Combining transcriptomic and TCRαβ repertoire analysis of circulating and tumor-infiltrating CD8 T cells from patients with metastatic melanoma, Lucca et al. identify a blood-based population with effector properties that reflect those of clonally related tumor-resident T cells.
Abstract
Understanding the relationship between tumor and peripheral immune environments could allow longitudinal immune monitoring in cancer. Here, we examined whether T cells that share the same TCRαβ and are found in both tumor and blood can be interrogated to gain insight into the ongoing tumor T cell response. Paired transcriptome and TCRαβ repertoire of circulating and tumor-infiltrating T cells were analyzed at the single-cell level from matched tumor and blood from patients with metastatic melanoma. We found that in circulating T cells matching clonally expanded tumor-infiltrating T cells (circulating TILs), gene signatures of effector functions, but not terminal exhaustion, reflect those observed in the tumor. In contrast, features of exhaustion are displayed predominantly by tumor-exclusive T cells. Finally, genes associated with a high degree of blood–tumor TCR sharing were overexpressed in tumor tissue after immunotherapy. These data demonstrate that circulating TILs have unique transcriptional patterns that may have utility for the interrogation of T cell function in cancer immunotherapy.
Introduction
Tumor infiltration by T cells, particularly CD8+ cells, is a significant prognostic factor in cancer (reviewed in Barnes and Amir, 2017). Cytotoxic T cells, characterized by lytic granule components and natural killer–associated receptors, play a pivotal role in tumor rejection by directly destroying tumor cells and associated structures in an antigen-specific fashion (Breart et al., 2008; Schietinger et al., 2013). In contrast, exhausted T cells are defined by sequential loss of effector functions, limited proliferative potential (Blank et al., 2019; Moskophidis et al., 1993), and increased, stable expression of coinhibitory receptors (reviewed in Blank et al., 2019; van der Leun et al., 2020). Inhibition of coinhibitory signals has proven highly successful in the treatment of solid malignancies (Ribas and Wolchok, 2018). Understanding the relationship between tumor and peripheral immune environments could allow longitudinal immune monitoring in situations in which tumor biopsies are unfeasible (Cohen and Buchbinder, 2019). Moreover, serial sampling for predicting and monitoring response to therapy is often necessary when weighing risk/benefit ratios for continuation of treatment.
Here, we analyzed paired transcriptome and TCRαβ repertoire of circulating and tumor-infiltrating T cells from matched tumor and blood samples from patients with metastatic melanoma at the single-cell level. We tested the hypothesis that circulating sister clones of clonally expanded tumor-infiltrating lymphocytes (TILs), i.e., clones with matching TCR sequences, can be interrogated to gain insight into the ongoing T cell response in the tumor. We found that the degree of cytotoxicity of the tumor infiltrate, but not exhaustion, can be inferred from circulating TILs. The most pronounced features of exhaustion are consistently displayed by tumor-exclusive clonal T cells. Thus, the degree of TCR repertoire sharing across tissues predicts the T cell phenotype, and genes associated with a high degree of blood–tumor TCR sharing are overexpressed by tissue-infiltrating T cells after successful immunotherapy.
Results and discussion
Detection of clonally expanded TILs in the circulation of melanoma patients
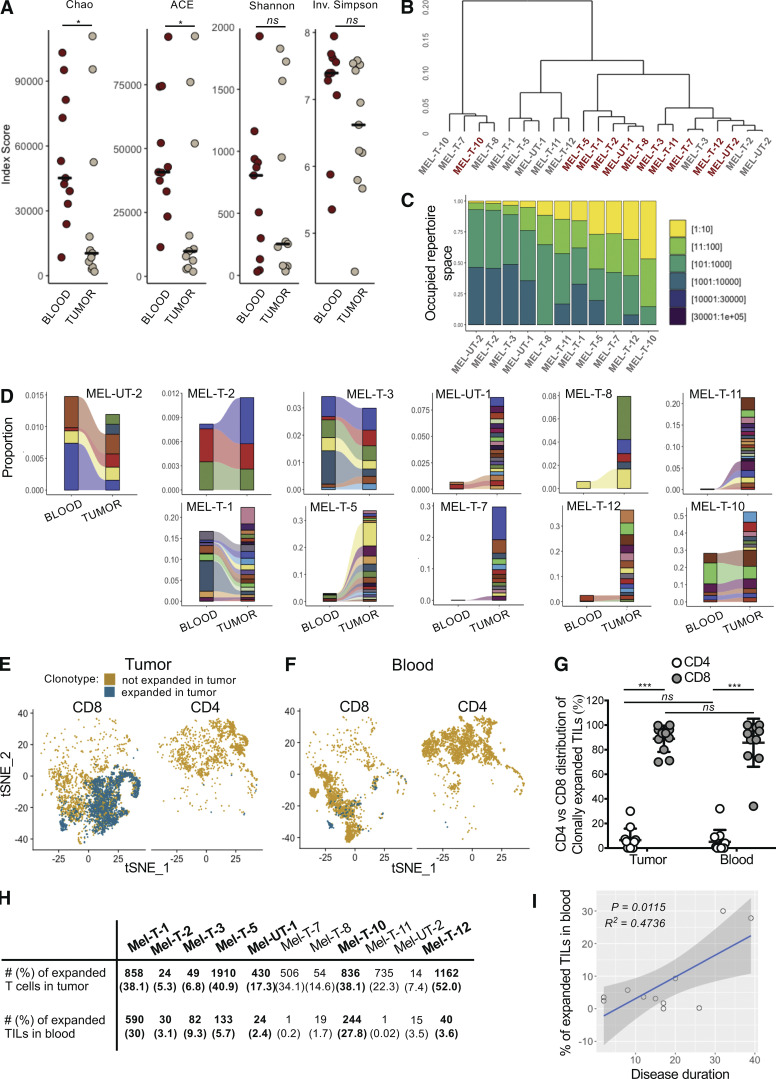
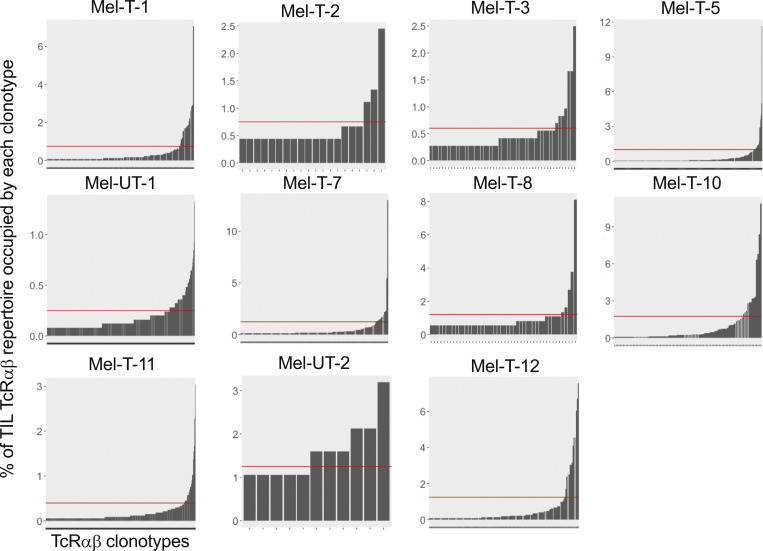
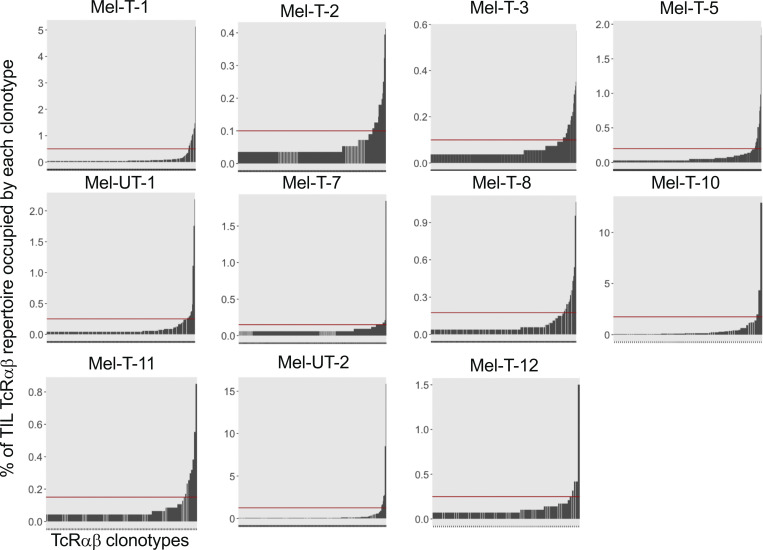
We analyzed paired transcriptomic and TCRαβ libraries from a total of 123,868 T cells derived from matched metastatic tumors and blood from 11 stage IV melanoma patients, selecting subjects with diverse treatment history (Table S1). A single TCRαβ clonotype (see Materials and methods) could be detected in 66.1% (48.5–76.6%) of the cells, and only these cells were retained for subsequent analyses. A reduction in the diversity of immune repertoires signals an ongoing immune response. We computed four estimates of repertoire diversity and compared tumor and blood repertoires across patients and observed a significant reduction in the richness of clonotype species in the tumor as calculated by the Chao (Chao et al., 1992) and ACE indexes (Laydon et al., 2014; Fig. 1 A). To identify patterns of similarity, we performed hierarchical clustering based on clonal size distribution and observed an overall separation of blood and tumor samples with two tumor samples, Mel UT-2 and Mel T-2, clustering closer to blood samples (Fig. 1 B). These two samples consistently ranked lowest when measuring the proportion of the repertoire occupied by the top 10 most frequent clonotypes (Fig. 1 C). Of note, Mel UT-2 was a mucosal melanoma and was therefore sun shielded (Table S1), consistent with the correlation between UV radiation exposure and immune infiltration (Wang et al., 2017). These analyses confirmed previous results showing restricted TCR repertoires in melanoma, indicative of ongoing antigen-specific responses (Kalaora et al., 2018). We then classified individual TIL clonotypes into clonally expanded or not based on the proportion of the repertoire occupied by each clonotype. Because of significant variability in the number of analyzed TILs across patients (1,127–5,404), for this classification we established individual percentage-based thresholds (Fig. S1). We next examined whether sister clones of expanded TILs could be detected in the blood based on exact clonotype matching. Comparing the proportion of tumor and blood repertoires occupied by the 20 most expanded TIL clonotypes highlights the existence of both shared and tumor-exclusive large clones (Fig. 1 D). Based on the largely mutually exclusive expression of the CD8A and CD4 genes, we found that both tumor-resident and circulating expanded TILs were disproportionally CD8 (Fig. 1, E–G). Finally, we observed a positive correlation between the frequency of circulating TILs in blood (average 7.5 ± 10.4%; Fig. 1 H) and the duration of metastatic disease (Fig. 1 I and Table S1).
Figure 1.
Detection of clonally expanded TILs in the circulation of melanoma patients. (A) Dot plots displaying indexes of diversity and species richness for tumor and blood TCRαβ repertoires of melanoma patients. Each dot represents one patient. Horizontal bars indicate medians. Statistical significance was determined using Wilcoxon rank sum test (ns, P ≥ 0.05; *, P < 0.05). (B) Hierarchical clustering of tumor and blood TCRαβ repertoires based on clonal size distribution. Red, blood; gray, tumor. (C) Bar graph displaying the proportion of TCRαβ repertoire space occupied by the top n clonotypes (bins in legend) for the tumor samples of individual patients. (D) Alluvial plots displaying the frequency of the top 20 most-expanded TIL clonotypes in blood and tumor for individual patients. (E and F) tSNE plot of TILs (E) and circulating T cells (F) divided by expression of CD8A (normalized counts >1) and CD4 (normalized counts >1) for one representative patient. T cells expressing TCRs expanded in the tumor are highlighted in blue. (G) Percentage of CD4+ (white) and CD8+ (gray) T cells among clonally expanded T cells in tumor and circulating TILs. Each data point represents the average for one patient. Data represent mean ± standard deviation. Statistical significance was determined using t test with Bonferroni-Dunn correction for multiple comparisons (ns, P ≥ 0.05; ***, P < 0.001). (H) Table displaying the absolute number and frequency of tumor-resident and blood-derived CD8 T cells expressing a TCR expanded in the tumor for each patient. Bold indicates patients in whom the absolute number of blood-derived T cells with a TCR expanded in the tumor (circulating TILs) was deemed large enough for subsequent analyses. (I) Scatter plots showing the relationship of the frequency of circulating TILs and disease duration. Adjusted R2 and F test P values are shown.
Figure S1.
Individual thresholds of clonal expansion in the tumor. Bar graphs displaying the frequency of TILs expressing a given TCRαβ clonotype on the y axis and individual TIL clonotypes ordered by increasing frequency on the x axis, for individual patients. Red horizontal lines represent individualized thesholds of expansion.
Cytotoxic profile of circulating TILs
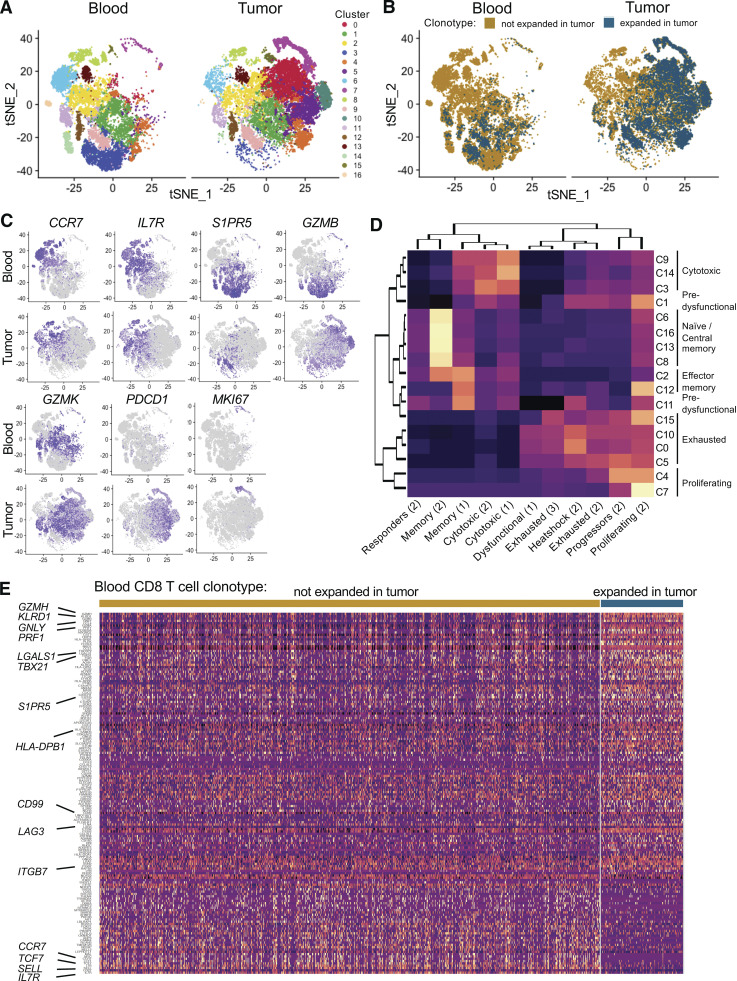
We then examined whether clonally expanded T cells in the tumor displayed specific functional traits by analyzing transcriptional profiles of individual CD8 T cells from all patients by grouping similar cells into clusters. We identified 17 clusters (Fig. 2 A), and clonally expanded TILs largely map onto clusters overrepresented in the tumor (Fig. 2 B). We further annotated each cluster by identifying cluster-specific gene markers (Table S2) and comparing these markers with gene sets derived from previous studies of the immune infiltrate in melanoma at the single-cell level (Li et al., 2019; Sade-Feldman et al., 2018; Tirosh et al., 2016). A large subgroup of blood-dominated clusters (6, 13, 16, and 8) corresponded to naive and central memory CD8 T cells, defined by the high expression of CCR7 and IL7R, overlapping with published signatures of central memory (Fig. 2, C and D). Clusters 2 and 12 displayed both signatures of central and effector memory, while clusters 3, 9, and 14 expressed high levels of cytotoxic molecules and clustered together with previously reported cytotoxic populations (Fig. 2, C and D). Tumor-dominated clusters 0, 5, 10, and 15 were enriched in signatures of exhaustion and terminal differentiation and are indeed marked by expression of PDCD1 (Fig. 2, C and D). Clusters 1 and 11 presented an intermediate profile, with both signatures of exhaustion and cytotoxicity, and higher expression of GZMK than GZMB (Zhang et al., 2019; Fig. 2, C and D), possibly representing predysfunctional T cells (Li et al., 2019). We also identified two clusters of actively proliferating cells (4 and 7) expressing high levels of MKI67 and predominantly containing tumor-derived, clonally expanded T cells.
Figure 2.
Cytotoxic profile of circulating TILs. (A) tSNE plot of CD8 T cells from 11 patients, split by tissue of origin and colored by cluster. Clustering was obtained with an SNN modularity optimization-based algorithm (k parameter = 5, resolution = 0.5). (B) tSNE plot of CD8 T cells from 11 patients, split by tissue of origin and colored by expression of a TCR expanded in the tumor (blue, threshold for expansion as in Fig. S1) or not (gold). (C) tSNE plots colored by scaled expression of indicated genes. (D) Clustered heatmap displaying the degree of overlap between lists of cluster markers (rows) and gene sets derived from published studies of melanoma TIL transcriptome (1 = Li et al., 2019; 2 = Sade-Feldman et al., 2018; 3 = Tirosh et al., 2016). (E) Heatmap of genes differentially expressed in circulating TILs and blood T cells with a TCR not expanded in the tumor with a delta percentage of expressing cells between the two groups of at least ±10%. Rows represent genes ordered by delta percentage of expressing cells. Columns represent individual cells grouped by identity class as indicated on the horizontal bars. Values are scaled and mean-centered log2-transformed gene counts. For visualization purposes, the dataset was randomly down sampled to include 1,000 cells.
In the blood, circulating TILs map to both cytotoxic and predysfunctional clusters. To further define transcriptional traits associated with this population, we compared transcription profiles of circulating TILs and other circulating T cells. We detected higher expression of multiple cytotoxic molecules such as granzymes, perforin, and natural killer–associated receptors as well as markers of tissue residence (LGALS1), migration (CD99), and tissue homing (ITGB7). Finally, LAG3, but not other coinhibitory receptors, was also significantly up-regulated by circulating TILs (Fig. 2 E).
Tumor signature of circulating TILs
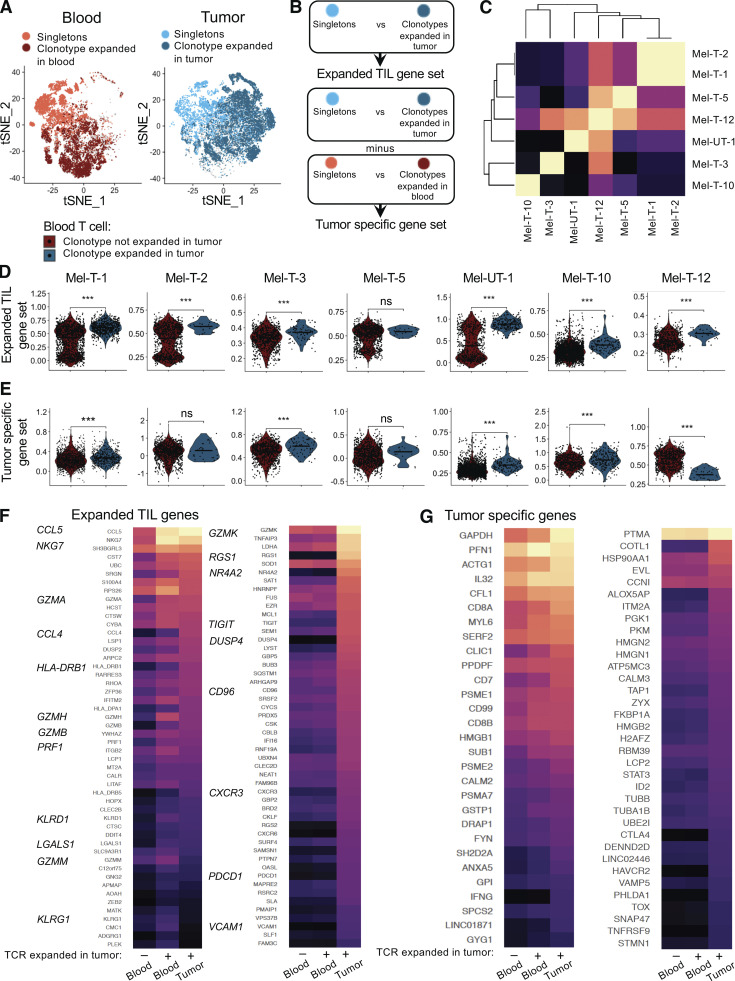
We then examined whether circulating TILs are enriched for features of tumor-resident, clonally expanded TILs. We focused on seven patients in whom a minimum of 20 circulating TILs could be detected (Fig. 1 H). For each patient, we defined expanded TIL gene sets consisting of genes differentially expressed between clonally expanded TILs and tumor-resident T cells with a unique TCR (singletons; Fig. 3 A). Similarly, we compared clonally expanded, blood-derived T cells (based on thresholds shown in Fig. S2) and blood-derived singletons (Fig. 3 A). To obtain a list of genes specifically associated with clonal expansion in the tumor, we incorporated a term for the interaction between clonal expansion and tissue into the linear model used for differential expression detection (Fig. 3 B). This analysis yielded two types of gene sets: those containing all genes relevant to clonal expansion in the tumor, including hallmarks of general activation (expanded TIL gene sets), and those containing only genes found exclusively when T cells expand in the tumor (tumor-specific gene sets; Table S3). We then calculated the overlap between expanded TIL gene sets from individual patients. The expanded TIL gene sets of Met-T-1 and Mel-T-2, the two patients with BRAF/NRAS/c-kit wild-type tumors, presented a high degree of overlap and shared multiple markers of cytotoxicity. A high degree of similarity was also observed for subjects Mel-T-5 and Mel-T-12, enriched for signatures of exhaustion (Fig. 3 C and Table S3).
Figure 3.
Circulating TILs reflect features of clonally expanded tumor-infiltrating T cells. (A) tSNE plots highlighting tumor-infiltrating cells from 11 patients with a TCR clonally expanded in the tumor (dark blue) or singletons (light blue) and blood-derived cells with a TCR clonally expanded in blood (dark red) or singletons (tomato red). (B) Diagram illustrating the definition of expanded TIL gene sets and TIL-specific gene sets. (C) Clustered heatmap of the similarity matrix between expanded TIL gene sets of individual patients. (D) Quantification of the expression of expanded TIL gene sets in circulating TILs (blue) versus other circulating T cells (red) for each patient. Statistical significance was determined using Wilcoxon rank sum test (ns, P ≥ 0.05; ***, P < 0.001). (E) Quantification of the expression of tumor-specific gene sets in circulating TILs (blue) versus other circulating T cells (red) for each patient. Statistical significance was determined using the Wilcoxon rank sum test (ns, P ≥ 0.05; ***, P < 0.001). (F and G) For selected genes belonging to the expanded TIL gene sets of at least three patients, expression is compared between tumor-resident clonally expanded TILs, circulating TILs, and other circulating CD8 T cells. (F) Heatmap displaying expression of expanded TIL genes differentially expressed (left panel) or not (right panel) between circulating TILs and other circulating T cells from 11 patients. (G) Heatmap displaying expression of tumor-specific genes differentially expressed (left panel) or not (right panel) between circulating TILs and other circulating T cells from 11 patients. For heatmaps, values are scaled and mean-centered log2-transformed gene counts. Statistical significance was determined with Wilcoxon rank sum tests, Bonferroni-corrected by the number of genes in the dataset (17.045) multiplied by the number of comparisons (3) with a P value threshold of 0.05.
Figure S2.
Individual thresholds of clonal expansion in blood. Bar graphs displaying the frequency of TILs expressing a given TCRαβ clonotype on the y axis and individual TIL clonotypes ordered by increasing frequency on the x axis, for individual patients.
We then used expanded TIL gene sets to score individual CD8 T cells from the blood and compared circulating TILs with other circulating T cells. Circulating TILs indeed had higher levels of aggregated expression of genes contained in expanded TIL gene sets in six out of seven patients (Fig. 3 D). Because expanded TIL gene sets contain multiple markers of activation and, as shown in Fig. 2, A and B, circulating TILs are an effector population, it is possible that this result may just reflect a difference in activation. Therefore, we also measured the expression of tumor-specific gene sets in circulating TILs. For four out of seven patients, circulating TILs were also enriched for tumor-specific gene sets (Fig. 3 E).
To visualize trends of expression of individual genes up-regulated by clonally expanded TILs, we selected genes contained in the expanded TIL modules of at least three patients and compared differential expression between tumor-resident expanded TILs, circulating TILs, and other circulating T cells (Table S4). Of 566 genes tested, 283 were more highly expressed in circulating TILs compared with other circulating T cells, including multiple markers of effector functions. In contrast, 200 genes had similarly lower expression in blood compared with tumor regardless of TCR sharing. These genes included inhibitory receptors and other markers of terminal differentiation (Fig. 3 F, top 50 genes). Likewise, of 105 genes induced in the tumor-specific sets of at least two patients, 29 were significantly up-regulated in circulating TILs, while 35 were not differentially expressed between blood populations. The tumor-specific genes reflected in circulating TILs included effector molecules (IL32 and IFNG), enzymes involved in carbohydrate metabolism (GAPDH, GPI, and GYG1), and genes involved in cytoskeleton dynamics (ACTG, MYL6, PFN1, and CFL1), possibly mediating adaptations required for tissue residency (Kumar et al., 2017). Tumor-specific genes not reflected in circulating TILs also included some regulators of cell structure (TUBA1B, TUBB, and COTL1) and additional inhibitory molecules (HAVCR2 and CTLA4) and transcription factors associated with exhaustion (TOX) and tissue residency (ID2; Fig. 3 G). Thus, these data not only indicate that circulating TILs reflect certain features of the ongoing T cell response in the tumor, including adaptations to tissue residency, but also reveal that key features of exhaustion in the tumor cannot be inferred from the periphery.
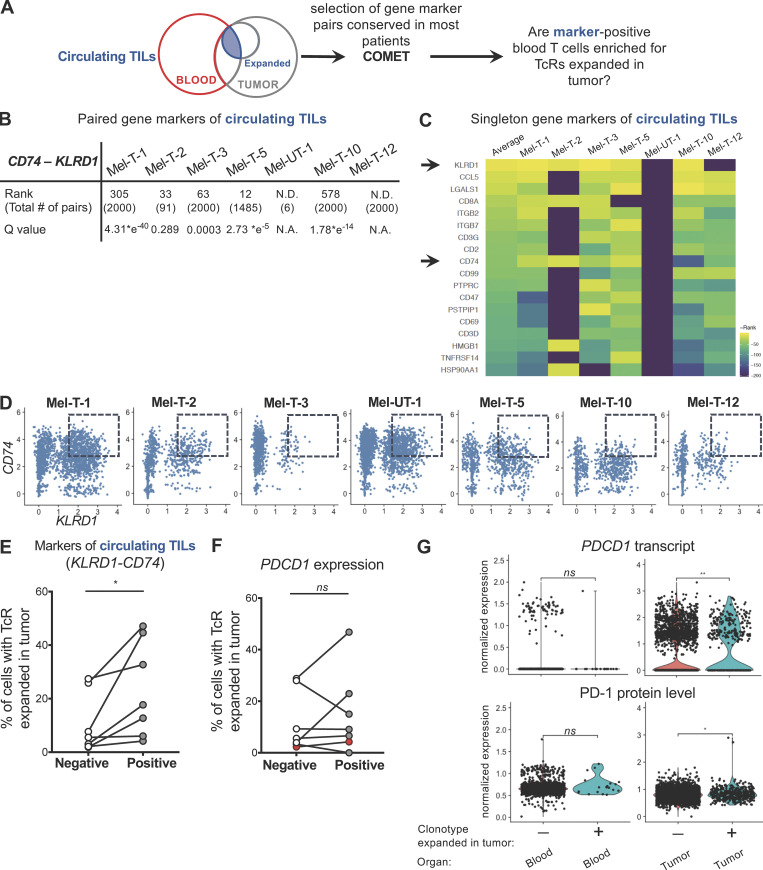
Coexpression of KLRD1 and CD74 enriches for circulating TILs
We then examined whether circulating TILs can be identified in the blood based on expression of gene markers. We classified circulating CD8 T cell populations based on sharing of TCRs expanded in the tumor. The transcriptomes of individual cells with this annotation were used as the input of COMET (combinatorial marker detection from single-cell transcriptomic data; Delaney et al., 2019 Preprint), a tool that uses the minimal hypergeometric test to rank single and paired gene marker panels from a curated list of surface-expressed genes (Chihara et al., 2018) for their ability to identify predefined populations (Fig. S3 A). Of 14 candidate gene marker pairs identified in five out of seven patients, the KLRD1-CD74 module had the highest average rank and a highly significant q value in most patients (Fig. S3 B). Moreover, KLRD1 and CD74 were also highly ranked as candidate gene markers of circulating TILs in the single-gene marker analysis for most patients (Fig. S3 C). We determined whether cells expressing high levels of the KLRD1-CD74 module were enriched for circulating TILs. Indeed, selecting for cells that coexpress KLRD1 and CD74 genes (Fig. S3 D) enriches for circulating TILs (Fig. S3 E). Interestingly, a significantly higher frequency of circulating TILs could not be found by classifying CD8 T cells by PDCD1 gene expression alone (Fig. S3 F). Analysis of PD-1 expression at the protein level consistently revealed that while tumor-resident expanded TILs display higher levels of both PDCD1 transcript and PD-1 protein than unexpanded cells, circulating TILs express similarly low levels of PD-1 as other circulating T cells (Fig. S3 G). Overall, these data suggest that it may be possible to identify a significant proportion of circulating T cells that are clonally related to tumor-resident T cells by measuring transcripts encoding for cell surface proteins in the absence of TCR sequence information.
Figure S3.
Coexpression of KLRD1 and CD74 enriches for circulating TILs. (A) Workflow for identification of gene markers of circulating TILs. Singleton and paired gene markers of circulating TILs were identified from the blood of each patient using COMET. Gene marker pairs detected in most patients were chosen, and blood T cells were classified based on coexpression of the selected gene markers. The percentages of circulating TILs in cells positive and negative for the selected gene markers were compared for each patient. (B) Table summarizing the rank of the gene marker pair KLRD1-CD74, the q value, and the total number of gene marker pairs identified by the COMET analysis. N.D. indicates that the gene marker pair KLRD1-CD74 was not detected in that patient. (C) Heatmap of the reverse rank (rank * −1, rows) of singleton gene markers of circulating TILs that were shared by at least five out of seven patients (columns), ordered by the average across patients. For pairs not detected in a specific patient, a reverse rank of −200 was assigned. Arrows indicate KLRD1 and CD74, detected by both the singleton and the paired gene marker analyses. (D) Scatter plots displaying the relationship between the integrated (Stuart et al., 2019) expression of KLRD1 and CD74 transcripts in each patient. Dashed boxes indicate double-positive cells, with expression of KLRD1 ≥ 1.5 and CD74 ≥ 3. (E) Percentage of cells expressing a TCR expanded in the tumor among cells that are positive or negative for the KLRD1-CD74 combination in each patient. (F) Percentage of cells expressing a TCR expanded in the tumor among cells that are positive or negative for PDCD1 expression in each patent. Red dots correspond to sample Mel-UT-1. Statistical significance was determined using paired t tests (ns, P ≥ 0.05; *, P < 0.05). (G) RNA-level (top) and protein level (bottom) expression of PD-1 in blood and tumor T cells from patient Mel-UT-1, grouped by clonotype expanded in the tumor or not. For RNA data, normalization was done by log transformation. For CITE-seq data, normalization was done by centered log-ratio. Statistical significance was determined using Wilcoxon rank sum test (ns, P ≥ 0.05; *, P < 0.05; **, P < 0.01).
Tumor–blood sharing as a predictor of cytotoxic phenotype
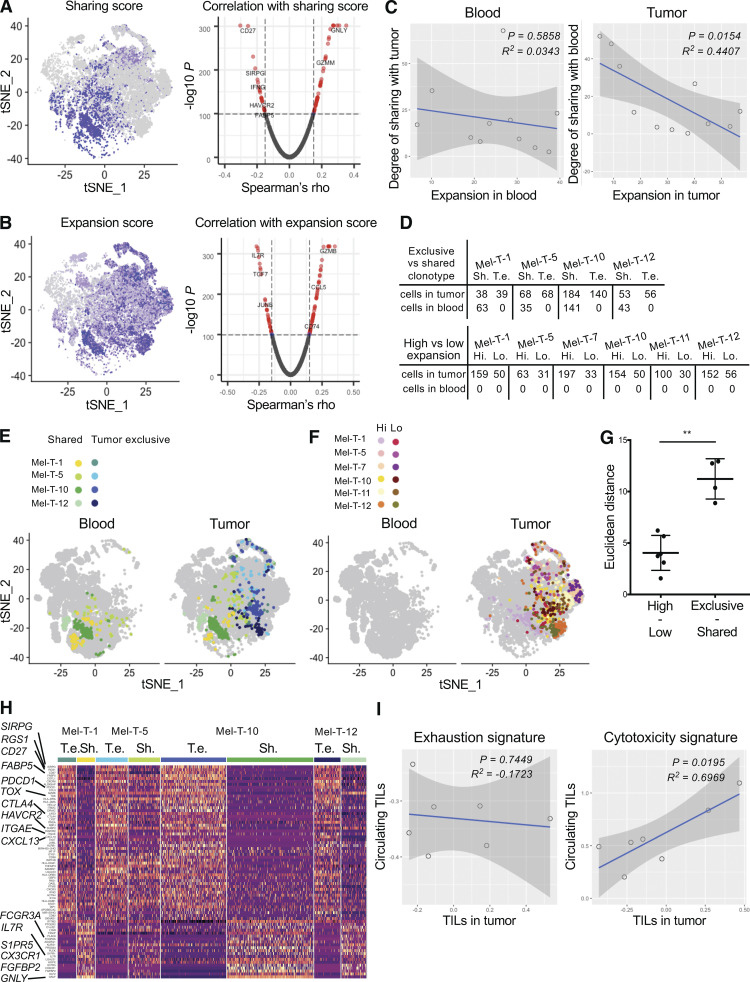
Because many markers of exhaustion could not be detected in the circulation, we hypothesized that these are predominantly expressed by tumor-exclusive clones. To this end, we grouped CD8 TILs and circulating CD8 T cells into clonal groups (i.e., groups of cells sharing the same TCR) and calculated expansion and blood–tumor sharing scores for each clonal group. Mapping the tumor expansion and sharing scores onto the TIL t-distributed stochastic neighbor embedding (tSNE) plot shows clearly that most highly expanded clonal groups are not highly shared with the blood (Fig. 4, A and B).
Figure 4.
Tumor–blood sharing as a predictor of cytotoxic phenotype. (A) tSNE plot of tumor CD8 T cells from 11 patients colored by expression of a blood–tumor sharing score (left) calculated for each clonal group in the tumor (cells sharing the same TCR) and volcano plot representing Spearman’s ρ coefficients and the adjusted P values of the correlations between sharing score and gene expression for CD8 TILs from 11 patients (right). (B) tSNE plot of tumor CD8 T cells from 11 patients colored by expression of an expansion score (left) and volcano plot representing the Spearman’s ρ coefficients and the adjusted P values of the correlations between expansion score and gene expression for CD8 TILs from 11 patients (right). (C) Scatter plots showing the relationship between expansion score in the blood (left) or in the tumor (right) and degree of sharing across the organ. Each dot represents the cumulative scores for each patient. Adjusted R2 and F test P values are shown. (D) Table summarizing the number of cells expressing selected tumor exclusive (T. e.) and shared (Sh.) clonotypes in tumor and blood for the indicated patient (top). Table summarizing the number of cells expressing selected highly expanded (Hi.) and lowly expanded (Lo.) tumor-exclusive clonotypes in tumor and blood for the indicated patient (bottom). (E and F) The indicated cells are mapped on a tSNE plot split by organ. (G) Euclidean distances between the average of the PC analysis embeddings of each cell in the tumor-exclusive versus shared clonal groups and highly expanded versus lowly expanded clonal groups. Dots represent clonal groups. Mean and standard deviation are displayed. Distances were calculated using the PCs that were determined to be statistically significant with an elbow plot (eight PCs). Statistical significance between groups was determined using the Mann-Whitney test (**, P < 0.01). (H) Heatmap displaying expression of genes differentially expressed between tumor exclusive versus shared clonal groups (rows, as in E). Cells are grouped and color-coded according to the clonal group to which they belong. Differential expression was established with the Wilcoxon rank sum test, retaining genes with an adjusted P value <0.05. Values are scaled and mean-centered log2-transformed gene counts. (I) Scatter plots showing the relationship of exhaustion signature (left) or cytotoxicity signature (right) between tumor-infiltrating CD8 T cells and blood-derived circulating TILs. Each dot represents one patient. Adjusted R2 and F test P values are shown.
We then determined whether gene expression in the tumor is associated with these metrics by calculating average Spearman’s ρ coefficients for each gene. The expansion score appears to be correlated with an activation signature, with markers of naive cells (IL7R, CCR7, and LEF1) displaying negative coefficients. Low values of the sharing score instead were associated with markers of exhaustion and terminal differentiation (HAVCR2, PDCD1, CXCL13, FABP5, and IFNG; Fig. 4, A and B; and Table S5). This approach did not allow us to separate the relative contribution of expansion and degree of sharing to these phenotypes. In fact, cumulative measures calculated for all the expanded clonal groups of each patient confirmed that these two metrics are not independent but are rather inversely correlated, specifically in the tumor (Fig. 4 C).
To separate the effects of expansion and degree of sharing, we selected clonal groups for which either of these parameters was constant. Specifically, we identified four clonal group pairs with similar expansion scores within pairs and opposite degrees of blood–tumor sharing. Similarly, we selected six clonal group pairs that were equally tumor exclusive and presented different degrees of clonal expansion (Fig. 4 D and Table S6). We noted that pairs that differ by degree of sharing occupy distinct areas of the cell space, whereas pairs with variable expansion scores all map to the same area (Fig. 4, E and F). This difference could be quantified by calculating the average Euclidean distances for each clonal group pair. Indeed, the distances between exclusive versus shared clonal groups were significantly larger than the ones between highly versus lowly expanded clonal groups (Fig. 4 G). These data indicate that in the tumor infiltrate, the degree of sharing with blood explains more variation than the degree of clonal expansion. Moreover, we identified differentially expressed genes among cells belonging to these clonal groups and were able to confirm that TILs with a TCR shared with blood T cells were highly enriched for signatures of central memory and cytotoxicity and presented fewer features of terminal exhaustion (Fig. 4 H).
As we demonstrated that circulating TILs express cytotoxic genes but low levels of exhaustion markers, we examined whether the cytotoxic score of circulating TILs was predictive of the cytotoxic score of the tumor infiltrate of the same patient. We scored individual tumor-derived CD8 TILs and circulating TILs for expression of a set of cytotoxicity markers previously reported in tumor-infiltrating T cells in single-cell RNA sequencing (scRNAseq)–based studies of multiple human cancers (see Materials and methods and van der Leun et al., 2020). We observed a significant direct correlation between cytotoxicity scores in the tumor versus the circulating TIL population (Fig. 4 I). Moreover, this was not the case when scoring tumor-derived and circulating TILs for an exhaustion signature (Fig. 4 I), indicating that the degree of cytotoxicity of the tumor infiltrate, but not exhaustion, can be inferred from the periphery by analyzing circulating TILs.
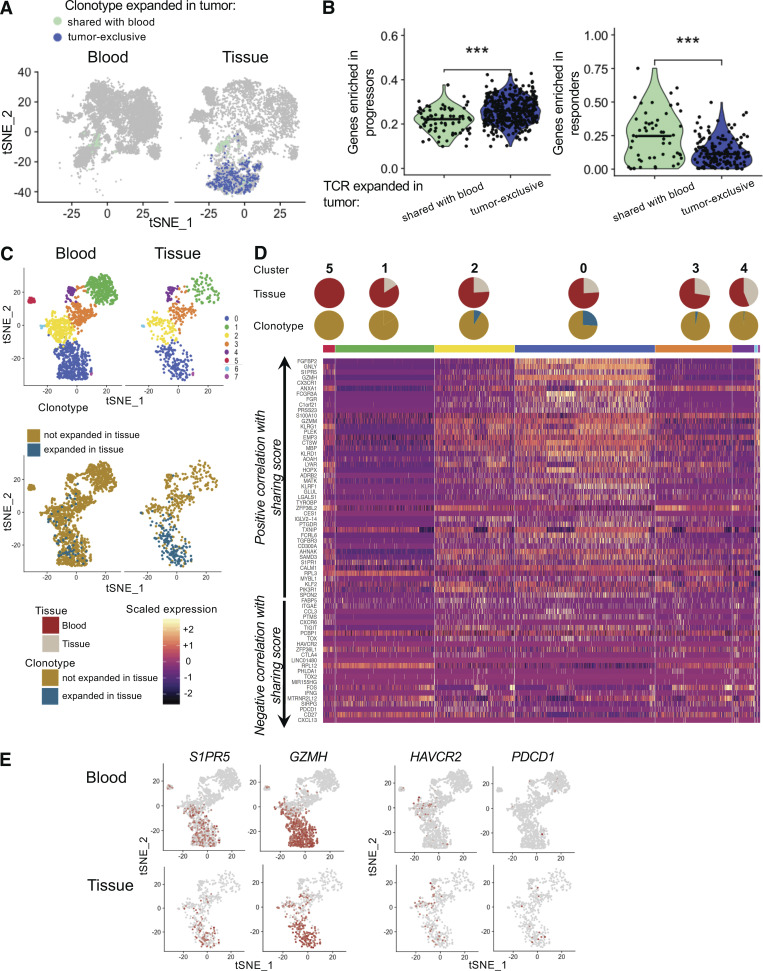
Signature of shared clones in the tissue of responders
It was recently reported that clinical response to immunotherapy is accompanied by an influx of new T cell clones from the periphery into the tumor microenvironment, suggesting that TILs with a high degree of TCR sharing with blood can be involved in tumor rejection (Yost et al., 2019). To address this, we classified expanded TILs from the immunotherapy naive patient Mel-T-1 as shared with blood or tumor exclusive and scored individual cells for gene sets associated with response or disease progression after checkpoint immunotherapy (Sade-Feldman et al., 2018). While tumor-exclusive TILs resembled TILs of progressors, genes predicting response were more represented in TILs shared with blood (Fig. 5, A and B). Furthermore, we examined whether defining sets of genes based on their correlation with the degree of blood–tumor sharing (Table S5) was sufficient to identify genes relevant to tumor rejection after immunotherapy. We analyzed residual tissue from two patients whose tumors responded clinically to checkpoint inhibitor immunotherapy. These samples did not contain any discernible tumor tissue, but rather focal aggregation of inflammatory cells including T cells and activated macrophages. We refer to these as tissue-infiltrating T cells. Similar to what we observed for tumor recurrent patients, in the two responder samples, <25% of tissue-infiltrating T cells carried a clonally expanded TCR (Fig. 5 C). We then analyzed the expression of genes that we had previously established to be associated with blood–tumor sharing or tumor exclusivity in the tissue (Fig. 4 A and Table S5). Genes positively correlated with blood–tumor sharing were highly expressed in the clusters dominated by clonally expanded tissue-infiltrating T cells (Fig. 5, D and E). These included cytotoxic effector molecules and receptors (GNLY, GZMH, GZMM, and KLRD1). Contrary to patients with recurrent disease, clonally expanded tissue-infiltrating T cells did not express high levels of coinhibitory receptors (TIGIT, HAVCR2, PDCD1, and CTLA4) and other markers of exhaustion (TOX, TOX2, and CXCL13) that we demonstrated to be negatively correlated with the blood–tumor sharing score (Fig. 5, D and E). Overall, these results suggest that genes positively correlated with the degree of blood–tumor sharing mark T cells that are involved in tumor rejection in response to immunotherapy.
Figure 5.
Features associated with blood–tumor sharing are highly expressed in the tissue of two patients who responded to immunotherapy. (A) tSNE plots of blood and tumor T cells from patient Mel-UT-1 highlighting tumor-infiltrating cells with a TCR clonally expanded in the tumor and tumor-exclusive (dark blue) or shared with blood (aqua). (B) Quantification of the expression of gene sets associated with response to immunotherapy or progression as described by Sade-Feldman et al. (2018) in tumor-resident cells indicated in A. Statistical significance was determined using Wilcoxon rank sum test (***, P < 0.001). (C) Top: tSNE plot of CD8 T cells from two patients, split by tissue of origin and colored by cluster. Clustering was obtained with an SNN modularity optimization-based algorithm (k parameter = 5, resolution = 0.5). Bottom: tSNE plot of CD8 T cells from two patients, split by tissue of origin and colored by expression of a TCR expanded in tissue (blue) or not (gold). (D) Heatmap of genes previously established to be positively and negatively correlated with TCR sharing score at an adjusted log (P value) less than or equal to −100 (as in Fig. 5 A). Rows represent genes grouped by correlation with TCR sharing. Columns represent individual cells grouped by cluster identity (horizontal bars), with clusters ordered by decreasing percentage of blood cells. Values are scaled and mean-centered log2-transformed gene counts. Pie charts stacked on top of the grouping color bar indicate tumor versus blood composition and expanded TIL composition for each cluster. (E) tSNE plot of CD8 T cells from two patients, split by tissue of origin and colored by scaled expression of the indicated genes.
Recent development in scRNAseq technology has significantly improved our understanding of the relationship between TCR identity and functional properties. A number of studies have analyzed the overlap between TCR repertoires of discrete TIL clusters to examine developmental continuity across functional states (Tirosh et al., 2016; Li et al., 2019; Azizi et al., 2018). Here, we show that the degree of TCR overlap between the distinct tumor and blood tissue compartments can be indicative of the functional state of TILs. Specifically, we show that tumor-derived T cells belonging to TCR clonal groups highly shared across tumor and blood display the highest cytotoxic signatures in the tumor. Of note, circulating sister clones of these TILs also display cytotoxic functions proportionally to what is observed in the tumor, suggesting that analysis of circulating TILs could be used to predict effector functions in the tumor. Previous work using similar approaches in various cancers indicate that cytotoxicity represents one of two main developmental states of CD8 TILs, the other being T cell exhaustion (Zheng et al., 2017; Guo et al., 2018; Zhang et al., 2018; Azizi et al., 2018). These two phenotypes are conceptualized as the extremes in a bifurcation developmental model, where T cells belonging to the same clonal group tend to follow one or the other differentiation pathway. We consistently observed that cytotoxic and exhausted phenotypes were also dichotomously associated with the pattern of TCR sharing across blood and tumor: while cytotoxic sister clones can be found in both tumor and blood, the most exhausted clonal groups appear to be tumor exclusive.
That the pattern of TCR sharing recapitulates CD8 T cell differentiation has a number of implications. In regard to the natural history of the antigen-specific response in the tumor, our data suggest that an exhausted phenotype is more likely to arise in T cells that have been primed locally in the tumor compared with central memory T cells seeded from the periphery. Indeed, tumor microenvironments contain antigen-presenting cells that are unfit for priming T cell responses, partly as a consequence of interactions with regulatory T cells (Wolf et al., 2015; Magnuson et al., 2018).
The most exhausted TILs belong to tumor-exclusive clonal groups, implying that features of exhaustion cannot be inferred by analyzing circulating TILs. Nevertheless, it is known that tumor-resident exhausted T cells have only modest potential for functional reinvigoration, partly due to hard-wired epigenetic changes (Jadhav et al., 2019; Ghoneim et al., 2017; Pauken et al., 2016). Recent work has established that in metastatic melanoma patients, clinical response to checkpoint inhibition immunotherapy is associated with peripheral expansion of large clonal groups expressing high levels of cytotoxic and effector markers, with strong overlap with genes that we found to be associated with a high degree of blood–tumor sharing (Fairfax et al., 2020). This observation suggests that circulating TILs contribute to response to immunotherapy, although the specificity of this population for tumor antigens remains to be demonstrated. Moreover, a study of basal cell carcinoma lesions sampled before and after anti-PD1 therapy found that preexisting exhausted TILs were largely replaced by novel expanded clonotypes, reinforcing the idea that exhausted clonal groups might be short-lived and unlikely to participate in long-term immune surveillance (Yost et al., 2019). We report that genes associated with a high degree of blood–tumor sharing are enriched for effector signatures. Consistently, a recent meta-analysis of four clinical trials of an anti–PD-L1 antibody (atezolizumab) in a variety of solid tumors found that in the tumor, gene signatures associated with clonotypes that were expanded in both tumor and normal adjacent tissue predicted a better response to PD-L1 blockade (Wu et al., 2020).
As we wished to examine circulating TILs in a variety of clinical settings, we did not select patients by prior exposure to immunotherapy or variability in clinical presentation. We thus were able to focus on findings that were reproducible in the patients analyzed, with the goal that these findings could be further corroborated in larger, uniformly treated patient cohorts. While we have further shown that it could be possible to enrich for circulating TILs by analyzing coexpression of genes encoding for surface markers, critical validation steps are required for implementation into a scalable immune-monitoring assay. Nevertheless, we note that in a companion paper by Pauken et al. (2021), circulating TILs could be identified by the expression of a set of gene markers partially overlapping with the ones that we found. The authors describe a combination of four negation markers (CCR7, FLT3LG, LTB, and GYPC) that allows for enrichment of circulating TILs in treatment naive metastatic melanoma patients. Indeed, three of these markers were also highly significantly associated with circulating TILs in our dataset in the two patients with the highest frequency of circulating TILs. Therefore, it appears that the approach of using the TCR as a molecular barcode for circulating TILs followed by an unbiased search for candidate markers can return the same genes across distinct patient cohorts and independent laboratories. Finally, we could not assess the directionality of circulating TILs with respect to their tumor-resident sister clones to determine whether circulating TILs include cells that have left the tumor bed and reentered the circulation. This possibility has been described for tissue-resident memory T cells in the skin: a subset of these cells can enter the circulation, where they remain transcriptionally representative of the skin-resident population (Klicznik et al., 2019). Additional work is required to assess if a similar phenomenon also occurs in cancer and whether related clonal T cells can sequentially patrol multiple metastases, including tissues to which immune cell trafficking is tightly regulated, such as the central nervous system.
In conclusion, we propose that in human cancer the pattern of TCR repertoire sharing across tissues recapitulates T cell differentiation states. Moreover, we report that it is possible to assess the ongoing immune response in the tumor by analyzing a circulating T cell population. Further work is necessary to better define circulating TILs as blood-based biomarkers to increase the precision of existing “liquid biopsy” assays.
Materials and methods
Human subjects
This study was approved by the Yale University Institutional Review Boards. All participants provided written informed consent for the study. All patients had histologically confirmed stage IV metastatic melanoma. Surgical resection was clinically indicated in all cases. Patients donated a portion of surgically resected tissue (for TIL isolation) and a time-matched blood sample (for peripheral blood mononuclear cell [PBMC] isolation).
Tumor dissociation and PBMC isolation
Freshly resected tumor samples were mechanically dissociated and digested in HBSS medium with collagenase IV (2.5 mg/ml) and DNase I (0.2 mg/ml; Worthington Biochemical Corporation) at 37°C for 30 min. TIL-enriched single-cell suspensions were then isolated using Lymphoprep gradient centrifugation. PBMCs were isolated from whole blood using Lymphoprep gradient centrifugation. For sample Mel-T-9, TILs and PBMCs were cryopreserved in GemCell human AB serum with 10% DMSO in liquid nitrogen. For sample Mel-T-11, cryopreserved TILs and PBMCs were kindly provided by the Advanced Cell Therapy Laboratory in the Department of Laboratory Medicine (Yale School of Medicine, New Haven, CT).
Cell sorting
All samples but Mel-T-9 and Mel-T-11 were sorted directly after PBMC and TIL isolation. For samples Mel-T-9 and Mel-T-11, cryopreserved TIL and PBMC aliquots were thawed following the protocol CG00039 from 10X Genomics (https://support.10xgenomics.com/). Samples were sorted on a BD FACS Aria II using a live-cell (Live/Dead Cell Viability Assay; Life Technologies), CD45+ (PerCP-Cy5.5 conjugated; eBioscience), TCRαβ+ (PE-Cy7 conjugated; eBioscience) gating strategy. At least 5,000 live T cells were sorted per sample for inclusion in the 10× library preparation.
Preparation of scRNAseq and single-cell TCR sequencing (scTCRseq) libraries
Sorted T cell samples were centrifuged at 300 rcf for 10 min at 4°C, resuspended in PBS with 0.04% BSA, and counted on a hemocytometer. Cell concentrations were adjusted to between 700 and 1,000 cells/µl. Paired single-cell gene expression and TCR libraries were prepared and sequenced at the Yale Center for Genome Analysis following standard protocols from 10× Genomics (Yale Center for Genome Analysis, 2021). Briefly, single cells were isolated in 1 nl of gel beads in emulsion using the GemCode technology. Depending on the initial cellularity of the sorted samples, between 5,000 and 10,000 cells were targeted. Cell barcoding, lysis, and reverse transcription of mRNA occurred within each gel bead in emulsion. cDNA libraries were then generated using next-generation sequencing PCR amplification with the 5-prime chemistry. Both scRNAseq libraries and scTCRseq libraries were generated for each sample.
CITE-seq staining
Sorted live/CD45+/TCRαβ+ cells derived from blood and tumor samples were incubated with 0.5 µg/sample of the following antibody mix: αCD4 clone RPA-T4, αCD8a clone RPA-T8, and αPD-1 clone EH12.2H7 (all from BioLegend, conjugated with TotalSeq oligos) in PBS:BSA 0.1% for 30 min on ice. Stained cells were centrifuged at 300 rcf for 10 min at 4°C, resuspended in PBS with 0.04% BSA, counted on a hemocytometer, and used for preparation of paired scRNAseq, scTCRseq, and single-cell CITE-seq libraries, as described above.
Sequencing
scRNAseq libraries were sequenced on an Illumina NovaSeq S4 instrument at a read length of 26 × 8 × 91 bp and at a depth of 300 million reads per sample. CITE-seq libraries were also sequenced at a read length of 26 × 8 × 91 bp and at a depth of 30 million reads per sample. scTCRseq libraries were sequenced on an Illumina NovaSeq S4 at a read length of 150 × 150 bp and at a depth of 20 million reads per sample. Data for all scRNAseq experiments are available through NCBI’s dbGaP at accession no. phs002289.v1.p1.
Data processing of scRNAseq and scTCRseq libraries
scRNAseq reads were aligned to the reference genome GRCh38 using the cellranger count pipeline from the CellRanger software v3.0.2 (10X Genomics). The mean number of reads per cell was 49,499 (28,471–103,776) for blood T cell samples and 92,357 (43,243–283,598) for tumor T cell samples. The median number of unique molecular identifiers per cell was 4,383 (2,624–5,820) for blood and 3,960 (2,440–4,836) for tumor samples. The median number of unique genes per cell was 1,444 (1,105–1,802) for blood and 1,465 (1,060–1,791) for tumor samples. Filtered gene-barcode matrices containing barcodes that had passed the threshold for cell detection were used for downstream analyses.
scTCRseq reads were processed using the cellranger vdj pipeline from CellRanger v3.0.2 (10X Genomics, reference genome GRCh38). The mean number of read pairs per cell was 4,792 (1,358–10,040) for blood-derived TCR libraries and 6,442 (1,765–27,411) for tumor-derived TCR libraries. The VDJ enrichment, defined as the percentage of reads mapping to any VDJ gene, was 89 ± 2.4% for blood and 90 ± 2.58% for tumor. TCR contigs called by cellranger vdj with high confidence, listed in the filtered_contig_annotations.csv output file, were retained for downstream analyses. Nucleotide and amino acid CDR3α and β sequences and count matrices of gene expression were matched for each cell based on barcode identities using custom R scripts and were imported into Seurat v3.0 (Satija Lab, 2021). When more than one CDR3α and β sequence was found for a given cell, additional sequences were appended, while NA (not applicable) values were assigned for missing sequences. For downstream analyses, all cells for which multiple CDR3α and β sequences existed or NA values were present were removed from the dataset.
Cluster analysis
Cell-level quality control was performed for individual patients and for blood and tumor gene expression matrices separately. Thresholds for removing low-quality cells were selected based on the distribution of the number of unique genes per cell and the percentage of genes that map to the mitochondrial genome. On average, cells that expressed between 258 and 3,896 unique genes and with a percentage of mitochondrial reads <13.75% were retained for downstream analyses.
For the cluster analysis, we filtered out T cell receptor genes and genes encoded by the Y chromosome and selected for cells with CD8A count ≥1 and CD3E + CD3D + CD3G ≥1. We then joined count matrices from individual patients to generate one merged Seurat object for blood-derived cells and one for tumor-derived cells. Counts were log normalized, and 2,000 variable genes were identified for blood and tumor separately using the Seurat function FindVariableFeatures with the vst method (variance stabilizing transformation). Tumor and blood datasets were subsequently integrated into a single dataset using the Seurat functions FindIntegrationAnchors and IntegrateData using 20 dimensions. For the resulting integrated object, scaled z-scores for each gene were computed using the ScaleData function and regressed against sequencing batch and fresh or frozen processing. Principal component (PC) analysis was then run to identify 30 PCs, of which the first eight were selected for tSNE dimensionality reduction using the elbow method on the distribution of the standard deviation of each PC. For clusters identification, we initially used the FindNeighbors function to build a shared nearest-neighbor (SNN) graph based on the k-nearest neighbor of each cell, with k = 5. Clusters were then determined with the FindClusters function, which uses the Louvain algorithm for modularity optimization (Blondel et al., 2008; resolution = 0.4). Markers of individual clusters were defined as the differentially expressed genes between cells belonging to each cluster versus every other cell using the Wilcoxon rank sum test as implemented in the function FindMarkers. Genes with an adjusted P value <0.05 were retained.
TCRαβ repertoire analysis
The TCRαβ repertoires of blood and tumor samples were analyzed using scRepertoire (Borcherding et al., 2020). Filtered contig annotation files from the output of cellranger vdj were used as input. Indexes of diversity (Shannon entropy, Inverted Simpson) and species richness (Chao, ACE) were calculated using the clonalDiversity function (cloneCall = “aa”), and the significance of group differences was tested with the Wilcoxon rank sum test. Hierarchical clustering of samples based on clone size distribution was performed with the function clonesizeDistribution (cloneCall = “aa”) derived from the package PowerTCR (Koch et al., 2018). The proportion of occupied repertoire space was plotted using the function clonalProportion (cloneCall = “aa”), and alluvial plots were generated with compareClonotypes after selecting the top 20 most expanded clonotypes in the tumor.
To obtain a binary definition of clonal expansion, thresholds were selected by inspection of histograms displaying the percentage of the tumor TCRαβ repertoire occupied by a given clonotype on the y axis, and individual TIL clonotypes were ordered by increasing the percentage of the tumor TIL repertoire on the x axis to identify a knee of the curve.
TIL expanded gene set analysis
Gene sets of clonally expanded TILs were calculated for each individual patient. To determine which genes are specifically associated with T cell expansion in the tumor, we modeled raw gene counts in a generalized linear model as implemented in DESeq2 (Love et al., 2014), including an interaction term between the expansion and the tissue variable: ~Tissue+Expansion+Tissue:Expansion. We ran our model on T cells from each patient separately, using a local fit. To generate expanded TIL gene sets, we filtered genes with log2FoldChange_tumor > 0 at an adjusted P value <0.1. To generate tumor-specific gene sets, we filtered genes with log2FoldChange_tumor > 0 at an adjusted P value <0.1 and log2FoldChange_interaction > 0 at an adjusted P value <0.1. We then used the function AddModuleScore with default parameters to calculate the average expression of each gene set on single cells from the blood for each patient. Blood cells were annotated based on TCR identity as circulating TILs (those with a TCR found to be above the threshold for clonal expansion in the tumor) or not. Statistical significance of the difference between the means of each group of cells was established for each patient using the Wilcoxon rank sum test.
COMET analysis
The input of the COMET analysis consisted of log-normalized counts of CD8 T cells from the blood of individual patients. Each cell was annotated as having or not having a TCR clonally expanded in the tumor. Expression matrices and cell identity annotations were uploaded to http://www.cometsc.com/comet (Singer, 2021), and COMET was run with default parameters. For each patient, we obtained an output file containing lists of gene pairs ranked by their power to identify circulating TILs. We joined these lists by the gene pair identity and filtered out all gene pairs that were not detected in at least five out of seven patients. For the remaining gene pairs, we calculated an average rank and ordered them by decreasing values of the average rank, the highest of which corresponded to the gene pair KLRD1-CD74.
Expansion and blood–tumor sharing score
At the clonotype level, the expansion score (Fig. 4 B) was defined as follows. For each clonotype, we calculated the percentage of the total tumor T cell infiltrate occupied by cells sharing that same clonotype (clonal groups). The patient-level expansion score, as displayed in Fig. 4 C, was calculated by adding up the expansion scores of individual clonal groups that were identified as clonally expanded, as exemplified in Fig. 1 A. Separate patient-level expansion scores were calculated for blood and tumor. The sharing score of individual tumor-expanded TCRs (Fig. 4 A) was defined as the percentage of cells in each clonal group that were also found in blood. For blood-expanded TCRs, the sharing score was a measure of the frequency of cells with that TCR that were also found in the tumor infiltrate. At the patient level, the degree of sharing was calculated by averaging the sharing score of individual clonal groups that were identified as clonally expanded as exemplified in Fig. 1 A. Separate patient-level sharing scores were calculated for blood and tumor.
Exhaustion and cytotoxicity gene signatures
Consensus markers of exhaustion and cytotoxicity were obtained from a recent review of CD8 states in human cancer (van der Leun et al., 2020). Specifically, the exhaustion gene set included HAVCR2, IFNG, ITGAE, PDCD1, CXCL13, LAYN, LAG3, TIGIT, CTLA4, and ENTPD1, while the cytotoxicity gene set consisted of CX3CR1, PRF1, GZMA, GZMH, GNLY, FGFBP2, KLRG1, GZMM, LYAR, TXNIP, FCRL6, NKG7, and KLRD1. We then used the function AddModuleScore with default parameters to calculate the average expression of each gene set on single cells. For each patient, we averaged the exhaustion and cytotoxicity scores of all tumor-infiltrating CD8 T cells and circulating TILs.
Online supplemental material
Fig. S1 contains individual thresholds of clonal expansion in the tumor. Fig. S2 contains individual thresholds of clonal expansion in the blood. Fig. S3 shows that coexpression of KLRD1 and CD74 enriches for circulating TILs. Table S1 presents demographics and clinical features of patients included in the study. Table S2 lists the top 10 marker genes for each cluster, with the percentage of positive cells in the cluster of interest, delta percentage of positive cells between that cluster and the rest of the dataset, and adjusted P value. Table S3 lists gene marker pairs of circulating TILs found in at least five of seven patients, with rank and q value for each patient. Table S4 shows the expanded TIL gene sets and tumor-specific gene sets for each patient. Table S5 lists the correlation coefficients between genes and expansion score, filtered for −logP ≥ 100, and the same for sharing score. Table S6 presents amino acid and nucleotide sequences of the TCRαβ CDR3 regions of clonotypes displayed in Fig. 4.
Supplementary Material
presents demographics and clinical features of patients included in the study.
lists the top 10 marker genes for each cluster, with percentage of positive cells in the cluster of interest, delta percentage of positive cells between that cluster and the rest of the dataset, and adjusted P value.
lists gene marker pairs of circulating TILs found in at least five out of seven patients, with rank and q value for each patient.
shows the expanded TIL gene sets and tumor-specific gene sets for each patient.
lists correlation coefficients between genes and expansion score, filtered for −logP ≥ 100, and the same for sharing score.
presents amino acid and nucleotide sequences of the TCRαβ CDR3 regions of clonotypes displayed in Fig. 4.
Acknowledgments
The authors thank Lesley Devine and Chao Wang for technical assistance with flow cytometry, Guilin Wang and the staff of the Yale Center for Genome Analysis for technical assistance with single-cell genomics, and Antonella Bacchiocchi from the Yale SPORE in Skin Cancer Biospecimen Core for assistance with sample collection. The authors would also like to thank Philip Coish and Jessica Wei for help with manuscript preparation and Matthew Lincoln and Rahul Dhodapkar for insightful comments on data analysis.
This work was supported by National Institutes of Health grant P50 CA121974 (to H.M. Kluger and M. Bosenberg) and National Institutes of Health grants P01 AI073748, U24 AI11867, R01 AI22220, UM 1HG009390, P01 AI039671, P50 CA121974, and R01 CA227473 (to D.A. Hafler).
Author contributions: L.E. Lucca planned and performed experiments, analyzed data, and wrote the manuscript with inputs from P.-P. Axisa, B. Lu, B. Harnett, S. Jessel, M. Singer, H.M. Kluger, and D.A. Hafler. P.-P. Axisa and B. Lu analyzed data. B. Harnett performed experiments. Le Zhang and K. Raddassi provided technical help with single-cell genomics. Lin Zhang provided technical help with sample acquisition. K. Olino and J. Clune harvested patient tissue samples. S. Jessel collected and reviewed patient clinical information. M. Singer oversaw data analysis. L.E. Lucca, M. Singer, H.M. Kluger, and D.A. Hafler conceptualized the study, and H.M. Kluger and D.A. Hafler planned and supervised the study and acquired funding.
References
- Azizi, E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. 2018. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 174:1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, T.A., and Amir E.. 2017. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br. J. Cancer. 117:451–460. 10.1038/bjc.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank, C.U., Haining W.N., Held W., Hogan P.G., Kallies A., Lugli E., Lynn R.C., Philip M., Rao A., Restifo N.P., et al. 2019. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19:665–674. 10.1038/s41577-019-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, V.D., Guillaume J-L., Lambiotte R., and Lefebvre E.. 2008. Fast unfolding of communities in large networks. J. Stat. Mech. 2008(10):P10008. [Google Scholar]
- Borcherding, N., Bormann N.L., and Kraus G.. 2020. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000 Res. 9:47. 10.12688/f1000research.22139.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breart, B., Lemaître F., Celli S., and Bousso P.. 2008. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest. 118:1390–1397. 10.1172/JCI34388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, A., Lee S.M., and Jeng S.L.. 1992. Estimating population size for capture-recapture data when capture probabilities vary by time and individual animal. Biometrics. 48:201–216. 10.2307/2532750 [DOI] [PubMed] [Google Scholar]
- Chihara, N., Madi A., Kondo T., Zhang H., Acharya N., Singer M., Nyman J., Marjanovic N.D., Kowalczyk M.S., Wang C., et al. 2018. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 558:454–459. 10.1038/s41586-018-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.V., and Buchbinder E.I.. 2019. The Evolution of Adjuvant Therapy for Melanoma. Curr. Oncol. Rep. 21:106. 10.1007/s11912-019-0858-3 [DOI] [PubMed] [Google Scholar]
- Delaney, C., Schnell A., Cammarata L.V., Yao-Smith A., Regev A., Kuchroo V.K., and Singer M.. 2019. Combinatorial prediction of gene-marker panels from single-cell transcriptomic data. bioRxiv. (Preprint posted September 24, 2019). 10.1101/655753 [DOI] [PMC free article] [PubMed]
- Eisenhauer, E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. 2009. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer. 45:228–247. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- Fairfax, B.P., Taylor C.A., Watson R.A., Nassiri I., Danielli S., Fang H., Mahé E.A., Cooper R., Woodcock V., Traill Z., et al. 2020. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 26:193–199. 10.1038/s41591-019-0734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenwald, J.E., Scolyer R.A., Hess K.R., Sondak V.K., Long G.V., Ross M.I., Lazar A.J., Faries M.B., Kirkwood J.M., McArthur G.A., et al. 2017. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67:472–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim, H.E., Fan Y., Moustaki A., Abdelsamed H.A., Dash P., Dogra P., Carter R., Awad W., Neale G., Thomas P.G., and Youngblood B.. 2017. De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation. Cell. 170:142–157.e19. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24:978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- Jadhav, R.R., Im S.J., Hu B., Hashimoto M., Li P., Lin J.-X., Leonard W.J., Greenleaf W.J., Ahmed R., and Goronzy J.J.. 2019. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. USA. 116:14113–14118. 10.1073/pnas.1903520116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaora, S., Wolf Y., Feferman T., Barnea E., Greenstein E., Reshef D., Tirosh I., Reuben A., Patkar S., Levy R., et al. 2018. Combined Analysis of Antigen Presentation and T-cell Recognition Reveals Restricted Immune Responses in Melanoma. Cancer Discov. 8:1366–1375. 10.1158/2159-8290.CD-17-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicznik, M.M., Morawski P.A., Höllbacher B., Varkhande S.R., Motley S.J., Kuri-Cervantes L., Goodwin E., Rosenblum M.D., Long S.A., Brachtl G., et al. 2019. Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 4:eaav8995. 10.1126/sciimmunol.aav8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, H., Starenki D., Cooper S.J., Myers R.M., and Li Q.. 2018. powerTCR: A model-based approach to comparative analysis of the clone size distribution of the T cell receptor repertoire. PLOS Comput. Biol. 14:e1006571. 10.1371/journal.pcbi.1006571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.-H., Lerner H., et al. 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laydon, D.J., Melamed A., Sim A., Gillet N.A., Sim K., Darko S., Kroll J.S., Douek D.C., Price D.A., Bangham C.R.M., and Asquith B.. 2014. Quantification of HTLV-1 clonality and TCR diversity. PLOS Comput. Biol. 10:e1003646. 10.1371/journal.pcbi.1003646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J.B.A.G., Blank C.U., et al. 2019. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 176:775–789.e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson, A.M., Kiner E., Ergun A., Park J.S., Asinovski N., Ortiz-Lopez A., Kilcoyne A., Paoluzzi-Tomada E., Weissleder R., Mathis D., and Benoist C.. 2018. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc. Natl. Acad. Sci. USA. 115:E10672–E10681. 10.1073/pnas.1810580115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis, D., Lechner F., Pircher H., and Zinkernagel R.M.. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 362:758–761. 10.1038/362758a0 [DOI] [PubMed] [Google Scholar]
- Pauken, K.E., Sammons M.A., Odorizzi P.M., Manne S., Godec J., Khan O., Drake A.M., Chen Z., Sen D.R., Kurachi M., et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 354:1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken, K.E., Shahid O., Lagattuta K.A., Mahuron K.M., Luber J.M., Lowe M.M., Huang L., Delaney C., Long J.M., Fung M.E., et al. 2021. Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J. Exp. Med. 10.1084/jem.20200920 [DOI] [PMC free article] [PubMed]
- Ribas, A., and Wolchok J.D.. 2018. Cancer immunotherapy using checkpoint blockade. Science. 359:1350–1355. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman, M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2018. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 175:998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija Lab . 2021. Seurat: R toolkit for single cell genomics. https://satijalab.org/seurat/
- Schietinger, A., Arina A., Liu R.B., Wells S., Huang J., Engels B., Bindokas V., Bartkowiak T., Lee D., Herrmann A., et al. 2013. Longitudinal confocal microscopy imaging of solid tumor destruction following adoptive T cell transfer. OncoImmunology. 2:e26677. 10.4161/onci.26677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. 2021. COMET: Combinatorial Marker Detection from Single-Cell Transcriptomic Data. http://www.cometsc.com/comet
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leun, A.M., Thommen D.S., and Schumacher T.N.. 2020. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 20:218–232. 10.1038/s41568-019-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Perry C.J., Meeth K., Thakral D., Damsky W., Micevic G., Kaech S., Blenman K., and Bosenberg M.. 2017. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30:428–435. 10.1111/pcmr.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, D., Sopper S., Pircher A., Gastl G., and Wolf A.M.. 2015. Treg(s) in Cancer: Friends or Foe? J. Cell. Physiol. 230:2598–2605. 10.1002/jcp.25016 [DOI] [PubMed] [Google Scholar]
- Wu, T.D., Madireddi S., de Almeida P.E., Banchereau R., Chen Y.-J.J., Chitre A.S., Chiang E.Y., Iftikhar H., O’Gorman W.E., Au-Yeung A., et al. 2020. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 579:274–278. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- Yale Center for Genome Analysis . 2021. 10X Genomics Single Cell RNA-Seq. https://medicine.yale.edu/keck/ycga/sequencing/10x/singcellsequencing/
- Yost, K.E., Satpathy A.T., Wells D.K., Qi Y., Wang C., Kageyama R., McNamara K.L., Granja J.M., Sarin K.Y., Brown R.A., et al. 2019. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25:1251–1259. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang, F., Wei K., Slowikowski K., Fonseka C.Y., Rao D.A., Kelly S., Goodman S.M., Tabechian D., Hughes L.B., Salomon-Escoto K., et al. Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium . 2019. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20:928–942. 10.1038/s41590-019-0378-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.-K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents demographics and clinical features of patients included in the study.
lists the top 10 marker genes for each cluster, with percentage of positive cells in the cluster of interest, delta percentage of positive cells between that cluster and the rest of the dataset, and adjusted P value.
lists gene marker pairs of circulating TILs found in at least five out of seven patients, with rank and q value for each patient.
shows the expanded TIL gene sets and tumor-specific gene sets for each patient.
lists correlation coefficients between genes and expansion score, filtered for −logP ≥ 100, and the same for sharing score.
presents amino acid and nucleotide sequences of the TCRαβ CDR3 regions of clonotypes displayed in Fig. 4.