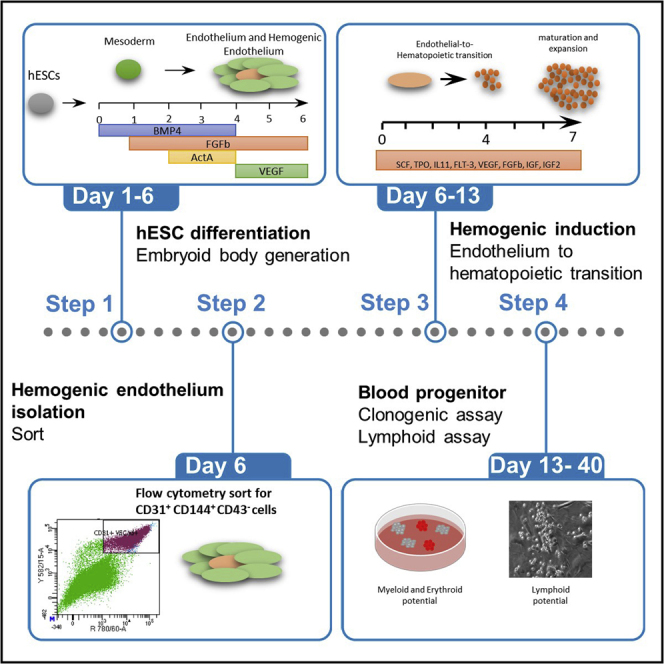
Summary
Little is known about the emergence of blood progenitors during human embryogenesis due to ethical reasons and restricted embryo access. The use of human embryonic stem cells (hESCs) as a model system offers unique opportunities to dissect human blood cell formation. Here, we describe a protocol allowing the differentiation of hESCs via embryoid bodies toward hemogenic endothelium and its subsequent differentiation to blood progenitors. This protocol relies on the formation of embryoid bodies, which is tricky if not carefully performed.
For complete details on the use and execution of this protocol, please refer to Garcia-Alegria et al. (2018).
Subject areas: Cell culture, Cell isolation, Flow cytometry/mass cytometry, Stem cells, Cell differentiation
Graphical Abstract
Highlights
Differentiation of hESCs to blood cell progenitors
Differentiation protocol via embryoid body formation
Isolation of hemogenic endothelium from day 6 embryoid bodies
Assays to determine hemogenic endothelium hematopoietic output
Little is known about the emergence of blood progenitors during human embryogenesis due to ethical reasons and restricted embryo access. The use of human embryonic stem cells (hESCs) as a model system offers unique opportunities to dissect human blood cell formation. Here, we describe a protocol allowing the differentiation of hESCs via embryoid bodies toward hemogenic endothelium and its subsequent differentiation to blood progenitors. This protocol relies on the formation of embryoid bodies, which is tricky if not carefully performed.
Before you begin
Note: This protocol has been successfully used with several hESC lines (Man5, Man7, Man9, H1).
Preparing reagents for hESC expansion on feeders
Timing: 1 day
-
1.Mitomycin C
-
a.The stock solution is prepared at a concentration of 2 mg/mL.
-
b.Add 1 mL of PBS to Mitomycin C vial to resuspend.
-
c.Transfer to a 1.5 mL Eppendorf tube and cover with foil to protect from light.
-
d.The reconstituted solution can be kept at +4°C for 2 weeks.
-
a.
CRITICAL: Mitomycin C is toxic, manipulate with care.
-
2.Gelatin 0.1%
-
a.Dissolve 0.5 g of gelatin in 500 mL of PBS for a final concentration of 0.1%.
-
b.Sterilize by autoclaving or by filtering through a 0.2 μm filter.
-
c.Aliquot in 50 mL screw-cap tubes and store at +4°C for up to 6 months.
-
a.
-
3.Plate coating
-
a.Dispense 1 mL of 0.1% gelatin solution per well of a 6-well plate.
-
b.Ensure that the surface is fully covered.
-
c.Maintain at +20°C for at least 20 min.
-
d.Aspirate the gelatin solution and replace with 1 mL of MEF medium.
-
a.
-
4.ROCK inhibitor (Y-27632, Chemdea cat# CD0141)
-
a.ROCK inhibitor is photosensitive, work quickly to avoid light exposure.
-
b.Resuspend the lyophilized compound in sterile H2O tissue culture grade for a final concentration of 100 mM.
-
c.Mix well by pipetting up and down.
-
d.Make aliquots of 20 μL and store at −80°C for up to 1 year.
-
a.
-
5.EDTA 0.5 mM
-
a.The 0.5 M stock solution is diluted in PBS Mg-/Ca2- at a final concentration of 0.5 mM.
-
b.Sterilize by filtering through a 0.2 μm filter and store at +4°C for up to 6 months.
-
c.Before use, aliquots are warmed up in a 37°C water bath.
-
a.
Note: Unless specified, PBS with calcium and magnesium is used elsewhere in the protocol
Preparing reagents for hESC expansion on feeder-free culture
Timing: 1 day
-
6.TeSR-E8 basal medium
-
a.Make a note of the lot number for each new bottle, in case of issues with hESC growth.
-
b.The supplement is stored at −20°C until needed and is thawed for 10–12 h at +4°C.
-
c.Add the supplement to the 500 mL bottle.
-
d.Add Penicillin/Streptomycin at a final concentration of 0.5%.
-
e.Make 40 mL aliquots in 50 mL screw-cap tubes and store at −20°C for up to 3 months.
-
f.Aliquots are thawed for 10–12 h at +4°C.
-
a.
-
7.Geltrex
-
a.Concentrated Geltrex is kept at −80°C.
-
b.Thaw Geltrex (5 mL) at +4°C for 10–12 h in a 50 mL Falcon tube containing ice.
-
c.Eppendorf tubes that are needed the next day for aliquoting are placed at −20°C for 10–12 h.
-
d.The following day prepare Geltrex aliquots by diluting the concentrated Geltrex with 20 mL of cold KO-DMEM (dilution 1:5).
-
e.Mix well and dispense in ice cold Eppendorf tubes (0.5 to 1 mL per tube) placed in cold rack (Corning CollRACK XT M24).
-
f.The aliquots of Geltrex are stored at −80°C for up to 1 year and thawed at +4°C for 10–12 h in a tube containing ice when needed.
-
a.
Preparing reagents for hESC differentiation
Timing: 1–3 days
-
8.Poly-HEMA plate coating
-
a.Poly-HEMA solution is made by dissolving 6 g of Poly-HEMA in 500 mL of 95% ethanol (25 mL of tissue culture grade H2O in 475 mL of biological grade ethanol) for 10–12 h on a heated stirrer at 30°C.
-
b.The solution is sterilized using a 0.2 μm filter (Nalgene) and kept at 20°C until needed.
-
c.10 cm tissue culture dishes are coated with 4 mL of Poly-HEMA solution and the dishes are allowed to dry completely for 24 h at 40°C in a ventilated oven. Similarly, 6-well plates are coated with 1 mL of Poly-HEMA solution per well. Plates and dishes are kept with their lids on during the drying step in the oven to maintain sterility.
-
d.Coated dishes and plates can be sealed with parafilm and stored at 20°C for 3 months.
-
a.
Alternatives: commercially available ultra-low adherent plates can be used (Corning Costar Ultra-Low Attachment plates, cat# 3471).
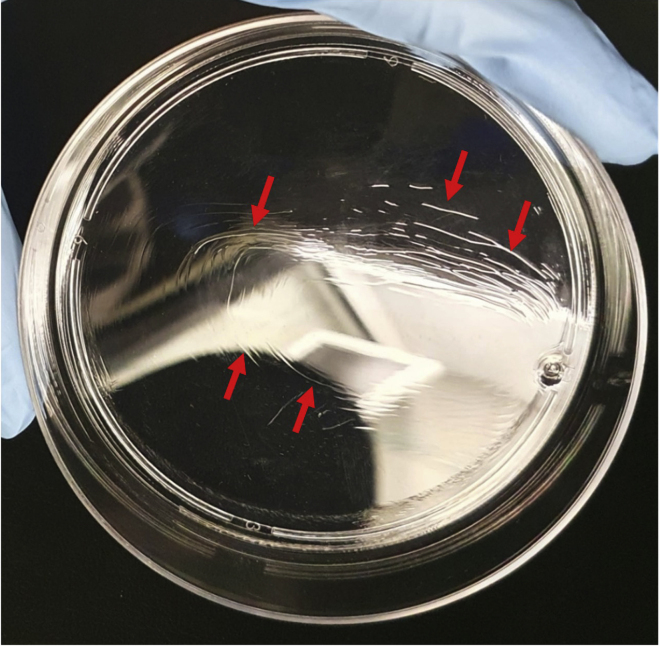
CRITICAL: Inspect plates visually to make sure that Poly-HEMA is coating the entire surface of the plate (see Figure 1 for suboptimal coating).
Figure 1.
Poly-HEMA coating
Photograph of suboptimal coating with Poly-HEMA showing ridge (arrows) and non-uniform coating.
-
9.L-ascorbic acid solution
-
a.Prepare a stock solution of 5 mg/mL in cold tissue culture grade H2O.
-
b.Leave on ice and vortex periodically until completely dissolved.
-
c.Filter sterilize, aliquot in Eppendorf tubes (0.2 to 1 mL per tube) and store at −20°C for up to 6 months.
-
d.Aliquots thawed need to be used on the day, unused solution is discarded.
-
a.
-
10.Monothioglycerol (MTG)
-
a.MTG should be aliquoted (1 mL) and stored at −20°C for up to 1 year.
-
b.When an aliquot is thawed, it can be kept for up to 1 month at +4°C.
-
c.Aliquoting MTG is strongly recommended as it minimizes the amount of oxidation due to repeated opening.
-
a.
CRITICAL: The amounts of MTG indicated in our protocols are the recommended concentrations. However, it is important to test each new batch of MTG as there are batch-to-batch variabilities.
-
11.Transferrin
-
a.Lyophilized transferrin is kept at −20°C.
-
b.Reconstitute 1 g of lyophilized transferrin with 50 mL of sterile tissue culture grade H2O warmed up at 37°C for a final concentration of 20 mg/mL.
-
c.Aliquot in 15 mL screw-cap tubes and store at −20°C for up to 6 months.
-
d.Working aliquots are stored at +4°C for up to 1 month.
-
a.
CRITICAL: The amounts of transferrin indicated in our protocols are recommended concentrations. However, it is important to test each new batch of transferrin as there is variabilityamong them.
-
12.Stempro-34 SFM
-
a.Stempro-34 is sold as a kit with 2 components.
-
b.The supplement is kept at −20°C and the liquid medium at +4°C.
-
c.Thaw the supplement at +4°C for 10–12 h.
-
d.Mix the supplement well with a pipette and add to the medium along with 1% Penicillin/Streptomycin.
-
e.Cover the bottle with foil if kept at +4°C. Store for a maximum of 2 weeks in the fridge or aliquot and keep at −20°C for up to 3 months.
-
a.
CRITICAL: When combined, the medium is unstable and therefore is kept for a maximum of 2 weeks +4°C. Alternatively, the reconstituted medium can be aliquoted and stored at −20°C for up to 3 months.
-
13.Cytokines (all stock concentrations are indicated in the tables)
-
a.All cytokines are stored lyophilized at −80°C.
-
b.Reconstitute according to the manufacturer’s recommendation and store at-80°C for 1 year.
-
c.Working aliquots are stored at +4°C and kept for up to 1 month.
-
a.
Note: Further dilutions are made in sterile PBS containing 0.1% BSA as recommended.
-
14.Methylcellulose
-
a.Weigh a 2 L glass flask with foil covering and magnetic stirring bar inside. Make a note of the weight.
-
b.Rinse the flask with water then boil approximately 100 mL water in the flask with the stirring bar inside and foil cover for 5–10 min. This is to sterilize the flask and stirring bar.
-
c.Pour out the remaining water carefully trying to maintain sterility.
-
d.Place the flask on a hot-plate stirrer and add 450 mL of tissue culture grade H2O. Replace the foil on top of the flask and bring the water to a boil.
-
e.Remove the flask from the hot-plate using heat resistant gloves. Add the methylcellulose powder (approximately 20 g depending on batch) being careful not to get any powder on the lip of the flask.
-
f.Place back on the hot-plate. Stir slowly and return to boil for approximately 5 min. Use a thermal glove to swirl flask as the water boils to prevent it boiling over. The methylcellulose is fully dissolved once no methylcellulose powder is seen upon visual inspection.
-
g.Turn off heat but leave stirring and allow to cool to 20°C.
-
h.Add 500 mL of 2× IMDM. Make sure IMDM is at 37°C, if it is cold when added to the methylcellulose then lumps will form.
-
i.Re-weigh the flask with the methylcellulose and IMDM.
-
j.Subtract weight of empty flask (obtained in step a).
-
k.Add sterile tissue culture grade H2O to make 1,000 × g. The final methylcellulose concentration is 20 g/L.
-
l.Place in a cold room checking and swirling every 5 min for 1 h to maintain a homogeneous solution as it cools down.
-
m.As the solution cools it will get thicker but swirling must be maintained throughout the process in order to avoid lumps. Leave 10–12 h in cold room.
-
n.The next day, aliquot approximately 100 mL into 125 mL bottles using a syringe without needle and place at −20°C for up to 1 year.
-
o.This last step clarifies the methylcellulose solution and makes the correct consistency.
-
a.
Note: 2× IMDM is made from powder and contains 2× pen/strep concentration. This solution is made by diluting the amount of IMDM powder necessary for 1 L of IMDM into 500 mL of tissue culture grade H2O, adding pen/strep at 2× the concentration and sterilizing by filtration (Nalgene).
Maintenance and expansion of hESCs onto an MEF layer
Timing: 2–3 weeks
hESCs need to be cultured onto mouse embryonic fibroblasts (MEFs) for maintenance and expansion prior to passage in feeder-free culture conditions for differentiation (Garcia-Alegria et al., 2018). MEFs are grown to confluency, mitotically inactivated with Mitomycin C and frozen in aliquots for later use. MEFs can be stored for up to 4 months at −80°C.
-
15.Inactivation of MEFs with Mitomycin C
-
a.Before starting, warm up TrypLE solution and MEF medium in a water bath at 37°C for at least 30 min.
-
b.Expand MEFs in 175 cm2 flasks at 37°C 5% O2 5% CO2. MEFs grow faster without differentiating in low O2 culture conditions. When MEF culture reaches 90% confluency, they are ready to be inactivated.
-
c.Aspirate medium from the flask.
-
d.Add 100 μl of Mitomycin C to 20 mL MEF medium to a final concentration of 10 μg/mL final and add to flask.
-
e.Swirl gently and incubate at 37°C for 3 h.
-
f.Aspirate medium and gently wash once with 20 mL MEF medium then twice with 20 mL PBS.
-
g.Add 12 mL TrypLE to the flask to cover the surface and incubate for 3 min at 37°C.
-
h.Collect the cells and place in 50 mL screw-cap tube and centrifuge at 1,800 × g for 5 min.
-
i.Aspirate the supernatant and resuspend in 10 mL of MEF medium.
-
j.Count the cells using a hemocytometer to determine total cell number and calculate the volume of freezing medium to use (generally 1–2 × 106 cells per mL per cryovial).
-
k.Centrifuge at 1,800 × g 5 min then aspirate the medium and resuspend the pellet in the appropriate amount of freezing medium.
-
l.Dispense 1 mL of cell suspension per cryovial. Place into a cryo-container and store at −80°C for 24–48 h.
-
m.Transfer to liquid nitrogen until required.
-
a.
-
16.Thawing hESCs
-
a.24 h before the hESCs are thawed, prepare the required number of wells with inactivated MEFs.
-
b.Work quickly when thawing the inactivated MEFs as DMSO is toxic to the cells when it is warm. Inactivated MEFs are plated on tissue culture dishes coated with gelatin which promotes better adherence. MEFs should cover the entire surface without being over-confluent. Typically, a vial containing 106 cells can be thawed onto a 10 cm dish or all the wells of a 6-well plate.
-
c.Remove a hESCs vial from the liquid nitrogen and thaw at 37°C in a water bath until a sliver of ice remains visible. Clean the vial with ethanol before opening to remove any contamination source.
-
d.Transfer the cells into a sterile screw-cap tube containing 10 mL hESC medium. Do not forget to add FGF2 at a concentration of 10 ng/mL to newly thawed hESC media before use. Centrifuge for 5 min at 1,800 × g.
-
e.Determine how much medium is required calculating 2 mL per well and add ROCK inhibitor at 10 μM final concentration to increase survival.
-
f.Remove MEF medium from the MEF-coated plates and add 1 mL of hESC medium containing rock inhibitor in each well.
-
g.Remove the supernatant from the pelleted hESCs and resuspend the pellet gently to keep cell clusters.
-
h.Place 1 mL of hESC suspension per MEF-coated well then gently move the plate up and down, left and right to evenly distribute the cells within the wells.
-
i.Carefully transfer the plate to an incubator at 37°C 5% CO2 in normoxia to avoid gathering of all cells in the middle of the wells.
-
j.Replace the medium every day, the addition of ROCK inhibitor is not required after the initial thawing. Maintain the culture until the colonies have reached a good size for passaging or 80% confluency (Figure 2). This takes usually 6 to 7 days after thawing, depending on the cell line and the quality of the frozen cells.
-
a.
-
17.Feeding hESC cultures
-
a.hESCs need to be fed with fresh medium every day. The hESC medium needs to be warmed up at 20°C for at least 30 min. Do not place in a 37°C water bath as it will affect the quality of the medium. Do not forget to add FGF2 at a concentration of 10 ng/mL to newly thawed hESC media before use.
-
b.Aspirate the medium from the wells without disturbing the adherent cell layer.
-
c.Add 1.5 mL of hESC medium warmed at 20°C per well. As the colonies get larger, add more medium when feeding daily (up to 3 mL by day 4 of culture) to avoid spontaneous differentiation and excessive acidification of the medium.
-
d.Incubate at 37°C 5% CO2 for 24 h.
-
a.
-
18.EDTA passage of hESC culture
-
a.24 h before passaging hESCs, plate inactivated MEFs.
-
b.On the day of passaging, leave hESC medium at 20°C for at least 30 min to warm up and place an aliquot of EDTA 0.5 mM solution in a 37°C water bath for at least 10 min prior to use.
-
c.Prepare the required volume of hESC medium, containing 10 μM ROCK inhibitor, for the number of wells in which the cells will be split, calculating 1.5 mL per well.
-
d.Check that MEFs cover the well and are healthy. Aspirate medium and dispense 1 mL of hESC medium containing ROCK inhibitor.
-
e.Carefully remove medium from the hESC colonies and wash quickly and gently with 0.5 mL of EDTA 0.5 mM to remove any trace of medium and debris.
-
f.Add 1 mL of EDTA 0. 5 mM and place cells in the incubator at 37°C for 2 min. Check the colonies under microscope, the cell-cell interactions should become more relaxed and cells within the colonies should become more refringent.
-
g.Carefully remove the EDTA solution without disturbing the cell layer. Let the cells dry by keeping the plate lid off for 1 min in the tissue culture hood.
-
h.To detach the hESCs, pipette 500 μl of hESC medium containing ROCK inhibitor directly over the colonies, aiming to detach small clumps of 5–6 cells, avoiding making a single cell suspension (Figure 3).
-
i.Repeat step h until reaching the desired volume of medium, calculating 500 μl per well in which the hESCs will be split (e.g., 2 mL of medium for a 1 in 4 split). This minimizes repeated pipetting of the cell clumps, minimizing cell death and single cell formation. Typically, hESCs are split at a ratio of 1 in 6 to 1 in 8.
-
j.Dispense 500 μl of hESC clump suspension dropwise to each new well. Gently move the plate up and down, left and right to evenly distribute the cell clumps within the wells.
-
k.Carefully transfer the plate to an incubator at 37°C 5% CO2 in normoxia to avoid gathering of all cell clumps to the middle of the wells.
-
a.
-
19.Freezing hESCs grown on MEFs
-
a.Before starting warm up an aliquot of EDTA solution in a water bath at 37°C for at least 10 min.
-
b.Use the protocol for EDTA passage.
-
c.Determine the volume of freezing medium needed calculating 1 mL per cryovial. Typically, 2 cryovials can be frozen per confluent well of a 6-well plate.
-
d.In step h of the EDTA passage protocol, use freezing medium instead of hESC medium.
-
e.Transfer the cell clumps into a cryovial, then place the cryovials in an appropriate freezing container (Isopropanol-filled or similar) to ensure a gradual decrease of the temperature.
-
f.Place the container at −80°C.
-
g.Transfer the cryovials to liquid nitrogen 24–48 h later.
-
a.
Note: Make sure you have frozen stocks of hESCs grown on MEFs prior to using a feeder-free culture system as hESCs are more likely to maintain a normal karyotype on feeders than in feeder-free culture conditions (Catalina et al., 2008; Kim et al., 2012).
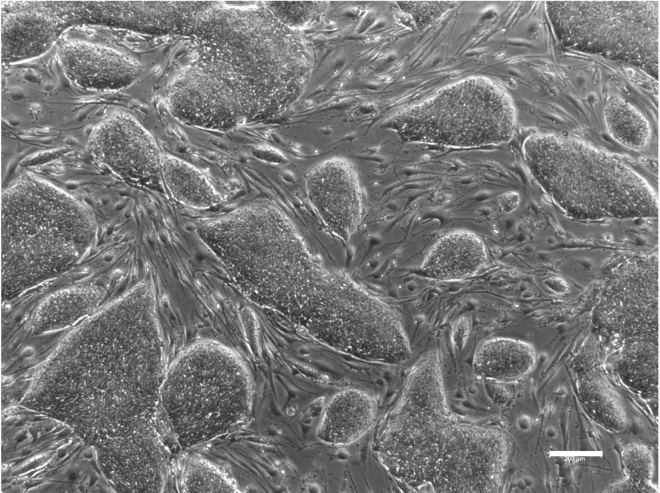
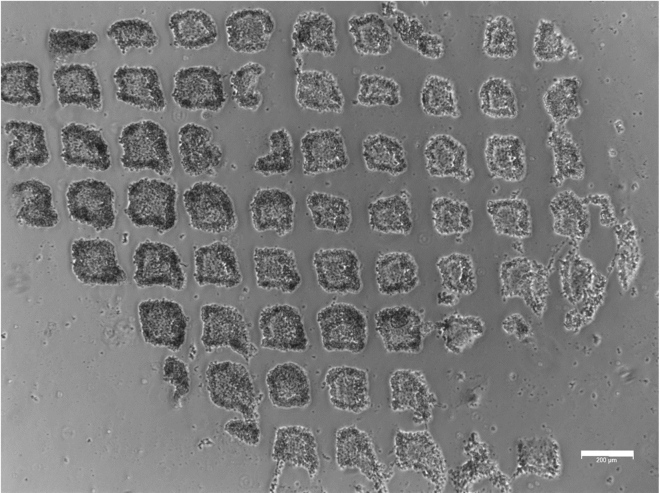
Figure 2.
hESC colonies morphology
Representative example of hESC colonies grown on mouse embryonic feeder (MEF) layer approaching 80% confluency. Scale bar, 200 μm.
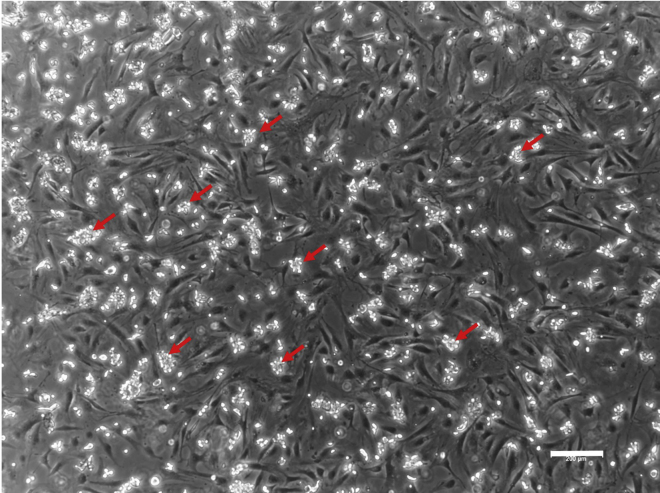
Figure 3.
hESC clumps
Small clumps of hESCs generated upon EDTA passaging. The clumps (arrows) are floating above the MEF layer on which they were just transferred. Scale bar, 200 μm.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Alpha-MEM | Lonza | BE02-002F |
| DMEM | Sigma-Aldrich | D6171-500mL |
| DMSO | Sigma-Aldrich | 276855 |
| EDTA, 0.5 M | Sigma-Aldrich | E7889 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | F9665 |
| Gelatin | Sigma-Aldrich | G1890 |
| Geltrex | Thermo Fisher Scientific | A1413302 |
| IMDM | Thermo Fisher Scientific | 12589059 |
| KO Serum Replacement | Thermo Fisher Scientific | 10828028 |
| KO-DMEM | Thermo Fisher Scientific | 10829018 |
| L-Ascorbic Acid | Sigma-Aldrich | A4544-100G |
| L-Glutamine, 200 mM | Thermo Fisher Scientific | 25030149 |
| Methylcellulose | Alfa Aesar | 36718 |
| Mitomycin C | Sigma-Aldrich | M4287 |
| Monothioglycerol (MTG) | Sigma-Aldrich | M-6145 |
| NEAA (MEM Non-essential Amino Acid Solution) | Merck | M7145-100ML |
| PBS | OXOID | BR0014G |
| PDS (plasma-derived serum) | Animal Technologies, Inc. | 11-131127-2 |
| Penicillin/streptomycin | Sigma-Aldrich | P0881-100ML |
| PFHM-II (protein-free hybridoma medium) | Thermo Fisher Scientific | 12040077 |
| Poly-HEMA | Sigma-Aldrich | P3932-25G |
| ProFreeze-CDM Medium | Lonza | 12-132A |
| ROCK inhibitor Y-27632 | Chemdea | CD0141 |
| β-Mercaptoethanol | Thermo Fisher Scientific | 11528926 |
| StemPro EZPassage tool | Thermo Fisher Scientific | 23181010 |
| STEMPRO-34 SFM | Thermo Fisher Scientific | 10639011 |
| StemSpan SFEM medium | Stemcell Technologies | 09650 |
| TeSR-E8 basal medium | Stemcell Technologies | 05990 |
| Transferrin | R&D systems | 2914-HT-001G |
| TrypLE Express Enzyme | Thermo Fisher Scientific | 12605036 |
| Water for cell culture applications | Lonza | LZBE17-724Q |
| Activin A (5 μg/mL) | Peprotech | 120-14 |
| BMP4 (10 μg/mL) | Peprotech | 120-05 |
| EPO (2,000 units/mL) | Peprotech | 100-64 |
| FGF2 (10 μg/mL) | Peprotech | 100-18C |
| FLT3-L (20 μg/mL) | Peprotech | 300-19 |
| GM-CSF (1 μg/mL) | Peprotech | 300-03 |
| IGF1 (50 μg/mL) | Peprotech | 100-11 |
| IGF2 (50 μg/mL) | Peprotech | 100-12 |
| IL-11 (10 μg/mL) | Peprotech | 200-11 |
| IL-2 (50 μg/mL) | Peprotech | 200-02 |
| IL-3 (100 μg/mL) | Peprotech | 200-03 |
| IL-6 (20 μg/mL) | Peprotech | 200-06 |
| IL-7 (10 μg/mL) | Peprotech | 200-07 |
| SCF (100 μg/mL) | Peprotech | 300-07 |
| TPO (100 μg/mL) | Peprotech | 300-18 |
| VEGF (10 μg/mL) | Peprotech | 100-20 |
| Experimental models: cell lines | ||
| Man5 | University of Manchester | n/a |
| Man9 | University of Manchester | n/a |
| H1 | WiCell | WA01 |
| Antibodies | ||
| CD117 PE-CY7 (1/100) | eBioscience | 25-1178-42 |
| CD144 PE (1/100) | BD Biosciences | 560410 |
| CD235a Biotin (1/100) | eBioscience | 13-9987-82 |
| CD309 AF647 (1/100) | BioLegend | 338909 |
| CD31 APC eF780 (1/100) | eBioscience | 47-0319-42 |
| CD34 PE (1/500) | Miltenyi Biotech | 130-081-002 |
| CD41a FITC (1/100) | eBioscience | 11-0419-42 |
| CD43 APC (1/100) | eBioscience | 17-0439-42 |
| CD45 PE CY5.5 (1/100) | eBioscience | 35-0459-42 |
| CD71 PE CY7 (1/5,000) | eBioscience | 25-0719-42 |
| CD73 PERCP eF710 (1/100) | eBioscience | 46-0739-42 |
| CD90 Biotin (1/100) | eBioscience | 13-0909-82 |
| Hoechst 33258 (1/100) | Abcam | Ab228550 |
| Streptavidin BV421 (1/100) | BioLegend | 405225 |
| Other | ||
| CollRACK XT M24 | Corning | 432040 |
| 0.2 μm filter | Nalgene | Z358223 |
| Costar ultra-low attachment plates | Corning | 3471 |
| 50 μm Filcon filter | BD Biosciences | 340630 |
| 5 mL snap cap tubes | Falcon | 352054 |
| Hematocytometer | Sigma-Aldrich | BR7 18605 |
| BD CompBeads | BD Biosciences | 552844 |
| FACSAria fusion flow cytometer | BD Biosciences | 656700 |
Materials and equipment
Antibody panel
| Laser | Channel | Antibody | Fluorochrome |
|---|---|---|---|
| 561 nm | Y780/60 | CD117 (cKIT) | PE-Cy7 |
| 561 nm | Y586/15 | CD144 | PE |
| 405 nm | V450/50 | CD235a (Glycophorin A) | Biotin |
| 640 nm | R670/14 | CD309 (VEGFR2) | AF647 |
| 640 nm | R780/60 | CD31 (PECAM-1) | APC-eFluor 780 |
| 561 nm | Y586/15 | CD34 | PE |
| 488 nm | B530/30 | CD41a | FITC |
| 640 nm | R670/14 | CD43 | APC |
| 561 nm | Y710/50 | CD45 | PE-Cy5.5 |
| 561 nm | Y780/60 | CD71 | PE-Cy7 |
| 488 nm | B710/50 | CD73 | PerCP-eFluor 710 |
| 405 nm | V450/50 | CD90 (Thy-1) | Biotin |
| 355 nm | U450/50 | Hoechst 33258 | n/a |
| 405 nm | V450/50 | Streptavidin | BV 421 |
Mouse embryonic feeders (MEFs) medium (filter sterilized and store at 4°C for up to 1 month)
| Reagent | Final concentration | Volume |
|---|---|---|
| IMDM | n/a | 442.5 mL |
| MTG (11.56 M) | 1.4 × 10−4 M | 0.063 mL of a 1/100 dilution |
| L-Glutamine 100× (200 mM) | 2 mM | 5 mL |
| FBS (heat inactivated at 56°C for 30 min) | 10% | 50 mL |
| Penicillin (10,000 units), streptomycin (10 mg/mL) | 50 units/mL, 0.05 mg/mL | 2.5 mL |
| Total | n/a | 500 mL |
OP9-DLL1 medium (Filter sterilized and store at 4°C for up to 1 month)
| Reagent | Final concentration | Volume |
|---|---|---|
| FBS (heat inactivated at 56°C for 30 min) | 20% | 20 mL |
| L-Glutamine 200 mM | 2 mM | 1 mL |
| α-MEM (pen/strep 1%) | n/a | 79 mL |
| Total | n/a | 100 mL |
Freezing medium for MEF and OP9-DLL1 cells (store at 4°C for up to 1 week)
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM | n/a | 40 mL |
| FBS (heat inactivated at 56°C for 30 min) | 50% | 50 mL |
| DMSO | 10% | 10 mL |
| Total | n/a | 100 mL |
hESC medium for culture on MEFs
| Reagent | Final concentration | Volume |
|---|---|---|
| KO-DMEM | n/a | 403 mL |
| NEAA | 1% | 5 mL |
| L-Glutamine (200 mM) | 2 mM | 5 mL |
| Penicillin (10,000 units), streptomycin (10 mg/mL) | 50 units/mL, 0.05 mg/mL | 2.5 mL |
| KO serum replacement | 16% | 80 mL |
| β-Mercapto ethanol (50 mM) | 0.1 mM | 1 mL |
| Total | 500 mL |
Note: The medium is sterilized by filtering through a 0.2μm filter, aliquoted into 50 mL screw-cap tubes and stored at −20°C for up to 4 months. Aliquots are thawed for 10–12 h at +4°C prior to use. FGF2 is added at a concentration of 10 ng/mL before use and store at +4°C for up to 2 weeks.
Freezing medium for hESCs (Store at 4°C for up to 1 week)
| Reagent | Final concentration | Volume |
|---|---|---|
| hESC medium | 34% | 3.4 mL |
| ProFreeze-CDM Medium | 50% | 5 mL |
| DMSO | 16% | 1.6 mL |
| Total | n/a | 10 mL |
Alternatives: You can use mFreSR medium (Stemcell Technologies, # 05855). This freezing medium is very efficient for high cell survival upon thawing but it is very expensive.
FACS wash buffer (Store at 4°C for up to 6 months)
| Reagent | Final concentration | Volume |
|---|---|---|
| FBS (heat inactivated at 56°C for 30 min) | 10% | 10 mL |
| PBS | n/a | 90 mL |
| Total | n/a | 100 mL |
Note: This buffer is sterilized by filtering through a 0.2 μm filter or autoclaved.
Embryoid body (EB) differentiation medium
| Reagent | Final concentration | IMDM 20% | Day 0 | Day 2 | Day 4 |
|---|---|---|---|---|---|
| FBS (heat inactivated at 56°C for 30 min) | 20% | 20 mL | n/a | n/a | n/a |
| L-Glutamine (200 mM) | 0.2 mM | 1 mL | 1 mL | 1 mL | 1 mL |
| Transferrin (20 mg/mL) | 0.15 mg/mL | 0.75 mL | 0.75 mL | 0.75 mL | 0.75 mL |
| MTG (0.150 M) | 4.5 × 10−4 M | 0.3 mL | 0.3 mL | 0.3 mL | 0.3 mL |
| Ascorbic acid (5 mg/mL) | 0.05 mg/mL | 1 mL | 1 mL | 1 mL | 1 mL |
| Geltrex (1:5 dilution) | 1:5,000 | 0.1 mL | 0.1 mL | n/a | n/a |
| ROCK inhibitor (100 mM) | 10 μM | 0.01 mL | 0.01 mL | n/a | n/a |
| BMP4 (10 μg/mL) | 10 ng/mL | n/a | 0.1 mL | 0.1 mL | n/a |
| FGF2 (10 μg/mL) | 5 ng/mL | n/a | n/a | 0.05 mL | 0.05 mL |
| Activin A (5 μg/mL) | 0.9 ng/mL | n/a | n/a | 0.018 mL | n/a |
| VEGF (10 μg/mL) | 12 ng/mL | n/a | n/a | n/a | 0.12 mL |
| IMDM | n/a | 76.95 mL | n/a | n/a | n/a |
| StemPro 34 (pen/strep 1%) | n/a | n/a | 96.95 mL | 96.95 mL | 96.95 mL |
| Total | n/a | 100 mL | 100 mL | 100 mL | 100 mL |
Note: Media for each step of the differentiation protocol is made on the day it is needed. For this media, MTG is first diluted at 26ul in 2ml of IMDM for more accurate pipetting of the exact volume to be added to the media as MTG is very viscous. 0.3mL of this dilution is added to the media.
Hemogenic endothelium (HE) inducing medium (prepared on the day it is needed)
| Reagent | Final concentration | Volume |
|---|---|---|
| FGF2 (10 μg/mL) | 5 ng/mL | 0.05 mL |
| IGF1 (50 μg/mL) | 25 ng/mL | 0.05 mL |
| IGF2 (50 μg/mL) | 25 ng/mL | 0.05 mL |
| SCF (100 μg/mL) | 50 ng/mL | 0.05 mL |
| TPO (100 μg/mL) | 50 ng/mL | 0.05 mL |
| IL11 (10 μg/mL) | 5 ng/mL | 0.05 mL |
| FLT3-L (20 μg/mL) | 20 ng/mL | 0.1 mL |
| StemSpan | n/a | 99.6 mL |
| Total | n/a | 100 mL |
CRITICAL: StemSpan is a very expensive medium. It must be aliquoted in small volumes (10 to 50 mL) and kept at −20°C. When needed, aliquots are thawed for 10–12 h at +4°C.
Semi-solid medium for clonogenic assay (prepared and used on the day it is needed)
| Reagent | Final concentration | Volume |
|---|---|---|
| Methylcellulose | 55% | 5.5 mL |
| PDS (plasma-derived serum) | 15% | 1.5 mL |
| PFHM-II (protein-free hybridoma medium) | 5% | 0.5 mL |
| L-Glutamine 200 mM | 2 mM | 0.1 mL |
| Transferrin (20 mg/mL) | 150 μg/mL | 0.075 mL |
| TPO (100 μg/mL) | 50 ng/mL | 0.005 mL |
| IL-3 (100 μg/mL) | 50 ng/mL | 0.005 mL |
| VEGF (5 μg/mL) | 10 ng/mL | 0.02 mL |
| SCF (100 μg/mL) | 100 ng/mL | 0.01 mL |
| EPO (2,000 units/mL) | 4 units/mL | 0.02 mL |
| IL-6 (20 μg/mL) | 10 ng/mL | 0.005 mL |
| IGF-1 (50 μg/mL) | 50 ng/mL | 0.01 mL |
| IL-11 (10 μg/mL) | 5 ng/mL | 0.005 mL |
| GM-CSF (1 μg/mL) | 1 ng/mL | 0.01 mL |
| IMDM (pen/strep 1%) | n/a | 2.235 mL |
| Total | n/a | 10 mL |
CRITICAL: Methylcellulose is very viscous, it needs to be warmed up at 37°C before use. Syringes and 18G blunt needles must be used to dispense methylcellulose. Do not use a pipette for dispensing. Extra volume must be taken into account as not all volume can be recovered from the tube in which the medium is prepared. Typically, an additional 5 mL must be calculated for the final volume when preparing this medium in a 50 mL tube and 0.5 mL when prepared in a 10 mL tube.
Medium for T lymphoid generation (prepared and used on the day it is needed)
| Reagent | Final concentration |
Week 1 and 2 | Week 3 and 4 | |
|---|---|---|---|---|
| Week 1 and 2 | Week 3 and 4 | |||
| FBS (heat inactivated at 56°C for 30 min) | 20% | 20% | 20 mL | 20 mL |
| L-Glutamine 200 mM | 2 mM | 2 mM | 1 mL | 1 mL |
| FLT3L (20 μg/mL) | 20 ng/mL | 10 ng/mL | 0.1 mL | 0.05 mL |
| SCF (100 μg/mL) | 100 ng/mL | n/a | 0.1 mL | n/a |
| IL2 (50 μg/mL) | 25 ng/mL | 25 ng/mL | 0.05 mL | 0.05 mL |
| IL7 (10 μg/mL) | 5 ng/mL | 5 ng/mL | 0.05 mL | 0.05 mL |
| α-MEM (pen/strep 1%) | n/a | n/a | 78.75 mL | 78.75 mL |
| Total | n/a | n/a | 100 mL | 100 mL |
Note: The addition of SCF is only required for the two first weeks of culture. The FLT3L concentration is decreased by half from week 3 onward.
Step-by-step method details
Differentiation of hESCs via embryoid body formation
Timing: 3–9 weeks
Prior to differentiation, hESCs are first amplified on Geltrex-coated dishes for at least one passage to remove MEFs which are inhibitory of differentiation toward mesoderm and blood cells.
-
1.Expansion of hESCs in feeder-free culture
-
a.Thaw a Geltrex aliquot on ice 10–12 h in a fridge. The following day prepare a further 1:40 dilution with cold KO-DMEM (total dilution factor from initial stock 1:200).
-
b.Dispense 1 mL of diluted Geltrex per well of a 6-well plate. Plates can be prepared in advance, sealed with parafilm, and kept in the fridge for up to one week.
-
c.Place the 6-well plate with Geltrex at 37°C for 15–20 min, check under the microscope that Geltrex pieces have attached, then aspirate the remaining Geltrex.
-
d.Wash gently with 1–2 mL KO-DMEM per well and add 1 mL of TeSR-E8 basal medium per well.
-
e.On the day of passaging, leave TeSR-E8 medium at 20°C for at least 30 min to warm up and place an aliquot of EDTA 0.5 mM solution in a 37°C water bath for at least 10 min prior to use.
-
f.Prepare the required volume of TeSR-E8 medium, containing 10 μM ROCK inhibitor, for the number of wells in which the cells will be split, calculating 1.5 mL per well.
-
g.Carefully remove medium from the hESC colonies and wash quickly and gently with 0.5 mL of EDTA 0.5 mM to remove any trace of medium and debris.
-
h.Add 1 mL of EDTA 0. 5 mM and place cells in the incubator at 37°C for 2 min. Check the colonies under microscope, the cell-cell interactions should become more relaxed and cells within the colonies should become more refringent.
-
i.Carefully remove the EDTA solution without disturbing the cell layer. Let the cells dry by keeping the plate lid off for 1 min in the tissue culture hood.
-
j.To detach the hESCs, pipette 500 μl of TeSR-E8 medium containing ROCK inhibitor directly over the colonies, aiming to detach small clumps of 5–6 cells, avoiding making a single cell suspension (Figure 3).
-
k.Repeat step j until reaching the desired volume of medium, calculating 500 μl per well in which the hESCs will be split (e.g., 2 mL of medium for a 1 in 4 split). This minimizes repeated pipetting of the cell clumps, minimizing cell death and single cell formation. Typically, hESCs are split at 1 in 6 or 1 in 8.
-
l.Dispense 500 μl of hESC clump suspension dropwise to each new well. Gently move the plate up and down, left and right to evenly distribute the cell clumps within the wells.
-
m.Carefully transfer the plate to an incubator at 37°C 5% CO2 in normoxia to avoid gathering of all cell clumps to the middle of the wells.
-
n.Replace medium without ROCK inhibitor after 24 h. The first passage on Geltrex may show spontaneous differentiation, more death, fewer attached clumps and more flattened morphology (Figure 4) than on feeders. Cells will recover after a few days and with further passaging on Geltrex.
-
o.Make a stock of hESCs grown on Geltrex before beginning an experiment if needed. Use the freezing protocol below.
-
a.
-
2.Freezing hESCs grown on Geltrex
-
a.Before starting, warm up an aliquot of EDTA solution in a water bath at 37°C for at least 10 min.
-
b.Use the protocol for EDTA passage.
-
c.Determine the volume of freezing medium needed calculating 1 mL per cryovial. Typically, 2 cryovials can be frozen per confluent well of a 6-well plate.
-
d.In step j of the EDTA passage protocol, use freezing medium instead of TeSR-E8 medium.
-
e.Transfer the cell clumps into a cryovial, then place the cryovials in an appropriate freezing container (Isopropanol-filled or similar) to ensure a gradual decrease of the temperature.
-
f.Place the container at −80°C.
-
g.Transfer the cryovials to liquid nitrogen 24–48 h later.
-
a.
-
3.
Differentiation of hESCs via Embryoid Body formation
Embryoid bodies (EBs) are made from hESCs grown on Geltrex when 80% confluence is reached. The formation of EBs is the first step in the differentiation process. This is best achieved by culturing small aggregates of hESCs in the presence of BMP4 for 24 h. At this stage, BMP4 promotes the survival of the hESCs. At day 0, it is important to add ROCK inhibitor to enhance survival and Geltrex to maintain a good aggregation of cells. At day 1, FGF2 is added to the EB medium then at day 2 and 4, the medium is changed.-
a.Remove the TeSR-E8 medium and wash quickly and gently with 1 mL of EDTA 0.5 mM per well of a 6-well plate.
-
b.Add 1 mL of EDTA 0.5 mM per well of a 6-well plate and incubate at 37°C for 2 min or more. The length of incubation with EDTA will depend on the cell line, so it is important to check under the microscope that the edges of the colonies are becoming refractive.
-
c.Aspirate EDTA and replace with 2 mL of IMDM 20% FBS EB harvesting medium.
-
d.Cut the hESC colonies into small clumps using a StemPro EZPassage tool, cutting up and down, then rotating the plate and cutting up and down in a perpendicular orientation to the previous cut. This is performed in a laminar flow hood (Figure 5).
-
e.Collect the clumps in a screw-cap tube, avoid making single cell suspension by pipetting too vigorously. Wash the plate thoroughly with extra medium to harvest all the clumps. A cell scraper can be used to reach the cells located at the edges of the well.
-
f.Let the clumps settle by gravity for 20 min. If the clumps are small, they can be centrifuged for 5 min at 600 × g.
-
g.Carefully aspirate the supernatant and gently add 2 mL (per well of 6-well plate) or 10 mL (per 10 cm dish) of Embryoid Body differentiation medium day 0.
-
h.Carefully transfer the clumps to Poly-HEMA coated plates. 6 wells of a 6-well plate are needed to seed a 10 cm dish of EBs or 1 well into 1 well of a 6-well plate. Incubate at 37°C, 5% O2, 5% CO2.
-
i.At day 1, add 0.5 mL per well or 2 mL per 10 cm dish of EB medium day 0, containing FGF2 for a final concentration of 5 ng/mL in the total volume.
-
j.At day 2, collect the EBs with a 10 mL serological pipette and let them settle by gravity for 15–20 min. Gently wash the dish with 6 mL of IMDM basal medium to collect all EBs. Resuspend in EB medium day 2. Replate the EBs in new Poly-HEMA-coated dishes or plates and incubate at 37°C 5% O2 5% CO2.
-
k.At day 4, collect the EBs (Figure 6), let them settle by gravity for 15–20 min and resuspend in EB medium day 4. Replate the EBs in new Poly-HEMA-coated dishes or plates and incubate at 37°C 5% O2 5% CO2 for 2 more days.
-
a.
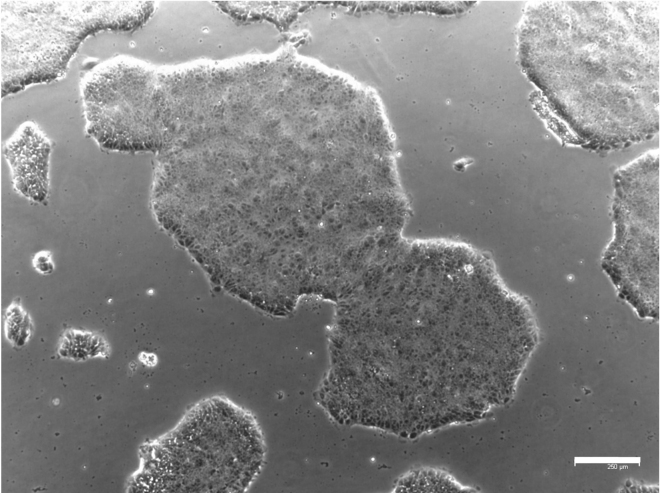
Figure 4.
hESC colonies morphology
Representative example of hESC colonies grown on Geltrex-coated dish. Scale bar, 250 μm.
Figure 5.
Square cutting for embryoid body seeding
hESC colonies grown on Geltrex-coated dish and cut in small squares for embryoid body seeding using a StemPro EZPassage tool. Scale bar, 200 μm.
Figure 6.
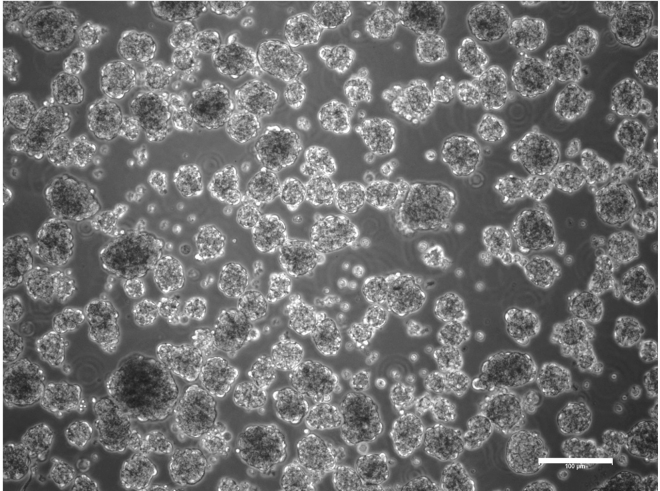
Embryoid bodies
Representative example of embryoid bodies at day 4 of differentiation. Scale bar, 100 μm.
Hemogenic endothelium isolation and culture
Hemogenic endothelium is an endothelial subset that give rise to all blood progenitors in specific culture conditions (Lacaud and Kouskoff, 2017). To determine the hemogenic potential of the culture, the endothelial population is isolated from EBs and further cultured in hematopoietic inducing conditions.
-
4.Isolation of hemogenic endothelium by cell sorting
-
a.Before starting the protocol, warm up the TrypLE solution in a water bath at 37°C.
-
b.Collect day 6 EBs in 50 mL screw-cap tubes and let them settle by gravity for 20 min. EBs from up to two 10 cm dishes are collected per 50 mL tube. Dishes can be washed with extra IMDM medium to ensure harvesting of all EBs. If the EBs are small, they can be centrifuged for 3 min at 400 × g.
-
c.Once the EBs are pelleted, carefully remove the supernatant and add 5 mL of warm TrypLE per 50 mL tube.
-
d.Incubate for 3 min at 37°C in a water bath, then vortex briefly. If EB clumps are still present in the suspension, return to the water bath for 2 min.
-
e.Add 5 mL IMDM 10% FBS and pipette up and down to disaggregate EBs. Allow any non-disaggregated EBs to settle and pass supernatant through a pre-wet 50 μm Filcon filter (BD bioscience, cat# 340630) into 5 mL snap cap tubes (Falcon, cat# 352054).
-
f.Add 2 mL IMDM 10% FBS to the remaining EBs and use a 5 mL syringe with a 18G blunt needle to disaggregate the remaining clumps. Allow any non-disaggregated EBs to settle and pass supernatant through a pre-wet Filcon filter into 5 mL snapcap tubes. Repeat until all EBs are collected.
-
g.Count cells using a hematocytometer.
-
h.Centrifuge for 3 min at 3,000 × g.
-
i.Resuspend cells in IMDM 10% FBS at 5 × 106 cells/mL and pass the cell solution through a pre-wet 50μm Filcon filter to remove any potential cell clumps. Set aside 200 μL of cell suspension for flow cytometry unstained control and single stained controls.
-
j.Stain the cell solution by adding anti-CD43, anti-CD144, and anti-CD31 antibodies directly to the cell solution. Hoechst 33258 is added as a cell live/death discriminator. The choice of fluorochromes will depend on the Flow Cytometer sorter filters and lasers settings. Stain between 20 × 106 and 50 ×106 cells to have enough cells after the sort for replating.
-
k.Incubate for 1 h at 4°C, mixing the cell solution after 30 min by vortexing gently.
-
l.Cells are washed once with 5 mL of IMDM 10% FBS, centrifuged for 5 min at 3,000 × g, the supernatant is discarded and the cells resuspended in IMDM 10% FBS at 5 × 106 cells/mL or less depending on the Flow Cytometer sorter used.
-
m.Compensation settings are performed using BD CompBeads (BD Bioscience) labeled with the antibodies used for staining the cells according to the manufacturer’s instruction (https://www.bdbiosciences.com/ds/pm/tds/552843.pdf).
-
n.Gating strategy is decided based on the unstained and single stained cell samples.
-
o.Live cells positive for CD144 and CD31 expression and negative for CD43 expression are sorted. Typically, the frequency of CD144+CD31+CD43− is 20% to 25%.
-
p.After the sort, cells are centrifuged for 5 min at 3,000 × g, the supernatant is discarded and the cells resuspended at 1 × 106 cells/mL in hemogenic endothelium inducing medium.
-
q.Cells are plated on gelatin-coated plates at 120,000 cells per well of a 6-well plate for analysis at day 4 and 80,000 cells for analysis at day 7. The media does not need to be changed during this period.
-
a.
-
5.Flow cytometry analysis at day 4 and 7 of hemogenic endothelium culture
-
a.Before starting warm up the TrypLE solution in a water bath at 37°C.
-
b.Collect the supernatant of the culture into a 15 mL screw-cap tube as it contains floating hematopoietic cells. The remaining attached cells which are mostly endothelial are trypsinized.
-
c.Add 1 mL of warm TrypLE to the well and incubate 3 min at 37°C in an incubator.
-
d.Cells are gently detached and resuspended by pipetting the TrypLE solution up and down. Cells are transferred to the tube containing the supernatant and the well is washed with 1 mL of IMDM 10% FBS
-
e.Centrifuge the cells for 3 min at 3,000 × g, aspirate the supernatant, and resuspend the pellet in 1 mL of FACS wash
-
f.Cells are stained for hematopoietic (CD43, CD235a) and endothelial markers (KDR, CD73) (Figure 7). For each set of markers, 1 × 105 to 5 × 105 cells are stained. Unstained control is also analyzed. Compensation settings are performed using BD CompBeads (BD Bioscience) labeled with the antibodies used for staining the cells according to the manufacturer’s instruction. Fluorescence minus one (FMO) controls should also be run to control the level of fluorescent spill over in the multicolor flow cytometry panels.
-
a.
-
6.Clonogenic assayNote: This assay measures the number and type of hematopoietic progenitors and is performed with cells from day 4 and day 7 hemogenic inducing cultures.
-
a.For each assay 3.5 mL of semi-solid medium is used, enough to seed three 35 mm dishes containing 1 mL each. 0.5 mL is prepared in excess as it is impossible to recover all the methylcellulose solution from the tube.
-
b.Warm up an aliquot of methylcellulose (MEC) at 37°C for at least 20 min.
-
c.Prepare the required amount of semi-solid medium, adding the methylcellulose last using a 5 mL syringe with 18G blunt needle.
-
d.Dispense 3.5 mL of semi-solid medium per 15 mL snapcap tube using a 5 mL syringe with 18G blunt needle.
-
e.Add 3.5 × 103 to 105 cells of day 4 or day 7 hemogenic culture to each 15 mL snapcap tube containing 3.5 mL of semi-solid medium and vortex vigorously briefly to homogenously mix the cells within the methylcellulose medium then let the air bubbles settle. The added volume of cells should not exceed 10% of the final volume.
-
f.Incubate for 5 min in a 37°C water bath to loosen up the methylcellulose and vortex vigorously to mix and allow the air bubbles to dissipate before dispensing.
-
g.Using a 1 mL syringe and 18G blunt needle, carefully distribute 1 mL of the solution per 35 mm sterile petri dish. Keep in mind it will go slowly due to the high density of this medium.
-
h.Place the dishes in a cell culture protective tray containing a dish with water to keep the appropriate humidity inside the tray and put in the incubator at 37°C, 5% CO2, 5% O2.
-
i.Ensure an appropriate level of water is maintained inside the tray for two weeks
-
j.Count the colonies after 14 days of culture.
-
a.
-
7.
T lymphocyte generation
The capacity to generate T lymphocytes from progenitors relies on a supportive stromal niche providing Notch signaling (Schmitt et al., 2004). To this end, cells are co-cultured on OP9 stromal cells expressing the human ligand DLL1 (obtained from the Zuniga-Pflucker group). These stroma cells are mitotically inactivated by Mitomycin C as described for the MEF above (section 15). It is advisable that these stromal cells express a fluorescence protein (GFP or similar) to exclude them in flow cytometry analysis. T cell potential is assessed either directly from sorted hemogenic cells or from cells obtained after 4 days of hemogenic inducing culture.-
a.One day before starting the assay, plate a vial containing 106 mitotically inactivated OP9-hDLL1-GFP mouse stromal cells in a 12-well plate or in a 6-well plate in OP9-DLL1 medium depending on the number of cell populations to be tested.
-
b.The next day, check that the OP9-hDLL1 cells are healthy and homogenously distributed (Figure 8).
-
c.Warm up an aliquot of the OP9-DLL1 medium at 20°C.
-
d.Hemogenic endothelium sorted from day 6 EBs can be directly used in this protocol. Alternatively, cells harvested after 4 days of hemogenic inducing culture can be seeded on OP9-DLL1 stroma layer (see step 5a–d for harvesting these cells).
-
e.The cells to be tested for T lymphocyte potential are seeded at minimum of 104 cells per cm2 in T lymphoid generation medium containing 20 ng/mL FLT3L, 100 ng/mL SCF, 25 ng/mL IL-2 and 5 ng/mL IL-7 human cytokines.
-
f.Cells must be transferred to a fresh inactivated OP9-hDLL1 cells at day 10 and every 5 days afterwards, keeping the culture for 1 month. The stroma cells must be thawed the day before passages
-
g.From day 15 onward, feed the cells with stroma medium containing 10 ng/mL FLT3L, 25 ng/mL IL-2 and 5 ng/mL IL-7 human cytokines
-
h.To passage the cells onto a fresh stroma cell layer, collect the supernatant into a 15 mL screw-cap tube as it contains floating hematopoietic cells.
-
i.Add 1 mL of warm TrypLE to the well and incubate 3 min at 37°C in an incubator.
-
j.Cells are gently detached and resuspended by pipetting the TrypLE solution up and down. Cells are transferred to the tube containing the supernatant previously collected and the well is washed with 1 mL of stroma medium and passed through a pre-wet 50μ m sterile Filcon to remove clumps of OP9 cells.
-
k.Centrifuge the cells for 5 min at 3,000 × g and add fresh OP9-DLL1 medium containing human cytokines according to the day of culture. Each well is transferred completely to another well, maintaining the culture without diluting the cell density.
-
l.After 1 month in culture, dissociate the culture as described for passaging and analyze the cells by flow cytometry for the expression of human CD45, CD8 and CD4 markers (Figure 9). Hoechst 33258 or any other suitable live dye is used for live/dead discrimination.
-
m.Use FMOs (fluorescent minus one), unstained sample, live single stained controls, and OP9-GFP stromal cells alone as controls for the flow cytometry data analysis.
-
n.The first gate must be done on the GFP negative CD45+ population to eliminate the OP9 cells that express high GFP.
-
a.
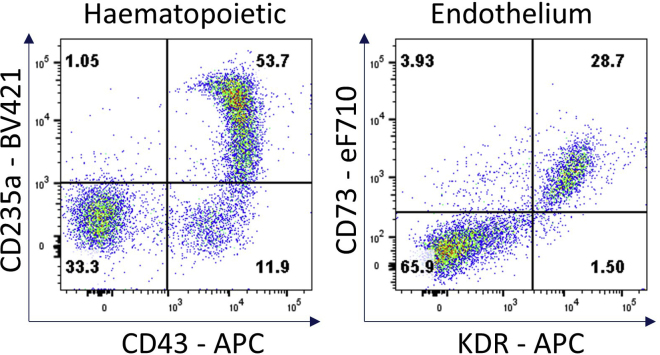
Figure 7.
Hematopoietic and endothelial cell characterization
Flow cytometry analysis of day 4 hemogenic culture for hematopoietic (CD43, CD235a) and endothelial (KDR, CD73) cell surface markers.
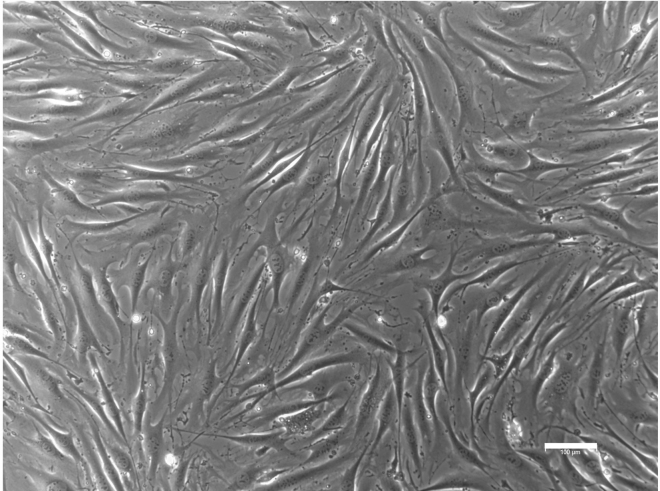
Figure 8.
OP9-DLL1 stroma
Representative example of a confluent layer of OP9-DLL1 stroma cells. Scale bar, 100 μm.
Figure 9.
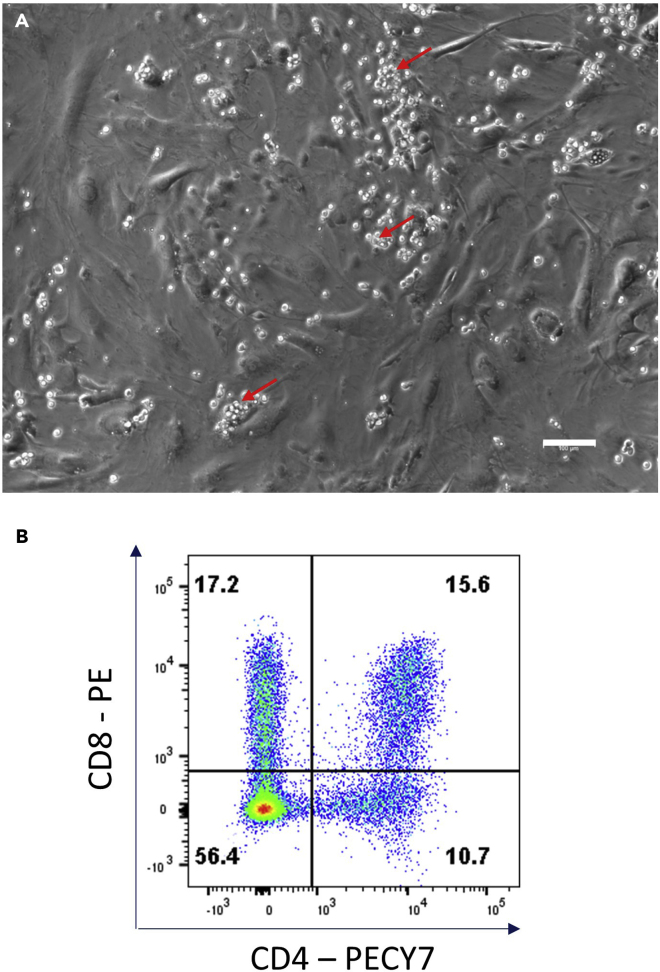
T lymphocyte characterization
(A) T lymphocyte culture on OP9-DLL1 showing T lymphocytes (red arrows) on top of the stroma layer. Scale bar, 100 μm.
(B) Flow cytometry staining for CD4 and CD8 cell surface markers.
Expected outcomes
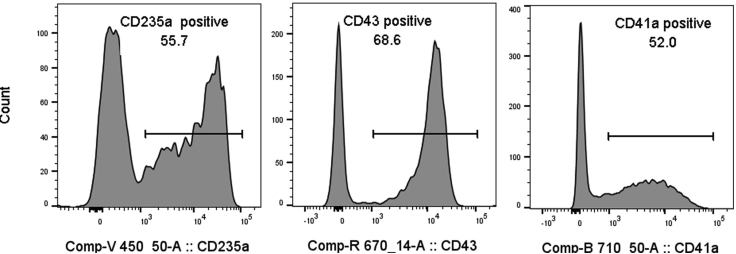
The successful differentiation of hESCs to blood progenitors will show a high frequency of cells stained for CD235a, CD43 and CD41a cell surface markers as measured by flow cytometry (Figure 10) and the generation of colonies with high proliferative and mixed potential in clonogenic assays (Figure 11).
Figure 10.
Hematopoiesis analysis
Flow cytometry analysis of the indicated cell surface markers at day 4 of hemogenic endothelium culture.
Figure 11.
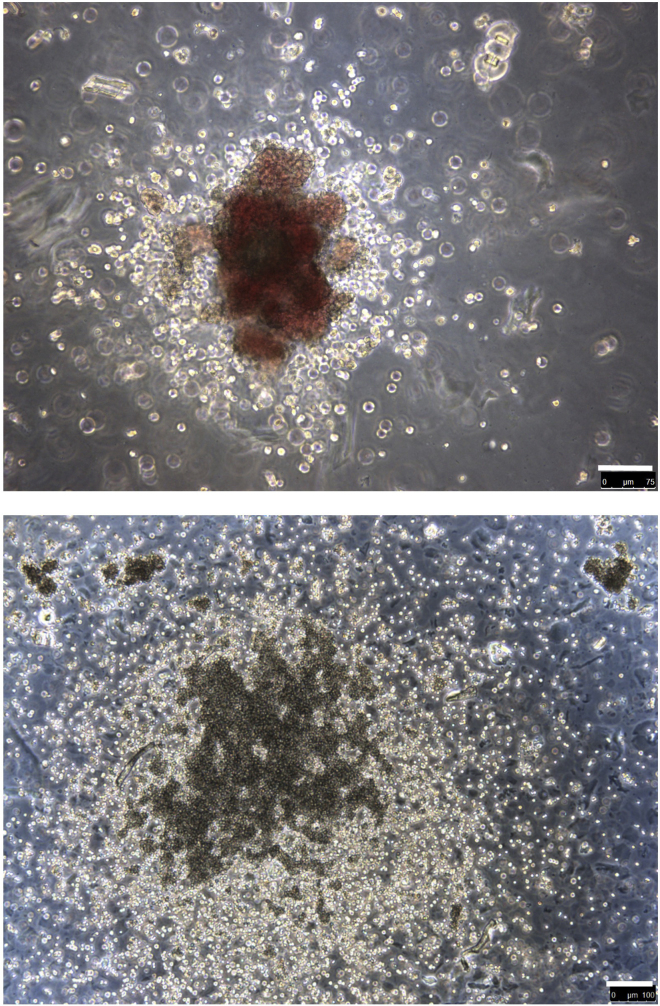
Hematopoietic colonies
Example of hematopoietic colonies in clonogenic assay, indicative of high proliferative and mixed potential. The upper panel shows a mixed erythroid-myeloid colony. Scale bar, 75 μm. The lower panel shows a mixed myeloid colony. Scale bar, 100 μm.
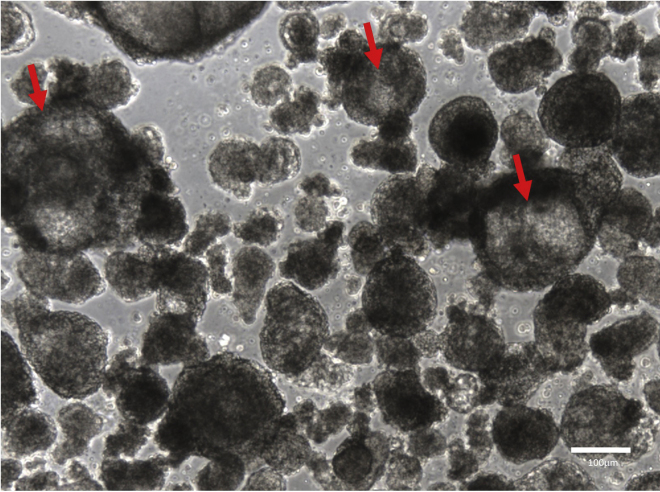
This is the final and successful result of a long process that can be monitored at different stages to ensure that the differentiation is proceeding as expected. hESC colonies should show minimal area of spontaneous differentiation on Geltrex prior to embryoid body formation. Once formed, EBs should increase in size over time and show limited amount of cell death. By day 6 of differentiation, a large fraction of the EBs should become cystic (Figure 12). The CD31+CD144+ endothelium population at day 6 of EB differentiation should represent 15%–25% of the total population which is the sign of an efficient differentiation process. Once replated in hemogenic endothelium culture, endothelial cells should give rise to floating blood cells within two days (Figure 13). These blood cells will increase in number over time in the culture.
Figure 12.
Embryoid bodies
Representative example of embryoid bodies at day 6 of differentiation showing cysts formation (red arrows). Scale bar, 100 μm.
Figure 13.
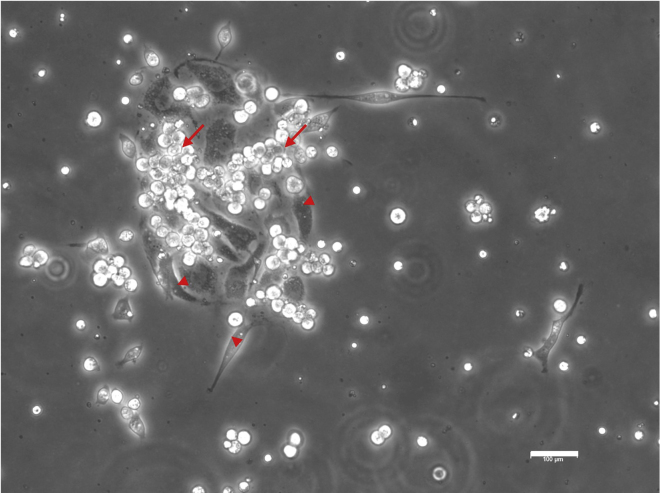
Endothelial to hematopoietic transition
Hemogenic endothelium culture at day 7 showing adherent endothelial cells (red arrow heads) and floating hematopoietic cells (red arrows). Scale bar, 100 μm.
Limitations
In this protocol, the differentiation of hESCs is driven in serum-free culture through the formation of 3D embryoid bodies. While all reagents and growth factors can be easily monitored for dosage or time of addition to the culture, the formation of the 3D structure gives rise to cell-cell interaction and activation/inhibition of signaling pathways that cannot be easily monitored or dissected. This is one of the main limitations of this differentiation protocol. These parameters are important in the differentiation process, but they cannot be controlled and therefore this introduces variation in the differentiation efficiency.
Other protocols using monolayer culture systems, either with stroma cells (Park et al., 2018) or with cell-free matrices (Niwa et al., 2011), have been described, but typically they are less efficient, generating more endothelium without hemogenic potential and scaling up monolayer culture for large experiments becomes difficult. A study comparing the different protocols was recently published (Tursky et al., 2020).
Troubleshooting
Problem 1
hESCs are spontaneously differentiating on MEFs (step 18: EDTA passage of hESC culture, in Maintenance and expansion of hESCs onto a MEF layer).
Potential solution
Usually, this is due to a poor or inadequate layer of feeders. If these are not valuable hESCs, discard the culture and start from a new frozen vial, making sure the feeder layer is healthy and covers the entire well. If these are precious hESCs, manually remove the differentiated areas of the culture, and passage on a healthy feeder layer.
Another potential cause could be that there were very few hESCs in the vial or that the freezing process was not well performed. If receiving a vial from another lab or from a company it may be advisable to thaw 1 vial in 1 unique well to ensure enough density and to enhance recovery of the cells. This is valid for problem 1 and 2; if cell density is too low, the cells struggle much more to survive and grow.
Also ensure that all the reagents, including ROCK inhibitor, are stored properly and are not expired.
Problem 2
hESCs are not growing or dying on MEFs (step 18: EDTA passage of hESC culture, in Maintenance and expansion of hESCs onto a MEF layer).
Potential solution
Make sure the hESC medium has not expired, has been kept at +4°C no longer than a week, that the FGF2 has been added before feeding the cells, and that the medium is changed daily. Check that the MEFs are still healthy and that no trace of Mitomycin C was left before freezing the inactivated MEFs. Make sure that the cells have been plated at a high enough density and as cell clumps, as hESCs struggle to grow at low confluency or from a single cell solution
Problem 3
Embryoid bodies (EBs) are not growing (step 3 in method details).
Potential solution
Make sure that EBs are not pipetted too vigorously or cut too small at the beginning, and that ROCK inhibitor and Geltrex have been added at day 0. If the EBs are not becoming cystic this is probably due to the VEGF not working correctly. Make sure that the StemPro medium is not expired and has been kept no more than 2 weeks at +4°C. It is important to keep track of batch and lot numbers for all reagents used and to record when a reagent is changed. Often, if the differentiation is very inefficient, it is due to a new reagent being suboptimal. In some instances, it is worth discussing with manufacturers to ask whether they changed a reagent specificity or whether problems have been reported by other users.
Problem 4
No hematopoiesis is produced from the hemogenic endothelium culture (step 5 in method details).
Potential solution
Make sure that the StemSpan medium is not expired and has been kept no more than 2 weeks at +4°C. Similar to problem 3, it is important to keep track of batch and lot numbers for all reagents used and to record when a reagent is changed. If the cytokines used in this step of the culture have been kept for too long at +4°C, their activities might decrease considerably. Refer to the manufacturer notice for expiration dates. Start a new experiment with freshly thawed cytokines.
Problem 5
Absence of T lymphocytes generation on OP9-DLL1 stroma layer (step 7 in method details).
Potential solution
Ensure that the stromal cells are healthy and correctly inactivated, that no trace of Mitomycin C was left before freezing, and that they are still expressing the Notch ligand.
Ensure that the cytokines and reagents are stored appropriately and are not expired.
Problem 6
Cells are reaggregating during the sort procedure (step 4 in method details).
Potential solution
Ensure that the cell concentration does not exceed 5 × 106 cells/mL. Either decrease the cell concentration further or add EDTA at 0.5 mM to prevent clump formation.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Valerie Kouskoff, Valerie.kouskoff@manchester.ac.uk.
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze datasets or code.
Acknowledgments
We thank the University of Manchester flow cytometry facility for help with sorts. This work was supported by the Medical Research Council (MR/P000673/1), the Biotechnology and Biological Sciences Research Council (BB/R007209/1), and Cancer Research UK (C5759/A20971).
Author contributions
Conceptualization: E.G.-A. and S.M.; Writing – Original Draft: E.G.-A., B.P., and V.K.; Writing – Review & Editing: E.G.-A., S.M., B.P., and V.K.; Funding Acquisition: V.K.; Supervision, E.G.-A. and V.K.
Declaration of interests
The authors declare no competing interests.
References
- Catalina P., Montes R., Ligero G., Sanchez L., de la Cueva T., Bueno C., Leone P.E., Menendez P. Human ESCs predisposition to karyotypic instability: is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol. Cancer. 2008;7:76. doi: 10.1186/1476-4598-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alegria E., Menegatti S., Fadlullah M.Z.H., Menendez P., Lacaud G., Kouskoff V. Early human hemogenic endothelium generates primitive and definitive hematopoiesis in vitro. Stem Cell Reports. 2018;11:1061–1074. doi: 10.1016/j.stemcr.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.E., Park J.A., Ha Y.W., Park S.K., Kim H.S., Oh S.K., Lee Y. Chromosomal modification in human embryonic stem cells cultured in a feeder-free condition after single cell dissociation using accutase. Dev. Reprod. 2012;16:353–361. doi: 10.12717/DR.2012.16.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud G., Kouskoff V. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp Hematol. 2017;49:19–24. doi: 10.1016/j.exphem.2016.12.009. [DOI] [PubMed] [Google Scholar]
- Niwa A., Heike T., Umeda K., Oshima K., Kato I., Sakai H., Suemori H., Nakahata T., Saito M.K. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS One. 2011;6:e22261. doi: 10.1371/journal.pone.0022261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.A., Kumar A., Jung H.S., Uenishi G., Moskvin O.V., Thomson J.A., Slukvin I.I. Activation of the arterial program drives development of definitive hemogenic endothelium with lymphoid potential. Cell Rep. 2018;23:2467–2481. doi: 10.1016/j.celrep.2018.04.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T.M., de Pooter R.F., Gronski M.A., Cho S.K., Ohashi P.S., Zuniga-Pflucker J.C. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Tursky M.L., Loi T.H., Artuz C.M., Alateeq S., Wolvetang E.J., Tao H., Ma D.D., Molloy T.J. Direct comparison of four hematopoietic differentiation methods from human induced pluripotent stem cells. Stem Cell Reports. 2020;15:735–748. doi: 10.1016/j.stemcr.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze datasets or code.