Abstract
Background
Intestinal digesta is commonly used for studying responses of microbiota to dietary shifts, yet evidence is accumulating that it represents an incomplete view of the intestinal microbiota. The present work aims to investigate the differences between digesta- and mucosa-associated intestinal microbiota in Atlantic salmon (Salmo salar) and how they may respond differently to dietary perturbations. In a 16-week seawater feeding trial, Atlantic salmon were fed either a commercially-relevant reference diet or an insect meal diet containing ~ 15% black soldier fly (Hermetia illucens) larvae meal. The digesta- and mucosa-associated distal intestinal microbiota were profiled by 16S rRNA gene sequencing.
Results
Regardless of diet, we observed substantial differences between digesta- and mucosa-associated intestinal microbiota. Microbial richness and diversity were much higher in the digesta than the mucosa. The insect meal diet altered the distal intestinal microbiota resulting in higher microbial richness and diversity. The diet effect, however, depended on the sample origin. Digesta-associated intestinal microbiota showed more pronounced changes than the mucosa-associated microbiota. Multivariate association analyses identified two mucosa-enriched taxa, Brevinema andersonii and Spirochaetaceae, associated with the expression of genes related to immune responses and barrier function in the distal intestine, respectively.
Conclusions
Our data show that salmon intestinal digesta and mucosa harbor microbial communities with clear differences. While feeding insects increased microbial richness and diversity in both digesta- and mucosa-associated intestinal microbiota, mucosa-associated intestinal microbiota seems more resilient to variations in the diet composition. To fully unveil the response of intestinal microbiota to dietary changes, concurrent profiling of digesta- and mucosa-associated intestinal microbiota is recommended whenever feasible. Specific taxa enriched in the intestinal mucosa are associated to gene expression related to immune responses and barrier function. Detailed studies are needed on the ecological and functional significance of taxa associated to intestinal microbiota dwelling on the mucosa.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-020-00071-3.
Keywords: Atlantic salmon, Diet, Black soldier fly, Microbiota, Digesta, Mucosa
Background
The global population is projected to reach 9.7 billion in 2050 [1], requiring an increase in the food supply by 25–70% [2]. Producing more safe and high-quality food in a sustainable way to meet the global population growth is a great challenge for our generation. Fish are considered as nutritionally valuable part of the human diet and play an important role in the global food supply [3, 4]. The average annual growth rate of world food fish consumption in the period 2019–2030 is projected to be 1.4%, reaching 28 million tonnes live weight in 2030 [5]. Atlantic salmon (Salmo salar) is the most produced marine fish species and one of the most economically important farmed fish worldwide [6]. While Atlantic salmon are strictly carnivorous in the wild, farmed Atlantic salmon have experienced a substantial shift in the diet composition due to a limited supply of marine ingredients. Marine ingredients used for Norwegian farmed Atlantic salmon have gradually been replaced by plant sources, decreasing from ~ 90% in 1990 to ~ 25% in 2016 [7]. Due to concerns on the economic, environmental and social sustainability of the current raw materials for Atlantic salmon farming [6], more sustainable alternative feed ingredients, such as insects [8] and single-cell organisms (bacteria, yeasts and microalgae) [9], have been developed and used. One of the insect species with the greatest potential as an alternative feed ingredient for salmon aquaculture is black soldier fly (BSF; Hermetia illucens), which is now produced at industrial scale in Europe. In terms of protein quality, BSF larvae have a favorable essential amino acid profile closer to fishmeal than that of soybean meal [10]. The nutritional value of BSF larvae meal has been extensively evaluated in various fish species including Atlantic salmon [11–21]. However, how dietary BSF larvae meal may influence the intestinal health, function and microbiota of fish remains largely unexplored.
It is now well established that intestinal microbiota plays a pivotal role in host development and physiology, from being an essential element for the development of normal gut functions and immunity [22, 23] to modulating lipid metabolism and energy balance [24, 25]. Recent advances in sequencing technologies have transformed our ability to study the composition and dynamics of fish intestinal microbiota, leading to increasing interest in selective manipulation of intestinal microbiota. Diet is one of the key factors in shaping the intestinal microbiota. While long-term dietary habits have a considerable effect on the structure and activity of host intestinal microbiota [26–28], short-term dietary change also alters the intestinal microbiota in a rapid and reproducible way [29]. Different dietary components selectively promote or suppress the growth of certain microbial clades, which in turn could inflict important effects on the host health and disease resistance [30, 31]. The use of alternative feed ingredients may not only affect the nutrient utilization, fish growth, health, welfare and product quality, but also intestinal microbiota in Atlantic salmon [32–34]. While studies in mammals and fish have revealed substantial differences between the digesta- and mucosa-associated intestinal microbiota [32, 35–38], most studies investigating diet effects on the intestinal microbiota of fish have sampled the digesta only or a mixture of digesta and mucosa. Evidence is accumulating that digesta- and mucosa-associated intestinal microbiota in fish respond differently to dietary changes [32, 39–42]. Profiling only one of or a mixture of digesta- and mucosa-associated microbiota may obscure the response of intestinal microbiota to dietary changes.
Characterizing intestinal microbiota and its associations with host responses is an essential step towards identifying key microbial clades promoting fish health and welfare. Ultimately, a milestone in the fish microbiota research would be knowing how to selectively manipulate the microbiota to improve the growth performance, disease resistance and health status of farmed fish. The main aims of the present study were (i) to compare distal intestinal microbiota of Atlantic salmon fed a commercially relevant diet or an insect meal diet, (ii) to further explore the dissimilarity between digesta- and mucosa-associated microbiota and the differences in their response to dietary changes, and (iii) to identify associations between microbial clades and host responses. This work was part of a larger study consisting of a freshwater and seawater feeding trial that aimed to investigate the nutritional value and possible health effects for Atlantic salmon of a protein-rich insect meal produced from BSF larvae. The results presented herein focus on the intestinal microbiota in seawater phase Atlantic salmon fed an insect meal diet containing ~ 15% BSF larvae meal for 16 weeks.
Results
To aid readers in interpreting the data we report here, results on the feed utilization, growth performance, fillet quality, intestinal histopathology and gene expression, which have been reported elsewhere [43–45], are summarized as the following. In brief, there was lack of evidence that the insect meal diet negatively affected the feed utilization, growth performance or fillet quality of Atlantic salmon. Profiling of genes related to lipid metabolism, immune responses, barrier functions and stress responses in the proximal and distal intestine showed little evidence of diet effect. Histopathological examination of intestinal segments showed enterocyte steatosis in the proximal and mid intestine in both diet groups, but it was less severe in the proximal intestine of fish fed the insect meal diet.
Hereafter, different sample groups are named based on the combination of diet (REF vs. IM) and sample origin (DID vs. DIM). Hence, in addition to the extraction blanks, library blanks and mock, we have four different sample types, i.e., REF-DID, REF-DIM, IM-DID and IM-DIM.
qPCR
Since Cq values of most mucosa DNA templates were out of the linear range of the standard curve, the raw Cq value was used as a proxy of 16S rRNA gene quantity in the diluted DNA templates (Fig. S1). On average, REF-DID showed the highest 16S rRNA gene quantities (mean Cq = 24.7), followed by the mocks (mean Cq = 26.1) and IM-DID (mean Cq = 28.4). Irrespective of diet, mucosa DNA templates (REF-DIM, IM-DIM) showed similar 16S rRNA gene quantities (mean Cq = 30) that were close to extraction blanks (mean Cq = 32.4).
Characteristics of the sequence data
The high-throughput sequencing generated a total number of 7.5 million raw reads for biological samples. The median of raw reads per sample was 96,231, with the minimum and maximum value being 14,940 and 151,916, respectively. After the sequence denoising and ASV filtering, a total number of 1620 unique ASVs was generated. The mean percentage of chloroplasts and mitochondria removed from the ASV table before filtering out contaminants was 7.4 and 0.2%, respectively. The mean percentage of chloroplast varied considerably between digesta (7.1%) and mucosa (0.2%) samples and between diets within digesta samples, e.g. REF-DID (24.9%) and IM-DID (4.4%). The number of effective sequences retained for the downstream data analysis was 3.6 million. The median of effective sequences per sample was 46,372, with the minimum and maximum value being 951 and 106,591, respectively.
Taxonomic composition
All the eight bacterial species included in the mock were successfully identified at genus level with E. faecalis, L. fermentum, L. monocytogenes and S. aureus further being annotated at the species level (Fig. S2A). At the genus level, the average Pearson’s r between the expected and observed taxonomic profile of the mock was 0.33, whereas the Pearson’s r between the observed taxonomic profile of the mock was 0.98. The relative abundance of most Gram-positive bacteria, L. monocytogenes and E. faecalis in particular, were underestimated. In contrast, the relative abundance of Gram-negative bacteria was overestimated. Most ASVs (97.5–99.9%) in the extraction and library blanks were classified as Pseudomonas (Fig. S2B), which was the main contaminating taxon removed from the biological samples. Other contaminating ASVs removed from the biological samples were classified as Curtobacterium, Jeotgalicoccus, Modestobacter, Cutibacterium, Hymenobacter, Brevundimonas, Micrococcus, Sphingomonas, Devosia, Sphingomonas aurantiaca and Marinobacter adhaerens. The exact sequence and taxonomy of the contaminating ASVs and their relative abundance in the extraction and library blanks are available in Table S1.
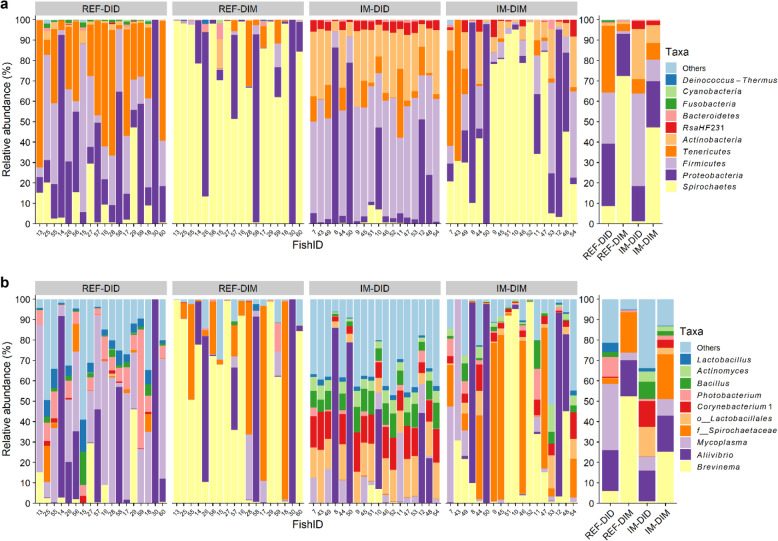
The taxonomic composition of mucosa samples showed higher similarity than that of the digesta samples, which were more diet-dependent (Fig. 1). At the phylum level, the dominant taxa of mucosa samples for both diets were Spirochaetes (REF-DIM, 72%; IM-DIM, 47%) (mean relative abundance), Proteobacteria (REF-DIM, 21%; IM-DIM, 23%), Firmicutes (REF-DIM, 1%; IM-DIM, 11%), Tenericutes (REF-DIM, 4%; IM-DIM, 8%) and Actinobacteria (REF-DIM, 1%; IM-DIM, 9%). For digesta samples, the dominant taxa of REF-DID were Tenericutes (33%), Proteobacteria (31%), Firmicutes (25%) and Spirochaetes (9%), whereas IM-DID was dominated by Firmicutes (45%), Actinobacteria (25%), Proteobacteria (17%), Tenericutes (7%) and RsaHF231 (4%) (Fig. 1a). At the genus level, the dominant taxa of mucosa samples for both diets were Brevinema (REF-DIM, 52%; IM-DIM, 25%), Spirochaetaceae (REF-DIM, 20%; IM-DIM, 22%), Aliivibrio (REF-DIM, 18%; IM-DIM, 18%) and Mycoplasma (REF-DIM, 4%; IM-DIM, 8%). For digesta samples, the dominant taxa of REF-DID were Mycoplasma (33%), Aliivibrio (20%), Photobacterium (10%), Brevinema (6%) and Lactobacillus (5%), whereas IM-DID was dominated by Aliivibrio (15%), Lactobacillales (14%), Corynebacterium 1 (13%), Bacillus (8%), Mycoplasma (7%) and Actinomyces (5%) (Fig. 1b).
Fig. 1.
Top 10 most abundant taxa of all samples at phylum (a) and genus (b) level. The samples are grouped by the sample type. The mean relative abundance of each taxon within the same sample type is displayed on the right side. o__, order; f__, family; REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa
Core ASVs
In total, 339 ASVs were identified as core ASVs based on their prevalence in each sample type (Fig. 2; Table S2). Three ASVs, classified as Aliivibrio, Brevinema andersonii, and Mycoplasma respectively, were identified as core ASVs in all the sample types. The Brevinema andersonii ASV was universally present in all the samples. Additionally, 11 ASVs were identified as core ASVs for digesta samples (REF-DID and IM-DID), which were classified as Geobacillus (1 ASV), Lactobacillus (3 ASVs), Mycoplasma (2 ASVs), Photobacterium (3 ASVs), Streptococcus (1 ASV) and Weissella (1 ASV). Two additional core ASVs were identified for the mucosa samples (REF-DIM and IM-DIM), which were classified as Brevinema andersonii and Spirochaetaceae, respectively. Six additional core ASVs were identified for fish fed the insect meal diet (IM-DID and IM-DIM), which were classified as Actinomyces, Corynebacterium 1, Corynebacterium aurimucosum ATCC 70097, Lactobacillales, RsaHF23 and Spirochaetaceae, respectively. No additional core ASVs were identified for fish fed the reference diet (REF-DID and REF-DIM). Lastly, 308 ASVs were found to be more prevalent in IM-DID than in any other sample type.
Fig. 2.
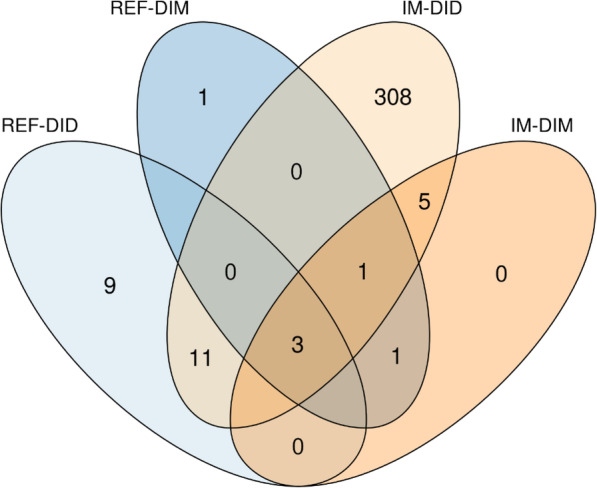
Venn’s diagram showing the shared and unique core ASVs in each sample type. The core ASVs were computed using a prevalence threshold at 80%. REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa
Alpha-diversity
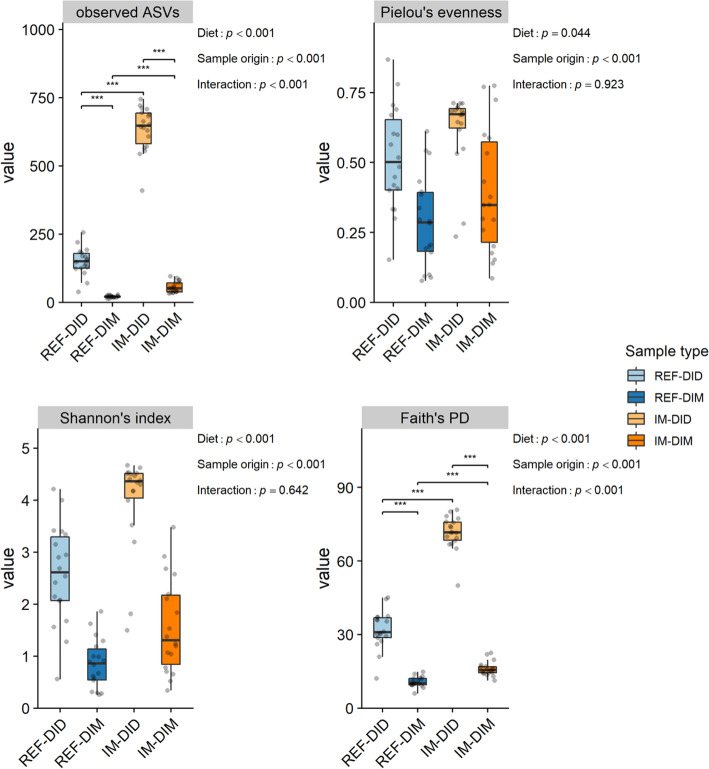
Regardless of diet, all the alpha-diversity indices were higher in digesta samples than mucosa samples (p < 0.05) (Fig. 3). Independent of sample origin, all the alpha-diversity indices were higher in fish fed the IM diet than those fed the REF diet (p < 0.05). A significant interaction between the diet and sample origin effect was detected for the observed ASVs (p < 0.001) and Faith’s phylogenetic diversity (p < 0.001), both of which showed a stronger diet effect in digesta samples than mucosa samples.
Fig. 3.
The sample origin and diet effects on the alpha-diversity of distal intestinal microbiota in seawater phase Atlantic salmon. The p value of the main effects and their interaction are displayed on the top-right corner of each sub-plot. Asterisks denote statistically significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001). PD, phylogenetic diversity; REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa
Beta-diversity
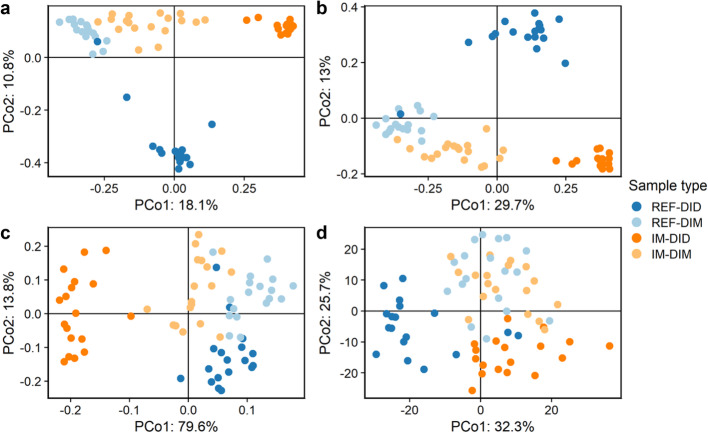
The PCoA plots built on the Jaccard and unweighted UniFrac distance matrix showed clear separations of samples belonging to different dietary groups and sample origins (Fig. 4a-b). However, the average distance between samples from different dietary groups was dependent on sample origin. Specifically, mucosa samples from different dietary groups formed clusters close to each other, whereas digesta samples from different dietary groups were far apart. The PCoA plots built on the Aitchison and PHILR transformed Euclidean distance matrix also showed separations of samples belonging to different dietary groups and sample origins (Fig. 4c-d). Again, the average distance between samples from different dietary groups was dependent on sample origin. Mucosa samples from different dietary groups formed clusters boarding (Fig. 4c) or overlapping (Fig. 4d) each other, whereas digesta samples from different dietary groups were more clearly separated.
Fig. 4.
The sample origin and diet effects on the beta-diversity of distal intestinal microbiota in seawater phase Atlantic salmon. The PCoA plots were built on Jaccard (a), unweighted UniFrac (b), Aitchison (c) and phylogenetic isometric log-ratio (PHILR) transformed Euclidean (d) distance matrix, respectively. PCo, principle coordinate; REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa
The PERMANOVA and its following conditional contrasts largely confirmed the PCoA results. Regardless of the distance matrix used, both main factors had significant effects on the beta-diversity and their interaction was significant as well (p < 0.05) (Table 2). Results on the tests of homogeneity of multivariate dispersions are shown in Table 3. For Jaccard distance, significant differences in the multivariate dispersions were observed between digesta and mucosa samples for both diets (REF-DID VS. REF-DIM, p = 0.045; IM-DID VS. IM-DIM, p = 0.002), and between diets for digesta samples (REF-DID VS. IM-DID, p = 0.002). For unweighted UniFrac distance, IM-DID showed lower multivariate dispersions than other sample types resulting in significant differences compared to REF-DID (p = 0.002) and IM-DIM (p = 0.002). For Aitchison distance, REF-DIM showed lower multivariate dispersions than other sample types resulting in significant differences compared to REF-DID (p = 0.046) and IM-DIM (p = 0.046). For PHILR transformed Euclidean distance, the differences in the multivariate dispersions among the sample types were not significant (p > 0.05).
Table 2.
PERMANOVA results and subsequent conditional contrasts
| Distance matrix | Main effects | Interaction | Conditional contrasts | ||||
|---|---|---|---|---|---|---|---|
| Diet | Sample origin | REF-DID VS. IM-DID |
REF-DIM VS. IM-DIM |
REF-DID VS. REF-DIM |
IM-DID VS. IM-DIM |
||
| Jaccard | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| Unweighted UniFrac | 0.001a | 0.001 | 0.001 | 0.001a | 0.001 | 0.001 | 0.001 |
| Aitchison | 0.001 | 0.003 | 0.004 | 0.002 | 0.004 | 0.004a | 0.002a |
| PHILR (Euclidean)b | 0.001 | 0.001 | 0.001 | 0.001 | 0.005 | 0.001 | 0.001 |
REF reference diet, IM insect meal diet, DID distal intestine digesta, DIM distal intestine mucosa
aMonte Carlo p value
bPhylogenetic isometric log-ratio transformed Euclidean distance
Table 3.
Test of homogeneity of multivariate dispersions among groups
| Distance matrix | Conditional contrasts | |||
|---|---|---|---|---|
| REF-DID VS. IM-DID |
REF-DIM VS. IM-DIM |
REF-DID VS. REF-DIM |
IM-DID VS. IM-DIM |
|
| Jaccard | 0.002 | 0.087 | 0.045 | 0.002 |
| Unweighted UniFrac | 0.002 | 0.711 | 0.200 | 0.002 |
| Aitchison | 0.453 | 0.046 | 0.046 | 0.369 |
| PHILR (Euclidean)a | 0.240 | 0.266 | 0.240 | 0.266 |
REF reference diet, IM insect meal diet, DID distal intestine digesta, DIM distal intestine mucosa
aPhylogenetic isometric log-ratio transformed Euclidean distance
Significant associations between microbial clades and sample metadata
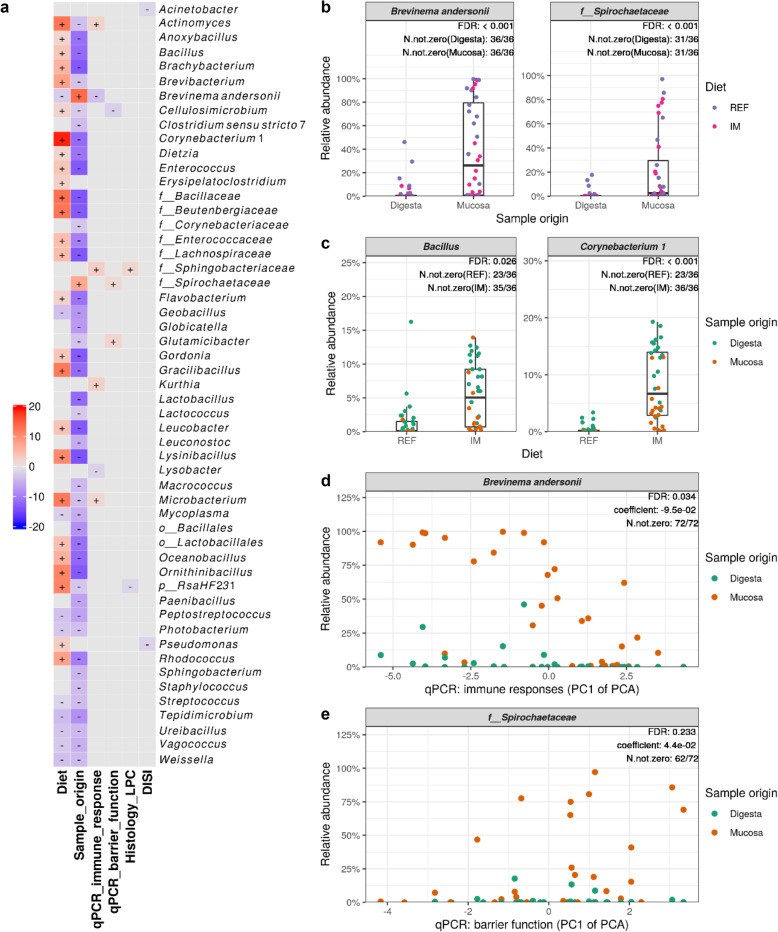
The multivariate association analysis identified 53 taxa showing significant associations with the metadata of interest (Fig. 5a). The diagnostic plots showing the raw data underlying the significant associations are shown in Figs. S3–8. Forty-seven differentially abundant taxa were identified for the sample origin effect, 45 of which, including Bacillus, Enterococcus, Flavobacterium, Lactobacillus, Lactococcus, Leuconostoc, Mycoplasma, Peptostreptococcus, Photobacterium, Staphylococcus, Streptococcus, Vagococcus and Weissella, showed lower relative abundances in the mucosa than the digesta (Fig. S3). In contrast, two taxa belonging to the Spirochaetes phylum, Brevinema andersonii and Spirochaetaceae, were enriched in the mucosa (Fig. 5b). Thirty-six differentially abundant taxa were identified for the diet effect, 26 of which showed increased relative abundances in fish fed the IM diet (Fig. S4). Among these 26 taxa, some were enriched in both intestinal digesta and mucosa which included Actinomyces, Bacillaceae, Bacillus, Beutenbergiaceae, Brevibacterium, Corynebacterium 1, Enterococcus, Lactobacillales, Microbacterium, Oceanobacillus and RsaHF231 (partially illustrated as Fig. 5c). For the histological scores, the relative abundance of Sphingobacteriaceae and RsaHF231 were found to increase and decrease, respectively, in fish scored abnormal regarding lamina propria cellularity (LPC) in distal intestine (Fig. S5). The relative abundance of Acinetobacter and Pseudomonas were negatively correlated with the distal intestine somatic index (DISI) (Fig. S6). Six taxa, including Actinomyces, Brevinema andersonii, Kurthia, Lysobacter, Microbacterium and the Sphingobacteriaceae, were found to associate with the expression of genes related to immune responses (Fig. S7). Notably, the relative abundance of Brevinema andersonii showed a clear positive correlation with the expression levels of immune genes (Fig. 5d), which decreased as the PC1 of the PCA increased. Furthermore, 3 taxa including Cellulosimicrobium, Glutamicibacter and the Spirochaetaceae were found to associate with the expression of genes related to barrier functions (Fig. S8). The relative abundance of the Spirochaetaceae showed a negative correlation with the expression levels of barrier function relevant genes (Fig. 5e), which decreased as the PC1 of the PCA increased.
Fig. 5.
Significant associations between microbial clades and sample metadata. a Heatmap summarizing all the significant associations between microbial clades and sample metadata. Color key: -log(q-value) * sign (coefficient). Cells that denote significant associations are colored (red or blue) and overlaid with a plus (+) or minus (−) sign that indicates the direction of association: Diet (+), higher abundance in salmon fed the IM diet; Sample_origin (+), higher abundance in mucosa samples; Histology_LPC (+), higher abundance in salmon scored abnormal regarding lamina propria cellularity (LPC) in the distal intestine; DISI (+), positive correlation between microbial clade abundance and distal intestine somatic index (DISI); qPCR_immune_response (+) / qPCR_barrier_function (+), negative correlation between microbial clade abundance and the gene expression levels. b Taxa that are more abundant in the intestinal mucosa than the digesta. c Representative taxa showing increased relative abundances in both intestinal digesta and mucosa of salmon fed the IM diet. d The positive correlation between the relative abundance of Brevinema andersonii and immune gene expression levels in the distal intestine. Note that the expression levels of the immune genes decreased as the PC1 of the PCA increased. e The negative correlation between the relative abundance of the Spirochaetaceae and the expression levels of barrier function relevant genes. Also note that the expression levels of the barrier function relevant genes decreased as the PC1 of the PCA increased. p__, phylum; o__, order; f__, family; FDR, false discovery rate; N.not.zero, number of observations that are not zero; REF, reference diet; IM, insect meal diet
Discussion
Core microbiota
In accordance with previous studies in Atlantic salmon [33, 46–51], Aliivibrio, Brevinema andersonii and Mycoplasma were identified as core microbiota in the present study. Aliivibrio is commonly found in the seawater phase Atlantic salmon intestine [48–50, 52–56] and has been identified as a core taxon of both wild and captive Atlantic salmon [47, 49, 50]. Provided its common presence in seawater, Aliivibrio may have originated from the surrounding water and colonized the intestinal mucosa as Atlantic salmon constantly drink seawater to prevent dehydration in a hyperosmotic environment. Currently, Aliivibrio comprises of four closely related species including Aliivibrio fischeri, Aliivibrio logei, Aliivibrio salmonicida and Aliivibrio wodanis, which were split from the Vibrio genus and reclassified as Aliivibrio in 2007 [57]. Strains of A. fischeri and A. logei have been described as bioluminescent symbionts of certain fishes and squids [58], whereas A. salmonicida and A. wodanis have been identified as pathogens for Atlantic salmon causing cold-water vibriosis [59] and ‘winter ulcer’ [60], respectively. We identified 7 Aliivibrio ASVs in this study, four of which, including the core Aliivibrio ASV, were closely related and clustered with unknow Aliivibrio species in the reference database. Among the known Aliivibrio species, A. logei is most closely related to the core Aliivibrio ASV, which was also the predominant Aliivibrio ASV found in the present study. One of the Aliivibrio ASVs, which was detected at very low abundances (< 0.00015%), was closely related to A. wodanis. These observations coincide with previous findings in Arctic seawater-farmed Atlantic salmon [54], suggesting that Aliivibrio in the salmon intestine mostly comprises of commensal species.
Though Spirochaetes has typically been found in low abundances in the Atlantic salmon intestine [32, 36, 40, 52, 61], two recent studies have identified Brevinema andersonii as a core taxon of both digesta- and mucosa-associated intestinal microbiota in seawater phase Atlantic salmon [48, 49]. Notably, Brevinema andersonii is also a predominant taxon in the digesta and mucosa in one of the studies [49]. Brevinema andersonii was initially isolated from short-tailed shrews (Blarina brevicauda) and white-footed mice (Peromyscus leucopus) as an infectious pathogen [62]. This taxon has also been found in the intestine and gill tissue of rainbow trout (Oncorhynchus mykiss) [63], and intestinal digesta of Senegalese sole (Solea senegalensis) [64].
Mycoplasma is widely distributed in nature and well known for its minute size and lack of cell wall. It seems to be particularly well-adapted to Atlantic salmon intestine [65]. Like Aliivibrio, it has been frequently identified as a core taxon of both wild and captive Atlantic salmon [33, 46, 48–51]. Notably, it was found to be more abundant in marine adults than in freshwater juvenile Atlantic salmon [50] and sporadically predominate intestinal microbial community in the digesta [33, 49, 50, 54, 66] and mucosa [48] reaching > 90% of total reads in extreme cases. Due to its small compact genome and limited biosynthesis capacities, Mycoplasma typically forms obligate parasitic or commensal relationships with its host to obtain necessary nutrients such as amino acids, fatty acids and sterols [67]. Recent shotgun-metagenomic sequencing of the Atlantic salmon Mycoplasma revealed that it is closely related to Mycoplasma penetrans [33, 68]. It was suggested that the presence of riboflavin encoding genes and lack of pathogenicity factors in the metagenome-assembled Mycoplasma genome is indicative of a symbiotic relationship between the Mycoplasma and Atlantic salmon [68].
Sample origin effect
In line with previous findings in mammals and fish [32, 35–38], we observed substantial differences between digesta- and mucosa-associated microbiota. The microbial richness and diversity were much higher in the digesta than the mucosa, as previously observed in seawater phase Atlantic salmon [32, 36, 49]. Furthermore, most of the bacterial taxa in the distal intestine, including those commonly found in the Atlantic salmon intestine such as Bacillus, Enterococcus, Flavobacterium, Lactobacillus, Lactococcus, Leuconostoc, Mycoplasma, Peptostreptococcus, Photobacterium, Staphylococcus, Streptococcus, Vagococcus and Weissella, were less abundant in the mucosa than in the digesta. These results are suggestive of a selection pressure from the host that determines which microbial clades colonize and flourish in the intestinal mucus layer [69]. In this study, two taxa belonging to the Spirochaetes phylum, Brevinema andersonii and Spirochaetaceae, were more abundant in the distal intestine mucosa than the digesta. As aforementioned, Spirochaetes were typically found in low abundances in the Atlantic salmon intestine. Yet a recent study also showed that irrespective of diets Brevinema andersonii seemed to be more abundant in the intestinal mucosa than the digesta of seawater phase Atlantic salmon [49]. Known for high motility and chemotactic attraction to mucin, some Spirochaetes can penetrate the mucus and associate with the intestinal mucosa [70–72]. Further work is required to confirm whether these taxa are consistently enriched in the intestinal mucus layer of seawater phase Atlantic salmon.
Diet effect
Diet is one of the key factors in shaping the fish intestinal microbiota. In agreement with previous findings in rainbow trout [42, 73, 74] and laying hens [75, 76], we found that the insect meal diet altered the distal intestinal microbiota assemblage resulting in higher microbial richness and diversity. Our findings, showing that the insect meal diet increased the relative abundance of Actinomyces, Bacillus, Brevibacterium, Corynebacterium 1 and Enterococcus, are in accord with recent studies in rainbow trout fed diets containing 30% BSF larvae meal [42, 74]. Importantly, these results were partly confirmed in other studies employing fluorescence in situ hybridization for targeted profiling of changes in the intestinal microbiota. Specifically, increased absolute abundance of Lactobacillus/Enterococcus was found in rainbow trout fed 20% dietary BSF larvae meal [77], whereas increased absolute abundance of Bacillus, Enterococcus and Lactobacillus was documented in Siberian sturgeon (Acipenser baerii) fed 15% BSF larvae meal [78].
The increases in the relative abundance of specific microbial clades in Atlantic salmon fed the insect meal diet may be explained by feed-borne microbiota and/or feed composition. Bacterial taxa, including Actinomyces, Bacillus, Brevibacterium, Corynebacterium, Enterococcus, Oceanobacillus and RsaHF231, have been found in BSF whole larvae or larvae intestine [79–82]. The fact that RsaHF231 has not been documented in fish before indicates that these bacterial taxa may have partially originated from BSF larvae meal. Our results from the freshwater feeding trial showed that these bacterial taxa were also enriched in the intestinal digesta and mucosa of Atlantic salmon smolts fed an insect meal diet containing 60% soldier fly larvae meal. Importantly, these bacterial taxa were also detected in the feed pellets which contained considerable amount of bacterial DNA (unpublished data). Given the hydrothermal treatments the feed pellets underwent during the extrusion, the feed-borne microbiota profiled by the DNA sequencing techniques could have largely originated from dead bacteria and bacterial spores rather than living bacteria. As sequencing-based methods cannot differentiate between living and dead cells, future studies should investigate to what extent the feed-borne microbiota may contribute to, or confound the observed diet effects on intestinal microbiota, using methods that distinguish living and dead bacteria such as viability PCR and RNA sequencing [83]. On the other hand, unique nutrients in the insect meal diet such as chitin, an essential component of the insect exoskeleton, may have selectively promoted the growth of certain intestinal microbes. Many bacterial species belonging to Bacillus can produce chitinase [84]. Bacillus and Lactobacillus were two of the predominant taxa in the intestinal mucosa of Atlantic salmon fed a 5% chitin diet, the former of which displayed the highest in vitro chitinase activity [85].
Significant interactions between diet and sample origin effect
We observed in the present study that the diet effect on the intestinal microbial community richness and structure was dependent on the sample origin, with mucosa-associated intestinal microbiota showing higher resilience to the dietary change. Our results corroborate previous findings in rainbow trout revealing that mucosa-associated intestinal microbiota was less influenced by dietary inclusion of 30% BSF larvae meal compared to digesta-associated intestinal microbiota [41, 42]. Results from molecular-based studies on salmonid intestinal microbiota hitherto suggest that diet modulates digesta- and mucosa-associated intestinal microbiota to varying degrees with the latter generally being more resilient to dietary interventions [32, 39–42, 48]. As such, current practices of profiling only one of or a mixture of digesta- and mucosa-associated microbiota may obscure the response of intestinal microbiota to dietary changes. To fully unveil the response of intestinal microbiota to dietary changes, we recommend concurrent profiling of digesta- and mucosa-associated intestinal microbiota whenever it is feasible.
Significant associations between microbial clades and sample metadata
To our knowledge, only a few studies have carried out association analysis between intestinal microbial clades and host responses in Atlantic salmon. As such, our results should be treated as preliminary observations and critically evaluated in later studies. Herein, we highlight the significant associations between two mucosa-enriched taxa and host gene expressions in the intestine. Specifically, Brevinema andersonii, part of the core microbiota, was associated with the expression of genes related to pro- and anti-inflammatory responses whereas the Spirochaetaceae was associated with the expression of genes related to barrier function. Intestinal microbiota is well known to modulate the local immune responses and intestinal epithelial barrier function [86]. Furthermore, it is hypothesized that mucosa-associated microbiota plays a more crucial role in shaping the host immunity in that it can interact both directly and indirectly with intestinal epithelial barrier whereas digesta-associated microbiota can only interact indirectly [69]. Taken together, further research should be undertaken to investigate the potential ecological and functional significance of these two taxa for seawater phase Atlantic salmon.
Quality control: use of mock and negative controls
As in any field of research, conducting a well-controlled microbiome study requires great care in the experiment design such as setting up appropriate experimental controls. The use of mock as a positive control allows for critical evaluation and optimization of microbiota profiling workflow. That all the bacterial taxa in the mock were correctly identified at the genus level indicates that the current workflow is reliable for the taxonomic profiling of intestinal microbiota. Furthermore, the taxonomic profile of mock from different DNA extraction batches was fairly similar, suggesting that the results generated by the current workflow are also largely reproducible. However, the low concordance between the expected and observed relative abundance of bacterial taxa in the mock is reminiscent of the fact that bias is introduced at different steps of the marker-gene survey [87–89], among which DNA extraction and PCR amplification are the two largest sources of bias due to preferential extraction and amplification of some microbial clades over others. In line with previous observations that Gram-positive bacteria may be more subjective to incomplete lysis during DNA extraction due to their tough cell walls [90, 91], the recovery of most Gram-positive bacteria in the mock was lower than the expected. The insufficient lysing of Gram-positive bacteria in the mock was largely mitigated in our later experiments by using a mixture of beads with different sizes for the bead beating during DNA extraction (unpublished data). The bias in the marker-gene sequencing experiments, as reflected in the observed taxonomic profile of the mock, highlights the necessity of validating such results by absolute quantification techniques such as cultivation (if possible), qPCR, flow cytometry and fluorescence in situ hybridization.
Reagent contamination is a common issue in molecular-based studies of microbial communities. The main contaminating taxon identified in this study is Pseudomonas, which has been reported as a common reagent contaminant in numerous studies [92–98]. Given the dominance of Pseudomonas in the negative controls of both DNA extraction and PCR, most of the observed contamination has likely derived from PCR reagents such as molecular-grade water [99–101]. Notably, Pseudomonas has also been isolated from intestinal digesta and mucosa of Atlantic salmon by traditional culturing approaches [85, 102–104], and reported as a member of Atlantic salmon core microbiota in culture-independent studies [32, 36, 46, 47, 51, 105]. Due to the low taxonomic resolution of amplicon sequencing, it is difficult to discern contaminating taxa from true signals solely based on taxonomic labels. The inclusion of negative controls, coupled with quantifications of microbial DNA concentration in the samples, has enabled fast and reliable identification of contaminating taxa in this study. Besides Pseudomonas, other common reagent contaminants, including Bradyrhizobium, Burkholderia, Comamonas, Methylobacterium, Propionibacterium, Ralstonia, Sphingomonas and Stenotrophomonas [97, 99, 101, 106–110], have also been frequently reported as members of Atlantic salmon intestinal microbiota, indicating that existing studies of Atlantic salmon intestinal microbiota may have been plagued with reagent contamination that is hard to ascertain due to lack of negative controls. As reagent contamination is unavoidable, study-specific and can critically influence sequencing-based microbiome analyses [99, 111, 112], negative controls should always be included and sequenced in microbiome studies especially when dealing with low microbial biomass samples like intestinal mucosa.
Conclusions
In summary, we confirmed previous findings in mammals and fish that intestinal digesta and mucosa harbor microbial communities with clear differences. Regardless of diet, microbial richness and diversity were much higher in the digesta than the mucosa. The insect meal diet altered the distal intestinal microbiota assemblage resulting in higher microbial richness and diversity. The diet effect was however dependent on the sample origin, with mucosa-associated intestinal microbiota being more resilient to the dietary change. To fully unveil the response of intestinal microbiota to dietary changes, concurrent profiling of digesta- and mucosa-associated intestinal microbiota is recommended whenever feasible. Lastly, we identified two mucosa-enriched taxa, Brevinema andersonii and Spirochaetaceae, which were associated with the expression in the distal intestine of genes related to immune responses and barrier function, respectively. As mucosa-associated microbiota could play a more critical role in shaping the host metabolism, their potential functional significance for seawater phase Atlantic salmon merits further investigations.
Methods
Experimental fish, diet and sampling
A 16-week seawater feeding trial with Atlantic salmon (initial body weight = 1.40 kg, S.D. = 0.043 kg) was conducted at the Gildeskål Research Station (GIFAS), Nordland, Norway. The experimental fish were randomly assigned into 6 adjacent square net pens (5 × 5 m) with a depth of 5 m, each containing 90 fish. The fish were fed, in triplicate net pens, either a commercially-relevant reference diet (REF) with a combination of fish meal, soy protein concentrate, pea protein concentrate, corn gluten and wheat gluten as the protein source, or an insect meal diet (IM) wherein all the fish meal and most of the pea protein concentrate were replaced by insect meal produced from BSF larvae. Formulation and proximate composition of the experimental diets are shown in Table 1. The diets were formulated to be isonitrogenous (39% crude protein), isolipidic (29% crude lipid) and isoenergetic (25 MJ/kg DM gross energy), and to meet the nutrient requirements of Atlantic salmon. The diets were extruded, dried and vacuum coated with oils, producing feed pellets with a diameter size of 3.5 mm (Cargill, Dirdal, Norway). The insect meal was produced by Protix Biosystems BV (Dongen, The Netherlands). The fly larvae were grown on feed substrates containing seaweed (Ascophyllum nodosum) and vegetable wastes (60:40). After 8 days of growing, the larvae were harvested and partially defatted before being dried and ground to make the insect meal. The insect meal contains about 52% crude protein and 18% crude lipid. Fish were fed by hand until apparent satiation once or twice daily depending on the duration of daylight. During the feeding trial, the water temperature was 8.3 ± 3.7 °C, dissolved oxygen 8.9 ± 1 mg/L and salinity 31.6 ± 0.8 ‰. Further details on the nutritional composition of the insect meal and diets have been reported elsewhere [45].
Table 1.
Formulation and proximate composition of the experimental diets
| REF | IM | |
|---|---|---|
| Ingredients (% wet-weight) | ||
| Fishmeal LT94 | 10 | 0 |
| Black soldier fly larva meala | 0 | 14,75 |
| Soy protein concentrate | 25 | 25 |
| Corn gluten meal | 7,5 | 7,5 |
| Wheat gluten meal | 3,35 | 6,88 |
| Pea protein concentrate 55 | 8,8 | 2,84 |
| Fish oil | 10,18 | 14,76 |
| Rapeseed oil | 20,95 | 14,73 |
| Binder | 12,32 | 11,24 |
| Additives | 1,89 | 2,29 |
| Chemical composition (wet-weight basis) | ||
| Dry matter (%) | 93 | 95 |
| Crude Protein (%) | 38 | 39 |
| Crude Lipid (%) | 29 | 29 |
| Ash (%) | 4,6 | 4,5 |
| Carbohydrates (%) | 11,6 | 11,4 |
| Gross energy (MJ/kg) | 24,6 | 25 |
| TBARS (nmol/g) | 3 | 4,9 |
REF reference diet, IM insect meal diet, TBARS Thiobarbituric acid reactive substances
aPartially defatted. Crude protein: 52%, crude lipid: 18%. Produced by the Protix Biosystems BV (Dongen, The Netherlands)
At the termination of the feeding trial, the average body weight of fish reached 3.7 kg. Six fish were randomly taken from each net pen, anesthetized with tricaine methanesulfonate (MS222®; Argent Chemical Laboratories, Redmond, WA, USA) and euthanized by a sharp blow to the head. After cleaning the exterior of each fish with 70% ethanol, the distal intestine, i.e., the segment from the increase in intestinal diameter and the appearance of transverse luminal folds to the anus, was aseptically removed from the abdominal cavity, placed in a sterile Petri dish and opened longitudinally. Only fish with digesta along the whole intestine were sampled to ensure that the intestine had been exposed to the diets. The intestinal digesta was gently scraped and collected into a 50 mL skirted sterile centrifuge tube and mixed thoroughly using a spatula. An aliquot of the homogenate was then transferred into a 1.5 mL sterile Eppendorf tube and snap-frozen in liquid N2 for the profiling of digesta-associated intestinal microbiota. A tissue section from the mid part of the distal intestine was excised and rinsed in sterile phosphate-buffered saline 3 times to remove traces of the remaining digesta. After rinsing, the intestinal tissue was longitudinally cut into 3 pieces for histological evaluation (fixed in 4% phosphate-buffered formaldehyde solution for 24 h and transferred to 70% ethanol for storage), RNA extraction (preserved in RNAlater solution and stored at − 20 °C) and profiling of mucosa-associated intestinal microbiota (snap-frozen in liquid N2), respectively. The collection of microbiota samples was performed near a gas burner to secure aseptic conditions. After the sampling of each fish, tools were cleaned and decontaminated by a 70% ethanol spray and flaming. Microbiota samples of the distal intestine digesta (DID) and mucosa (DIM) were transported in dry ice and stored at − 80 °C until DNA extraction.
DNA extraction
Total DNA was extracted from ~ 200 mg distal intestine digesta or mucosa using the QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany; catalog no., 51,504) with some modifications to the manufacturer’s specifications as described before [32], except that 2 mL prefilled bead tubes (Qiagen; catalog no., 13,118–50) were used for the bead beating. For quality control purposes, a companion “blank extraction” sample was added to each batch of sample DNA extraction by omitting the input material, whereas an additional microbial community standard (ZymoBIOMICS™, Zymo Research, California, USA; catalog no., D6300), i.e. mock, was included for each DNA extraction kit as a positive control. The mock consists of 8 bacteria (Pseudomonas aeruginosa, Escherichia coli, Salmonella enterica, Lactobacillus fermentum, Enterococcus faecalis, Staphylococcus aureus, Listeria monocytogenes, Bacillus subtilis) and 2 yeasts (Saccharomyces cerevisiae, Cryptococcus neoformans).
Amplicon PCR
The V1–2 hypervariable regions of the bacterial 16S rRNA gene were amplified using the primer set 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 338R (5′-GCW GCC WCC CGT AGG WGT-3′) [113]. The PCR was run in a total reaction volume of 25 μL containing 12.5 μL of Phusion® High-Fidelity PCR Master Mix (Thermo Scientific, CA, USA; catalog no., F531L), 10.9 μL molecular grade H2O, 1 μL DNA template and 0.3 μL of each primer (10 μM). The amplification program was set as follows: initial denaturation at 98 °C for 3 min; 35 cycles of denaturation at 98 °C for 15 s, annealing decreasing from 63 °C to 53 °C in 10 cycles for 30 s followed by 25 cycles at 53 °C for 30 s, and extension at 72 °C for 30 s; followed by a final extension at 72 °C for 10 min. For samples with faint or invisible bands in the agarose gel after PCR, the PCR condition was optimized by applying serial dilutions to the DNA templates to reduce the influence of PCR inhibitors. All the digesta samples were diluted 1:2 in buffer ATE (10 mM Tris-Cl, pH 8.3, with 0.1 mM EDTA and 0.04% NaN3) whereas all the mucosa samples were diluted 1:32. The formal amplicon PCR was run in duplicate incorporating two negative PCR controls, which were generated by replacing the template DNA with molecular grade H2O. The duplicate PCR products were then pooled and examined by a 1.5% agarose gel electrophoresis.
Quantification of 16S rRNA gene by qPCR
To assist in identifying contaminating sequences, the 16S rRNA gene quantity in the diluted DNA templates used for the amplicon PCR was measured by qPCR. The qPCR assays were performed using a universal primer set (forward, 5′-CCA TGA AGT CGG AAT CGC TAG-3′; reverse, 5′-GCT TGA CGG GCG GTG T-3′) that has been used for bacterial DNA quantification in previous studies [114, 115]. The assays were carried out using the LightCycler 96 (Roche Applied Science, Basel, Switzerland) in a 10 μL reaction volume, which contained 2 μL of PCR-grade water, 1 μL diluted DNA template, 5 μL LightCycler 480 SYBR Green I Master Mix (Roche Applied Science) and 1 μL (3 μM) of each primer. Samples, together with the extraction blanks and mock, were run in duplicate in addition to Femto™ bacterial DNA standards (Zymo Research; catalog no., E2006) and a no-template control of the qPCR assay. The qPCR program encompassed an initial enzyme activation step at 95 °C for 2 min, 45 three-step cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 15 s, and a melting curve analysis at the end. Quantification cycle (Cq) values were determined using the second derivative method [116]. The specificity of qPCR amplification was confirmed by evaluating the melting curve of qPCR products and the band pattern on the agarose gel after electrophoresis. The inter-plate calibration factor was calculated following the method described in [117], using the bacterial DNA standards as inter-plate calibrators.
Sequencing
The sequencing was carried out on a Miseq platform following the Illumina 16S metagenomic sequencing library preparation protocol [118]. Briefly, the PCR products were cleaned using the Agencourt AMPure XP system (Beckman Coulter, Indiana, USA; catalog no., A63881), multiplexed by dual indexing using the Nextera XT Index Kit (Illumina, California, USA; catalog no., FC-131-1096) and purified again using the AMPure beads. After the second clean-up, representative libraries were selected and analyzed using the Agilent DNA 1000 Kit (Agilent Technologies, California, USA; catalog no., 5067–1505) to verify the library size. Cleaned libraries were quantified using the Invitrogen Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, California, USA; catalog no., Q32854), diluted to 4 nM in 10 mM Tris (pH 8.5) and finally pooled in an equal volume. Negative controls with library concentrations lower than 4 nM were pooled in equal volume directly. Due to the low diversity of amplicon library, 15% Illumina generated PhiX control (catalog no., FC-110-3001) was spiked in by combining 510 μL amplicon library with 90 μL PhiX control library. The library was loaded at 6 pM and sequenced using the Miseq Reagent Kit v3 (600-cycle) (Illumina; catalog no., MS-102-3003).
Sequence data processing
The raw sequence data were processed by the DADA2 1.14 in R 3.6.3 [119] to infer amplicon sequence variants (ASVs) [120]. Specifically, the demultiplexed paired-ended reads were trimmed off the primer sequences (forward reads, first 20 bps; reverse reads, first 18 bps), truncated at the position where the median Phred quality score crashed (forward reads, at position 290 bp; reverse reads, at position 248 bp) and filtered off low-quality reads. After trimming and filtering, the run-specific error rates were estimated and the ASVs were inferred by pooling reads from all the samples sequenced in the same run. The chimeras were removed using the “pooled” method after merging the reads. The resulting raw ASV table and representative sequences were imported into QIIME2 (version, 2020.2) [121]. The taxonomy was assigned by a scikit-learn naive Bayes machine-learning classifier [122], which was trained on the SILVA 132 99% OTUs [123] that were trimmed to only include the regions of 16S rRNA gene amplified by our primers. ASVs identified as chloroplasts or mitochondria were excluded from the ASV table. The ASV table was conservatively filtered to remove ASVs that had no phylum-level taxonomic assignment or appeared in only one biological sample. Contaminating ASVs were identified based on two suggested criteria: contaminants are often found in negative controls and inversely correlate with sample DNA concentration [98]. The ASVs filtered from the raw ASV table were also removed from the representative sequences, which were then inserted into a reference phylogenetic tree built on the SILVA 128 database using SEPP [124]. The alpha rarefaction curves and the core metrics results were generated with a sampling depth of 10,000 and 2047 sequences per sample, respectively (Fig. S9). For downstream data analysis and visualization, QIIME2 artifacts were imported into R using the qiime2R package [125] and a phyloseq [126] object was assembled from the sample metadata, ASV table, taxonomy and phylogenetic tree. The core ASVs were calculated using a prevalence threshold at 80% and visualized by the Venn’s diagram. The alpha-diversity indices, including observed ASVs, Pielou’s evenness, Shannon’s index and Faith’s phylogenetic diversity (PD), were computed via the R packages microbiome [127] and picante [128]. For beta-diversity analyses, we used distance matrices including Jaccard distance, unweighted UniFrac distance, Aitchison distance and phylogenetic isometric log-ratio (PHILR) transformed Euclidean distance. Since rarefying remains to be the best solution for unweighted distance matrices [129], the Jaccard distance and unweighted UniFrac distance were computed in QIIME2 using the rarefied ASV table. The compositionality-aware distance matrices, Aitchison distance and PHILR transformed Euclidean distance, were calculated using the unrarefied ASV table. The Aitchison distance was computed by the DEICODE plugin in QIIME2, a form of Aitchison distance that is robust to high levels of sparsity by using the matrix completion to handle the excessive zeros in the microbiome data [130]. The PHILR transform of the ASV table was performed in R using the philr package [131]. The selected distance matrices were explored and visualized by the principal coordinates analysis (PCoA).
Multivariate association analysis
To reduce the multiple testing burden, the ASV table was collapsed at the genus level before running the multivariate association analysis. Bacterial taxa of very low abundance (< 0.01%) or low prevalence (present in < 25% of samples) were removed from the feature table. The microbial clades were then tested for significant associations with metadata of interest by MaAsLin2 (version, 0.99.12) (https://huttenhower.sph.harvard.edu/maaslin2) in R, using the default parameters. The results of the analysis are the associations of specific microbial clades with metadata, deconfounding the influence of other factors included in the model. Association was considered significant when the q-value was below 0.25. Metadata included in the multivariate association testing are fixed factors Diet + Sample origin + distal intestine somatic index (DISI) + lamina propria cellularity (histological scores) + immune response (qPCR) + barrier function (qPCR), and random factors FishID + NetPen. FishID was nested in NetPen, and NetPen nested in Diet. The methodological approach to these parameters was reported in a previous study [44]. Lamina propria cellularity reflects the severity of inflammation in the distal intestine. Based on the degree of cellular infiltration within the lamina propria, a value of normal, mild, moderate, marked or severe was assigned. To make the data appropriate for the association testing, the highly skewed five-category scores were collapsed into more balanced binary data, i.e., normal and abnormal. The immune-related genes included for the association testing were myeloid differentiation factor 88 (myd88), interleukin 1β (il1β), interleukin 8 (il8), cluster of differentiation 3 γδ (cd3γδ), transforming growth factor β1 (tgfβ1), interferon γ (ifnγ), interleukin 17A (il17a), fork-head box P3 (foxp3) and interleukin 10 (il10), whose expression levels were higher in the distal intestine of fish assigned abnormal regarding lamina propria cellularity. Since the expression levels of immune-related genes were highly correlated, we ran a principal component analysis (PCA) and extracted the first principle component (PC1) for the association testing to avoid multicollinearity and reduce the number of association testing. For genes relevant to the barrier function, which included claudin-15 (cldn15), claudin-25b (cldn25b), zonula occludens 1 (zo1), E-cadherin / cadherin 1 (cdh1) and mucin-2 (muc2), we also used the PC1 of the PCA for the association testing based on the same considerations.
Statistics
All the statistical analyses were run in R except for the PERMANOVA, which was run in PRIMER (version 7; PRIMER-e). The differences in the alpha-diversity indices were compared using linear mixed-effects models via the lme4 package [132]. Predictor variables in the models included the fixed effects Diet + Sample origin + Diet x Sample origin, and the random effects FishID + NetPen. The models were validated by visual inspections of residual diagnostic plots generated by the ggResidpanel package [133]. The statistical significance of fixed predictors was estimated by Type III ANOVA with Kenward-Roger’s approximation of denominator degrees of freedom via the lmerTest package [134]. When the interaction between the main effects was significant, conditional contrasts for the main effects were made via the emmeans package [135]. To compare the differences in beta-diversity, we performed the PERMANOVA [136] using the same predictors included in the linear mixed-effects models. Terms with negative estimates for components of variation were sequentially removed from the model via term pooling, starting with the one showing the smallest mean squares. At each step, the model was reassessed whether more terms needed to be removed or not. Conditional contrasts for the main effects were constructed when their interaction was significant. Monte Carlo p values were computed as well when the unique permutations for the terms in the PERMANOVA were small (< 100). The homogeneity of multivariate dispersions among groups was visually assessed with boxplots and was formally tested by the permutation test, PERMDISP [137], via the R package vegan [138]. Multiple comparisons were adjusted by the Benjamini-Hochberg procedure where applicable. Differences were regarded as significant when p < 0.05.
Supplementary Information
Additional file 1: Figure S1. Quantification of bacterial 16S rRNA gene in different sample types using qPCR. Since the Cq values of most mucosa-associated samples were out of the linear range of the standard curve, the Cq value was used as a proxy of 16S rRNA gene quantity which is reliable for the screening of contaminant sequences. Data are presented as mean ± 1 standard deviation overlaying the raw data points. Abbreviations: REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa. Figure S2. Taxonomic profile of the mock (A) and contaminating features in the negative controls (B). The lowest level of taxonomic ranks was displayed for each taxon. EB, extraction blank; LB, library blank. Figure S3. Microbial clades showing significant associations with sample origin. p__, phylum; o__, order; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations; REF, reference diet; IM, insect meal diet. Figure S4. Microbial clades showing significant associations with diet. p__, phylum; o__, order; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations; REF, reference diet; IM, insect meal diet. Figure S5. Microbial clades showing significant associations with histological scores on lamina propria cellularity in the distal intestine. p__, phylum; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S6. Microbial clades showing significant associations with distal intestine somatic index (DISI). FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S7. Microbial clades showing significant associations with immune gene expressions in the distal intestine. Since the expression levels of immune genes were highly correlated, we ran a principle component analysis (PCA) and used the first principle component (PC1) for the association testing to avoid multicollinearity and reduce the number of association testing. Note that the expression levels of immune genes decrease as the PC1 increases from left to right. Hence, a positive correlation coefficient denotes a negative association between the microbial clade and immune gene expressions, and vice versa. f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S8. Microbial clades showing significant associations with expressions of barrier function related genes in the distal intestine. Since the expression levels of barrier function related genes were highly correlated, we ran a principle component analysis (PCA) and used the first principle component (PC1) for the association testing to avoid multicollinearity and reduce the number of association testing. Note that the expression levels of barrier function related genes decrease as the PC1 increases from left to right. Hence, a positive correlation coefficient denotes a negative association between the microbial clade and barrier function related gene expressions, and vice versa. f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S9. Rarefaction curves based on Observed ASVs for the different sample types. The rarefaction analysis showed that mucosa samples (REF-DIM, IM-DIM) reached the saturation phase at a subsampling depth of 2000 sequences whereas digesta samples (REF-DID, IM-DID) reached the saturation phase at a subsampling depth of 9500 sequences. To preserve a maximum number of samples for the downstream data analysis, we rarefied the ASV table to 2047 sequences per sample which left out 2 samples. To ensure that the subsampling depth of 2047 sequences per sample produced reliable comparisons of microbial communities, we computed compositionality-aware distance matrices, the Aitchison distance and PHILR transformed Euclidean distance, which do not require rarefying and use all the sequences in the samples.
Additional file 2: Table S1. Contaminating features removed from the ASV table.
Additional file 3: Table S2. The prevalence of core ASVs in different sample types.
Acknowledgments
The authors wish to thank Ellen K. Hage for organizing the sampling and technicians at the GIFAS for their committed animal care and supports during the sampling.
Abbreviations
- ASVs
amplicon sequence variants
- BSF
black soldier fly
- Cq
quantification cycle
- DID
distal intestine digesta
- DIM
distal intestine mucosa
- DISI
distal intestine somatic index
- IM
insect meal
- LPC
lamina propria cellularity
- PC1
principal component one
- PCA
principal component analysis
- PCoA
principal coordinates analysis
- PD
phylogenetic diversity
- PERMANOVA
permutational multivariate analysis of variance
- PHILR
phylogenetic isometric log-ratio
- REF
reference
Authors’ contributions
TMK and ÅK concepted and designed the study. YL, LB and KG participated in the sample collection. YL, LB and AJ-T carried out the laboratory works. YL performed the bioinformatics, statistical analyses and data visualization. YL and LB completed the first draft of the manuscript. All the authors read, revised and approved the final version of the manuscript.
Funding
YL was granted a scholarship from the China Scholarship Council to pursue his PhD degree at Norwegian University of Life Sciences. This work was a spin-off of the “AquaFly” project (grant number, 238997), funded by the Research Council of Norway and managed by the Institute of Marine Research. Costs related to this study were covered by Norwegian University of Life Sciences. The funding agencies had no role in study design, data collection and interpretation, decision to publish or preparation of the manuscript.
Availability of data and materials
The raw 16S rRNA gene sequencing data are deposited at the NCBI SRA database under the BioProject PRJNA555355. Other raw data and code for reproducing our results are available from the GitHub repository (https://github.com/yanxianl/Li_AqFl2-Microbiota_ASM_2020).
Ethics approval and consent to participate
The experiment was conducted in compliance with the Norwegian Animal Welfare Act 10/07/2009 and the EU Directive on the Protection of Animals used for Scientific Purposes (2010/63/EU).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanxian Li and Leonardo Bruni contributed equally to this work.
References
- 1.United Nations. World population prospects 2019: Highlights [https://population.un.org/wpp/Publications/. Accessed 3 Oct 2020.
- 2.Hunter MC, Smith RG, Schipanski ME, Atwood LW, Mortensen DA. Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience. 2017;67:386–391. doi: 10.1093/biosci/bix010. [DOI] [Google Scholar]
- 3.Tacon AGJ, Metian M. Fish matters: importance of aquatic foods in human nutrition and global food supply. Rev Fish Sci. 2013;21:22–38. doi: 10.1080/10641262.2012.753405. [DOI] [Google Scholar]
- 4.Khalili Tilami S, Sampels S. Nutritional value of fish: lipids, proteins, vitamins, and minerals. Rev Fish Sci Aquac. 2018;26:243–253. doi: 10.1080/23308249.2017.1399104. [DOI] [Google Scholar]
- 5.FAO . The state of world fisheries and aquaculture. Rome: FAO; 2020. [Google Scholar]
- 6.FAO . The state of world fisheries and aquaculture. Rome: FAO; 2018. [Google Scholar]
- 7.Aas TS, Ytrestøyl T, Åsgård T. Utilization of feed resources in the production of Atlantic salmon (Salmo salar) in Norway: an update for 2016. Aquac Rep. 2019;15:100216. doi: 10.1016/j.aqrep.2019.100216. [DOI] [Google Scholar]
- 8.Sánchez-Muros M-J, Barroso FG, Manzano-Agugliaro F. Insect meal as renewable source of food for animal feeding: a review. J Clean Prod. 2014;65:16–27. doi: 10.1016/j.jclepro.2013.11.068. [DOI] [Google Scholar]
- 9.Glencross BD, Huyben D, Schrama JW. The application of single-cell ingredients in aquaculture feeds—a review. Fishes. 2020;5:22. doi: 10.3390/fishes5030022. [DOI] [Google Scholar]
- 10.Barroso FG, de Haro C, Sánchez-Muros M-J, Venegas E, Martínez-Sánchez A, Pérez-Bañón C. The potential of various insect species for use as food for fish. Aquaculture. 2014;422-423:193–201. doi: 10.1016/j.aquaculture.2013.12.024. [DOI] [Google Scholar]
- 11.Bondari K, Sheppard D. Soldier fly, Hermetia illucens L., larvae as feed for channel catfish, Ictalurus punctatus (Rafinesque), and blue tilapia, Oreochromis aureus (Steindachner) Aquac Fish Manage. 1987;18:209–220. [Google Scholar]
- 12.Devic E, Leschen W, Murray F, Little DC. Growth performance, feed utilization and body composition of advanced nursing Nile tilapia (Oreochromis niloticus) fed diets containing black soldier fly (Hermetia illucens) larvae meal. Aquac Nutr. 2017;24:416–423. doi: 10.1111/anu.12573. [DOI] [Google Scholar]
- 13.Hu JR, Wang GX, Huang YH, Sun YP, He F, Zhao HX, Li NN. Effects of substitution of fish meal with black soldier fly (Hermetia illucens) larvae meal, in yellow catfish (Pelteobagrus fulvidraco) diets. Isr J Aquac. 2017;6:1382–90.
- 14.Kroeckel S, Harjes AGE, Roth I, Katz H, Wuertz S, Susenbeth A, Schulz C. When a turbot catches a Fly: evaluation of a pre-pupae meal of the black soldier fly (Hermetia illucens) as fish meal substitute — growth performance and chitin degradation in juvenile turbot (Psetta maxima) Aquaculture. 2012;364-365:345–352. doi: 10.1016/j.aquaculture.2012.08.041. [DOI] [Google Scholar]
- 15.Li SL, Ji H, Zhang BX, Zhou JS, Yu HB. Defatted black soldier fly (Hermetia illucens) larvae meal in diets for juvenile Jian carp (Cyprinus carpio var. Jian): growth performance, antioxidant enzyme activities, digestive enzyme activities, intestine and hepatopancreas histological structure. Aquaculture. 2017;477:62–70. doi: 10.1016/j.aquaculture.2017.04.015. [DOI] [Google Scholar]
- 16.Lock ER, Arsiwalla T, Waagbo R. Insect larvae meal as an alternative source of nutrients in the diet of Atlantic salmon (Salmo salar) postsmolt. Aquac Nutr. 2016;22:1202–1213. doi: 10.1111/anu.12343. [DOI] [Google Scholar]
- 17.Magalhaes R, Sanchez-Lopez A, Leal RS, Martinez-Llorens S, Oliva-Telesa A, Peres H. Black soldier fly (Hermetia illucens) pre-pupae meal as a fish meal replacement in diets for European seabass (Dicentrarchus labrax) Aquaculture. 2017;476:79–85. doi: 10.1016/j.aquaculture.2017.04.021. [DOI] [Google Scholar]
- 18.Renna M, Schiavone A, Gai F, Dabbou S, Lussiana C, Malfatto V, Prearo M, Capucchio MT, Biasato I, Biasibetti E, et al: Evaluation of the suitability of a partially defatted black soldier fly (Hermetia illucens L.) larvae meal as ingredient for rainbow trout (Oncorhynchus mykiss Walbaum) diets. J Anim Sci Biotechnol. 2017;8:57. [DOI] [PMC free article] [PubMed]
- 19.Sealey WM, Gaylord TG, Barrows FT, Tomberlin JK, McGuire MA, Ross C, St-Hilaire S. Sensory analysis of rainbow trout, Oncorhynchus mykiss, fed enriched black soldier fly prepupae, Hermetia illucens. J World Aquac Soc. 2011;42:34–45. doi: 10.1111/j.1749-7345.2010.00441.x. [DOI] [Google Scholar]
- 20.St-Hilaire S, Sheppard C, Tomberlin JK, Irving S, Newton L, McGuire MA, Mosley EE, Hardy RW, Sealey W. Fly prepupae as a feedstuff for rainbow trout, Oncorhynchus mykiss. J World Aquac Soc. 2007;38:59–67. doi: 10.1111/j.1749-7345.2006.00073.x. [DOI] [Google Scholar]
- 21.Vargas A, Randazzo B, Riolo P, Truzzi C, Gioacchini G, Giorgini E, Loreto N, Ruschioni S, Zarantoniello M, Antonucci M. Rearing zebrafish on black soldier fly (Hermetia illucens): biometric, histological, spectroscopic, biochemical, and molecular implications. Zebrafish. 2018;15(4):404–19. [DOI] [PubMed]
- 22.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falcinelli S, Picchietti S, Rodiles A, Cossignani L, Merrifield DL, Taddei AR, Maradonna F, Olivotto I, Gioacchini G, Carnevali O. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci Rep. 2015;5:9336. doi: 10.1038/srep09336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol. 2018;3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajardo K, Jaramillo-Torres A, Kortner TM, Merrifield DL, Tinsley J, Bakke AM, Krogdahl Å. Alternative protein sources in the diet modulate microbiota and functionality in the distal intestine of Atlantic salmon (Salmo salar) Appl Environ Microbiol. 2017;83:e02615–e02616. doi: 10.1128/AEM.02615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Angell IL, Rod Sandve S, Snipen LG, Olsen Y, Rudi K. Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma-dominated gut microbiota at sea. Aquac Environ Interact. 2019;11:31–39. doi: 10.3354/aei00297. [DOI] [Google Scholar]
- 34.Schmidt V, Amaral-Zettler L, Davidson J, Summerfelt S, Good C. Influence of fishmeal-free diets on microbial communities in Atlantic salmon (Salmo salar) recirculation aquaculture systems. Appl Environ Microbiol. 2016;82:4470–4481. doi: 10.1128/AEM.00902-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajardo K, Rodiles A, Kortner TM, Krogdahl Å, Bakke AM, Merrifield DL, Sorum H. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci Rep. 2016;6:30893. doi: 10.1038/srep30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Looft T, Allen HK, Cantarel BL, Levine UY, Bayles DO, Alt DP, Henrissat B, Stanton TB. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014;8:1566. doi: 10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyons PP, Turnbull JF, Dawson KA, Crumlish M. Exploring the microbial diversity of the distal intestinal lumen and mucosa of farmed rainbow trout Oncorhynchus mykiss (Walbaum) using next generation sequencing (NGS) Aquac Res. 2017;48:77–91. doi: 10.1111/are.12863. [DOI] [Google Scholar]
- 39.Huyben D, Sun L, Moccia R, Kiessling A, Dicksved J, Lundh T. Dietary live yeast and increased water temperature influence the gut microbiota of rainbow trout. J Appl Microbiol. 2018;124:1377–1392. doi: 10.1111/jam.13738. [DOI] [PubMed] [Google Scholar]
- 40.Jaramillo-Torres A, Rawling MD, Rodiles A, Mikalsen HE, Johansen LH, Tinsley J, Forberg T, Aasum E, Castex M, Merrifield DL. Influence of Dietary Supplementation of Probiotic Pediococcus acidilactici MA18/5M During the Transition From Freshwater to Seawater on Intestinal Health and Microbiota of Atlantic Salmon (Salmo salar L.) Front Microbiol. 2019;10:2243. doi: 10.3389/fmicb.2019.02243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimoldi S, Gini E, Iannini F, Gasco L, Terova G. The effects of dietary insect meal from Hermetia illucens Prepupae on autochthonous gut microbiota of rainbow trout (Oncorhynchus mykiss). Animals. 2019;9(4):143. [DOI] [PMC free article] [PubMed]
- 42.Terova G, Rimoldi S, Ascione C, Gini E, Ceccotti C, Gasco L. Rainbow trout (Oncorhynchus mykiss) gut microbiota is modulated by insect meal from Hermetia illucens prepupae in the diet. Rev Fish Biol Fish. 2019;29:465–486. doi: 10.1007/s11160-019-09558-y. [DOI] [Google Scholar]
- 43.Bruni L, Belghit I, Lock E-J, Secci G, Taiti C, Parisi G. Total replacement of dietary fish meal with black soldier fly (Hermetia illucens) larvae does not impair physical, chemical or volatile composition of farmed Atlantic salmon (Salmo salar L.) J Sci Food Agric. 2020;100:1038–1047. doi: 10.1002/jsfa.10108. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Kortner TM, Chikwati EM, Belghit I, Lock E-J, Krogdahl Å. Total replacement of fish meal with black soldier fly (Hermetia illucens) larvae meal does not compromise the gut health of Atlantic salmon (Salmo salar) Aquaculture. 2020;520:734967. doi: 10.1016/j.aquaculture.2020.734967. [DOI] [Google Scholar]
- 45.Belghit I, Liland NS, Gjesdal P, Biancarosa I, Menchetti E, Li Y, Waagbø R, Krogdahl Å, Lock E-J. Black soldier fly larvae meal can replace fish meal in diets of sea-water phase Atlantic salmon (Salmo salar) Aquaculture. 2019;503:609–619. doi: 10.1016/j.aquaculture.2018.12.032. [DOI] [Google Scholar]
- 46.Dehler CE, Secombes CJ, Martin SA. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.) Sci Rep. 2017;7:13877. doi: 10.1038/s41598-017-13249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dehler CE, Secombes CJ, Martin SAM. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.) Aquaculture. 2017;467:149–157. doi: 10.1016/j.aquaculture.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Feckaninova A, Lokesh J, Koscova J, Sorensen M, Femandes J, Kiron V. Lactobacillus dominate in the intestine of Atlantic salmon fed dietary probiotics. Front Microbiol. 2019;9:3247. [DOI] [PMC free article] [PubMed]
- 49.Gupta S, Lokesh J, Abdelhafiz Y, Siriyappagouder P, Pierre R, Sorensen M, Fernandes JMO, Kiron V. Macroalga-derived alginate oligosaccharide alters intestinal bacteria of Atlantic salmon. Front Microbiol. 2019;10. [DOI] [PMC free article] [PubMed]
- 50.Llewellyn MS, McGinnity P, Dionne M, Letourneau J, Thonier F, Carvalho GR, Creer S, Derome N. The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. ISME J. 2015;10:1280–1284. doi: 10.1038/ismej.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster TMU, Consuegra S, Hitchings M, de Leaniz CG. Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity. Appl Environ Microbiol. 2018;84:e00691–e00618. doi: 10.1128/AEM.00691-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fogarty C, Burgess CM, Cotter PD, Cabrera-Rubio R, Whyte P, Smyth C, Bolton DJ. Diversity and composition of the gut microbiota of Atlantic salmon (Salmo salar) farmed in Irish waters. J Appl Microbiol. 2019;127:648–657. doi: 10.1111/jam.14291. [DOI] [PubMed] [Google Scholar]
- 53.Godoy FA, Miranda CD, Wittwer GD, Aranda CP, Calderon R. High variability of levels of Aliivibrio and lactic acid bacteria in the intestinal microbiota of farmed Atlantic salmon Salmo salar L. Ann Microbiol. 2015;65:2343–2353. doi: 10.1007/s13213-015-1076-3. [DOI] [Google Scholar]
- 54.Karlsen C, Ottem K, Brevik ØJ, Davey M, Sørum H, Winther-Larsen H. The environmental and host-associated bacterial microbiota of Arctic seawater-farmed Atlantic salmon with ulcerative disorders. J Fish Dis. 2017;40:1645–1663. doi: 10.1111/jfd.12632. [DOI] [PubMed] [Google Scholar]
- 55.Zarkasi KZ, Abell GCJ, Taylor RS, Neuman C, Hatje E, Tamplin ML, Katouli M, Bowman JP. Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J Appl Microbiol. 2014;117:18–27. doi: 10.1111/jam.12514. [DOI] [PubMed] [Google Scholar]
- 56.Zarkasi KZ, Taylor RS, Abell GC, Tamplin ML, Glencross BD, Bowman JP. Atlantic salmon (Salmo salar L.) gastrointestinal microbial community dynamics in relation to digesta properties and diet. Microb Ecol. 2016;71:589–603. doi: 10.1007/s00248-015-0728-y. [DOI] [PubMed] [Google Scholar]
- 57.Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol. 2007;57:2823–2829. doi: 10.1099/ijs.0.65081-0. [DOI] [PubMed] [Google Scholar]
- 58.Dunlap PV, Ast JC, Kimura S, Fukui A, Yoshino T, Endo H. Phylogenetic analysis of host–symbiont specificity and codivergence in bioluminescent symbioses. Cladistics. 2007;23:507–532. doi: 10.1111/j.1096-0031.2007.00157.x. [DOI] [Google Scholar]
- 59.Egidius E, Wiik R, Andersen K, Hoff K, Hjeltnes B. Vibrio salmonicida sp. nov., a new fish pathogen. Int J Syst Evol Microbiol. 1986;36:518–520. [Google Scholar]
- 60.Lunder T, Sørum H, Holstad G, Steigerwalt AG, Mowinckel P, Brenner DJ. Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with 'winter ulcer'. Int J Syst Evol Microbiol. 2000;50:427–450. doi: 10.1099/00207713-50-2-427. [DOI] [PubMed] [Google Scholar]
- 61.Webster TMU, Rodriguez-Barreto D, Castaldo G, Gough P, Consuegra S. Garcia de Leaniz C: environmental plasticity and colonisation history in the Atlantic salmon microbiome: a translocation experiment. Mol Ecol. 2020;29:886–898. doi: 10.1111/mec.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson JF, Johnson RC, Magnarelli LA, Hyde FW, Andreadis TG. New infectious spirochete isolated from short-tailed shrews and white-footed mice. J Clin Microbiol. 1987;25:1490–1494. doi: 10.1128/JCM.25.8.1490-1494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown RM, Wiens GD, Salinas I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2019;86:497–506. doi: 10.1016/j.fsi.2018.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tapia-Paniagua S, Vidal S, Lobo C, Prieto-Álamo M, Jurado J, Cordero H, Cerezuela R, de la Banda IG, Esteban M, Balebona M. The treatment with the probiotic Shewanella putrefaciens Pdp11 of specimens of Solea senegalensis exposed to high stocking densities to enhance their resistance to disease. Fish Shellfish Immunol. 2014;41:209–221. doi: 10.1016/j.fsi.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 65.Heys C, Cheaib B, Busetti A, Kazlauskaite R, Maier L, Sloan WT, Ijaz UZ, Kaufmann J, McGinnity P, Llewellyn MS. Neutral processes dominate microbial community assembly in Atlantic salmon, Salmo salar. Appl Environ Microbiol. 2020;86(8):e02283–19. [DOI] [PMC free article] [PubMed]
- 66.Holben W, Williams P, Saarinen M, Särkilahti L, Apajalahti J. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb Ecol. 2002;44:175–185. doi: 10.1007/s00248-002-1011-6. [DOI] [PubMed] [Google Scholar]
- 67.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/MMBR.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheaib B, Yang P, Kazlauskaite R, Lindsay E, Heys C, De Noa M, Schaal P, Dwyer T, Sloan W, Ijaz UZ, Llewellyn MS: Unpicking the mysterious symbiosis of Mycoplasma in salmonids. Preprint at https://wwwbiorxivorg/content/101101/20200717209767v1 2020.
- 69.Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol Rev. 2011;35:681–704. doi: 10.1111/j.1574-6976.2011.00270.x. [DOI] [PubMed] [Google Scholar]
- 70.Kennedy M, Rosnick D, Ulrich R, Yancey R., Jr Association of Treponema hyodysenteriae with porcine intestinal mucosa. J Gen Microbiol. 1988;134:1565–1576. doi: 10.1099/00221287-134-6-1565. [DOI] [PubMed] [Google Scholar]
- 71.Milner J, Sellwood R. Chemotactic response to mucin by Serpulina hyodysenteriae and other porcine spirochetes: potential role in intestinal colonization. Infect Immun. 1994;62:4095–4099. doi: 10.1128/IAI.62.9.4095-4099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Witters NA, Duhamel GE: Motility-regulated mucin association of Serpulina pilosicoli, the agent of colonic spirochetosis of humans and animals. In Advances in Experimental Medicine and Biology. Volume 473: Springer; 1999: 199–205. [DOI] [PubMed]
- 73.Bruni L, Pastorelli R, Viti C, Gasco L, Parisi G. Characterisation of the intestinal microbial communities of rainbow trout (Oncorhynchus mykiss) fed with Hermetia illucens (black soldier fly) partially defatted larva meal as partial dietary protein source. Aquaculture. 2018;487:56–63. doi: 10.1016/j.aquaculture.2018.01.006. [DOI] [Google Scholar]
- 74.Huyben D, Vidaković A, Hallgren SW, Langeland M. High-throughput sequencing of gut microbiota in rainbow trout (Oncorhynchus mykiss) fed larval and pre-pupae stages of black soldier fly (Hermetia illucens) Aquaculture. 2019;500:485–491. doi: 10.1016/j.aquaculture.2018.10.034. [DOI] [Google Scholar]
- 75.Borrelli L, Coretti L, Dipineto L, Bovera F, Menna F, Chiariotti L, Nizza A, Lembo F, Fioretti A. Insect-based diet, a promising nutritional source, modulates gut microbiota composition and SCFAs production in laying hens. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawasaki K, Hashimoto Y, Hori A, Kawasaki T, Hirayasu H, Iwase S, Hashizume A, Ido A, Miura C, Miura T, et al. Evaluation of black soldier fly (Hermetia illucens) larvae and pre-pupae raised on household organic waste, as potential ingredients for poultry feed. Animals. 2019;9(3):98. [DOI] [PMC free article] [PubMed]
- 77.Jozefiak A, Nogales-Merida S, Mikolajczak Z, Rawski M, Kieronczyk B, Mazurkiewicz J. The utilization of full-fat insect meal in rainbow trout (Oncorhynchus mykiss) nutrition: the effects on growth performance, intestinal microbiota and gastrointestinal tract histomorphology. Ann Anim Sci. 2019;19:747–765. doi: 10.2478/aoas-2019-0020. [DOI] [Google Scholar]
- 78.Jozefiak A, Nogales-Merida S, Rawski M, Kieronczyk B, Mazurkiewicz J: Effects of insect diets on the gastrointestinal tract health and growth performance of Siberian sturgeon (Acipenser baerii Brandt, 1869). BMC Microbiol. 2019;15(1):348. [DOI] [PMC free article] [PubMed]
- 79.Jiang CL, Jin WZ, Tao XH, Zhang Q, Zhu J, Feng SY, Xu XH, Li HY, Wang ZH, Zhang ZJ. Black soldier fly larvae (Hermetia illucens) strengthen the metabolic function of food waste biodegradation by gut microbiome. Microb Biotechnol. 2019;12:528–543. doi: 10.1111/1751-7915.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bruno D, Bonelli M, De Filippis F, Di Lelio I, Tettamanti G, Casartelli M, Ercolini D, Caccia S. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions. Appl Environ Microbiol. 2019;85:e01864–e01818. doi: 10.1128/AEM.01864-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wynants E, Frooninckx L, Crauwels S, Verreth C, De Smet J, Sandrock C, Wohlfahrt J, Van Schelt J, Depraetere S, Lievens B. Assessing the microbiota of black soldier fly larvae (Hermetia illucens) reared on organic waste streams on four different locations at laboratory and large scale. Microb Ecol. 2019;77:913–930. doi: 10.1007/s00248-018-1286-x. [DOI] [PubMed] [Google Scholar]
- 82.Zheng L, Crippen TL, Singh B, Tarone AM, Dowd S, Yu Z, Wood TK, Tomberlin JK. A survey of bacterial diversity from successive life stages of black soldier fly (Diptera: Stratiomyidae) by using 16S rDNA pyrosequencing. J Med Entomol. 2013;50:647–658. doi: 10.1603/ME12199. [DOI] [PubMed] [Google Scholar]
- 83.Emerson JB, Adams RI, Roman CMB, Brooks B, Coil DA, Dahlhausen K, Ganz HH, Hartmann EM, Hsu T, Justice NB, et al. Schrodinger's microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome. 2017;5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cody R. Distribution of chitinase and chitobiase in Bacillus. Curr Microbiol. 1989;19:201–205. doi: 10.1007/BF01570162. [DOI] [Google Scholar]
- 85.Askarian F, Zhou ZG, Olsen RE, Sperstad S, Ringo E. Culturable autochthonous gut bacteria in Atlantic salmon (Salmo salar L.) fed diets with or without chitin. Characterization by 16S rRNA gene sequencing, ability to produce enzymes and in vitro growth inhibition of four fish pathogens. Aquaculture. 2012;326:1–8. doi: 10.1016/j.aquaculture.2011.10.016. [DOI] [Google Scholar]
- 86.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLaren MR, Willis AD, Callahan BJ. Consistent and correctable bias in metagenomic sequencing experiments. Elife. 2019;8:e46923. [DOI] [PMC free article] [PubMed]
- 88.Brooks JP, Edwards DJ, Harwich MD, Rivera MC, Fettweis JM, Serrano MG, Reris RA, Sheth NU, Huang B, Girerd P. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung F-E. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35:1069. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 91.Santiago A, Panda S, Mengels G, Martinez X, Azpiroz F, Dore J, Guarner F, Manichanh C. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. doi: 10.1186/1471-2180-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lazarevic V, Gaia N, Girard M, Schrenzel J. Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. 2016;16:73. doi: 10.1186/s12866-016-0689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jousselin E, Clamens AL, Galan M, Bernard M, Maman S, Gschloessl B, Duport G, Meseguer AS, Calevro F. Coeur d'acier a: assessment of a 16S rRNA amplicon Illumina sequencing procedure for studying the microbiome of a symbiont-rich aphid genus. Mol Ecol Resour. 2016;16:628–640. doi: 10.1111/1755-0998.12478. [DOI] [PubMed] [Google Scholar]
- 95.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 96.Jervis-Bardy J, Leong LEX, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, Nosworthy E, Morris PS, O'Leary S, Rogers GB, Marsh RL. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016;8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O'Hanlon JF. Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol. 2002;68:1548–1555. doi: 10.1128/AEM.68.4.1548-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett. 2003;219:87–91. doi: 10.1016/S0378-1097(02)01190-4. [DOI] [PubMed] [Google Scholar]
- 102.Cantas L, Fraser TWK, Fjelldal PG, Mayer I, Sorum H. The culturable intestinal microbiota of triploid and diploid juvenile Atlantic salmon (Salmo salar) - a comparison of composition and drug resistance. BMC Vet Res. 2011;7:71. doi: 10.1186/1746-6148-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Navarrete P, Fuentes P, De la Fuente L, Barros L, Magne F, Opazo R, Ibacache C, Espejo R, Romero J. Short-term effects of dietary soybean meal and lactic acid bacteria on the intestinal morphology and microbiota of Atlantic salmon (Salmo salar) Aquac Nutr. 2013;19:827–836. doi: 10.1111/anu.12047. [DOI] [Google Scholar]
- 104.Hatje E, Neuman C, Stevenson H, Bowman JP, Katouli M. Population dynamics of Vibrio and Pseudomonas species isolated from farmed Tasmanian Atlantic salmon (Salmo salar L.): a seasonal study. Microb Ecol. 2014;68:679–687. doi: 10.1007/s00248-014-0462-x. [DOI] [PubMed] [Google Scholar]
- 105.He XP, Chaganti SR, Heath DD. Population-specific responses to interspecific competition in the gut microbiota of two Atlantic salmon (Salmo salar) populations. Microb Ecol. 2018;75:140–151. doi: 10.1007/s00248-017-1035-6. [DOI] [PubMed] [Google Scholar]
- 106.Laurence M, Hatzis C, Brash DE. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One. 2014;9:e97876. doi: 10.1371/journal.pone.0097876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barton HA, Taylor NM, Lubbers BR, Pemberton AC. DNA extraction from low-biomass carbonate rock: an improved method with reduced contamination and the low-biomass contaminant database. J Microbiol Methods. 2006;66:21–31. doi: 10.1016/j.mimet.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 109.Weyrich LS, Farrer AG, Eisenhofer R, Arriola LA, Young J, Selway CA, Handsley-Davis M, Adler CJ, Breen J, Cooper A. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019;19:982–996. doi: 10.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanner MA, Goebel BM, Dojka MA, Pace NR. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/AEM.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 112.de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, Peacock SJ, Smith GCS, Parkhill J. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 113.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vandeputte D, Kathagen G, D'Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 115.Ramseier CA, Kinney JS, Herr AE, Braun T, Sugai JV, Shelburne CA, Rayburn LA, Tran HM, Singh AK, Giannobile WV. Identification of pathogen and host-response markers correlated with periodontal disease. J Periodontol. 2009;80:436–446. doi: 10.1902/jop.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rasmussen R: Quantification on the LightCycler. In Rapid cycle real-time PCR. Springer; 2001: 21–34.
- 117.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Illumina I: 16S Metagenomic sequencing library preparation. Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System 2013:1–28.
- 119.R Core Team . R: A language and environment for statistical computing. 2013. [Google Scholar]
- 120.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Janssen S, McDonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, Winker K, Kado DM, Orwoll E, Manary M. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3:e00021–e00018. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bisanz JE. qiime2R: Importing QIIME2 artifacts and associated data into R sessions. 2019. [Google Scholar]
- 126.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lahti L, Shetty S, Blake T, Salojarvi J. Microbiome r package. Tools Microbiome Anal R. 2017.
- 128.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 129.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, Zengler K. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019;4:e00016–e00019. doi: 10.1128/mSystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Silverman JD, Washburne AD, Mukherjee S, David LA. A phylogenetic transform enhances analysis of compositional microbiota data. Elife. 2017;6:e21887. doi: 10.7554/eLife.21887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 133.Goode K, Rey K: ggResidpanel: panels and interactive versions of diagnostic plots using 'ggplot2'. 2019.
- 134.Kuznetsova A, Brockhoff PB, Christensen RHB: lmerTest package: tests in linear mixed effects models. J Stat Softw 2017, 82.
- 135.Lenth R. emmeans: estimated marginal means, aka least-squares means. 2019. [Google Scholar]
- 136.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 137.Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 138.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, et al. vegan: community ecology package. 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Quantification of bacterial 16S rRNA gene in different sample types using qPCR. Since the Cq values of most mucosa-associated samples were out of the linear range of the standard curve, the Cq value was used as a proxy of 16S rRNA gene quantity which is reliable for the screening of contaminant sequences. Data are presented as mean ± 1 standard deviation overlaying the raw data points. Abbreviations: REF, reference diet; IM, insect meal diet; DID, distal intestine digesta; DIM, distal intestine mucosa. Figure S2. Taxonomic profile of the mock (A) and contaminating features in the negative controls (B). The lowest level of taxonomic ranks was displayed for each taxon. EB, extraction blank; LB, library blank. Figure S3. Microbial clades showing significant associations with sample origin. p__, phylum; o__, order; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations; REF, reference diet; IM, insect meal diet. Figure S4. Microbial clades showing significant associations with diet. p__, phylum; o__, order; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations; REF, reference diet; IM, insect meal diet. Figure S5. Microbial clades showing significant associations with histological scores on lamina propria cellularity in the distal intestine. p__, phylum; f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S6. Microbial clades showing significant associations with distal intestine somatic index (DISI). FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S7. Microbial clades showing significant associations with immune gene expressions in the distal intestine. Since the expression levels of immune genes were highly correlated, we ran a principle component analysis (PCA) and used the first principle component (PC1) for the association testing to avoid multicollinearity and reduce the number of association testing. Note that the expression levels of immune genes decrease as the PC1 increases from left to right. Hence, a positive correlation coefficient denotes a negative association between the microbial clade and immune gene expressions, and vice versa. f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S8. Microbial clades showing significant associations with expressions of barrier function related genes in the distal intestine. Since the expression levels of barrier function related genes were highly correlated, we ran a principle component analysis (PCA) and used the first principle component (PC1) for the association testing to avoid multicollinearity and reduce the number of association testing. Note that the expression levels of barrier function related genes decrease as the PC1 increases from left to right. Hence, a positive correlation coefficient denotes a negative association between the microbial clade and barrier function related gene expressions, and vice versa. f__, family; FDR, false discovery rate; N.not.zero, number of non-zero observations. Figure S9. Rarefaction curves based on Observed ASVs for the different sample types. The rarefaction analysis showed that mucosa samples (REF-DIM, IM-DIM) reached the saturation phase at a subsampling depth of 2000 sequences whereas digesta samples (REF-DID, IM-DID) reached the saturation phase at a subsampling depth of 9500 sequences. To preserve a maximum number of samples for the downstream data analysis, we rarefied the ASV table to 2047 sequences per sample which left out 2 samples. To ensure that the subsampling depth of 2047 sequences per sample produced reliable comparisons of microbial communities, we computed compositionality-aware distance matrices, the Aitchison distance and PHILR transformed Euclidean distance, which do not require rarefying and use all the sequences in the samples.
Additional file 2: Table S1. Contaminating features removed from the ASV table.
Additional file 3: Table S2. The prevalence of core ASVs in different sample types.
Data Availability Statement
The raw 16S rRNA gene sequencing data are deposited at the NCBI SRA database under the BioProject PRJNA555355. Other raw data and code for reproducing our results are available from the GitHub repository (https://github.com/yanxianl/Li_AqFl2-Microbiota_ASM_2020).