Abstract
All current vaccines for COVID-19 utilize ancestral SARS-CoV-2 spike with the goal of generating protective neutralizing antibodies. The recent emergence and rapid spread of several SARS-CoV-2 variants carrying multiple spike mutations raise concerns about possible immune escape. One variant, first identified in the United Kingdom (B.1.1.7, also called 20I/501Y.V1), contains eight spike mutations with potential to impact antibody therapy, vaccine efficacy, and risk of reinfection. Here, we show that B.1.1.7 remains sensitive to neutralization, albeit at moderately reduced levels (∼sim;2-fold), by serum samples from convalescent individuals and recipients of an mRNA vaccine (mRNA-1273, Moderna) and a protein nanoparticle vaccine (NVX-CoV2373, Novavax). A subset of monoclonal antibodies to the receptor binding domain (RBD) of spike are less effective against the variant, while others are largely unaffected. These findings indicate that variant B.1.1.7 is unlikely to be a major concern for current vaccines or for an increased risk of reinfection.
Keywords: SARS-CoV-2 variants, COVID-19, neutralizing antibodies, vaccines, monoclonal antibodies, B.1.1.7, Moderna, Novavax
Graphical abstract
The increasing prevalence and diversity of SARS-CoV-2 spike variants raises concerns for potential immune escape. Using a validated pseudovirus neutralization assay, Shen et al. show that the B.1.1.7 variant escapes a subset of monoclonal antibodies but remains susceptible to vaccine-elicited antibodies and serum samples from people who recovered from COVID-19.
Introduction
Genetic evolution in the SARS-CoV-2 virus (severe acute respiratory syndrome coronavirus 2) is an increasing concern for the COVID-19 (coronavirus disease 2019) pandemic. Continued high infection rates are providing opportunities for the virus to acquire mutations that contribute to virus spread and possible immune evasion. Mutations in the viral spike are a particular concern because this glycoprotein mediates virus attachment and entry (Ou et al., 2020) and is the major target for neutralizing antibodies (Piccoli et al., 2020). The D614G spike variant that spread rapidly during March and April of 2020 (Biswas and Majumder, 2020; Isabel et al., 2020) was found in most sequences globally by June 2020 and is the earliest evidence for adaptive evolution of this virus in humans. The D614G mutation imparts increased infectivity in vitro (Hou et al., 2020; Korber et al., 2020), accelerated transmission in hamsters (Hou et al., 2020), and shows a modest increase in neutralization susceptibility (Weissman et al., 2021), all of which are explained by a more open conformation of the receptor binding domain (RBD) (Weissman et al., 2021; Yurkovetskiy et al., 2020). The mutation does not appear to increase disease severity despite an association with higher virus loads in respiratory secretions (Korber et al., 2020). Notably, several vaccines proved highly efficacious in phase 3 trials conducted while D614G was the dominant variant in the global pandemic (Baden et al., 2020; Polack et al., 2020; Voysey et al., 2021).
Newer variants with additional mutations are spreading rapidly in the UK (variant B.1.1.7, also called 20I/501Y.V1), South Africa (variant B.1.351, also called 20H/501Y.V2), Brazil (variant B.1.1.248, also called P.1 and 20J/501Y.V3), and California (variant B.1.429, also called Cal.20C and 452R.V1) (Figure S1; for daily updates of the global sampling of these variants, see GISAID’s “Tracking of Variants” page: https://www.gisaid.org/hcov19-variants/) (Tegally et al., 2020, Zhang et al., 2021, Naveca et al., 2021, Rambaut et al., 2020). Among them, the B.1.1.7 lineage of SARS-CoV-2 has caused public health concern because of its high rate of transmission in the UK (Rambaut et al., 2020). This variant contains 17 non-synonymous mutations, including the D614G mutation and 8 additional mutations in spike: ΔH69-V70, ΔY144, N501Y, A570D, P681H, T716I, S982A, and D1118H. Three B.1.1.7 spike mutations are of particular concern: a two-amino-acid deletion at position 69-70 of the N-terminal domain (NTD); N501Y, located in the receptor binding motif (RBM); and P681H, proximal to the furin cleavage site (Rambaut et al., 2020). Each of these three mutations are also found in other variants of interest. Epidemiological evidence and mathematical modeling data suggest the variant is more transmissible than the SARS-CoV-2 variants that were circulating prior to its introduction (Figure 1 ) (Davies et al., 2020; Public Health England, 2021; Galloway et al., 2021; Volz et al., 2021) and, though initially reported as not more pathogenic (Public Health England, 2020), evidence of increased mortality rate has also been reported (NERVTAG, 2021). As mutations in spike have potential to alter virus infectivity and/or susceptibility to neutralizing antibodies, one critical question is whether this B.1.1.7 variant will evade current vaccines, all of which are based on ancestral spike.
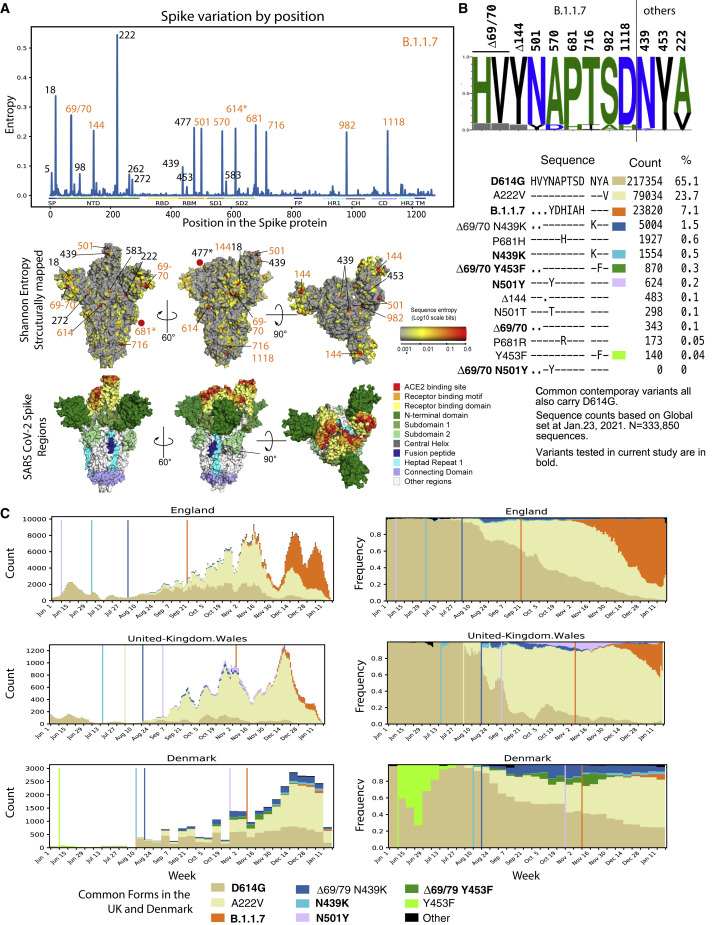
Figure 1.
Epidemiology tracing of mutations in B.1.1.7 and co-circulating relevant mutations in the UK and Danish SARS-CoV-2 epidemics
(A) Entropy scores summarizing the level of diversity found in positions in spike. These scores are dependent on sampling, and recent sampling from the UK and Denmark has been particularly intense relative to other regions of the world (Figure S1). B.1.1.7 mutations are highlighted in orange. The subset of B.1.1.7 sites with greater entropy scores (69/70, 681, and 501) are also often found in the context of other variants. The most variable site in spike is at 222 and is indicative of the GV clade. G614 has dominated global sampling since June 2020, and the entropy at 614 reflects presence of the ancestral form, D614, sampled in the early months of the pandemic. These same entropy scores are first mapped by linear position in the protein and then mapped onto the spike structure below the graph. Regions of spike are indicated by the same colors in the linear and structural maps.
(B) Frequencies of variants in relevant positions. Using the Analyze Align (AA) tool at cov.lanl.gov, we extracted the columns of interest for the B.117 spike mutations, and the additional sites of interest at 439, 453, and 222, out of a 333,850-sequence set extracted from GISAID on January 23, 2021. The logo at the top indicates the AA frequency in the full dataset; the gray boxes indicate deletions. All common forms of combinations of mutations at these sites of interest are shown, followed by their count and percentage. The forms that were common in the UK and Denmark are each assigned a color and used to map transition in frequencies of these forms over time in (C).
(C) Weekly running averages for each of the major variants in the UK and Denmark, based on the variants shown in (B), are plotted; the actual counts are on the left, and relative frequencies on the right. Some windows in time are very poorly sampled, some very richly. The vertical lines indicate when a variant is first sampled in a region. Note the lavender N501Y in Wales; this is N501Y found out of the context of B.1.1.7 and transient. The shifts in relative prevalence from the G clade (beige, D614G) to the GV clade (cream, A222V) to the B.1.1.7 variants (orange) are shown.
Here, we assessed the neutralization phenotype of the B.1.1.7 variant using convalescent sera, monoclonal antibodies (mAbs), and serum samples from phase 1 trials of an mRNA-based vaccine (mRNA-1273, Moderna) and a protein nanoparticle vaccine (NVX-CoV2373, Novavax). In addition, we characterized another two RBD mutations, N439K and F453Y, that showed limited circulation in both Denmark and the UK preceding the circulation of the B.1.1.7 variant; these RBD mutations are each most often found coupled with the same ΔH69-V70 that is in B.1.1.7.
Results
Rationale for testing B.1.1.7 variant and select subvariants
B.1.1.7 contains eight mutations in spike (Figure 1), and the lineage is associated with many additional mutations throughout the SARS-CoV-2 genome (Figure S2). Among the spike mutations, N501Y is suggested to increase RBD interaction with its cellular receptor, angiotensin-converting enzyme 2 (ACE2) (Santos and Passos, 2021), and has been shown to be critical for adaptation of SARS-CoV-2 to infect mice (Gu et al., 2020). N501Y has twice reached frequencies between 10%–20% in local populations as a single mutation in a D614G spike backbone (once in Wales, Figures 1C and S1, and also once in Victoria, Australia), but in these cases it did not persist. N501Y is also evident in a distinctive variant that is increasing in frequency in South Africa, 501Y.V2 (B.1.351), and accompanies other mutations in spike that can confer partial resistance to convalescent sera (Wang et al., 2021; Wibmer et al., 2021) and vaccine sera (Wang et al., 2021; Wu et al., 2021). A double deletion of amino acids H69-V70 in the N-terminal domain (NTD) of spike often co-occurs with one of three mutations in RBD: N501Y, N439K, or Y453F (Kemp et al., 2020). Y453F is associated with a mink farm outbreak in Denmark, with and without the presence of a ΔH69-V70 deletion (Kemp et al., 2020; van Dorp et al., 2020), but is also found in people in Denmark and the UK (Figure 1). N439K mutation usually occurs with ΔH69-V70, but occurs frequently without the ΔH69-V70 mutation as well. Y453F and N439K have been reported to escape neutralization by REGN10933 (Baum et al., 2020) and REGN10987 (Thomson et al., 2020) respectively, the two mAbs that comprise the REGN-COV2 cocktail regimen (Weinreich et al., 2021). Neither N439K nor Y453F is resistant to both of the REGN-COV2 mAbs, indicating involvement of the two sites in interactions with different RBD mAb species. N439K has also been reported to resist neutralization while maintaining virus fitness/infectivity (Li et al., 2020; Thomson et al., 2020). Another mutation of obvious concern in B.1.1.7 is P681H, proximal to the furin cleavage site (Figures 1C, S2, and S3) that has arisen many times independently (Figure S1) and has come to dominate the local epidemic in Hawaii.
Neutralization of variant B.1.1.7 by serum from convalescent individuals and vaccine recipients
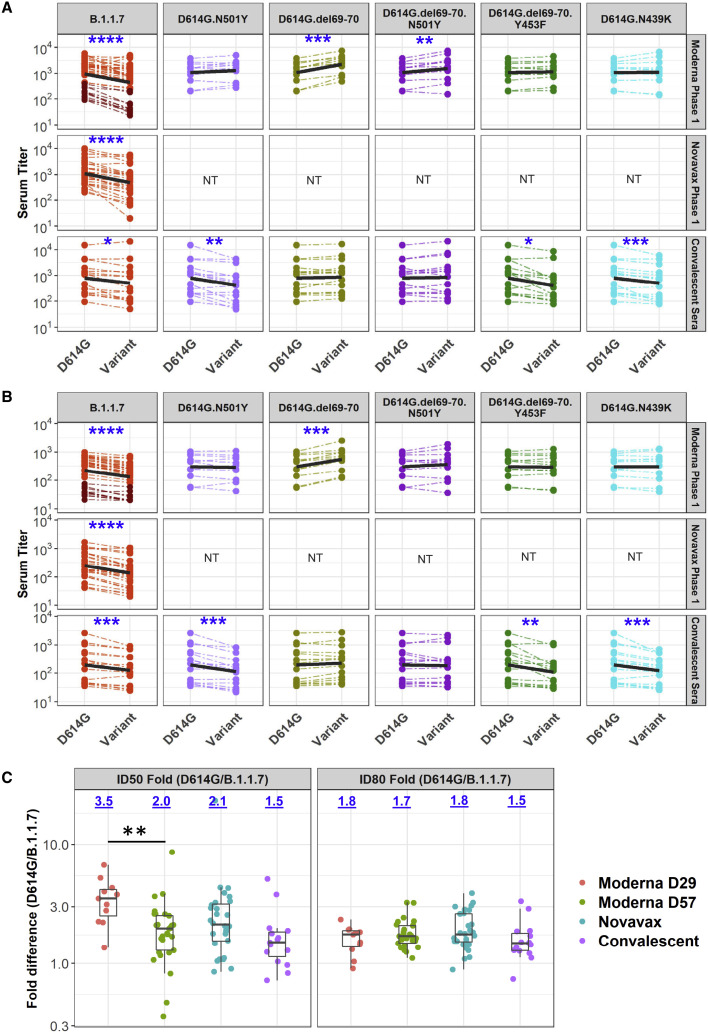
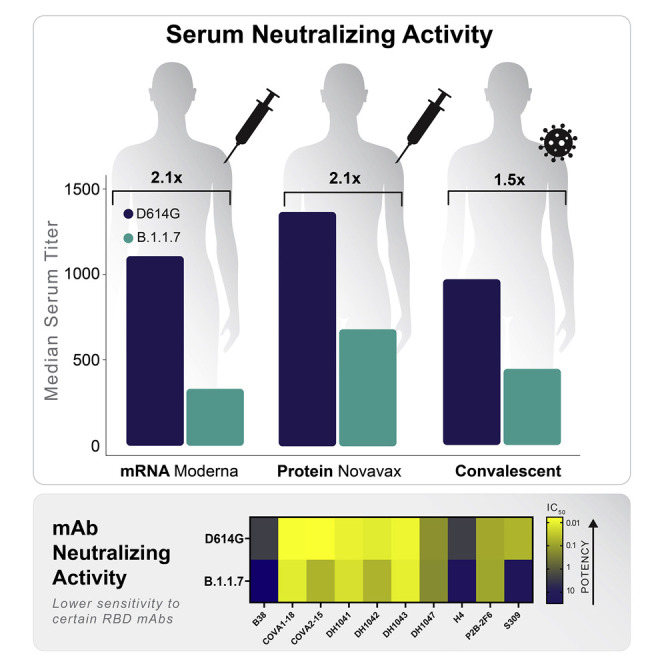
SARS-CoV-2 variant B.1.1.7 was compared to the D614G variant in neutralization assays with serum samples from 15 COVID-19 convalescent individuals, 40 recipients of the Moderna mRNA-1273 vaccine (11 samples from 29 days post-first inoculation, day 29; 29 samples from 28 days post-second inoculation, day 57), and 28 recipients of the Novavax spike protein nanoparticle vaccine NVX-CoV2373 (2 weeks post-second inoculation). Selection of NVX-CoV2373 vaccine serum samples was random and not pre-selected based on any selection criterion of anti-spike or neutralizing titers. The B.1.1.7 variant was neutralized by all vaccine sera, although with modestly diminished susceptibility compared to the D614G variant (Figures 2A and 2B). A modest decrease in neutralization susceptibility was also seen with convalescent sera, although not to the same extent seen with vaccine sera. Median ID50 titers of sera from both phase 1 vaccine trials were on average 2.1-fold lower against B.1.1.7 than against D614G (Table S1). The fold difference in ID50 titer ranged from 0.36 to 8.62 for Moderna sera, with an interquartile range (IQR) of 1.6 to 2.9. The fold difference in ID50 titers ranged from 0.85 to >20 for Novavax sera, with an IQR of 1.5 to 3.0. Median ID80 titers of sera from both phase 1 trials were on average 1.7-fold lower against B.1.1.7 than against D614G (Table S1), with a tighter range of fold difference compared to ID50. The fold difference in ID80 ranged from 0.91 to 3.21 for Moderna sera, with an IQR of 1.4 to 1.9. The fold difference in ID80 titer ranged from 0.89 to 3.98 for Novavax, with an IQR of 1.5 to 2.6. Convalescent sera showed an average of 1.5-fold (group median) lower ID50 titer against the B.1.1.7 variant (range 0.7 to 5.5; IQR = 1.1 to 1.8) and 1.5-fold (group median) lower ID80 titer (range 0.7 to 3.3; IQR = 1.3 to 1.8). The fold differences were statistically significant with p < 0.0001 for both ID50 and ID80 for Moderna and Novavax phase 1 sera, and p < 0.001 for ID80 of both sets of vaccine sera and the convalescent sera (Wilcoxon signed-rank test, paired, two-tailed; false discovery rate [FDR] corrected q values <0.1, corresponding to p < 0.064 in this study, were considered as significant) (Figures 2A and 2B; Table S2).
Figure 2.
Neutralization of variants by vaccine and convalescent sera
(A and B) Serum ID50 (A) and ID80 (B) titers of neutralization of each variant relative to D614G by vaccine sera (top 2 rows) and convalescent sera. Dashed thin lines represent individual samples; thick black lines represent geometric means of each sample group as indicated on the right. NT, not tested. Samples in dark and light red colors in the Moderna panel against B.1.1.7 are D29 and D57 samples, respectively. See also Table S1.
(C) Fold decline of ID50 (left) and ID80 (right) titers for each variant over D614G (D614G/variant) for each serum sample set as identified. Numbers on top of each plot show median fold differences. Upper and lower border of each box represent IQR of the fold differences, respectively, and the middle bars in boxes represent group median. Statistical significance of comparisons are indicated in all panels as ∗p < 0.05 (p < 0.064 corresponds to q < 0.1), ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Wilcoxon signed-rank paired test for (A and B); Wilcoxon rank-sum test for (C). See also Table S2.
Notably, sera with weaker neutralizing activity from the Moderna trial exhibited a more substantial reduction in activity against the B.1.1.7 variant. Because most low-titer samples in this trial were from day 29 (single inoculation), we compared the change in neutralization titers for day 29 and day 57 samples from the Moderna trial, as well as all samples from the Novavax trial, and from convalescent individuals (Figure 2C). The day 29 samples from the Moderna trial exhibited the greatest decrease in ID50 titer among the sample sets, and the decrease was significantly larger than the decrease for day 57 samples against the B.1.1.7 variant (Table S1; p = 0.0007), suggesting that antibody maturation can alleviate neutralization resistance.
Neutralization of additional variants by serum from convalescent individuals and vaccine recipients
To gain insights into whether the reduced neutralization susceptibility of the B.1.1.7 variant was due to a single spike mutation or a combination of two or more spike mutations, we characterized B.1.1.7 subvariants containing either N501Y alone, ΔH69-V70 alone, and a combination of N501Y+ΔH69-V70, each in the D614G background. We also tested ΔH69-V70+Y453F and N439K in the D614G background because ΔH69-V70 is commonly shared with these mutations; they were first identified in association with recent outbreaks in minks and zoonotic transmission to humans in Denmark. Due to limited supplies, sera from the Novavax phase 1 trial were not included in these assays. Interestingly, the ΔH69-V70 mutation rendered the virus more susceptible to neutralization by Moderna (mRNA-1273 vaccine) sera but not convalescent sera (Figure 2). Median ID50 and ID80 titers for Moderna sera were 2-fold higher against D614G.ΔH69-V70 than against D614G (p < 0.001), while titers of convalescent sera were comparable to D614G (Figures 2A and 2B; Table S2). N501Y had no significant impact on susceptibility to Moderna sera but did impart modest resistance to convalescent sera (Figure 2). Median ID50 and ID80 titers for convalescent sera against the N501Y variant were 2.2- and 1.8-fold lower (p < 0.01 and p < 0.001 for ID50 and ID80), while titers for Moderna sera were comparable to D614G (Figures 2A and 2B; Table S2). When both the ΔH69-V70 deletion and the N501Y mutation were present, the increase in susceptibility caused by ΔH69-V70 was diminished but still significant. No significant difference in neutralization titers was observed when both the ΔH69-V70 deletion and the N501Y mutation were present, except for a statistically significant (p < 0.01) but minimal 1.18-fold increase in ID50 titer of Moderna sera against D614G.ΔH69-V70.N501Y compared to D614G (Figures 2A and 2B; Table S2). The variant with both the ΔH69-V70 deletion and the Y453F mutation showed decreased neutralization susceptibility to convalescent sera, but not Moderna sera. Median ID50 and ID80 titers for convalescent sera were 1.7- and 1.5-fold lower against D614G.ΔH69-V70.Y453F than against D614G (p = 0.012 [q = 0.026] and p < 0.001, respectively) (Figures 2A and 2B; Table S2). When neutralization titers for variants with and without the Y453F mutation were compared, median ID50 titers of Moderna and convalescent sera were 2.1- and 3.6-fold lower, respectively, against D614G.ΔH69-V70.Y453F than against D614G.ΔH69-V70 (p < 0.001 and p < 0.0001, respectively) (Table S2). Median ID80 titers also were significantly lower for Moderna and convalescent sera against D614G.ΔH69-V70.Y453F than against D614G.ΔH69-V70 (1.8- and 2.5-fold; p < 0.01 and p < 0.001, respectively) (Table S2). Therefore, Y453F mutation can reverse the increased susceptibility conferred by the ΔH69-V70 mutation, demonstrating cooperative interactions between the RBD and NTD of spike. The D614G.N439K variant showed neutralization titers comparable to D614G for Moderna sera, and slight (1.6- and 1.4-fold) but significant (p < 0.001) decrease in ID50 and ID80 titers for convalescent sera (Figures 2A and 2B; Table S2).
Neutralization by mAbs
Neutralization of the B.1.1.7 variant and corresponding subvariants was assessed with a panel of RBD-targeting mAbs: DH1041, DH1042, DH1043, DH1047, B38, H4, P2B-2F6, COVA1-18, COVA2-15, and S309. The B.1.1.7 variant showed greatest resistance to mAbs B38, COVA2-15, and S309 (>10-fold difference in either IC50 or IC80 concentration compared to D614G) (Table 1 ). Resistance to B38 could not be fully explained by N501Y, ΔH69-V70, or the combination of these two mutations, whereas resistance to COVA2-15 and COVA1-18 was largely due to N501Y. Resistance to S309 was associated with N501Y, although this mutation alone accounted for only part of the resistance seen with the complete B.1.1.7 variant. The complete set of mutations and subsets of mutations in B.1.1.7 tested here had little (4.7-fold reduction in IC50 for DH1042 and H4) or no impact on other RBD antibodies (Table 1).
Table 1.
Neutralization of variants by mAbs
| Virus | Valuea | mAbs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B38 | COVA1-18 | COVA2-15 | DH1041 | DH1042 | DH1043 | DH1047 | H4 | P2B-2F6 | S309 | ||
| D614G | IC50 | 2.10 | 0.0060 | 0.0058 | 0.0094 | 0.011 | 0.0081 | 0.14 | 2.10 | 0.071 | 0.048 |
| IC80 | 12.00 | 0.031 | 0.028 | 0.036 | 0.068 | 0.040 | 0.54 | 18.00 | 1.20 | 0.69 | |
| D614G.B117 | IC50 | 30 (14.2x) | 0.011 (1.8x) | 0.05 (8.7x) | 0.015 (1.6x) | 0.052 (4.7x) | 0.01 (1.3x) | 0.17 (1.2x) | 9.9 (4.7x) | 0.075 (1.1x) | 9.2 (191.4x) |
| IC80 | >50 (> 4x) | 0.16 (5.1x) | 0.95 (34.5x) | 0.039 (1.1x) | 0.13 (1.9x) | 0.05 (1.3x) | 0.64 (1.2x) | >50 (> 3x) | 2.2 (1.8x) | >50 (> 72x) | |
| D614G.N501Y | IC50 | 4.7 (2.2x) | 0.032 (5.2x) | 0.084 (14.6x) | 0.01 (1.1x) | 0.012 (1.1x) | 0.015 (1.8x) | 0.072 (0.5x) | 3.7 (1.7x) | 0.043 (0.6x) | 0.15 (3.1x) |
| IC80 | >50 (> 4x) | 0.19 (6.1x) | 0.93 (33.8x) | 0.034 (1x) | 0.06 (0.9x) | 0.058 (1.5x) | 0.43 (0.8x) | 46 (2.5x) | 0.93 (0.8x) | >50 (> 72x) | |
| D614G.del69-70 | IC50 | 1.5 (0.7x) | 0.0042 (0.7x) | 0.0051 (0.9x) | 0.007 (0.8x) | 0.0095 (0.9x) | 0.0077 (0.9x) | 0.1 (0.7x) | 2.5 (1.2x) | 0.1 (1.4x) | 0.018 (0.4x) |
| IC80 | 9.9 (0.8x) | 0.026 (0.8x) | 0.026 (0.9x) | 0.029 (0.8x) | 0.053 (0.8x) | 0.033 (0.8x) | 0.5 (0.9x) | 26 (1.4x) | 1.1 (0.8x) | 18 (26.8x) | |
| D614G.del69-70.N501Y | IC50 | 2.5 (1.2x) | 0.026 (4.3x) | 0.051 (8.8x) | 0.0044 (0.5x) | 0.0061 (0.6x) | 0.0074 (0.9x) | 0.049 (0.4x) | 3.1 (1.5x) | 0.033 (0.5x) | 0.11 (2.2x) |
| IC80 | 40 (3.3x) | 0.18 (5.8x) | 11.6 (56.4x) | 0.025 (0.7x) | 0.039 (0.6x) | 0.035 (0.9x) | 0.28 (0.5x) | 30 (1.6x) | 1.0 (0.8x) | >50 (> 72x) | |
| D614G.del69-70.Y453F | IC50 | 1.2 (0.5x) | 0.0083 (1.4x) | 0.0055 (0.9x) | 0.0046 (0.5x) | 0.004 (0.4x) | 0.011 (1.4x) | 0.048 (0.3x) | 0.64 (0.3x) | 0.094 (1.3x) | 0.098 (2x) |
| IC80 | 15 (1.3x) | 0.043 (1.4x) | 0.03 (1.1x) | 0.027 (0.8x) | 0.03 (0.5x) | 0.05 (1.3x) | 0.24 (0.4x) | 5.7 (0.3x) | 1.3 (1x) | 11 (15.5x) | |
| D614G.N439K | IC50 | 1.3 (0.6x) | 0.0061 (1x) | 0.011 (1.9x) | 0.0075 (0.8x) | 0.0063 (0.6x) | 0.017 (2.1x) | 0.016 (0.1x) | 3.6 (1.7x) | 0.15 (2.2x) | 0.046 (0.9x) |
| IC80 | 19 (1.6x) | 0.049 (1.6x) | 0.1 (3.8x) | 0.035 (1x) | 0.07 (1x) | 0.059 (1.5x) | 0.33 (0.6x) | 42 (2.3x) | 1.6 (1.3x) | 0.38 (0.6x) | |
Numbers in parentheses followed by “x” represent fold differences over D614G. NA signifies data not available. >3-fold increase in IC50 or IC80 concentration over D614G represents moderate decrease in susceptibility. >10-fold increase in IC50 or IC80 concentration over D614G represents substantial decrease in susceptibility. <0.33-fold of the IC50 or IC80 of D614G represents moderate increase in susceptibility.
Unit of IC50 and IC80 concentrations is μg/mL.
The mAbs were largely unaffected by the Y453F and N439 mutations (Table 1). A modest increased sensitivity was seen in two cases: DH1047 assayed against D614G.N439K, and H4 assayed against D614G.ΔH69-V70.Y453F. The latter variant also exhibited partial resistance to S309, which was mostly seen at IC80, and the level of change was similar to that caused by the ΔH69-V70 deletion alone, indicating that the deletion is the cause of the decreased susceptibility rather than the Y453F.
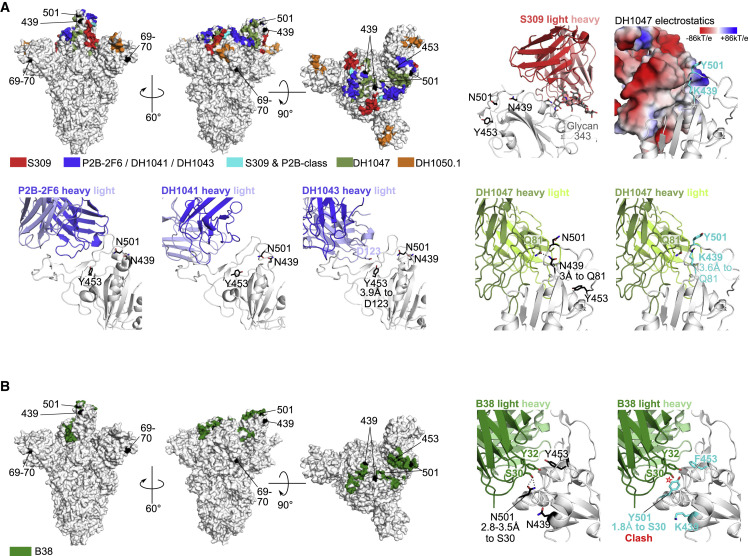
We used structural analyses to understand the molecular mechanisms of mAb neutralization resistance or lack thereof. Mapping of the mAb epitopes (<4Å) on the spike trimer showed that the RBD mutations were within, or close to, the epitopes of all RBD antibodies, while ΔH69-V70 is in the NTD (Figure 3 ). N501Y and Y453F are in close proximity to the B38 paratope. Modeling of the N501Y mutation showed a potential clash between Y501 in spike and S-30 in B38 light chain, consistent with the neutralization resistance of N501Y to B38. mAbs P2B-2F6, DH1041, and DH1043 had very similar epitopes and were grouped together (P2B-class in Figure 3A; also similar to class 2 RBD mAbs) (Barnes et al., 2020). The RBD mutations were further from the P2B-2F6 and DH1041 paratopes, explaining the lack of impact of these mutations. Y453 is close (3.9Å) to the DH1043 paratope but with no predicted polar interactions. Structural modeling of N439K suggests a potentially more favorable interaction as Lys interacts with a negatively charged patch on the DH1047 surface. Although N501 is close to the DH1047 paratope, its side chain is oriented away from the mAb, suggesting no substantial impact due to N501Y. Y453 is also in close proximity to the B38 paratope, but structural modeling showed no substantial impact.
Figure 3.
Structural analyses for antibody resistance mutations
(A) Three top left panels: full spike trimer with antibody epitopes for S309, P2B-2F6, DH1041, DH1043, DH1047, and DH1050.1. Epitopes for P2B-2F6, DH1041, and DH1043 are similar and are grouped together. Top row, second from right panel shows location of spike sites 439, 453, and 501 with respect to S309. These spike sites are not close to S309 (>11Å). Top row rightmost panel shows the DH1047 antibody colored according to vacuum electrostatic potential and the modeled mutations at spike sites Lys-439 and Tyr-501. Bottom row, rightmost two panels: DH1047 interaction with sites 439, 453, and 501 using wild-type amino acids (second from right) and modeled mutations (rightmost). Bottom row, three left panels: the location of spike sites 439, 453, and 501 with P2B-2F6, DH1041, and DH1043. Polar interactions between antibody and spike residues of interest are shown with dotted black lines.
(B) Similar to (A), except with B38 antibody. The modeled Tyr-501 is predicted to clash with light chain Ser-30 (~1.8Å, red star).
The decreased neutralization observed for S309 by ΔH69-V70 and N501Y could not be explained by structural analyses. Both the RBD mutations and ΔH69-V70 are distal from S309 (>11Å), suggesting allosteric interactions. Notably, S309 is the only mAb in this study that interacts strongly with a glycan (at site 343), and changes in spike dynamics or conformations can impact glycan processing (Wagh et al., 2020).
Discussion
Recent months have seen the emergence of a growing number of novel SARS-CoV-2 variants that can rapidly and repeatedly shift in prevalence in local populations and even globally (Hodcroft et al., 2020; Korber et al., 2020). Newer variants carry multiple spike mutations (Naveca et al., 2021; Rambaut et al., 2020; Tegally et al., 2020) that are a potential concern for immune escape. The B.1.1.7 variant studied here was first detected in England in September 2020, where it rapidly came to dominate the regional pandemic and has now been detected in over 70 countries. Variants in the UK and Denmark followed a shifting dynamic, starting with the emergence of the G clade as the dominant form, followed by increasing prevalence of the GV clade that mirrored across Europe (Hodcroft et al., 2020), and then regional appearance of variants that carried combinations of ΔH69-V70 with Y453F and N439K, finally to be followed by the introduction of the B.1.1.7 variant, which rapidly rose to dominance in the UK and is now beginning to increase in frequency in Denmark. The serial waves of variant prevalence in these two countries suggest complex dynamics that may come into play as SARS-CoV-2 continues to evolve. Furthermore, co-circulation of major variants in a geographically local region may enable recombination (Varabyou et al., 2020), bringing together mutations that enhance fitness either through infectivity or immunological resistance.
Prior to the emergence of this variant, two SARS-CoV-2 vaccines based on ancestral spike proved highly effective and recently received emergency use authorization, including the Moderna mRNA-1273 vaccine studied here (Baden et al., 2020) and a similar mRNA vaccine developed by Pfizer/BioNTek (Polack et al., 2020). Another vaccine based on ancestral spike nanoparticles developed by Novavax (Keech et al., 2020) is currently undergoing phase 3 testing in the UK, USA, and Mexico, with phase 2 a/b testing ongoing in South Africa. In addition to vaccines, several potent RBD-specific mAbs have received emergency use authorization for treatment of mild-to-moderate COVID-19 in the USA (FDA press release, November 21, 2020 and November 9, 2020), while still other therapeutic mAbs are in development (also see recent announcement) (Tuccori et al., 2020).
Here, we show in a lentivirus-based pseudovirus assay that variant B.1.1.7 exhibits only modestly reduced neutralization susceptibility in the presence of convalescent sera (1.5-fold average) and sera from the Moderna and Novavax phase 1 studies (2-fold average after two inoculations) using the prototypic D614G variant as comparator. Our data on the Moderna mRNA vaccine sera are in agreement with a recent study testing B.1.1.7 using sera from a similar mRNA vaccine produced by Pfizer (Muik et al., 2021). While it is not known for certain what level of neutralization is required for the remarkable efficacy in phase 3 studies completed to date, it is noteworthy that both the Moderna and Pfizer/BioNTech mRNA vaccines demonstrated substantial efficacy prior to the second (final) dose (Baden et al., 2020; Polack et al., 2020). Neutralization titers have been shown to increase by approximately 10-fold after the second dose for both vaccines (Anderson et al., 2020; Jackson et al., 2020; Walsh et al., 2020), suggesting that a 2-fold reduction in neutralization will have minimal impact on vaccine efficacy in people who receive both doses of vaccine. The recent finding that the Novavax vaccine was 95.6% effective against the common variant and 85.6% effective against B.1.1.7 (Callaway and Mallapaty, 2021) is consistent with our results. Receiving the second dose in a timely manner is encouraged for maximum efficacy in regions where the B.1.1.7 variant circulates. In addition, the three RBD mutations N439K, Y453F, and N501Y showed greater impact on neutralization by convalescent sera than by vaccine sera (only Moderna sera were tested against partial variants containing single RBD mutations), suggesting that the mRNA vaccine is more robust in tolerating isolated RBD mutations than natural infections.
In contrast to our findings with polyclonal sera from convalescent individuals and vaccine recipients, the B.1.1.7 variant exhibited markedly reduced susceptibility to a subset of RBD-specific mAbs. Partial escape from four mAbs (COVA1-18, COVA2-15, S309, and to a lesser extent, B38) was associated with the N501Y mutation. Modest escape from two additional mAbs (DH1042 and H4) could not be mapped with the specific mutations tested. Notably, B.1.1.7 exhibited no escape from four RBD-specific mAbs tested here (DH1041, DH1043, DH1047, and P2B-2F6).
In summary, our findings indicate that B.1.1.7 is not a neutralization escape variant of concern for vaccine efficacy and the risk of reinfection. In addition, although the variant is considerably less susceptible to certain mAbs, other RBD-specific mAbs retain full activity. While this is encouraging, it is becoming increasingly clear that SARS-CoV-2 continues to evolve and that new variants may arise that pose a greater risk for immune escape. Early identification and characterization of newly emerging variants requires robust genetic surveillance coupled with rapid laboratory and clinical investigation to facilitate the timely design and testing of next generation vaccines and therapeutic mAbs, should they be needed.
Limitations of study
Potential drawbacks to our studies include the following. (1) we do not have age and gender information for all samples due to the need for remaining blinded for ongoing work. Nevertheless, we do have age and sex information for the set of Moderna samples used for testing B.1.1.7. We observed comparable level of change in neutralization titers among the age groups and between genders against the variant (Table S1). (2) The neutralization assay we performed utilizes pseudoviruses rather than live viruses. Pseudovirus neutralization assay is deemed a valuable assay for evaluating clinical study samples due to its high throughput, level of formal qualification/validation, ease of incorporating spike variants, and biosafety advantages. Highly significant correlations have been reported between pseudovirus and live virus neutralization assays in evaluating antibody responses against SARS-CoV-2 (Schmidt et al., 2020).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DH1041, DH1042, DH1043, and DH1047 | Kevin Saunders & Barton Haynes, Duke | Li et al., 2021 |
| B38 and H4 | Peter Kwong, VRC/NIH | Wu et al., 2020 |
| P2B-2F6 | Peter Kwong, VRC/NIH | Ju et al., 2020 |
| S309 | Peter Kwong, VRC/NIH | Pinto et al., 2020 |
| Critical commercial assays | ||
| Luciferase Cell Culture Lysis 5x Reagent | Promega | Cat# E1531 |
| Bright-Glo Luciferase Assay System | Promega | Cat# #2650 |
| Deposited data | ||
| Tables S1, S2, and S3 | This paper | https://data.mendeley.com/datasets/ty33r9g972/1 |
| Additional dilution data | This paper | https://data.mendeley.com/datasets/stwvcrkswf/1 |
| Experimental models: Cell lines | ||
| 293T/ACE2 cells | Drs. Mike Farzan and Huihui Mu at Scripps | 293T/ACE2 cells |
| HEK293T/17 | ATCC | CRL-11268 |
| Oligonucleotides | ||
| Primers for site-direct mutegenesis, see Table S3. | This paper | N/A |
| Software and algorithms | ||
| R (Version 3.6.1) | The R Foundation for Statistical Computing | http://www.r-project.org/ |
| R packages: tidyverse (Version 1.2.1); dplyr (Version 0.8.5); ggplot2 (Version 3.3.0); coin (1.3-1) | The R Foundation for Statistical Computing | https://cran.r-project.org/ |
| Python (Version 3.4.2) package | Python | https://github.com/nfusi/qvalue |
| PyMOL (Version 2.0) | PyMOL Molecular Graphics System | Pymol.org |
| Sequencher (Version 5.4.6) | Gene Codes Corporation | www.Genecodes.com |
| SnapGene (Version 5.2.4) | GSL Biotech LLC | www.snapgene.com |
Resource availability
Lead contact
Further information and requests for supporting data, resources, and reagents should be directed to and will be fulfilled upon request by the Lead Contact: David Montefiori (monte@duke.edu).
Materials availability
Reagents from this study are available upon request.
Data and code availability
Neutralization dilution data have been deposited to Mendeley Data: https://data.mendeley.com/datasets/stwvcrkswf/1.
Experimental model and subject details
Human
Moderna phase 1 study (NCT04283461) is a phase I, open-label, dose-ranging clinical trial in healthy males and non-pregnant females, starting at 18 years of age testing the mRNA-1273 vaccine. mRNA-1273 is a lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine that encodes for a full-length, prefusion stabilized spike (S) protein of SARS-CoV-2. Trial subjects received 2 vaccine immunization on day 1 and day 29 of the study.
Novavax phase 1 study (NCT04368988) is a 2-part, randomized, observer-blinded, placebo-controlled, Phase 1/2 trial. The study tested a SARS-CoV-2 recombinant nanoparticle vaccine with or without Matrix-M adjuvant (Keech et al., 2020) in health volunteered age between 18 and 84 years.
HVTN 405/HPTN 1901 is an observational cohort study HVTN 405/HPTN 1901 (NCT04403880). Participants were followed starting at 1-8 weeks post resolution of COVID-19 or 2-10 weeks post most recent positive SARS-CoV-2 test, if asymptomatic, and then 2 months, 4 months, and 1 year later (optional).
Ethics statement
Clinical trials described in this manuscript were approved by the appropriate Institutional Review Boards (IRBs). Informed consent was obtained from all subjects in the trials.
Method details
Serum samples
Sera for the mRNA-1273 phase 1 study (NCT04283461) were obtained from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases for the mRNA-1273 phase 1 study team and Moderna Inc. The phase 1 study protocols and results are reported previously (Anderson et al., 2020; Jackson et al., 2020). The phase 1 trial tested the identical vaccine (mRNA-1273), dose (100 μg) and schedule as used in the Moderna phase 3 (NCT04470427). mRNA-1273 is a lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine that encodes for a full-length, prefusion stabilized spike protein of SARS-CoV-2 (Baden et al., 2020). Samples tested against the B.1.1.7 variant (together with D614G as control) were collected at day 29 (4 weeks post 1st inoculation) or day 57 (4 weeks post 2nd inoculation). Samples tested against the subvariants (together with D614G as control) were all from day 57.
Novavax phase 1 sera were obtained from Novavax. The phase 1 study (NCT04368988) tested a 5 μg dose of SARS-CoV-2 recombinant nanoparticle vaccine with or without 50 μg of Matrix-M adjuvant (Keech et al., 2020). Serum samples (N = 28) tested here were from the vaccine arm with the Maxtrix-M adjuvant, which is the identical vaccine in the ongoing Novavax global phase 3 study (NC04611802). Samples tested represent the entire Phase 1 vaccine cohort and were not pre-selected for higher titer responses at 2 weeks post 2nd inoculation (day 35).
Convalescent sera were collected in an observational cohort study conducted by the HIV Vaccine Trial Network and the HIV Prevention Trials Network (protocol HVTN 405/HPTN 1901; NCT04403880). Samples were collected from the first visit of the study, scheduled at 1-8 weeks post resolution of COVID-19, or 2-10 weeks post most recent positive SARS-CoV-2 test, if asymptomatic. The subset of samples included in this study were pre-selected as representing high, medium and low neutralization titers against the D614G variant of SARS-CoV-2.
MAbs
Antibodies B38, H4, P2B-2F6, and S309 (Ju et al., 2020; Pinto et al., 2020; Wu et al., 2020), were provided by Dr. Peter Kwong. Antibodies DH1041, DH1042, DH1043, and DH1047 were provided by Drs. Kevin Saunders, Dapeng Li, and Barton Haynes (Li et al., 2021). Antibodies COVA1-18 and COVA2-15 were provided by Dr. Rogier Sanders (Brouwer et al., 2020).
Cells
HEK293T/17 cells (ATCC cat. no. CRL-11268) and 293T/ACE2.MF (provided by Drs. Michael Farzan and Huihui Mu) were maintained in 12 mL of growth medium (DMEM, 10% heat-inactivated fetal bovine serum, 50 μg gentamicin/mL, 25mM HEPES) in T-75 culture flasks in a humidified 37°C, 5% CO2 environment. Puromycin (3 μg/mL) was added to the growth medium for maintaining 293T/ACE2.MF cells. Cells were split at confluency using TrypLE Select Enzyme solution (Thermo Fisher Scientific).
Pseudotyped virus production
SARS-CoV-2 spike-pseudotyped viruses were prepared and titrated for infectivity essentially as described previously (Korber et al., 2020). An expression plasmid encoding codon-optimized full-length spike of the Wuhan-1 strain (VRC7480), was provided by Drs. Barney Graham and Kizzmekia Corbett at the Vaccine Research Center, National Institutes of Health (USA). Mutations were introduced into VRC7480 by site-directed mutagenesis (Weissman et al., 2021) using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (Catalog # 210518) using primers as listed in Table S3. All mutations were confirmed by full-length spike gene sequencing by Sanger Sequencing, using Sequencher and SnapGene for sequence analyses. Pseudovirions were produced in HEK293T/17 cells (ATCC cat. no. CRL-11268) by transfection using Fugene 6 (Promega Cat#E2692) and a combination of spike plasmid, lentiviral backbone plasmid (pCMV ΔR8.2) and firefly Luc reporter gene plasmid (pHR’ CMV Luc) (Naldini et al., 1996) in a 1:17:17 ratio in Opti-MEM (Life Technologies). Transfection mixtures were added to pre-seeded HEK293T/17 cells in T-75 flasks containing 12 mL of growth medium and incubated for 16-20 h at 37°C. Medium was removed and 15 mL of fresh growth medium added. Pseudovirus-containing culture medium was collected after an additional 2 days of incubation and clarified of cells by low-speed centrifugation and 0.45 μm micron filtration.
TCID50 assays were performed prior to freezing aliquots of the viruses at −80°C. Viruses were serially diluted 3-fold or 5-fold in quadruplicate for a total of 11 dilutions in 96-well flat-bottom poly-L-lysine-coated culture plates (Corning Biocoat). An additional 4 wells served as background controls; these wells received cells but no virus. Freshly suspended 293T/ACE2.MF cells were added (10,000 cells/well) and incubated for 66-72 h. Medium was removed by gentle aspiration and 30 μl of Promega 1X lysis buffer was added to all wells. After a 10 min incubation at room temperature, 100 μl of Bright-Glo luciferase reagent was added to all wells, mixed, and 105 μl of the mixture was added to a black/white plate (Perkin-Elmer). Luminescence was measured using a GloMax Navigator luminometer (Promega). TCID50 was calculated using the method of Reed and Muench as described (Johnson and Byington, 1990).
Neutralization assay
Neutralization was measured in a formally validated assay that utilized lentiviral particles pseudotyped with SARS-CoV-2 spike and containing a firefly luciferase (Luc) reporter gene for quantitative measurements of infection by relative luminescence units (RLU). A pre-titrated dose of virus was incubated with 8 serial 5-fold dilutions of serum samples in duplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottom poly-L-lysine-coated culture plates. Cells were detached using TrypLE Select Enzyme solution, suspended in growth medium (100,000 cells/mL) and immediately added to all wells (10,000 cells in 100 μL of growth medium per well). One set of 8 wells received cells + virus (virus control) and another set of 8 wells received cells only (background control). After 66-72 h of incubation, medium was removed by gentle aspiration and 30 μl of Promega 1X lysis buffer was added to all wells. After a 10 min incubation at room temperature, 100 μl of Bright-Glo luciferase reagent was added to all wells. After 1-2 min, 110 μl of the cell lysate was transferred to a black/white plate. Luminescence was measured using a GloMax Navigator luminometer (Promega). Neutralization titers are the inhibitory dilution (ID) of serum samples, or the inhibitory concentration (IC) of mAbs at which RLUs were reduced by either 50% (ID50/IC50) or 80% (ID80/IC80) compared to virus control wells after subtraction of background RLUs. Serum samples were heat-inactivated for 30 min at 56°C prior to assay. This pseudotyped neutralization assay has been formally validated and reviewed by FDA for evaluation of phase 3 clinical trial samples. In addition, all assays were performed in compliance with GCLP guidelines.
Phylogenetic trees
The tree in Figure S1 is based on the GISAID data sampled on Jan. 17th, 2021, and passed through a quality control filter, and presented as the “tree of the day” at the cov.lanl.gov website https://cov.lanl.gov/components/sequence/COV/rainbow.comp. The “full” alignment was used as described previously (Korber et al., 2020). Only the mutations of interest for this study are tracked in this tree for clarity. The tree is rooted using the Wuhan-Hu-1 isolate (GenBank accession NC_045512).
All Phylogenetic trees are constructed using parsimony, TNT version 1.5 (Goloboff and Catalano, 2016), with 5 or 10 random-sequence addition replicates with TBR (tree-bisection-reconnection) branch swapping (command: “mult = rep REPS tbr hold 1 wclu 1000,” where REPS equals 5 or 10, with the bbreak cluster value set to 40).
Structural analyses
We used PDB: 7C2L (Chi et al., 2020) for the full trimeric spike structure, and antibody spike complex structures from Li et al. (Li et al., 2021) for DH1041-DH1047 antibodies, PDB: 6WPS (Pinto et al., 2020) for S309, PDB: 7BWJ (Ju et al., 2020) for P2B-2F6, and PDB: 6XDG for REGN antibodies (Hansen et al., 2020). Antibody epitopes were defined as spike amino acids with any heavy atoms within 4Å antibody heavy atoms (Barnes et al., 2020). Antibody epitope, electrostatics and polar bonds calculations as well as mutation modeling were performed in PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). Mutations were modeled with spike or RBD in isolation and the rotamers with least predicted strain in PyMOL were used. For B38, to identify the most amenable Y-501 rotamers, the N501Y mutation was modeled with the antibody-RBD complex; however, all identified rotamers induced substantial clashes and the rotamer with the least clash was retained. PyMOL was also used for structural renderings for all figures.
Quantification and statistical analysis
Neutralization ID50 titers or IC50 concentrations between each variant and D614G, or between other pairs of variants with and without the N501Y or ΔH69-V70 were compared using the Wilcoxon signed-rank 2-tailed test. Decrease in ID50 and ID80 titer for Moderna Day 29 and Day 57 samples against B.1.1.7 were compared using the Wilcoxon rank-sum test, 2-tailed. To correct for multiple test corrections, false discovery rates (FDR or q values) were calculated as in (Storey and Tibshirani, 2003) implemented in a Python package (https://github.com/nfusi/qvalue) for Python version 3.4.2. All tests with q < 0.1 were considered as significant, which corresponded to p < 0.042. Wilcoxon signed-rank test and Wilcoxon rank-sum test were performed using the coin package (version 1.3-1) with R (version 3.6.1). Wilcoxon signed-rank test and Wilcoxon rank-sum test were performed using the coin package (version 1.3-1) with R (version 3.6.1).
Acknowledgments
We thank the HIV Vaccine Trials Network and HIV Prevention Trial Network for serum samples from COVID-19 convalescent individuals. We thank Peter Kwong for generously sharing the mAbs B38, H4, P2B-2F6, and S309 produced at the Vaccine Research Center, NIH. We also thank Jin Tong, Elize Domin, Wenhong Feng, and Miroslawa Bilska for excellent technical assistance. We thank Francesca Suman for assistance with graphic design. Original data and specimens for Protocol 20-0003 were supported by the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases. K.W. and B.K. were supported by LANL LDRD 20190441ER. D.C.M., X.S., H.T., and C.M. were supported by the COVID-19 Prevention Network (CoVPN) and the National Institutes of Health. D.L., B.F.H., and K.O.S. were supported by a grant from the State of North Carolina from federal CARES Act funds, and NIAID grant AI142596.
Author contributions
D.C.M. designed the study, coordinated assays, data analysis, and manuscript preparation, and helped write and edited the manuscript. X.S. helped with study design, coordinated the study, performed data analysis and visualization, and wrote the manuscript. H.T. participated in study design, site-directed mutagenesis, data generation, and manuscript writing and editing. C.M. participated in data generation and reviewing, and manuscript review and editing. D.L. and K.O.S. produced, purified, and provided mAbs, and edited the manuscript. B.F.H. provided mAbs and edited the manuscript. N.H. contributed data, data visualizations, and to manuscript preparation. B.K. helped with study design, generated data visualizations, and contributed to data interpretation and manuscript preparation. J.T., H.Y., and W.F. also helped generate data visualizations. K.W. provided structural analyses and helped with data interpretation and manuscript preparation. .
Declaration of interests
R.P. is an employee of Moderna, Inc. G.S. and G.M.G. are employees of Novavax, Inc.
Published: March 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.03.002.
Supplemental information
References
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas N.K., Majumder P.P. Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J. Med. Res. 2020;151:450–458. doi: 10.4103/ijmr.IJMR_1125_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Mallapaty S. Novavax offers first evidence that COVID vaccines protect people against variants. Nature. 2021;590:17. doi: 10.1038/d41586-021-00268-9. [DOI] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J., Pearson C.A.B., Russell T.W., Tully D.C., Abbott S., Gimma A., et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv. 2020 doi: 10.1101/2020.12.24.20248822. [DOI] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff P., Catalano S. TNT, version 1.5, with a full implementation of phylogenetic morphometrics. Cladistics. 2016 doi: 10.1111/cla.12160. [DOI] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D., Vaughan T.G., Comas I., Candelas F.G., Stadler T., et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel S., Graña-Miraglia L., Gutierrez J.M., Bundalovic-Torma C., Groves H.E., Isabel M.R., Eshaghi A., Patel S.N., Gubbay J.B., Poutanen T., et al. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci. Rep. 2020;10:14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.A., Byington R.E. In: Techniques in HIV Research. Aldovani A., Walker B.D., editors. Stockton Press; New York, NY: 1990. Infectivity assay (virus yield assay) pp. 71–76. [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S., Datir R., Collier D., Ferreira I., Carabelli A., Harvey W., Robertson D., Gupta R. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion DH69/V70. bioRxiv. 2020 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. bioRxiv. 2021 doi: 10.1101/2020.12.31.424729. [DOI] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci O., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021 doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Naveca F., Nascimento V., Souza V., Corado A., Nascimento F., Silva G., Costa A., Duarte D., Pessoa K., Gonçalves L., et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. 2021. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585
- NERVTAG NERVTAG paper on COVID-19 variant of concern B.1.1.7. 2021. https://www.gov.uk/government/publications/nervtag-paper-on-covid-19-variant-of-concern-b117
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 strain in Southern California, USA. medRxiv. 2021 doi: 10.1101/2021.01.18.21249786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (2020). Public Health England: Investigation of novel SARS-COV-2 variant Variant of Concern 202012/01, P.H. England, ed. (Technical briefing 2: December 28, 2020). https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959361/Technical_Briefing_VOC202012-2_Briefing_2.pdf.
- Public Health England (2021). Investigation of novel SARS-COV-2 variant Variant of Concern 202012/01 (Technical briefing 3: January 8, 2021). P.H. England, ed. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959360/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3.pdf
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- Santos J.C., Passos G.A. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv. 2021 doi: 10.1101/2020.12.29.424708. [DOI] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. bioRxiv. 2020 doi: 10.1101/2020.11.04.355842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuccori M., Ferraro S., Convertino I., Cappello E., Valdiserra G., Blandizzi C., Maggi F., Focosi D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. MAbs. 2020;12:1854149. doi: 10.1080/19420862.2020.1854149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Tan C.C., Lam S.D., Richard D., Owen C., Berchtold D., Orengo C., Balloux F. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv. 2020 doi: 10.1101/2020.11.16.384743. [DOI] [Google Scholar]
- Varabyou A., Pockrandt C., Salzberg S.L., Pertea M. Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie. bioRxiv. 2020 doi: 10.1101/2020.09.21.300913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh K., Hahn B.H., Korber B. Hitting the sweet spot: exploiting HIV-1 glycan shield for induction of broadly neutralizing antibodies. Curr. Opin. HIV AIDS. 2020;15:267–274. doi: 10.1097/COH.0000000000000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Lihong L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.-G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., van den Berg K., Rossouw T., Boswell M., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. [DOI] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Neutralization dilution data have been deposited to Mendeley Data: https://data.mendeley.com/datasets/stwvcrkswf/1.