Abstract
Backgroundː
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), within few months of being declared as a global pandemic by WHO, the number of confirmed cases has been over 75 million and over 1.6 million deaths since the start of the Pandemic and still counting, there is no consensus on factors that predict COVID-19 case progression despite the diversity of studies that reported sporadic laboratory predictive values predicting severe progression. We review different biomarkers to systematically analyzed these values to evaluate whether are they are correlated with the severity of COVID-19 disease and so their ability to be a predictor for progression.
Methods
The current meta-analysis was carried out to identify relevant articles using eight different databases regarding the values of biomarkers and risk factors of significance that predict progression of mild or moderate cases into severe and critical cases. We defined the eligibility criteria using a PICO model.
Results
Twenty-two relevant articles were selected for meta-analysis the following biomarkers C-reactive protein, interleukin-6, LDH, neutrophil, %PD-1 expression, D-dimer, creatinine, AST and Cortisol all recorded high cut-off values linked to severe and critical cases while low lymphocyte count, and low Albumin level were recorded. Also, we meta- analyzed age and comorbidities as a risk factors of progression as hypertension, Diabetes and chronic obstructive lung diseases which significantly correlated with cases progression (p < 0.05).
Conclusions
ː The current meta-analysis is the first step for analysing and getting cut-off references values of significance for prediction COVID-19 case progression. More studies are needed on patients infected with SARS-CoV-2 and on a larger scale to establish clearer threshold values that predict progression from mild to severe cases. In addition, more biomarkers testing also help in building a scoring system for the prediction and guiding for proper timely treatment.
Keywords: Meta-analysis, COVID-19, SARS-CoV-2, Prediction of severe cases, Prediction of critical cases, Biomarkers of risk for COVID-19 case progression, Comorbidity of risk for COVID-19 case progression
Introduction
Coronaviruses (CoV) are a group of viruses that lead to diseases variable from mild to many serious diseases such as Middle Eastern respiratory syndrome (MERS-CoV) and severe acute respiratory syndrome (SARS-CoV). Dec. 31, 2019, the Chinese Health Authority notified the World Health Organization (WHO) of numerous cases of pneumonia of unknown etiology in the city of Wuhan, in the area of Hubei, in central China [1]. On January 7, a new coronavirus, originally abbreviated as 2019-nCoV by WHO, was identified by a patient's throat swab sample, has not been recognized before in humans [2,3]. Subsequently, this pathogen was renamed as coronavirus 2 severe acute respiratory syndrome (SARS-CoV-2) and the coronavirus designated by WHO 2019 (COVID-19) [4] by January 30, in China 7736 cases recorded, 12,167 suspected and 82 cases had been perceived in 18 countries. Accordingly, WHO stated the SARS-CoV-2 epidemic as an international public health emergency [5]. On 11 March 2020, the World Health Organization (WHO) stated that COVID-19 as a pandemic [6]. While writing this review, worldwide cases over 75 million and over 1.6 million deaths which are increasing with the upward curve [7]. The dependence on CT scan findings to detect and diagnose cases of coronavirus affection although accurate, does not estimate or predict which patient will go for a severe or lethal course [8,9]. Therefore, it is mandatory to identify mild/moderate patients who can pass to severe/critical condition and give them efficient treatment to prevent deterioration as the early proper treatment of cases prone to severe/critical malignant progression are important ways to reduce mortality [10]. Unfortunately, most investigators focused on cross-sectional description and the comparison of clinical, laboratory, and CT imaging results [[11], [12], [13], [14]]. Some have focused on finding risks for death [15,16]. Few have presented their data and mentioned the prediction of progression of mild COVID-19 cases, but still, the work is separate and there is no review to gather and filter the information of significance. So our purpose is to fill the gap by doing meta-analysis of published researches based on criteria to obtain reference figures of statistically significant biomarkers levels (p < 0.05) and getting mean number of all means of the analyzed papers to be easily consulted to detect cases that expected to deteriorate into severe/critical condition as a first step to start building a scoring system that can be objectively expect COVID-19 case progression which is best known to our knowledge this is the first time to be done. This will help doctors to begin efficient aggressive treatment without delay by adequately predicting severe/critical cases which will improve survival rates and reduce both illness and fatality in the COVID-19 pandemic.
Methods
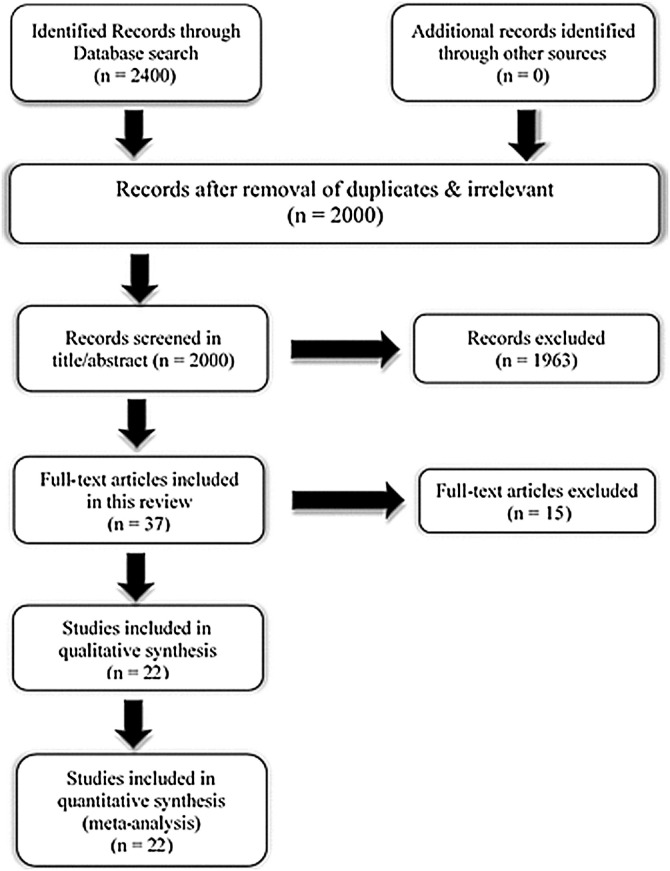
Using an online database, to conduct a meta-analysis on the early prediction of COVID-19 cases prone to deterioration. The main articles are mainly taken from PubMed, Google Scholar, MEDLINE, UpToDate, Medscape, Embase, Web of Science, and the preprint server medRxiv using the terms “COVID-19”, “SARS-CoV-2”, “prediction of severe/critical cases”, “Vulnerability Index”, “Risk factors mortality”, “prediction of survival”, “prediction of malignant progression”, “risk score” are the keywords for our search. Publications were left out if they had inappropriate data and were not appropriate to the specific purpose of our review. All relevant study reference lists have been selected to identify missing publications. After duplicates & irrelevant were removed, we identified 2000 papers. Search, of titles and abstracts were conducted by 6 independent working researchers by applying eligibility criteria and filter the relevant ones. Disagreements were resolved by discussion. We defined the eligibility criteria using a PICO model as follows. Population: no specific reference population. Intervention: Noninvasive lab investigations and biomarkers assay. Comparison: Imaging investigations, clinical symptoms progression or intra-individual pre-post comparison. Outcome: Patients with certain biomarkers levels progressed from mild/moderate to severe/critical. All eligible deemed articles have been retrieved for full-text review [Fig. 1]. We included studies for meta-analysis those show cohort, case-series case-control; studies on patients with severe or mild illness, also those had death groups and survivors. We considered severity of the disease as “mild” for those patients described in the studies as mild or moderate cases, while considered ‘severe’ for described as having severe disease, ICU cases or required mechanical ventilation. Only studies of ≥30 patients were included. Only studies of score greater than 6 were considered of high quality of total score 9 according to Newcastle-Ottawa Scale (NOS) guidelines that was used by 4 independent reviewers, disagreement was resolved by additional reviewer [17].
Fig. 1.
Systematic review flow chart for literature refinement.
Statistical analysis
Meta-analyses were done using the metacount and metabin functions for continuous and binary variables respectively from the R package meta in R Studio [18]. For each study with more than one values of the biomarkers, a single value was calculated using the method described in Higgins and Green [19]. An overall weighted mean (SD) of the biomarkers was estimated within the mild and severe groups using standard weighted approach with built-in R functions. The overall difference in means or events between severe and mild patients were quantified by pooling the differences provided by the original studies using Random effect models, and the results represented as forest plots. Results for age and biomarkers were given as Mean Difference, MD (95%CI) while those of comorbidities as Relative Risks, RR (95%CI). Statistical heterogeneity between studies was assessed using I². For results with more than 2 studies, publication bias was assessed using the Egger regression [20].
Results
Literature screening and assessment
A total of 2400 records were identified from the databases. After a detailed assessment, 22 studies involving 4138 COVID-19 patients were included in the meta-analysis (Fig. 1).
Characteristics of included studies
19 studies included in the meta-analysis were conducted in China examined Chinese patients, the remaining three were conducted in Iran, Italy and Cameron. All included studies were published in 2020. A large proportion of these studies (n = 20) were single center data collected. All included studies received quality scores of 6–9, indicating high quality (Table 1 ).
Table 1.
Characteristics of the included studies in meta-analysis.
| Author | Study design | Country | Cohort size | Biomarkers studied/comorbidity | Comments |
|---|---|---|---|---|---|
| Wang et al. [11] | Retrospective cohort, single center | China | 138 | Neutrophil cnt, Lymphocyte cnt, LDH | Higher Neutrophil count, LDH and lower lymphocyte count and significantly correlate this relation to severe critical cases |
| Yang et al. [12] | Retrospective cohort, multi center | China | 149 | Neutrophil cnt, Lymphocyte cnt, D-dimer, albumin, AST, creatinine, LDH, CRP | CT scan cannot exclude the diagnosis of COVID-19 as some patients with COVID-19 can present with normal chest finding however high biomarkers levels |
| Zhou et al. [15] | Retrospective cohort, multi center | China | 191 | Lymphocyte cnt, albumin, D-dimer, IL-6, creatinine, hypertension, Diabetes chronic obstructive lung disease | Considered D-dimer > 1 μg/mL could help clinicians to identify patients with poor prognosis at an early stage. |
| Diao et al. [24] | Retrospective cohort, multi center | China | 522 | Lymphocyte cnt, IL-6 | Recorded significant reduction in T cell counts in COVID-19 patients, and the surviving T cells appear functionally exhausted. Also, they negatively corrected T cells to IL-6. Considered Non-ICU patients with total T cells counts < 800/μL still require urgent intervention, even in the immediate absence of more severe symptoms due to a high risk for further deterioration in condition. |
| Liu et al. [34] | Retrospective cohort, single center | China | 40 | Neutrophil cnt, Lymphocyte cnt, AST, LDH, creatinine, D-dimer, CRP, hypertension, Diabetes | Associated higher degree of lymphopenia and a proinflammatory cytokines with COVID-19 disease severity. |
| Feng et al. [36] | Retrospective cohort, single center | China | 132 | Lymphocyte cnt, Neutrophil cnt, CRP, IL-6, hypertension, Diabetes, chronic obstructive lung disease | Proposed CT scan as early screening could not satisfy every patient in COVID-19 outbreak and considered use of machine-learning algorithms to analyze clinical symptoms, biomarkers and other clinical information as a good tool for diagnosis and early prediction of cases prognosis before further CT examination |
| Qin et al. [37] | Retrospective cohort, single center | China | 452 | CRP, IL-6, Neutrophil cnt, Lymphocyte cnt | Compared different inflammatory biomarkers higher levels in severe and non-severe COVID-19 cases |
| Liu et al. [39] | Retrospective cohort, single center | China | 140 | IL-6, lymphocytes, neutrophils, AST, CRP, Creatinine, D-Dimer | They measured different biomarkers and correlated them with disease progression |
| Wu et al. [41] | Retrospective cohort, single center | China | 201 | IL-6 | Significantly correlated higher IL-6 levels in severe cases |
| Chen et al. [43] | Retrospective cohort, single center | China | 99 | IL-6 | Considered high IL-6 levels one of the measures that may detect COVID-19 severity. |
| Ji et al. [51] | Retrospective cohort, single center | China | 33 | CRP | Statistically significant increase in CRP with increase severity of the disease and considered it one of the measures can be used to judge severity |
| Etoga et al. [53] | Cross sectional single center | Cameroon | 80 | Cortisol | This study recorded higher levels of cortisol among COVID-19 cases who need further oxygen therapy than those of mild condition |
| Ramezani et al. [54] | Cross sectional single center | Iran | 30 | Cortisol | This study significantly correlated higher levels of cortisol in non-survived patients of Covid-19 in comparison with surviving patients. |
| Li et al. [55] | Retrospective cohort, single center | China | 132 | CRP | This study recorded noticeable difference of CRP between mild and severe critical cases |
| Tang et al. [56] | Retrospective cohort, single center | China | 183 | D-Dimer | Recorded higher levels of D-Dimer among non survivors COVID-19 cases |
| Zhang et al. [57] | Retrospective cohort, single center | China | 343 | D-Dimer | They study considered D-dimer level on admission > 2.0 μg/mL could eff ;ectively predict hospital mortality in patients with COVID-19 |
| Huang et al. [58] | Prospective cohort, single center | China | 41 | IL-6, D-Dimer | Recorded higher levels of IL-6 and D-dimer among severe cases |
| Cheng et al. [60] | Prospective cohort, single center | China | 701 | Creatinine | They correlated high level of creatinine with severity and worse outcome in COVID-19 cases |
| Luo et al. [61] | Retrospective cohort, single center | China | 35 | LDH | Considered higher levels of LDH may indicate severity of the disease by their recorded levels of LDH in severe cases |
| Li et al. [62] | Retrospective cohort, single center | China | 134 | Lymphocyte cnt, Neutrophils cnt, D-dimer, albumin, AST, creatinine, IL-6, CRP, hypertension | Reached cut off value for decrease in albumin levels with the progression of the disease even they considered it as an independent predictor (cut-off point: 35.1 g/L) of the risk of non survivors among critical COVID-19 cases |
| Ferrari et al. [63] | Retrospective cohort, single center | Italy | 207 | LDH | LDH higher level among COVID-19 cases and considered it may help in diagnosis of such cases |
| Mo et al. [64] | Retrospective cohort, single center | China | 155 | LDH | Recorded higher levels among complicated cases and correlated LDH biomarker with the development of the disease. |
Publication bias
The calculated p values of Egger’s and the Begg’s test for all analyzed studies outcomes showed no publication bias (Table 2 ). The recorded p values were >0.05 which indicates no publication bias.
Table 2.
Egger’s test of funnel plot asymmetry (publication bias).
| Variable | Bias | Statistics | P value |
|---|---|---|---|
| Age | −1.5 | −1.3 | 0.28 |
| Lymphocytes | −0.3 | −0.1 | 0.92 |
| D-Dimer | 3.1 | 3.1 | 0.05 |
| IL-6 | 4.7 | 0.9 | 0.42 |
| Neutrophils count | 0.03 | 0.01 | 0.99 |
| Creatinine | 0.2 | 0.1 | 0.92 |
| CRP | 0.3 | 0.1 | 0.94 |
| LDH | 6 | 1.3 | 0.33 |
| Hypertension | 1.4 | 1.0 | 0.50 |
| Diabetes | 0.06 | 0.07 | 0.95 |
Bias: the intercept from the Egger’s regression; p value of <0.05 signifies that the intercept is different from 0 and implying significant publication bias.
Age distribution
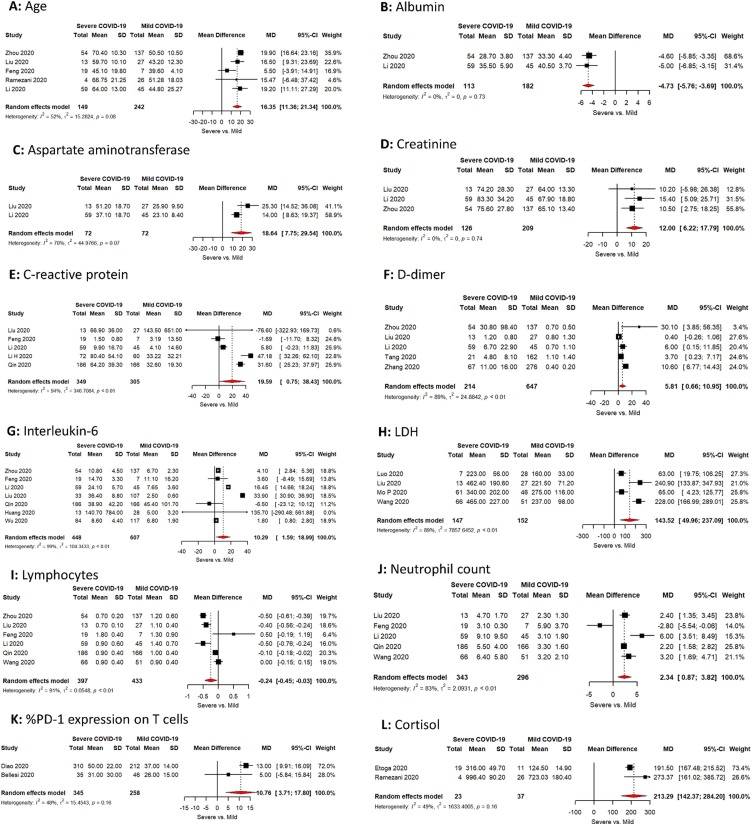
Severe cases were on average 16 years significantly older than those with Mild cases: Age difference 16.4 years (95%CI; 11.4, 21.3). There are no major differences in the study estimates and publication bias (I2 is <75%, Bias = −1.4, p value = 0.28). The mean age of severe group cases was 66.9 (STD 15.4) (Fig. 2 A).
Fig. 2.
(A–F): Meta-analysis of the difference between COVID-19 patients with severe vs mild disease in: (A) Mean age (B) Albumin level (C) Aspartate aminotransferase (D) Creatinine (E) C-reactive protein (F) D-Ddimer. (G–L): Meta-analysis of the difference between COVID-19 patients with severe vs mild disease in: (G) Interleukin-6 (H) LDH (I) Lymphocytes (J) Neutrophil count (K) %PD-1 expression on T cells (L) Cortisol.
Albumin
Albumin level was significantly lower in severe cases vs mild cases. The difference was of −4.7 g/L (95% CI; −5.8, −3.7). The mean Albumin level in severe group cases was 30.4 g/L (STD 6.1) (Fig. 2B).
Aspartate aminotransferase
Severe cases recorded significantly higher level of AST vs mild cases with difference of 18.6 IU/L (95% CI; 7.8–29.5), with no major difference in in the study estimates (I2 is <75%). The recorded mean AST level in severe group cases was 42.4 IU/L (STD 19.5) (Fig. 2C).
Creatinine
Creatinine level showed significantly higher levels in severe vs mild cases with difference of 12.00 μmol/L (95% CI; 6.2; 17.8), with no major difference in the study estimates (I2 is <75%). Mean creatinine level in severe cases was 77.1 μmol/L (STD 31.2) (Fig. 2D).
C-reactive protein
C-reactive protein level was higher in severe vs mild cases with difference of 19.6 mg/L (95% CI; 0.8; 38.4), however there was a major difference in the study estimates (I2 is >75%) with no significant publication bias (Bias = 0.3, p value = 0.9) Mean C-reactive recorded level in severe cases was 58.2 mg/L (STD 47) (Fig. 2E).
D-dimer
D-Dimer recorded higher levels in severe vs mild cases with mean difference of 5.8 μg/mL (95% CI; 0.7; 10.9), however was a major difference in the study estimates (I2 is >75%). Publication bias was insignificant publication (Bias= -3.4, p value = 0.5). Mean D-Dimer level in severe cases was 12.9 μg/mL (STD 52.7) (Fig. 2F).
Interleukin-6
IL-6 showed higher levels in severe vs mild cases with mean difference of 10.3 pg/mL (95% CI; 1.6; 19.0), there was a major difference in the study estimates (I2 is >75%). No significant publication bias (Bias = 4.7, p value = 0.4). Mean IL-6 level in severe cases was 29.6 pg/mL (STD 138) (Fig. 2G).
LDH
LDH level was higher in severe vs mild cases with mean difference of 143.52 U/L (95% CI; 50; 237.1), major difference was recorded in study estimates (I2 is >75%). No significant publication bias (Bias = 6, p value = 0.3). Mean LDH level in severe cases was 382 U/L (STD 221) (Fig. 2H).
T-Lymphocytes count
T-Lymphocytes count was lower in severe vs mild cases, the mean difference was -0.24 × 109/L (95% CI; −0.45; −0.03), the study estimate recorded major difference estimates (I2 is >75%). No significant publication bias (Bias = −0.3, p value = 0.9). Mean lymphocyte count in severe cases was 0.8 × 109/L (STD 0.46) (Fig. 2I).
Neutrophil count
Neutrophil count recorded higher levels in severe vs mild cases, the mean difference was 2.3 × 109/L (95% CI; 0.9; 3.8), the study estimate recorded major difference estimates (I2 is >75%). No significant publication bias (Bias = 0.03, p value = 0.9). Mean neutrophil count in severe cases was 6.1 × 109/L (STD 5.8) (Fig. 2J).
%PD-1 expression on T cells
%PD-1 expression on T-Lymphocytes was higher in severe vs mild cases, with mean difference of 10.8 (95% CI; 3.7; 17.8), there was no significant heterogeneity in the studies used (I2 is <75%). Mean of %PD-1 expression on T lymphocytes (the marker of T-cell exhaustion) in severe cases was 47 (STD 24) (Fig. 2K).
Cortisol
Although there are only two published studies about cortisol level among COVID-19 cases we included in our meta-analysis to check the significance of this biomarker and accordingly to recommend its importance for further studies. Cortisol recorded significant higher level in severe vs mild cases with mean difference of 213.3 nmol/L (95% CI; 142.4; 284.2), No significant heterogeneity was recorded in study estimates (I2 is >75%). Mean cortisol level in severe cases was 794 nmol/L (STD 264) (Fig. 2L).
Hypertension
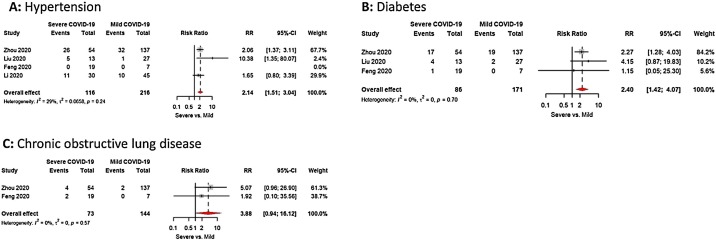
Co-morbidity with hypertension was twice much more common in severe cases vs mild cases with RR 2.1(95% CI; 1.5; 3.0), No significant heterogeneity recorded in study estimates (I2 is < 75%) or publication bias (Bias = 1.4, p value = 0.5) (Fig. 3 A).
Fig. 3.
(A–C): Meta-analysis of the difference between COVID-19 patients with severe vs mild disease in: (A) Hypertension (B) Diabetes (C) Chronic obstructive lung disease.
Diabetes
Co-morbidity with diabetes was twice and half much more common in severe cases vs mild cases with RR 2.4 (95% CI; 1.4; 4.0), No significant heterogeneity recorded in study estimates (I2 is <75%) or publication bias (Bias = 0.06, p value = 0.9) (Fig. 3B).
Chronic obstructive lung disease
Co-morbidity with chronic obstructive pulmonary disease was about four times much more common in severe cases vs mild cases with RR 3.9 (95% CI; 0.9; 16.1), No significant heterogeneity recorded in study estimates (I2 is <75%) (Fig. 3C).
Study limitation
Our meta-analysis included a large number of COVID-19 patients, but there are some limitations due to heterogeneity found in some studies, as well as most of the studies were from China and single center, so geographical and ethnic variations could not be studied well.
Discussion
COVID-19 The disease
COVID-19 is a respiratory infection with common signs including respiratory symptoms, fever, cough, and difficulty in breathing. In severe cases, the infection can cause pneumonia, severe acute respiratory syndrome, renal failure, and death [21,22]. Based on the staging of the infectious disease: The prodromal period is a phase in which the host begins to manifest general signs and symptoms. The openly symptomatic period is a phase in which the disease signs or symptoms are more evident, with positive laboratory results and chest imaging. For ICU patients, the ICU period is a phase in which the symptoms are more evident and severe. The period of decline is a phase in which clinical symptoms begin to decrease, laboratory results and chest imaging improve, and saturation of oxygen returns to normal [23,24]. The Chinese National Health Commission classified clinical types into mild/moderate/severe/critical [25].
Factors that favor disease severity and worsen the prognosis
Factors that favor disease severity and complications: Selecting cases with risk factors that could progress through a serious and complicated COVID-19 disease course is like selecting between death and life. Several researchers have investigated many risk factors, some findings were statistically significant (p < 0.05).
Age and comorbidity
Severe cases are those prone to serious complications and are at risk of death. The danger of serious complications from COVID-19 is greater for some vulnerable populations, particularly the elderly, the weak, or persons with multiple chronic diseases [10,21,[26], [27], [28], [29], [30]]. Some studies reported advanced age and comorbidity with hypertension, as the most important risk factors for malignant progression [31]. The hazard of death was difficult to calculate, but some studies on COVID-19 patients in Wuhan reported that the risk of death rises with age and for those with diabetes, heart disease, clotting disorders. Since the fatality rate has changed from an average of 1%, to be 6% for persons with cancer, hypertension or chronic respiratory diseases, 7% for diabetes, and 10% for heart diseases. There was also a strong age gradient; the fatality rate for those over 80 was > 15% [15].
Our meta-analysis confirmed older age at more risk for case progression as we recorded age was higher by 16 years in COVID-19 severe cases than mild, with overall mean age of analyzed severe cases 66.9 (STD 15.4). Also, we recorded comorbidity of hypertension increase risk of progression from mild to severe twice, Diabetes twice and half, and Chronic obstructive lung disease showed four times which is the highest risk of progression among our analyzed studies.
Biomarkers of prediction for case progression
Inflammatory markers
Lymphopenia is a common feature in COVID-19 patients and could be a critical factor associated with disease severity and mortality [24,32,33,[35], [36], [37]]. Even some studies considered low lymphocyte count is a biomarker is of >90% accuracy or of vital role for prediction of case progression that may aid as a possible therapeutic target [27]. Diao et al. studied lymphocytes function by measuring exhaustion markers and recorded increased expression of PD-1 and Tim-3 in T cells in cases proceeded from prodromal to symptomatic phases, further indicating T-cell depletion. The authors stated that these counts even in patients without intensive care may require aggressive intervention even in the absence of serious symptoms as liable to worsening of their condition [24]. Our meta-analysis on %PD-1 expression on T-Lymphocytes as one of exhaustion markers confirmed being higher in severe vs mild cases. The mean of %PD-1 expression in severe cases in studies analyzed was 47 (STD 24).
Many studies found a close and significant correlation between IL-6 high levels and case progression and even mortality. Some are negatively correlated it with low T-lymphocyte count and higher expression of PD-1 among severe cases [16,24,27,38,39]. Our meta-analysis confirmed higher levels of IL-6 in severe vs mild cases with overall mean level in severe cases 29.6 pg/mL (STD 138). Several studies recorded neutrophil count as an important biomarker that can predict progression of mild cases into severe/critical one either alone or compared to lymphocytes count. Some considered it useful in surveillance and may predict critically ill patients early [11,16,37,40,41]. Our results revealed higher neutrophil count in severe vs mild cases, with the overall mean neutrophil count in analyzed studies is 6.1 × 109/L (STD 5.8). C-reactive protein was like a marker of common use several studies recorded higher levels in severe/critical than mild COVID-19 cases and some considered it to be of help for detection of case progression [10,27,35,39,[41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51]]. Our analysis confirmed higher levels of CRP among severe vs mild COVID-19 cases with overall mean level in severe cases 58.2 mg/L (STD 47).
Cortisol level
Three published studies on cortisol level in COVID-19 cases recorded higher level of cortisol among severe vs mild COVID-19 cases [[52], [53], [54]]. Our meta-analysis confirmed its higher levels in severe vs mild cases with over all mean of the studied severe cases is 794 nmol/L (STD 264).
Abnormal coagulation
Several studies recorded high levels of D-Dimer which reflect abnormal coagulation results among severe vs mild COVID-19 cases. Some considered certain level could effectively predict in-hospital mortality in patients although the recorded levels were different [11,15,27,39,[55], [56], [57]].
Our analysis confirmed D-Dimer higher levels in severe vs mild cases with overall mean level in severe cases is 12.9 μg/mL (STD 52.7).
Multiorgan injury
COVID-19 severe cases not only related to respiratory tract infection but also progression into severe complicated cases and increased mortality could be due to multiorgan injury. Some studied worked on biomarkers those could reflect organs injury as AST, creatinine, LDH and Albumin on which they reported higher levels of the first three biomarkers and lower levels of the fourth one among severe/critical cases. Some strongly corrected decrease Albumin level with disease progression [11,16,27,39,41,[46], [47], [48], [49],[58], [59], [60], [61], [62], [63], [64]]. Our analysis confirmed higher levels of AST, creatinine and LDH among severe vs mild covid-19 cases with overall mean levels in severe cases 42.4 IU/L (STD 19.5), 77.1 μmol/L (STD 31.2), and 382 U/L (STD 221) respectively. Albumin level was significantly lower among severe cases with overall mean level in severe cases is 30.4 g/L (STD 6.1).
Reference keys and prediction of severity
We collected the overall mean levels of our biomarkers of study those all included studies in our meta-analysis to seal the gap, best known to our knowledge not done before in any published meta-analysis on COVID-19, and reach a possible prediction references keys with their levels that can be used as a reference that can predict sunsets for mild or moderate COVID-19 cases who are vulnerable to progress into a severe course, entailing aggressive interventions in absence of severe symptoms which will be of great value to reduce the burden on intensive care and reduce complications and mortality (Table 3, Table 4) . We considered it as the first step to start building a scoring system that can be used for the prediction that guides for proper timely treatment which will be of help to reduce mortality. We are working on another review article that will explain the immunopathogenesis of COVID-19 and explaining why these predictors are keys in predicting progression of the cases to severe/critical, we also propose the possible solutions for effective treatment.
Table 3.
The collected biomarkers of all meta-analyzed studies of statistical significance with calculated mean of all recorded means in the studies of analysis to be a help key levels for prediction of progression from a mild/moderate case of COVID-19 into severe/critical case.
| Positive COVID-19 patient | |||
|---|---|---|---|
| Biomarkers of prediction progression of cases from mild/moderate to severe/critical | |||
| Non-high-risk groupa | |||
| A-Indicators of COVID-19 progression into severe/critical condition | B-Indicators of multiorgan injury | ||
| C-reactive protein | ≥58.2 mg/L (STD 47) | LDH | ≥382 U/L (STD 221) |
| Aspartate aminotransferase (AST), U/L | ≥42.4 IU/L (STD 19.5) | ||
| Neutrophil count | ≥ 6.1 × 109/L (STD 5.8). | Albumin | ≤30.4 g/L (STD 6.1) |
| T-Lymphocytes count | ≤ 0.8 × 109/L (STD 0.46). | ||
| D-Dimer | ≥12.9 μg/mL (STD 52.7) | ||
| T Cell Function: % PD-1 expression on T cells | Creatinine | ≥77.1 μmol/L (STD 31.2) | |
| % PD-1 On T-Cell | ≥47 (STD 24) | ||
| Cytokines: | |||
| IL-6 | ≥29.6 pg/mL (STD 138) | ||
| Cortisol level: | ≥794 nmol/L (STD 264) | ||
Patient younger than 66.9 years (STD 15.4) with no comorbidity if show any indicator in this group considered a risk for progression into severe/ critical condition which necessitates aggressive treatment even if CT findings not clear yet and the patient condition still apparently mild.
Table 4.
The collected risk factors of meta-analyzed studies of statistical significance to help as a key for prediction of progression from a mild/moderate case of COVID-19 into severe/critical case.
| Positive COVID-19 patient | |
|---|---|
| Risk factors of prediction progression of cases from Mild/moderate to severe/critical | |
| High-risk group | |
| Risk factor | |
| A-Age ≥ 66.9 (STD 15.4) | |
| B-Any age with Comorbidity: | Risk degree for progression |
| 1-Chronic obstructive lung disease | Four times |
| 2-Hypertension | Twice |
| 3-Diabetes | Twice and half |
Funding
Researchers would like to thank the Deanship of Scientific Research, Qassim University for funding publication of this project.
Competing interests
None declared.
Ethical approval
Not required.
Authors’ contributions
First two authors contributed equally to the manuscript and read and approved the final version of the manuscript. Other authors main work in reading and applying selection criteria final discussion and approval of final version of manuscript.
Acknowledgment
Thanks to all health care workers: Doctors, nursing, intensive care, laboratories, Radiology, technicians, and volunteers who are in the front lines of the pandemic and making a great effort in fighting the disease all over the world. Special thanks for those who sacrificed their lives to save the life of thousands.
References
- 1.Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H., et al. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13(May (5)):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO | Novel Coronavirus — China [Internet]. World Health Organization [cited Jul 1, 2020]. Available from: http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/.
- 4.Gorbalenya Alexander E., Baker Susan C., Baric Ralph S., de Groot Raoul J., Drosten C., Gulyaeva Anastasia A., et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(April (4)):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burki T.K. Coronavirus in China. Lancet Respir Med. 2020;8(March (3)):238. doi: 10.1016/S2213-2600(20)30056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. 1 May 2020. allAfrica.com (English). https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-.11-march-2020.
- 7.WHO. Coronavirus disease 2019 (COVID-19) “Weekly Epidemiological Update.” 2020 December 22. https://www.who.int/publications/m/item/weekly-epidemiological-update---22-december-2020.
- 8.Dangis A., Gieraerts C., Bruecker Y.D., Janssen L., Valgaeren H., Obbels D., et al. Accuracy and reproducibility of low-dose submillisievert chest CT for the diagnosis of COVID-19. Radiol Cardiothorac Imaging. 2020;2(April (2)) doi: 10.1148/ryct.2020200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pianura E., Di Stefano F., Cristofaro M., Petrone A., Albarello F., Fusco N., et al. COVID-19: a review of the literature and the experience of INMI Lazzaro Spallanzani two months after the epidemic outbreak. J Radiol Rev. 2020;7:196–207. doi: 10.23736/S2723-9284.20.00022-4. [DOI] [Google Scholar]
- 10.Bai X., Fang C., Zhou Y., Bai S., Liu Z., Xia L., et al. Predicting COVID-19 malignant progression with AI techniques. medRxiv. 2020 https://www.medrxiv.org/content/10.1101/2020.03.20.20037325v2.supplementary-material?versioned=true [Google Scholar]
- 11.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA J Am Med Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W., Cao Q., Qin L., Wang X., Cheng Z., Pan A., et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): a multi-centre study in Wenzhou city, Zhejiang, China. J Infect. 2020;80(April (4)):388–393. doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H. Outbreak of novel coronavirus (COVID-19): what is the role of radiologists? Eur Radiol. 2020;30(June (6)):3266–3267. doi: 10.1007/s00330-020-06748-2. https://link.springer.com/article/10.1007/s00330-020-06748-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(April (4)):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(May (5)):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 18.Team R.C. R Foundation for Statistical Computing; Vienna: 2018. R: a language and environment for statistical computing.https://www.R-project.org [Google Scholar]
- 19.Higgins P.T., Green S. Wiley; England: 2008. Cochrane handbook for systematic reviews of interventions: version 5.1.0. [Google Scholar]
- 20.Egger M., Smith G.D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeCaprio D., Gartner J., Burgess T., Kothari S., Sayed S., McCall C.J. Building a COVID-19 vulnerability index. medRxiv. 2020 doi: 10.1101/2020.03.16.20036723. https://www.medrxiv.org/content/10.1101/2020.03.16.20036723v2 [DOI] [Google Scholar]
- 22.Tuttolomondo D., Frizzelli A., Aiello M., Bertorelli G., Majori M., Chetta A. Beyond the lung involvement in COVID-19 patients. A review. Minerva Med. 2020 doi: 10.23736/S0026-4806.20.06719-1. In press. [DOI] [PubMed] [Google Scholar]
- 23.Parker N., Schneegurt M., Thi Tu A.-H., Lister P., Forster B.M. Microbiology. OpenStax; Houston, Texas: 2016. Characteristics of infectious disease.https://openstax.org/books/microbiology/pages/15-1-characteristics-of-infectious-disease Available from: [Google Scholar]
- 24.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Health Commission of the People’s Republic of China . 2020. The notice of launching guideline on diagnosis and treatment of the novel coronavirus pneumonia (NCP)http://www.nhc.gov.cn/yzygj/s7653p/202002/3b09b894ac9b4204a79db5b8912d4440/files/7260301a393845fc87fcf6dd52965ecb.pdf . [Accessed 18 February 2020] [in Chinese] [Google Scholar]
- 26.Why is it so hard to calculate how many people will die from covid-19? [Internet]. [cited Jul 1, 2020]. Available from: https://www.newscientist.com/article/mg24532733-700-why-is-it-so-hard-to-calculate-how-many-people-will-die-from-covid-19/.
- 27.Yan L., Zhang H.-T., Xiao Y., Wang M., Sun C., Liang J., et al. Prediction of criticality in patients with severe Covid-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan. medRxiv. 2020 doi: 10.1101/2020.02.27.20028027. https://www.medrxiv.org/content/10.1101/2020.02.27.20028027v2 [DOI] [Google Scholar]
- 28.Song C.-Y., Xu J., He J.-Q., Lu Y.-Q. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020 doi: 10.1101/2020.03.05.20031906. https://www.medrxiv.org/content/10.1101/2020.03.05.20031906v1 [DOI] [Google Scholar]
- 29.Shi Y., Yu X., Zhao H., Wang H., Zhao R., Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24(1):108–112. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velavan T.P., Meyer C.G. Estimation of risk factors for COVID-19 mortality - preliminary results. Trop Med Int Health. 2020;25(March (3)):278–280. doi: 10.1101/2020.02.24.20027268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J., Tong Z., Guan X., Du B., Qiu H., Slutsky A.S. Critical care crisis and some recommendations during the COVID-19 epidemic in China. Intensive Care Med. 2020;46(May (5)):837–840. doi: 10.1007/s00134-020-05979-7. https://link.springer.com/article/10.1007/s00134-020-05979-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA J Am Med Assoc. 2020;323(April (13)):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y., Tu M., Wang S., Chen S., Zhou W., Chen D., et al. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: a retrospective single center analysis. Travel Med Infect Dis. 2020;36 doi: 10.1016/j.tmaid.2020.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55(May) doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(March (1)) doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng C., Huang Z., Wang L., Chen X., Zhai Y., Zhu F., et al. A novel triage tool of artificial intelligence assisted diagnosis aid system for suspected COVID-19 pneumonia in fever clinics. medRxiv. 2020 doi: 10.21037/atm-20-3073. https://www.medrxiv.org/content/10.1101/2020.03.19.20039099v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;27(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genevieve M., Crowther N.J., et al. Endothelial Dysfunction as a Primary Consequence of SARS-CoV-2 Infection. Adv Exp Med Biol. 2021;1321:33–43. doi: 10.1007/978-3-030-59261-5_3. [DOI] [PubMed] [Google Scholar]
- 39.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., et al. Prognostic value of interleukin-6, Creactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127(April) doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T., Zhang J., Yang Y., Ma H., Li Z., Zhang J., et al. The potential role of IL-6 in monitoring severe case of coronavirus disease 2019. medRxiv. 2020;(March) doi: 10.15252/emmm.202012421. https://www.medrxiv.org/content/10.1101/2020.03.01.20029769v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. medRxiv. 2020;(February) doi: 10.1186/s12879-020-05681-5. https://www.medrxiv.org/content/10.1101/2020.02.25.20025643v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(April (1)):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J., Wu X., Zeng W., Guo D., Fang Z., Chen L., et al. Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Invest Radiol. 2020;55(May (5)):257–261. doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan C., Huang Y., Shi F., Tan K., Ma Q., Chen Y., et al. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J Med Virol. 2020;92(7):856–862. doi: 10.1002/jmv.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020;50(4):332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W., Tao Z.-W., Wang L., Yuan M.-L., Liu K., Zhou L., et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J. 2020;133(May (9)):1032–1038. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(February (5)):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(March (15)):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji W., Bishnu G., Cai Z., Shen X. Analysis clinical features of COVID-19 infection in secondary epidemic area and report potential biomarkers in evaluation. medRxiv. 2020;(March) 2020.03.10.20033613. [Google Scholar]
- 52.Tan T., Khoo B., Mills E.G., Phylactou M., Patel B., Eng P.C., et al. Association between high serum total cortisol concentrations and mortality from COVID-19. Lancet Diabetes Endocrinol. 2020;8(8):659–660. doi: 10.1016/S2213-8587(20)30216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Etoga M.C.E., Inna A.H., Guewo-Fokeng M., Dehayem M., Boli A.O., Manga J.A.N., et al. [DOI]
- 54.Ramezani M., Simani L., Karimialavijeh E., Rezaei O., Hajiesmaeili M., Pakdaman H. The role of anxiety and cortisol in outcomes of patients with Covid-19. BCN. 2020;11(2):179–184. doi: 10.32598/bcn.11.covid19.1168.2. http://bcn.iums.ac.ir/article-1-1771-en.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Xiang X., Ren H., Xu L., Zhao L., Chen X., et al. Serum Amyloid A is a biomarker of severe Coronavirus Disease and poor prognosis. J Infect. 2020;80(6):646–655. doi: 10.1016/j.jinf.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z, et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost [Internet]. [cited 2020 Apr 25];n/a(n/a). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jth.14859. [DOI] [PMC free article] [PubMed]
- 58.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(February (10223)):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(February (18)):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97(May (5)):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo W, Lin Y, Yao X, Shi Y, Lu F, Wang Z, et al. Clinical findings of 35 cases with novel coronavirus pneumonia outside of Wuhan. Preprint from Research Square, 17 Apr 2020, PPR: PPR151913 10.21203/rs.3.rs-22554/v1. [DOI]
- 62.Li J., Li M., Zheng S., Li M., Zhang M., Sun M., et al. Plasma albumin levels predict risk for nonsurvivors in critically ill patients with COVID-19. Biomark Med. 2020;14(July (10)):827–837. doi: 10.2217/bmm-2020-0254. Epub 2020 Jun 3. PMID: 32490680; PMCID: PMC7273900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med (CCLM) 2020;58(7):1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- 64.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020;(March) doi: 10.1093/cid/ciaa270. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]