technique
Sputum and bronchoalveolar lavage (BAL) samples were collected for quantitative bacterial culture. Specimens were streaked on agar plates, including blood agar, MacConkey's, and chocolate agar, and then were incubated at 37 °C (chocolate agar with 5% CO2). After incubation for 24 and 48 h, the bacteria were identified by a Phoneix-100 Automatic Microbiology System (BD, Maryland, USA). The results were reported according to the standard of the National Guide to Clinical Laboratory Procedures of China (third edition) [11].
Sputum and nasopharyngeal swab samples were used in a multiple real-time PCR (RT-PCR) assay using the Multiple Combined Real-time PCR Detection Kit (Uninovo Biological Technology, Jiangsu, China) for 15 respiratory pathogens including Streptococcus pyogenes, Staphylococcus aureus, Klebsiella pneumoniae, Haemophilus influenzae, Legionella pneumophila, Mycobacterium tuberculosis, Acinetobacter baumannii, Chlamydia pneumoniae (CP), Mycoplasma pneumoniae (MP), Moraxella catarrhalis (M. catarrhalis), Escherichia coli (E. coli), S. maltophilia, Streptococcus pneumoniae, P. aeruginosa, and Pneumocystis carinii (PC). Multiple RT-PCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, USA) and reported according to the manufacturer's instructions.
Fungal infection was determined by serological assays, including a (1,3)-β-D-glucan (G) test and a galactomannan (GM) test. Serum samples, including the early stage (1–5 days), middle stage (5–12 days), and late stage (≥12 days) after ICU admission, were collected and stored at −80 °C. The kits for the G and GM tests were purchased from Dynamiker Biotechnology Co., Ltd (Tianjin, China). The tests were performed using a Dynamiker Automatic ELISA Workstation (A200; Tianjin, China) and results are reported according to the manufacturer's instructions.
1.3. Statistical analyses
All statistical analyses were performed in SPSS Version 22.0 (IBM, New York, USA). Figures were generated using GraphPad Prism Version 6.0 (GraphPad Software, San Diego, Canada) and Illustrator Version CS6 (Adobe, San Jose, Canada). Abnormally distributed continuous variables are presented as median and interquartile range (IQR), and were compared with a Mann–Whitney test. Independent binomial variables are presented as percentages, and were analyzed by either Pearson's chi-square test or Fisher's exact test. A P value < 0.05 was considered statistically significant.
2. Results
A total of 20 COVID-19 patients admitted to the ICU from January 30, 2020 to April 13, 2020 were included in this study. There were 7 patients (35.0%) in the severe group with the median age of 39 years (interquartile range, IQR, 36–67 years). Thirteen patients (65.0%) were in the critical group with a median age of 69 years (IQR, 64–80 years). Compared to patients in the severe group, patients in the critical group was significantly older than in severe group (P < 0.01), and the length of stay in ICU of critical group was significantly (P < 0.01) longer than that of severe group. The critical group including 4 cases (30.8%) had significantly (P = 0.005) higher rate of patients supported by extracorporeal membrane oxygenation (ECMO) (see Table 1 ).
Table 1.
Demographic and clinical characteristics of critically ill COVID-19 patients upon ICU admission.
| Characteristic | Severe Group (n = 7) | Critical Group (n = 13) | P |
|---|---|---|---|
| Age (years), median (IQR) | 39 (36–67) | 69 (64–80) | <0.001 |
| Gender, n (%) | |||
| Female | 5 (71.4) | 6 (46.2) | 0.374 |
| Male | 2 (28.6) | 7 (53.8) | |
| Comorbidities, n (%) | |||
| Hypertension | 7 (53.8) | 1 (14.3) | 0.158 |
| Diabetes | 1 (7.7) | 1 (14.3) | 1.0 |
| Cardiovascular disease | 1 (7.7) | 0 | 1.0 |
| Chronic lung disease | 4 (30.8) | 0 | 0.249 |
| Tumor | 2 (15.4) | 0 | 0.521 |
| Antibiotics use, n (%) | 5 (71.4) | 12 (92.3) | 0.270 |
| Antifungal drugs use, n (%) | 3 (42.9) | 9 (69.2) | 0.359 |
| Oxygen therapy, n (%) | |||
| HFNO | 7 (100.0) | 1 (7.7) | 1.0 |
| Invasive ventilator | 0 | 12 (92.3) | 1.0 |
| ECMO | 0 | 4 (30.8) | 0.005 |
| Length of stay (days), median (IQR) | |||
| Hospitalized | 37 (32–47) | 43 (42–50) | 0.168 |
| ICU admitted | 12 (6–12) | 22 (21–44) | <0.001 |
IQR, interquartile range; HFNO, high-flow oxygen/non-invasive ventilator; ECMO, extracorporeal membrane oxygenation.
2.1. Identification of respiratory bacterial infections
In these 13 critical patients, 23 BAL samples and 73 sputum samples were obtained for bacterial cultures. A total of 56 respiratory samples (58.3%) were identified to have respiratory bacterial pathogens (Table 2 ). The two most common pathogens in the critical group were Burkholderia cepacia (18/96, 18.8%) and S. maltophilia (15/96, 15.6%).
Table 2.
Microbial culture and multiple RT-PCR assay results from critically ill COVID-19 patients.
| Characteristic | Total | Severe Group | Critical Group |
|---|---|---|---|
| Culture test undertaken, n (%) | |||
| BAL | 23 | 0 | 23 |
| Sputum | 76 | 3 | 73 |
| Culture results, n (%) | |||
| B. cepacia | 18 | 0 | 18 (18.8) |
| S. maltophilia | 15 | 0 | 15 (15.6) |
| S. aureus | 7 | 0 | 7 (7.3) |
| A. baumannii | 6 | 0 | 6 (6.3) |
| M. morganii | 5 | 0 | 5 (5.2) |
| E. cloacae | 4 | 0 | 4 (4.2) |
| K. pneumoniae | 1 | 0 | 1 (1.0) |
| No growth | 43 | 3 (100.0) | 40 (41.7) |
| RT-PCR test undertaken, n (%) | |||
| Nasopharyngeal swab | 75 | 20 | 55 |
| Sputum | 41 | 30 | 11 |
| RT-PCR results, n (%) | |||
| S. maltophilia | 44 | 14 (28.0) | 30 (45.5) |
| S. aureus | 36 | 8 (16.0) | 28 (42.4) |
| P. aeruginosa | 32 | 14 (28.0) | 18 (27.3) |
| A. baumannii | 21 | 10 (20.0) | 11 (16.7) |
| E. coli | 11 | 4 (8.0) | 7 (10.6) |
| M. catarrhalis | 9 | 5 (10.0) | 4 (6.1) |
| K. pneumoniae | 7 | 3 (6.0) | 4 (6.1) |
| H. influenzae | 2 | 1 (2.0) | 1 (1.5) |
| S. pneumoniae | 2 | 2 (4.0) | 0 |
| S. pyogenes | 1 | 0 | 1 (1.5) |
| Non-detected | 20 | 12 (24.0) | 8 (12.1) |
RT-PCR assay detected at least one bacterial pathogen in 38 of the 50 samples collected from the severe group, which yielded a positivity rate of 76.0% (Table 2). The most abundant pathogens were S. maltophilia (14, 28.0%), P. aeruginosa (14, 28.0%), and A. baumannii (10, 20.0%). Multiple RT-PCR assay detected significantly more pathogens in 58 of 66 specimens from the critical group, yielding a higher positivity rate of 87.9%. The most prevalent bacteria were S. maltophilia (30, 45.5%). The RT-PCR assay detected 31 more respiratory pathogens, including P. aeruginosa, E. coli, M. catarrhalis, H. influenzae, S. pneumoniae, and S. pyogenes, than the culture method. We note, however, that our RT-PCR assay was not designed to detect B. cepacia, M. morganii, and Escherichia cloacae. L. pneumophila, M. tuberculosis, CP, MP, and PC were not detected in either the severe group or the critical group.
As shown in Table 3 , in the 96 specimens with a positive result from the multiple RT-PCR assay, more than half (50, 52.1%) of the specimens had only one bacterium detected. Twenty-seven (28.1%), 12 (12.5%), and 7 (7.3) specimens had two, three, and four types of bacteria detected, respectively. There were no statistically significant differences in the frequency of different bacteria detected between the severe and critical groups (Table 3).
Table 3.
Bacteria detected from critically ill COVID-19 patients using a multiple RT-PCR assay.
| Characteristic | Total | Severe Group (n = 38) | Critical Group (n = 58) | P |
|---|---|---|---|---|
| Any bacteria | 96 (100.0) | 38 (100.0) | 58 (100.0) | 0.136 |
| Single bacterium | 50 (52.1) | 21 (55.3) | 29 (50.0) | 0.679 |
| Dual bacteria | 27 (28.1) | 12 (31.6) | 15 (25.9) | 0.542 |
| Triple bacteria | 12 (12.5) | 3 (7.9) | 9 (15.5) | 0.269 |
| Quadruple bacteria | 7 (7.3) | 2 (5.3) | 5 (8.6) | 0.700 |
2.2. Characteristics of bacterial pathogens through the ICU course
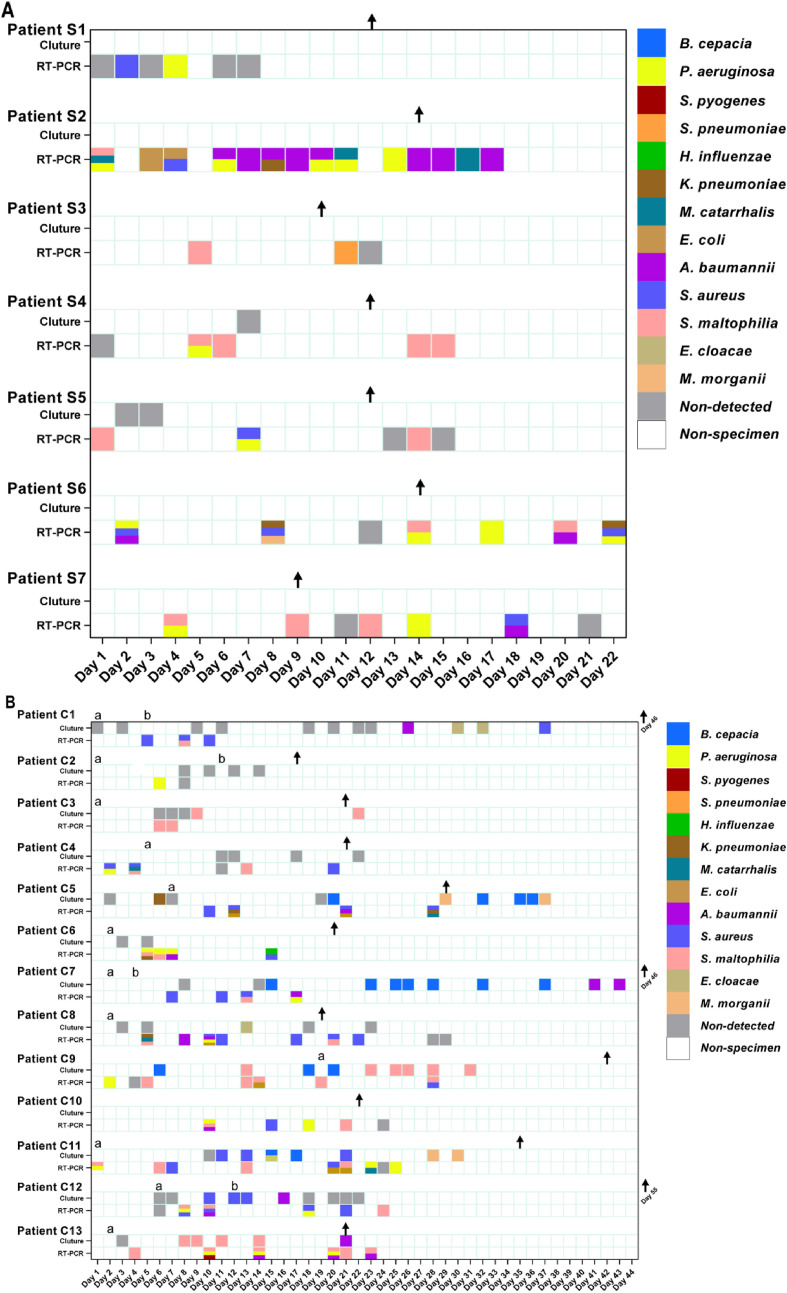
In the severe group, at least one respiratory bacterial pathogen was detected in all 7 patients via multiple RT-PCR assay during the first five days after ICU admission, with the most common bacterium being P. aeruginosa (5/7, 71.4%) (Fig. 1 A). Pathogens from the first specimens of 5 patients, including patients S2, S4, S5, S6, and S7, were still detected positive during the subsequent tested specimens. However, patients S1 and S3 had different pathogens detected in each subsequent tested specimen. Except patient S1, all other 6 patients had pathogens identified in the respiratory specimens after they were transferred to the general ward. In the severe group, the longest length of time for a positive respiratory bacterium, including K. pneumoniae, S. aureus and P. aeruginosa, was 22 days from patient S6. Bactria were still able to be identified from specimens from the 7 patients after their SARS-CoV-2 RT-PCR tests were negative.
Fig. 1.
Respiratory pathogens from each specimen in critically ill COVID-19 patients. Respiratory pathogens were detected by microbial culture and a multiple RT-PCR assay. Detected bacteria in individual specimens of each patient in the severe and critical groups are shown in A and B, respectively. The results from specimens were classified as the date of patients admitted to the ICU. Patients S1 – 7 had severe illness and patients C1 – 13 had critical illness. Note. a, the date of receiving mechanical ventilation; b, the date of supporting ECMO; ↑, the date of being transferred to the general ward.
In the critical group, bacteria were cultured from the respiratory samples from 9 out of 12 patients (except patients C2, C4 and C6). As shown in Fig. 1B, positive culture results were only obtained 6 days after ICU admission. Only 2 patients (patients C3 and C8) had only one bacterium identified (S. maltophilia and E. cloacae, respectively). At least two types of bacteria were cultured from the specimens of the other 7 patients. The specimen collected in patient C7 42 days after ICU admission was positive for A. baumannii, which was the longest time for a positive culture result. A positive result was obtained much earlier by RT-PCR than that obtained via the microbial culture test, especially in patients with an invasive ventilator. Samples obtained from patients, including patients C5, C6, C8, C11, and C12, a few days after the invasive ventilator application provided a positive result via multiple RT-PCR assay. P. aeruginosa was the most prevalent bacterium collected from the critical group during the early stages and was detected in the samples first collected from the 7 patients, including patients C2, C4, C6, C9, C10, C11, and C12. A. baumannii and S. aureus were more likely to be detected during the later stage of ICU admission, which was observed in patients C1, C5, C8, C9, and C13. In all of the collected specimens at different time points for multiple RT-PCR assay, P. aeruginosa persisted the longest, up to 25 days in Patient C11. Bacteria were still detected in specimens collected from 8 patients via multiple RT-PCR, even after their SARS-CoV-2 RT-PCR tests were negative.
2.3. Identification of fungal pathogens infections
There was no significant difference in the positivity rates of the G test and GM test from specimens collected from patients in the severe group during the early, middle, and late stages of ICU admission (P > 0.05) (Table 4 ). In the severe group, the positive rate of G tests increased to 57.1% during the middle stage and dropped to 50.0% in the late stage since only 4 patients stayed more than 12 days in the ICU. In the critical group, the rates of positive G tests and GM tests from the early and late stages were higher than those of the severe group. The positive G-test and GM-test rates tended to increase with the length of ICU stay, since the positive G-test rates from the late stage (84.6%) were much higher than that of the samples from the middle stage (69.2%) and early stage (46.2%).
Table 4.
G test and GM test results from critically ill COVID-19 patients according to disease course.
| ICU admission | No. of tested | Positive cases in severe group (%) |
No. of tested | Positive cases in critical group (%) |
||
|---|---|---|---|---|---|---|
| G test | GM test | G test | GM test | |||
| Early stage (1–5 days) | 7 | 2 (28.6) | 1 (14.3) | 13 | 6 (46.2) | 3 (23.1) |
| Middle stage (5–12 days) | 7 | 4 (57.1) | 3 (42.9) | 13 | 9 (69.2) | 4 (30.8) |
| Late stage (≥12 days) | 4 | 2 (50.0) | 2 (50.0) | 13 | 11 (84.6) | 7 (53.8) |
3. Discussion
In this study, we described the characteristics of co-infections in critically ill ICU patients with COVID-19. Culture results from sputum specimens or BAL specimens were available for 2 severe patients and 9 critical patients; however, there were no pathogens isolated in the early-stage specimens from the severe group, which might be due to the limited volume of the collected samples. In the critical group, B. cepacia and S. maltophilia were the two most common bacteria that were cultured. The majority of specimens that yielded a cultured organism were collected until 6 days after ICU admission. Most of the specimens collected during the late stage of ICU admission had a positive result from the bacterial culture. Thus far, previous studies have reported that the positive culture rates of respiratory bacterial pathogens from hospitalized patients with COVID-19 ranged from 2.0% to 17.2% [6,8,9]. The rate of bacterial co-infection in our study was higher than previous studies. This difference might be explained by the critical category, as well as the multiple times that the specimens were collected in our study [[12], [13], [14]]. This suggests that multiple attempts should be made to collect specimens from critically ill COVID-19 patients at different time points in order to identify pathogenic opportunistic bacteria. Additionally, a review, which included 806 COVID-19 cases, reported that a large percentage of COVID-19 patients (72%) received the broad-spectrum antimicrobial therapy. Our results showed that most of the cultured bacteria were detected during the middle and late stages of ICU admission. Thus, the selections and recommendations of empiric antimicrobial therapy for respiratory bacterial co-infection should not be only based on the results from early cultured specimens in critically ill patients with COVID-19 [15].
We used a commercially available kit for the multiple respiratory pathogens RT-PCR assay, which is able to detect 15 respiratory pathogens. Compared with the microbial culture test, the multiple RT-PCR assay identified more bacteria in specimens from patients both in the severe group and critical group. The distribution of positive results from the multiple RT-PCR assay tended to appear during the early-stage and middle-stage of ICU admittance. S. maltophilia, S. aureus, P. aeruginosa, and A. baumannii were the most common bacteria detected in this study. As these are frequently identified pathogens attributed to hospital-acquired pneumonia (HAP) in the ICU, it was difficult to distinguish colonization from infection due to the high sensitivity of the multiple RT-PCR assay [[16], [17], [18], [19]]. We also analyzed the distribution of pathogens identified using multiple RT-PCR assay throughout the ICU course. Our results showed that the multiple tests performed at different time points could improve the positive rates. A previous study showed that COVID-19 patients were more susceptible to developing VAP due to the increased duration of ventilation [20]. In our study, the majority of identified bacteria, either by culturing or multiple RT-PCR assay, were identified during the treatments without mechanical ventilation. Therefore, multiple RT-PCR assay in combination with a sensitive and rapid diagnostic test might shorten the time to pathogen identification and thereby decrease the risk of false-negative cultures, thus optimizing antimicrobial therapy in critically ill patients with COVID-19.
Respiratory viral infections, such as influenza, are recognized as an independent risk factor for co-infection with Aspergillus, which wis associated with a high mortality rate in critically ill patients admitted to the ICU. Positive (1,3)-β-D-glucan and galactomannan serum antigens in the critical group were more common than those in the severe group. However, Aspergillus was not identified in any specimens by culturing, which might be due to the frequent use of antifungal prophylaxis in these patients. However, the positive rate of fungal antigenemia was high, indicating that fungal co-infection among critically ill patients with COVID-19 should not be ignored [21]. Further studies on fungal co-infection need to address a more rapid serological assay or antifungal prophylaxis among critically ill patients with COVID-19.
Limitations of our study include a small sample size in a single study center. Our retrospective design also brought biases into the sample collection and comparisons. For example, some patients received multiple RT-PCR assays but other patients received culture analysis. Additionally, we did not have the results for the antimicrobial resistance patterns. More investigations should be performed to further study the co-infections and antimicrobial resistance patterns in COVID-19 patients.
We demonstrated a high frequency of respiratory bacterial and fungal co-infections in critically ill COVID-19 patients admitted to the ICU. These co-infections could be detected by culture from sputum and BAL samples, multiple RT-PCT from the sputum or nasopharyngeal swab samples, or serological fungal antigen tests. Careful examinations and necessary tests should be performed to exclude co-infections in order to appropriately treat critically ill ICU COVID-19 patients.
Funding
This study was supported by the Beijing Municipal Science and Technology Commission (Z201100005420012), Beijing Municipal Administration of Hospital Clinical Medicine Development of Special Funding Support (ZYLX201802), and the Beijing Traditional Chinese Medicine Technology Development Fund Project (YJ2020-01).
Access to data
Not available.
Contributions
YSY and LJY performed all conception and design of the study. YSY and LXZ performed the experiments. PL and XP contributed clinical samples and clinical data collection. HMX and DCJ analyzed the data. LJY and WLH wrote the manuscript. All authors reviewed the manuscript.
Declaration of competing interest
All authors declare that they have no conflicts of interest.
Acknowledgements
We would like to thank the frontline medical providers at Beijing Ditan Hospital for their bravery and efforts in the prevention and control of SARS-CoV-2.
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C.P., Adhi F., Highland K. Recognition and management of respiratory coinfection and secondary bacterial pneumonia in patients with COVID-19. Cleve Clin J Med. 2020 doi: 10.3949/ccjm.87a.ccc015. [DOI] [PubMed] [Google Scholar]
- 5.Rawson T.M., Moore L., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes S., Troise O., Donaldson H., Mughal N., Moore L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai C.C., Wang C.Y., Hsueh P.R. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents. J Microbiol Immunol Infect. 2020;53:505–512. doi: 10.1016/j.jm.ii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharifipour E., Shams S., Esmkhani M., Khodadadi J., Fotouhi-Ardakani R., Koohpaei A. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:646. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Vidal C., Sanjuan G., Moreno-García E., Puerta-Alcalde P., Garcia-Pouton N., Chumbita M. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Health Commission The people's Republic of China. National clinical guidance for COVID-19 diagnosis and treatment of China. http://www.nhc.gov.cn/yzygj/s7653p/202008/0a7bdf12bd4b46e5bd28ca7f9a7f5e5a.shtml (eighth edition). Available at:
- 11.Ying-wu Ye, Wang Yu-san, Shen zi-yu. 3rd ed. Southeast University Press; Nanjing, China: 2006. National guide to clinical laboratory procedures. [Google Scholar]
- 12.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A. Clinical characteristics of Covid-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerci P., Bellut H., Mokhtari M., Gaudefroy J., Mongardon N., Charpentier C. Outcomes of Stenotrophomonas maltophilia hospital-acquired pneumonia in intensive care unit: a nationwide retrospective study. Crit Care. 2019;23:371. doi: 10.1186/s13054-019-2649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agodi A., Barchitta M., Cipresso R., Giaquinta L., Romeo M.A., Denaro C. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intensive Care Med. 2007;33:1155–1161. doi: 10.1007/s00134-007-0671-6. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Chetrit E., Wiener-Well Y., Lesho E., Kopuit P., Broyer C., Bier L. An intervention to control an ICU outbreak of carbapenem-resistant Acinetobacter baumannii: long-term impact for the ICU and hospital. Crit Care. 2018;22:319. doi: 10.1186/s13054-018-2247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paling F.P., Wolkewitz M., Bode L., Klein Klouwenberg P., Ong D., Depuydt P. Staphylococcus aureus colonization at ICU admission as a risk factor for developing S. aureus ICU pneumonia. Clin Microbiol Infect. 2017;23:49. doi: 10.1016/j.cmi.2016.09.022. e9-49. e14. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H., Zhang Y., Wu J., Li Y., Zhou X., Li X. Risks and features of secondary infections in severe and critical ill COVID-19 patients. Emerg Microb Infect. 2020;9:1958–1964. doi: 10.1080/22221751.2020.1812437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei Y., Song Y., Shu Y., Zhao Y., Huo X., Wang H. Fungal antigenemia in patients with severe Coronavirus disease 2019 (COVID-19): the facts and challenges. J Microbiol Immunol Infect. 2020;53:657–659. doi: 10.1016/j.jmii.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]