To The Editor: The duration of transmissibility of coronavirus disease 2019 (Covid-19) and the associated level of contagion have been uncertain. We cultured severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in serial respiratory samples obtained from hospitalized patients with Covid-19 to assess the duration of shedding of viable virus.
The data reported here represent all the patients with Covid-19, as confirmed by positive real-time reverse transcriptase–polymerase chain reaction (RT-PCR) testing, who were hospitalized at Chung-Ang University Hospital in Seoul, South Korea, between February and June 2020. The Allplex 2019-nCoV Assay (Seegene) for nasopharyngeal and oropharyngeal samples was used for real-time RT-PCR testing.1 Patients were isolated until two consecutive negative or inconclusive results on real-time RT-PCR were documented, at least 24 hours apart.2,3 We endeavored to obtain samples at approximately 2-day intervals, but this was not always possible. Viral RNA was quantitated with the use of the cycle-threshold value for the N gene of SARS-CoV-2.4 Viral cultures were conducted by means of a plaque assay until at least two consecutive cultures showed no growth.
We compared the time from the onset of illness to viral clearance in culture with the time to clearance in real-time RT-PCR tests.5 Detailed methods and sensitivities of the culture and real-time RT-PCR assay and the definition and estimation of the time to viral clearance are described in the Supplementary Appendix, available with the full text of this letter at NEJM.org.
A total of 21 patients with Covid-19 were enrolled. Their clinical characteristics are shown in Table S1 in the Supplementary Appendix. The median age of the patients was 62 years, and 76% of the patients were men. A total of 71% of the patients had pneumonia, and 38% were receiving supplemental oxygen therapy. The median Sequential Organ Failure Assessment (SOFA) score was 0 (scores range from 0 to 24, with higher scores indicating more severe organ dysfunction and a higher risk of death), and the median Acute Physiology and Chronic Health Evaluation (APACHE) II score was 5 (scores range from 0 to 71, with higher scores indicating more severe disease and a higher risk of death); these scores indicated mild-to-moderate illness. A total of 165 samples were tested by means of real-time RT-PCR at intervals of 1 to 5 days (median, 2). Of these 165 samples, 89 were cultured for SARS-CoV-2. The timing of the tests, the kinetics of the viral loads, and the clinical course in each patient are shown in Table S2.
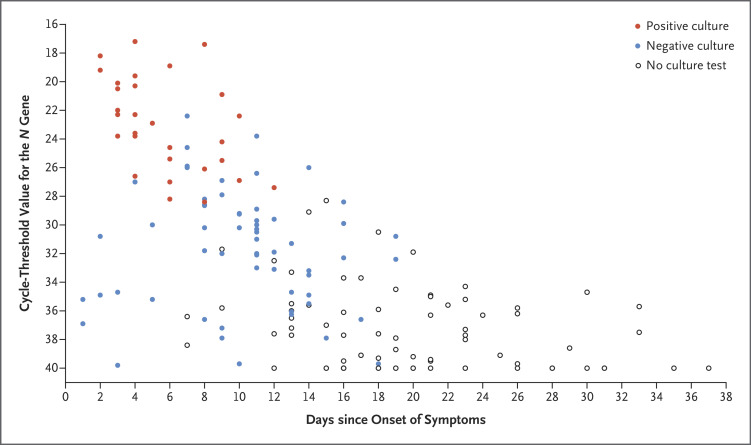
SARS-CoV-2 was cultured in 29 of the 89 samples (33%) (Figure 1). The median time from symptom onset to viral clearance in culture was 7 days (95% confidence interval [CI], 5 to 10), and the median time from symptom onset to viral clearance on real-time RT-PCR was 34 days (lower boundary of the 95% CI, 24 days) (Fig. S1 and Table S4). The latest positive viral culture was 12 days after symptom onset (in Patient 6). Viable virus was identified until 3 days after the resolution in fever (in Patient 14). Viral culture was positive only in samples with a cycle-threshold value of 28.4 or less. The incidence of culture positivity decreased with an increasing time from symptom onset and with an increasing cycle-threshold value (Table S3).
Figure 1. Timing of Presence or Absence of Viable SARS-CoV-2 on Viral Culture and Cycle-Threshold Values for 165 Serial Samples Obtained from 21 Consecutive Patients Hospitalized with Covid-19.
Viral loads were determined with the cycle-threshold value for the N gene of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).4 Sampling intervals ranged from 1 to 5 days (median, 2). Each circle represents a sample obtained on the specified day. Viral culture was positive only in samples with a cycle-threshold value of 28.4 or less and in those that were obtained as long as 12 days after symptom onset. Covid-19 denotes coronavirus disease 2019.
Our findings may be useful in guiding isolation periods for patients with Covid-19 and in estimating the risk of secondary transmission among close contacts in contract tracing. Given the small sample size, inconsistent timing of sampling, and relatively mild illness of the enrolled patients, our results should be verified in larger and more diverse groups of patients.
Supplementary Appendix
Disclosure Forms
This letter was published on January 27, 2021, at NEJM.org.
Footnotes
Supported by Chung-Ang University Research Grants in 2020 (to Dr. Kim) and by a grant (NRF-2018M3A9H4056537, to Dr. Park) from the National Research Foundation of Korea, funded by the Ministry of Science and Information and Communication Technology.
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Allplex 2019-nCoV assay: instructions for use. Seoul, South Korea: Seegene, 2020. (Cat. no. RP10250X/RP10252W.) [Google Scholar]
- 2.Response guidelines to prevent the spread of COVID-19. 8-1 Ed. Korea Disease Control and Prevention Agency, May 20, 2020. (In Korean) (http://ncov.mohw.go.kr).
- 3.Hong KH, Lee SW, Kim TS, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med 2020;40:351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020;26:672-675. [DOI] [PubMed] [Google Scholar]
- 5.Reich NG, Lessler J, Cummings DAT, Brookmeyer R. Estimating incubation period distributions with coarse data. Stat Med 2009;28:2769-2784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.