Abstract
Chromodomain helicase/ATPase DNA binding protein 1-like gene (CHD1L) is a multifunctional protein participated in diverse cellular processes, including chromosome remodeling, cell differentiation and development. CHD1L is a regulator of chromosomal integrity maintenance, DNA repair and transcriptional regulation through its bindings to DNA. By regulating kinds of complex networks, CHD1L has been identified as a potent anti-apoptotic and pro-proliferative factor. CHD1L is also an oncoprotein since its overexpression leads to dysregulation of related downstream targets in various cancers. The latest advances in the functional molecular basis of CHD1L in normal cells will be described in this review. As the same time, we will describe the current understanding of CHD1L in terms of structure, characteristics, function and the molecular mechanisms underlying CHD1L in tumorigenesis. We inference that the role of CHD1L which involve in multiple cellular processes and oncogenesis is well worth further studying in basic biology and clinical relevance.
Keywords: CHD1L, ALC1, SNF2, Chromatin remodeling, Tumorigenesis
Introduction
The CHD1L gene (Chromodomain helicase/ATPase DNA binding protein 1-like gene), also called the ALC1 gene (amplified in liver cancer 1), locates on chromosome 1q21 region of human hepatoma cells and is cloned by Guan using the comparative genomic hybridization (CGH) technique [1–3]. Because CHD1L has a consistent helicase sequence motif found in helicase superfamily 2 proteins [1], it is classified as a sucrose non-fermentation 2 like (SNF2-like) subfamily of the SNF2 family [4–6]. Most SNF2-like proteins can utilize the energy, which is released from their DNA-dependent ATPase activity, to stabilize or interfere with protein-DNA interactions [7] and participate in a variety of nuclear activities, such as transcriptional inhibition or activation, DNA recombination and repair [8, 9]. Like most SNF-like proteins, CHD1L is a regulator of chromosome integrity, transcriptional regulation and DNA repair through its bindings to DNA. The functional diversity of CHD1L has always been related to the characteristics of helicases or chromatin remodeling enzymes that interact with PAR and catalyze the sliding of nucleosomes stimulated by PAR polymerase 1 (PARP1) [10, 11].
CHD1L plays an important role as a transcription and translation activator of its target genes [1, 6, 12–19]. CHD1L can promote cell proliferation, enhance cell migration and inhibit apoptosis by regulating various complex networks [6, 12, 13]. For example, CHD1L is a well-known activator of ARHGEF9, TCTP, SPOCK1 and NTKL [14–17] and can also lead to deregulation of p53, TCTP and Nur77 [1, 15, 18]. Importantly, CHD1L shows carcinogenicity in the process of malignant transformation. CHD1L overexpression in cancer cells is considered as a biomarker of short tumor-free survival time and poor prognosis [12, 20–28].
We will provide an overview of current understanding about CHD1L on structure, characteristics, function and the molecular mechanisms underlying CHD1L in cancer development in this review. As the same time, we will describe the latest development of CHD1L functions in normal cells. Finally, we will conclude that CHD1L is an attractively clinical target in the future molecular therapy of cancer.
Molecular and structural features of CHD1L
SNF2 superfamily
CHD1L protein belongs to the SNF2 superfamily proteins, which was identified by Ma [1]. The SNF2 superfamily proteins include ATP-dependent chromatin remodeling enzymes and play key roles in the organization of genomic DNA in the natural chromatin state [7, 29]. The SNF2 superfamily proteins are further divided into ISWI (simulated switches), INO80 (inositol), CHD (chromosomal domain helicase DNA binding) and SWI/SNF (mating switch/non-sucrose fermentation) families [29]. In the mammalian, the ISWI family comprises SNF2H and SNF2L. The SWI/SNF family proteins contain brahma (BRM) and brahma-related gene 1 (BRG1). Based on the existence or nonexistence of additional domains, the CHD family contains three subfamilies: Chd1-Chd2, Chd3-Chd4, and Chd5-Chd9 [29]. The CHD family has the characteristics of two signature sequence motifs. One is the conserved SNF2_N domain at the N-terminal tandem chromodomain, and the other is the SNF2-like ATPase domain at the center, also known as the helicase superfamily c-terminal domain (HELICc) [9]. The SNF2_N domains (containing 280 amino acids (aa)) of CHD1L and CHD1 have 45% identity, while their HELICc domains (containing 107 amino acids) have 59% identity [1].
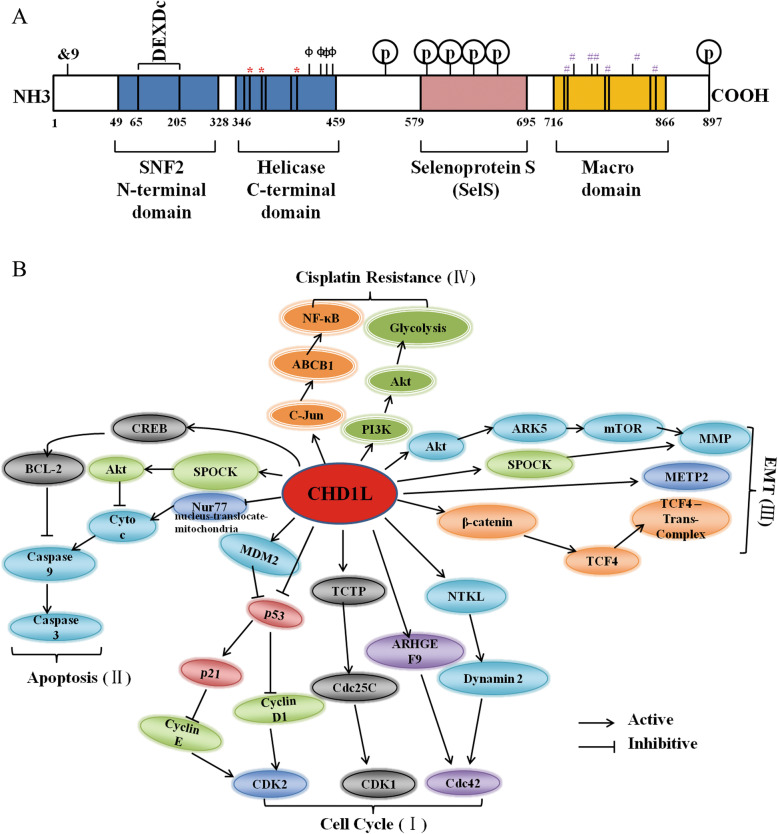
CHD1L is mapped to chromosome 1q21 [30]. The full-length messenger RNA of CHD1L (NM_004284.6) contains 3036 base pairs with a presumptive open reading frame encoding an 897 aa protein [1]. As shown in Fig. 1a, CHD1L mainly contains four domains: a conserved SNF2_N domains, a helicase superfamily domains (HELICc), selenoprotein S (SelS) and a Macro domains [1]. The upstream of CHD1L gene is flavin containing monooxygenase 5 (FMO5) gene, while the downstream of which have a long intergenic non-coding RNA 624 (LINC00624) and a prostaglandin reductase pseudo gene (LOC100130018) [30]. In addition, human CHD1L has a Selenoprotein S (SelS) region (579–695 aa), which is a plasma membrane protein that exists in many cell types and tissues [31]. Schematic representation of human CHD1L protein is shown in Fig. 1a.
Fig. 1.
Structural representation, transcriptional effects and regulatory pathways of CDH1L. a Schematic representation of CHD1L protein. CHD1L encompasses four domains, SNF2 family N-terminal domain (49–328 aa, blue color) containing the ATP-binding region in N-terminal, helicase C-terminal domain (346–459 aa, blue color) containing nucleotide binding region, Selenoprotein S (SelS) (579–695 aa, pink color) consisting of several mammalian SelS sequences and a macro domain (716–866 aa, yellow color) containing ADP-ribose binding region in C-terminal. &9:Omega-N-methylarginine, no additional details recorded. DEXDc (65–205 aa):DEAD-like helicases superfamily. A diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. This domain contains the ATP-binding region (74–78 aa, ATP binding site) and DEAH box (174–177 aa, putative Mg++ binding site). *: nucleotide binding region (371–374, 394–395, 421–423 aa). #: putative ADP-ribose binding sites (723–724, 741, 748, 750–751, 844, 846–848 aa). ɸ: ATP-binding sites (429, 450, 454, 457 aa). There are six putative phosphorylation sites: phospho-serine at 540, 607, 618, 628, 636 and 891 amino acids, respectively. b The transcriptional effects and regulatory pathways of CDH1L. (I) CHD1L accelerate cell cycle transition, which directly binds to the promoter regions of MDM2, P53, TCTP, ARHGEF9 and NTKL. (II) CHD1L promotes cell apoptosis death through activing caspase pathway, which enhances the phosphorylation of CREB, increases SPOCK expression and inhibits nuclear to mitochondrial translocation of Nur77. (III) CHD1L activate the transcription of Akt, METP2, TCF4 genes, leading to EMT (include invasion and metastasis). (IV) CHD1L regulates NF-κB and glycolysis pathways to facilitate cisplatin resistance
Characteristics and experssion pattern
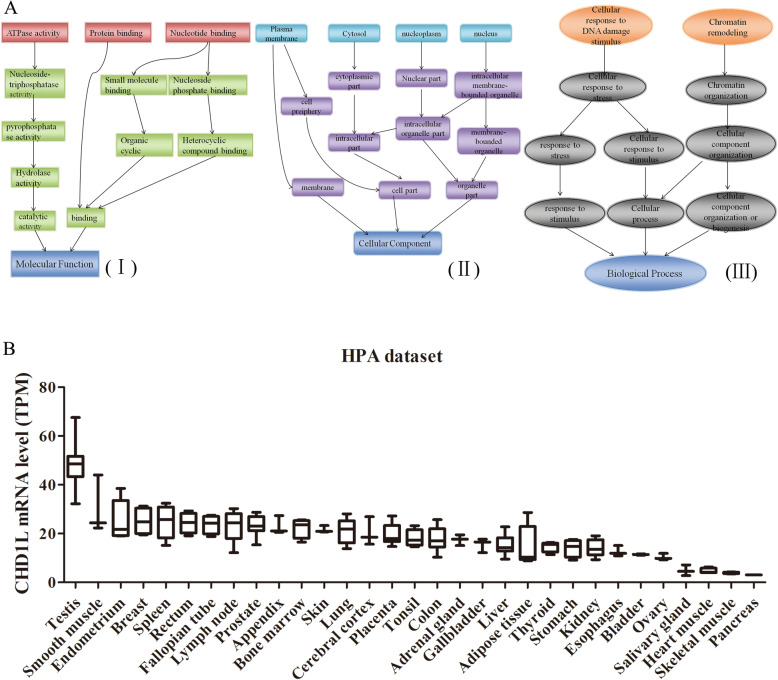
CHD1L is a highly conserved gene among species in nature (https://www.ncbi.nlm.nih.gov/homologene/?term=11590). Human CHD1L is more than 87, 73, and 66% similar to mammals, the less-evolved vertebrate chicken and zebrafish, respectively (Table 1). As conserved gene in eukaryota (HomoloGene ID: 11590), CHD1L is involved in molecular function, cellular component, biological process (Fig. 2a, Table 2) (http://www.informatics.jax.org/homology/GOGraph/11590#Annotations). Since CHD1L is expressed in many species including invertebrates, it appears that CHD1L exists in the common ancestor of vertebrates. In human, CHD1L can be found in various tissues and exhibit different expression patterns in space and time [22] (Fig. 2b). For example, in the male reproductive system (testis), CHD1L is less expressed in mature cells than progenitor cells [32, 33].
Table 1.
conserved gene CHD1L homology in Eukaryota (HomoloGene:11590) [https://www.ncbi.nlm.nih.gov/homologene/?term=11590]
| HomoloGene:11590. Gene conserved in Eukaryota | ||||||
|---|---|---|---|---|---|---|
| Species | Symbol | Genetic location | Protein Acc. | Protein length | Identity(%)a | |
| protein | DNA | |||||
| Human (H.sapiens) | CHD1L | Chr1 q21.1 | NP_004275.4 | 897 aa | ||
| Mouse (M.musculus) | Chd1l | Chr3 42.17 cM | NP_080815.1 | 900 aa | 87.6 | 85.3 |
| Rat (R.norvegicus) | Chd1l | Chr2 q34 | XP_006233080.1 | 926 aa | 88.2 | 86.1 |
| Chimpanzee (P.troglodytes) | CHD1L | Chr1 | XP_001158033.1 | 896 aa | 99.0 | 99.1 |
| Cattle (B.taurus) | CHD1L | Chr3 | NP_001032909.1 | 897 aa | 91.6 | 90.3 |
| Dog (C.lupus) | CHD1L | Chr17 | XP_005630931.1 | 918 aa | 88.0 | 88.5 |
| Chicken (G.gallus) | CHD1L | Chr1 | XP_004938249.1 | 900 aa | 73.4 | 72.6 |
| Zebrafish (D.rerio) | chd1l | Chr6 | NP_956607.1 | 1026 aa | 66.5 | 64.1 |
aVS Human (H.sapiens)
Fig. 2.
Characteristics and experssion pattern of CHD1L. a Conserved gene CHD1L in eukaryota (HomoloGene ID:11590) involves in molecular function (I), cellular component (II), biological process (III)., cellular component, biological process. b CHD1L mRNA expression overview of tissue category from HPA dataset. TPM: Transcript Per Million
Table 2.
Function of CHD1L in HomoloGene:11590 (http://www.informatics.jax.org/homology/GOGraph/11590#Annotations)
| Category | Classification term | Gene Ontology IDa | Reference | |
|---|---|---|---|---|
| Full set of experimental annotations for Human-Mouse-Rat CHD1L genes in HomoloGene:11590 | Molecular Function | ATPase activity | GO:0016887 | [11] |
| nucleotide binding | GO:0000166 | |||
| protein binding | GO:0005515 | |||
| Cellular Component | plasma membrane | GO:0005886 | b | |
| cytosol | GO:0005829 | |||
| nucleoplasm | GO:0005654 | |||
| nucleus | GO:0005634 | [11] | ||
| Biological Process | cellular response to DNA damage stimulus | GO:0006974 | [11] | |
| chromatin remodeling | GO:0006338 |
Biological function
Studies have described five spliced variants of the CHD1L protein, and even found six spliced transcribed variants of the CHD1L gene [30]. Interestingly, CHD1L is a conserved protein with four conserved protein domains, namely SNF2_N domain, HELICc, SelS and Macro domain (Fig. 1a).
SNF2_N domain is an important part of many proteins that involved in a variety of cell biological processes including DNA repair, chromatin unwinding, DNA recombination, and transcription regulation, but also is composed of some proteins with little functional information [9, 34].
HELICc is a component of multiple helicases and helicase-related proteins and DEAD-, DEXDc-, DEAH-box associated proteins [35, 36], hepatitis C virus NS3, ski2p and yeast initiation factor 4A [37]. The HELICc is not an autonomous folding unit, but is an essential part of the helicases that utilize the energy from nucleoside triphosphate hydrolysis to provide fuel for their translocation along DNA and unwinding double-stranded DNA in the process [36, 38]. The HELICc also contains DEAD-like helicase superfamily (ATP-binding) region which participates in ATP-dependent DNA or RNA unwinding.
SelS family contains several mammalian SelS sequences. SelS is a disordered protein which has a seleno sulfide bond (between Cys-174 and Sec-188) and a redox potential (− 234 mV) [31]. SelS is an efficient reductase that can catalyze the reduction of hydrogen peroxide [39]. SelS also has the ability to resist hydrogen peroxide inactivation and may have an evolutionary advantage compared to cysteine-containing peroxidases [40].
Macro domain is a high-affinity ADP-ribose binding module, which exists in various proteins in the form of independent domains or complexed with other domains (such as poly ADP-ribose polymerase (PARPs) and histone macroH2A). Poly ADP-ribose can be recognized as a ligand by some macro domains. Initially, the macro domain was identified as performing ADP-ribose-1″-monophosphate (Appr-1″-p) processing activity. Besides, the macro domain also play important roles in different ADP-ribose pathways, including DNA transcription, chromatin biology, DNA repair and long-term memory formation [34].
Roles of CHD1L in chromatin remodeling
The CHD1L protein includes highly conserved helicase motifs that exist in other SNF2 family members such as CHD1, ISWI and Snf2 [41]. However, unlike CHD1, CHD1L contains a macro domain that can recognize poly ADP-ribose (PAR), but not a chromo domain that recognizes methylated histone tails [11, 42]. PAR is produced by PAR polymerase (PARP), and single-stranded DNA breaks (SSB) and gaps can activate PARP [43, 44]. The occurrence of SSB is during base-excision repair (BER) [45] and can eliminate base damage, including alkylation and oxidation [45, 46]. The formation of PAR on chromatin protein near SSB promotes the recruitment of CHD1L and other BER factors to damaged bases [47]. CHD1L promotes BER by spreading chromatin at the sites of DNA damage [48]. PARP1 stimulates the chromatin-repositioning activity of CHD1L [10, 49]. CHD1L contributes to PARP-dependent BER without affecting the recruitment of DNA damage by XRCC1 or Polβ [48]. Chromatin relaxation is one of the earliest responses of cells to DNA damage, PARP1 triggers a conformational change that activates CHD1L to drive chromatin relaxation [50–52]. The co-operation between CHD1L and PARP is also related to the nucleotide excision repair of UV-induced DNA damage [53]. Another important role of CHD1L is involved in the transcriptional control for DNA damage responses, which supports the fact that CHD1L interacts with Tripartite Motifcontaining 33, a multifunctional protein involved in transcriptional regulation [54]. In addition, CHD1L promotes cell tolerance by slowing the site of DNA damage (as a chromatin remodeler) induced by replication forks of camptothecin, a topoisomerase I toxin that produces single strand breaks and causes the breakdown of replication forks [55]. CHD1L-dependent nucleosome remodeling is required for the efficient handover between PARP1/2, DNA glycosylases, and APEX1 downstream of lesion excision. Loss of ALC1 confers methyl-methanesulfonate, PARP inhibitors and formyl-dU sensitivity, which is synthetic lethal with homologous recombination deficiency (HRD) [56]. Therefore, CHD1L not only has a PAR-dependent chromatin remodeling activity, but also promotes DNA repair reactions within the chromatin range.
The critical role of CHD1L in development and differentiation
The developmental processes of the mammalian embryo are routinely analyzed according to their potential genetic components. Among many genes which are characterized by their roles in the early embryonic development, especially in pre-implantation [57], CHD1L is a key developmental regulator and is necessary for the early stages of development [58–60]. Inhibiting the production of CHD1L can arrest embryo at the pre-blastocyst stage in mice embryonic stem (ES) cells by microinjecting antisense morpholinone [58]. CHD1L regulates stem cell pluripotency through interplaying with PARP1 during early developmental stage [59]. However, CHD1L is not necessary for the survival, pluripotency and differentiation of cultured ES cells [58]. CHD1L is essential for embryonic events that are distinct from events in ES cells [58].
CHD1L is developmentally regulated and expressed in human fetuses, with the highest expression of CHD1L in the brain, followed by the kidney, then muscle, liver, thymus, lung, heart, and spleen [61]. CHD1L is indispensable for the development of human embryonic neuroepithelium. CHD1L overexpression in hESCs promoted neuroepithelial differentiation in both self-renewal and directional differentiation conditions. CHD1L knockdown impaired hESC differentiation into neuroepithelium. CHD1L overexpression dramatically upregulated PAX6 expression which is a key regulatory gene in eye and brain development. Interestingly, CHD1L highly expressed in cells of the ventricular (germinal) zone of E14 mouse embryos, and it colocalized with PAX6-positive cells [60]. The expression of CHD1L is higher in the kidney of fetal than in adult (4:1). This fact means that CHD1L is particularly important in kidney development [61]. CHD1L also plays a key role in congenital anomalies of the kidneys and the urinary tract (CAKUT). Three different heterozygous missense variants of CHD1L (variant Gly700Arg, variant Ile765Met and variant Ile827Val) were revealed by sequencing the entire coding region of the CHD1L gene in 61 CAKUT patients and exons 18, 19 and 21 in 24 CAKUT patients. The interaction between all three CHD1L variants and PARP1 decreased compared with the wild-type CHD1L. Therefore, chromatin remodeling and ATPase activities of CHD1L are low in CAKUT [61].
In adult, CHD1L expression is the highest in the testis, and it is mainly localized in undifferentiated spermatogonia, suggesting the role of CHD1L in spermatogenesis [32, 61]. Spermatogenic stem cells (SSCS) are adult stem cells, which are parthenogenetic cells based on spermatogenesis and male fertility [33]. CHD1L is a novel and intrinsic regulator of SSCs self-renewal and survival, which is at least partially mediated by the GDNF signaling pathway [32].
Molecular mechanism of CHD1Lin tumorigenesis
Multiple studies have shown that CHD1L plays important roles in cell proliferation, cell metastasis, cell apoptosis, cell cycle transition and drug resistance through a variety of mechanisms. CHD1L can promote G1/S transition and DNA synthesis by upregulating cyclins, CDK2, 4 and downregulating P27, Rb and p53 in transgenic mouse models [62]. In the development of HCC, CHD1L has been shown to participate in many ways, such as CHD1L-ARHGEF9-CDC42-EMT axis [14], CHD1L-TCTP-CDC25C-CDK1 pathway [15], CHD1L-SPOCK1-Akt signaling pathway [16]. In breast cancer, CHD1L promoted cell metastasis and invasion through the PI3K/AKT/ARK5/mTOR/MMP pathway [4]. CHD1L might promote cell motility and cell cycle progression through the MDM2/p53 pathway [63]. CHD1L could induce G1/S transition by the dysregulation of p53-cyclinE-CDK2 pathway in glioma [64]. CHD1L influenced cell proliferation by activating the Wnt/β-catenin/TCF pathway in pancreatic cancer [65]. The dysregulation of p53-cyclin D1-CDK2 pathway might be related to CHD1L-induced G1/S transition, while CHD1L might drive EMT and MET and cause metastasis of cholangiocarcinoma cells [20]. In terms of drug resistance, the upregulation of CHD1L could promote cisplatin resistance of NSCLC cells through c-Jun/ABCB1/NF-κB axis [66]. However, the downregulation of CHD1L enhanced cisplatin cytotoxicity of esophageal squamous cell carcinoma cells by inhibiting the glycolysis of PI3K/AKT pathway [67]. Taken together, inappropriate expression of CHD1L target genes and deregulation of CHD1L system may link CHD1L to tumorigenesis by several mechanisms (Fig. 1b).
Transcriptional effects of CHD1L on target genes
ARHGEF9
ARHGEF9, also called Collybistin, is one of the guanine nucleotide exchange factor (GEF) superfamily, which catalyzes GDP-GTP exchange in small GTPases of Rho family [68, 69]. An important molecular mechanism of cancer metastasis is the activation of the Rho family of small GTPases, which leads to the rearrangement of the actin cytoskeleton and regulates cadherin-dependent cell-to-cell contacts [70–72]. Most mammalian GEFs targeting Rho GTPases can accelerate cancer cell invasion by enhancing GTP loading on Rho proteins [73]. ARHGEF9 can encode a special guanine nucleotide exchange factor (GEF) for the Rho small GTPase Cdc42 [74]. ARHGEF9 is a target gene of the transcriptional regulator CHD1L [14]. CHD1L can upregulate transcription of ARHGEF9, which then increases Cdc42 activity, causing EMT and finally invasion and metastasis of HCC. CHD1L-ARHGEF9-Cdc42-EMT might be a novel pathway to participate in the progression and metastasis of HCC [14].
SPOCK1
Sparc/osteoectin, cwcv and kazal-like domain proteoglycan 1 (SPOCK1) is a secreted protein and can encode a Ca2 + −binding matricellular glycoprotein, which belongs to acidic and rich in cysteine (SPARC) family [75]. The SPARC family plays a role in cell migration, cell proliferation and cell apoptosis of certain types of cancer [76]. As a member of the family, SPOCK1 also plays a key role in proliferation, adhesion and migration of cancer cells [77–79]. CHD1L can activate transcription of SPOCK1 by binding to the 5’upstream region of SPOCK1, and promote anti-apoptotic effect by activating the AKT pathway and inhibiting the cytochrome c/caspase-9/caspase-3 pathway [16].
TCTP
Translationally controlled tumor protein (TCTP) expresses in almost all mammalian tissues as a housekeeping gene. TCTP is a pro-survival factor by inhibiting apoptosis and promoting cell cycle as a tubulin-binding protein [80]. CHD1L protein can bind directly to the 5′ upstream region (nt − 733/− 1027) of TCTP and activate its transcription [15]. Then, TCTP could promote the ubiquitin proteasome degradation of CDC25C, and downregulate CDK1 activity by inhibiting the dephosphorylation of CDK1 at Tyr15 and lead to a faster mitotic exit during mitotis [15]. The CHD1L-TCTP-CDC25C-CDK1 pathway could cause malignant transformation of hepatocytes, and its phenotype accelerated the mitotic process and produced aneuploidy [15].
MDM2
MDM2 is characterized by dynamic negative regulation of the tumor suppressor p53 [81], which can promote cell cycle transition in p53-dependent [82] and p53-independent [83] manner. Increased levels of MDM2 could promote the ubiquitination and degradation of E-cadherin, which in turn drove cancer cell invasion [84]. CHD1L might facilitate the progress of breast cancer cells via the MDM2/p53 signaling pathway [63].
NTKL
N-terminal kinase like protein gene (NTKL) locates on 11q13 and encodes a 808 amino acid protein [85], which exhibited in golgi apparatus, centrosomes, cytoplasm and nucleus of subcellular localizations [86]. Golgi NTKL was reported to interact with Cop1 vesicles and regulate golgi morphology [87], while centrosome NTKL played an important role in cell division [88]. NTKL was frequently upregulated by CHD1L in primary HCC cases and exhibited a strong oncogenic ability [17].
ABCB1
The ATP-Binding Cassette Sub-Family B Member 1 (ABCB1), also called the plasma membrane glycoprotein (P-glycoprotein), locates on the chromosome 7q21.12, which encodes a 170 KD trans-membrane glycoprotein and belongs the ATP-binding cassette (ABC) transporters family [89]. The ABC transporter family has a transport effect on chemotherapeutic agents, which causes the occurrence and development of multidrug resistance (MDR). ABCB1 is the most important resistance-inducing protein. ABCB1 was a potential downstream target gene of CHD1L in NSCLC cells [66]. The up-regulation of ABCB1 by CHD1L depended on the transcription of c-Jun. ABCB1 knockdown coupled with CHD1L ectopic expression enhanced the effect of cisplatin on apoptosis of NSCLC cells [66].
β-Catenin
β-catenin is a typical influencer of Wnt signaling pathway and a component of cell-cell adhesion complex, which participates in cell proliferation, metastasis, differentiation and tumorigenesis [90, 91]. When the Wnt signaling pathway is activated, β-catenin accumulates in the cytoplasm and then enters the nucleus to form a complex with the nuclear transcription factor Tcf/Lef. Then, β-catenin can activate a series of downstream target genes, which participate in the transcription process in different ways [92]. CHD1L overexpression significantly increased β-catenin expression in pancreatic cancer [65]. However, elevated β-catenin expression exhibited in the CHD1L-KD group in glioma [64]. Therefore, β-catenin may play different roles in different tumors as a target gene of CHD1L.
p53
p53 protein is essential for effective suppression of human tumors [93]. Overexpression of CHD1L inhibits the expression of p53 in HCC [1], breast cancer [63] and glioma [64], makes p53 lose its anti-cancer effect. p53 protein can upregulate p21 expression, and the latter acts as a Cdk2 inhibitor, inactivating cyclinE-CDk2 complex to control the S phase entry [62, 94, 95]. CHD1L overexpression could promote cell proliferation by downregulating the p53-p21-cyclinE-Cdk2 pathway in HCC [1]. However, the loss of CHD1L resulted in increased expression of p53 and p21, while decreased expression of cyclinE and Cdk2 in glioma [64].
Nur77
Nur77, also named NR4A1, is a unique transcriptional factor and belongs to the orphan nuclear receptor superfamily [96, 97]. Nur77 is a key member of the p53-independent apoptotic pathway, which directly targets Bcl-2 and induces the latter to adopt a pro-apoptotic conformation, thereby triggering the release of cytochrome c and the activation of caspase-9 and caspase-3 [98–100]. CHD1L can inhibit the nuclear translocation of Nur77 to mitochondria. Macro domain of CHD1L acts to interact with Nur77, while the CHD1L mutants lacking residues 600–897 cannot interact with Nur77 and prevents Nur77-mediated apoptosis of hepatocellular carcinoma [18].
hMLH1
Human mutL homolog 1 gene (hMLH1) is located on the chromosome 3p21.3–23, which encodes a protein of 756 amino acids. hMLH1 is an important member of DNA mismatch repair (MMR) protein family. MMR is a common DNA repair process which helps maintain the stability and integrity of genetic substances [101]. It has been observed that hMLH1 protein interacts with mismatched genes and DNA repair enzymes, and enhances DNA repair [102]. Downregulation of hMLH1 could lead to the loss of DNA MMR repair function and promote tumor progress [102]. CHD1L can inhibit hMLH1 expression in cholangiocarcinoma cells, which was related to the malignant progression and prognosis of cholangiocarcinoma [103].
CHD1L related disease
The homology and difference between CHD1L and CHD1
CHD1 locates in 5q15 and encodes a protein composed of 1710 amino acids (hCHD1: NP_001261), belongs to the Chd family proteins that play an important role in transcriptional regulation and developmental processes [29]. CHD1 is characterized by two N-terminal tandem chromodomains (yellow rectangles), a central SNF2-like ATPase domain (green oval), and a C-terminal DNA binding domain (blue oval) (Fig. 3a) [104]. Chromodomains are modules implicated in the recognition of lysine-methylated histone tails and nucleic acids. Lysine methylation at a specific site on the histone H3 tails is associated with transcriptional regulation. Methylation at H3 K4, H3 K36 and H3 K79 is linked to transcriptional activity, whereas methylation at H3 K9, H3 K27 and H4 K20 serves as a marker for epigenetic silencing [105]. The SNF2-like ATPase domain defines the ATP-dependent chromatin remodeling proteins. The Chd1 DNA-binding domain is consist of a SANT and SLIDE domain, which is required for efficient nucleosome sliding and believed to be essential for sensing the length of DNA flanking the nucleosome core [106].
Fig. 3.
The homology and difference between CHD1L and CHD1. a Structural diagram of CHD1L and CHD1. CHD1 contains two N-terminal tandem chromodomains (yellow rectangles), a central SNF2-like ATPase domain (green oval), and a C-terminal DNA binding domain (blue oval). CHD1L contains a SNF2_N domain (green oval), HELICc (blue oval), Sels (black rectangle) and a Macro domain (red rectangle). b Multiple Alignment Results of CHD1L and CHD1 Protein Sequence. The sequence homology between the SNF2_N domain of CHD1L (280 aa) and SNF2-like ATPase domain of CHD1 have 45% identity (126/280, yellow indicator). The sequence homology between the HELICc domain of CHD1L (107 aa) and DNA binding domain of CHD1 have 59% identity (63/107, green indicator)
Sequence homology analysis showed that CHD1L (initially called ALC1, hCHD1L: NM_004284.6) contains a conserved SNF2_N domain (green oval), a helicase superfamily c-terminal domain (HELICc) (blue oval), Sels domain (black rectangle) and a Macro domain (red rectangle) in human by Ma et al. (Fig. 3a) [1]. The SNF2_N domain is composed of 280 amino acids, and the sequence homology between the SNF2_N domain of CHD1L and SNF2-like ATPase domain of CHD1 have 45% identity (126/280, yellow indicator). The HELICc domain is composed of 107 amino acids, and the sequence homology between the HELICc domain of CHD1L and DNA binding domain of CHD1 have 59% identity (63/107, green indicator) (Fig. 3b). Therefore, the name of CHD1L was given.
The difference structures between CHD1L and CHD1 are that CHD1 has two tandem chromodomians that affect DNA functions, including transcription, replication, recombination and repair, while CHD1L uses the Marco domain to perform biological functions by binding PAR. The Macro domain proteins also recognize poly-ADP-ribose as a ligand, and the ADP-ribosylation of proteins is an important posttranslational modification that occurs in a variety of biological processes, including DNA repair, transcription, chromatin biology and long-term memory formation [42].
Studies have shown that CHD1 plays an important role in embryonic stem cell differentiation, hematopoietic stem cells emergence and tumorigenesis. Full length CHD1 is required in embryonic stem cell differentiation, and loss of the serine-rich region (SRR) renders CHD1 unable to support normal differentiation of ESCs into the three germ layers [107]. Endothelial-specific deletion of CHD1 leads to loss of definitive hematopoietic progenitors, anemia, and lethality by embryonic day (E)15.5 [108]. CHD1 is the 5q21 tumor suppressor gene, and inactivation of CHD1 abolishes recruitment of androgen receptor (AR) to result in downregulation of AR-responsive genes (eg. FOXO1, NKX3–1 and PPARγ) in prostate cancer [109]. Recurrent deletion of CHD1 is a driver of prostate cancer cell invasiveness [110]. CHD1 drives immune suppression in PTEN-deficient prostate cancer [111]. a novel CHD1-RUNX1 fusion collaborated with FLT3-ITD mutation in the development of acute myeloid leukemia [112]. However, CHD1L is an oncoprotein, which is overexpression in various cancers.
Abnormal expression and pan-cancer analysis of CHD1L
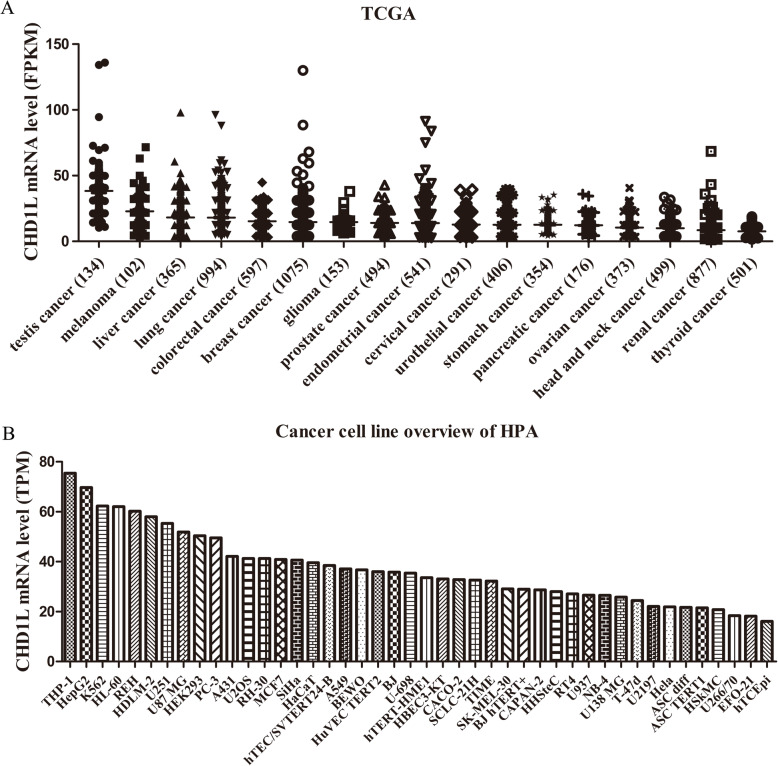
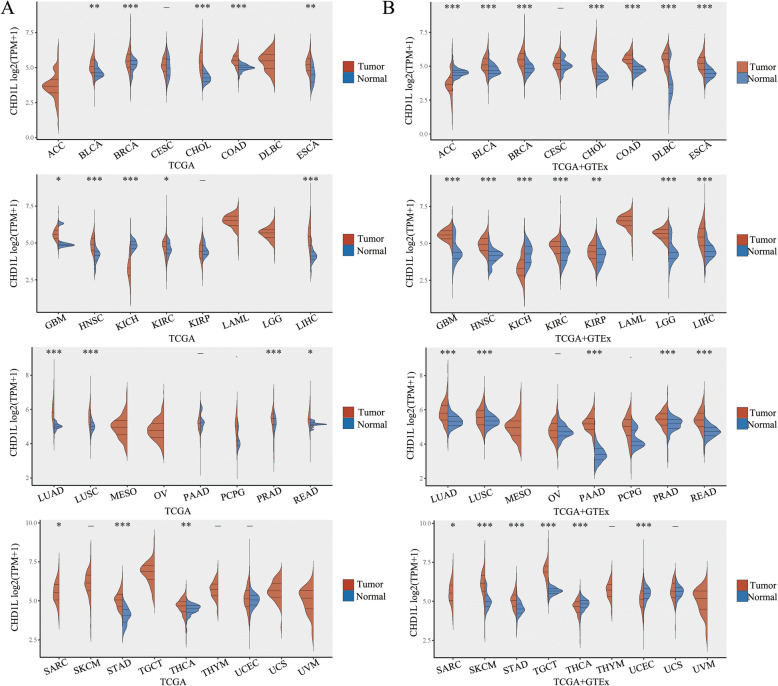
Several studies have found that CHD1L has a strong carcinogenic ability, including promoting tumor cell proliferation, invasion, migration, metastasis and inhibiting tumor cell apoptosis [1, 6, 12, 13]. In mESCs, overexpression of CHD1L increases tumor susceptibility [58]. It is documented that CHD1L is deregulation in various neoplastic diseases. CHD1L is an important oncoprotein that is overexpressed in various malignant tumors, including HCC [1, 14–18, 21], ovarian cancer [23], gastric cancer [22], colorectal carcinoma [24], bladder cancer [25], breast cancer [4, 26, 63], nasopharyngeal carcinoma [27], glioma [64], NSCLC [12, 66], myeloma [6], pancreatic cancer [65], esophageal carcinoma [28, 67] and cholangiocarcinoma [20, 103] (Fig. 4 and Table 3). Further, pan-cancer analysis of CHD1L expression was determined in The Cancer Genome Atlas (TCGA) dataset (https://tcga-data.nci.nih.gov/tcga/) and The Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/). In TCGA dataset, the expression of CHD1L in BLCA, BACA, CHOL, COAD, ESCA, GBM, HNSC, KIRC, LIHC, LUAD, LUSC, PRAD, READ, SARC, STAD and THCA was higher than the normal tissues, while in KICH was lower than normal tissues, and the differences are statistically significant (Fig. 5a). In the TCGA combined GTEx database, CHD1L is expressed in BLCA, BACA, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, PAAD, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA and UCEC are all higher than normal tissues, while in ACC, KICH is lower than normal tissues, and the difference is statistically significant (Fig. 5b).
Fig. 4.
CHD1L mRNA expression overview of cancer category and cell lines from TCGA (a) and HPA (b) dataset. FPKM: Fragments Per Kilobase of exon per Million. TPM:Transcript Per Million
Table 3.
Summary of the current literature on CHD1L deregulation in solid cancers
| Cancer | Deregulation | Downstream targets | Phenotypic effect | clinical impact | References |
|---|---|---|---|---|---|
| HCC | CHD1L↑ | P53↓, p21↓, cyclinE↑, CDK2↑ | G1/S phase transition↑, apoptosis↓, proliferation↑ | [1] | |
| CHD1L↑ | Nur77↓ | apoptosis↓ | [18] | ||
| CHD1L↑ | ARHGEF9↑, Cdc42↑ | migration↑, invasion↑, metastasis↑ | [14] | ||
| CHD1L↑ | TCTP↑, Cdc25c↓, Cdk1↓ | Mitotic progression↑, aneuploidy↑ | [15] | ||
| CHD1L↑ | SPOCK1↑, Akt↑ | apoptosis↓, invasion↑, metastasis↑ | [16] | ||
| CHD1L↑ | NTKL↑ | cell growth↑, colony formation↑, G1/S transition↑ | [17] | ||
| CHD1L↑ | NA | Poor Prognosis | [21] | ||
| Ovarian carcinoma | CHD1L↑ | NA | metastasis↑ | Prognostic biomarker | [23] |
| CHD1L↑ | METAP2↑ | invasion↑, metastasis↑ | [19] | ||
| Gastric cancer | CHD1L↑ | NA | Prognostic biomarker | [22] | |
| Colorectal carcinoma | CHD1L↑ | NA | G1/S phase transition↑, apoptosis↓ | Prognostic biomarker | [24] |
| Bladder cancer | CHD1L↑ | NA | Prognostic biomarker | [25] | |
| Breast cancer | CHD1L↑ | NA | Prognostic biomarker | [27] | |
| CHD1L↑ | MDM2↑, p53↓ | Cell cycle↑, cell motility↑ | [63] | ||
| CHD1L↑ | PI3K↑, Akt↑, ARK5↑, mTOR↑, MMP2↑, MMP9↑ | chemotaxis↑, invasion↑, lung colonization↑ | [4] | ||
| Nasopharyageal carcinoma | CHD1L↑ | NA | Prognostic biomarker | [27] | |
| Glioma | CHD1L↑ | PCNA↑, β-catenin ↓, cyclinD1↑, p53↓, p21↓, cyclinE↑, Cdk2↑, c-capase3↓, Bcl2↑ | G1/S phase transition↑, proliferation↑, apoptosis↓, migration↑, invasion↑ | [64] | |
| NSCLC | CHD1L↑ | NA | Prognostic biomarker | [12] | |
| CHD1L↑ | ABCB1↑, c-Jun↑, NF-kB↑ | Cisplatin resistance | [66] | ||
| Myeloma | CHD1L↑ | c-capase9↓, capase3↓ | Anti-apoptosis, cell adhesion-mediated drug resistance↑ | [6] | |
| Pancreatic cancer | CHD1L↑ | β-catenin↑ | Cell proliferation↑ | Poor prognosis | [65] |
| Esophageal carcinoma | CHD1L↑ | NA | proliferation↑, apoptosis↓, metastasis↑, invasion↑ | Poor prognosis | [28] |
| CHD1L↑ | PI3K/Akt pathway↑ | viability↑, apoptosis↓, cisplatin cytotoxicity↓, glycolysis↑ | [67] | ||
| Cholangiocarcinoma | CHD1L↑ | hMLH1↓ | Prognostic biomarker | [103] | |
| CHD1L↑ | P53↓, cyclinD1↑, CDK2↑, E-cadherin↓, N-cadherin↑, Vimentin↑ | EMT↑, G1/S transition↑, Cell proliferation↑ | Poor prognosis | [20] |
Fig. 5.
Pan-cancer analysis of CHD1L expression in TCGA dataset (a) and the TCGA combined GTEx dataset (b)
Prognostic biomarker and prognosis analysis of CHD1L
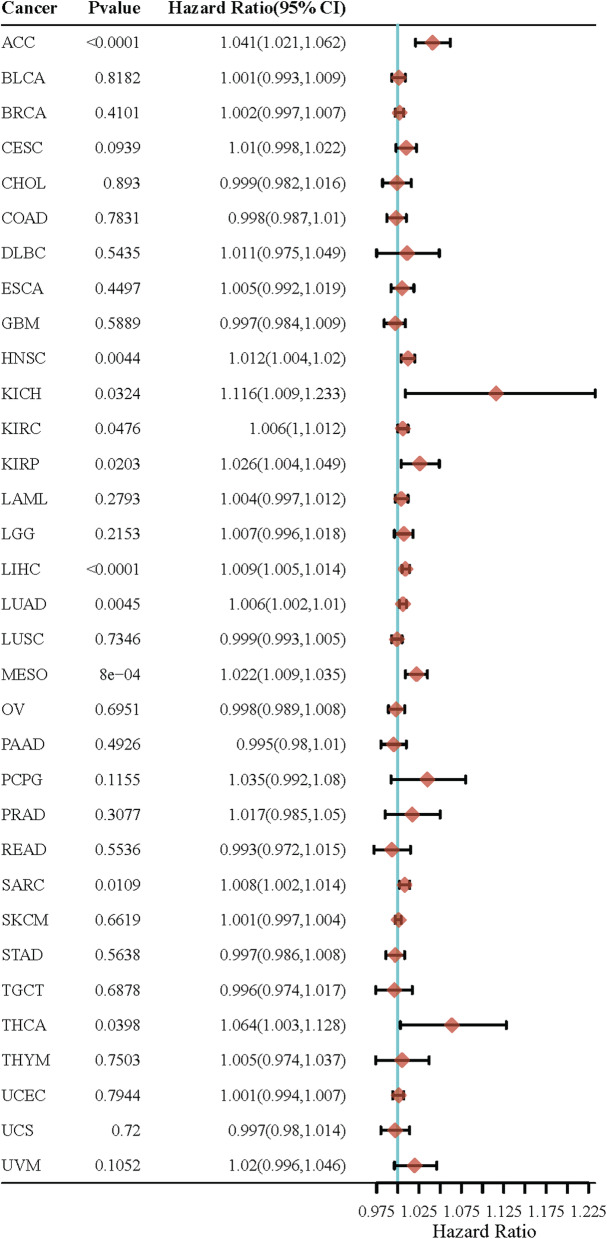
CHD1L is considered a cancer or prognostic biomarker, because its overexpression is usually associated with poor prognosis, such as in HCC [21], gastric cancer [22], bladder cancer [25], breast cancer [26], NSCLC [12], cholangiocarcinoma [20], and nasopharyngeal carcinoma [27]. CHD1L expression is correlated with tumor size and stage in bladder cancer [25], nasopharyngeal carcinoma [27] and pancreatic cancer [65] in contrast to HCC [17] and ovarian cancer [23]. Further, the prognostic analysis of CHD1L was obtained from the TCGA database. The abnormal expression of CHD1L may be prognostic factor of ACC, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, MESO, SARC and THCA (Fig. 6).
Fig. 6.
Prognostic analysis of CHD1L in TCGA database
CHD1L and drug resistance
CHD1 as tumor suppressor gene, is currently rare in tumor resistance mechanisms. CHD1L as a potent anti-apoptotic and pro-proliferative factor, CHD1L overexpression is correlated with the increase of chemotherapy resistance in myeloma [6], NSCLC [66] and ESCC [67]. Bortezomib decreased the protein levels of CHD1L, but cell adhesion increased the expression levels of CHD1L, overexpression of CHD1L contributes to cell adhesion-mediated drug resistance (CAM-DR) in multiple myeloma cells [6]. Overexpression of CHD1L increased the transcription of c-Jun which targeted directly to the promoter of ABCB1 (initially isolated in drug-resistant Chinese hamster ovary cancer cells), and the upregulated ABCB1 phosphoried the NF-κB downstream Iκ-Bα and p65, therefore, CHD1L could induce cisplatin resistance via c-Jun-ABCB1-NF-κB axis in NSCLC [66]. Knockdown of CHD1L enhanced cisplatin cytotoxicity of ESCC cells by inhibition of glycolysis through inactivation of the PI3K/Akt pathway [67]. CHD1L results in cell malignant change and chemotherapy resistance through complex mechanisms in several solid tumors (Fig. 1b). Therefore, for patients with increased expression of CHD1L, effective inhibition of CHD1L may represent a promising therapeutic direction.
The pharmacological inhibition of CHD1L
Since CHD1L was discovered in 2008, it acts as an oncogene in a variety of tumors, but relatively few reported CHD1L as a druggable target and establishes a novel therapeutic strategy for the treatment of cancers. CHD1L possesses intrinsic poly (ADP)-ribose (PAR) chains via a C-terminal macro domain, which rapidly recruited to sites of DNA damage by PAR chains synthesized by PARP1/2 [11]. Therefore, PARP inhibitors may be applied to the target of CHD1L. Blessing et al. reveal CHD1L manipulation impacts the single-strand DNA break repair response and potentiates PARPi-induced cancer killing through PARP2 trapping [113]. PARP inhibitor Olaparib suppressed the DNA damage repair signaling and repressed the global pluripotent transcriptional network through CHD1L-mediated condensation of the chromatin structure in HCC [114]. Lead CHD1L inhibitors (Compounds 5–7) display potent antitumor activity by reversing TCF-driven EMT and induction of cleaved Ecadherin mediated extrinsic apoptosis in colorectal cancer [115]. The pharmacological inhibition of CHD1L might represent a promising therapeutic strategy for patients with decrease CHD1L expression. An overview of the CHD1L inhibitors is described in Table 4.
Table 4.
Description of CHD1L-inhibitors identified so far
| Compound | Chemical structure | Molecular Weight | Mechanism | Effect on CHD1L target genes | IC50 | References |
|---|---|---|---|---|---|---|
| 2-(4-Methoxyphenyl)-5-(methylsulfonyl)-4-(phenylsulfonyl)-1,3-oxazole | 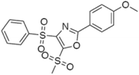 |
393.43 | Inhibit that CHD1L binds the TCF complex, reverse TCF-driven EMT | TCF complex WNT response elements (WRE) (eg. c-Myc, vimentin, slug, LEF1, and N-cadherin) | 3 μmol/L | [115] |
| N-(4-{[6-Methyl-2-(1-pyrrolidinyl)-4-pyrimidinyl]amino}phenyl)-2-(2-thienyl)acetamide | 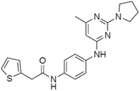 |
393.51 | 5.5 μmol/L | [115] | ||
| 2-(4-{4-[(3-Chloro-4-methylphenyl)amino]-2-pteridinyl}-1-piperazinyl)ethanol | 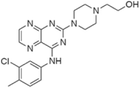 |
399.88 | 4 μmol/L | [115] | ||
| Olaparib | 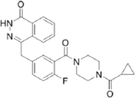 |
434.46 | suppresses the DNA damage repair signaling, repress the key pluripotent transcriptional factors | DNA damage repair genes (eg. SSRP1,ERCC3,CHD1L,TP53BP1,TRIP13), the key pluripotent transcriptional factors (eg. SOX2, OCT4, c-MYC) |
5–50 μM depending on Assays (HCC cells), 10–50 mg/kg (xenograft mouse model) |
[113] [114] |
| Niraparib | 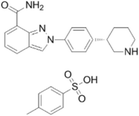 |
492.59 | suppresses the DNA damage repair signaling | DNA damage repair genes (eg. SSRP1,ERCC3,CHD1L,TP53BP1,TRIP13) | Not available (10 μM treated HCC cells) | [114] |
Conclusions
CHD1L is a multifunctional protein that plays important roles in normal or pathological conditions. However, how does CHD1L manage all those diverse functions? This may be related to its three main domains: the N-terminal domain of SNF2 family, HELICc and Macro domain. In normal cells, CHD1L mainly functions through the Macro domain. First, CHD1L recognizes PAR through the macro domain. PARP activates the chromatin-repositioning enzyme of CHD1L, which in turn activates PAR-dependent chromatin remodeling and promotes DNA repair. In addition, under pathological conditions, CHD1L is usually abnormally expressed in a variety of tumors. The mechanism of CHD1L involved in tumorigenesis may be the activation of certain genes (such as ARHGEF9, SPOCK1, TCTP) and/or suppression of other genes (such as p53, Nur77, hMLH1). The overexpression of CHD1L is also related to the clinical characteristics and prognosis of patients, so the expression level of CHD1L can be considered as a potential prognostic factor for cancer.
In conclusion, CHD1L recognizes several activities that play key roles in many biological processes. The significance of CHD1L in cellular and pathological effects have not been fully understood and remain to be explored. Therefore, this review suggests that CHD1L may be a novel biomarker for cancers and represents a fascinating target for molecular cancer therapy in the future.
Acknowledgements
Not applicable.
Abbreviations
- CHD1L
Chromodomain helicase/ATPase DNA binding protein 1-like gene
- ALC1
Amplified in liver cancer 1
- SNF2
Sucrose non-fermenting 2
- CGH
Comparative genomic hybridization
- ARHGEF9
Rho guanine nucleotide exchange factor 9
- SPOCK1
Sparc/osteonectin, cwcv, and kazal-like domains proteoglycan 1
- TCTP
Translationally controlled tumor protein
- ABCB1
ATP-Binding Cassette Sub-Family B Member 1
- hMLH1
Human mutL homolog 1 gene
- NTKL
N-terminal kinase like protein
- TPM
Transcript Per Million
- FPKM
Fragments Per Kilobase of exon per Million
- ACC
Adrenocortical carcinoma
- BLCA
Bladder Urothelial Carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid Neoplasm Diffuse Large B-cell Lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney Chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute Myeloid Leukemia
- LGG
Brain Lower Grade Glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and Paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcomav
- SKCM
Skin Cutaneous Melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular Germ Cell Tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine Corpus Endometrial Carcinoma
- UCS
Uterine Carcinosarcoma
- UVM
Uveal Melanoma
Authors’ contributions
X.X., X.L., Z.L. and N.M. conceived and designed the review. X.X., X.L., A.L. conducted the literature search and participated in the analysis and interpretation of data. X.X., Z.L. and N.M. drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded in part by National Natural Science Foundation of China (No.81902802), Medical Science and Technology Foundation of Guangdong Province (No.A2018063), Traditional Chinese Medicine Bureau of Guangdong Province (No.20191260 & 20181206) and Technology Project of Guangzhou Municipal Health Commission (No.20181A010017 & 20201A011020). The Science and Technology Planning project of Guangzhou (No.2018040100160).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xifeng Xiong and Xudong Lai contributed equally to this work.
Contributor Information
Aiguo Li, Email: Liaiguo7161@ext.jnu.edu.cn.
Zhihe Liu, Email: zliu0731@ext.jnu.edu.cn.
Ningfang Ma, Email: nfma@gzhmu.edu.cn.
References
- 1.Ma NF, Hu L, Fung JM, Xie D, Zheng BJ, Chen L, et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47(2):503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- 2.Guan XY, Zhang H, Bittner M, Jiang Y, Meltzer P, Trent J. Chromosome arm painting probes. Nat Genet. 1996;12(1):10–11. doi: 10.1038/ng0196-10. [DOI] [PubMed] [Google Scholar]
- 3.Guan XY, Fang Y, Sham J, Kwong D, Zhang Y, Liang Q, et al. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30(1):110. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1063>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Mu QJ, Li HL, Yao Y, Liu SC, Yin CG, Ma XZ. Chromodomain helicase/ATPase DNA-binding protein 1-like gene (CHD1L) expression and implications for invasion and metastasis of breast Cancer. PLoS One. 2015;10(11):e0143030. doi: 10.1371/journal.pone.0143030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, Chow E, et al. Understanding the impact of 1q21.1 copy number variant. Orphanet J Rare Dis. 2011;6:54. doi: 10.1186/1750-1172-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, He Y, Miao X, Wu Y, Han J, Wang Q, et al. Cell adhesion induces overexpression of chromodomain helicase/ATPase DNA binding protein 1-like gene (CHD1L) and contributes to cell adhesion-mediated drug resistance (CAM-DR) in multiple myeloma cells. Leuk Res. 2016;47:54–62. doi: 10.1016/j.leukres.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23(14):2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88(6):737–740. doi: 10.1016/S0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 9.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A. 1997;94(21):11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, et al. Poly (ADP-ribosyl) ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106(33):13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly (ADP-ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science (New York, NY) 2009;325(5945):1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He LR, Ma NF, Chen JW, Li BK, Guan XY, Liu MZ, et al. Overexpression of CHD1L is positively associated with metastasis of lung adenocarcinoma and predicts patients poor survival. Oncotarget. 2015;6(31):31181–31190. doi: 10.18632/oncotarget.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Chen L, Ma NF, Chow RK, Li Y, Song Y, et al. CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors. Hepatology. 2016;63(5):1544–1559. doi: 10.1002/hep.28437. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Chan TH, Yuan YF, Hu L, Huang J, Ma S, et al. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J Clin Invest. 2010;120(4):1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan TH, Chen L, Liu M, Hu L, Zheng BJ, Poon VK, et al. Translationally controlled tumor protein induces mitotic defects and chromosome missegregation in hepatocellular carcinoma development. Hepatology. 2012;55(2):491–505. doi: 10.1002/hep.24709. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Chen L, Chan TH, Liu M, Kong KL, Qiu JL, et al. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology. 2013;144(1):179–91 e4. doi: 10.1053/j.gastro.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Liu M, Chen L, Chan TH, Jiang L, Yuan YF, et al. Overexpression of N-terminal kinase like gene promotes tumorigenicity of hepatocellular carcinoma by regulating cell cycle progression and cell motility. Oncotarget. 2015;6(3):1618–1630. doi: 10.18632/oncotarget.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Hu L, Chan TH, Tsao GS, Xie D, Huo KK, et al. Chromodomain helicase/adenosine triphosphatase DNA binding protein 1-like (CHD1l) gene suppresses the nucleus-to-mitochondria translocation of nur77 to sustain hepatocellular carcinoma cell survival. Hepatology. 2009;50(1):122–129. doi: 10.1002/hep.22933. [DOI] [PubMed] [Google Scholar]
- 19.He WP, Guo YY, Yang GP, Lai HL, Sun TT, Zhang ZW, et al. CHD1L promotes EOC cell invasiveness and metastasis via the regulation of METAP2. Int J Med Sci. 2020;17(15):2387–2395. doi: 10.7150/ijms.48615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Chai Y, Ding Y, Yuan T, Wu C, Huang C. CHD1L is associated with poor survival and promotes the proliferation and metastasis of intrahepatic cholangiocarcinoma. Oncol Rep. 2019;42(2):657–669. doi: 10.3892/or.2019.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyeon J, Ahn S, Park CK. CHD1L is a marker for poor prognosis of hepatocellular carcinoma after surgical resection. Korean J Pathol. 2013;47(1):9–15. doi: 10.4132/KoreanJPathol.2013.47.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z, Zhao J, Xian G, Geng W, Rong Z, Wu Y, et al. CHD1L is a novel independent prognostic factor for gastric cancer. Clin Transl Oncol. 2014;16(8):702–707. doi: 10.1007/s12094-013-1136-8. [DOI] [PubMed] [Google Scholar]
- 23.He WP, Zhou J, Cai MY, Xiao XS, Liao YJ, Kung HF, et al. CHD1L protein is overexpressed in human ovarian carcinomas and is a novel predictive biomarker for patients survival. BMC Cancer. 2012;12:437. doi: 10.1186/1471-2407-12-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji X, Li J, Zhu L, Cai J, Zhang J, Qu Y, et al. CHD1L promotes tumor progression and predicts survival in colorectal carcinoma. J Surg Res. 2013;185(1):84–91. doi: 10.1016/j.jss.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Tian F, Xu F, Zhang ZY, Ge JP, Wei ZF, Xu XF, et al. Expression of CHD1L in bladder cancer and its influence on prognosis and survival. Tumour Biol. 2013;34(6):3687–3690. doi: 10.1007/s13277-013-0951-4. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Zong Y, Fei X, Chen X, Huang O, He J, et al. Presence of CHD1L over-expression is associated with aggressive tumor biology and is a novel prognostic biomarker for patient survival in human breast cancer. PLoS One. 2014;9(8):e98673. doi: 10.1371/journal.pone.0098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su FR, Ding JH, Bo L, Liu XG. Chromodomain helicase/ATPase DNA binding protein 1-like protein expression predicts poor prognosis in nasopharyngeal carcinoma. Exp Ther Med. 2014;8(6):1745–1750. doi: 10.3892/etm.2014.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu ZH, Zhang Q, Ding YJ, Ren YH, Yang HP, Xi Q, et al. Overexpression of CHD1L is associated with poor survival and aggressive tumor biology in esophageal carcinoma. Oncotarget. 2017;8(43):74178–74187. doi: 10.18632/oncotarget.18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618(1–2):30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W, Su Y, Xu F. CHD1L: a novel oncogene. Mol Cancer. 2013;12(1):170. doi: 10.1186/1476-4598-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, et al. Characterization of mammalian selenoproteomes. Science. 2003;300(5624):1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 32.Liu SS, Bai YS, Feng L, Dong WW, Li Y, Xu LP, et al. Identification of CHD1L as an important regulator for Spermatogonial stem cell survival and self-renewal. Stem Cells Int. 2016;2016:4069543. doi: 10.1155/2016/4069543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu SS, Maguire EM, Bai YS, Huang L, Liu Y, Xu L, et al. A Novel Regulatory Axis, CHD1L-MicroRNA 486-Matrix Metalloproteinase 2, Controls Spermatogonial Stem Cell Properties. Mol Cell Biol. 2019;39(4):e00357–18. [DOI] [PMC free article] [PubMed]
- 34.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D2D3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bork P, Koonin EV. An expanding family of helicases within the 'DEAD/H' superfamily. Nucleic Acids Res. 1993;21(3):751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24(5):192–198. doi: 10.1016/S0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 37.Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274(22):15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 38.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12(1):123–133. doi: 10.1016/S0959-440X(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Li F, Rozovsky S. The intrinsically disordered membrane protein selenoprotein S is a reductase in vitro. Biochemistry. 2013;52(18):3051–3061. doi: 10.1021/bi4001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Rozovsky S. Contribution of selenocysteine to the peroxidase activity of selenoprotein S. Biochemistry. 2013;52(33):5514–5516. doi: 10.1021/bi400741c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croset A, Cordelieres FP, Berthault N, Buhler C, Sun JS, Quanz M, et al. Inhibition of DNA damage repair by artificial activation of PARP with siDNA. Nucleic Acids Res. 2013;41(15):7344–7355. doi: 10.1093/nar/gkt522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Silva I, Pelletier JD, Lagueux J, D'Amours D, Chaudhry MA, Weinfeld M, et al. Relative affinities of poly (ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta. 1999;1430(1):119–126. doi: 10.1016/S0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 45.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31(2):101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, Selby CP, Adar S, Adebali O, Sancar A. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J Biol Chem. 2017;292(38):15588–15597. doi: 10.1074/jbc.R117.807453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanzlikova H, Gittens W, Krejcikova K, Zeng Z, Caldecott KW. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017;45(5):2546–2557. doi: 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuda M, Cho K, Ooka M, Shimizu N, Watanabe R, Yasui A, et al. ALC1/CHD1L, a chromatin-remodeling enzyme, is required for efficient base excision repair. PLoS One. 2017;12(11):e0188320. doi: 10.1371/journal.pone.0188320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottschalk AJ, Trivedi RD, Conaway JW, Conaway RC. Activation of the SNF2 family ATPase ALC1 by poly (ADP-ribose) in a stable ALC1.PARP1.Nucleosome intermediate. J Biol Chem. 2012;287(52):43527–43532. doi: 10.1074/jbc.M112.401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, et al. The poly (ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell. 2016;27(24):3791–3799. doi: 10.1091/mbc.E16-05-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh HR, Nardozza AP, Moller IR, Knobloch G, Kistemaker HAV, Hassler M, et al. A poly-ADP-ribose trigger releases the auto-inhibition of a chromatin remodeling oncogene. Mol Cell. 2017;68(5):860–71 e7. doi: 10.1016/j.molcel.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Lehmann LC, Hewitt G, Aibara S, Leitner A, Marklund E, Maslen SL, et al. Mechanistic insights into autoinhibition of the oncogenic chromatin remodeler ALC1. Mol Cell. 2017;68(5):847–59 e7. doi: 10.1016/j.molcel.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199(2):235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulkarni A, Oza J, Yao M, Sohail H, Ginjala V, Tomas-Loba A, et al. Tripartite motif-containing 33 (TRIM33) protein functions in the poly (ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with amplified in liver Cancer 1 (ALC1) protein. J Biol Chem. 2013;288(45):32357–32369. doi: 10.1074/jbc.M113.459164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ooka M, Abe T, Cho K, Koike K, Takeda S, Hirota K. Chromatin remodeler ALC1 prevents replication-fork collapse by slowing fork progression. PLoS One. 2018;13(2):e0192421. doi: 10.1371/journal.pone.0192421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hewitt G, Borel V, Segura-Bayona S, Takaki T, Ruis P, Bellelli R, et al. Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol Cell. 2021 ;81(4):767–83.e11. [DOI] [PMC free article] [PubMed]
- 57.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6(1):133–144. doi: 10.1016/S1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- 58.Snider AC, Leong D, Wang QT, Wysocka J, Yao MW, Scott MP. The chromatin remodeling factor Chd1l is required in the preimplantation embryo. Biol Open. 2013;2(2):121–131. doi: 10.1242/bio.20122949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang B, Chen W, Li H, Chien Y, Chang W, Hsieh P, et al. CHD1L regulated PARP1-driven Pluripotency and chromatin remodeling during the early-stage cell reprogramming. Stem Cells. 2015;33(10):2961–2972. doi: 10.1002/stem.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dou D, Zhao H, Li Z, Xu L, Xiong X, Wu X, et al. CHD1L promotes neuronal differentiation in human embryonic stem cells by Upregulating PAX6. Stem Cells Dev. 2017;26(22):1626–1636. doi: 10.1089/scd.2017.0110. [DOI] [PubMed] [Google Scholar]
- 61.Brockschmidt A, Chung B, Weber S, Fischer DC, Kolatsi-Joannou M, Christ L, et al. CHD1L: a new candidate gene for congenital anomalies of the kidneys and urinary tract (CAKUT) Nephrol Dial Transplant. 2012;27(6):2355–2364. doi: 10.1093/ndt/gfr649. [DOI] [PubMed] [Google Scholar]
- 62.Chen M, Huang JD, Hu L, Zheng BJ, Chen L, Tsang SL, et al. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS One. 2009;4(8):e6727. doi: 10.1371/journal.pone.0006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, Wu J, Fei X, Chen W, Li Y, Shen K, et al. CHD1L promotes cell cycle progression and cell motility by up-regulating MDM2 in breast cancer. Am J Transl Res. 2019;11(3):1581–1592. [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Zhang L, Zhao H, Qiu X, Chen W, Wang D, et al. CHD1L regulates cell cycle, apoptosis, and migration in Glioma. Cell Mol Neurobiol. 2016;36(4):565–576. doi: 10.1007/s10571-015-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu C, Fu X, Zhong Z, Zhang J, Mou H, Wu Q, et al. CHD1L expression increases tumor progression and acts as a predictive biomarker for poor prognosis in pancreatic Cancer. Dig Dis Sci. 2017;62(9):2376–2385. doi: 10.1007/s10620-017-4641-8. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, He LR, Gao Y, Zhou NN, Liu Y, Zhou XK, et al. CHD1L contributes to cisplatin resistance by upregulating the ABCB1-NF-kappaB axis in human non-small-cell lung cancer. Cell Death Dis. 2019;10(2):99. doi: 10.1038/s41419-019-1371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li F, Zhang Z, Wang P, Wen P, Xu Q, Wang Y, et al. ALC1 knockdown enhances cisplatin cytotoxicity of esophageal squamous cell carcinoma cells by inhibition of glycolysis through PI3K/Akt pathway. Life Sci. 2019;232:116679. doi: 10.1016/j.lfs.2019.116679. [DOI] [PubMed] [Google Scholar]
- 68.Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3(1):22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- 69.Wherlock M, Mellor H. The rho GTPase family: a Racs to Wrchs story. J Cell Sci. 2002;115(Pt 2):239–240. doi: 10.1242/jcs.115.2.239. [DOI] [PubMed] [Google Scholar]
- 70.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 71.Braga VM. Cell-cell adhesion and signalling. Curr Opin Cell Biol. 2002;14(5):546–556. doi: 10.1016/S0955-0674(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 72.Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via rho family GTPases. J Cell Biol. 2000;150(3):567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582(14):2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 74.Reid T, Bathoorn A, Ahmadian MR, Collard JG. Identification and characterization of hPEM-2, a guanine nucleotide exchange factor specific for Cdc42. J Biol Chem. 1999;274(47):33587–33593. doi: 10.1074/jbc.274.47.33587. [DOI] [PubMed] [Google Scholar]
- 75.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44(3):480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tai IT, Tang MJ. SPARC in cancer biology: its role in cancer progression and potential for therapy. Drug Resist Updat. 2008;11(6):231–246. doi: 10.1016/j.drup.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao Y, et al. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol Cancer. 2015;14:12. doi: 10.1186/s12943-014-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen D, Zhou H, Liu G, Zhao Y, Cao G, Liu Q. SPOCK1 promotes the invasion and metastasis of gastric cancer through slug-induced epithelial-mesenchymal transition. J Cell Mol Med. 2018;22(2):797–807. doi: 10.1111/jcmm.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Veenstra VL, Damhofer H, Waasdorp C, Steins A, Kocher HM, Medema JP, et al. Stromal SPOCK1 supports invasive pancreatic cancer growth. Mol Oncol. 2017;11(8):1050–1064. doi: 10.1002/1878-0261.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, et al. Translationally controlled tumor protein is a target of tumor reversion. Proc Natl Acad Sci U S A. 2004;101(43):15364–15369. doi: 10.1073/pnas.0406776101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 82.Deb SP. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol Cancer Res. 2003;1(14):1009–1016. [PubMed] [Google Scholar]
- 83.Girnita L, Shenoy SK, Sehat B, Vasilcanu R, Vasilcanu D, Girnita A, et al. Beta-arrestin and Mdm2 mediate IGF-1 receptor-stimulated ERK activation and cell cycle progression. J Biol Chem. 2007;282(15):11329–11338. doi: 10.1074/jbc.M611526200. [DOI] [PubMed] [Google Scholar]
- 84.Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26(19):7269–7282. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kato M, Yano K, Morotomi-Yano K, Saito H, Miki Y. Identification and characterization of the human protein kinase-like gene NTKL: mitosis-specific centrosomal localization of an alternatively spliced isoform. Genomics. 2002;79(6):760–767. doi: 10.1006/geno.2002.6774. [DOI] [PubMed] [Google Scholar]
- 86.Burman JL, Bourbonniere L, Philie J, Stroh T, Dejgaard SY, Presley JF, et al. Scyl1, mutated in a recessive form of spinocerebellar neurodegeneration, regulates COPI-mediated retrograde traffic. J Biol Chem. 2008;283(33):22774–22786. doi: 10.1074/jbc.M801869200. [DOI] [PubMed] [Google Scholar]
- 87.Chafe SC, Mangroo D. Scyl1 facilitates nuclear tRNA export in mammalian cells by acting at the nuclear pore complex. Mol Biol Cell. 2010;21(14):2483–2499. doi: 10.1091/mbc.e10-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang SX, Garcia-Gras E, Wycuff DR, Marriot SJ, Kadeer N, Yu W, et al. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J Biol Chem. 2005;280(19):19115–19126. doi: 10.1074/jbc.M413793200. [DOI] [PubMed] [Google Scholar]
- 89.Sharom FJ. The P-glycoprotein multidrug transporter. Essays Biochem. 2011;50(1):161–178. doi: 10.1042/bse0500161. [DOI] [PubMed] [Google Scholar]
- 90.Bamji SX, Rico B, Kimes N, Reichardt LF. BDNF mobilizes synaptic vesicles and enhances synapse formation by disrupting cadherin-beta-catenin interactions. J Cell Biol. 2006;174(2):289–299. doi: 10.1083/jcb.200601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129(7):1614–1627. doi: 10.1038/jid.2008.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 93.Green DR, Chipuk JE. p53 and metabolism: inside the TIGAR. Cell. 2006;126(1):30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 94.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 95.Zolota V, Sirinian C, Melachrinou M, Symeonidis A, Bonikos DS. Expression of the regulatory cell cycle proteins p21, p27, p14, p16, p53, mdm2, and cyclin E in bone marrow biopsies with acute myeloid leukemia. Correlation with patients' survival. Pathol Res Pract. 2007;203(4):199–207. doi: 10.1016/j.prp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 96.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 97.Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl):S57–S66. doi: 10.1016/S0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- 98.Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116(4):527–540. doi: 10.1016/S0092-8674(04)00162-X. [DOI] [PubMed] [Google Scholar]
- 99.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25(34):4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 100.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479–489. doi: 10.1016/S0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 101.Yan S, Sorrell M, Berman Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mole Life Sci. 2014;71(20):3951–3967. doi: 10.1007/s00018-014-1666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 103.Hua J, Li S, Huang C. Clinical significance of chromodomain helicase/ATPase DNA binding protein 1-like and human mutL homolog 1 gene expression in cholangiocarcinoma. Oncol Lett. 2018;16(3):2989–2994. doi: 10.3892/ol.2018.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 2011;30(13):2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okuda M, Horikoshi M, Nishimura Y. Structural polymorphism of Chromodomains in Chd1. J Mol Biol. 2007;365(4):1047–1062. doi: 10.1016/j.jmb.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 106.Sharma A, Jenkins KR, Héroux A, Bowman GD. Crystal structure of the Chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA*. J Biol Chem. 2011;286(49):42099–42104. doi: 10.1074/jbc.C111.294462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Piatti P, Lim CY, Nat R, Villunger A, Geley S, Shue YT, et al. Embryonic stem cell differentiation requires full length Chd1. Sci Rep. 2015;5(1):8007. [DOI] [PMC free article] [PubMed]
- 108.Fong Ming Koha b c, Lizamad e CO, Wonga,b,c P, Hawkinsd,e JS, Zoveind,e AC, Ramalho-Santosa aM. Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. PNAS. 2015;23:E1734–E1E43. doi: 10.1073/pnas.1424850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate Cancer. Cancer Res. 2013;73(9):2795–2805. doi: 10.1158/0008-5472.CAN-12-1342. [DOI] [PubMed] [Google Scholar]
- 110.Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD, Pollack JR. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2011;31(37):4164–4170. doi: 10.1038/onc.2011.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao D, Cai L, Lu X, Liang X, Li J, Chen P, et al. Chromatin Regulator CHD1 Remodels the Immunosuppressive Tumor Microenvironment in PTEN-Deficient Prostate Cancer. Cancer Discov. 2020;10(9):1374–87. [DOI] [PMC free article] [PubMed]
- 112.Yao H, Pan J, Wu C, Shen H, Xie J, Wang Q, et al. Transcriptome sequencing reveals CHD1 as a novel fusion partner of RUNX1 in acute myeloid leukemia with t(5;21)(q21;q22). Mol Cancer. 2015;14(1):81. [DOI] [PMC free article] [PubMed]
- 113.Blessing C, Mandemaker IK, Gonzalez-Leal C, Preisser J, Schomburg A, Ladurner AG. The Oncogenic Helicase ALC1 Regulates PARP Inhibitor Potency by Trapping PARP2 at DNA Breaks. Mol Cell. 2020;80(5):862–75.e6. doi: 10.1016/j.molcel.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 114.Yang XD, Kong FE, Qi L, Lin JX, Yan Q, Loong JHC, et al. PARP inhibitor Olaparib overcomes Sorafenib resistance through reshaping the pluripotent transcriptome in hepatocellular carcinoma. Mol Cancer. 2021;20(1):20. [DOI] [PMC free article] [PubMed]
- 115.Abbott JM, Zhou Q, Esquer H, Pike L, Broneske TP, Rinaldetti S, et al. First-in-class inhibitors of oncogenic CHD1L with preclinical activity against colorectal Cancer. Mol Cancer Ther. 2020;19(8):1598–1612. doi: 10.1158/1535-7163.MCT-20-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.