Abstract
Various circular RNAs (circRNAs) have been shown to exert vital functions in tongue squamous cell carcinoma (TSCC). However, their roles in TSCC progression remain to be elucidated. This research aimed to investigate the role and mechanism of hsa_circ_0000003 (circ_0000003) in TSCC progression. Here, we found that circ_0000003 expression was upregulated in TSCC tissues and cell lines, and high circ_0000003 expression was correlated with advanced TNM stage, increased tumor size and poor patient survival. Circ_0000003 was revealed to facilitate cell proliferation, migration and invasion of TSCC cells. Mechanistically, we found that circ_0000003 acted as a competing endogenous RNA (ceRNA) that sponged miR-330-3p, thereby elevating glutaminase (GLS) expression. Accordingly, cell invasion, migration, glutamine consumption, α-ketoglutarate (α-KG) production and ATP production were significantly decreased by circ_0000003 knockdown in TSCC cells, and these effects were reversed by miR-330-3p inhibition. In conclusion, circ_0000003 facilitates TSCC cell proliferation, migration, invasion and glutamine catabolism by regulating the miR-330-3p/GLS pathway.
Keywords: circ_0000003, tongue squamous cell carcinoma, miR-330-3p, glutamine metabolism, glutaminase
Introduction
Tongue squamous cell carcinoma (TSCC) is the most prevalent malignancy of oral cancer and accounts for 25–40% of all cases (1). Despite significant advancements in surgery, radiotherapy, chemotherapy and other therapies, the 5-year survival rate for TSCC patients has remained poor during the past few decades (1,2). Even though the pathogenesis of TSCC has been investigated extensively over the past few decades, the mechanisms underlying the occurrence and development of TSCC have not been fully clarified (2). Therefore, elucidation of the mechanisms of TSCC progression may aid in devising novel strategies for the diagnosis, therapy and prognosis of TSCC.
Circular RNAs (circRNAs) are a class of newly discovered non-coding RNAs (ncRNAs) with covalently closed loops, which are more stable than conventional linear RNAs (3). Increasing evidence suggests that circRNAs exert a crucial role in tumorigenesis (4–6). In addition, a number of circRNAs have been implicated in the progression of TSCC (7–10). For instance, hsa_circ_0001742 was found to promote TSCC progression by regulating the miR-431-5p/ATF3 or miR-634/RAB1A axis (9,10). Interestingly, hsa_circ_0000003 (circ_0000003) is a poorly conserved circRNA that has been reported to facilitate cell proliferation, invasion and migration of non-small cell lung cancer (NSCLC) cells by regulating the miR-338-3p/insulin receptor substrate 2 (IRS2) axis (11). Although the tumor-promoting role of circ_0000003 has been reported in NSCLC (11), its role in other tumors has not yet been reported.
Recent studies have demonstrated that glutamine metabolism plays a vital role in the progression of various tumors, including lung adenocarcinoma, prostate cancer and B-lymphoma (12,13). In these tumors, not only tumor cell growth, but also tumor invasion and metastasis are promoted by dysregulated glutamate metabolism (12,13). Interestingly, some circRNAs have been shown to be involved in the regulation of glutamine metabolism (14–16). Among them are circHMGCS1, which was demonstrated to promote cell proliferation and glutaminolysis in hepatoblastoma cells by facilitating the expression of glutaminase (GLS), one of the key enzymes in glutamine metabolism (15), and circ_0016418, which was shown to facilitate glutamine catabolism in melanoma cells by upregulating GLS expression via interaction with miR-605-5p (16). The above studies suggest that some circRNAs may affect the occurrence and development of tumors by regulating glutamate metabolism. However, the mechanism of how glutamate metabolism influences tumor physiology remains unclear.
In the present study, the expression level of circ_0000003 in TSCC and its roles in TSCC progression were investigated. Furthermore, whether circ_0000003 exerts its functions by regulating glutamate metabolism via the miR-330-3p/GLS axis in TSCC was assessed.
Materials and methods
Clincical tissues
The clinical research was conducted with the approval of the Medical Ethics Committee of Taizhou University Hospital (approval no. 2018-092), and 40 TSCC patients at Taizhou University Hospital (Taizhou, Zhejiang, China) provided written consent to donate their cancerous tissues for this research before surgery. All patients were adults with no history of other cancers, and had not undergone any targeted therapeutic treatments prior to surgery. The 40 paired TSCC tissues and their paracancerous nontumor samples (>1 cm away from the edge of TSCC tissues) were collected between January 2015 and December 2017, and diagnosed by more than two pathologists. All the specimens were immediately snap-frozen in liquid nitrogen and preserved at −80°C before use.
Cell culture
Human TSCC cell lines (SCC25, SCC4, Cal27 and SCC1) were obtained from the American Type Culture Collection (ATCC), and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) and 1% Penicillin-Streptomycin Solution. Human oral keratinocytes (HOKs) were obtained from the ScienCell Research Laboratories (cat. no. 2610), and cultured in Oral Keratinocyte Medium (ScienCell Research Laboratories).
Construction of plasmid and cell transfection
For overexpression, circ_0000003 was cloned into mammalian expression vector pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) to construct overexpressing plasmid pcDNA3.1-circ_0000003 (p-circ_0000003). Empty pcDNA3.1 vector was used as a vector control. Short hairpin RNAs (shRNAs) targeting circ_0000003 (pLKO.1-sh-circ_0000003, sh-circ_0000003), negative control shRNAs (sh-NC), miR-330-3p inhibitors (UCUCUGCAGGCCGUGUGCUUUGC) (17), miRNA inhibitor negative control (miRNA inhibitor-NC; CAGUACUUUUGUGUAGUACAA) (18), miR-330-3p mimics (GCAAAGCACACGGCCUGCAGAGA) (17) and miRNA scrambled control (control mimics; UUCUCCGAACGUGUCACGUTT) (18) were all synthesized by GenePharma. All the obtained constructs were subsequently transfected via Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers protocol.
Cell proliferation assay
A total of 5×103 transfected TSCC cells were seeded into 96-well plates. After incubation for 0, 24, 48, 72 and 96 h, the medium was discarded, and the fresh medium containing 10 µl Cell Counting Kit-8 (CCK-8) solution (Beyotime Institute of Biotechnology) was mixed and pipetted into the TSCC cells. After reaction for 4 h, the absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.).
Cell invasion and migration assays
Transwell chambers (Corning Inc.) precoated with Matrigel (BD Biosciences) were used to detect the TSCC cell invasion ability. For cell invasion assay, TSCC cells were seeded into the upper chamber of Matrigel-coated inserts and incubated for 48 h. The invasive TSCC cells were fixed in 70% ethanol at room temperature and stained with 0.1% crystal violet for 30 min. Then the stained cells were lysed with 200 µl of lysis reagent, and 100 µl of lysate was pipetted into a 96-well plate to detect the absorbance value at 560 nm using a microplate reader (550; Bio-Rad).
Transwell assay without Matrigel was performed to detect the TSCC cell migration ability. For cell migration detection, TSCC cells were resuspended into a single-cell suspension in 250 µl DMEM containing 1% FBS, and then 5×104 cells per well were pipetted into uncoated 8-µm transwell filter inserts (Corning Inc.) in 24-well plates in triplicate. DMEM (500 µl) containing 15% FBS was added into the lower chambers as a chemoattractant. The TSCC cells were incubated for 16 h, and the non-migratory TSCC cells in the upper chamber were wiped away gently with a cotton swab. Then the migratory TSCC cells on the bottom side of the upper chamber were fixed in 100% methanol at room temperature and stained with 0.5 µg/ml 4,6-diamidino-2-phenylindole (DAPI) for 5 min. Finally the stained cells were counted with a fluorescence microscope (original magnification, ×200; Eclipse 80i; Nikon Corp.) in five random fields.
Glutamine, α-ketoglutarate (α-KG) and ATP assays
Glutamine consumption, α-KG and ATP production were tested as detailed previously (19). Glutamine consumption levels were analyzed using the glutamine assay kit (BioVision Inc.). α-KG production levels were detected with the fluorescence-based lactate assay kit (BioVision Inc.). ATP production levels were detected with the enhanced ATP assay kit (Beyotime Institute of Biotechnology). All the above kits were operated according to the manufacturers protocol.
Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
Total RNA of TSCC cells and tissues was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific Inc.), and then the first-strand cDNA was synthesized using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara). RT-qPCR was performed using the SYBR Green qPCR Kit (Thermo Fisher Scientific Inc.) with a StepOne™ Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific Inc.). The thermocycling conditions were: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 55°C for 30 sec, and 72°C for 30 sec. Relative gene expression was calculated using the 2−ΔΔCq method (20). GAPDH and U6 were respectively used as control references for circRNA/mRNA and miRNA. Their primers were purchased from Sangon Biotech, and the sequences as described in previous studies (11,21–25) are listed in Table I.
Table I.
RT-qPCR primer sequences.
| Name | Primer sequences |
|---|---|
| circ_0000003 | F: 5′-TTGACTGGGCCAATACCAGC-3′ |
| R: 5′-ACAGGAAGGCGTTCAGTGTG-3′ | |
| miR-31 | F: 5′-UAGCAGCACAGAAAUAUUGGC-3′ |
| R: 5′-CAAUAUUUCUGUGCUGCUAUU-3′ | |
| miR-197 | F: 5′-CGGTAGTCTGATACTGTAA-3′ |
| R: 5′-GTGCTCCGAAGGGGGT-3′ | |
| miR-210 | F: 5′-TAATATAGCCCCTGCCCACC-3′ |
| R: 5′-TATGCTTGTTCTCGTCTCTGTGTC-3′ | |
| miR-330-3p | F: 5′-ACACTCCAGCTGGGAGAGACGTCCGGCACAC-3′ |
| R: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGTTTCGT-3′ | |
| miR-338-3p | F: 5′-GTGTGGCTCTGAGGTCCTTG-3′ |
| R: 5′-ATCCGTGAAGTTGTTCGTGG-3′ | |
| GLS | F: 5′-TTCCAGAAGGCACAGACATGGTTG-3′ |
| R: 5′-GCCAGTGTCGCAGCCATCAC-3′ | |
| GAPDH | F: 5′-GCACCGTCAAGGCTGAGAAC-3′ |
| R: 5′-GCCTTCTCCATGGTGGTGAA-3′ | |
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
F, forward; R, reverse; GLS, glutaminase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blot analysis
RIPA buffer (Beyotime Institute of Biotechnology) was used to extract the total protein from the TSCC cells, which was then quantified using the BCA Kit (Pierce; Thermo Fisher Scientific Inc.). Then the protein samples were separated by 10% SDS-PAGE electrophoresis, and transferred onto a PVDF membrane (Millipore Corp.). After blocking with 5% defatted milk powder, the membrane was incubated overnight at 4°C with specific primary antibodies (both from Abcam) against glutaminase (dilution 1:1,000, cat. no. ab156876), and GAPDH (dilution 1:1,000, cat. no. ab181602). Next, the membranes were incubated with horseradish peroxidase-conjugated rabbit secondary antibody (dilution 1:2,000, cat. no.7074, Cell Signaling Technology) for 1 h at room temperature. The immunoreactive bands were visualized by enhanced chemiluminescence ECL kit, and then scanned by a LAS-4000 imaging system (Fujifilm).
Bioinformatics analysis
The interaction between circ_0000003 and miRNAs was predicted using Circular RNA Interactome (https://circinteractome.nia.nih.gov/). miR-330-3p that may interact with GLS mRNA 3-untranslated region (UTR) was searched using miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/).
Luciferase reporter assay
The partial sequences of circ_0000003 or GLS 3′-UTR, which contain the putative miR-330-3p-binding site, were amplified via PCR and constructed into the pmirGLO Luciferase vector (Promega Corp.) to generate wild-type circ_0000003 reporter (circ_0000003-WT) or GLS reporter (GLS-WT). The GeneArt™ Site-Directed Mutagenesis System (Thermo Fisher Scientific, Inc.) was used to produce miR-330-3p target site-mutation circ_0000003 (circ_0000003-MUT) reporter or miR-330-3p target site-mutation GLS 3′-UTR (GLS-MUT) reporter. All constructs were verified via DNA sequencing. Subsequently, the luciferase reporters and miR-330-3p mimic or control mimic were co-transfected into the cells using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were collected at 48 h after transfection, and then the relative firefly luciferase activities were measured using a Dual-Glo luciferase assay system (Promega Corp.) following the manufacturers instructions. Firefly luciferase activities were normalized to Renilla luciferase activities.
Pull-down assay with biotinylated miR-330-3p
TSCC cell lysates were collected using RIPA buffer plus RNase inhibitor (Promega Corp.), followed by transfection with biotin-labeled wild-type (WT) miR-330-3p (Bio-miR-330-3p-WT), mutated (MUT) miR-330-3p (Bio-miR-330-3p-MUT) or antagonistic miR-330-3p probe (Bio-NC-probe), which were all designed and synthesized by GenePharma. Then, the TSCC cell lysates were mixed with M-280 streptavidin magnetic beads (Sigma-Aldrich; Merck KGaA) for 3 h at 4°C. The pull-down products were subjected to RT-qPCR for circ_0000003 expression.
RNA immunoprecipitation (RIP) assay
RIP assay was carried out using the RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore Corp.). In brief, TSCC cells were lysed in RIP lysis buffer, followed by incubation with anti-Ago2 antibody (catalog no.ab57113, Abcam) and protein G magnetic beads. After 6 washes, the immunocomplexes bound by Ago2 were eluted, and then incubated with proteinase K at 55°C for 30 min to digest the proteins, followed by RNA extraction and RT-qPCR analysis for the expression of circ_0000003 or GLS mRNA.
Statistical analysis
All data were statistically analyzed via SPSS 25.0 (IBM Corp.). Paired t-tests were used in the comparisons between TSCC tissues and their adjacent normal tissues. Unpaired t-tests were used in the comparisons between two groups of TSCC cells. Differences among ≥3 groups were assessed using one-way ANOVA followed by Tukeys post-hoc test. The overall survival curve was generated and assessed by Kaplan-Meier and log-rank tests. The associations between circ_0000003 expression and clinicopathological parameters of TSCC patients were assessed using the χ2 test (for analyzing sex and age) or Fishers exact test (for analyzing TNM stage and tumor size) in Table II. Among circ_0000003, miR-330-3p and GLS mRNA, their linear correlations were assessed using Pearsons correlation analysis and two-tailed t-test.
Table II.
Correlation between circ_0000003 and the clinicopathological features of the TSCC patients (N=40).
| Clinicopathological | |||||
|---|---|---|---|---|---|
| Circ_0000003 factors | All patients | Low level | High level | χ2 | P-value |
| Age (years) | |||||
| ≤55 | 17 | 9 | 8 | 0.102 | 0.749 |
| >55 | 23 | 11 | 12 | ||
| Sex | |||||
| Male | 24 | 11 | 13 | 0.417 | 0.519 |
| Female | 16 | 9 | 7 | ||
| TNM stage | |||||
| I+II | 18 | 13 | 5 | 6.465 | 0.011a |
| III+IV | 22 | 7 | 15 | ||
| Tumor size (cm) | |||||
| ≤2 | 21 | 14 | 7 | 4.912 | 0.027a |
| >2 | 19 | 6 | 13 | ||
The χ2 test was used for comparison between groups
P<0.05 indicates a statistically significant result. TSCC, tongue squamous cell carcinoma; TNM, Tumor-Node-Metastasis.
Results
Circ_0000003 is upregulated in TSCC and promotes TSCC cell proliferation
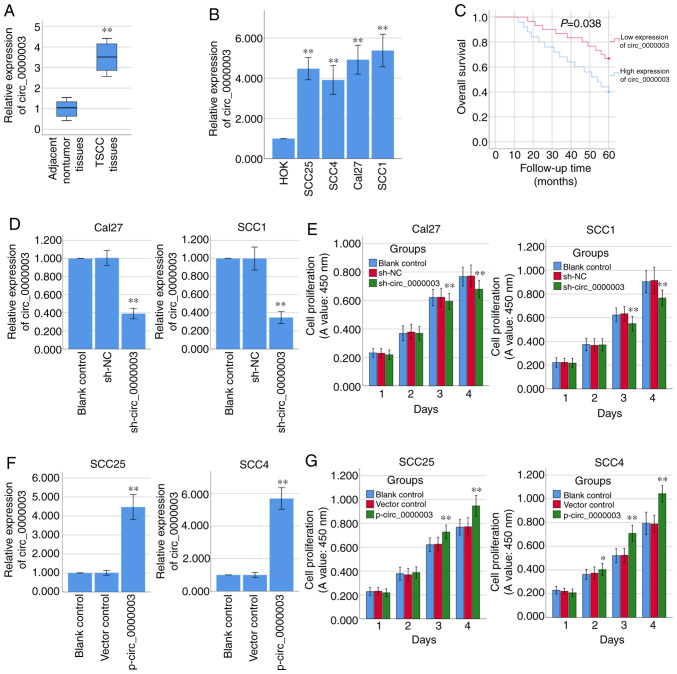
To determine the role of circ_0000003 in TSCC, we tested circ_0000003 expression levels in 40 pairs of TSCC tissues and their paired adjacent normal tissues. As shown in Fig. 1A, circ_0000003 expression was significantly increased in the TSCC tissues compared with that noted in the paired adjacent normal tissues. We also examined circ_0000003 expression levels in HOKs and TSCC cell lines (SCC25, SCC4, Cal27 and SCC1), and found that circ_0000003 expression was significantly increased in the TSCC cells when compared with the HOKs cells (Fig. 1B). The above data revealed that circ_0000003 is highly expressed in TSCC tissues and cell lines. Furthermore, the clinicopathological characteristics of the 40 TSCC patients are shown in Table II. High expression of circ_0000003 was found to be significantly correlated with advanced TNM stage and increased tumor size. In addition, high expression of circ_0000003 predicted a poor patient prognosis (Fig. 1C).
Figure 1.
Circ_0000003 is upregulated in TSCC tissues and cell lines and promotes TSCC cell proliferation in vitro. (A) Circ_0000003 level was examined in TSCC tissues (n=40) and their paired adjacent nontumor tissues (n=40). **P<0.01 vs. the nontumor tissues. (B) Circ_0000003 level was measured in HOK and TSCC cells (SCC25, SCC4, Cal27 and SCC1). **P<0.01 vs. the HOK. (C) Survival rates of TSCC patients with high and low circ_0000003 expression were analyzed via Kaplan-Meier survival analysis. (D) Circ_0000003 shRNA (sh-circ_0000003) transfection silenced the circ_0000003 expression in Cal27 and SCC1 cells. (E) CCK-8 assay was conducted to detect the cell proliferative ability of Cal27 and SCC1 cells with circ_0000003 knockdown. (F) p-circ_0000003 transfection enforced the circ_0000003 expression in SCC25 and SCC4 cells. (G) CCK-8 assay was conducted to detect the cell proliferative ability of SCC25 and SCC4 cells with circ_0000003 overexpression. *P<0.05 and **P<0.01 vs. controls. circ_0000003, hsa_circ_0000003; TSCC, tongue squamous cell carcinoma; HOK, human oral keratinocytes; sh-circ_0000003, circ_0000003 shRNA; CCK-8, Cell Counting Kit-8; p-circ_0000003, pcDNA3.1-circ_0000003; sh, shRNA; NC, negative control.
Loss-of-function and gain-of-function experiments were performed to determine the biological functions of circ_0000003. RT-qPCR analysis confirmed that circ_0000003 was significantly knocked down via sh-circ_0000003 in the Cal27 and SCC1 cells (Fig. 1D), and CCK-8 assay showed that circ_0000003 knockdown significantly repressed the cell proliferation of Cal27 and SCC1 cells (Fig. 1E). Meanwhile, circ_0000003 was overexpressed via p-circ_0000003 in SCC25 and SCC4 cells (Fig. 1F), and circ_0000003 overexpression significantly promoted the cell proliferation of SCC25 and SCC4 cells (Fig. 1G).
Circ_0000003 regulates the tumor phenotype of TSCC cells by targeting miR-330-3p/GLS axis
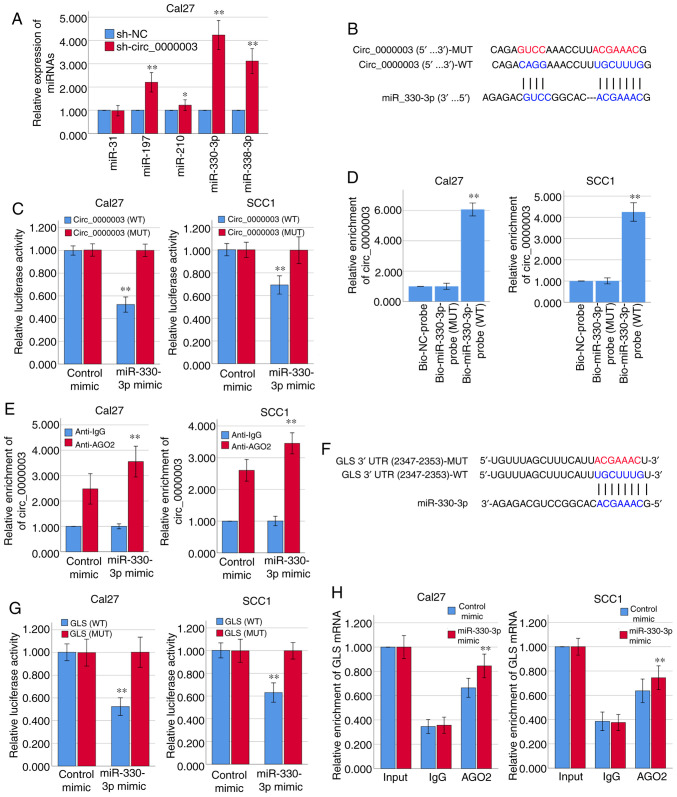
Bioinformatics analysis via Circular RNA Interactome (https://circinteractome.nia.nih.gov/) indicated that the top five predicted potential target miRNAs of circ_0000003 are miR-31, miR-197, miR-210, miR-330-3p and miR-338-3p (Table III). The results of RT-qPCR demonstrated that circ_0000003 knockdown in Cal27 cells facilitated the expression of miR-197, miR-210, miR-330-3p and miR-338-3p, respectively, among which the increase in miR-330-3p expression was the most obvious (Fig. 2A). The binding sites that miR-330-3p may share with circ_0000003 are showed in Fig. 2B. Subsequently, the results from the dual-luciferase reporter assay verified that miR-330-3p could bind with circ_0000003 at the molecular level in Cal27 and SCC1 cells (Fig. 2C).
Table III.
Predicted targets of circ_0000003.
| CircRNA Mirbase ID | CircRNA (Top)-miRNA (Bottom) pairing | Site type | CircRNA start | CircRNA end | 3′ pairing | Local AU | Position | TA | SPS | Context + score | Context + score percentile |
|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0000003 (5′ … 3′) | 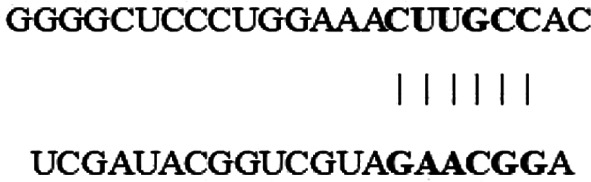 |
7mer-1a | 16762 | 16768 | 0.001 | 0.062 | 0.114 | 0.003 | −0.047 | 0.059 | 5 |
| hsa-miR-31 (3′ … 5′) | |||||||||||
| hsa_circ_0000003 (5′ … 3′) | 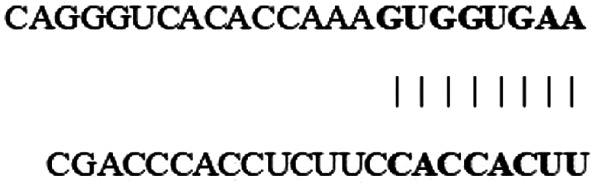 |
8mer-1a | 19032 | 19039 | −0.008 | −0.016 | 0.265 | 0.018 | −0.068 | −0.056 | 71 |
| hsa-miR-197 (3′ … 5′) | |||||||||||
| hsa_circ_0000003 (5′ … 3′) | 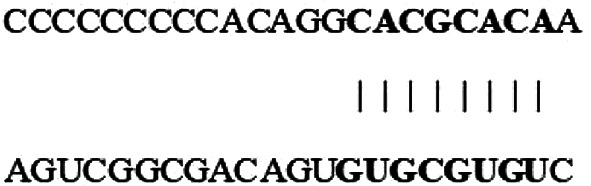 |
8mer-1a | 23553 | 23560 | 0.024 | 0.184 | 0.265 | −0.116 | −0.058 | 0.052 | 3 |
| hsa-miR-210 (3′ … 5′) | |||||||||||
| hsa_circ_0000003 (5′ … 3′) | 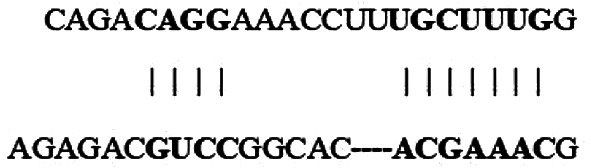 |
7mer-m8 | 1836 | 1842 | −0.025 | 0.058 | 0.151 | 0.020 | 0.023 | 0.107 | 14 |
| hsa-miR-330-3p (3′ … 5′) | |||||||||||
| hsa_circ_0000003 (5′ … 3′) |  |
7mer-m8 | 128 | 134 | −0.016 | 0.032 | −0.044 | 0.011 | −0.032 | −0.169 | 90 |
| hsa-miR-338-3p (3′ … 5′) |
Figure 2.
Circ_0000003 modulates the tumor phenotype of TSCC cells via the miR-330-3p/GLS axis. (A) After transfection of sh-circ_0000003, expression levels of miR-31, miR-197, miR-210, miR-330-3p and miR-338-3p were detected via RT-qPCR. *P<0.05 and **P<0.01 vs. sh-NC. (B) Bioinformatics analysis suggested that miR-330-3p may share binding sites with circ_0000003. (C) Luciferase reporter assay indicated the molecular level combination of miR-330-3p and circ_0000003. **P<0.01 vs. circ_0000003(MUT). (D) Circ_0000003 was pulled down by biotinylated miR-330-3p-WT. **P<0.01 vs. Bio-miR-330-3p-probe(MUT). (E) AGO2-RIP followed by RT-qPCR to detect circ_0000003 enrichment level after miR-330-3p overexpression via miR-330-3p mimics. **P<0.01 vs. Anti-IgG. (F) Bioinformatics analysis suggested the associability within miR-330-3p and GLS. (G) Luciferase reporter assay indicated the covalent targeting of miR-330-3p with GLS mRNA 3′-UTR. **P<0.01 vs. GLS(MUT). (H) RIP assays using antibodies against AGO2 or IgG were performed in cellular lysates from Cal27 and SCC1 cells, and RT-qPCR demonstrated the relative enrichment of GLS mRNA in Cal27 and SCC1 cells transfected with miR-330-3p or control mimics. **P<0.01 vs. control mimic. circ_0000003, hsa_circ_0000003; TSCC, tongue squamous cell carcinoma; sh-circ_0000003, circ_0000003 shRNA; qRT-PCR, Reverse transcription and quantitative polymerase chain reaction; WT, wild-type; RIP, RNA immunoprecipitation; GLS, glutaminase; 3′-UTR, 3-untranslated region; MUT, mutant type.
For further verification of the interaction between miR-330-3p and circ_0000003, RNA pull-down assay was conducted using biotinylated miR-330-3p-WT and miR-330-3p-MUT probes. As shown in Fig. 2D, only miR-330-3p-WT could bind and precipitate circ_0000003 in the cell lysates of Cal27 and SCC1 cells. However, there was no interaction between circ_0000003 and miR-330-3p-MUT or biotin-labeled antagonistic miR-330-3p probe (Bio-NC-probe). To determine whether circ_0000003 binds miR-330-3p in an AGO2-dependent manner, we conducted anti-AGO2 RIP in Cal27 and SCC1 cells overexpressing miR-330-3p via miR-330-3p mimics, and found that circ_0000003 enrichment was significantly increased after overexpression of miR-330-3p in Cal27 and SCC1 cells (Fig. 2E).
Furthermore, the two online bioinformatics databases, miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/), suggested that GLS mRNA 3′-UTR had the high associability with miR-330-3p (Fig. 2F; Table IV). Luciferase reporter assay verified that miR-330-3p covalently targeted GLS mRNA 3′-UTR in Cal27 and SCC1 cells (Fig. 2G). Moreover, RIP assay revealed a higher level of GLS mRNA enrichment in the AGO2 group after overexpression of miR-330-3p in Cal27 and SCC1 cells (Fig. 2H). Therefore, we here found that circ_0000003 has a direct effect on miR-330-3p, and miR-330-3p has a direct effect on GLS mRNA 3′-UTR.
Table IV.
Prediction of binding sites of GLS 3′-UTR and mir-330-3p.
| Predicted consequential pairing of target region (top) and miRNA (bottom) | Site type | Context++ score | Context++ score percentile | Weighted context++ score | Conserved branch length | PCT | |
|---|---|---|---|---|---|---|---|
| Position 2347–2353 of GLS 3′-UTR | 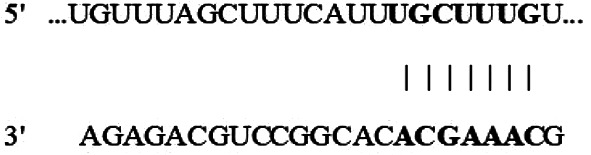 |
7mer-m8 | −0.11 | 92 | −0.11 | 3.834 | N/A |
| hsa-miR-330-3p |
Association of circ_0000003, miR-330-3p and GLS in TSCC
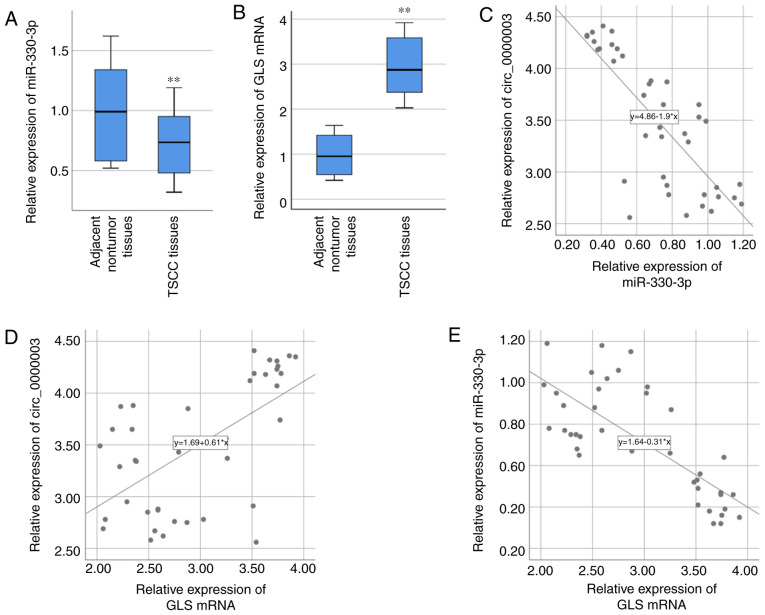
Based on the above bioinformatics analyses and related verification experiments, there may be certain interactions among circ_0000003, miR-330-3p and GLS. Therefore, miR-330-3p and GLS were chosen for our further studies. miR-330-3p expression in the 40 specimens of TSCC tissues was examined via RT-qPCR, and the result demonstrated that miR-330-3p expression was significantly lower in TSCC tissues compared with that noted in the adjacent nontumor tissues (Fig. 3A). GLS mRNA expression in the 40 specimens of TSCC tissues was also examined via RT-qPCR, and the result demonstrated that GLS mRNA expression was significantly higher in TSCC tissues compared with that found in the paired adjacent normal tissues (Fig. 3B). After statistically analyzing the associations of their expressions, circ_0000003 expression was found to be negatively correlated with miR-330-3p expression (R=−0.783, P<0.01, Fig. 3C), while positively correlated with GLS mRNA expression (R=0.606, P<0.01, Fig. 3D). Moreover, miR-330-3p expression was negatively correlated with GLS mRNA expression (R=−0.753, P<0.01, Fig. 3E).
Figure 3.
Correlations between circ_0000003, miR-330-3p and GLS in TSCC. (A) miR-330-3p expression and (B) GLS mRNA expression were detected by RT-qPCR in the TSCC tissues and compared with that in the adjacent nontumor tissues. **P<0.01 vs. the nontumor tissues. (C-E) Bivariate correlation analysis of (C) circ_0000003 and miR-330-3p, (D) circ_0000003 and GLS mRNA, and (E) miR-330-3p and GLS mRNA. circ_0000003, hsa_circ_0000003; GLS, glutaminase; TSCC, tongue squamous cell carcinoma; RT-qPCR, reverse transcription and quantitative polymerase chain reaction.
Circ_0000003 downregulates miR-330-3p expression and upregulates GLS expression in TSCC cells
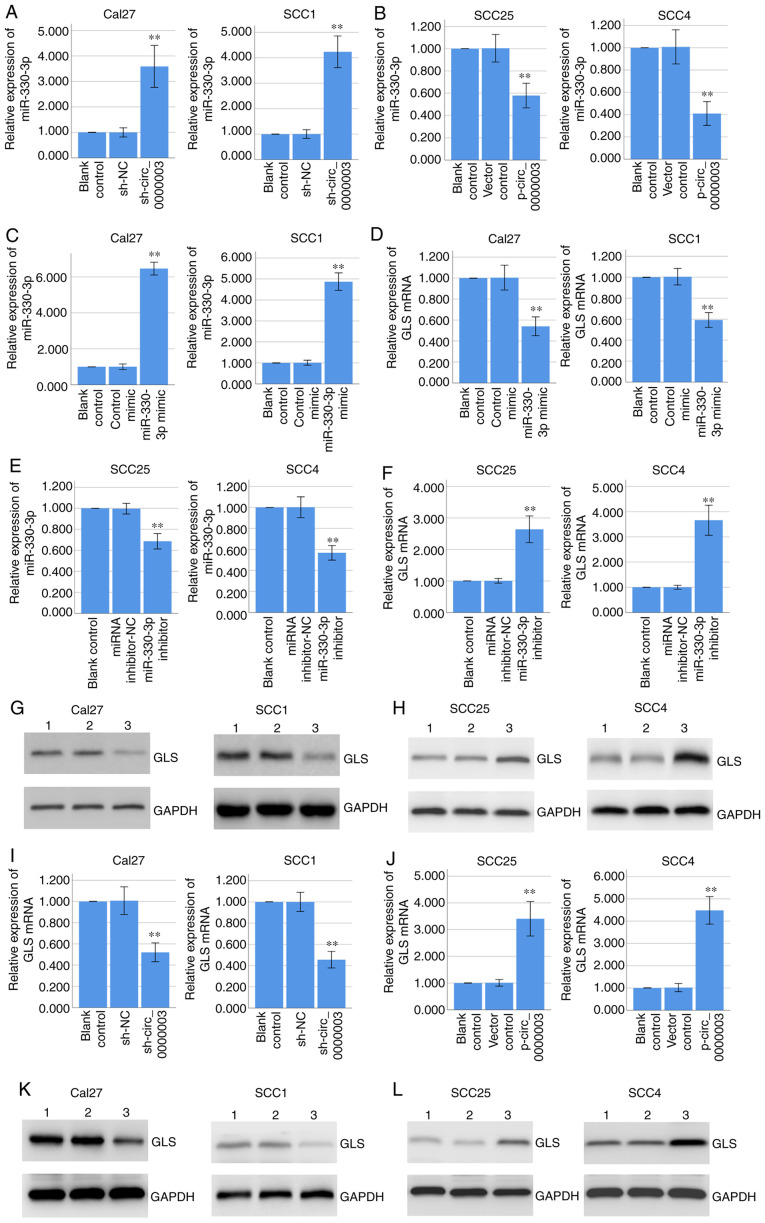
RT-qPCR assay demonstrated that knockdown of circ_0000003 significantly promoted miR-330-3p expression in Cal27 and SCC1 cells (Fig. 4A), while overexpression of circ_0000003 significantly inhibited miR-330-3p expression in the SCC25 and SCC4 cells (Fig. 4B). Furthermore, RT-qPCR assay indicated that overexpression of miR-330-3p via miR-330-3p mimics significantly decreased GLS mRNA expression in Cal27 and SCC1 cells (Fig. 4C and D), while inhibition of miR-330-3p via miR-330-3p inhibitors significantly increased GLS mRNA expression in the SCC25 and SCC4 cells (Fig. 4E and F). Coincidentally, western blot analysis demonstrated that overexpression of miR-330-3p decreased GLS protein expression in Cal27 and SCC1 cells (Fig. 4G), while inhibition of miR-330-3p increased GLS protein expression in SCC25 and SCC4 cells (Fig. 4H).
Figure 4.
Circ_0000003 regulates miR-330-3p and GLS expressions, and miR-330-3p regulates GLS expression. (A and B) RT-qPCR determined the miR-330-3p expression in Cal27 and SCC1 cells transfected with sh-NC or sh-circ_0000003, and in SCC25 and SCC4 cells transfected with empty vector or p-circ_0000003. (C and D) RT-qPCR determined the miR-330-3p and GLS mRNA expression in Cal27 and SCC1 cells transfected with control mimic or miR-330-3p mimic. (E and F) RT-qPCR determined the miR-330-3p and GLS mRNA expression in SCC25 and SCC4 cells transfected with miRNA inhibitor-NC or miR-330-3p inhibitor. (G) Western blot analysis determined the GLS protein expression in Cal27 and SCC1 cells transfected with control mimic or miR-330-3p mimic. Lane 1, blank control; lane 2, control mimic; lane 3, miR-330-3p mimic. (H) Western blot analysis determined the GLS protein expression in SCC25 and SCC4 cells transfected with miRNA inhibitor-NC or miR-330-3p inhibitor. Lane 1, blank control; lane 2, miRNA inhibitor-NC; lane 3, miR-330-3p inhibitor. (I and J) RT-qPCR determined the GLS mRNA expression in Cal27 and SCC1 cells transfected with sh-NC or sh-circ_0000003, and in SCC25 and SCC4 cells transfected with empty vector or p-circ_0000003. (K) Western blot analysis determined the GLS protein expression in Cal27 and SCC1 cells transfected with sh-NC or sh-circ_0000003. Lane 1, blank control; Lane 2, sh-NC; Lane 3, sh-circ_0000003. (L) Western blot analysis determined the GLS protein expression in SCC25 and SCC4 cells transfected with empty vector or p-circ_0000003. Lane 1, blank control; lane 2, vector control; lane 3, p-circ_0000003. **P<0.01 vs. controls. circ_0000003, hsa_circ_0000003; GLS, glutaminase; RT-qPCR, reverse transcription and quantitative polymerase chain reaction; sh, shRNA; miRNA, microRNA; NC, negative control; sh-circ_0000003, circ_0000003 shRNA; p-circ_0000003, pcDNA3.1-circ_0000003.
Coincidentally, RT-qPCR assay indicated that knockdown of circ_0000003 significantly decreased GLS mRNA expression in Cal27 and SCC1 cells (Fig. 4I), while overexpression of circ_0000003 significantly increased GLS mRNA expression in the SCC25 and SCC4 cells (Fig. 4J). Coincidentally, western blot analysis demonstrated that circ_0000003 knockdown decreased GLS protein expression in Cal27 and SCC1 cells (Fig. 4K), while circ_0000003 overexpression increased GLS protein expression in SCC25 and SCC4 cells (Fig. 4L). Overall, our results indicated that circ_0000003 downregulated miR-330-3p expression and upregulated GLS expression, and miR-330-3p downregulated GLS expression, suggesting circ_0000003 may promote GLS expression by restraining miR-330-3p.
Circ_0000003 regulates cell invasion and migration in TSCC cells
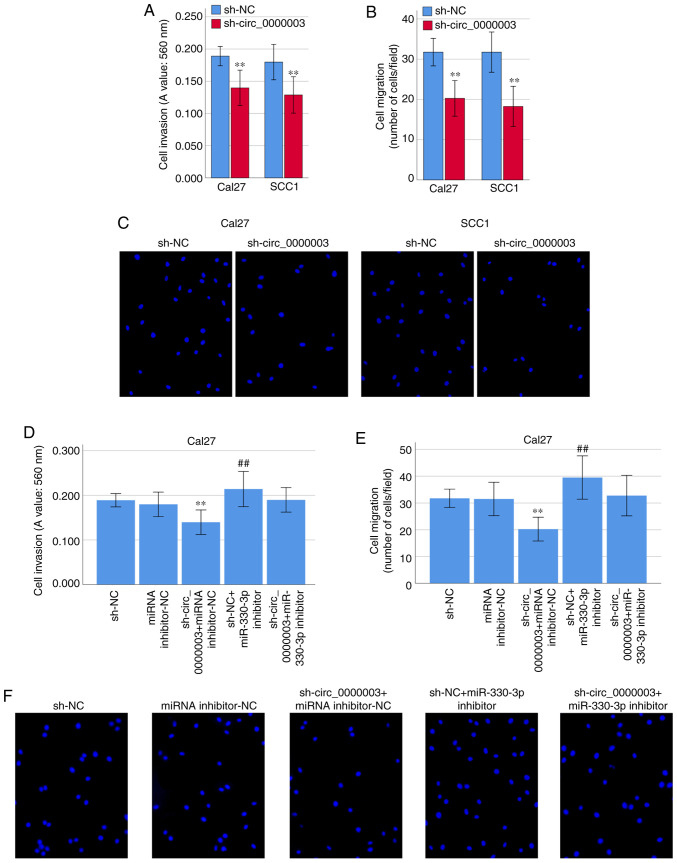
Interestingly, transfection of sh-circ_0000003 caused a dramatic reduction in cell invasion (Fig. 5A) and migration (Fig. 5B and C). To clarify whether miR-330-3p was involved in the inhibition of cell invasion and migration capability elicited by circ_0000003 knockdown, co-transfection experiments were performed in Cal27 cells. As shown in Fig. 5D-F, co-transfection of sh-NC + miR-330-3p inhibitor resulted in significantly increased cell invasion and migration capability. Meanwhile, the transfection of miR-330-3p inhibitor reversed the decrease in cell invasion and migration induced by sh-circ_0000003 (Fig. 5D-F). These results indicated that the cell invasion and migration were significantly restrained by circ_0000003 knockdown, and then it was reversed after transfection with miR-330-3p inhibitor in TSCC cells.
Figure 5.
Circ_0000003 facilitates cell invasion and migration capability in TSCC by inhibiting miR-330-3p. (A) Cell invasion levels in TSCC cells after circ_0000003 knockdown. (B) Cell migration levels in TSCC cells after circ_0000003 knockdown. (C) Representative images of the cell migration in Cal27 and SCC1 cells after circ_0000003 knockdown (original magnification, ×200). (D) Cell invasion and (E) migration capability in Cal27 cells transfected with sh-NC, miRNA inhibitor-NC, sh-circ_0000003 + miRNA inhibitor-NC, sh-NC + miR-330-3p inhibitor, and sh-circ_0000003 + miR-330-3p inhibitor. **P<0.01 vs. sh-NC; ##P<0.01 vs. miRNA inhibitor-NC. (F) Representative images of the cell migration in Cal27 cells after the transfections (original magnification, ×200). circ_0000003, hsa_circ_0000003; TSCC, tongue squamous cell carcinoma; sh-circ_0000003, circ_0000003 shRNA; sh, shRNA; NC, negative control.
Circ_0000003 regulates glutamine metabolism in TSCC cells
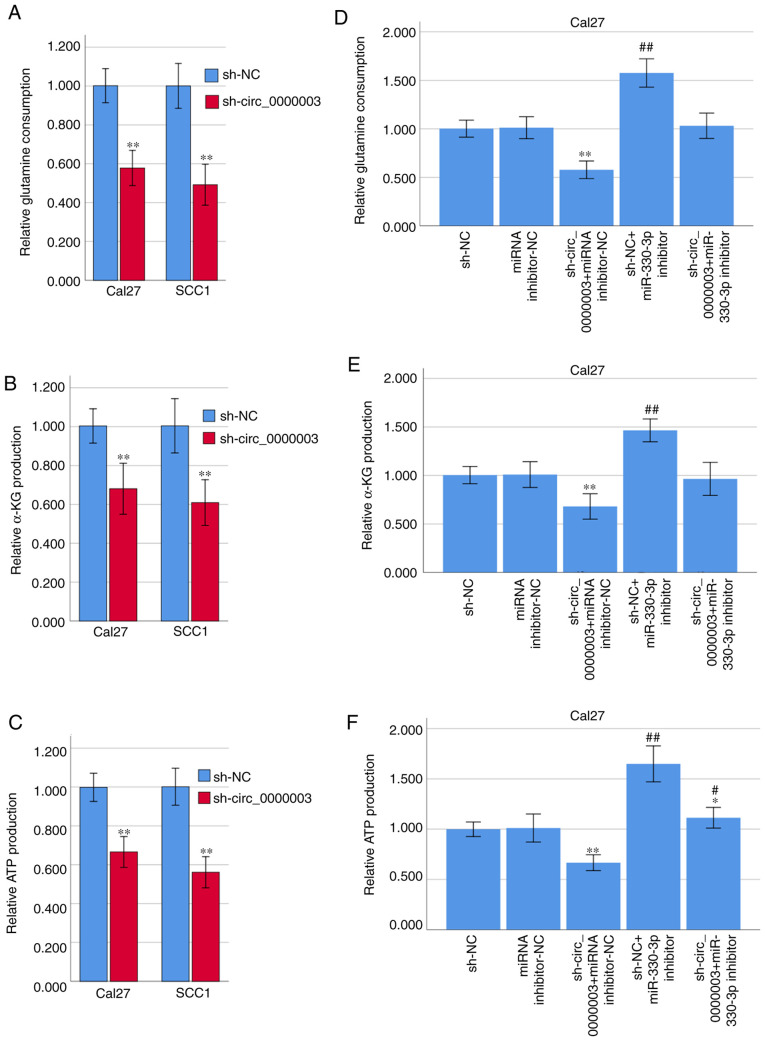
Since some circRNAs have been found to be involved in the progression of glutamine metabolism (14), we asked whether circ_0000003 could facilitate TSCC progression by regulating glutamine metabolism. Interestingly, transfection of sh-circ_0000003 caused a dramatic reduction in glutamine consumption, α-KG production and ATP production in TSCC cells (Fig. 6A-C). To clarify whether miR-330-3p was involved in the inhibition of glutamine metabolism elicited by circ_0000003 knockdown, co-transfection experiments were performed in Cal27 cells. As shown in Fig. 6D-F, co-transfection of sh-NC + miR-330-3p inhibitor resulted in significantly increased glutamine consumption, α-KG production, and ATP production. Meanwhile, miR-330-3p inhibitor transfection reversed the decrease in glutamine consumption, α-KG production and ATP production induced by sh-circ_0000003 transfection (Fig. 6D-F). These results indicated that the glutamine consumption, α-KG production and ATP production were significantly restrained by circ_0000003 knockdown, and then it was reversed after transfection with miR-330-3p inhibitor in TSCC cells.
Figure 6.
Circ_0000003 facilitates glutamine metabolism in TSCC by inhibiting miR-330-3p. (A) Glutamine consumption, (B) cellular α-KG production, and (C) ATP production levels in TSCC cells after circ_0000003 knockdown. (D) Glutamine consumption, (E) α-KG production, and (F) ATP production levels in Cal27 cells transfected with sh-NC, miRNA inhibitor-NC, sh-circ_0000003 + miRNA inhibitor-NC, sh-NC + miR-330-3p inhibitor, and sh-circ_0000003 + miR-330-3p inhibitor. *P<0.05, **P<0.01 vs. sh-NC; #P<0.05, ##P<0.01 vs. miRNA inhibitor-NC. circ_0000003, hsa_circ_0000003; TSCC, tongue squamous cell carcinoma; α-KG, α-ketoglutarate; ATP, adenosine triphosphate; sh-circ_0000003, circ_0000003 shRNA; sh, shRNA; NC, negative control.
Discussion
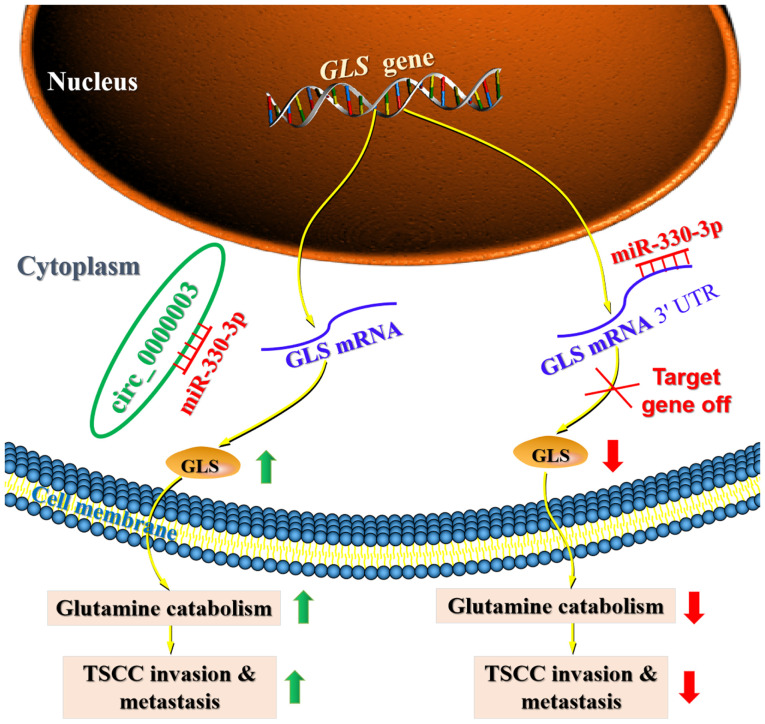
Recently, a number of circulating RNAs (circRNAs) have been revealed to participate in the progression of tongue squamous cell carcinoma (TSCC) by sponging miRNAs to regulate protein-coding target gene expression (7–9). Moreover, some previous studies have demonstrated that circRNAs play a regulatory role in tumor progression by regulating cell metabolism, including glucose metabolism, glutamine catabolism and lipid metabolism (14). However, to date, there is no report on how circRNAs affect TSCC progression by regulating cell metabolism. Our current findings provide evidence demonstrating the essential biological relevance of circRNAs in TSCC, and uncover that dysregulation of circ_0000003 may be responsible for TSCC progression. Importantly, the current research demonstrated that circ_0000003 elevated glutaminase (GLS) expression by sponging miR-330-3p, leading to promotion of TSCC glutamine catabolism, invasion and metastasis (Fig. 7).
Figure 7.
Proposed functional model demonstrating the oncogenic role of the circ_0000003/miR-330-3p/GLS axis in TSCC glutamine catabolism and tumorigenesis. The green upward arrows represent upregulation, and the red downward arrows depict downregulation. circ_0000003, hsa_circ_0000003; GLS, glutaminase; TSCC, tongue squamous cell carcinoma.
Circ_0000003 has been demonstrated to facilitate the cell proliferation, invasion and migration of NSCLC cells via targeting the miR-338-3p/IRS2 axis (11). However, circ_0000003 has not been systematically studied in other tumors. Because there are very few articles concerning circ_0000003, we deduced the hypothesis about circ_0000003 mainly through bioinformatics analysis and the study about circ_0000003 in NSCLC (11). Thus, we detected circ_0000003 expression in TSCC tissues and cell lines, and found that circ_0000003 was upregulated in TSCC tissues and cell lines, and high circ_0000003 expression was associated with poor patient prognosis and advanced tumor progression, suggesting that circ_0000003 has an oncogenic function in TSCC. Here, we used bioinformatics online analysis software to speculate possible targets for circ_0000003 to elucidate the potential mechanism of circ_0000003 in TSCC. Then, we found that miR-330-3p expression was strongly increased by circ_0000003 knockdown in TSCC cells compared with other potential targeting miRNAs. Therefore, miR-330-3p was chosen as a candidate for further research.
Several studies have revealed that miR-330-3p exerts a tumor-suppressive effect in various tumors (23,26–28). For instance, circ-0016068 restrained miR-330-3p expression and facilitated insertion region-1 (BMI-1) expression to promote the cell proliferation, invasion and migration of prostate cancer cells (23). Moreover, upregulation of miR-330-3p restricted the cell proliferation, colony formation and invasion of laryngeal squamous cell carcinoma (LSCC) cells by targeting transformer-2 protein homolog β (Tra2β) to repress Akt activation (27). In our current research, miR-330-3p expression was markedly reduced in TSCC tissues, and was negatively correlated with circ_0000003 expression in the TSCC tissues. More importantly, our present study revealed that circ_0000003 was a decoy for miR-330-3p, and circ_0000003 participated in the progression of TSCC by sponging miR-330-3p.
Increasing studies indicate that circRNAs contain miRNA binding sites and function as competing endogenous RNAs (ceRNAs) that sponge downstream miRNAs and restrain their expression and functions (11,29). For instance, circ_0000003 has been shown to sponge miR-338-3p and restrain miR-338-3p expression, leading to promotion of cell proliferation and invasion of NSCLC cells (11). In the current research, our gain- and loss-of-function experiments indicated that circ_0000003 knockdown facilitated miR-330-3p expression, whereas circ_0000003 overexpression repressed miR-330-3p expression. Moreover, our rescue experiments demonstrated that circ_0000003 facilitated cell proliferation, migration and invasion by counteracting the miR-330-3p-mediated effects on TSCC cells. Therefore, our current study presents strong evidence for a tumor-promoting role of the circ_0000003/miR-330-3p interaction in TSCC.
Increasing evidence has revealed that miRNAs can bind to the 3′-UTR of their downstream target genes, thereby inhibiting the target gene expression at the post-transcriptional levels (30). Thus, we here used two bioinformatics databases (miRDB and TargetScan) to speculate potential target genes for miR-330-3p, and found GLS may be a target gene of miR-330-3p. GLS is a key enzyme involved in the progression of glutamine metabolism, and functions as a tumor-promoting factor in various tumors (31). The knockdown of GLS repressed cell proliferation and induced apoptosis by inhibiting the Wnt/β-catenin pathway in prostate cancer cells (32). Moreover, circ_0016418 facilitated melanoma progression and glutamine catabolism by sponging miR-605-5p to promote GLS expression (16). In the current research, we verified that miR-330-3p directly bound to GLS 3′-UTR and negatively regulated GLS expression.
Aberrant energy metabolism, such as increased glutamine catabolism, is one of critical hallmarks of cancer (33,34). In addition to abnormal glycolysis, several previous studies revealed that enhanced glutamine metabolism was correlated with tumor metastasis and poor prognosis in patients with cancer (12,35). GLS is a rate-limiting enzyme for glutamine catabolism, catalyzing the hydrolysis of glutamine to glutamate (31,36). Interestingly, GLS has multiple effects in cancer cells, including maintaining mitochondrial metabolism, activating cell signaling, and promoting cancer cell growth (31,35,36). Moreover, dysfunction of glutamine metabolism has been found in oral squamous cell carcinoma (OSCC) and head and neck squamous cell carcinoma (HNSCC) (37,38). However, it is still unclear whether glutamine metabolism exerts a vital regulatory role in TSCC progression, and whether the glutamine metabolism-related molecules can be used as potential biomarkers or targets for TSCC diagnosis, treatment and prognosis. Therefore, glutamine metabolism may be a good entry point for future diagnosis and treatment of TSCC.
Several circRNAs have, interestingly, been shown to be involved in the progression of cell metabolism in various tumors (14). Recently, accumulating evidence has illuminated that altered glutamine catabolism exerts a critical effect on cell proliferation in tumor cells (39). Moreover, circHMGCS1 was found to regulate glutamine metabolism in hepatoblastoma cells by targeting the miR-503-5p/IGF/PI3K/Akt axis (15). However, there is no systematic study on the correlation between circRNAs and glutamine metabolism in TSCC. Therefore, we here explored how circ_0000003 regulated glutamine metabolism in TSCC. Herein, our current study demonstrated that circ_0000003 knockdown significantly repressed glutamine consumption, α-KG production and ATP production of TSCC cells, indicating that circ_0000003 knockdown impeded glutamine catabolism in TSCC cells. Moreover, in the current rescue experiments, miR-330-3p inhibitor reversed the reduction in glutamine consumption, α-KG production and ATP production induced by sh-circ_0000003 transfection in Cal27 cells, suggesting circ_0000003 knockdown may exert the inhibitory effects on the glutamine metabolism by facilitating miR-330-3p expression and function in TSCC.
In summary, this research work suggests a vital role for circ_0000003 in glutamine metabolism and tumor progression by binding and restraining miR-330-3p to facilitate GLS expression in TSCC. This work provides a novel mechanism for understanding cancer metabolism, and suggests that the circ_0000003/miR-330-3p/GLS axis may provide novel strategies for TSCC diagnosis, therapy and prognosis. However, there are some limitations that exist in the present study. First, the limited TSCC tissue samples might not fully substantiate the accuracy of the present results. Secondly, there is a lack of parallel studies on HOK cells as a control to detect cell proliferation and invasion. Furthermore, the role of circ_0000003 in TSCC was explored only in TSCC cell lines in vitro, but not in a TSCC mouse model in vivo. Finally, but perhaps extremely important, the relationships between circ_0000003 and other potential targeting miRNAs should be explored in future work.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY16H160033), the Basic Public Welfare Research Project of Zhejiang Province (2016C33189), the National Natural Science Foundation of China (81902138), the Zhejiang Provincial Medical Science and Technology Project of China (2019RC314), the National College Students Innovation and Entrepreneurship Training Program of China (202010350053), and the Science and Technology Project of Taizhou City (20ywa61).
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY16H160033), the Basic Public Welfare Research Project of Zhejiang Province (2016C33189), the National Natural Science Foundation of China (81902138), the Zhejiang Provincial Medical Science and Technology Project of China (2019RC314), the National College Students Innovation and Entrepreneurship Training Program of China (202010350053), and the Science and Technology Project of Taizhou City (20ywa61).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors contributions
JY designed the study. CQ, SC and SL contributed to the cell transfection, RT-qPCR, western blotting, RIP, RNA pull-down and luciferase assays. CQ performed the CKK-8 and Transwell assays. SC performed the glutamine, α-KG and ATP assays. YW collected the clinical data and follow-up data. JY, CQ and SC participated in writing and revising the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The research protocol conformed to the principles outlined in the Declaration of Helsinki. All patients provided written informed consent and the protocol of the study was approved by the Medical Ethics Committee of Taizhou University Hospital (Taizhou, Zhejiang, China) (approval no. 2018-092).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cohen N, Fedewa S, Chen AY. Epidemiology and demographics of the head and neck cancer population. Oral Maxillofac Surg Clin North Am. 2018;30:381–395. doi: 10.1016/j.coms.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Hussein AA, Forouzanfar T, Bloemena E, de Visscher J, Brakenhoff RH, Leemans CR, Helder MN. A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br J Cancer. 2018;119:724–736. doi: 10.1038/s41416-018-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Agostino S, Riccioli A, De Cesaris P, Fontemaggi G, Blandino G, Filippini A, Fazi F. Circular RNAs in embryogenesis and cell differentiation with a focus on cancer development. Front Cell Dev Biol. 2020;8:389. doi: 10.3389/fcell.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajappa A, Banerjee S, Sharma V, Khandelia P. Circular RNAs: Emerging role in cancer diagnostics and therapeutics. Front Mol Biosci. 2020;7:577938. doi: 10.3389/fmolb.2020.577938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei T, Ye P, Yu GY, Zhang ZY. Circular RNA expression profiling identifies specific circular RNAs in tongue squamous cell carcinoma. Mol Med Rep. 2020;21:1727–1738. doi: 10.3892/mmr.2020.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Bi L, Zhang C. Circular RNA_0001742 has potential to predict advanced tumor stage and poor survival profiles in tongue squamous cell carcinoma management. J Clin Lab Anal. 2020;34:e23330. doi: 10.1002/jcla.23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu YT, Li XX, Zeng LW. Circ_0001742 promotes tongue squamous cell carcinoma progression via miR-431-5p/ATF3 axis. Eur Rev Med Pharmacol Sci. 2019;23:10300–10312. doi: 10.26355/eurrev_201912_19668. [DOI] [PubMed] [Google Scholar]
- 10.Shao B, He L. Hsa_circ_0001742 promotes tongue squamous cell carcinoma progression via modulating miR-634 expression. Biochem Biophys Res Commun. 2019;513:135–140. doi: 10.1016/j.bbrc.2019.03.122. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Niu X, Li H, Liang Y, Sun Z, Yan Y. Circ_0000003 promotes the proliferation and metastasis of non-small cell lung cancer cells via miR-338-3p/insulin receptor substrate 2. Cell Cycle. 2019;18:3525–3539. doi: 10.1080/15384101.2019.1690883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kery M, Papandreou I. Emerging strategies to target cancer metabolism and improve radiation therapy outcomes. Br J Radiol. 2020;93:20200067. doi: 10.1259/bjr.20200067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. CircRNAs in cancer metabolism: A review. J Hematol Oncol. 2019;12:90. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhen N, Gu S, Ma J, Zhu J, Yin M, Xu M, Wang J, Huang N, Cui Z, Bian Z, et al. CircHMGCS1 promotes hepatoblastoma cell proliferation by regulating the IGF signaling pathway and glutaminolysis. Theranostics. 2019;9:900–919. doi: 10.7150/thno.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Zhang X, Li X, Wan X. Circ_0016418 promotes melanoma development and glutamine catabolism by regulating the miR-605-5p/GLS axis. Int J Clin Exp Pathol. 2020;13:1791–1801. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H, Hu Y, Wang C, Zhang X, He D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death Dis. 2020;11:284. doi: 10.1038/s41419-020-2447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Sun YY, Zhang ZX, Li YH, Xu ZM, Fu WN. Transcriptional suppression of microRNA-27a contributes to laryngeal cancer differentiation via GSK-3β-involved Wnt/β-catenin pathway. Oncotarget. 2017;8:14708–14718. doi: 10.18632/oncotarget.14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen G, Chen X, Xin H, Tang C, Zeng J. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8:113650–113661. doi: 10.18632/oncotarget.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Xu L, Yang T. miR-31 Modulates liver cancer HepG2 cell apoptosis and Invasion via ROCK1/F-Actin Pathways. Onco Targets Ther. 2020;13:877–888. doi: 10.2147/OTT.S227467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Wang W, Zhang M, Sun W, Shi W, Li F. Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:827. doi: 10.3389/fcell.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Zhang Y, Zhou YM, Zhang ZJ, Li X. miR-210 is a serological biomarker for predicting recurrence and prognosis of colon carcinoma patients with liver metastases after radiofrequency ablation treatment. Cancer Manag Res. 2020;12:9077–9085. doi: 10.2147/CMAR.S267731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Wang L, Mao S, Liu M, Zhang W, Zhang Z, Guo Y, Huang B, Yan Y, Huang Y, Yao X. miR-1-3p contributes to cell proliferation and invasion by targeting glutaminase in bladder cancer cells. Cell Physiol Biochem. 2018;51:513–527. doi: 10.1159/000495273. [DOI] [PubMed] [Google Scholar]
- 26.He D, Yang X, Kuang W, Huang G, Liu X, Zhang Y. The Novel Circular RNA Circ-PGAP3 promotes the proliferation and invasion of triple negative breast cancer by regulating the miR-330-3p/Myc axis. Onco Targets Ther. 2020;13:10149–10159. doi: 10.2147/OTT.S274574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Y, Zhu H, Gao W. MicroRNA-330-3p represses the proliferation and invasion of laryngeal squamous cell carcinoma through downregulation of Tra2β-mediated Akt signaling. Mol Cell Probes. 2020;52:101574. doi: 10.1016/j.mcp.2020.101574. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Liu G, Li T, Wang N, Wu J, Zhi H. MiR-330-3p functions as a tumor suppressor that regulates glioma cell proliferation and migration by targeting CELF1. Arch Med Sci. 2020;16:1166–1175. doi: 10.5114/aoms.2020.95027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren S, Lin P, Wang J, Yu H, Lv T, Sun L, Du G. Circular RNAs: Promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol Ther Methods Clin Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanan R, Schratt G. miRNA regulation of social and anxiety-related behaviour. Cell Mol Life Sci. 2020;77:4347–4364. doi: 10.1007/s00018-020-03542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masisi BK, El Ansari R, Alfarsi L, Rakha EA, Green AR, Craze ML. The role of glutaminase in cancer. Histopathology. 2020;76:498–508. doi: 10.1111/his.14014. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Mao S, Guo Y, Wu Y, Yao X, Huang Y. Inhibition of GLS suppresses proliferation and promotes apoptosis in prostate cancer. Biosci Rep. 2019;39:BSR20181826. doi: 10.1042/BSR20181826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo HC, Yu YC, Sung Y, Han JM. Glutamine reliance in cell metabolism. Exp Mol Med. 2020;52:1496–1516. doi: 10.1038/s12276-020-00504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JH, Pyun WY, Park HW. Cancer Metabolism: Phenotype, signaling and therapeutic targets. Cells. 2020;9:2308. doi: 10.3390/cells9102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matés JM, Campos-Sandoval JA, Santos-Jiménez JL, Márquez J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett. 2019;467:29–39. doi: 10.1016/j.canlet.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Shah R, Chen S. Metabolic Signaling Cascades Prompted by Glutaminolysis in Cancer. Cancers (Basel) 2020;12:2624. doi: 10.3390/cancers12092624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa T, Washio J, Takahashi T, Echigo S, Takahashi N. Glucose and glutamine metabolism in oral squamous cell carcinoma: insight from a quantitative metabolomic approach. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;118:218–225. doi: 10.1016/j.oooo.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Liu R, Shuai Y, Huang Y, Jin R, Wang X, Luo J. ASCT2 (SLC1A5)-dependent glutamine uptake is involved in the progression of head and neck squamous cell carcinoma. Br J Cancer. 2020;122:82–93. doi: 10.1038/s41416-019-0637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matés JM, Di Paola FJ, Campos-Sandoval JA, Mazurek S, Márquez J. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin Cell Dev Biol. 2020;98:34–43. doi: 10.1016/j.semcdb.2019.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.