Abstract
Objective
The nosocomial pathogen, Acinetobacter baumannii, has acquired clinical significance due to its ability to persist in hospital settings and survive antibiotic treatment, which eventually resulted in the rapid spread of this bacterium with antimicrobial resistance (AMR) phenotypes. This study used a multidrug-resistant A. baumannii (strain ATCC BAA1605) as a model to study the genomic features of this pathogen.
Results
One circular chromosome and one circular plasmid were discovered in the complete genome of A. baumannii ATCC BAA1605 using whole-genome sequencing. The chromosome is 4,039,171 bp long with a GC content of 39.24%. Many AMR genes, which confer resistance to major classes of antibiotics (beta-lactams, aminoglycosides, tetracycline, sulphonamides), were found on the chromosome. Two genomic islands were predicted on the chromosome, one of which (Genomic Island 1) contains a cluster of AMR genes and mobile elements, suggesting the possibility of horizontal gene transfer. A subtype I-F CRISPR-Cas system was also identified on the chromosome of A. baumannii ATCC BAA1605.
This study provides valuable genome data that can be used as a reference for future studies on A. baumannii. The genome of A. baumannii ATCC BAA1605 has been deposited at GenBank under accession no. CP058625 and CP058626.
Keywords: Acinetobacter baumannii, Antimicrobial resistance, Crispr/cas system, Hybrid genome assembly, Nanopore MinION, Illumina miSeq
Introduction
Acinetobacter baumannii, a gram-negative opportunistic human pathogen, has currently been recognized as one of the most challenging nosocomial pathogens. Ventilator-associated-pneumonia (VAP) is the major outcome of A. baumannii infections, with an overall in-hospital mortality rate of 63.3% [1]. Other presentations may include meningitis, urinary tract infection, bacteremia and skin or wound infections [2]. Multidrug-resistant (MDR) A. baumannii was thought to have first emerged in the United States of America in military treatment facilities during the 2003–2004 outbreak MDR A. baumannii which caused serious infections to the injured military personnel returned from the war. This subsequently resulted in the spread of MDR A. baumannii to other areas and increased the rate of distribution of resistance genes [3]. Carbapenem-resistant A. baumannii has recently been categorized by the World Health Organization (WHO) as a top priority pathogen requiring urgent development of novel antibiotics [4]. Several studies have reported a higher than the national average (50–60%) carbapenem resistance rates of A. baumannii isolates from individual hospitals in Malaysia. For example, carbapenem resistance rate of > 70% was reported for the clinical isolates collected from Universiti Kebangsaan Malaysia Medical Centre (UKMCC Kuala Lumpur, Malaysia) between 2010 and 2011 [5]. Resistance to other major classes of antibiotics (such as cephalosporins, aminoglycosides, fluoroquinolones, etc.) was also reported in Malaysia [6].
A. baumannii tolerates unfavourable environmental conditions, such as nutrient limitation and desiccation. It colonizes almost any surfaces including medical equipment. The ability of A. baumannii to form biofilm enhances its survival under stress conditions [7]. This could increase the chances of transmission and MDR development attained by either mutations or genetic elements. Clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR-Cas) is a responsive immune system that could play a role in the exchange of bacterial genetic information, colonization and biofilm production [8]. CRISPR-Cas system can be classified into two main classes, which includes 6 major types and 33 different subtypes [9]. Subtype I-Fb is reported as the most common CRISPR-Cas in A. baumannii [10].
Understanding the genome characteristics of A. baumannii, especially the MDR strains, could provide useful information in dealing with this pathogen such as drug development. In this study, we aimed to sequence the MDR A. baumannii strain ATCC BAA1605 which was originally isolated from a sputum sample of military personnel returning from Afghanistan and admitted to a Canadian hospital in 2006. This strain has been known for its MDR phenotype; however, its genomic features remain poorly studied. To our knowledge, this is the first reported complete genome of A. baumannii strain ATCC BAA1605.
Materials and methods
Sample preparation
A. baumannii ATCC BAA1605 (ATCC® BAA-1605™) was purchased from American Type Culture Collection (ATCC, USA). This strain was originally isolated from a sputum sample of a hospitalized patient according to ATCC (USA). A. baumannii was cultured on Mueller–Hinton broth (Oxoid, UK) and incubated overnight at 37℃ with continuous shaking at 200 rpm. DNA was isolated using phenol–chloroform phase-separation method according to Sambrook and Russell [11]. Leeds Acinetobacter medium (LAM) supplemented with antibiotics (HiMedia, India) was used to selectively isolate MDR Acinetobacter. Selective supplement composed of 3 antibiotics: vancomycin, cefsulodin and cefradine. Antibiotic-sensitive A. baumannii strain 65 (isolated from Segamat, Malaysia, unpublished data) was used as a control. Antibiotic susceptibility test (AST) was performed using disk diffusion method according to Clinical and Laboratory Standard Institute (CLSI) [12]. Classes of antibiotics tested were listed in Additional file 1: Figure S2C.
De novo whole-genome sequencing and assembly
A hybrid short- and long-read based-WGS was performed to construct the complete genome of A. baumannii ATCC BAA1605. Briefly, short-read sequencing data was generated by Nextera XT library preparation kit and sequenced on the Illumina Miseq using a 2 × 250 bp paired-end configuration. DNA libraries for long-reads sequencing were prepared using the Ligation Sequencing Kit protocol (SQK-LSK109) and long-reads sequencing data was generated on a MinION FLO-MIN106 flow cell and MinION MK1B sequencing device (Oxford Nanopore Technologies). Base-calling was conducted using Guppy v3.2.10 through MinKnow v3.6.17, using fast base calling configuration. Quality of short Illumina reads was assessed using FastQC v0.11.5 (https://github.com/s-andrews/FastQC), followed by adapter trimming using Trimmomatic v0.36 [13]. Chromosomal assembly was performed using Flye v2.7 [14]. Plasmid recovery was done by checking short-reads sequencing data assembled de novo using SPAdes v3.13.0 [15]. The whole-genome sequence was later corrected and polished using Pilon v1.23 [16]. Quality of the corrected assembly was evaluated using BUSCO v4.0.6 [17], using pseudomonales_odb10 as database.
Genome annotation, genome map and plasmid identification
The whole genome was annotated using Prokka v1.13 [18]. Genome map was plotted using BLAST Ring Image Generator (BRIG) v0.95 [19]. Plasmid identification was determined using BLASTn [20] against NCBI database, and sourmash v3.3.0 search-containment method [21] against PLSDB database [22]. Top 10 plasmids from sourmash results were aligned with A. baumannii ATCC BAA1605 plasmid (CP058626) using Mauve v2.4.0 progressive alignment [23].
Genome analysis
Whole genome of A. baumannii ATCC BAA1605 was compared with two reference strains: A. baumannii ATCC BAA-1790 (the only A. baumannii ATCC BAA strain with complete genome available in NCBI database) and A. baumannii ASM211692v1 (the representative strain of A. baumannii in NCBI). Whole genome alignment of the 3 strains was constructed using Mauve v2.4.0 progressive alignment [23]. AMR genes were identified by Abricate v1.0.1 (https://github.com/tseemann/abricate), using Comprehensive Antibiotic Resistance Database (CARD) [24]. Prophage regions and CRISPR-Cas proteins were detected using the web-tools, PHASTER [25] and CRISPRCasFinder [26], respectively. CRISPR-Cas with evidence levels of 3 and 4 represented highly likely candidates according to Couvin, Bernheim [26], and was selected for further analysis. Core genomes of different strains of A. baumannii that carry CRISPR-Cas (downloaded from the NCBI database) were extracted using Roary v3.13.0 [27]. The phylogenetic tree of CRISPR-Cas of A. baumannii was constructed with the concatenated core genome sequences using FastTree [28] and visualized using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Genomic islands were predicted using IslandViewer 4 [29] and schematic representation of the genes was constructed using DNA Features Viewer v3.0.1 [30].
Genome accession numbers
The SRA accession numbers of long- and short-reads sequence data are SRX8666155 and SRX8666156, respectively. The sequences of the complete annotated genome of A. baumannii ATCC BAA1605 has been deposited at GenBank with accession CP058625 (https://www.ncbi.nlm.nih.gov/nuccore/CP058625.1) and CP058626 (https://www.ncbi.nlm.nih.gov/nuccore/CP058626.1).
Results
Genome properties
A. baumannii ATCC BAA1605 contains one circular chromosome (CP058625) and one circular plasmid (CP058626) with the sizes of 4,039,171 bp (GC content = 39.24%) and 8,731 bp (GC content = 34.37%), respectively (Fig. 1a, Additional file 1: Table S1). A high degree of completeness was obtained for this genome with a BUSCO score of 98.9%, of which 774 genes were complete, 6 were fragmented, and 2 were missing BUSCO orthologs out of the 782 BUSCO groups searched. Genome annotation is summarized in Table S2 and Table S3. Comparing to the two reference strains (A. baumannii ATCC BAA-1790 and A. baumannii ASM211692v1), A. baumannii ATCC BAA1605 has a slightly larger number of genes and chromosome size but carry a smaller plasmid (Additional file 1: Table S4). The chromosomes and plasmids for the three strains share similar GC content. Whole genome alignments of the 3 strains showed some genome rearrangement (Additional file 1: Figure S1).
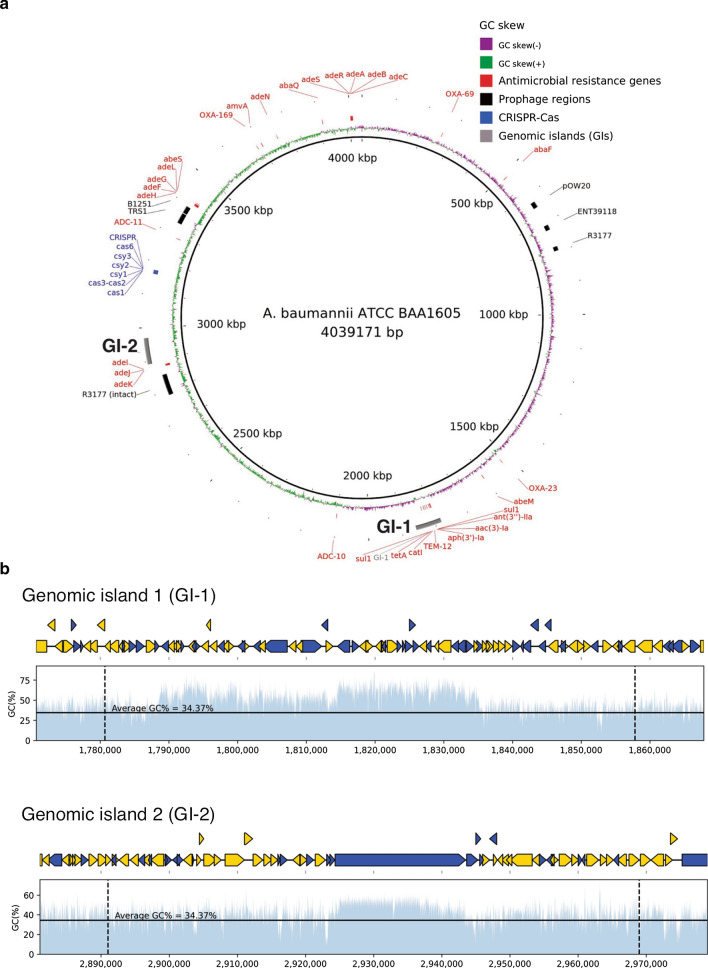
Fig. 1.
Genomic features of A. baumannii ATCC BAA1605 chromosome (CP058625). a Genome map. The innermost colored circle denotes the GC skew of genomic sequences (purple: negative; green: positive), followed by distributions of antimicrobial resistance genes (red), prophage regions (black), CRISPR-Cas system (blue) and genomic islands (GIs) predicted by IslandViewer 4 (grey). b Schematic representation of the genes present in the predicted GIs from IslandViewer 4 and their GC contents. The upper panel depicts the genes and their GC contents found in the GI-1 (approximately 1.78 Mb to 1.86 Mb), while lower panel depicts the genes and their GC contents found in the GI-2 (approximately 2.89 Mb to 2.97 Mb). Yellow arrows: Annotated CDS; Blue arrows: Unannotated/hypothetical proteins
Antimicrobial resistance
Thirty AMR genes were detected on the chromosome of A. baumannii ATCC BAA1605 and their distribution is shown in Fig. 1a. No AMR genes were detected on the plasmid. Identification of these AMR genes suggests the resistance of this pathogen to few major classes of antibiotics, including beta-lactams, fluoroquinolone, tetracycline, aminoglycoside, and sulphonamide (Additional file 1: Table S5). A. baumannii ATCC BAA1605 grew on LAM supplemented with antibiotics that select for MDR Acinetobacter (Additional file 1: Figure S2A and S2B). This finding is further supported by the results of AST (Additional file 1: Figure S2C), where A. baumannii ATCC BAA1605 showed resistance to all classes of antibiotics tested.
Prophage regions and genomic islands
One intact prophage region was identified on the A. baumannii ATCC BAA1605 chromosome with a length of 65.5 Kb (Fig. 1a, Additional file 1: Table S6). There are 79 open reading frames (ORFs) present in this intact prophage region. The best hit for this region corresponds to the Acinetobacter phage YMC11/11/R3177. There are five additional incomplete prophage regions scattered throughout the genome (Fig. 1a, Additional file 1: Table S6).
Two clear genomic islands (GI-1 and GI-2) were identified on the chromosome of this bacterium (Fig. 1a). Genes found within these GIs are listed in Additional file 1: Table S7 and Table S8. A cluster of AMR genes, such as bla and tet, and DNA recombination enzymes including resolvase (tnpR) and DNA-invertase (hin) were found in GI-1. The GC content in these GIs is generally higher than the average GC content of the genome (34.37%) (Fig. 1b).
Plasmid identification and verification
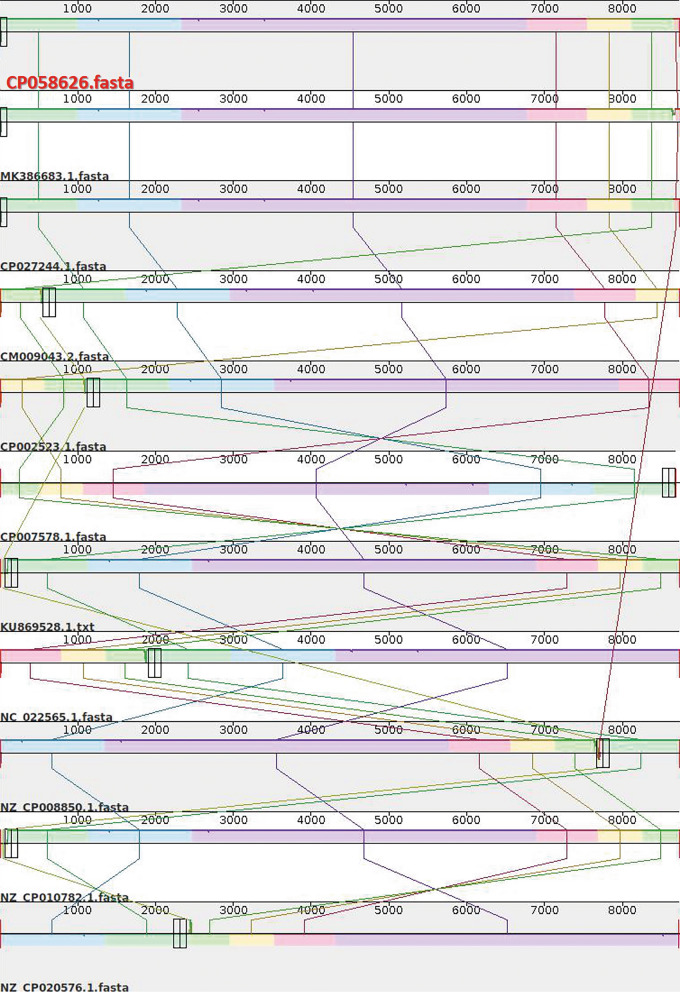
Plasmid sequence of A. baumannii ATCC BAA1605 was aligned with the top 10 plasmids obtained from sourmash results (Additional file 1: Table S9) and shown in Fig. 2. BLASTn results revealed 100% identity and coverage of A. baumannii ATCC BAA1605 plasmid with the plasmids of other A. baumannii strains in the NCBI database. Comparison against PLSDB database using sourmash showed 100% similarity with A. baumannii strain ABAY14012 plasmid pABAY14012_4D (MK386683.1), followed by A. baumannii strain WCHAB005078 plasmid p2_005078 (CP027244.1) with a similarity of 99.8% (Fig. 2, Additional file 1: Table S9).
Fig. 2.
Plasmid sequence alignments of A. baumannii ATCC BAA1605 plasmid (CP058626) with top 10 plasmids from sourmash results using Mauve. The plasmid of A. baumannii ATCC BAA1605 strain used in this study is shown on top and highlighted in red. Each coloured blocks depicts the homologous sites of sequence that aligned to part of another genome
CRISPR-Cas system
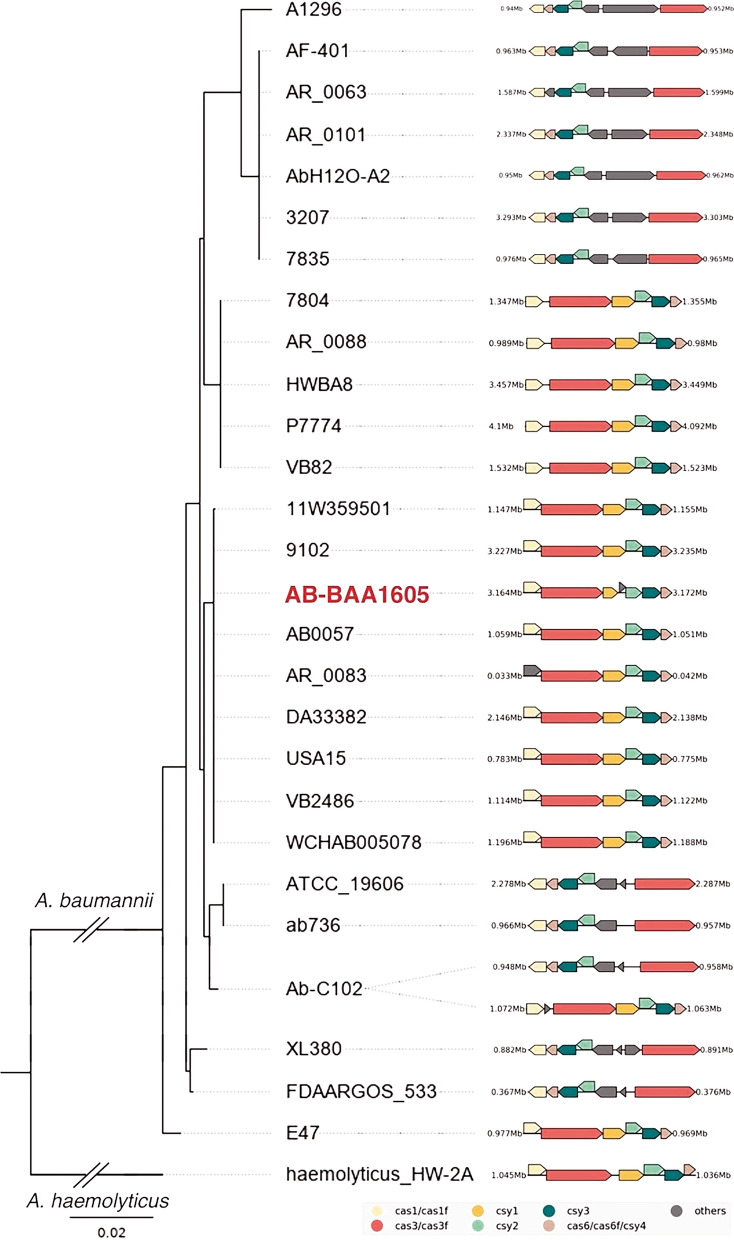
A subtype I-F CRISPR-Cas was identified on the chromosome of A. baumannii ATCC BAA1605, with a 3150 bp CRISPR array containing 52 spacers (Fig. 3, Additional file 1: Table S10). The spacers are flanked by 28 bp CRISPR repeats. The Cas proteins are listed in Additional file 1: Table S11. Phylogenetic tree of A. baumannii carrying CRISPR-Cas was shown in Fig. 3. It is observed that strains with close phylogenetic relationships have similar genes and share gene synteny on their CRISPR-Cas system.
Fig. 3.
Phylogenetic tree of CRISPR-Cas system of different strains of A. baumannii. Organization of the CRISPR-Cas systems in the genomes are shown on the right. Yellow, cas1/cas1f; Red, cas3/cas3f; Orange, csy1; Light green, csy2; Dark green, csy3; Brown, cas6/cas6f/cas4; Gray, CDS not related to CRISPR. The A. baumannii ATCC BAA1605 strain used in this study is highlighted in red. A. haemolyticus strain HW-2A with CRISPR-Cas system was used as an outgroup
Discussion
WGS reveals one circular chromosome and one circular plasmid in this MDR A. baumannii ATCC BAA1605. High degree of completeness from BUSCO score indicates successful genome assembly and accurate analysis. There are many AMR genes identified on the chromosome of this pathogen, indicating its resistance to a few major classes of antibiotics. This has been further evidenced by culturing A. baumannii ATCC BAA1605 on LAM added with selective supplement that contained antibiotics selectively for MDR Acinetobacter, and AST results. For example, multiple bla genes that encode carbapenem-hydrolyzing class D β-lactamases, including the highly prevalent blaOXA-23 gene [31], were detected on the chromosome of this pathogen, suggested its resistance to carbapenems which then confirmed by AST. One intact, complete prophage region was detected on the chromosome with high similarity to Acinetobacter phage YMC11/11/R3177. Genes, such as prophage integrase intA and intS, and virulence regulon transcriptional activator virF, were found within the intact prophage region, suggesting their roles in regulating many phage-encoded virulence factors.
GI-1 that was detected on the chromosome contains a cluster of AMR genes and mobile genetic elements. This could suggest the possibility of acquired AMR via horizontal gene transfer since no AMR genes were found on the plasmid. It is reported that the genomic islands of pathogenic A. baumannii generally possess genes such as heavy metal resistance genes, AMR genes and competence proteins, that facilitate their survival under unfavourable conditions [32]. The findings of this study are consistent with this notion.
A subtype I-F CRISPR-Cas system was identified on the chromosome of A. baumannii ATCC BAA1605. Since the spacers in the CRISPR array do not change over time, the high number of spacers identified in the CRISPR loci of this pathogen suggests that it might have encountered a high number of phages attacks. The presence of CRISPR-Cas system could explain the low number of plasmid found within this genome.
Conclusion
Multidrug-resistant A. baumannii has become an emerging threat to public health, especially to immunocompromised patients. Yet, there are many drug-resistant A. baumannii that have not been fully explored in terms of their genomes. This is the first reported complete genome of a MDR A. baumannii (strain ATCC BAA1605). This study provides data that can be used as a reference for future studies on MDR A. baumannii and improves our understanding of the genomic features of this reference strain.
Limitations
The virulent determinants identified in the genome of A. baumannii ATCC BAA1605 could serve as preliminary data; however, future experiments, such as PCR, can be conducted to validate the presence of these virulent genes.
Supplementary Information
Additional file 1: Table S1. Summary of the genome of A. baumannii ATCC BAA1605: one chromosome and one plasmid. Table S2. Summary of annotation of A. baumannii ATCC BAA1605 chromosome using Prokka. Table S3. Summary of annotation of A. baumannii ATCC BAA1605 plasmid using Prokka. Table S4. Comparisons of the chromosome and plasmid of A. baumannii strain ATCC BAA1605 with A. baumannii strain ATCC BAA-1790 and A. baumannii ASM211692v1. Table S5. Antibiotic resistance profiles of A. baumannii ATCC BAA1605 identified by CARD. Table S6. Predicted prophage regions in A. baumannii ATCC BAA1605 using PHASTER. Table S7. Genes found in Genomic Island 1 (1.78 Mb to 1.86 Mb) predicted by IslandViewer 4 in A. baumannii ATCC BAA1605 and their coordinates. Table S8. Genes found in Genomic Island 2 (2.89 Mb to 2.97 Mb) predicted by IslandViewer 4 in A. baumannii ATCC BAA1605 and their coordinates. Table S9. Top 10 plasmids and their similarity with A. baumannii ATCC BAA1605 plasmid using sourmash search-containment method against PLSDB database. Table S10. Summary of CRISPR array identified in chromosome of A. baumannii ATCC BAA1605. Table S11. Subtype I-F cas genes identified in the chromosome of A. baumannii ATCC BAA1605 and their coordinates. Figure S1. Whole genome alignments of A. baumannii ATCC BAA1605 (top), A. baumannii ATCC BAA-1790 (middle) and A. baumannii ASM211692v1 (bottom) using Mauve. Each coloured blocks depicts the homologous sites of sequence that aligned to part of another genomes. Figure S2. Identification and antibiotic resistance profiling of A. baumannii ATCC BAA1605. MDR A. baumannii ATCC BAA1605 and antibiotic-sensitive A. baumannii strain 65 (control) cultured on Leeds Acinetobacter medium either without selective supplement (A) or with antibiotics supplementation (B). (C) Antibiotic susceptibility profiling of A. baumannii ATCC BAA1605 using disk diffusion method.
Acknowledgements
This work was supported by the Tropical Medicine & Biology Multidisciplinary Platform and School of Science, Monash University Malaysia.
Abbreviations
- AMR
Antimicrobial resistance
- Cas
CRISPR-associated protein
- CDS
Coding sequence
- CRISPR
Clustered regularly interspaced short palindromic repeats
- GI
Genomic Island
- MDR
Multidrug-resistant
- ORF
Open reading frame
- WGS
Whole-genome sequencing
- PCR
Polymerase chain reaction
- LAM
Leeds Acinetobacter medium
Authors’ contributions
KET isolated the DNA of A. baumannii ATCC BAA1605 and performed the genome analysis, elucidation of the data and drafted the manuscript. MZHMZ carried out the whole-genome sequencing and helped with the bioinformatic analysis. MZHMZ was supported by Monash Malaysia R&D Sdn Bhd. QA and HST supervised the project and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The complete sequences of the genome of A. baumannii ATCC BAA1605 are available in GenBank with the accession numbers: CP058625 and CP058626 (Bioproject ID: PRJNA643902, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA643902/).
Ethics approval and consent to participate
Not applicable.
Consent for publication.
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kah Ern Ten, Email: kah.ten@monash.edu.
Muhammad Zarul Hanifah Md Zoqratt, Email: muhammad.zarulhanifah@monash.edu.
Qasim Ayub, Email: qasim.ayub@monash.edu.
Hock Siew Tan, Email: tan.hocksiew@monash.edu.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-021-05493-z.
References
- 1.Čiginskienė A, Dambrauskienė A, Rello J, Adukauskienė D. Ventilator-associated pneumonia due to drug-resistant Acinetobacter baumannii: risk factors and mortality relation with resistance profiles, and independent predictors of in-hospital mortality. Medicina. 2019;55(2):49. doi: 10.3390/medicina55020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergogne-Berezin E, Towner K. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9(2):148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott P, Deye G, Srinivasan A, Murray C, Moran K, Hulten E, et al. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis. 2007;44(12):1577–1584. doi: 10.1086/518170. [DOI] [PubMed] [Google Scholar]
- 4.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Biglari S, Alfizah H, Ramliza R, Rahman MM. Molecular characterization of carbapenemase and cephalosporinase genes among clinical isolates of Acinetobacter baumannii in a tertiary medical centre in Malaysia. J Med Microbiol. 2015;64(1):53–58. doi: 10.1099/jmm.0.082263-0. [DOI] [PubMed] [Google Scholar]
- 6.Mohd Rani F, Rahman NIA, Ismail S, Alattraqchi AG, Cleary DW, Clarke SC, et al. Acinetobacter spp. infections in Malaysia: a review of antimicrobial resistance trends, mechanisms and epidemiology. Front Microbiol. 2017;8:2479. doi: 10.3389/fmicb.2017.02479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaddy JA, Actis LA. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009;4(3):273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162(1):27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karah N, Samuelsen O, Zarrilli R, Sahl J, Wai S, Uhlin B. CRISPR-Cas subtype I-Fb in Acinetobacter baumannii: evolution and utilization for strain subtyping. PLoS ONE. 2015;10(2):e0118205. doi: 10.1371/journal.pone.0118205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006 doi: 10.1101/pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- 12.CLSI. Performance standards for antimicrobial susceptibility testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30(15):2114. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 15.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 18.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 19.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Pierce NT, Irber L, Reiter T, Brooks P, Brown CT. Large-scale sequence comparisons with sourmash. F1000Res. 2019;8:1006. doi: 10.12688/f1000research.19675.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galata V, Fehlmann T, Backes C, Keller A. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 2019;47(D1):D195–D202. doi: 10.1093/nar/gky1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46(W1):W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zulkower V, Rosser S. DNA Features Viewer: a sequence annotation formatting and plotting library for Python. Bioinformatics. 2020;36(15):4350–4352. doi: 10.1093/bioinformatics/btaa213. [DOI] [PubMed] [Google Scholar]
- 31.Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16(1):35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakkala H, Samantarrai D, Gribskov M, Siddavattam D. Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains (Research Article) (Report) PLoS ONE. 2019;14(6):e0218204. doi: 10.1371/journal.pone.0218204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary of the genome of A. baumannii ATCC BAA1605: one chromosome and one plasmid. Table S2. Summary of annotation of A. baumannii ATCC BAA1605 chromosome using Prokka. Table S3. Summary of annotation of A. baumannii ATCC BAA1605 plasmid using Prokka. Table S4. Comparisons of the chromosome and plasmid of A. baumannii strain ATCC BAA1605 with A. baumannii strain ATCC BAA-1790 and A. baumannii ASM211692v1. Table S5. Antibiotic resistance profiles of A. baumannii ATCC BAA1605 identified by CARD. Table S6. Predicted prophage regions in A. baumannii ATCC BAA1605 using PHASTER. Table S7. Genes found in Genomic Island 1 (1.78 Mb to 1.86 Mb) predicted by IslandViewer 4 in A. baumannii ATCC BAA1605 and their coordinates. Table S8. Genes found in Genomic Island 2 (2.89 Mb to 2.97 Mb) predicted by IslandViewer 4 in A. baumannii ATCC BAA1605 and their coordinates. Table S9. Top 10 plasmids and their similarity with A. baumannii ATCC BAA1605 plasmid using sourmash search-containment method against PLSDB database. Table S10. Summary of CRISPR array identified in chromosome of A. baumannii ATCC BAA1605. Table S11. Subtype I-F cas genes identified in the chromosome of A. baumannii ATCC BAA1605 and their coordinates. Figure S1. Whole genome alignments of A. baumannii ATCC BAA1605 (top), A. baumannii ATCC BAA-1790 (middle) and A. baumannii ASM211692v1 (bottom) using Mauve. Each coloured blocks depicts the homologous sites of sequence that aligned to part of another genomes. Figure S2. Identification and antibiotic resistance profiling of A. baumannii ATCC BAA1605. MDR A. baumannii ATCC BAA1605 and antibiotic-sensitive A. baumannii strain 65 (control) cultured on Leeds Acinetobacter medium either without selective supplement (A) or with antibiotics supplementation (B). (C) Antibiotic susceptibility profiling of A. baumannii ATCC BAA1605 using disk diffusion method.
Data Availability Statement
The complete sequences of the genome of A. baumannii ATCC BAA1605 are available in GenBank with the accession numbers: CP058625 and CP058626 (Bioproject ID: PRJNA643902, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA643902/).