Abstract
Transcriptomics in Parkinson’s disease offers insights into the pathogenesis of Parkinson’s disease but obtaining brain tissue has limitations. In order to bypass this issue, we profile and compare differentially expressed genes and enriched pathways (KEGG) in two peripheral tissues (blood and skin) of 12 Parkinson’s disease patients and 12 healthy controls using RNA-sequencing technique and validation with RT-qPCR. Furthermore, we compare our results to previous Parkinson’s disease post mortem brain tissue and blood results using the robust rank aggregation method. The results show no overlapping differentially expressed genes or enriched pathways in blood vs. skin in our sample sets (25 vs. 1068 differentially expressed genes with an FDR ≤ 0.05; 1 vs. 9 pathways in blood and skin, respectively). A meta-analysis from previous transcriptomic sample sets using either microarrays or RNA-Seq yields a robust rank aggregation list of cortical gene expression changes with 43 differentially expressed genes; a list of substantia nigra changes with 2 differentially expressed genes and a list of blood changes with 1 differentially expressed gene being statistically significant at FDR ≤ 0.05. In cortex 1, KEGG pathway was enriched, four in substantia nigra and two in blood. None of the differentially expressed genes or pathways overlap between these tissues. When comparing our previously published skin transcription analysis, two differentially expressed genes between the cortex robust rank aggregation and skin overlap. In this study, for the first time a meta-analysis is applied on transcriptomic sample sets in Parkinson’s disease. Simultaneously, it explores the notion that Parkinson’s disease is not just a neuronal tissue disease by exploring peripheral tissues. The comparison of different Parkinson’s disease tissues yields surprisingly few significant differentially expressed genes and pathways, suggesting that divergent gene expression profiles in distinct cell lineages, metabolic and possibly iatrogenic effects create too much transcriptomic noise for detecting significant signal. On the other hand, there are signs that point towards Parkinson’s disease-specific changes in non-neuronal peripheral tissues in Parkinson’s disease, indicating that Parkinson’s disease might be a multisystem disorder.
Keywords: Biomarkers, brain, neurodegeneration, blood, transcriptomics, skin
Impact statement
This work is relevant because a meta-analysis of transcriptomic studies of Parkinson’s disease (PD) has not been attempted yet. Furthermore, two peripheral and easily obtainable tissues are being analyzed in this study, paving a way for future large-scale studies from these in vivo accessible tissues in PD. This work maps changes that are consistent between previous work and the current one and, particularly, maps the gene expression changes that might be of specific interest, because they are differentially changed even after applying robust and rigorous statistical methods. This aims to prompt readers to investigate into the qualities of specific gene expressions or pathways regarding Parkinson’s disease.
Introduction
Parkinson’s disease (PD) has an appreciable rate of clinical misdiagnosis. The diagnosis made by experienced clinicians is ∼90% concordant with the following pathological diagnosis.1,2 Furthermore, PD is not a uniform disease where next to causal monogenetic mutations (<10% of all PD patients), many low-risk loci account for much of the genetic risk in sporadic PD.3 Although the effect size of each locus is small, the combined effect increases the risk of PD substantially.4 These loci change gene expression levels rather than changing the coded protein qualities.5 Both monogenetic and sporadic changes in PD result in pathognomic loss of neurons in substantia nigra (SN) and in accumulation of misfolded α-synuclein in Lewy bodies. The incidence of PD increases with aging, which reflects the accumulation of changes caused by altered gene expression.6 Therefore, mapping differentially expressed genes (DEGs) in PD is useful in discovering these multiple changes and could offer diagnostic and prognostic markers and possible treatment options.
However, there are substantial obstacles. PD is canonically thought to be a disease affecting a specific population of neurons. Obtaining tissue for transcriptomic analyses from brain has some limitations, since it is performed post mortem on advanced PD patients. Another factor is that RNA-molecule is instable and the length of post mortem interval and RNA integrity must be considered. A possibility to bypass these obstacles is to sample peripheral tissues, especially the blood, for gene expression. There is evidence that blood gene expression profile does not differ significantly from the brain.7,8 Thus, analyzing blood for easily obtainable biomarkers in PD has become widespread.9–17
Another issue with gene expression studies in PD is the lack of reproducibility of the findings. For example, from a set of 22 DEGs identified with microarray from PD whole blood by Scherzer et al.,18 only three were found overlapping from 1367 DEGs in a subsequent study by Kauczynska et al.9 Lack of reproducibility between datasets could come from different analytic procedures and some from the inherent genetic and transcriptional heterogeneity of PD. PD has been associated with aberrant splicing19 and using microarrays or polymerase chain reaction (PCR) limits mapping this heterogeneity. Unlike arrays, RNA sequencing (RNA-Seq) does not require probes but can detect alternatively spliced or novel transcripts.20,21 RNA-Seq has also been shown to be highly accurate for quantifying expression levels and has less background noise. This enables a smaller sample size, while still having more statistical power compared to microarrays.22,23
In this study, we compare two peripheral tissues in PD—whole blood and skin. We identified the DEGs using RNA-Seq with subsequent validation with RT-qPCR. In addition to describing DEGs, our second goal is to describe enriched functional pathways in PD whole blood and skin. Thirdly, we aim to compare our results with previous datasets from central nervous system (CNS) and blood. The usage of different methods has limited comparing and integrating previous data and thus far enabled only descriptive reviews of PD transcriptomics.24 Meta-analysis could possibly offer a more reproducible set of DEGs or functional pathways for PD, but the use of different methods, annotations, and the publication of incomplete DEG lists hampers integrating the results of previous work. In order to bypass this issue, we use the robust rank aggregation method25 to analyze the aggregated results from our study and previous datasets. The aim here is to determine which DEGs and enriched pathways are ranked the highest within a specific tissue and whether CNS tissue changes overlap with those from heterogenous peripheral tissue populations.
Materials and methods
Study subjects
The study was conducted in concordance with the Declaration of Helsinki and approved by the local Ethics Committee. All subjects gave their written consent to participate in the study. The RNA-Seq from whole blood included 12 patients with clinically diagnosed idiopathic PD and 12 matched healthy controls (HC). Patient characteristics chosen for skin RNA-Seq have been published previously.26 The inclusion of patients was based on: (1) a diagnosis of idiopathic PD according to the QSBB criteria27; (2) on standard medical treatment for PD; and (3) no other severe diagnoses based on medical interview. The demographic data, history of disease, and clinical data were documented, and a summary of participant characteristics and comparison with the previously published skin dataset is presented in Table 1. Clinical disease markers were assessed using validated instruments including the Movement Disorders Unified Parkinson’s Disease Rating Scale (MDS-UPDRS),28 the Hoehn and Yahr Scale (HY),29 the Schwab and England Activities of Daily Living Scale (SE-ADL),30 and the Mini Mental State Examination (MMSE).31
Table 1.
Clinical characteristics of blood and skin RNA-Seq patients.
| Subj. no | Gender (% of males) | Agea | Disease onset agea | Duration of diseasea | 1st degree relatives with PD (%) | HYa | SE-ADLa | MMSEa | MDS-UPDRSa |
|---|---|---|---|---|---|---|---|---|---|
| Blood RNA-Seq patients | |||||||||
| 1 | M | 73 | 65 | 9 | No | 3 | 80 | 30 | 77 |
| 2 | F | 82 | 74 | 9 | No | 4 | 60 | 26 | 79 |
| 3 | M | 67 | 50 | 17 | No | 4 | 70 | 28 | 103 |
| 4 | M | 69 | 68 | 1.2 | No | 3 | 80 | 30 | 56 |
| 5 | F | 85 | 70 | 15 | No | 2.5 | 60 | 23 | 44 |
| 6 | M | 85 | 67 | 18 | No | 4 | 40 | 27 | 159 |
| 7 | F | 68 | 65 | 3 | Yes | 1.5 | 100 | 30 | 26 |
| 8 | F | 71 | 66 | 5 | No | 3 | 70 | 24 | 73 |
| 9 | F | 48 | 47 | 1.1 | No | 1.5 | 90 | 30 | 33 |
| 10 | F | 69 | 67 | 2 | No | 2.5 | 80 | 29 | 72 |
| 11 | M | 76 | 75 | 1.7 | No | 2 | 90 | 25 | 36 |
| 12 | M | 73 | 72 | 1 | No | 1 | 95 | 29 | 22 |
| Total | 50% | 72.2 (±10.0) | 65.5 (±8.6) | 6.9 (±6.5) | 8% | 2.7 (±1) | 76.3 (±17.2) | 27.6 (±2.5) | 65 (±38.7) |
| Skin RNA-Seq patients | |||||||||
| 1 | M | 60 | 46 | 14 | No | 2.5 | 80 | 28 | 83 |
| 2 | F | 82 | 74 | 9 | No | 4 | 60 | 26 | 79 |
| 3 | F | 77 | 68 | 9 | No | 3 | 70 | 30 | 68 |
| 4 | M | 63 | 58 | 4 | No | 2.5 | 90 | 30 | 40 |
| 5 | F | 76 | 73 | 3 | No | 3 | 80 | 27 | 77 |
| 6 | M | 81 | 80 | 1 | No | 2.5 | 90 | 30 | 43 |
| 7 | M | 67 | 64 | 3 | No | 2 | 90 | 29 | 65 |
| 8 | F | 68 | 65 | 3 | No | 1.5 | 100 | 30 | 26 |
| 9 | M | 75 | 64 | 11 | No | 4 | 70 | 30 | 105 |
| 10 | F | 69 | 67 | 2 | No | 2.5 | 80 | 29 | 72 |
| 11 | F | 80 | 76 | 4 | No | 2 | 95 | 25 | 28 |
| 12 | F | 65 | 55 | 10 | No | 4 | 60 | 30 | 114 |
| Total | 42% | 71.9 (±7.5) | 65.8 (±9.6) | 6.1 (±4.3) | 0% | 3.1 (±0.9) | 80.4 (±13.2) | 28.7 (±1.8) | 66.7 (±28.1) |
| P-value | 0.70 | 0.95 | 0.93 | 0.71 | 0.33 | 0.75 | 0.51 | 0.24 | 0.91 |
aAverage (± standard deviation).
PD patients selected for the whole blood RNA-Seq had a mean age of 72.2 (±10.0) years with a mean disease duration of 6.9 (±6.5) years and mean HY stage of 2.7 (±1). The results were validated with qRT-PCR with further 59 PD and 33 HC blood samples (including the 12 + 12 from RNA-Seq), and a breakdown of clinical characteristics can be found in Supplementary Table 1.
Sampling and RNA sequencing
At the time of the medical interview, venous blood of all study subjects was collected into Tempus Blood RNA Tubes (Thermo Fisher Scientific Inc., CA, USA) for the whole blood RNA-Seq. The tubes were immediately frozen and stored in liquid nitrogen at −80°C. The RNA was extracted applying Tempus Spin RNA Isolation Kit (Thermo Fisher Scientific Inc., CA, USA) combined with DNase treatment (RNase-Free DNase Set, Qiagen, Hilde, Germany), according to the manufacturers’ protocols. The globin mRNA was removed from the extracted total RNA using GLOBINclear Kit, human (Thermo Fisher Scientific Inc, CA, USA). The lowest acceptable concentration of globin clear RNA was 36.5 ng/µL for PD patients and 39.5 ng/µL for the control group; 50 ng of each RNA sample was amplified with Ovation RNA-Seq System V2 Kit (NuGen Technologies Inc., CA, USA) and the output double stranded DNA was used to prepare SOLiD 5500 W System DNA fragment libraries according to the manufacturers’ protocols (Thermo Fisher Scientific Inc, CA, USA). Barcoding adapters were applied, and the 12 libraries were pooled prior to sequencing. Fragment (single-end) sequencing chemistry was applied with SOLiD 5500 W XL platform resulting in reads with length of 75 bp. Methods of skin RNA-Seq preparation are described in detail in our previous paper.26
RNA sequencing data analysis
LifeScope software was used for aligning RNA-Seq reads to the hg19 reference genome. The alignment was performed with recommended default parameters. Gene-level read counts were obtained from LifeScope alignment summary statistics and DeSeq2 package in R32 was used to test the differential expression of genes. Detected differential expression of genes was considered statistically significant at a false discovery rate (FDR) ≤ 0.05. Followingly, a pathway enrichment analysis was performed using the hypergeometric test implemented by ClusterProfiler in R33 and KEGG pathway annotations and pathway results were considered significant at P-adjusted ≤ 0.05. A reanalysis from the previously published skin data26 was done with the Clusterprofiler package for R (Input DEGs with an FDR of ≤0.05, cut-off for pathways at P-adjusted ≤0.05).
Validation with RT-qPCR
In a larger group of 59 PD patients and 33 controls (including the RNA-Seq samples), validation top DEGs of whole blood RNA sequencing data with RT-qPCR was conducted. Total-RNA was converted to cDNA using random primers and High Capacity cDNA Reverse Transcription Kit with an RNase Inhibitor (Applied Biosystems). TaqMan Gene Expression Assay with VIC-label was used as the housekeeping gene (Actin) and FAM as the gene of interest. The samples were prepared using the TaqMan® Gene Expression Master Mix (Applied Biosystems). The genes that statistically had the largest change in gene expression were OGT; UBE2J1, KIR2DL3, LCE5A, ENOSF1, FAM219B, CTSL, IL8R1, MIAT, SBSN, ETV1, SPRRE2, CTSD, KRTDAP, and S100A2. Followingly, duplex quantitative real-time PCR (qRT-PCR) analysis was performed in three replicates to minimize technical errors. The data were analyzed using the 2−ΔCT method.
Meta-analysis of previously published gene expression datasets from blood, cortex, and SN
For the comparison of our work with previously published transcription studies’ results, a search was conducted on PubMed. Minimal criteria for including these gene expression results in our analysis were: (1) the list of DEGs is openly accessible (2) the list is original data, (3) list did not contain selectively presented gene expressions, lists with statistical significance cut-offs were allowed, (4) idiopathic PD vs. control comparison, (5) there are enough lists per tissue type for further analysis. The search yielded three lists from cortex,34–36 five from SN,37–41 and five from blood.13,16,42–44 Details of the input lists can be found in Supplementary Table 2. The meta-analysis of the gene expression datasets published by the authors of this paper and other previously published PD gene expression studies was conducted using the Robust rank aggregation (RRA) method by Kolde et al.25 This method summarizes the ranked DEGs lists reported by the authors of these studies by assigning significance scores for the genes that appear more frequently in the top of the ranked lists of differentially expressed genes. This method allows for incomplete ranked gene lists to be included as well, therefore we also included gene rankings where only DEGs with significant P-values were published or a fold change threshold was applied. We conducted the meta-analysis separately in each type of tissue (three in total: cortex, SN, and blood). All gene identifiers were converted to Entrez IDs prior to analysis with RRA. The N parameter was calculated as N(i) + N(avg. u), where N(i) denotes the number of genes present on all the different microarray chips used by the studies included in our meta-analysis and N(avg. u) denotes the average number of genes per microarray that were not included in the intersected N(i). The N was calculated separately for each set of ranked gene lists in different tissues. DEGs were considered statistically significant at FDR ≤ 0.05. For pathway analysis that was conducted with ClusterProfiler KEGG package for R DEGs with a score/P-value ≤0.05 were included and the results were considered statistically significant at FDR ≤ 0.05.
Results
Differential expression of genes and pathways in PD blood
The RNA-Seq yielded 21.4 ± 3.3 million reads (mean ± SD) per sample, out of which 73.6 ± 0.8% aligned to the genome at least once. No cut-offs to reads were applied; >20 million reads per sample in a 12-sample set should provide optimal statistical power for this analysis. Sequencing deeper than 10 M reads and having a sample size >12 does not significantly improve statistical power and precision for detecting DEGs.45–47
Raw data were subjected to differential expression testing with DeSeq2. Differential expression testing of whole blood RNA-Seq data resulted in 25 DEGs between control and PD samples (Table 2). Most of DEGs in blood are upregulated, but notably UBE2J1 (also known as HIP2) is downregulated, with a foldchange of 0.63. Significantly upregulated genes include A2M, LINC00612, FAM219B, KIR2DL3, OGT, ENOSF1, MIAT, IL18R1 and the only other downregulated gene CUTALP. Pathway analysis from PD blood shows one enriched pathway–cholesterol metabolism.
Table 2.
Differentially expressed genes from PD blood.
| Gene | P-adjusted |
|---|---|
| UBE2J1 | 0.002456533032969 |
| OGT | 0.004525065433154 |
| C15orf17 | 0.005583830092225 |
| LUC7L3 | 0.005583830092225 |
| C12orf33 | 0.008624302294215 |
| STAT4 | 0.008624302294215 |
| PTPN4 | 0.008624302294215 |
| A2M | 0.00966346623602 |
| ENOSF1 | 0.011203502577878 |
| MIAT | 0.012084815414542 |
| CD36 | 0.016921618832212 |
| IL18R1 | 0.018943723956247 |
| PYHIN1 | 0.018943723956247 |
| PPT1 | 0.018943723956247 |
| LOC253039 | 0.018943723956247 |
| SYTL2 | 0.023012888462431 |
| EPM2AIP1 | 0.02323506201148 |
| PCGF3 | 0.025439081994619 |
| PKM2 | 0.025439081994619 |
| ZNF767 | 0.032474874139179 |
| RAB32 | 0.037059204955581 |
| PRPF4B | 0.037487943308261 |
| STARD9 | 0.038488470301337 |
| AGPAT4 | 0.048712754929204 |
| GMPPA | 0.048712754929204 |
Differential expression of genes and pathways in PD skin and comparison with blood
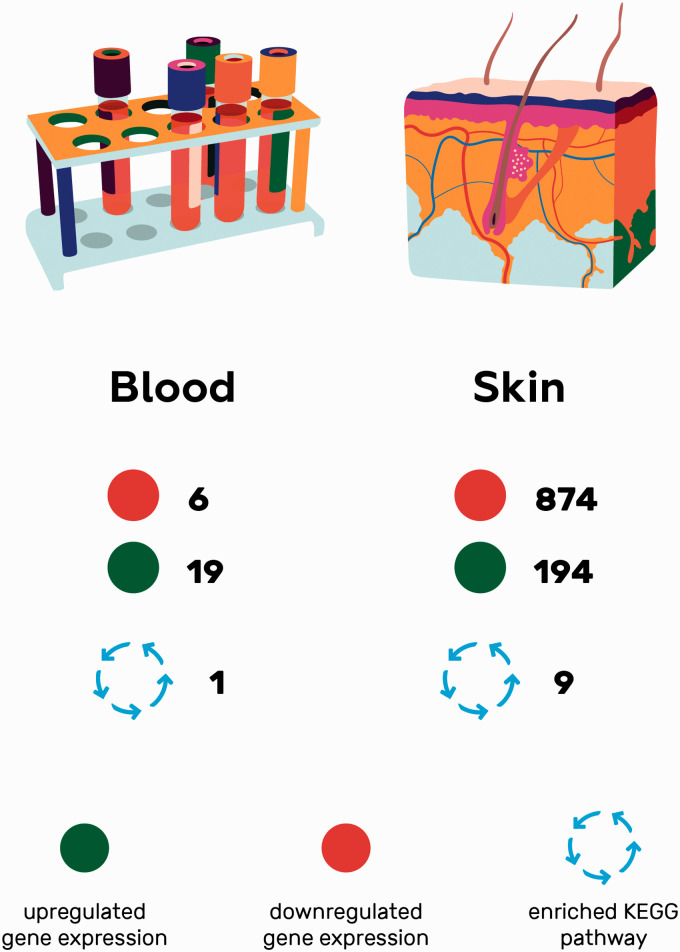
Skin RNA-Seq yielded 1068 DEGs (data available upon request). A clear pattern of global downregulation appears, with 874 of total DEGs being downregulated.26 Nine enriched pathways from PD skin were detected (Table 3). If blood and skin gene expressions are compared using the standard cut-off values for significance, no overlap in DEGs or pathways is to be found (Figure 1.)
Table 3.
Enriched KEGG pathways from PD skin.
| ID | Description | GeneRatio | BgRatio | P value | P. adjust | q value | Count |
|---|---|---|---|---|---|---|---|
| hsa05016 | Huntington disease | 44/434 | 193/7528 | 9.3451827163023e-16 | 2.68206743957876e-13 | 2.59697709168822e-13 | 44 |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 36/434 | 149/7528 | 6.6323638782285e-14 | 9.5174421652579e-12 | 9.21549507290697e-12 | 36 |
| hsa05012 | Parkinson disease | 33/434 | 142/7528 | 2.46947683273819e-12 | 2.36246616998621e-10 | 2.28751538190485e-10 | 33 |
| hsa00190 | Oxidative phosphorylation | 31/434 | 133/7528 | 1.07724856969573e-11 | 7.72925848756688e-10 | 7.48404269472824e-10 | 31 |
| hsa03010 | Ribosome | 31/434 | 153/7528 | 4.8176225916021e-10 | 2.38471885093517e-08 | 2.3090620819618e-08 | 31 |
| hsa05010 | Alzheimer disease | 33/434 | 171/7528 | 4.98547494969025e-10 | 2.38471885093517e-08 | 2.3090620819618e-08 | 33 |
| hsa04714 | Thermogenesis | 36/434 | 231/7528 | 3.27933904006457e-08 | 1.34452900642647e-06 | 1.30187294222112e-06 | 36 |
| hsa04723 | Retrograde endocannabinoid signaling | 24/434 | 148/7528 | 3.35738799540263e-06 | 0.000120446294335069 | 0.000116625056682407 | 24 |
| hsa04260 | Cardiac muscle contraction | 13/434 | 78/7528 | 0.000458898234034117 | 0.0146337547964213 | 0.0141694893315798 | 13 |
Figure 1.
A schematic comparison of the original RNA-Seq data from Parkinson’s disease blood and skin samples visualizing the differences in number of DEGs and KEGG pathways and showing no overlap.
Results of the meta-analysis
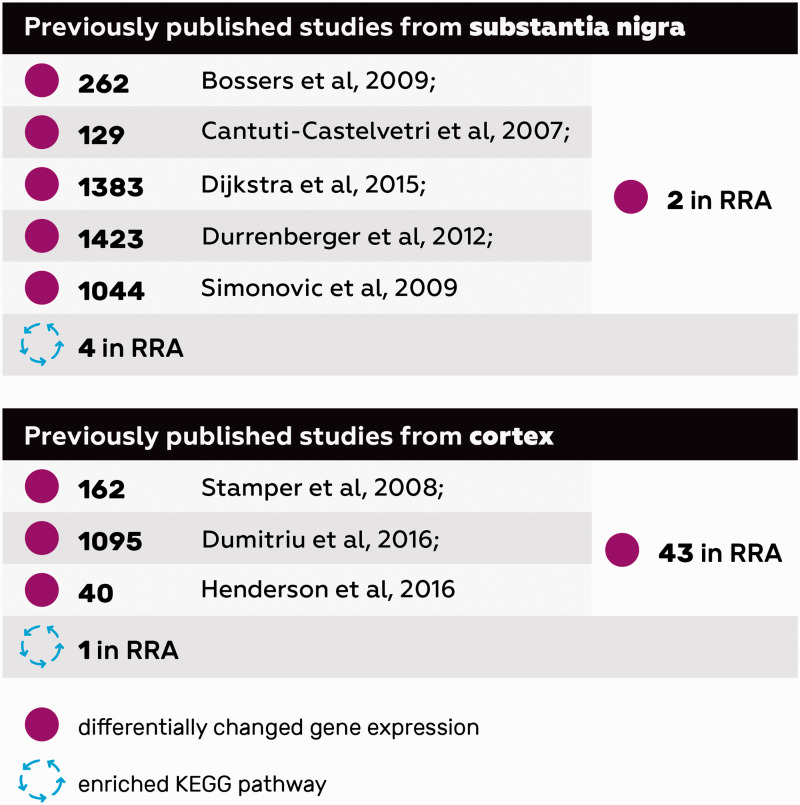
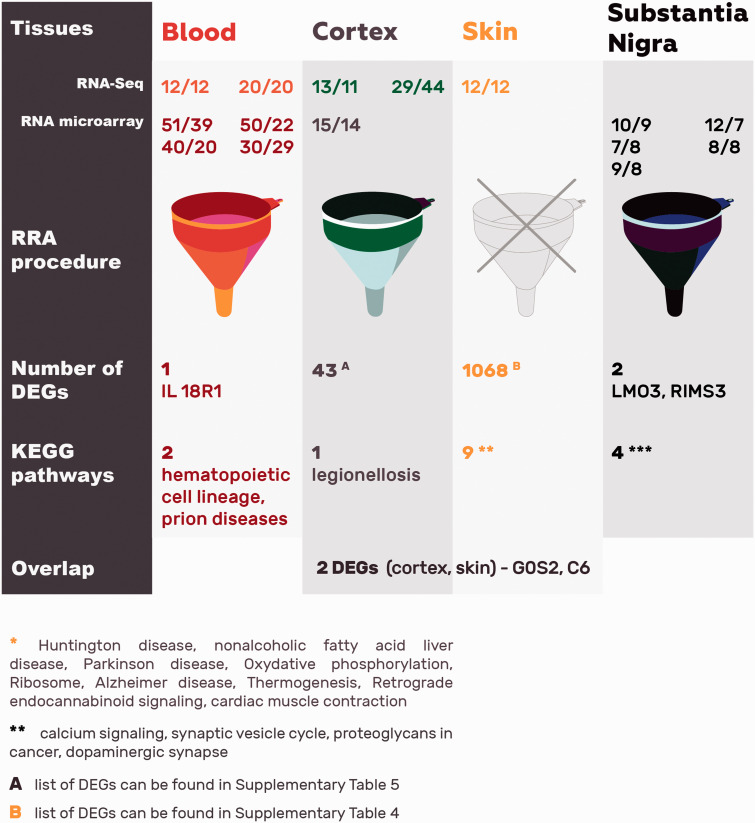
When five previously published sets of DEGs from PD blood13,16,42–44 and the current work were integrated into an RRA list, only one gene remained significant: IL18R1. It is an interleukin receptor linked with proinflammatory responses that belongs to the immunoglobulin superfamily. Pathway enrichment analysis of the top 163 DEGs from blood RRA list found two enriched KEGG pathways: hematopoietic cell lineage (FDR = 0.001) and prion diseases (FDR = 0.037).
Thereafter, we decided to investigate how these findings from peripheral tissues in PD correlate to transcriptomic profiles from CNS. In order to do that, aggregated ranked gene list was created based on the results of three previous studies on the genes expression in cortex34–36 and five studies on SN37–41 (Figure 2). In case of differentially expressed genes in the cortex of PD patients, RRA resulted in 43 significant genes (data available upon request) despite differences in the location and exact cell composition of cortical samples. Topmost significant DEG is PENK, which is elevated in the brain during oxidative stress.48 Furthermore, heat shock proteins HSPA1B, HSPA6, and SERPINH1 and growth factors BDNF, VGF, and CSF3 were elevated. KEGG pathway analysis based on 475 DEGs yielded one enriched pathway: legionellosis. In the case of differentially expressed genes in the SN of PD patients, RRA yielded two significant genes: LMO3 and RIMS3. LMO3 is an oncogene predominantly expressed in the brain, while RIM3 is involved in vesicle trafficking. Pathway analysis from the 390 DEGs yielded four KEGG pathways to be enriched: calcium signaling pathway, synaptic vesicle cycle, proteoglycans in cancer and dopaminergic synapse.
Figure 2.
A schematic overview of previously published cortex and substantia nigra RNA-Seq and microarray studies showing the number DEGs in each single study, overlapping DEGs and KEGG pathways in the RRA analysis.
Comparing the DEGs from three different tissues (blood, cortex, and SN) yielded no overlap (Table 4). Also, reanalysis of enriched pathways from these tissues using KEGG database yielded no overlapping pathways (Table 5). When comparing all RRA lists with the currently only skin DEGs list, only two genes overlap between cortex and skin: G0S2 and C6. No statistically significant pathway was found comparing four different tissues in PD.
Table 4.
Significantly changed gene expressions from blood, cortex, substantia nigra after RRA analysis.
| Tissue | Gene ID | Score | FDR | SYMBOL |
|---|---|---|---|---|
| Blood | 8809 | 1.28802794373959e-05 | 0.050954385454338 | IL18R1 |
| Substantia nigra | 55885 | 3.93808672219532e-06 | 0.00859806402721747 | LMO3 |
| Substantia nigra | 9783 | 5.21727186117565e-06 | 0.00859806402721747 | RIMS3 |
| Cortex | 5179 | 2.27636804825777e-05 | 0.026952197691372 | PENK |
| Cortex | 4548 | 4.8484098221707e-05 | 0.0274016611662194 | MTR |
| Cortex | 56261 | 6.94298847117046e-05 | 0.0274016611662194 | GPCPD1 |
| Cortex | 1440 | 0.00016460604283402 | 0.0324822591192467 | CSF3 |
| Cortex | 29102 | 0.00016460604283402 | 0.0324822591192467 | DROSHA |
| Cortex | 3310 | 0.00016460604283402 | 0.0324822591192467 | HSPA6 |
| Cortex | 3627 | 0.000329206064413617 | 0.0389772851187413 | CXCL10 |
| Cortex | 50486 | 0.000329206064413617 | 0.0389772851187413 | G0S2 |
| Cortex | 8289 | 0.000329206064413617 | 0.0389772851187413 | ARID1A |
| Cortex | 627 | 0.000392790400009593 | 0.0389772851187413 | BDNF |
| Cortex | 203523 | 0.00043728447392925 | 0.0389772851187413 | ZNF449 |
| Cortex | 54431 | 0.000441864529348627 | 0.0389772851187413 | DNAJC10 |
| Cortex | 23332 | 0.000493800064848919 | 0.0389772851187413 | CLASP1 |
| Cortex | 3303 | 0.000493800064848919 | 0.0389772851187413 | HSPA1A |
| Cortex | 57822 | 0.000493800064848919 | 0.0389772851187413 | GRHL3 |
| Cortex | 3315 | 0.000658388044250058 | 0.041505605825661 | HSPB1 |
| Cortex | 729 | 0.000658388044250058 | 0.041505605825661 | C6 |
| Cortex | 7874 | 0.000658388044250058 | 0.041505605825661 | USP7 |
| Cortex | 57542 | 0.000683883429288061 | 0.041505605825661 | KLHL42 |
| Cortex | 151613 | 0.000701108206514544 | 0.041505605825661 | TTC14 |
| Cortex | 486 | 0.000822970002727162 | 0.0423650644882157 | FXYD2 |
| Cortex | 6401 | 0.000822970002727162 | 0.0423650644882157 | SELE |
| Cortex | 85369 | 0.000822970002727162 | 0.0423650644882157 | STRIP1 |
| Cortex | 23704 | 0.000987545940390364 | 0.0449713228239304 | KCNE4 |
| Cortex | 283742 | 0.000987545940390364 | 0.0449713228239304 | FAM98B |
| Cortex | 7425 | 0.000987545940390364 | 0.0449713228239304 | VGF |
| Cortex | 3304 | 0.00115211585734979 | 0.0470381094862812 | HSPA1B |
| Cortex | 5304 | 0.00115211585734979 | 0.0470381094862812 | PIP |
| Cortex | 5469 | 0.00115211585734979 | 0.0470381094862812 | MED1 |
| Cortex | 58517 | 0.00123118103210076 | 0.0472408735878559 | RBM25 |
| Cortex | 4009 | 0.00131667975371558 | 0.0472408735878559 | LMX1A |
| Cortex | 6415 | 0.00131667975371558 | 0.0472408735878559 | SELENOW |
| Cortex | 6539 | 0.00131667975371558 | 0.0472408735878559 | SLC6A12 |
| Cortex | 126308 | 0.00144191115876224 | 0.0473996041471313 | MOB3A |
| Cortex | 4922 | 0.00148123762959785 | 0.0473996041471313 | NTS |
| Cortex | 871 | 0.00148123762959785 | 0.0473996041471313 | SERPINH1 |
| Cortex | 96610 | 0.00148123762959785 | 0.0473996041471313 | BMS1P20 |
| Cortex | 2168 | 0.00164578948510674 | 0.0487153687591595 | FABP1 |
| Cortex | 3337 | 0.00164578948510674 | 0.0487153687591595 | DNAJB1 |
| Cortex | 91748 | 0.00164578948510674 | 0.0487153687591595 | ELMSAN1 |
| Cortex | 51116 | 0.00181033532035238 | 0.0498473725417957 | MRPS2 |
| Cortex | 54541 | 0.00181033532035238 | 0.0498473725417957 | DDIT4 |
| Cortex | 56116 | 0.00181033532035238 | 0.0498473725417957 | PCDHB@ |
Table 5.
Enriched KEGG pathways from blood, cortex, substantia nigra after RRA analysis.
| Tissue | ID | Description | GeneRatio | BgRatio | P value | P. adjust | q value | geneID | Count |
| Blood | hsa04640 | Hematopoietic cell lineage | 14/181 | 97/7914 | 7.7189077408129e-06 | 0.00129677650045657 | 0.00124315040457302 | 933/2208/2322/7850/948/930/3552/2323 | 8 |
| Blood | hsa05020 | Prion diseases | 8/181 | 35/7914 | 0.000481311510902407 | 0.0404301669158022 | 0.0387582427200359 | 712/714/3552/713 | 4 |
| Substantia nigra | hsa04020 | Calcium signaling pathway | 14/181 | 193/7914 | 0.00012816581859865 | 0.0305034648264787 | 0.0273870117637115 | 4923/5027/2066/6543/8913/5159/489/3708/185/80228/2065/3710/444/23236 | 14 |
| Substantia nigra | hsa04721 | Synaptic vesicle cycle | 8/181 | 78/7914 | 0.000383587904674655 | 0.0456469606562839 | 0.0409833392889236 | 6857/4905/5864/535/23025/440279/22999/6571 | 8 |
| Substantia nigra | hsa05205 | Proteoglycans in cancer | 13/181 | 204/7914 | 0.000779054186583262 | 0.0479601005770201 | 0.043060152220854 | 286/2066/7480/5962/3708/1499/10451/6198/6383/2065/3710/7473/2549 | 13 |
| Substantia nigra | hsa04728 | Dopaminergic synapse | 10/181 | 131/7914 | 0.000806052110538154 | 0.0479601005770201 | 0.043060152220854 | 10681/2932/1813/6323/3708/6571/5528/2893/3710/23236 | 10 |
| Cortex | hsa05134 | Legionellosis | 10/207 | 56/7914 | 1.50285900432062e-06 | 0.00039074334112336 | 0.000367013988423561 | 3310/3303/3304/4791/5970/7100/840/4792/10767/3305 | 10 |
Discussion
RNA-seq results from blood and skin yielded very different number of significantly changed DEGs (25 vs. 1068), but no overlap. In the blood, a gene was found which has previously shown to have altered expression in PD venous blood—UBE2J19,18,49 The protein encoded by this gene is a ubiquitin-conjugating enzyme located in the membrane of the endoplasmic reticulum. UBE2J1 is involved in ubiquitin-proteasome system, which targets proteins for degradation.50 In PD, the ubiquitin-proteasome system is impaired and contributes to aberrant protein degradation. UBE2J1 is also linked with HD, where it ubiquitinylates aberrant protein huntingtin.51 Another gene, A2M encodes for a protease inhibitor and cytokine transporter that is linked with AD where it contributes to the degradation of A-beta.52 The only statistically significant pathway in the blood was cholesterol metabolism. Interestingly, epidemiological studies demonstrate an association between higher levels of cholesterol and lower risk of PD.53 A study showing higher plasma levels of (S)24-OH-cholesterol, a metabolite possibly reflecting brain cholesterol metabolism, was linked to lower odds of having PD.54 In the PD brain, aberrant alpha-synuclein impacts cholesterol metabolism which neurons depend on in synapse formation. Altered levels of cholesterol in neuronal cultures lead to impaired formation of synapses and less neurotransmitter release.55 DEGs in the skin reflect the PD-related dermal changes.26 A reanalysis of this data yielded three KEGG pathways that were associated with neurodegeneration. Huntington disease (HD) pathway contains genes that are involved in vesicular transport, Ca2+ signaling and (through disrupted Ca2+ signaling), mitochondrial dysfunction. Notably, a study56 compared post mortem brain tissues between PD and HD and found an overlap of 25% of DEGs. The second neurodegenerative pathway enriched in PD skin was Alzheimer’s disease (AD) pathway which contains genes that increase in the production of amyloidogenic A-beta peptides or affect posttranslational processing of proteins. Lastly, KEGG Parkinson’s disease pathway itself, which characterizes the intracellular accumulation of alpha-synuclein and impaired mitochondrial function, is enriched in PD skin. The fact that major neurodegenerative pathways are mapped in a peripheral tissue in PD suggests two things. Firstly, that there are global pan-neurodegenerative mechanisms involved in PD. Secondly, these pan-neurodegenerative mechanisms are detectable from a peripheral tissue. A group of enriched pathways emerge from skin that are linked to metabolic changes in PD, for example oxidative phosphorylation pathway, which is another key player in neurodegenerative diseases.57 Non-alcoholic fatty liver disease (NAFLD) is a pathway of lipid accumulation which is affected by insulin resistance and leads to elevated oxidative stress though mitochondrial beta-oxidation of fatty acids in the liver. NAFLD has already been linked to AD,58 the implication in PD needs to be investigated. Another pathway related to lipid metabolism affected in PD skin is thermogenesis pathway in which chemical energy is converted into heat in brown adipose tissue that is controlled by sympathetic nervous system. This has also been shown to be implicated in murine models of HD,56 a key regulator in this pathway is peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α) was upregulated in PD skin samples.26 Its activation was demonstrated to improve the phenotype in mouse models of PD.59 Retrograde endocannabinoid signaling pathway affects synaptic plasticity and neurotransmission in the brain. It is suggested dyskinesias in PD might be caused through disrupted endocannabinoid signaling among other effectors.60,61 Initially, it was thought that endocannabinoids are exclusive to the CNS, but further research has described their involvement in peripheral tissues. In the skin, changes in endocannabinoid system are linked to melanoma,62 which is interesting considering the epidemiological bidirectional link between PD and melanoma.63
When comparing our sample sets from skin and blood it is clear that skin yields more significant changes. Low DEG count from blood might be due to higher number of different cell lineages, metabolic, and possibly iatrogenic effects that create too much transcriptomic noise for detecting significant signal. The lack of overlap between results of skin and blood lead us to investigate comparability with previous studies. As the output of individual experiments can be noisy, data integration could increase the signal and lessen false positive findings. Therefore, we performed meta-analysis using the RRA method25 of the lists of DEGs from previously published open-access studies and the present study in hopes of finding robust PD-specific changes (Figure 3). Another significant reason that lead us to perform this meta-analysis was to allow for a comparison between tissues of different origin (especially the CNS and the periphery).
Figure 3.
A schematic overview of the studies involved in RRA analysis (51/39 is the number of Parkinson’s disease patients and healthy controls in the original study respectively), the output DEGs and KEGG pathways, and the overlap between the investigated studies.
In a descriptive review of transcriptomic studies of PD, whole blood shows significant changes in pathways involved in inflammation, mitochondrial function, immune function, protein chaperones, RNA processing, and programmed cell death.24 In our analysis using the RRA method, the only significant DEG in blood was IL18R1. It is an interleukin receptor linked with proinflammatory responses of the immunoglobulin superfamily. PD is characterized by a dysregulated inflammatory environment. In a PD model of MPTP-treated mice, GM-CSF administration induced IL18R1, which was argued to be due to GM-CSF induced neuroprotective mechanisms.64 IL18R1 has also been associated with neuroinflammation in multiple sclerosis.65 Altered levels of IL18R1 found in PD blood underline the relevance of neuroinflammatory processes in neurodegeneration. In the enrichment analysis of PD blood, RRA lists two pathways which were significant: hematopoietic cell lineage and prion diseases pathway. Hematopoietic cell lineage pathway is a part of the immune system, and changes in this lineage have previously been published in PD.44,66 Prion diseases pathway is possibly linked to PD prion-like propagating qualities.67 In order to compare the transcriptomic profiles in PD of our current peripheral tissues and the aggregated results from peripheral blood with previous transcriptomic results from CNS, aggregated lists of these sample sets needed to be made. The two specific localizations in the CNS that had enough transcriptomic sample sets for an RRA analysis—SN and cortex showed no overlap in DEGs or pathways. There were multiple DEGs in cortex RRA list (Table 4) with only two (G0S2 and C6) showing an overlap with the skin RNA-Seq results. G0/G1 switch gene 2 (G0S2) is a major regulator of lipid metabolism in adipocytes.68 Complement component 6 (C6) is a protein in the lytic membrane attack complex (MAC) macromolecule C5b-9. The MAC inserts itself into foreign cells, causing lysis. It has been shown immunohistochemically in the brains of AD and PD69 where its abundant staining is one of the signs of active neuroinflammation in neurodegeneration. The only significantly enriched pathway in the cortex of PD was legionellosis pathway; how this relates to PD remains to be known. A component of this pathway TLR/MYD88 signaling has previously been associated with neurodegenerative diseases.70 In the RRA of SN, four enriched pathways were significant: calcium signaling, synaptic vesicle cycle, proteoglycans in cancer, and dopaminergic synapse pathway. Genes that give rise to PD have a known causal role in Ca2+ homeostasis which is regulated in the calcium signaling pathway.71 Synaptic vesicle cycle pathway has already been associated with PD previously.72 Proteoglycans in cancer pathway might be altered in PD due to neuroinflammation, modulating the fibrotic process in the extracellular matrix as previously reported in neurodegeneration.73,74 Enriched dopaminergic synapse pathway in an RRA list from SN describes the pathognomonic changes of PD.
Due to the novelty of using RRA in analyzing transcriptome data, the optimal number of studies that need to be integrated for the results to be relevant still needs to be determined. If feasible, integrating studies with small numbers could be an alternative to increasing sample size per study in obtaining more reproducible and robust results. Another aspect that should be considered in future transcriptomic studies of PD that might yield more reproducible results, is dividing PD patients into endophenotypes. Also, in the current work and, indeed, in many of the previous studies, the effects of dopaminergic medication and natural disease course remain undistinguishable, as most of the samples are from patients already receiving dopaminergic therapy. However, so far it is not been clearly shown that therapy naïve patients have very different transcriptomic profiles.9
Taken together, the differential expression of very few genes remained significant in direct comparison between PD blood and skin and in ranked aggregation lists and there was virtually no overlap between tissues. There are multiple pathways that lead to the pathognomonic sign of dopaminergic neuron death in PD, for an example alpha-syn aggregation and mitochondrial stress. There is a possibility that changes in adjacent DEGs in a pathway that converge into having the same metabolic effect is why transcriptomic studies so far have failed to yield robust reproducible results. Reproducing the exact DEGs even in the same tissue type has proven to be difficult and thus the comparison between tissues of different origin is hampered, especially when taking the current meta-analysis into regard. However, there are signs that point towards changes in non-neuronal tissues in PD, indicating that PD might be a multisystem disorder.
Key points:
RNA-Seq analysis from PD whole blood and skin yielded very different results, blood being a lot more heterogeneous. There were no exact overlapping DEGs or enriched KEGG pathways.
Integrating previous studies from blood, cortex, and SN into robust ranked aggregation lists yields virtually no significantly changed DEGs and no overlap between tissues. Notable exceptions are IL18R1 from blood, LMO3 and RIMS3 from SN, PENK from cortex and G0S2, and C6 from cortex and skin.
Since the DEGs show great interstudy variability, there are also only a few significantly enriched pathways and no direct overlap between tissues in PD.
Peripheral tissues should be used in transcriptomic studies of PD, because of their accessibility in vivo and moderate comparability to PD specific changes in CNS.
Of the two peripheral tissues we investigated, skin shows more PD specific changes. The epidemiological link between PD and melanoma indicates that there might be some pathomechanistic similarities in these diseases. This warrants more transcriptomic studies from PD skin.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220967325 for Transcriptomic profiles in Parkinson’s disease by Lille Kurvits, Freddy Lättekivi, Ene Reimann, Liis Kadastik-Eerme, Kristjan M Kasterpalu, Sulev Kõks, Pille Taba and Anu Planken in Experimental Biology and Medicine
Authors’ contributions: LK drafted the manuscript and was involved in analyzing and validation processes. SK analyzed the raw data and revised the manuscript. PT and AP revised the manuscript. LKE gathered the clinical data. FL contributed in the RRA and pathway analysis. ER contributed in the transcriptomic analysis. KMK ran the validating tests.
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ETHICAL APPROVAL: Ethical approval was received from Research Ethics Committee of the University of Tartu.
Funding: This work was supported by Estonian Research Council Personal research funding [PUT1239].
Supplemental material: Supplemental material for this article is available online.
ORCID iDs
Lille Kurvits https://orcid.org/0000-0002-5027-3539
Freddy Lättekivi https://orcid.org/0000-0003-0121-1056
Kristjan M Kasterpalu https://orcid.org/0000-0002-8852-5850
Sulev Kõks https://orcid.org/0000-0001-6087-6643
References
- 1.Hughes AJ, Daniel SE, Lees AJ. Improved accuracy of clinical diagnosis of Lewy body Parkinson’s disease. Neurology 2001; 57:1497–9 [DOI] [PubMed] [Google Scholar]
- 2.Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, Litvan I, Lang AE, Halliday G, Goetz CG, Gasser T, Dubois B, Chan P, Bloem BR, Adler CH, Deuschl G. MDS clinical diagnostic criteria for Parkinson’s disease: MDS-PD clinical diagnostic criteria. Mov Disord 2015; 30:1591–601 [DOI] [PubMed] [Google Scholar]
- 3.Verstraeten A, Theuns J, Van Broeckhoven C. Progress in unraveling the genetic etiology of parkinson disease in a genomic era. Trends Genet 2015; 31:140–9 [DOI] [PubMed] [Google Scholar]
- 4.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R; International Parkinson's Disease Genomics Consortium (IPDGC); Parkinson's Study Group (PSG) Parkinson's Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson's Disease Consortium; Alzheimer Genetic Analysis Group, Ikram MA, Ioannidis JP, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB.. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet 2014; 46:989–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron 2010; 68:201–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadastik-Eerme L, Taba N, Asser T, Taba P. Incidence and mortality of Parkinson’s disease in Estonia. Neuroepidemiology 2019; 53:63–72 [DOI] [PubMed] [Google Scholar]
- 7.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 2006; 141B:261–8 [DOI] [PubMed] [Google Scholar]
- 8.Cooper-Knock J, Kirby J, Ferraiuolo L, Heath PR, Rattray M, Shaw PJ. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol 2012; 8:518–30 [DOI] [PubMed] [Google Scholar]
- 9.Kauczynska KM, Praveen S, Toft JA, Skogar M, Sæbø O, Lönneborget S. A. Found in transcription: accurate Parkinson's disease classification in peripheral blood. J Park Dis 2013; 3:19–29 [DOI] [PubMed] [Google Scholar]
- 10.Aguiar Pm de C, Severino P. Biomarkers in Parkinson disease: global gene expression analysis in peripheral blood from patients with and without mutations in PARK2 and PARK8. Einstein São Paulo 2010; 8:291–7 [DOI] [PubMed] [Google Scholar]
- 11.Shehadeh LA, Yu K, Wang L, Guevara A, Singer C, Vance J. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson’s disease. PLoS One 2010; 5:e9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alieva AK, Shadrina MI, Filatova EV, Karabanov AV, Illarioshkin SN, Limborska SA, Slominsky PA. Involvement of endocytosis and alternative splicing in the formation of the pathological process in the early stages of Parkinson’s disease. BioMed Res Int 2014; 2014:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Infante J, Prieto C, Sierra M, Sánchez-Juan P, González-Aramburu I, Sánchez-Quintana C, Berciano J, Combarros O, Sainz J. Identification of candidate genes for Parkinson’s disease through blood transcriptome analysis in LRRK2-G2019S carriers, idiopathic cases, and controls. Neurobiol Aging 2015; 36:1105–9 [DOI] [PubMed] [Google Scholar]
- 14.Infante J, Prieto C, Sierra M, Sánchez-Juan P, González-Aramburu I, Sánchez-Quintana C, Berciano J, Combarros O, Sainz J. Comparative blood transcriptome analysis in idiopathic and LRRK2 G2019S-associated Parkinson’s disease. Neurobiol Aging 2016; 38:214.e1–e5 [DOI] [PubMed] [Google Scholar]
- 15.Pinho R, Guedes LC, Soreq L, Lobo PP, Mestre T, Coelho M, Rosa MM, Gonçalves N, Wales P, Mendes T, Gerhardt E, Fahlbusch C, Bonifati V, Bonin M, Miltenberger-Miltényi G, Borovecki F, Soreq H., Ferreira JJ, Outeiro TF. Gene expression differences in peripheral blood of Parkinson’s disease patients with distinct progression profiles. PLoS One 2016; 11:e0157852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calligaris R, Banica M, Roncaglia P, Robotti E, Finaurini S, Vlachouli C, Antonutti L, Iorio F, Carissimo A, Cattaruzza T, Ceiner A, Lazarevic D, Cucca A, Pangher N, Marengo E, di Bernardo D, Pizzolato G, Gustincich S. Blood transcriptomics of drug-naïve sporadic Parkinson’s disease patients. BMC Genomics 2015; 16:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potashkin JA, Santiago JA, Ravina BM, Watts A, Leontovich AA. Biosignatures for Parkinson’s disease and atypical parkinsonian disorders patients. PLoS One 2012; 7:e43595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherzer CR, Eklund AC, Morse LJ, Liao Z, Locascio JJ, Fefer D, Schwarzschild MA, Schlossmacher MG, Hauser MA, Vance JM, Sudarsky LR, Standaert DG, Growdon JH, Jensen RV, Gullans SR. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc Natl Acad Sci U S A 2007; 104:955–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu RH, Liu SP, Huang SJ, Chen HJ, Chen PR, Lin YH, Ho JC, Chang WL, Tsai CH, Shyu WC, Linet SZ. Aberrant alternative splicing events in Parkinson’s disease. Cell Transplant 2013; 22:653–61 [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014; 9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Iterson M, ’t Hoen P, Pedotti P, Hooiveld GJEJ, den Dunnen JT, van Ommen GJB, Boer JM, Menezes RX. Relative power and sample size analysis on gene expression profiling data. BMC Genomics 2009; 10:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Zhao S, Li C-I, Sheng Q, Shyr Y. RNAseqPS: a web tool for estimating sample size and power for RNAseq experiment. Cancer Inform 2014; 13:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borrageiro G, Haylett W, Seedat S, Kuivaniemi H, Bardien S. A review of genome-wide transcriptomics studies in Parkinson’s disease. Eur J Neurosci 2018; 47:1–16 [DOI] [PubMed] [Google Scholar]
- 25.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 2012; 28:573–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planken A, Kurvits L, Reimann E, Kadastik-Eerme L, Kingo K, Kõks S, Taba P. Looking beyond the brain to improve the pathogenic understanding of Parkinson’s disease: implications of whole transcriptome profiling of patients’ skin. BMC Neurol 2017; 17:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet 2009; 373:2055–66 [DOI] [PubMed] [Google Scholar]
- 28.Goetz CG, Tilley BC, Shaftman SR, Stebbins GT, Fahn S, Martinez-Martin P, Poewe W, Sampaio C, Stern MB, Dodel R, Dubois B, Holloway R, Jankovic J, Kulisevsky J, Lang AE, Lees A, Leurgans S, LeWitt PA, Nyenhuis D, Olanow CW, Rascol O, Schrag A, Teresi JA, van Hilten JJ, LaPelle N, Movement Disorder Society UPDRS Revision Task Force. Movement disorder society-sponsored revision of the unified Parkinson’s disease rating scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov Disord 2008; 23:2129–70 [DOI] [PubMed] [Google Scholar]
- 29.Hoehn MM, Yahr MD. Parkinsonism: onset, progression and mortality. Neurology 1967; 17:427–42 [DOI] [PubMed] [Google Scholar]
- 30.Schwab RS, Eaigf, Donaldson IML. Projection technique for evaluating surgery in Parkinson’s disease. In: ■ (eds) 3rd symposium on surgery in Parkinson’s disease. Edinburgh: Livingstone, 1969, pp.152–157.
- 31.Folstein MF, Folstein SE, McHugh PR. Mini-mental state. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975; 12:189–98 [DOI] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol 2012; 16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson-Smith A, Corneveaux JJ, De Both M, Cuyugan L, Liang WS, Huentelman M, Adler C, Driver-Dunckley E, Beach TG, Dunckley TL. Next-generation profiling to identify the molecular etiology of parkinson dementia. Neurol Genet 2016; 2:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumitriu A, Golji J, Labadorf AT, Gao B, Beach TG, Myers RH, Longo KA, Latourelle JC. Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in parkinson disease. BMC Med Genomics 2015; 9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamper C, Siegel A, Liang WS, Pearson JW, Stephan DA, Shill H, Connor D, Caviness JN, Sabbagh M, Beach TG, Adler CH, Dunckley T. Neuronal gene expression correlates of Parkinson’s disease with dementia. Mov Disord 2008; 23:1588–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bossers K, Meerhoff G, Balesar R, van Dongen JW, Kruse CG, Swaab DF, Verhaagen J. Analysis of gene expression in Parkinson’s disease: possible involvement of neurotrophic support and axon guidance in dopaminergic cell death. Brain Pathol 2009; 19:91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, Andersen SL, Stephens RM, Benes FM, Sonntag KC. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain 2009; 132:1795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantuti-Castelvetri I, Keller-McGandy C, Bouzou B, Asteris G, Clark TW, Frosch MP, Standaert DG. Effects of gender on nigral gene expression and parkinson disease. Neurobiol Dis 2007; 26:606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dijkstra AA, Ingrassia A, de Menezes RX, van Kesteren RE, Rozemuller AJM, Heutink P, van de Berg WDJ. Evidence for immune response, axonal dysfunction and reduced endocytosis in the substantia nigra in early stage Parkinson’s disease. PLoS One 2015; 10:e0128651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durrenberger PF, Grünblatt E, Fernando FS, Monoranu CM, Evans J, Riederer P, Reynolds R, Dexter TD. Inflammatory pathways in Parkinson’s disease; a BNE microarray study. Park Dis 2012; 2012:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santiago JA, Potashkin JA. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc Natl Acad Sci U S A 2015; 112:2257–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kedmi M, Bar-Shira A, Gurevich T, Giladi N, Orr-Urtreger A. Decreased expression of B cell related genes in leukocytes of women with Parkinson’s disease. Mol Neurodegener 2011; 6:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soreq L, Israel Z, Bergman H, Soreq H. Advanced microarray analysis highlights modified neuro-immune signaling in nucleated blood cells from Parkinson’s disease patients. J Neuroimmunol 2008; 201-202:227–36 [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 2014; 30:301–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ching T, Huang S, Garmire LX. Power analysis and sample size estimation for RNA-Seq differential expression. RNA 2014; 20:1684–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schurch NJ, Schofield P, Gierliński M, Cole C, Sherstnev A, Singh V, Wrobel N, Gharbi K, Simpson GG, Owen-Hughes T, Blaxter M, Barton1 GJ. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016; 22:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenberger J, Petrovics G, Buzas B. Oxidative stress induces proorphanin FQ and proenkephalin gene expression in astrocytes through p38- and ERK-MAP kinases and NF-κB: oxidative stress and proorphanin FQ. J Neurochem 2008; 79:35–44 [DOI] [PubMed] [Google Scholar]
- 49.Molochnikov L, Rabey JM, Dobronevsky E, Bonuccelli U, Ceravolo R, Frosini D, Grünblatt E, Riederer P, Jacob C, Aharon-Peretz J, Bashenko Y, Youdim MBH, Mandel SA. A molecular signature in blood identifies early Parkinson’s disease. Mol Neurodegener 2012; 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith SB, Dampier W, Tozeren A, Brown JR, Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One 2012; 7:e33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem 1996; 271:19385–94 [DOI] [PubMed] [Google Scholar]
- 52.Varma VR, Varma S, An Y, Hohman TJ, Seddighi S, Casanova R, Beri A, Dammer EB, Seyfried NT, Pletnikova O, Moghekar A, Wilson MR, Lah JJ, O'Brien RJ, Levey AI, Troncoso JC, Albert MS, Thambisetty M. Alpha-2 macroglobulin in Alzheimer’s disease: a marker of neuronal injury through the RCAN1 pathway. Mol Psychiatry 2017; 22:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin U, Park SJ, Park SM. Cholesterol metabolism in the brain and its association with Parkinson’s disease. Exp Neurobiol 2019; 28:554–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, Sterling NW, Du G, Sun D, Stetter C, Kong L, Zhu Y, Neighbors J, Lewis MM, Chen H, Hohl RJ, Mailman RB. Brain cholesterol metabolism and Parkinson’s disease. Mov Disord 2019; 34:386–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alecu I, Bennett SAL. Dysregulated lipid metabolism and its role in α-Synucleinopathy in Parkinson’s. Disease Front Neurosci 2019; 13:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory And metabolic defects in Huntington’s disease transgenic mice implicate PGC-1α in Huntington’s disease neurodegeneration. Cell Metab 2006; 4:349–62 [DOI] [PubMed] [Google Scholar]
- 57.Singh A, Kukreti R, Saso L, Kukreti S. Oxidative stress: a key modulator in neurodegenerative diseases. Molecules 2019; 24:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Estrada LD, Ahumada P, Cabrera D, Arab JP. Liver dysfunction as a novel player in alzheimer’s progression: looking outside the brain. Front Aging Neurosci 2019; 11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin HL, Mounsey RB, Mustafa S, Sathe K, Teismann P. Pharmacological manipulation of peroxisome proliferator-activated receptor γ (PPARγ) reveals a role for anti-oxidant protection in a model of Parkinson’s disease. Exp Neurol 2012; 235:528–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giuffrida A, Martinez A. The endocannabinoid system and parkinson disease. In: ■ (eds) The endocannabinoid system. Amsterdam: Elsevier, 2017, pp.63–81
- 61.Castillo PE, Younts TJ, Chávez AE, Hashimotodani Y. Endocannabinoid signaling and synaptic function. Neuron 2012; 76:70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Río C, del Millán E, García V, Appendino G, DeMesa J, Muñoz E. The endocannabinoid system of the skin. A potential approach for the treatment of skin disorders. Biochem Pharmacol 2018; 157:122–33 [DOI] [PubMed] [Google Scholar]
- 63.Liu R, Gao X, Lu Y, Chen H. Meta-analysis of the relationship between parkinson disease and melanoma. Neurology 2011; 76:2002–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosloski LM, Kosmacek EA, Olson KE, Mosley RL, Gendelman HE. GM-CSF induces neuroprotective and anti-inflammatory responses in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine intoxicated mice. J Neuroimmunol 2013; 265:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gillett A, Thessen Hedreul M, Khademi M, Espinosa A, Beyeen AD, Jagodic M, Kockum I, Harris RA, Olsson T. Interleukin 18 receptor 1 expression distinguishes patients with multiple sclerosis. Mult Scler J 2010; 16:1056–65 [DOI] [PubMed] [Google Scholar]
- 66.Schlachetzki JCM, Prots I, Tao J, Chun HB, Saijo K, Gosselin D, Winner B, Glass CK, Winkler J. A monocyte gene expression signature in the early clinical course of Parkinson’s disease. Sci Rep 2018; 8:10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brundin P, Melki R. Prying into the prion hypothesis for Parkinson’s disease. J Neurosci 2017; 37:9808–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta 2013; 1831:276–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGeer PL, McGeer EG. Inflammation and neurodegeneration in Parkinson’s disease. Parkinsonism Relat Disord 2004; 10:S3–S7 [DOI] [PubMed] [Google Scholar]
- 70.Xiang W, Chao ZY, Feng DY. Role of toll-like receptor/MYD88 signaling in neurodegenerative diseases. Rev Neurosci 2015; 26:407–14. [DOI] [PubMed] [Google Scholar]
- 71.Zaichick SV, McGrath KM, Caraveo G. The role of Ca 2+ signaling in Parkinson’s disease. Dis Model Mech 2017; 10:519–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esposito G, Ana Clara F, Verstreken P. Synaptic vesicle trafficking and Parkinson’s disease. Dev Neurobiol 2012; 72:134–44 [DOI] [PubMed] [Google Scholar]
- 73.Heindryckx F, Li JP. Role of proteoglycans in neuro-inflammation and Central nervous system fibrosis. Matrix Biol 2018; 68-69:589–601 [DOI] [PubMed] [Google Scholar]
- 74.Labadorf A, Choi SH, Myers RH. Evidence for a pan-neurodegenerative disease response in Huntington’s and Parkinson’s disease expression profiles. Front Mol Neurosci 2018; 10:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220967325 for Transcriptomic profiles in Parkinson’s disease by Lille Kurvits, Freddy Lättekivi, Ene Reimann, Liis Kadastik-Eerme, Kristjan M Kasterpalu, Sulev Kõks, Pille Taba and Anu Planken in Experimental Biology and Medicine