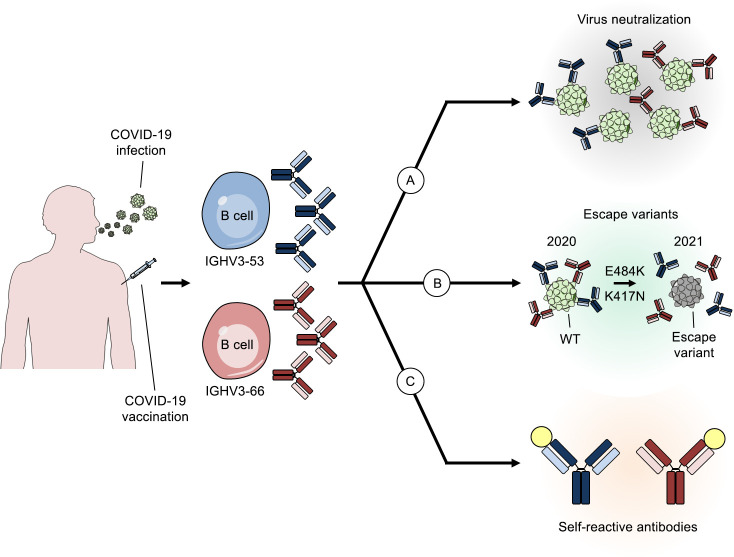
Rappuoli and Andreano highlight that, in humans, neutralizing antibodies deriving from germlines IGHV3-53/IGHV3-66 dominate the immune response to SARS-CoV-2 infection and discuss potential correlations with self-reacting antibodies in COVID-19.
Abstract
The neutralizing antibody response to SARS-CoV-2 is dominated by antibodies deriving from germlines IGHV3-53/IGHV3-66, which are also associated with self-reacting antibodies. Could vaccines avoid the expansion of this immunodominant response, decrease the risk of autoimmunity, and still protect against emerging SARS-CoV-2 variants?
Infection and vaccination with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the infectious agent of coronavirus disease 2019 (COVID-19), is characterized by an immunodominant antibody response against the spike protein receptor binding domain (RBD). This response is mediated by the immunoglobulin G heavy-chain variable (IGHV) germline region IGHV3-53 and the closely related IGHV3-66, which constitute >10% of SARS-CoV-2–neutralizing antibodies targeting the RBD compared with 0.5% and 2.6% in the naive repertoire of healthy individuals (Fagiani et al., 2020; Wang et al., 2021 Preprint; Robbiani et al., 2020). These germlines provide an optimal framework of protection, as they interfere with the interaction between the RBD and human angiotensin-converting enzyme 2 (ACE2; Hoffmann et al., 2020), which is the receptor used by the virus to infect host cells (Fig. 1 A). Very potent SARS-CoV-2–neutralizing antibodies can be generated by expanding preexisting B cell germlines or by adding very few somatic mutations without the need of extensive affinity maturation. For instance, Qiang et al. described SARS-CoV-2–neutralizing antibodies generated from a human antibody library constructed 20 yr before the COVID-19 pandemic, while Tan et al. observed that single point mutations, like Y58F on the heavy chain, can increase the germline-binding affinity to the RBD by 10 to 1,000-fold (Tan et al., 2021 Preprint; Qiang et al., 2020 Preprint). The binding of germline IGHV3-53/IGHV3-66 antibodies to the SARS-CoV-2 RBD seems to leverage on germline-conserved motives such as 32NY33, which binds to RBD residues L455 and A475 in several of the described antibodies, and 53SGGS56 that seem to be central to the recognition of several RBD residues (Barnes et al., 2020). Indeed, conservative mutations of this latter motif to 53TGGT56 increases the binding affinity to the RBD of 50-fold forming a salt bridge with residue E484 (Wu et al., 2020; Yuan et al., 2020). Another important conserved germline residue of IGHV3-53/IGHV3-66 antibodies is the D/E97, which in some antibodies makes a salt bridge with the RBD residue K417 (Wu et al., 2020; Yuan et al., 2020).
Figure 1.
The cryptic role of IGHV3-53 and IGHV3-66 antibodies in COVID-19. Summary of the roles of two predominant antibody germlines elicited by COVID-19 infection. (A–C) The figure shows different functions of IGHV3-53 and IGHV3-66 antibodies after infection which are virus neutralization (A), evolution of escape variants (B), and possible autoimmunity (C).
Interestingly, while most of the interactions between the 25 RBD residues and IGHV3-53/IGHV3-66 neutralizing antibodies are mediated by hydrogen bonds, only K417 and E484 have been described to form a salt bridge resulting in a stronger interaction and higher pressure on these residues (Qiang et al., 2020 Preprint; Wu et al., 2020; Yuan et al., 2020 Preprint).
Given that predominant neutralizing antibodies in the human population derive from these germlines, most of the emerging variants arise to escape this immune response. Indeed, emerging variants, such as 501Y.V2, B.1.1.28.1, and B.1.1.248, isolated in South Africa, Brazil, and Japan, respectively, showed mutations in residues K417 and E484, which are involved in the interaction between the RBD and IGHV3-53/IGHV3-66 antibodies (Wu et al., 2020; Yuan et al., 2020). These two mutations are currently among the most mutated residues on the spike protein according to the Global Initiative on Sharing All Influenza Data (GISAID) database, which gathers >463,000 complete SARS-CoV-2 genome sequences (Elbe and Buckland-Merrett, 2017). In fact, residues E484 and K417 on the RBD are mutated in 0.63% (n = 2,959) and 0.33% (n = 1,531) of the cases, respectively. In addition, the S477 RBD residue, which has also been described to interact with the heavy chain of IGHV3-53/IGHV3-66 antibodies, is currently mutated in N477 in 5.35% (n = 24,819) of sequenced genomes according to the GISAID database (Yuan et al., 2020; Elbe and Buckland-Merrett, 2017).
In conclusion, the mutations on the RBD residues K417 and E484, which significantly reduce the neutralization activity of sera from convalescent and vaccinated subjects and abolish the neutralization activity of many monoclonal antibodies, have been clearly selected to escape the immune pressure of predominant antibody germlines (Fig. 1 B).
Interestingly, severe COVID-19 disease has been also associated with a number of difficult-to-explain pathologies in multiple organs in addition to the respiratory tract. These are associated with dramatic increases of autoantibodies against phospholipids with prothrombotic properties, autoantibodies against cytokines and other immunoregulatory proteins, and autoantibodies against cell surface proteins (Bastard et al., 2020; Wang et al., 2020 Preprint; Zuniga et al., 2021 Preprint; Zuo et al., 2020). It has been proposed that the autoantibodies may be responsible for some of the delay in the onset of severe symptoms in COVID-19 patients (Khamsi, 2021).
In particular, >52% of subjects in a cohort of 172 hospitalized COVID-19 patients showed detectable antiphospholipid autoantibodies in their serum, which could result in life-threatening thrombophilia (Zuo et al., 2020), while another recent study showed that autoantibodies against Annexin A2 were elevated in hospitalized COVID-19 patients and these levels predicted mortality (Zuniga et al., 2021 Preprint). In addition, >10% of 987 individuals with severe COVID-19 had autoantibodies that blocked the action of type 1 IFN molecules (Bastard et al., 2020).
While there is no evidence yet that the autoantibodies described in the above studies derive all or in part from the IGHV3-53/IGHV3-66 germlines, there is evidence that neutralizing antibodies against SARS-CoV-2 deriving from these germlines also bind self-antigens. Several studies (Qiang et al., 2020 Preprint; Kreer et al., 2020; Andreano et al., 2021) have shown that IGHV3-53/IGHV3-66–derived neutralizing antibodies cross-react with self-antigens. Other studies report that IGHV3-53/IGHV3-66–derived antibodies are associated with autoimmune disorders such as Kawasaki syndrome (Johnson et al., 2020). Therefore, it is tempting to speculate that the IGHV3-53/IGHV3-66–mediated immunodominant response against the SARS-CoV-2 virus RBD may contribute to the exacerbation of the COVID-19 disease (Fig. 1 C).
If our hypothesis is correct, second generation vaccines against COVID-19 should be carefully designed to avoid the immunodominant response mediated by IGHV3-53/IGHV3-66 in order to avoid propagating the most common escape variants and the generation of potentially harmful autoantibodies. Structure-based antigen design has been successfully used in the HIV field to change immunogen/germline interaction. In this case, the antigen was engineered to bind the rare B cells BG18-like precursors in order to generate broadly neutralizing antibodies against HIV (Steichen et al., 2019). Here we propose using structure-based antigen design for the opposite purpose; that is, to eliminate the immunodominant interaction between the SARS-CoV-2 spike protein and the IGHV3-53/IGHV3-66 germlines.
Acknowledgments
This work was funded by the European Research Council advanced grant agreement number 787552 (vAMRes).
R. Rappuoli is an employee of the GSK group of companies. E. Andreano and R. Rappuoli are listed as inventors of full-length human monoclonal antibodies described in Italian patent applications nos. 102020000015754 (filed on June 30, 2020) and 102020000018955 (filed on August 3, 2020).
References
- Andreano, E., et al. 2021. Cell. 10.1016/j.cell.2021.02.035 [DOI] [Google Scholar]
- Barnes, C.O., et al. 2020. Nature. 10.1038/s41586-020-2852-1 [DOI] [Google Scholar]
- Bastard, P., et al. 2020. Science. 10.1126/science.abd4585 [DOI] [Google Scholar]
- Elbe, S., and Buckland-Merrett G.. 2017. Glob. Chall. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiani, F., et al. 2020. Signal Transduct. Target. Ther. 10.1038/s41392-020-00287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., et al. 2020. Cell. 10.1016/j.cell.2020.02.052 [DOI] [Google Scholar]
- Johnson, T.A., et al. 2020. J. Hum. Genet. 10.1038/s10038-020-00864-z [DOI] [Google Scholar]
- Khamsi, R. 2021. Nature. 10.1038/d41586-021-00149-1 [DOI] [Google Scholar]
- Kreer, C., et al. 2020. Cell. 10.1016/j.cell.2020.06.044 [DOI] [Google Scholar]
- Qiang, M., et al. 2020. bioRxiv. 10.1101/2020.11.06.370676 (Preprint posted November 6, 2020) [DOI]
- Robbiani, D.F., et al. 2020. Nature. 10.1038/s41586-020-2456-9 [DOI] [Google Scholar]
- Steichen, J.M., et al. 2019. Science. 10.1126/science.aax4380 [DOI] [Google Scholar]
- Tan, T.J.C., et al. 2021. bioRxiv. 10.1101/2021.01.26.428356 (Preprint posted January 27, 2021) [DOI]
- Wang, E.Y., et al. 2020. medRxiv. 10.1101/2020.12.10.20247205 (Preprint posted February 1, 2021) [DOI]
- Wang, Z., et al. 2021. bioRxiv. 10.1101/2021.01.15.426911 (Preprint posted January 30, 2021) [DOI]
- Wu, N.C., et al. 2020. Cell Rep. 10.1016/j.celrep.2020.108274 [DOI] [Google Scholar]
- Yuan, M., et al. 2020. Science. 10.1126/science.abd2321 [DOI] [Google Scholar]
- Zuniga, M., et al. 2021. medRxiv. 10.1101/2020.12.28.20248807 (Preprint posted January 4, 2021) [DOI]
- Zuo, Y., et al. 2020. Sci. Transl. Med. 10.1126/scitranslmed.abd3876 [DOI] [Google Scholar]