Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive subtype of leukemia with poor prognosis, and biomarkers and novel therapeutic targets are urgently needed for this disease. Our previous studies have found that inhibition of the B-cell leukemia/lymphoma 11B (BCL11B) gene could significantly promote the apoptosis and growth retardation of T-ALL cells, but the molecular mechanism underlying this effect remains unclear. This study intends to investigate genes downstream of BCL11B and further explore its function in T-ALL cells. We found that PTK7 was a potential downstream target of BCL11B in T-ALL. Compared with the healthy individuals (HIs), PTK7 was overexpressed in T-ALL cells, and BCL11B expression was positively correlated with PTK7 expression. Importantly, BCL11B knockdown reduced PTK7 expression in T-ALL cells. Similar to the effects of BCL11B silencing, downregulation of PTK7 inhibited cell proliferation and induced apoptosis in Molt-4 cells via up-regulating the expression of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and p27. Altogether, our studies suggest that PTK7 is a potential downstream target of BCL11B, and downregulation of PTK7 expression via inhibition of the BCL11B pathway induces growth retardation and apoptosis in T-ALL cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40364-021-00270-3.
To the Editor:
The B-cell leukemia/lymphoma 11B (BCL11B) gene plays an important role in the development of T-cell acute lymphoblastic leukemia (T-ALL) [1, 2]. Our previous studies have shown that down-regulation of BCL11B effectively inhibits proliferation and induces apoptosis of T-ALL cells [3, 4]. PTK7 (protein tyrosine kinase 7), the target protein of sgc8 DNA aptamer, has been identified as a potential biomarker for T-ALL [5]. However, the detailed role and downstream molecular mechanisms of BCL11B and relationship between BCL11B and PTK7 remain undefined. In this study, we determined the BCL11B target genes in T-ALL patients using the Gene Expression Omnibus (GEO) database and used specific siRNAs (small interfering ribonucleic acid) to down-regulate the expression of this gene in T-ALL cell lines to explore the mechanism.
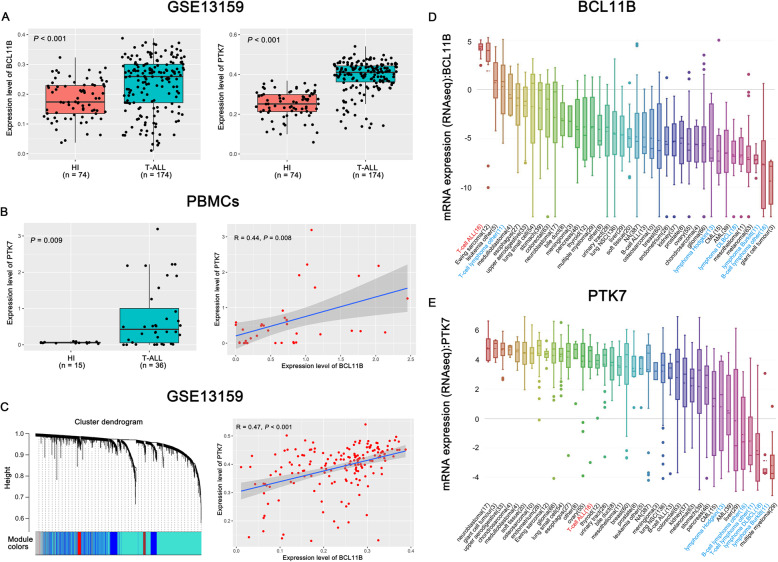
A total of 220 de novo T-ALL patients from the GEO database and 36 peripheral blood mononuclear cells (PBMCs) of T-ALL from our center were used for analysis and validation. In GSE13159 and GSE28497 datasets, we found that the BCL11B was highly expressed in T-ALL (P < 0.001, Fig. 1a and S1a). These results were consistent with our previous study [6]. Moreover, similar results of PTK7 were found in GSE13159, GSE28497 and PBMCs (P < 0.05, Fig. 1a-b and S1b), which was also in line with previous report [7]. Next, to identify genes downstream of BCL11B, the BCL11B co-expression network was further characterized by weighted gene co-expression network analysis (WGCNA). Interestingly, Bioinformatics analysis [8] showed a significant positive correlation between the expression of BCL11B and PTK7 in GSE13159, GSE28497 and PBMCs (P < 0.05, Fig. 1b-c and S1c). Furthermore, we studied the expression of BCL11B and PTK7 in different cell lines from the Cancer Cell Line Encyclopedia (CCLE). Previous studies have shown that BCL11B is overexpressed in the acute type of adult T-cell leukemia/lymphoma (ATLL), and it is under-expressed in other lymphoma type. Consistently, BCL11B and PTK7 was highly expressed in the T-ALL cells line but had low expression in the lymphoma cell lines (Fig. 1d-e). Based on these findings, we proposed that PTK7 might be an important gene downstream of BCL11B in T-ALL.
Fig. 1.
Positive correlation between BCL11B and PTK7 in T-cell acute lymphoblastic leukemia (T-ALL). (a) The expression levels of BCL11B (left panel) and PTK7 (right panel) in T-ALL patients and healthy individuals (HIs) in the GSE13159 dataset. (b) Validation of the correlation between BCL11B and PTK7. Expression levels of the PTK7 gene in PBMCs from T-ALL patients and His (left panel). Linear correlation analysis of the BCL11B and PTK7 genes in T-ALL samples (right panel). (c) BCL11B and PTK7 are positively correlated in the GSE13159 dataset. Left panel: co-expression modules were obtained by weighted gene co-expression network analysis (WGCNA). The colored row beneath the dendrogram shows the module assignment determined by the Dynamic Tree Cut. A cluster dendrogram demonstrated that 10,459 differentially expressed genes were enriched in 11 co-expression network modules, while BCL11B and PTK7 were located in the same blue module. Right panel: a positive correlation was detected between BCL11B and PTK7. Both BCL11B (d) and PTK7 (e) are highly expressed in T-ALL compared to lymphoma cell lines. Data were obtained through the Cancer Cell Line Encyclopedia (CCLE). Detailed methods are available in Materials and Methods in Additional file 1
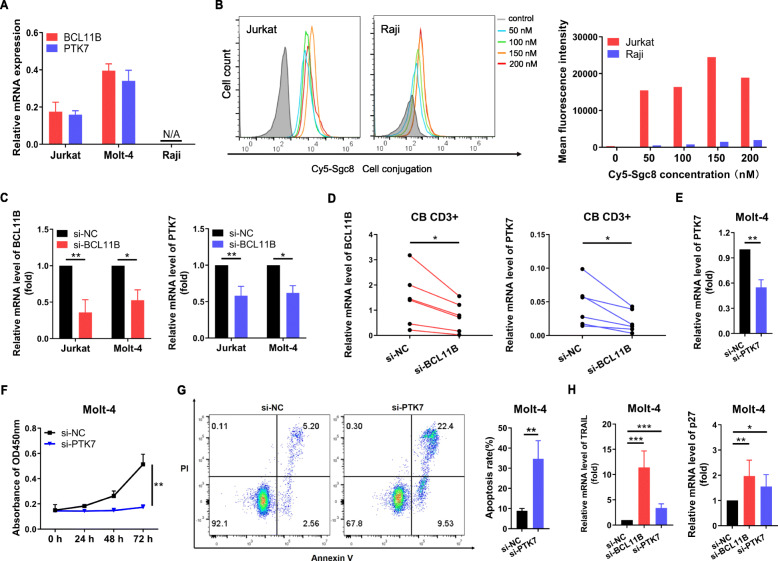
We sought to further verify the association between BCL11B and PTK7 in both T-ALL and non-T-ALL cell lines at the mRNA and protein levels. As shown in Fig. 2a, BCL11B and PTK7 mRNA were highly expressed in T-ALL cell lines (Jurkat and Molt-4), but almost absent in BCL11B-negative Burkitt lymphoma cell line (Raji). Next, an aptamer was used to determine the cell surface protein expression of PTK7 in Jurkat and Raji cells. Sgc-8, the PTK7-specific aptamer, was labeled with a Cy5 fluorescent reporter and incubated with Jurkat and Raji cells under different concentrations, which revealed that Cy5-Sgc8 specifically bound to Jurkat cells but did not react with Raji cells (Fig. 2b). Excitingly, there was a significant decrease in PTK7 mRNA expression after silencing BCL11B expression in the Jurkat and Molt-4 cells and cord blood (CB) CD3+ T cells (Fig. 2c-d). These results confirmed that PTK7 might be regulated by the BCL11B signaling pathway in both T-ALL cell lines and human CD3+ T cells. Based on the above results, we attempted to further understand the role of PTK7 in T-ALL cells. Compared to the negative control, the proliferation of Molt-4 cells transfected with PTK7-siRNA was significantly decreased (P < 0.01, Fig. 2e-f). Meanwhile, the Annexin V/PI-positive cells demonstrated a significant increase for PTK7-siRNA transfected Molt-4 cells, reaching 34.66 ± 5.21% (P = 0.008, Fig. 2g). In addition, recent reports have shown that tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and p27 are found to be involved in the BCL11B pathway to regulate the cell cycle and apoptosis [3, 9, 10]. Interestingly, significant increases in the expression levels of TRAIL and p27 were detected in the PTK7-siRNA group, which was consistent with the trend exhibited in the BCL11B-siRNA group (Fig. 2h). These data suggested that PTK7 was a downstream BCL11B target gene in T-ALL cell growth and apoptosis.
Fig. 2.
Downregulation of PTK7 induces cell growth retardation and apoptosis. (a) The BCL11B and PTK7 mRNA expression levels in T-ALL (Jurkat and Molt-4) and non-T-ALL (Raji) cell lines. GAPDH was used as an endogenous reference. (b) Cultured Jurkat and Raji cells were treated with Cy5-Sgc8 for 1 h at different concentrations. Flow cytometry analysis of the specific cell binding of Cy5-Sgc8 to Jurkat cells. (c) Expression of PTK7 in T-ALL cell lines 48 h after BCL11B siRNA transfection. (d) Effect of BCL11B siRNA on the BCL11B mRNA expression level by real-time quantitative polymerase chain reaction (qRT-PCR) 48 h after transfection in CB CD3+ T cells. Negative control siRNA-treated cells were used for comparison. (e) Knockdown efficiency in Molt-4 cells were analyzed after knocking down the PTK7 gene. (f) Molt-4 cells were transfected with si-PTK7 and then cell proliferation was assessed by Cell counting kit-8 (CCK-8) assay. (g) Apoptosis in Molt-4 cells transfected with si-PTK7 was measured by flow cytometry after 48 h. (h) Expression of the TRAIL and p27 genes in Molt-4 cells 48 h after transfected with BCL11B siRNA and PTK7 siRNA. Non-specific siRNA treated cells were used as negative control. Asterisks signify statistically significant differences (***, P < 0.001; **, P < 0.01; *, P < 0.05)
In summary, this is the first report demonstrating that PTK7 is an important functional downstream target gene of BCL11B in T-ALL. Our study provides rationale for targeting BCL11B/PTK7 in the development of therapeutics for T-cell malignancies.
Supplementary Information
Additional file 1. Materials and Methods.
Additional file 2: Figure S1. Expression patterns of BCL11B and PTK7 in the GSE28497 dataset. High expression of BCL11B (A) and PTK7 (B) in T-ALL. (C) BCL11B and PTK7 had a positive correlation.
Acknowledgements
Not applicable.
Abbreviations
- BCL11B
B-cell leukemia/lymphoma 11B
- CCLE
Cancer cell line encyclopedia
- GEO
Gene expression omnibus database
- HI
Healthy individual
- PTK7
Protein tyrosine kinase receptor 7
- PBMCs
Peripheral blood mononuclear cells
- siRNA
Small interfering ribonucleic acid
- T-ALL
T-cell acute lymphoblastic leukemia
- TRAIL
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand
- WGCNA
Weighted gene co-expression network analysis
Authors’ contributions
Kehan Li, Cunte Chen and Rili Gao performed the experiments, wrote the paper, and analyzed the data. Xibao Yu, Youxue Huang, Zheng Chen, Zhuandi Liu, Shaohua Chen and Xin Huang helped analyze the data. Gengxin Luo provided primary cells and patient information. Grzegorz K. Przybylski, Yangqiu Li, and Chengwu Zeng designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from Intergovernmental International Cooperation on Scientific and Technological Innovation project of Chinese Ministry of Science and Technology (No. 2017YFE0131600), the Guangdong Science and Technology Project (No. 2020A0505100042), the National Science Center, Poland (No. DEC-2013/09/B/NZ1/01867) (GKP.), the Polish National Centre for Research and Development (No. WPC1/BCL/2019) (GKP), the National Natural Science Foundation of China (No.81770158), and the Pearl River S&T Nova Program of Guangzhou, China (No. 201906010002).
Availability of data and materials
Data available on request.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of the affiliated hospitals of Jinan University. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kehan Li, Cunte Chen and Rili Gao contributed equally to this work.
Contributor Information
Grzegorz K. Przybylski, Email: grzegorz.przybylski@igcz.poznan.pl
Yangqiu Li, Email: yangqiuli@hotmail.com.
Chengwu Zeng, Email: bio-zcw@163.com.
References
- 1.Ha VL, Luong A, Li F, Casero D, Malvar J, Kim YM, et al. The T-ALL related gene BCL11B regulates the initial stages of human T-cell differentiation. Leukemia. 2017;31(11):2503–2514. doi: 10.1038/leu.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329(5987):89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabarczyk P, Przybylski GK, Depke M, Völker U, Bahr J, Assmus K, et al. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26(26):3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Chen S, Shen Q, Chen S, Yang L, Grabarczyk P, et al. Down regulation of BCL11B expression inhibits proliferation and induces apoptosis in malignant T cells by BCL11B-935-siRNA. Hematology. 2011;16(4):236–242. doi: 10.1179/102453311X13025568941961. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Li M, Gao X, Chen Y, Liu T. Nanotechnology in cancer diagnosis: progress, challenges and opportunities. J Hematol Oncol. 2019;12(1):137. doi: 10.1186/s13045-019-0833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X, Chen S, Shen Q, Yang L, Li B, Zhong L, et al. Analysis of the expression pattern of the BCL11B gene and its relatives in patients with T-cell acute lymphoblastic leukemia. J Hematol Oncol. 2010;3(1):44. doi: 10.1186/1756-8722-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang G, Zhang M, Yue B, Yang M, Carter C, Al-Quran SZ, et al. PTK7: a new biomarker for immunophenotypic characterization of maturing T cells and T cell acute lymphoblastic leukemia. Leuk Res. 2012;36(11):1347–1353. doi: 10.1016/j.leukres.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Liang C, Wang S, Chio C, Zhang Y, Zeng C, et al. Expression patterns of immune checkpoints in acute myeloid leukemia. J Hematol Oncol. 2020;13(1):28. doi: 10.1186/s13045-020-00853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamimura K, Mishima Y, Obata M, Endo T, Aoyagi Y, Kominami R. Lack of Bcl11b tumor suppressor results in vulnerability to DNA replication stress and damages. Oncogene. 2007;26(40):5840–5850. doi: 10.1038/sj.onc.1210388. [DOI] [PubMed] [Google Scholar]
- 10.Nie Y, Lu W, Chen D, Tu H, Guo Z, Zhou X, et al. Mechanisms underlying CD19-positive ALL relapse after anti-CD19 CAR T cell therapy and associated strategies. Biomark Res. 2020;8:18. doi: 10.1186/s40364-020-00197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Materials and Methods.
Additional file 2: Figure S1. Expression patterns of BCL11B and PTK7 in the GSE28497 dataset. High expression of BCL11B (A) and PTK7 (B) in T-ALL. (C) BCL11B and PTK7 had a positive correlation.
Data Availability Statement
Data available on request.