Abstract
Objectives:
The study aimed at detecting the prevailing hepatitis B virus (HBV) genotypes and the presence of clinically relevant mutations in the precore/core gene of the HBV DNA, among patients with chronic infection in South-eastern, Nigeria.
Methods:
A total of 72 participants with chronic HBV infection were enrolled into the study. Plasma samples from those with detectable HBV DNA were subjected to nested Polymerase Chain Reaction amplification using the precore/core specific primers. This resulted to the successful amplification and sequencing of the HBV precore/core region DNA from 16 participants. Mutation analysis on the precore/core region detected the presence of certain HBV precore/core gene mutations. Genotyping was carried out by phylogenetic analysis.
Results:
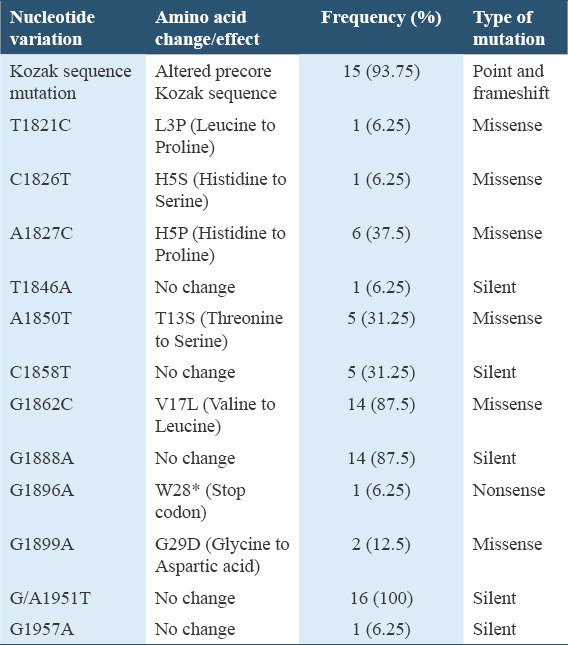
The precore region mutation at nucleotide position 1896, which is a G to A change resulting to a nonsense mutation, was detected in 6.25% of the participants. Other HBV precore region mutations that were detected include: G1899A, T1846A, G1862C, G1888A, T1821C, C1826T, A1827C, A1850T, C1858T, precore start codon Kozak sequence mutations and some novel core region mutations such as G/A1951T and G1957A. Genotyping revealed the existence of HBV genotype/subgenotype A1 (87.5%) and D (12.5%) among the participants. There was no significant difference in the occurrence of specific precore/core mutations among the HBV/hepatitis C virus dually infected and HBV mono-infected participants.
Conclusion:
The data suggest the likelihood of a more severe outcome of hepatitis caused by HBV in South-eastern Nigeria due to the occurrence of a variety of precore/core mutation, which resulted to HBeAg-negative chronic HBV infection among the participants.
Keywords: Hepatitis B Virus, mutation, precore/core gene
Introduction
Hepatitis B virus (HBV) infection is a major cause of acute and chronic liver disease and affects approximately two billion people worldwide. More than 240 million people worldwide are chronic carriers and may develop serious complications in the future.[1] In 2015, the World Health Organization (WHO) estimated that HBV infection resulted in about 887,000 deaths due to cirrhosis and hepatocellular carcinoma (HCC). The prevalence of HBV infection is highest in the WHO Western Pacific Region and the WHO African Region, where 6.2% and 6.1% of the adult population are infected respectively.[2] In Nigeria, HBV is hyperendemic with the seroprevalence of hepatitis B surface antigen (HBsAg) estimated to range from 10% to 40%.[3-8]
The therapeutic response and pathogenicity of different HBV genotypes vary. According to the overall nucleotide sequence variations of the entire genome, HBV is classified into at least ten different genotypes (A-J) differing by at least 8% of the DNA sequence. These genotypes are furthermore divided into different subgenotypes that differ by at least 4%. Genotype and subgenotype distributions vary with geographic location. Genotype E occurs in West and Central Africa.[9,10]
HBV mutations have been found both in acute and chronic patients and in all the four HBV open reading frames (preS/S, polymerase, precore/core, and X). Mutations in the precore/core open reading frame mainly results to HBeAg negative hepatitis because this region controls the transcription of precore mRNA. The precore mRNA is used exclusively for expression of the precore protein, which is a precursor of secreted HBeAg.[11] The e antigen is an important target for antibody mediated and cell-mediated immune responses. Hence, the loss of e antigen production by precore mutants may help the virus to evade the host immune response. The occurrence of the precore mutation seems to be a strategy of viral selection secondary to the immunological pressure against HBV.[12] A variety of these mutations have been observed. They usually result by developing a stable Watson–Crick base pair match so as to stabilize a convoluted area of the HBV genome, known as the e encapsidation signal, which is very important for productive HBV replication. These mutations have a role in ensuring the perpetuation of viral replication by enhancing the stability of the secondary structure of the pregenomic RNA through the disruption of base pairing within the secondary structure.[11,13]
Resistance to drugs for HBV treatment as a result of HBV mutation has been observed.[14,15] Resistance to the immunomodulators, interferon (IFN) alpha-2b and pegylated IFN (PEG IFN) alpha-2a have been reported to be caused by precore mutations which appear to influence response to treatment.[16,17] The precore stop codon mutation (G1896A) is resistant to IFN treatment.[17,18] Viral mutations have also been reported to be associated with reactivation. The HBV precore G1896A mutation is known to be associated with HBV reactivation.[19-23] Fatal cases of HBV reactivation harboring this mutation have been reported.[24,25] The emergence of this mutant might predict a possible occurrence of HBV reactivation.
This study aimed at determining the HBV genotypes and frequency of occurrence of known precore/core region mutations in HBV DNA sequences from chronically infected patients in South-eastern, Nigeria.
Methods
The participants are HBsAg positive patients diagnosed with chronic HBV infection, who visited the Polymerase Chain Reaction (PCR) Laboratory in Nnamdi Azikiwe University Teaching Hospital, Nnewi, Anambra State, Nigeria. A total of 72 participants were enrolled into the study. They were males and females aged between 20 and 67 years.
The study is a cross-sectional study, which was carried out between January 2018 and June 2019. The participants were recruited by consecutive sampling method during their visit to the laboratory. The consenting participants who met the inclusion criteria were recruited.
Inclusion criteria
The inclusion criteria were a diagnosis of chronic HBV infection, which is defined as HBsAg positivity for more than 6 months. Hence, patients from the South-eastern part of Nigeria whose laboratory request forms indicated a diagnosis of chronic HBV infection and were treatment naïve, were enrolled into the study.
Exclusion criteria
The patients diagnosed with cirrhosis or HCC and pregnant women were excluded from the study. The participants that tested positive to human immunodeficiency virus (HIV I and II) antibodies were also excluded from the study.
Ethical statement
An ethical clearance was issued by the Health Research Ethics Committee of the Nnamdi Azikiwe University Teaching Hospital, Nnewi, Anambra State (Reference number: NAUTH/CS/66/VOL.10/33/2017/028) on January 12, 2018. Informed consent was taken from all individual participants.
Serology
Five milliliters of whole blood were collected from the participants in tubes containing potassium-ethylenediaminetetra-acetic acid and 2 ml was also collected in plain tubes. The plasma and serum were separated within 24 h. The samples were assayed for HIV I and II using Determine test kit (Alere, Belgium). The samples that tested positive were excluded from the study.
The plasma samples were tested for HBeAg, HBeAb, HBsAg, HBcAb, and HBsAb using a lateral flow chromatographic immunoassay (Combo Cassette HBV panel immunoassay manufactured by Lusys Laboratories Inc U.S.A) which has a 99.9% sensitivity and 99.75% specificity. The interpretation of test results was performed according to the manufacturer’s specification. Assay was also carried out for hepatitis C virus (HCV) antibodies using the SD Bioline immunochromatographic rapid test kit (Standard Diagnostics Inc., Korea). The serum samples were used to assay for hepatitis D virus (HDV) total antibody and serum alpha-fetoprotein using the enzyme linked immunosorbent assay technique.
Quantitative assay of HBV DNA
The HBV DNA quantification was done using the Cobas AmpliPrep/Cobas TaqMan system (Roche Molecular Systems, Inc., Branchburg, NJ, USA)
Viral DNA Isolation
The HBV DNA was extracted from the samples with detectable HBV DNA by the phenol-chloroform method. Briefly, 250 μl of plasma treated proteinase K (Qiagen, UK) was added to 750 μl of TRIzol reagent (Thermofisher, UK). The DNA was subsequently precipitated with 300 μl of absolute ethanol. The DNA pellets were washed twice with 0.1M sodium citrate in 10% ethanol and rinsed in 75% ethanol. The DNA pellets were air dried in a heating block at 37°C. Finally, 100 μl of DNAse/RNAse-free water was added to each sample to dissolve the DNA.
Amplification of the Precore/Core Gene
Nested PCR for the amplification of the HBV precore/core gene was performed on the samples. The ABI 2730 thermal cycler (Applied Biosystems, UK) was used for this purpose. For the first-stage PCR, 25 μl of reaction mixture, containing 2 μl of the DNA sample, 2.5 μl of 10× PCR buffer, 1.25 μl of 5 mM MgCl2, 2 μl of 2.5mM dNTPs, 1 μl of each outer primer (10 μM), 0.2 μl of DNA Taq polymerase, and 15.05 μl of DNAse/RNase-free water, was amplified for 35 cycles (95°C for 60 s, 55°C for 30 s, 72°C for 60 s) with a final extension step at 72°C for 10 min. After the first round amplification, 2 μl of the PCR product was re-amplified for another 35 cycles with 1 μl of each inner primer and under the same condition as the first round PCR. The outer sense primer was 5’ -CTGGGAGGAGTTGGGGGA-3’, nucleotide positions 1770–1787; the outer antisense primer was 5’ - CAATGCTCAGGAGACTCTAA-3’, nucleotide positions 2476–2495; the inner sense primer was 5’ -GGTCTTTGTACTCGGAGGCT-3’, nucleotide positions 1788–1808; and the inner antisense primer was 5’ -GTCAGAAGGCAAAAAAGAGA-3’, nucleotide positions 2467–2486.[26]
The PCR products were electrophoresed on 1.5% agarose gel. To prevent cross-contamination, DNA extraction and PCR reaction mixture preparation were done in separate rooms from that in which the amplified samples were handled. The WHO international standard for HBV DNA containing 103 IU/ml which was obtained from the National Institute for Biological standards and controls (Hertfordshire, UK) was used as HBV DNA positive plasma sample for the optimization of the method. It was also used as a positive control for the assay. The negative control used for the PCR assay was DNAse/RNAse-free water.
HBV DNA sequencing, mutation analysis, and phylogenetic analysis
The PCR products were purified and DNA sequencing was performed using the inner primers, in the forward and reverse directions. This was done using the Applied Biosystems genetic automated sequencer (ABI Prism 3130X1 Forster City, CA 94404, USA) with the BigDye Terminator Cycle sequencing kit v1.1 (Applied-Biosystems). The obtained HBV precore/core gene sequences were subjected to the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) search for initial identification of the HBV so as to ensure that the sequences obtained matched only HBV sequences (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?PAGE=Nucleotides). GenBank sequences showing the highest matching scores were retrieved and retained for multiple sequence alignment. HBV genotypes were determined by phylogenetic analysis using the sequences with the highest BLAST hit from the NCBI website and the published HBV sequences available from the genotyping reference set on the NCBI website (http://www.ncbi.nlm.nih.gov/projects/genotyping/view.cgi?db=2) and a few other sequences of genotypes not represented in the reference set. The sequences and all the reference sequences were used for multiple sequence alignment using the MUSCLE program[27] implemented in the Molecular Evolutionary Genetics Analysis X software[28] which was also used to perform the phylogenetic analysis. A phylogenetic tree was constructed by the neighbor-joining method[29] and evolutionary distances were determined using the Kimura 2-parameter method.[30] Bootstrap resampling and reconstruction were carried out 1000 times to confirm the reliability of the phylogenetic tree.[31] GenBank accession numbers of all reference sequences, followed by the country of origin of the strains, as well as the corresponding genotype/subgenotype, are indicated. The sequences from this study are indicated by a black-colored triangular shape. Bootstrap values of 70 and above are shown along the nodes. The sequences appearing on the same node with the reference sequences were interpreted to be from the same genotype/subgenotype. For mutation analysis, the HBV precore/core gene sequences were re-aligned according to their genotypes using their genotype specific reference sequences and the nucleotide variations with the reference sequences were noted. The sequences were subsequently translated into amino acid residues and the amino acid variations with the reference sequences were also noted.
A total of 32 reference sequences of HBV genotypes A to J isolates were obtained from the NCBI database and were used for comparison with the sequences of the isolates in this study for the purpose of phylogenetic and mutational analysis. The accession numbers of the sequences are as follows: Genotype A: A1-KT327902, AF090842, KY810139; A2-X51970; A3-AM180624; A4-AY934764; A5-FJ692554; A6-GQ331047; genotype B: AB602818, AB073846, D00329; genotype C: AB014381, X04615; genotype D: X85254, X65259, M32138, AB674414, AF043593, X72702; genotype E: AB032431, X75657; genotype F: X69798, AF223965, AB036910; genotype G: AB064310, AF405706, AF160501; genotype H: AY090460, AY090457, AY090454; genotype I: AB231908; and genotype J: AB486012.
Data were stored using Microsoft excel and statistical analysis was carried out using Stata statistical package, version 16 (Stata Statistical Software, Stata Corporation, College Station, TX, USA). The categorical variables were compared using Pearson’s Chi-square or Fisher’s exact test while the median (Interquartile Range) of continuous variables were compared by Mann–Whitney U-test. A P < 0.05 was considered as statistically significant.
Results
All the participants were positive for HBsAg and HBcAb. HBV DNA was detected in a total of 45 samples by the Cobas AmpliPrep/Cobas TaqMan system. The complete HBV precore region and the partial core region were successfully amplified by nested PCR as a 500bp product in 16 participants and were sequenced. The agarose gel electrophoresis result showing some amplified samples is shown in Figure 1. The phylogenetic tree obtained from multiple sequence alignment with reference sequences is shown in Figure 2. A total of 14 (87.5%) sequences clustered around the subgenotype A1 reference sequences while 2 (12.5%) sequences clustered around the genotype D reference sequences.
Figure 1.

Agarose gel electrophoresis result of the precore/core region. Lane A=1000 bp DNA ladder, lane B= 100bp DNA ladder, lane 1= Hepatitis B virus (HBV) positive control sample, lane 2= HBV negative control, lanes 5, 8, 13, 14, 15, and 19 are the amplified DNA samples. Lanes 3, 4, 6, 7, 9, 10, 11, 12, 16, 17, 18, 20, 21, 22, 23, and 24 are DNA samples that were not successfully amplified.
Figure 2.
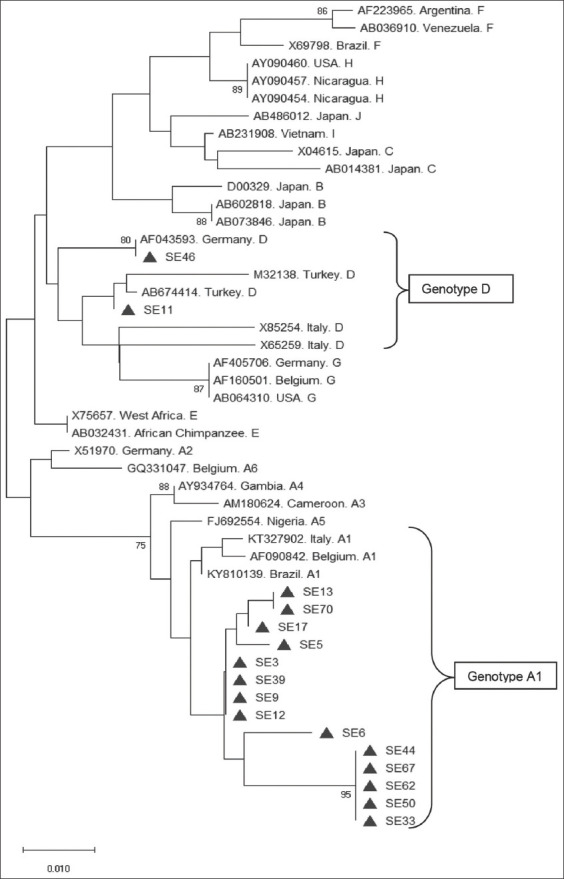
Phylogenetic tree showing the 16 hepatitis B virus precore/core gene sequences obtained in this study and the reference sequences
Seven of the HBV DNA precore/core gene sequences obtained in this study were submitted to the NCBI GenBank database and were assigned the accession numbers; MT893925 to MT893931.
For mutation analysis, a cross-section of the multiple sequence alignment with reference sequences of genotype A and genotype D is shown in Figure 3. The translated amino acid sequences can be seen in Figure 4. The HBeAg was negative in all the participants whose samples were successfully amplified and sequenced. The frequency of occurrence of the HBV precore/core gene mutations is shown in Table 1.
Figure 3.
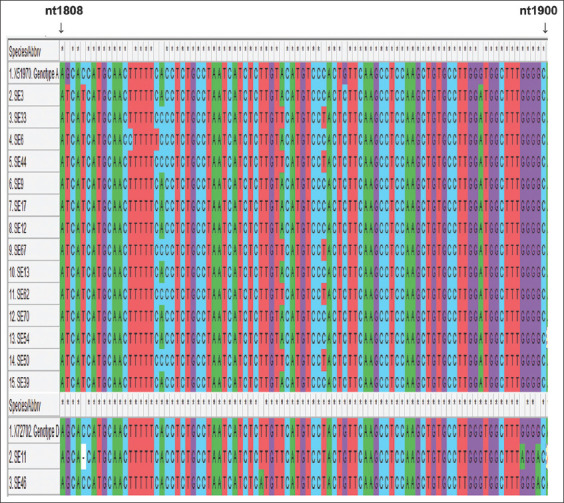
Multiple sequence alignment of 16 hepatitis B virus DNA sequences and genotype specific wild-type references showing the Kozak sequence in the basal core promoter (nt1809 to nt1813) and the complete precore gene (nt1814 to nt1900). The asterisks symbol represents areas of nucleotide similarity while its absence signifies nucleotide variation
Figure 4.
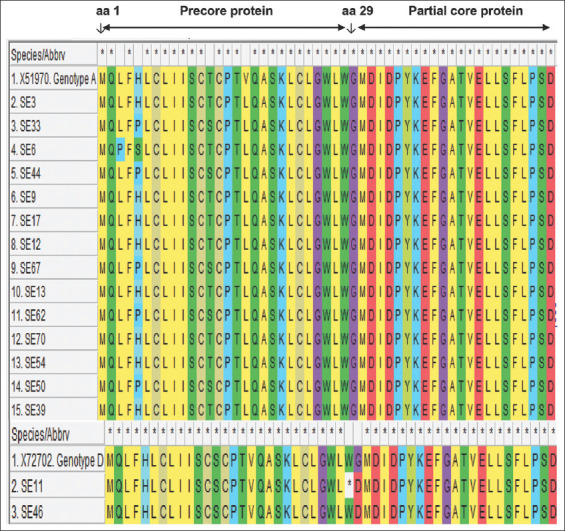
Translated protein sequences of the complete precore gene (amino acid 1 to 29) and the partial core gene (amino acid 1 to 21). The precore translational stop codon mutation at codon 28 observed in SE11 is designated by “*”
Table 1.
Frequency of occurrence and types of HBV precore/core gene mutation
An assessment of the distribution of HBV precore/core region specific mutations among the HBV genotypes in Table 2 showed that there exists a significant statistical difference between the occurrence of G1862C, G1888A, and G1899A specific mutations among the HBV subgenotype A1 and genotype D infected participants (P < 0.05, Fisher’s exact test). There was no significant statistical difference in the occurrence of other specific mutations among the HBV subgenotype A1 and genotype D infected participants (P > 0.05, Fisher’s exact test). On comparing the HBV DNA and serum AFP levels among the genotypes, the median HBV DNA and AFP levels among participants infected with HBV genotype D were higher than that of genotype A and a trend toward statistical significance was observed as illustrated in the box plots shown in Figure 5.
Table 2.
Distribution of HBV precore/core region specific mutations among the HBV genotypes
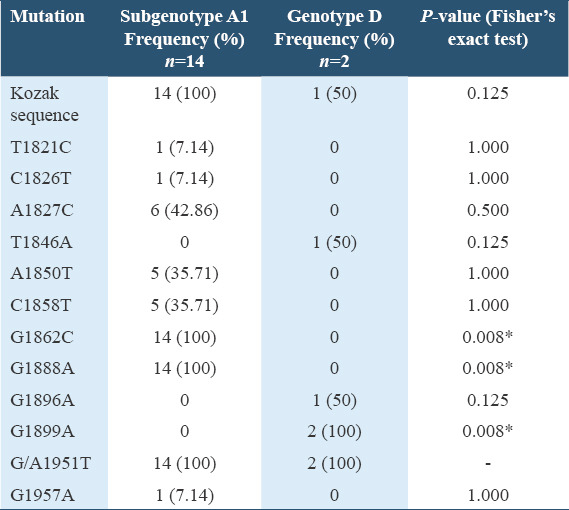
Figure 5.
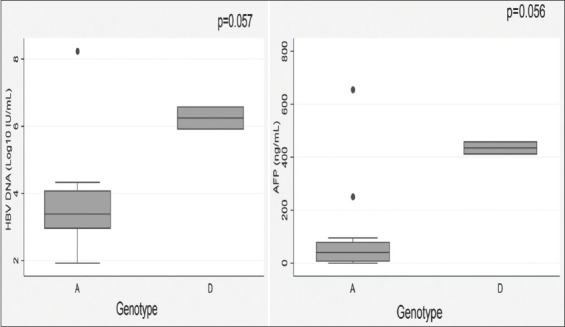
Graphical representation of the median and interquartile ranges of hepatitis B virus (HBV) DNA and serum AFP levels across HBV genotypes A and D
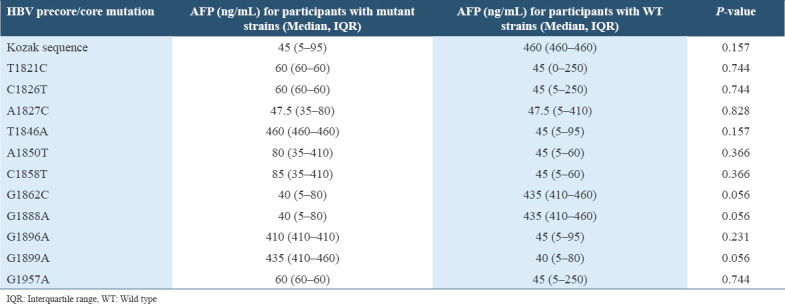
On assessing the levels of HBV DNA and serum AFP in participants with the specific HBV precore/core gene mutation and those with the wild type (WT) strains, the median viral load and AFP level was higher among the participants with certain HBV specific mutations than those with the WT strains and vice versa but the difference was not of statistical significance. However, a trend toward statistical significance was observed in the difference between the median of HBV DNA and AFP levels among the participants with the G1899A mutation (P = 0.057 and P = 0.056, respectively), where a higher median value was observed among those with the mutant strains for both HBV DNA and AFP levels [Tables 3 and 4].
Table 3.
The HBV DNA levels of the participants with specific HBV precore/core mutation compared with those with the WT strain
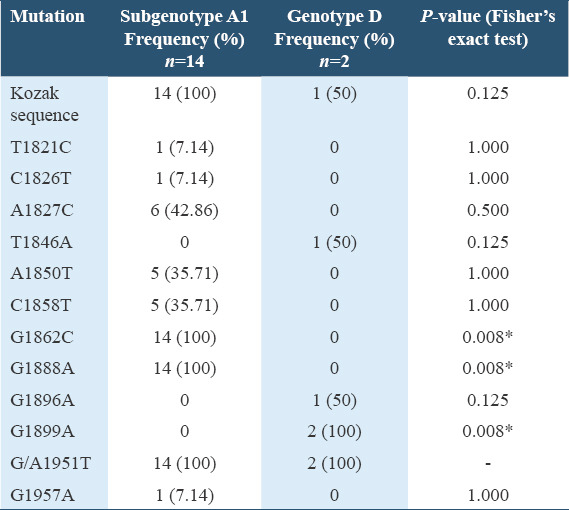
Table 4.
The serum AFP levels of the participants with specific HBV precore/core mutation compared with those with the WT strains
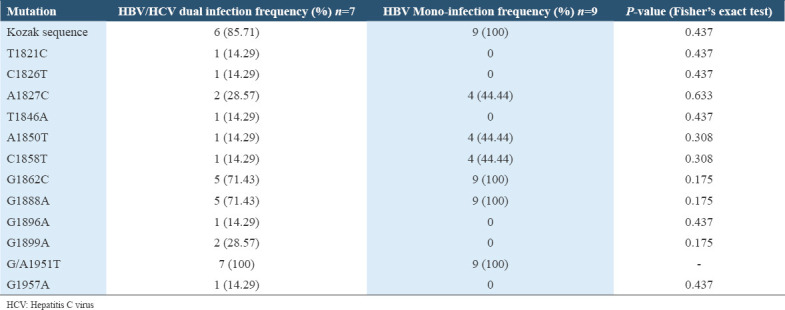
No case of HBV/HDV dual infection was observed among the participants whose HBV DNA was successfully sequenced. There was no statistically significant difference between the frequency of occurrence of HBV precore/core specific mutation among the HBV/HCV dually infected participants and among the HBV mono-infected participants (P > 0.05, Fisher’s exact test) as elucidated in Table 5.
Table 5.
The occurrence of HBV precore/core mutation among HBV/HCV dually infected participants
Discussion
Certain HBV precore/core region mutations reported in this study are indicators that patients who have chronic HBV infection may develop serious chronic illness, which could be a serious public health problem in Nigeria. These HBV mutations were present in all the successfully sequenced samples and may have caused the absence of HBeAg, which was observed in all of them. The mutations include the nonsense, missense, silent, and frameshift mutations occurring on the precore/core regions of the HBV DNA.
The G1896A mutation is a nonsense mutation occurring in the HBV precore region leading to a G-to-A shift, which causes a conversion of codon 28 from TGG (tryptophan) to a premature stop codon TAG, and subsequent termination of the expression of HBeAg.[32] This mutation has been reported to be significantly associated with the development of cirrhosis and HCC.[33] It was observed in this study at a frequency of 6.25% [Table 1], in a participant infected with HBV genotype D. The report from this study is in concordance with a report from India where the G1896A mutation was not detected among the patients infected with HBV subgenotype A1 but in the genotype D infected patients.[34] The G1896A mutation has previously been reported among HIV/HBV co-infected Nigerians in a study done in Jos, North-central, Nigeria as 57%.[35] It has also been reported as 34.1% in Iran,[36] 54.9% in France,[37] and 16.7% in India.[38] These reports are higher than the report from this study. The low prevalence of this mutation in this study is probably because of the different geographical location and the genotype distribution of the HBV sequences obtained in this study, as majority of the sequences were genotype A. The occurrence of this mutation is dependent upon the nucleotide (cytosine or thymine) at position 1858 because it forms a base pair with the nucleotide at position 1896 in the pregenomic RNA loop, at the ε encapsidation sign. The presence of a cytosine at position 1858 prevents the occurrence of G-to-A mutation at nucleotide position 1896, as it would destabilize the stem-loop structure of the RNA encapsidation signal.[39] The G1896A mutation occurs frequently in genotypes/subgenotypes which have 1858T, such as C1, D, E, and F[11,40] and was reported to be least frequently observed in genotype A.[40]
A missense mutation observed in this study is the G1899A mutation which occurred in 12.5% of the participants [Table 1]. This mutation caused a change from guanine to adenine at nucleotide 1899 which resulted to a glycine to aspartic acid amino acid substitution at residue 29 of the precore region. The G1899A precore mutation causing the amino acid variation G29D have also been reported to be significantly associated with the development of cirrhosis and HCC.[33,41,42] The report from this study is higher than the report from India which recorded a prevalence rate of 6.1%[38] for G1899A precore mutation. However, it is lower than the reports from North-central, Nigeria where a prevalence rate of 34% was observed[35] and France where a prevalence rate of 29.3% was reported.[37] These differences may be due to the different geographical regions.
Another missense mutation observed in this study is at position 1862 and it affects the bulge of the RNA encapsidation signal. It has been reported to occur most frequently in subgenotype A1 and may affect HBeAg expression at the post-translational level.[43-45] In this study, this mutation occurred in 87.5% of the participants [Table 1] and caused a nucleotide change from guanine to cytosine (G1862C), leading to a change in the translated amino acid from valine to leucine at position 17 of the precore region. This mutation has previously been reported in Zimbabwe among 44% of HBV isolates from blood donors infected with subgenotype A1.[46] The report from this study is in agreement with the assertion that this mutation occurs frequently in subgenotype A1 sequences, as the mutation occurred in all the subgenotype A1 sequences and did not occur in any of the genotype D sequences [Table 2]. However, in contrast, a study done in Cameroun reported the occurrence of the G1862C mutation in patients infected with HBV genotype/subgenotype E and A3.[47]
The G1888A HBV mutation is a silent mutation of clinical relevance which was detected in 87.5% of the participants [Table 1]. This is higher than the report from a previous study where it was detected in 66% of sequenced strains from Rwanda.[48] This may be due to the fact that the majority of the participants in this study were infected with subgenotype A1 HBV strain and this specific mutant has been reported to occur specifically among the HBV subgenotype A1 strains.[11] It was interesting to note that this mutation occurred in all the subgenotype A1 sequences obtained in this study but none occurred in the genotype D sequences [Table 2].
The Kozak sequence which is located upstream from the precore translational start codon (1809–1813) was observed to be different in some of the sequences obtained in this study. It was not the regular wild-type GCACC sequence, but TCATC, as a result of a double mutation (G1809T and C1812T) which occurred in all the participants infected with HBV subgenotype A1 [Table 2]. Mutations in the Kozak sequence, which interfere with the translation of HBeAg is typical of subgenotype A1 and do not occur in subgenotype A2.[40,49] It was interesting to note that this peculiar double mutation was observed in this study only among the subgenotype A1 sequences. This may be the reason for its high occurrence in this study as subgenotype A1 sequences were observed to be predominant. However, this did not corroborate a report from Ethiopia which recorded this mutation among patients infected with HBV genotypes A, D, and E.[50] Another Kozak sequence mutation observed in this study was a deletion of cytosine (C) at nucleotide 1812 (frameshift mutation) which occurred in one of the genotype D sequences. This novel mutation may hamper the translation of HBeAg.
Mutations occurring at nucleotide positions 1821–1828, the region important for pregenomic RNA initiation and synthesis,[51] which have not been widely reported such as T1821C, C1826T, and A1827C mutations were observed in this study. Some novel mutations observed in this study which may also be of clinical importance include the G/A1951T, G1957A, and nucleotide 1812 deletion of cytosine (C). Frameshift mutations in the precore region have been reported to result to enhanced HBV replication and might be linked with the development of fulminant hepatitis.[52]
Some notable mutations of the HBV precore/core region which have been described elsewhere were not observed in this study. The precore start codon was conserved in all the sequenced strains obtained in this study. However, variations of the precore start codon have rarely been described but have been reported among a few genotype A1 strains isolated from South Africa and also from some untyped strains from Japan and Europe.[53,54] Furthermore, the core gene mutations previously reported to be associated with liver disease progression in chronic patients and particularly in HCC patients such as C1913A/G,[55] C1914G,[41] and G1915T[56] were not identified in any of the sequences from this study.
Data on the prevalence of HBV genotypes in South-eastern, Nigeria, are scarce. The available data on the prevalence of HBV genotypes in other parts of Nigeria reported that genotype E is the predominant HBV genotype.[7,35,57-59] HBV genotype E has also been reported to be predominant in Sub-Saharan Africa in general.[60,61] Interestingly, this was not the case with the present study conducted in the South-eastern part of the country as genotype E was not detected. The predominant HBV genotype/subgenotype detected in this study is subgenotype A1, which occurred in 14/16 sequences as shown in Figure 2. The HBV subgenotype A1 has been reported to circulate in South Africa, Malawi, Somalia, Uganda, Tanzania, Yemen, Nepal, Philippines, India, and Brazil.[49] It was proposed that genotype A originated from Africa and subgenotype A1 was reported to be prevalent in sub-Saharan Africa.[62] However, there is paucity of data available on the occurrence of HBV subgenotype A1 in Nigeria. The detection of genotype A in this study agrees with the reports from two different studies carried out among subjects from South-West, Nigeria where HBV genotype A was detected in 15/18 subjects[63] and in 8/49 subjects.[64] The detection of HBV genotype D (2/16) in this study is in agreement with a report about the genotypic distribution of HBV in HBV/HIV infected subjects in North-central, Nigeria where genotype D was detected in one of the subjects.[35] HBV genotype D has also been reported in a recent study in the northern part of the country where HBV genotypes A, B, C, D, and E were detected through multiplex PCR.[65]
Genotypes have a significant influence on the HBV DNA levels of chronic HBV carriers.[66,67] The features of subgenotype A1 which occur as a result of the presence of mutations include: Lower HBV DNA levels, rapid disease progression, and high rate of developing HCC, even in the absence of cirrhosis.[11,34,43,68] It was evident in this study when comparing between genotypes A and D infected participants in Figure 5 that a lower median HBV DNA level was observed among those infected with HBV subgenotype A1 than those infected with HBV genotype D. It was also observed in Figure 5 that the median serum AFP level which is the most widely used biomarker for HCC[69] was higher in genotype D than subgenotype A1 infected participants, and the difference indicated a trend toward statistical significance (P = 0.056). However, this finding may need to be verified with a larger sample size, as it has been previously reported that individuals infected with HBV subgenotype A1, developed HCC 6.5 years earlier than individuals infected with other genotypes/subgenotypes.[49]
The occurrence of some HBV precore/core mutations has been reported to result to an increase in HBV DNA load. High HBV DNA loads are associated with the progression of chronic liver disease and an increased risk of HCC.[70] Furthermore, high serum AFP level in adults generally indicates a high possibility of HCC in patients with chronic hepatitis or cirrhosis.[71] In this study, as elucidated in Tables 3 and 4, the median HBV DNA and serum AFP levels were higher in participants infected with HBV strains which have some specific precore/core gene mutations, when compared with those of the participants infected with the WT strains. Although there were no statistically significant differences in the median HBV DNA load or serum AFP levels between the participants with the different specific mutations and those with the WT strains, the occurrence of G1899A mutation was marked with a higher level of HBV DNA and serum AFP, and a trend toward statistical significance for both parameters (P = 0.057 and P = 0.056, respectively) was evident. The case was different with the G1888A and G1862C mutations, in which lower HBV DNA and serum AFP levels were observed in participants infected with the mutant strains than the WT strains, and the differences also showed a trend toward statistical significance. This may be explained by the fact that these two mutations occurred specifically in HBV subgenotype A1 sequences which have been reported to have lower HBV DNA levels and high rate of developing HCC even in the absence of cirrhosis, as a result of the presence of mutations.[34,43,68,72] Low level HBV DNA load is not always an indication of improved conditions and in some patients it indicates advanced disease.[73,74]
Precore/core mutations have been reported to occur less frequently in HBV/HCV co-infected than in HBV mono-infected patients because of the inhibitory effect of HCV on HBV.[75,76] This was observed in most cases in this study as the frequency of occurrence of most specific precore/core mutants were higher among the HBV mono-infected participants than the HBV/HCV dually infected participants but the differences were not statistically significant [Table 5].
The limitation of this study is its inability to successfully amplify all the DNA samples, most likely caused by several genetic variations that resulted to a mismatch in the PCR assay. There is need for future studies to explore other optimized techniques that will increase the rate of amplification of HBV precore/core gene within the study area.
Conclusion
This study is the first of its kind to report the occurrence of HBV precore/core gene mutations in South-eastern, Nigeria where HBV is highly endemic. This spectrum of mutation suggests the likelihood of a more severe outcome of chronic HBV infection within the study area. The detection of HBV subgenotype A1 and genotype D in this study suggests the predominance of HBV genotypes other than E in the South-eastern part of the country. HBV subgenotype A1, which was the most common genotype detected in this study, is known to result to severe consequences such as rapid disease progression and a high rate of developing HCC. Consequently, determining HBV genotypes and mutations in the precore/core gene will provide valuable information for the management of patients with chronic HBV infection within the region.
Authors’ Declaration Statements
Ethical statement
The study was approved by the Health Research Ethics Committee of the Nnamdi Azikiwe University Teaching Hospital, Nnewi, Anambra State (Reference number: NAUTH/CS/66/VOL.10/33/2017/028) on January 12, 2018 and informed consent was taken from all individual participants.
Availability of Data and Material
Data will be made available by the corresponding author upon request. Seven of the HBV DNA precore/core gene sequences obtained in this study were deposited to the NCBI database and were assigned the following accession numbers; MT893925, MT893926, MT893927, MT893928, MT893929, MT893930, and MT893931.
Competing Interests
None declared.
Funding Statement
No funding was available for this study.
Authors’ Contribution
Conception and design: Chinenye Mbamalu, Ifeoma Ekejindu, and Ifeoma Enweani; Collection of data and laboratory analysis: Chinenye Mbamalu, David Igwe, Stephen Kalu, and Gloria Akaeze; Data analysis and interpretation: Chinenye Mbamalu, Ifeoma Ekejindu, Ifeoma Enweani, and David Igwe; Manuscript writing: All authors participated; Final approval of manuscript: All authors participated.
Acknowledgments
None.
ORCID link of the corresponding author: 0000-0001-8521-8321
References
- 1.Caligiuri P, Cerruti R, Icardi G, Bruzzone B. Overview of hepatitis B virus mutations and their implications in the management of infection. World J Gastroenterol. 2016;22:145–54. doi: 10.3748/wjg.v22.i1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Hepatitis B Fact Sheets. Geneva: World Health Organization; 2019. [Last accessed on 2020 Jun 30]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b . [Google Scholar]
- 3.Olayinka AT, Oyemakinde A, Balogun MS, Ajudua A, Nguku P, Aderinola M, et al. Seroprevalence of hepatitis B infection in Nigeria:A national survey. Am J Trop Med Hyg. 2016;95:902–7. doi: 10.4269/ajtmh.15-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musa B, Bussell S, Borodo MM, Samaila AA, Femi OL. Prevalence of hepatitis B virus infection in Nigeria, 2000-2013:A systematic review and meta-analysis. Niger J Clin Pract. 2015;18:163–72. doi: 10.4103/1119-3077.151035. [DOI] [PubMed] [Google Scholar]
- 5.Fasola FA, Kotila TR, Akinyemi JO. Trends in transfusion-transmitted viral infections from 2001 to 2006 in Ibadan, Nigeria. Intervirology. 2008;51:427–31. doi: 10.1159/000209671. [DOI] [PubMed] [Google Scholar]
- 6.Forbi JC, Onyemauwa N, Gyar SD, Oyeleye AO, Entonu P, Agwale SM. High prevalence of hepatitis B virus among female sex workers in Nigeria. Rev Inst Med Trop SP. 2008;50:219–21. doi: 10.1590/s0036-46652008000400006. [DOI] [PubMed] [Google Scholar]
- 7.Odemuyiwa SO, Mulders MN, Oyedele OI, Ola SO, Odaibo GN, Olaleye DO, et al. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J Med Virol. 2001;65:463–9. [PubMed] [Google Scholar]
- 8.Olubuyide IO, Ola SO, Aliyu B, Dosumu OO. Prevalence and epidemiological characteristics of Hepatitis B and C infections among doctors and dentists in Nigeria. East Afr Med J. 1997;74:357–61. [PubMed] [Google Scholar]
- 9.Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–50. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 10.Shi W, Zhang Z, Ling C, Zheng W, Zhu C, Carr MJ, et al. Hepatitis B virus subgenotyping:History, effects of recombination, misclassifications, and corrections. Infect Genet Evol. 2013;16:355–61. doi: 10.1016/j.meegid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Kramvis A. Molecular characterization of the genotypes and mutants of hepatitis B virus from South Africa. South Afr J Epidermiol Infect. 2008;23:29–1. [Google Scholar]
- 12.Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen-negative chronic hepatitis B. J Hepatol. 2001;34:617–24. doi: 10.1053/jhep.2001.27834. [DOI] [PubMed] [Google Scholar]
- 13.Jong-Keun J, Gye-Soon Y, Wang-Shick R. Evidence that the 5'-End cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J Virol. 2000;74:5502–8. doi: 10.1128/jvi.74.12.5502-5508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, et al. Ultra-deep pyrosequencing of hepatitis b virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J Infect Dis. 2009;199:1275–85. doi: 10.1086/597808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoulim F, Fournier C, Marion-Audibert A. Hepatitis B virus drug resistance and combination therapy. Hepatology Rev. 2007;4:34–47. [Google Scholar]
- 16.Keeffe EB, Dieterich DT, Han SH, Jacobson IM, Martin P, Schiff ER, et al. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States:an update. Clin Gastroenterol Hepatol. 2006;4:936–62. doi: 10.1016/j.cgh.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Wei L, Jiang D, Cong X, Fei R, Xiao J, et al. In vitro resistance to interferon of hepatitis B virus with precore mutation. World J Gastroenterol. 2005;11:649–55. doi: 10.3748/wjg.v11.i5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonneveld MJ, Rijckborst V, Zwang L, Zeuzem S, Heathcote EJ, Simon K, et al. Hepatitis B e antigen levels and response to peginterferon:Influence of precore and basal core promoter mutants. Antivir Res. 2013;97:312–7. doi: 10.1016/j.antiviral.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Steinberg JL, Yeo W, Zhong S, Chan JY, Tam JS, Chan PK, et al. Hepatitis B virus reactivation in patients undergoing cytotoxic chemotherapy for solid tumours:precore/core mutations may play an important role. J Med Virol. 2000;60:249–55. [PubMed] [Google Scholar]
- 20.Yeo W, Zhong S, Chan PK, Ho WM, Wong HT, Chan AS, et al. Sequence variations of precore/core and precore promoter regions of hepatitis B virus in patients with or without viral reactivation during cytotoxic chemotherapy. J Viral Hepat. 2000;7:448–58. doi: 10.1046/j.1365-2893.2000.00257.x. [DOI] [PubMed] [Google Scholar]
- 21.Dai MS, Lu JJ, Chen YC, Perng CL, Chao TY. Reactivation of precore mutant hepatitis B virus in chemotherapy-treated patients. Cancer. 2001;92:2927–32. doi: 10.1002/1097-0142(20011201)92:11<2927::aid-cncr10109>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 22.Chen PM, Yao NS, Wu CM, Yang MH, Lin YC, Hsiao LT, et al. Detection of reactivation and genetic mutations of the hepatitis B virus in patients with chronic hepatitis B infections receiving hematopoietic stem cell transplantation. Transplantation. 2002;74:182–8. doi: 10.1097/00007890-200207270-00007. [DOI] [PubMed] [Google Scholar]
- 23.Alexopoulou A, Theodorou M, Dourakis SP, Karayiannis P, Sagkana E, Papanikolopoulos K, et al. Hepatitis B virus reactivation in patients receiving chemotherapy for malignancies:Role of precore stop-codon and basic core promoter mutations. J Viral Hepat. 2006;13:591–6. doi: 10.1111/j.1365-2893.2006.00728.x. [DOI] [PubMed] [Google Scholar]
- 24.Sugauchi F, Tanaka Y, Kusumoto S, Matsuura K, Sugiyama M, Kurbanov F, et al. Virological and clinical characteristics on reactivation of occult hepatitis B in patients with hematological malignancy. J Med Virol. 2011;83:412–8. doi: 10.1002/jmv.21995. [DOI] [PubMed] [Google Scholar]
- 25.Marusawa H, Imoto S, Ueda Y, Chiba T. Reactivation of latently infected hepatitis B virus in a leukemia patient with antibodies to hepatitis B core antigen. J Gastroenterol. 2001;36:633–6. doi: 10.1007/s005350170049. [DOI] [PubMed] [Google Scholar]
- 26.Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, et al. Hepatitis B virus BCP, precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol. 2010;82:1115–25. doi: 10.1002/jmv.21774. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE:Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1987;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Croagh CM, Desmond PV, Bell SJ. Genotypes and viral variants in chronic hepatitis B:A review of epidemiology and clinical relevance. World J Hepatol. 2015;7:289–03. doi: 10.4254/wjh.v7.i3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Qahtani AA, Al-Anazi MR, Nazir N, Abdo AA, Sanai FM, Al-Hamoudi WK, et al. The correlation between hepatitis B virus precore/core mutations and the progression of severe liver disease. Front Cell Infect Microbiol. 2018;8:355. doi: 10.3389/fcimb.2018.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopalakrishnan D, Keyter M, Shenoy KT, Leena KB, Thayumanavan L, Thomas V, et al. Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J Gastroenterol. 2013;19:9294–306. doi: 10.3748/wjg.v19.i48.9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant J, Agbaji O, Kramvis A, Yousif M, Auwal M, Penugonda S, et al. Hepatitis B virus sequencing and liver fibrosis evaluation in HIV/HBV co-infected Nigerians. Trop Med Int Health. 2017;22:744–54. doi: 10.1111/tmi.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kheirabad AK, Farshidfar G, Nasrollaheian S, Gouklani H. Prevalence and characteristics of precore mutation in Iran and its correlation with genotypes of Hepatitis B. Electron Phys. 2017;9:4114–23. doi: 10.19082/4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducancelle A, Pivert A, Bertrais S, Boursier J, Balan V, Veillon P, et al. Different precore/core mutations of hepatitis B interact with, limit, or favor liver fibrosis severity. J Gastroenterol Hepatol. 2016;31:1750–6. doi: 10.1111/jgh.13338. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharya H, Bhattacharya D, Nagarajan M, Reesu R, Roy S, Attayur PS. Prevalence of mutations in basal core promoter and precore region of hepatitis B virus in vaccinated and nonvaccinated individuals of the aboriginal nicobarese tribe of car nicobar Island, India. Intervirology. 2014;57:357–64. doi: 10.1159/000365756. [DOI] [PubMed] [Google Scholar]
- 39.Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trepo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant:Possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–10. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revill P, Yuen L, Walsh R, Perrault M, Locarnini S, Kramvis A. Bioinformatic analysis of the hepadnavirus e-antigen and its precursor identifies remarkable sequence conservation in all orthohepadnaviruses. J Med Virol. 2010;82:104–15. doi: 10.1002/jmv.21645. [DOI] [PubMed] [Google Scholar]
- 41.Malik A, Singhal DK, Albanyan A, Husain SA, Kar P. Hepatitis B virus gene mutations in liver diseases:A report from New Delhi. PLoS One. 2012;7:e39028. doi: 10.1371/journal.pone.0039028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng JX, Zeng Z, Zheng YY, Ying SJ, Zhang DY, Yu YY, et al. Role of hepatitis B virus base core and precore/core promoter mutations on hepatocellular carcinoma in untreated older genotype C Chinese patients. J Viral Hepat. 2011;18:423–31. doi: 10.1111/j.1365-2893.2011.01458.x. [DOI] [PubMed] [Google Scholar]
- 43.Sugauchi F, Kumada H, Acharya SA, Shrestha SM, Gamutan MT, Khan M, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol. 2004;85:811–20. doi: 10.1099/vir.0.79811-0. [DOI] [PubMed] [Google Scholar]
- 44.Inoue J, Ueno Y, Nagasaki F, Wakui Y, Kondo Y, Fukushima K, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis. Virol J. 2009;395:202–9. doi: 10.1016/j.virol.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 45.Bhoola NH, Kramvis A. Expression of wild-type or G1862T mutant HBe antigen of subgenotype A1 of hepatitis B virus and the unfolded protein response in Huh7 cells. J Gen Virol. 2017;98:1422–33. doi: 10.1099/jgv.0.000793. [DOI] [PubMed] [Google Scholar]
- 46.Gulube Z, Chirara M, Kew M, Tanaka Y, Mizokami M, Kramvi A. Molecular characterization of hepatitis B virus isolates from Zimbabwean blood donors. J Med Virol. 2011;83:235–44. doi: 10.1002/jmv.21954. [DOI] [PubMed] [Google Scholar]
- 47.Amougou MA, Marchio A, Bivigou-Mboumba B, Noah DN, Bana R, Atangana PJ, et al. Enrichment in selected genotypes, basal core and precore mutations of hepatitis B virus in patients with Hepatocellular carcinoma in Cameroun. J Viral Hepat. 2019;26:1086–93. doi: 10.1111/jvh.13131. [DOI] [PubMed] [Google Scholar]
- 48.Norder H, Twagirumugabe T, Said J, Tian Y, Tang KW, Lindh M. High frequency of either altered pre-core start codon or weakened Kozak sequence in the core promoter region in hepatitis B virus A1 strains from Rwanda. Genes. 2019;10:182. doi: 10.3390/genes10030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramvis A, Kew MC. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol Res. 2007;37:27–32. doi: 10.1111/j.1872-034X.2007.00100.x. [DOI] [PubMed] [Google Scholar]
- 50.Belyhun Y, Liebert UG, Maier M. Analysis of HBV basal core promoter/precore gene variability in patients with HBV drug resistance and HIV infection in Nortewst Ethiopia. PLoS One. 2018;13:e0191970. doi: 10.1371/journal.pone.0191970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen IH, Huang CJ, Ting LP. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647–57. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue J, Ueno Y, Wakui Y, Fukushima K, Kondo Y, Kakazu E, et al. Enhanced replication of hepatitis B virus with frameshift in the precore region found in fulminant hepatitis patients. J Infect Dis. 2011;204:1017–25. doi: 10.1093/infdis/jir485. [DOI] [PubMed] [Google Scholar]
- 53.Ochwoto M, Chauhan R, Gopalakrishnan D, Chen CY, Ng'ang'a Z, Okoth F, et al. Genotyping and molecular characterization of hepatitis B virus in liver disease patients in Kenya. Infect Genet Evol. 2013;20:103–10. doi: 10.1016/j.meegid.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Raimondo G, Schneider R, Stemler M, Smedile V, Rodino G, Will H. A new hepatitis B virus variant in a chronic carrier with multiple episodes of viral reactivation and acute hepatitis. Virol J. 1990;179:64–8. doi: 10.1016/0042-6822(90)90274-u. [DOI] [PubMed] [Google Scholar]
- 55.Zhang A, Wan Z, You S, Liu H, Zhu B, Chen J, et al. Association of hepatitis B virus mutations of A1846T and C1913A/g with acute-on-chronic liver failure development from different underlying chronic liver diseases. Hepat Mon. 2013;13:e12445. doi: 10.5812/hepatmon.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahyuni RM, Utsumi T, Juniastuti Yano Y, Murti IS, Amin M, et al. Analysis of hepatitis B virus genotype and gene mutation in patients with advanced liver disease in East Kalimantan, Indonesia. Biomed Rep. 2019;10:303–10. doi: 10.3892/br.2019.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forbi JC, Vaughan G, Purdy MA, Campo DS, Xia GL, Ganova-Raeva LM, et al. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS One. 2010;5:e11615. doi: 10.1371/journal.pone.0011615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faleye TO, Adewumi MO, Ifeorah IM, Omoruyi EC, Bakarey SA, Akere A, et al. Detection of hepatitis B virus isolates with mutations associated with immune escape mutants among pregnant women in Ibadan, Southwestern Nigeria. Springerplus. 2015;4:43. doi: 10.1186/s40064-015-0813-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oladeinde BH, Ekejindu IM, Omoregie R, Odia I, Aguh OD, Okwu UM. New strains of hepatitis B virus genotype E circulating in Nigeria. Int J Health Sci (Qassim) 2018;12:25–9. [PMC free article] [PubMed] [Google Scholar]
- 60.Forbi JC, Ben-Ayed Y, Xia GL, Vaughan G, Drobeniuc J, Switzer WM, et al. Disparate distribution of hepatitis B virus genotypes in four sub-Saharan African countries. J Clin Virol. 2013;58:59–66. doi: 10.1016/j.jcv.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Candotti D, Diarra B, Bisseye C, Tao I, Quang KP, Sanou M, et al. Molecular characterization of hepatitis B virus in blood donors from Burkina Faso:Prevalence of quasi-subgenotype A3, genotype E, and mixed infections. J Med Virol. 2016;88:2145–56. doi: 10.1002/jmv.24589. [DOI] [PubMed] [Google Scholar]
- 62.Hannoun C, Soderstrom A, Norkrans G, Lindh M. Phylogeny of African complete genomes reveals a West African genotype A subtype of hepatitis B virus and relatedness between Somali and Asian A1 sequences. J Gen Virol. 2005;86:2163–67. doi: 10.1099/vir.0.80972-0. [DOI] [PubMed] [Google Scholar]
- 63.Akintule OA, Olusola BA, Odaibo GN, Olaleye DO. Occult HBV Infection in Nigeria. Arch Bas App Med. 2018;6:87–3. [PMC free article] [PubMed] [Google Scholar]
- 64.Olinger CM, Venard V, Njayou M, Oyefolu AB, Maïga I, Kemp AJ, et al. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa:New subtypes, mixed infections and recombinations. J Gen Virol. 2006;87:1163–73. doi: 10.1099/vir.0.81614-0. [DOI] [PubMed] [Google Scholar]
- 65.Ahmad AE, Bakari AG, Musa BO, Mustapha SK, Jamoh BY, Abdullahi IN, et al. Pattern of prevalent Hepatitis B virus genotypes in Zaria, Nigeria. Niger Postgrad Med J. 2019;26:80–6. doi: 10.4103/npmj.npmj_59_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du H, Li T, Zhang HY, He ZP, Dong QM, Duan XZ, et al. Correlation of hepatitis B virus (HBV) genotypes and mutations in basal core promoter/precore with clinical features of chronic HBV infection. Liver Int. 2007;27:240–6. doi: 10.1111/j.1478-3231.2006.01400.x. [DOI] [PubMed] [Google Scholar]
- 67.Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype:Recent advances. J Gastroenterol Hepatol. 2011;26:123–30. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- 68.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. J Hepatol. 2006;44:915–24. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- 69.Lou J, Zhang L, Lv S, Zhang C, Jiang S. Biomarkers for hepatocellular carcinoma. Biomark Cancer. 2017;9:1–9. doi: 10.1177/1179299X16684640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghabeshi S, Sharifi Z, Hosseini SM, Shooshtari MM. Correlation between viral load of HBV in chronic hepatitis B patients and precore and Basal core promoter mutations. Hepat Mon. 2013;13:e7415. doi: 10.5812/hepatmon.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou YM, Yang JM, Li B, Yin ZF, Xu F, Wang B, et al. Clinicopathologic characteristics of intrahepatic cholangiocarcinoma in patients with positive serum a-fetoprotein. World J Gastroenterol. 2008;14:2251–4. doi: 10.3748/wjg.14.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kramvis A, Kew MC, Bukofzef S. Hepatitis B virus precore mutants in serum and liver of Southern African blacks with hepatocellular carcinoma. J Hepat. 1998;28:132–41. doi: 10.1016/s0168-8278(98)80212-2. [DOI] [PubMed] [Google Scholar]
- 73.Ledesma MM, Galdame O, Bouzas B, Tadey L, Livellara B, Giuliano S, et al. Characterization of the basal core promoter and precore regions in anti-HBe-positive inactive carriers of hepatitis B virus. Int J Infect Dis. 2011;15:314–20. doi: 10.1016/j.ijid.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Alam S, Ahmad N, Alam K. Correlation between hepatitis B viral DNA load and extent of liver pathology in patients with chronic hepatitis B. Hepat Mon. 2008;8:4. [Google Scholar]
- 75.De Mitri MS, Morsica G, Cassini R, Bagaglio S, Andreone P, Bianchi G, et al. Low replication and variability of HBV pre-core in concomitant infection with hepatitis B and hepatitis C viruses. Arch Virol. 2007;152:395–4. doi: 10.1007/s00705-006-0836-6. [DOI] [PubMed] [Google Scholar]
- 76.Hung CH, Chen CH, Lu SN, Wang JH, Hu TH, Huang CM, et al. Precore/core promoter mutations and hepatitis B virus genotype in hepatitis B and C dually infected patients treated with interferon-based therapy. Antiviral Res. 2012;93:55–63. doi: 10.1016/j.antiviral.2011.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available by the corresponding author upon request. Seven of the HBV DNA precore/core gene sequences obtained in this study were deposited to the NCBI database and were assigned the following accession numbers; MT893925, MT893926, MT893927, MT893928, MT893929, MT893930, and MT893931.


