Abstract
The novel coronavirus (SARS-CoV-2) outbreak has started taking away the millions of lives worldwide. Identification of known and approved drugs against novel coronavirus disease (COVID-19) seems to be an urgent need for the repurposing of the existing drugs. So, here we examined a safe strategy of using approved drugs of SuperDRUG2 database against modeled membrane protein (M-protein) of SARS-CoV-2 which is essential for virus assembly by using molecular docking-based virtual screening. A total of 3639 drugs from SuperDRUG2 database and additionally 14 potential drugs reported against COVID-19 proteins were selected. Molecular docking analyses revealed that nine drugs can bind the active site of M-protein with desirable molecular interactions. We therefore applied molecular dynamics simulations and binding free energy calculation using MM-PBSA to analyze the stability of the compounds. The complexes of M-protein with the selected drugs were simulated for 50 ns and ranked according to their binding free energies. The binding mode of the drugs with M-protein was analyzed and it was observed that Colchicine, Remdesivir, Bafilomycin A1 from COVID-19 suggested drugs and Temozolomide from SuperDRUG2 database displayed desirable molecular interactions and higher binding affinity towards M-protein. Interestingly, Colchicine was found as the top most binder among tested drugs against M-protein. We therefore additionally identified four Colchicine derivatives which can bind efficiently with M-protein and have better pharmacokinetic properties. We recommend that these drugs can be tested further through in vitro studies against SARS-CoV-2 M-protein.
Keywords: SARS-CoV-2, Membrane protein, Docking, Molecular dynamics simulations, Pharmacokinetics, MM-PBSA
Introduction
The coronavirus disease (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 , 2 The disease was first reported in December 2019 at Wuhan (China) and currently spread all around the world.3 As of 1st November 2020, approximately 46 million cases and 1.2 million deaths have been reported globally according to World Health Organization (WHO) (https://www.who.int/). The most common symptoms associated with disease reported are fever, cough, fatigue, breathing difficulties and pneumonia.4 , 5 The SARS-CoV-2 was identified as the member of the family Coronaviridae which is subdivided into two subfamilies and have four genera: α, β, γ and Δ coronaviruses. The SARS-CoV-2 belongs to β-coronavirus together with SARS-CoV and MERS-CoV viruses.6 The virus have largest single stranded RNA genome of size 27–34 kb.7 , 8 The current therapeutic options under investigation for treating COVID-19 were antiviral, convalescent plasma and hyper immune immunoglobulin.9 , 10 There is still insufficient clinical data to recommend the use of these agents for the treatment. The urgent need of developing potential diagnostic, therapeutic, and preventive strategies till the vaccination arrives is drug repurposing. The main viral therapeutic proteins identified are spike protein (S-protein), 3C-like protease (3CLpro), papain like protease (PLpro), RNA-dependent RNA polymerase (RdRp), nucleocapsid protein (N-protein), transmembrane protease serine‑2, envelope protein (E-protein) and the membrane (M) protein.11 The spike protein is responsible for the interaction of virus to host cell receptor angiotensin-converting enzyme 2 (ACE2) and after the entry of the virus into the host cell, viral polyproteins are processed by 3CLpro and PLpro resulting in the release of nonstructural proteins (NSPs). The NSPs further forms a replication-transcriptase complex which is then assembled by RdRp and helicase leading to the production of mRNA. Simultaneously, sub genomic proteins translate structural and other accessory proteins S, M, N and E protein.12, 13, 14 As stated earlier, S-protein helps in the entry of the virus inside the host cell while E-protein is an integral membrane protein responsible for envelop formation and assembly of the virus.15 The M-protein is present in greater amounts in coronaviruses and conserved among β-coronaviruses.15 The M-protein from SARS-CoV-2 shares over 98% sequence identity with Bat and Pangolin.16 M-protein plays an important role in maintaining the shape of the virus envelop and also interacts with E-protein to form virions.12 Furthermore, M-protein also appears to affect the immune responses by inhibiting nuclear factor kappa B (NF- κB) therefore resulting in the proliferation of the virus.17 Additionally, M-protein inhibits the interaction of 3-phosphoinositide-dependant protein kinase 1 (PDK1) and protein kinase B (PKB) which resulted in release of caspases which eventually causes cell death.18 This literature survey revealed that M-protein can be a potential target for limiting and targeting the formation of virions and preventing inflammation in host cells.11 , 19 Recent studies have repurposed numerous drug-like candidates against well-known targets of SARS-CoV-2 viz. S-protein, 3CLpro, PLpro, RdRp and helicase.9 , 20 Few of the drugs such as Umifenovir, Lopinavir, Ritonavir, Hydroxychloroquine, Remdesivir and Favipiravir are under clinical trials against COVID-19.21 Only Remdesivir, a RdRp inhibitor has been found most effective till date and therefore approved by FDA for emergency use.22, 23, 24 Researchers around the world are still testing and searching for effective solutions against COVID-19. Apart from the well-known protein targets (3CLpro, PLpro, RdRp), researchers are now targeting other structural (E, N and M-protein) and accessory proteins of the virus.25, 26, 27 Among these proteins the M-protein whose role could be vital for viral entry, replication, assembly and maintenance of the virus envelop along with N, E and S proteins can be a potential targeting strategy for COVID-19 further seeks our attention.11 , 12 , 19 Unfortunately, the 3 dimensional (3D) crystal structure of M-protein is not reported till date in protein data bank (PDB). Therefore, a homology modelled structure of SARS-CoV-2 M-protein was obtained from DeepMind algorithm AlphaFold system's project for COVID-19 structures prediction (https://deepmind.com/). The structure was validated using standard validation methods. Thereafter, the molecular docking-based virtual screening of 3639 drugs from SuperDRUG2 database along with 14 previously reported COVID-19 drugs was performed and the selected drugs were subjected to molecular dynamics simulations.21 , 28, 29, 30 Additionally, the selected drugs were scrutinized for their binding affinity with M-protein by rigorous binding free energy calculations using MM-PBSA.
Materials and methods
Modeling of M-protein
Homology modeled three-dimensional (3D) structure of M-protein was downloaded from DeepMind algorithm AlphaFold system (https://deepmind.com/). The AlphaFold system uses convolutional neural network for protein structure prediction.31 In brief, this neural networks approach uses protein sequence to predict distances and angles between chemical bonds which connects amino acids. Predicted properties were then combined into a score and these scores were then used to screen proteins database to find structures that match the prediction.31 Recently Pandey et al., 2020 published a paper using drug repurposing approach for COVID-19 using the homology model of a membrane protein Nsp6 from DeepMind algorithm AlphaFold system.32 For present study, server uses SARS-CoV-2 M-protein sequence (UniProtKB id VME1_SARS2) as input for the membrane protein model generation. The generated model was energy minimized using Discovery Studio (DS) v18 (www.accelrys.com Accelrys Inc. San Diego, USA) to remove the steric clashes and lowest energy minimized structure was selected. To confirm the reliability of the SARS-CoV-2 M-protein model structure was then aligned with SARS-CoV sequence (UniProtKB id Q19QW6_SARS). The model was further evaluated by PROCHECK and ERRAT analysis using SAVES server (https://saves.mbi.ucla.edu/). The PROCHECK analysis evaluates the stereochemical quality of the protein backbone residues, while the ERRAT analysis provides details about protein folding.33 , 34 The validated model was then selected and prepared using the Clean Protein protocol in DS for molecular docking. During the preparation of the M-protein, the hydrogen atoms were added, bonding order was checked and terminal residues were adjusted. The binding site of M-protein for further process of molecular docking was identified by using CASTp 3.0 online server (http://cast.engr.uic.edu). CASTp 3.0 detects the global topological and protein dimensions to identify the active site and its volume.35 The top ranked site with larger surface area was considered as the active site of M-protein.36 The selected ligand binding site have amino acids Ile48, Leu51, Leu52, Trp55, Phe96, Met109, Trp110, Phe112, Asn113 and Pro114.
Ligand selection and preparation
The drug repurposing strategy was performed by using 3639 drugs from SuperDRUG2 database.28 , 29 We additionally selected 13 drugs (Lopinavir, Ritonavir, Hydroxychloroquine, Chloroquine, Ivermectin, Azithromycin, Favipiravir, Colchicine, Remdesivir, Tamiflu, Nitazoxanide, Toremifine, Umifenovir) reported active against other therapeutic target proteins of COVID-19 and downloaded them from PubChem database (https://pubchem.ncbi.nlm.nih.gov/).21 , 30 Interestingly, Gordon et al. identified that Bafilomycin A1 can bind with SARS-CoV-2 M-protein, we therefore also included this drug in our dataset for binding mode analysis.37 The drugs were saved in a single coordinated SDF format file. For each drug molecule, different possible conformers were generated and energy minimization was performed by using universal force field of PyRx software (https://pyrx.sourceforge.io/).
Molecular docking
To carry out the molecular docking studies, virtual screening software PyRx was employed.38 , 39 PyRx works based on empirical-based free energy scoring function and Lamarckian Genetic Algorithm. Molecular docking was performed in the grid box generated based on the binding site information provided by the CASTp 3.0 online server.35 The binding site residues inside the grid box with X, Y and Z axis and dimensions were adjusted to 13.49 Å × 0.41 Å × 2.45 Å with an exhaustiveness of 8, and the calculations were conducted in such a manner that only lowest energy pose was obtained as an output.
Molecular dynamics simulations
Molecular dynamics simulation technique is widely used in drug discovery to study the behavior of protein-ligand complexes at atomic level.40 , 41 In the present investigation, top ranked compounds retrieved from the molecular docking calculations were subjected to molecular dynamics simulations with M-protein. Topology parameters of the potential drug candidates were generated by using SwissParam server.42 Thereafter, these complexes were simulated for 50 ns using GROningen MAchine for Chemical Simulations (GROMACS v5.1.5),43 following the same protocol as described in our previous report.44
Binding free energy calculations
Protein-ligand complexes obtained via molecular dynamics simulations were further subjected to Molecular Mechanics-based Poisson-Boltzmann Surface Area (MM-PBSA) analysis for the calculation of their binding free energies.45 Based on the RMSD plots, last 10 ns simulation trajectories were selected and a total of 40 snapshots were taken at a regular interval. The g_mmpbsa tool for GROMACS was employed to calculate the different parameters of binding free energies with the methodologies described in previous reports.46, 47, 48, 49
Results
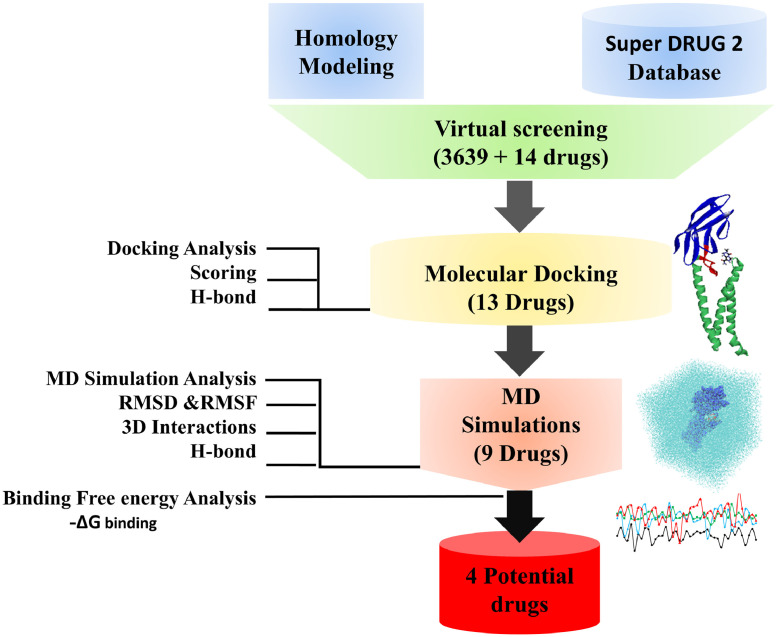
The present study provides a comprehensive details on targeting the M-protein of novel coronavirus using homology modeling, molecular docking-based virtual screening, binding mode analyses using molecular dynamics simulations with SARS-CoV-2 M-protein and free energy calculations. The schematic representation of the workflow has been described (Fig. 1 ).
Fig. 1.
Schematic representation of the workflow implemented in the current study.
M-protein modeling
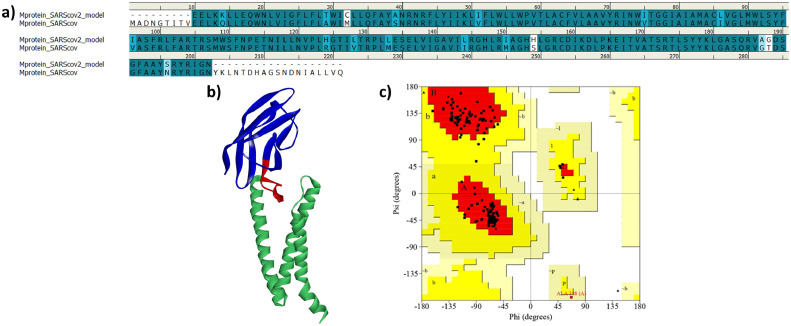
Homology modeled structure of M-protein was downloaded from DeepMind algorithm AlphaFold system's project for COVID-19 structure prediction. The server uses deep neural network learning algorithm AlphaFold system (https://deepmind.com/). The server uses SASR-CoV-2 M-protein sequence (UniProtKB id VME1_SARS2) was used as input for the construction of 3D model. The obtained modeled structure was first aligned with SARS-CoV sequence (UniProtKB id Q19QW6_SARS). It was observed that the obtained model has 87.1% sequence identity and 93.1% sequence similarity with the SARS-CoV M-protein (Fig. 2 a). This analysis is parallel with the reported sequence identity between both the viruses SARS-CoV and SARS-CoV-2.16 , 50 The model was then submitted to SAVES server for the generation of Ramachandran plot to predict the distribution of residues.51 , 52 The analysis revealed that 92.5% residues are in the allowed region (Fig. 2c). In addition, the model was also analyzed by ERRAT web server and the analysis revealed that the quality factor of our model was 92.04%. Generally, models with quality factor more that 50% is of acceptable quality.50 , 53 The above detailed analysis of selected model using sequence alignment, PROCHECK and ERRAT revealed that the model is of reliable quality and can be used in further studies (Fig. 2b).
Fig. 2.
(a) Sequence alignment of the M-protein of SARS-CoV-2 with the SARS-CoV M-protein. Dark blue color and light blue color represents amino acid identity and similarity respectively, (b) 3D structure of the selected model with the transmembrane and C-terminal domain shown in blue and light green color respectively while the C-terminal conserved region is shown as red (c) Validation of the selected homology model of SARS-CoV-2 M-protein by Ramachandran plot. Plot displays 92.5% residues in the allowed region.
Molecular docking
The energy minimized drug database was used for docking-based virtual screening using PyRx software. PyRx works on empirical-based free energy scoring function and Lamarckian Genetic Algorithm. Molecular docking was performed in the grid box generated as stated above based on the binding site information provided by the CASTp 3.0 online server and the docking results were analyzed on the basis of binding energy scores. A total of 13 drugs from SuperDRUG2 database displayed acceptable binding energy score lower than −7.0 kcal/mol.38 , 54 , 55 Additionally the drugs were also docked on two more possible sites predicted by CASTp 3.0 server. The results confirms that drugs have high binding affinity toward previously selected Top1binding site Table S2. The molecular interactions of selected drugs with were Top1 binding site were further analyzed and it was observed that five drugs displayed better molecular interactions with the active site residues of M-protein. The selected potential drugs formed hydrogen bonds with the active site residues Ala40, Asn41, Arg44, Asn113, and Glu115 of M-protein (Table S1). It has been observed that the drug Temozolomide from SuperDRUG2 database displayed highest binding affinity of −8.9 kcal/mol among selected drugs. The three drugs (Colchicine, Remdesivir and Bafilomycin A1) were also selected from the 14 COVID-19 reported drugs suggested on the basis of binding energy scores and molecular interactions (Table S1). The selected drug were additionally, docked with known drug targets. It was observed that drugs displayed comparable binding affinity towards SASR-CoV-2 M-protein complete details were provided in Table S3.
Molecular dynamics simulations
The selected drugs from molecular docking were subjected for molecular dynamics simulation for 50 ns run using GROMACS. The drugs were then analyzed for RMSD, potential energy, RMSF and formation of hydrogen bonds with M-protein. Furthermore, the binding free energies for selected drugs were calculated using MM-PBSA tool and drugs were ranked accordingly. Drugs which displayed acceptable binding were considered for detailed analyses (Table S1).
Stability analyses of simulated systems
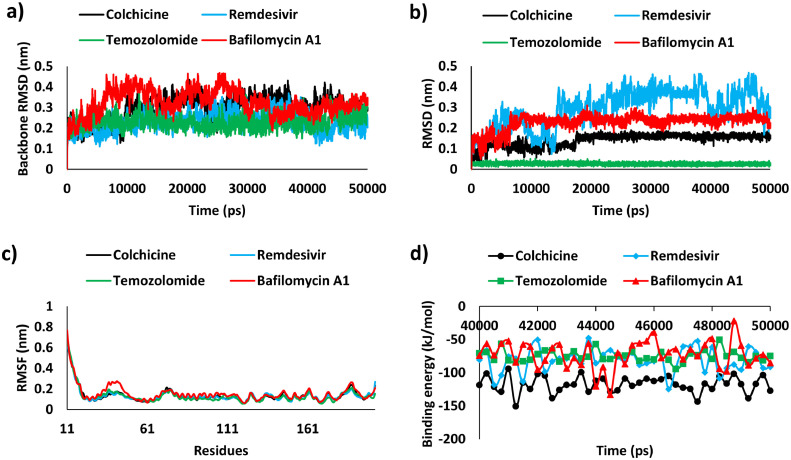
To examine the structural fluctuations of the docked complexes, the RMSD value for the protein backbone and ligand was calculated for the time duration of 50 ns.56 , 57 Except Remdesivir that showed stable RMSD from 5 ns till the end of the simulation run, the backbone RMSD of the Bafilomycin A1, Colchicine and Temozolomide initially increased between 10 and 25 ns and thereafter attained steadiness till the end of the simulation. (Fig. 3 a). The mean RMSD value for M-protein backbone bound with Colchicine, Remdesivir, Temozolomide and Bafilomycin A1 was observed to be 0.29, 0.23, 0.23 and 0.32 nm, respectively. The RMSD pattern of M-protein bound drugs (Fig. 3b) revealed that Temozolomide showed stable RMSD from beginning till the end of the simulation run whereas Bafilomycin A1, Colchicine, Remdesivir complexed with M-protein reached equilibrium in the time duration of 15 ns. In drug discovery the RMSD values below 0.3 nm were generally considered as stable interestingly, our simulated systems were found within acceptable RMSD range.44 , 58, 59, 60 The RMSF analysis showed minimal residual flexibility for all the M-protein and ligand complexes and were found stable throughout the simulation. The functionally important residues Ala40 and Asn41 displayed stable behavior and no fluctuations were observed towards the start of N-terminal and the end of C-terminal region of the SARS-CoV-2 M-protein. The mean RMSF value for M-protein with Colchicine, Remdesivir, Temozolomide and Bafilomycin A1 complexes was observed to be 0.13, 0.13, 0.13 and 0.15 nm, respectively (Fig. 3c).
Fig. 3.
Molecular dynamics simulation analysis. a) RMSD profile of the backbone atoms of M-protein, b) RMSD profile of bound drugs, c) RMSF analyses and d) MM-PBSA estimated binding free energy of selected potential drugs (Colchicine, Remdesivir, Temozolomide and Bafilomycin A1) from last 10 ns trajectories of simulation run.
Additionally, stability of the simulated complexes was also analyzed by potential energy plots.61 , 62 The selected protein-ligand complexes attained equilibrium and were found to be stabilized throughout the period of simulation (Figure S2a). The binding stability of M-protein with Colchicine, Remdesivir, Temozolomide and Bafilomycin A1 was also estimated by hydrogen bonding (H-bond) analysis. H-bond analyses revealed that Temozolomide forms the highest number of H-bonds with M-protein followed by Remedesivir, Bafilomycin A1 and Colchicine (Figure S2b). The average values of RMSD, RMSF, potential energy and H-bond analyses has been displayed (Table 1 ). Additionally, all the above mentioned parameters were compared with SARS-CoV-RdRp-Remedesivir and were found satisfactory (Figure S3). The analysis revealed that the RMSD values of protein backbone atoms and ligand were found <0.3 nm which is very similar with our results with M-protein. RMSF analysis revealed the proteins showed residual fluctuations but the average values were found <0.3 nm. The stable potential energy and comparable number of hydrogen bonds were observed in both the protein.
Table 1.
Molecular dynamics simulation analysis of selected drugs against SARS-CoV-2 M-protein.
| Sr. No. | Drugs Name | Binding Free energy (kJ/mol) | RMSD backbone (nm) | RMSD ligand (nm) | RMSF backbone (nm) | Potential energy (kJ/mol) | No. of Hydrogen Bonds |
|---|---|---|---|---|---|---|---|
| 1 | Colchicine | −117.62 | 0.29 | 0.14 | 0.13 | −1,056,643.76 | 0.75 |
| 2 | Remdesivir | −81.17 | 0.23 | 0.29 | 0.13 | −1,057,314.45 | 2.17 |
| 3 | Temozolomide | −74.94 | 0.23 | 0.02 | 0.13 | −1,057,813.98 | 2.48 |
| 4 | Bafilomycin A1 | −72.31 | 0.32 | 0.22 | 0.15 | −1,056,947.96 | 1.29 |
Binding free energy calculations
The MM-PBSA method was used to estimate the binding affinity of ligands from the last 10 ns simulation trajectories (Table S1). Previous studies confirmed that binding free energy values lower than −30 kJ/mol can be considered for binding, however lower binding free energy values more favorable for interactions.63 , 64 In present investigation, four drugs were able to display acceptable binding free energy values therefore other drugs were removed from further analysis. The predicted binding free energies (ΔGbind) for selected drugs Colchicine, Remdesivir, Temozolomide and Bafilomycin A1 were −117.62, −81.17, −74.94 and −72.31 kJ/mol, respectively (Fig. 3d and Table 1). Our analyses confirmed that Colchicine was observed to show lowest ΔGbind values among all drugs, therefore can bind more tightly to the M-protein this is followed by Remdesivir, Temozolomide and Bafilomycin A1. To further understand the binding in detail free energy values were decomposed in individual components (Table 2 ). Analysis revealed that the major favorable contributors for protein-ligand complexes were van der Waals interactions (∆EvdW) and solvent energy surface area SASA non-polar energy (∆Gnp). The greater electrostatic contribution (∆Eele = −69.88, −36.28, −47.62 kJ/mol) of Remdesivir, Temozolomide, Bafilomycin A1 in comparison to ∆Eele = 18.88 kJ/mol of Colchicine was observed. The polar solvation energies (∆Gpol) contributed positively to the total binding free energies and therefore opposes the complex formation.65 The binding affinity of the drug Remedesivir was also analyzed with SARS-CoV-RdRp (Figure S3). As expected, Remedesivir displayed better binding affinity for its known target RdRp (−104 kJ/mol) but interestingly, showed considerable binding affinity toward M-protein (−81.17 kJ/mol) confirms the ability of the drug to target multi-targets.
Table 2.
Binding free energies of simulated protein-ligand complexes through MM-PBSA method.
| Ligand | ∆EvdW (kJ/mol) | ∆Eele (kJ/mol) | ∆Gpol (kJ/mol) | ∆Gnp (kJ/mol) | ΔGbind (kJ/mol) |
|---|---|---|---|---|---|
| Colchicine | −148.40±9.55 | 18.88±8.41 | 27.39±9.63 | −15.49±0.77 | −117.62±12.20 |
| Remdesivir | −177.51±14.43 | −69.88±14.05 | 186.21±24.68 | −19.98±2.00 | −81.17±17.84 |
| Temozolomide | −132.06± 8.25 | −36.28±7.67 | 104.34±8.23 | −10.94±0.44 | −74.94±8.39 |
| Bafilomycin A1 | −155.44±20.09 | −47.62±16.02 | 150.89±23.65 | −20.13±2.01 | −72.31±20.44 |
Molecular interaction analysis
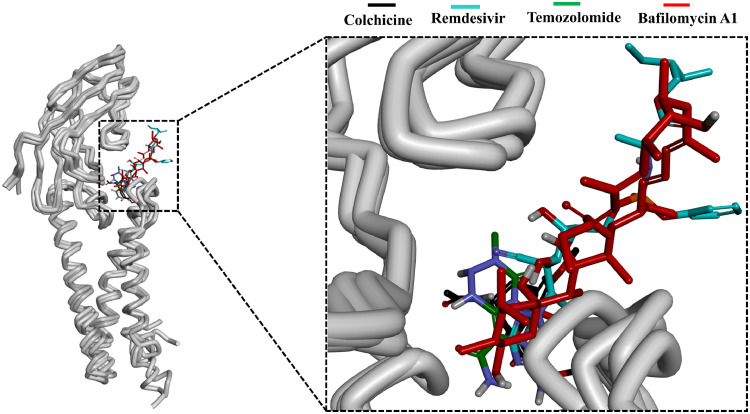
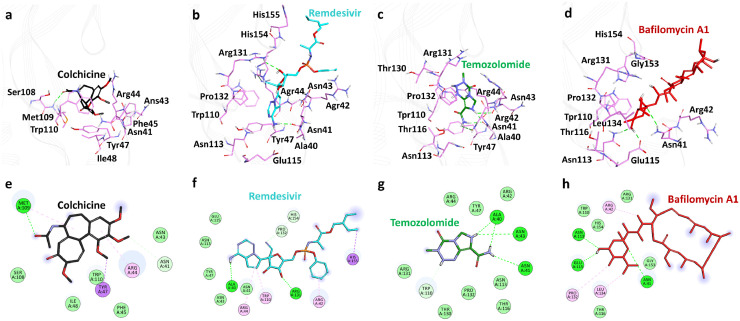
The binding site of the M-protein was identified in between the conserved C-terminal and transmembrane domain (Fig. 2b). The average structure for all the selected complexes were calculated from the last 10 ns of stable MD trajectories and superimposed. All the selected drugs were observed to bind inside the defined binding site (Fig. 4 ). A deep insight on molecular interaction pattern revealed that Colchicine formed hydrogen bond interaction with the amino-acid Met109, Remdesivir showed hydrogen bonding with the amino-acids Ala40 and Arg131, Temozolomide displayed hydrogen bonding with amino-acids Ala40, Asn41 and Asn43 and Bafilomycin A1 showed hydrogen bonding with the amino-acid residues Asn41, Asn113 and Glu115 of SARS-CoV-2 M-protein (Fig. 5 a, b, c and d). On further noticing the bonding distance between the mentioned drugs and their respective interacting amino-acids, it is clearly visible that the drugs have shown the bonding in less than 3.5 Å distance (Table 3 ).44 , 61 , 66 The protein-drug molecular interactions were also stabilized by non-polar interactions within the active site residues of the protein. Colchicine was observed to form van der Waals interactions with active site residues of M-protein Asn41, Asn43, Ile48, Ser108, Trp110 and π-alkyl interactions with Arg44, Tyr47 whereas Remdesivir displayed van der Waals interactions with Asn41, Asn43, Tyr47, Asn113, Glu115, Pro132, His154 and π-alkyl interactions with residues Arg42, Arg44, Trp110, His155. Temozolomide formed van der Waals interactions with Arg42, Arg44, Tyr47, Trp110, Asn113, Thr116, Thr130, Arg131, Pro132 of M-protein and Bafilomycin A1 formed van der Waals interactions with Trp110, Thr116, Arg131, Gly153, His154 and π-alkyl interactions with Arg42, Pro132, Leu134 (Table 3). Recently, Bhowmik et al. identified through an in silico study that Caffeic acid and Ferulic acid can be effective against M-protein.25 The identified compounds were reported to target Asn43, Tyr47 Lys51 through hydrogen bond and residues Leu46, Leu51, and Leu54 through hydrophobic interactions. It is noteworthy to mention here that our identified drugs target SARS-CoV-2 M-protein through hydrogen bonds and hydrophobic interactions hence providing enough support as future COVID-19 inhibitors (Table 3).
Fig. 4.
Binding patterns of the potential drugs in the active site of SARS-CoV-2 M-protein. Superimposition of the selected drugs (left) and enlarged view (right). M-protein is shown as gray tube whereas Colchicine, Remdesivir, Temozolomide and Bafilomycin A1 are in black, cyan, green and red respectively.
Fig. 5.
Molecular interaction analysis. Upper panel displaying 3D interaction patterns of the potential drugs in the active site of SARS-CoV-2 M-protein. Protein in background is shown as line ribbon (gray) whereas residues involved in interactions are shown as stick representation (light pink). Drugs are displayed as sticks with a) Colchicine (black) b) Remdesivir (cyan) c) Temozolomide (green) and d) Bafilomycin A1 (red). Hydrogen bonds were displayed in green dashed lines. The lower panel displaying 2D interactions of potential drugs in the active site SARS-CoV-2 M-protein. e) Colchicine f) Remdesivir g) Temozolomide and h) Bafilomycin A1. Residues shown in dark green color represent conventional hydrogen bonds, light green color represents van der Waals interactions and pink or purple color are alkyl interactions.
Table 3.
Molecular interactions of potential drugs with SARS-CoV-2 M-protein.
| Name | Hydrogen Bond Interactions |
van der Waals Interactions | π -π /π-alkyl interactions | |||
|---|---|---|---|---|---|---|
| Amino acid | Amino acid atom | Ligand atom | Distance (<3.5 Å) | |||
| Colchicine | Met109 | HN | O6 | 2.67 | Asn41, Asn43, Ile48, Ser108, Trp110 | Arg44, Tyr47 |
| Remdesivir | Ala40 | O | H20 | 2.08 | Asn41, Asn43, Tyr47, Asn113, Glu115, Pro132, His154 | Arg42, Arg44, Trp110, His155 |
| Arg131 | HH21 | O3 | 2.67 | |||
| Temozolamide | Ala40 | O | HN | 1.98 | Arg42, Arg44, Tyr47, Trp110, Asn113, Thr116, Thr130, Arg131, Pro132 | |
| Asn41 | OD1 | H1 | 3.07 | |||
| Asn43 | O | HN | 3.07 | |||
| Bafilomycin A1 | Asn41 | HD22 | O1 | 3.01 | Trp110, Thr116, Arg131, Gly153, His154 | Arg42, Pro132, Leu134 |
| Asn113 | HD22 | O3 | 2.04 | |||
| Glu115 | OG1 | H53 | 2.10 | |||
The repurposed drug Colchicine is an alkaloid-based drug derived from Colchicum autumnale and has been used against gout flare. The drug is currently under randomized clinical trials against COVID-19.67 Colchicine is reported to target mitosis and microtubule assembly.68 It is also reported to have antiviral effects against Dengue and Zika viruses and its derivatives were reported to reduce HIV virus load.69 , 70 The drug Remdesivir which is a nucleotide analog is a well-known inhibitor of SARS-CoV-2 RdRp.71, 72, 73, 74 Interestingly, Remdesivir is the first drug got an emergency FDA approval against COVID-19.75 Recently, in silico drug repurposing studies also identified that the drug can bind SARS-CoV-2 3CLpro.76 , 77 These observations conclude that Remdesivir can bind both the proteins efficiently and hence found most effective against SARS-CoV-2 infections. The concept of multi-target drugs which can target multiple proteins simultaneously has been already in use and found effective and hence Remdesivir can be selected as a hit against M-protein.78 The Bafilomycin A1 is reported to inhibit SARS-CoV-2 NSP6 and M-protein and is in pre-clinical studies.37 The drug was also reported to inhibit SARS-CoV-2 in vitro studies and RdRp.79 Here, our study revealed the binding mode of Bafilomycin A1 inside the active site of M-protein. In addition, Temozolomide from SuperDRUG2 database was found effective against SARS-CoV-2 M-protein as well. The drug is an anti-cancer agent approved by the FDA for use in the first-line treatment of glioblastoma. It is classified as an alkylating agent and is supposed to stop replication of cells.80
Substructure search for colchicine
According to binding free energy and binding mode analyses, out of the 14 COVID-19 reported drugs, Colchicine was found ranked on the top (Table 2). The drug is derived from Colchicum autumnale and has been approved by FDA for the treatment of gout flares and Familial Mediterranean fever.81 Intriguingly, Colchicine is currently under randomized clinical trials against COVID-19.67 It is hypothesized that Colchicine can interfere with SARS-CoV-2 spike protein function and therefore may block viral entry. The detailed mechanism of this is not fully reported yet. In the present study, we have observed that Colchicine binds M-protein with high affinity (−117.62 kJ/mol) with polar and non-polar interactions. Colchicine is reported to be a substrate of P-glycoprotein (P-gp) and CYP3A4 due to this drugs like Lopinavir and Ritonavir can increase the potential for Colchicine toxicity.67 To avoid this toxicity effect we searched for similar compounds using PubChem database (https://pubchem.ncbi.nlm.nih.gov/) to obtained Colchicine-like substructures. A total of 683 compounds were retrieved and used for molecular docking with M-protein. The docking analysis revealed 10 compounds binding M-protein with better binding affinity than Colchicine and also formed more number of hydrogen bonds than Colchicine (Table S4). These compounds were further subjected for calculation of their pharmacokinetic properties with online web tool SwissADME (http://www.swissadme.ch/index.php).82 Four compounds were observed to display comparable pharmacokinetic properties with Colchicine (Table 4 ). Interestingly, the compound with PubChem ID 6,711,380 was found better among all selected derivatives in terms of pharmacokinetic properties predicted.
Table 4.
Pharmacokinetic properties analyses for Colchicine substructures.
| Pharmacokinetic Properties | Compound name/PubChem ID |
||||
|---|---|---|---|---|---|
| Colchicine | 24,988,904 | 118,725,563 | 146,048,789 | 6,711,380 | |
| GI absorption | High | Low | Low | Low | Low |
| BBB permeant | No | No | No | No | No |
| P-gp substrate | Yes | Yes | Yes | Yes | Yes |
| CYP1A2 inhibitor | No | No | No | No | No |
| CYP2C19 inhibitor | No | No | No | No | No |
| CYP2D6 inhibitor | Yes | No | No | No | No |
| CYP3A4 inhibitor | Yes | Yes | Yes | Yes | No |
| Water solubility | Soluble | Moderately soluble | Moderately soluble | Moderately soluble | Soluble |
Conclusions
The membrane glycoprotein is conserved across the β-coronaviruses. The multiple sequence alignment shows a remarkable similarity over 96% among the SARS-CoV M variants. Membrane protein exists as a Homomultimer and interacts with envelope protein and nucleocapsid protein. In this study, four commercially available drugs Colchicine an alkaloid drug used in the management of gout, Remdesivir an investigational drug currently under trial against coronavirus, Temozolomide an oral alkylating agent used for the treatment of refractory anaplastic astrocytoma and Bafilomycin A1 an experimental toxic macrolide antibiotic is shown to be important and highly potent for the inhibition of SARS‐CoV‐2 M-protein. Moreover, Colchicine derivatives were also found effective against M-protein and displayed better pharmacokinetic properties. The lead drug candidates showed molecular interactions with binding pocket residues of the modelled M-protein of SARS‐CoV‐2. However further studies are required to authenticate the effectiveness of these purposed drugs.
Author contributions
K.A.P. and V.K. conceived the idea of the project. K.A. P. performed Molecular docking and contributed in writing and V.K. performed Molecular Dynamics simulation, binding free energy analyses and contributed in writing. K.A. P., V.K., K.W.L. and T.C.V., analyzed the results. All the authors have read and approved the manuscript for submission.
Declaration of Competing Interest
The authors report no conflicts of interest in this work.
Acknowledgements
The authors acknowledge to Vignan's Foundation for Science, Technology and Research (Deemed to be university), Vadlamudi-522213 and DST-FIST networking facility to carry out this work. Author also thank Gyeongsang National University to carry out Molecular dynamics simulation work. This research was partially supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (No. NRF-2018M3A9A70–57263).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.xphs.2021.03.004.
Appendix. Supplementary materials
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020;55(6) doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant M.C., Geoghegan L., Arbyn M. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): a systematic review and meta-analysis of 148 studies from 9 countries. PLoS ONE. 2020;15(6) doi: 10.1371/journal.pone.0234765. Hirst JA, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harapan H., Itoh N., Yufika A. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sexton N.R., Smith E.C., Blanc H., Vignuzzi M., Peersen O.B., Denison M.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J Virol. 2016;90(16):7415–7428. doi: 10.1128/jvi.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valencia D.N. Brief review on COVID-19: the 2020 pandemic caused by SARS-CoV-2. Cureus 2020;12(3). doi:10.7759/cureus.7386. [DOI] [PMC free article] [PubMed]
- 9.Kaddoura M., AlIbrahim M., Hijazi G. COVID-19 therapeutic options under investigation. Front Pharmacol. 2020;11:1196. doi: 10.3389/fphar.2020.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Wang G., Cai X peng. An overview of COVID-19. J Zhejiang Univ Sci B. 2020;21(5):343–360. doi: 10.1631/jzus.B2000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlachakis D., Papakonstantinou E., Mitsis T. Molecular mechanisms of the novel coronavirus SARS-CoV-2 and potential anti-COVID19 pharmacological targets since the outbreak of the pandemic. Food Chem Toxicol. 2020;146 doi: 10.1016/j.fct.2020.111805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prajapat M., Sarma P., Shekhar N. Update on the target structures of SARS-CoV-2: a systematic review. Indian J Pharmacol. 2020;52(2):142–149. doi: 10.4103/ijp.IJP_338_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65(2):69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol. 2007;81(7):3597–3607. doi: 10.1128/jvi.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed Res Int. 2020;2020 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang X., Gao J., Zheng H. The membrane protein of SARS-CoV suppresses NF-κB activation. J Med Virol. 2007;79(10):1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsoi H., Li L., Chen Z.S., Lau K.F., Tsui S.K.W., Chan H.Y.E. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1PKB/Akt signalling. Biochem J. 2014;464(3):439–447. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- 19.Rajarshi K., Khan R., Singh M.K., Ranjan T., Ray S., Ray S. Essential functional molecules associated with SARS-CoV-2 infection: potential therapeutic targets for COVID-19. Gene. 2020 doi: 10.1016/j.gene.2020.145313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dotolo S., Marabotti A., Facchiano A., Tagliaferri R. A review on drug repurposing applicable to COVID-19. Brief Bioinform. 2020;2020(0):1–16. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drożdżal S., Rosik J., Lechowicz K. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist Updat. 2020;53 doi: 10.1016/j.drup.2020.100719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C., Tian L., Liu Y. A promising antiviral candidate drug for the COVID-19 pandemic: a mini-review of remdesivir. Eur J Med Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko W.C., Rolain J.M., Lee N.Y. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. 2020. doi:10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed]
- 26.Das G., Das T., Chowdhury N., Chatterjee D., Bagchi A., Ghosh Z. Repurposed drugs and nutraceuticals targeting envelope protein: a possible therapeutic strategy against COVID-19. Genomics. November 2020 doi: 10.1016/j.ygeno.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borkotoky S., Banerjee M. A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem) J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1774419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siramshetty V.B., Eckert O.A., Gohlke B.O. SuperDRUG2: a one stop resource for approved/marketed drugs. Nucleic Acids Res. 2018;46(D1):D1137–D1143. doi: 10.1093/nar/gkx1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goede A., Dunkel M., Mester N., Frommel C., Preissner R. Super drug: a conformational drug database. Bioinformatics. 2005;21(9):1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 30.Wu R., Wang L., Kuo H.C.D. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Reports. 2020;6(3):56–70. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senior A.W., Evans R., Jumper J. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 32.Pandey P., Prasad K., Prakash A., Kumar V. Insights into the biased activity of dextromethorphan and haloperidol towards SARS-CoV-2 NSP6: in silico binding mechanistic analysis. J Mol Med. 2020;98(12):1659–1673. doi: 10.1007/s00109-020-01980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- 34.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(W1):W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helal M.A., Shouman S., Abdelwaly A. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J Biomol Struct Dyn. 2020:1. doi: 10.1080/07391102.2020.1822208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon D., Jang G., Bouhaddou M. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020;19:4. doi: 10.1101/2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni S.A., Nagarajan S.K., Ramesh V., Palaniyandi V., Selvam S.P., Madhavan T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J Mol Struct. 2020;1221 doi: 10.1016/j.molstruc.2020.128823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 40.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9(1):71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 43.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 44.Kumar R., Kumar V., Lee K.W. A computational drug repurposing approach in identifying the cephalosporin antibiotic and anti-hepatitis C drug derivatives for COVID-19 treatment. Comput Biol Med. 2021;130 doi: 10.1016/j.compbiomed.2020.104186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar R., Parameswaran S., Bavi R. Investigation of novel chemical scaffolds targeting prolyl oligopeptidase for neurological therapeutics. J Mol Graph Model. 2019;88:92–103. doi: 10.1016/j.jmgm.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 48.Bavi R., Kumar R., Rampogu S. Novel virtual lead identification in the discovery of hematopoietic cell kinase (HCK) inhibitors: application of 3D QSAR and molecular dynamics simulation. J Recept Signal Transduct. 2017;37(3):224–238. doi: 10.1080/10799893.2016.1212376. [DOI] [PubMed] [Google Scholar]
- 49.Zeb A., Kim D., Alam S.I. Computational simulations identify pyrrolidine-2,3-dione derivatives as novel inhibitors of Cdk5/p25 complex to attenuate Alzheimer's pathology. J Clin Med. 2019;8(5):746. doi: 10.3390/jcm8050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas S. The structure of the membrane protein of sars-cov-2 resembles the sugar transporter semisweet. Pathog Immun. 2020;5(1):342–363. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J Biomol Struct Dyn. April 2020:1–13. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bavi R., Kumar R., Rampogu S. Molecular interactions of UvrB protein and DNA from Helicobacter pylori: insight into a molecular modeling approach. Comput Biol Med. 2016;75:181–189. doi: 10.1016/j.compbiomed.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Messaoudi A., Belguith H., Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor Biol Med Model. 2013;10(1):22. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J A. Francis D., C.S S. K.G A. C S. Variyar E.J. Repurposing simeprevir, calpain inhibitor IV and a cathepsin F inhibitor against SARS-CoV-2 and insights into their interactions with Mpro. J Biomol Struct Dyn. September 2020:1–12. doi: 10.1080/07391102.2020.1813200. [DOI] [PubMed] [Google Scholar]
- 55.Jordaan M.A., Ebenezer O., Damoyi N., Shapi M. Virtual screening, molecular docking studies and DFT calculations of FDA approved compounds similar to the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz. Heliyon. 2017:e04642. doi: 10.1016/j.heliyon.2020.e04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys J. 2010;98(5):861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao K.H., Chen K.-.B., Lee W.-.Y., Sun M.-.F., Lee C.-.C., Chen C.Y.-.C. Ligand-based and structure-based investigation for Alzheimer's disease from traditional Chinese medicine. Evidence-Based Complement Altern Med. 2014;2014:1–16. doi: 10.1155/2014/364819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh W., Karabencheva-Christova T.G., Black G.W., Ainsley J., Dover L., Christov C.Z. Conformational dynamics, ligand binding and effects of mutations in NirE an S-Adenosyl-l-methionine dependent methyltransferase. Sci Rep. 2016;6(1):1–9. doi: 10.1038/srep20107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rangwala H., Karypis G. fRMSDPred: predicting local RMSD between structural fragments using sequence information. Proteins Struct Funct Genet. 2008;72(3):1005–1018. doi: 10.1002/prot.21998. [DOI] [PubMed] [Google Scholar]
- 61.Zeb A., Son M., Yoon S., Kim J.H., Park S.J., Lee K.W. Computational simulations identified two candidate inhibitors of Cdk5/p25 to abrogate tau-associated neurological disorders. Comput Struct Biotechnol J. 2019;17:579–590. doi: 10.1016/j.csbj.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian Y., Gao Y., Chen Y., Liu G., Ju X. Identification of the fipronil resistance associated mutations in nilaparvata lugens GABA receptors by molecular modeling. Molecules. 2019;24(22):4116. doi: 10.3390/molecules24224116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rifai E.A., Van Dijk M., Vermeulen N.P.E., Yanuar A., Geerke D.P. A comparative linear interaction energy and MM/PBSA Study on SIRT1-ligand binding free energy calculation. J Chem Inf Model. 2019;59(9):4018–4033. doi: 10.1021/acs.jcim.9b00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poli G., Granchi C., Rizzolio F., Tuccinardi T. Application of MM-PBSA methods in virtual screening. Molecules. 2020;25(8):1971. doi: 10.3390/molecules25081971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh S., Sk M.F., Sonawane A., Kar P., Sadhukhan S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: an in-silico analysis. J Biomol Struct Dyn. July 2020:1–16. doi: 10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raschka S., Wolf A.J., Bemister-Buffington J., Kuhn L.A. Protein–ligand interfaces are polarized: discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J Comput Aided Mol Des. 2018;32(4):511–528. doi: 10.1007/s10822-018-0105-2. [DOI] [PubMed] [Google Scholar]
- 67.Schlesinger N., Firestein B.L., Brunetti L. Colchicine in COVID-19: an old drug, new use. Curr Pharmacol Reports. 2020;6(4):137–145. doi: 10.1007/s40495-020-00225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niel E., Scherrmann J.M. Colchicine today. Jt Bone Spine. 2006;73(6):672–678. doi: 10.1016/j.jbspin.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Worachartcheewan A., Songtawee N., Siriwong S., Prachayasittikul S., Nantasenamat C., Prachayasittikul V. Rational design of colchicine derivatives as anti-HIV agents via QSAR and molecular docking. Med Chem (Los Angeles) 2018;15(4):328–340. doi: 10.2174/1573406414666180924163756. [DOI] [PubMed] [Google Scholar]
- 70.Richter M., Boldescu V., Graf D. Synthesis, biological evaluation, and molecular docking of combretastatin and colchicine derivatives and their hCE1-Activated prodrugs as antiviral agents. Chem Med Chem. 2019;14(4):469–483. doi: 10.1002/cmdc.201800641. [DOI] [PubMed] [Google Scholar]
- 71.Koulgi S., Jani V., Uppuladinne M.V.N., Sonavane U., Joshi R. Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2. RSC Adv. 2020;10(45):26792–26803. doi: 10.1039/d0ra04743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J Phys Chem B. 2020;124(32):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 73.Gordon C.J., Tchesnokov E.P., Woolner E., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. doi:10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed]
- 74.Yin W., Mao C., Luan X. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science (80-) 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.FDA Approves First Treatment for COVID-19 | FDA.https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19. Accessed December 10, 2020.
- 76.Naik V.R., Munikumar M., Ramakrishna U. Remdesivir (GS-5734) as a therapeutic option of 2019-nCOV main protease–in silico approach. J Biomol Struct Dyn. 2020:1. doi: 10.1080/07391102.2020.1781694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen H.L., Thai N.Q., Truong D.T., Li M.S. Remdesivir strongly binds to both RNA-dependent RNA polymerase and main protease of SARS-COV-2: Evidence from molecular simulations. J Phys Chem B. 2020;124(50):11337–11348. doi: 10.1021/acs.jpcb.0c07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trezza A., Iovinelli D., Santucci A., Prischi F., Spiga O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-70863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piplani S., Singh P., Winkler D.A., Petrovsky N. November 2020. Computationally Repurposed Drugs and Natural Products Against RNA Dependent RNA Polymerase As Potential COVID-19 Therapies.http://arxiv.org/abs/2011.14241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., F.G. Stevens M, D. Bradshaw T. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2011;5(1):102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 81.Colchicine - StatPearls - NCBI Bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK431102/. Accessed December 10, 2020.
- 82.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.