Abstract
The pregnane X receptor (PXR, NR1I2) is a xenobiotic-sensing nuclear receptor that defends against toxic agents. We have shown that PXR promotes chronic ethanol (EtOH)–induced steatosis. Therefore, we examined the role of PXR in binge EtOH-induced hepatotoxicity. Male wild-type (WT) and Pxr-null mice were orally administered three binge doses of EtOH (4.5 g/kg, every 12 hours) and were euthanized 4 hours after the final dose. Pxr-null mice displayed higher basal mRNA levels of hepatic lipogenic transcription factor sterol regulatory element binding protein 1c (Srebp-1c) and its target stearoyl-CoA desaturase 1 and the lipid peroxide–detoxifying aldo-keto reductase 1b7 (Akr1b7) and higher protein levels of EtOH-metabolizing alcohol dehydrogenase 1 (ADH1). In both genotypes, binge EtOH-induced triglyceride accumulation was associated with inhibition of fatty acid β-oxidation and upregulation of Srebp-1c–regulated lipogenic genes and hepatic CYP2E1 protein. Unexpectedly, gene expression of Cyp2b10, a constitutive androstane receptor target gene implicated in EtOH hepatotoxicity, was PXR-dependent upregulated by binge EtOH. In addition, in WT mice, binge EtOH-induced inhibition of hepatic Akr1b8 mRNA, and protein levels of aldehyde dehydrogenase 1A1 and antiapoptotic Bcl-2 but increased proapoptotic Bax protein expression, leading to increases in residual EtOH concentration and the cellular oxidative stress marker, malondialdehyde. In contrast, Pxr-null mice displayed increased Akr1b7 gene and ADH1 protein expression and hypertriglyceridemia after binge EtOH exposure. Taken together, this study demonstrates that PXR ablation prevents EtOH-induced upregulation of Cyp2b10 and that PXR potentiates binge EtOH-induced oxidative stress and inhibition of EtOH catabolism but protects against alcoholic hyperlipidemia.
Introduction
Alcoholic liver disease (ALD) is a common pathology associated with excessive alcohol use. Despite intense research efforts to unravel the molecular basis of ethanol (EtOH)–induced liver damage, there are currently no Food and Drug Administration–approved treatments for ALD. Interestingly, published reports have implicated the involvement of ligand-activated transcription factors called nuclear receptors in ALD (Pang et al., 2009; Gyamfi and Wan, 2010; Chen et al., 2011; Wu et al., 2014).
The pregnane X receptor (PXR, NR1I2) is a xenobiotic-sensing nuclear receptor that coordinates the metabolic response to drugs and toxic agents and is highly expressed in the liver and intestine (Pavek and Dvorak, 2008). A recent report established that loss of PXR is protective against chronic EtOH-induced hepatic triglyceride accumulation and steatosis, whereas PXR signaling aggravates hepatotoxicity (Choi et al., 2018). Chronic EtOH-induced hepatotoxicity in wild-type (WT) mice was associated with gene induction of transcription factors involved in lipogenesis and upregulation of Pxr, constitutive androstane receptor (Car), and CAR target gene Cyp2b10 (Choi et al., 2018). Importantly, PXR deficiency suppressed these changes. Similar to the effect of chronic EtOH on WT mice, activation of mouse CAR by 1,4-bis[2-(3,5-dichloropyridyloxy)] benzene and EtOH infusion enhanced both acute and chronic EtOH-induced hepatotoxicity (Chen et al., 2011; Wang et al., 2017). Furthermore, EtOH can increase both PXR and its target Cyp3a11/CYP3A gene in rodents, in human primary hepatocytes, and in the liver of human alcoholics (Kostrubsky et al., 1995; Niemelä et al., 2000; He et al., 2006). Taken together, it appears that PXR/CAR crosstalk mediates and exacerbates EtOH-induced steatosis and ALD pathogenesis.
Intriguingly, both ligand PXR activation and PXR knockdown in mice, human hepatoma HepG2 cells, and human hepatocytes led to hepatic lipid accumulation (Nakamura et al., 2007; Moreau et al., 2009; Spruiell et al., 2014b; Bitter et al., 2015). However, the underlying mechanisms involved in the dual role of PXR activation and ablation in steatosis are still being unraveled. Interestingly, steatosis induced by PXR activation in mice implicated the involvement of sterol regulatory element binding protein (SREBP)–independent activation of the free fatty acid uptake transporter CD36 and several lipogenic enzymes, including stearoyl-CoA desaturase 1 (SCD1) and long-chain free fatty acid elongase, as well as suppression of peroxisome proliferator–activated receptor a (PPARa) target genes (Zhou et al., 2006; Nakamura et al., 2007). In contrast, human PXR activation by rifampicin in HepG2 cells was reported to involve the SREBP1-dependent lipogenic pathway (Bitter et al., 2015), suggesting species differences in PXR ligand-induced steatosis. However, ligand-dependent PXR activation and steatosis induction in primary human hepatocytes did not involve CD36, but upregulation of the thyroid hormone responsive SPOT 14 homolog, which activates lipogenic fatty acid synthase (FAS) expression (Moreau et al., 2009).
On the other hand, PXR ablation results in increased hepatic lipid accumulation in untreated Pxr-null mice, possibly involving upregulation of the Srebp1c target Scd1 (Nakamura et al., 2007; Spruiell et al., 2014b). Furthermore, PXR knockdown in HepG2 cells also resulted in lipid accumulation; however, it was not associated with SCD1 upregulation but instead increased expression of aldo-keto reductase 1B 10 (AKR1b10), which has been implicated in steatosis and steatohepatitis (Ma et al., 2008; Tsuzura et al., 2014; Bitter et al., 2015). Understanding the mechanisms connecting PXR and EtOH-induced steatosis will provide important insights into potential therapeutic targets.
Binge (acute) alcohol drinking is a more common form of alcohol consumption than chronic alcoholism, with more than 75% of all alcohol consumed by binging (Sarkar and Chang, 2013; Shukla et al., 2013). Binge drinking contributes significantly to the incidence of cirrhosis seen in patients with ALD (Parrish et al., 1993). Furthermore, binge EtOH administration to mice produced behavioral abnormalities and changes in blood EtOH concentration (BEC), recapitulating some of the changes in human binge drinkers (Carson and Pruett, 1996). Acute EtOH exposure to both humans and rodent models also produced steatosis, lipid peroxidation (LPO), and mitochondrial dysfunction (Meagher et al., 1999; Shukla et al., 2013; Spruiell et al., 2015). Compared with chronic EtOH exposure, relatively little is known regarding the molecular mechanisms of binge EtOH-induced hepatotoxicity. PXR signaling promotes chronic EtOH-induced steatosis (Choi et al., 2018). However, whether PXR is also involved in binge EtOH-induced hepatotoxicity is unknown.
Therefore, we binge dosed male WT and Pxr-null mice and evaluated them for hepatotoxicity, lipid profiles, and changes in lipid, alcohol metabolism, and PXR target gene expression.
Materials and Methods
Animal Care and Treatment.
This study used male C57BL/6J mice (which served as the WT) and Pxr-null mice that were developed by Staudinger et al. (2001) and characterized previously (Staudinger et al., 2001; Spruiell et al., 2014b). Mice (three to five per cage) were housed at the Animal Resources Complex at North Carolina Central University in polycarbonate cages on racks directly vented via the facility’s exhaust system at 22°C with a 12-hour/12-hour light/dark cycle. Age-matched (aged 10–12 weeks) male WT and Pxr-null mice were each randomly separated into two groups (n = 8 to 9 for each group) and gavaged with three doses of 4.5 g/kg EtOH or saline solution every 12 hours at 9:00 am, 9:00 pm, and 9:00 am the next day. Four hours after the final dose, mice were anesthetized with isoflurane and euthanized. The liver was rapidly dissected, weighed, snap-frozen in liquid nitrogen, and stored at –80°C. Blood samples collected by cardiac puncture from anesthetized mice were centrifuged at 3000 rpm for 15 minutes to collect serum and were stored at −80°C until they were assayed for EtOH concentration, liver enzymes, triglycerides, and cholesterol. All procedures were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the North Carolina Central University Institutional Animal Care and Use Committee.
Hematoxylin and Eosin Staining of Liver Sections.
Liver tissues were fixed in 10% formalin/phosphate-buffered saline and used for hematoxylin and eosin staining and for histologic examination.
Quantification of Alanine Aminotransferase, Aspartate Aminotransferase, Cholesterol, and Triglyceride in Serum.
Serum was prepared from whole blood after centrifugation and stored at −80°C. Levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in the serum were determined using the Cholestech LDX analyzer (Cholestech Corporation, Hayward, CA) as reported previously (Spruiell et al., 2014a,b). Serum triglyceride and cholesterol levels were measured using commercially available kits (Wako Pure Chemical Industries, Richmond, VA).
Determination of Serum Alcohol Concentration.
Blood samples were collected after three doses of binge EtOH administration and were centrifuged at 3000 rpm for 15 minutes to collect serum for the measurement of BEC. The EtOH L3K assay kit for quantitative measurement of EtOH concentration (Sekisui Diagnostics P.E.I. Inc., Charlottetown, PE, Canada) was used according to the manufacturer’s instructions as described previously (Spruiell et al., 2015). The reaction is based on the enzymatic conversion of EtOH by alcohol dehydrogenase (ADH) to acetaldehyde and NADH. EtOH concentration in the serum was quantified as the rate of increase in NADH absorbance due to the reduction of NAD+ at 380 nm.
Determination of Hepatic Triglyceride, Cholesterol, and Nonesterified Fatty Acid Levels.
Total hepatic lipids were isolated from 100 mg liver tissue as previously described (Gyamfi et al., 2008). Hepatic triglyceride, cholesterol, and nonesterified fatty acid levels were measured using commercially available test kits (Wako Pure Chemical Industries).
Determination of LPO in Liver Tissues.
LPO in liver was measured as the thiobarbituric acid reactive product, malondialdehyde (MDA), using a thiobarbituric acid reactive substances kit (ZeptoMetrix, Buffalo, NY) according to the manufacturer’s instructions. Protein contents in liver homogenates were determined by the BCA protein assay kit (Thermo Scientific, Rockford, IL).
Western Blot Analyses from Liver Extracts.
Western blot analysis was performed using frozen livers as described previously (Spruiell et al., 2014a). The protein concentrations in liver homogenates were quantified by the BCA protein assay kit (Thermo Scientific). Briefly, 40 μg liver homogenates was mixed with Laemmli loading buffer containing β-mercaptoethanol and denatured at 100°C. Proteins were separated on 10% or 15% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. The membranes were incubated overnight with rabbit polyclonal primary antibody for CYP2E1 (1:1000, ab28146, lot no. GR182611-20; Abcam, Cambridge, MA). Primary antibodies from Santa Cruz Biotechnology Inc. (Dallas, TX) were all used at a concentration of 1:200 for mouse monoclonal ADH1 (Sc-137078 lot no. L3015), rabbit polyclonal antibody for glucose-regulated protein GRP78 (Sc-13968, lot no. H1809), and goat polyclonal antibody for CY3A11 (SC-30621, lot no. D2109). Rabbit monoclonal primary antobody for aldehyde dehydrogenase ALDH1A1 (1:1000, NB110-55451, lot no. GR182706-7) were from Novus Biologicals (Littleton, CO). Rabbit polyclonal primary antibody for CYP2B10 (1:5000, AB9916, lot no. 2794496) was from Sigma-Aldrich (St. Louis, MO). Rabbit polyclonal primary antibodies for Bax (no. 2772, lot no. 10), caspase-12 (no. 2202, lot no. 8), Bcl-2 (no. 2870, lot no. 5), and phosphorylated-elF2α (Ser51) (no. 9721, lot no. 12) were from Cell Signaling Technology (Danvers, MA) and were all used at a concentration of 1:1000. Blots were then incubated with the appropriate peroxidase–conjugated anti-rabbit (no. 7074, lot no. 26) or anti-mouse (no. 7076, lot no. 32) IgG secondary antibodies (Cell Signaling Technology) or donkey anti-goat (Sc-2020, lot no. 2113) IgG secondary antibody (Santa Cruz Biotechnology Inc.) diluted at 1:5000 in Tris-buffered saline/Tween 20 plus 1% milk for 60 minutes at room temperature. After initial probing, blots were stripped and reprobed with α-tubulin antibody (1:1000, no. 2125, lot no. 9; Cell Signaling Technology). Enhanced chemiluminescence was used to identify protein bands. ImageJ software (National Institutes of Health, Bethesda, MD) was used to quantify band intensity.
Total RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction.
Total RNA isolation, cDNA synthesis, and real-time quantitative polymerase chain reaction (PCR) were performed as previously described (Spruiell et al., 2014b). mRNA expression of the following genes was quantified: Ppara, carnitine palmitoyltransferase 1 (Cpt1), liver fatty acid binding protein 1 (Lfabp1), apolipoprotein B-1 (Apob1), Srebp1c, Scd1, Fas, acetyl-CoA carboxylase 1a (Acc1a), diacylglycerol acyltransferase 1 (Dgat1), Dgat2, Srebp2, 3-hydroxy-3-methyl-glutaryl CoA reductase (Hmgcr), Pxr, Car, Cyp2b10, farnesoid X receptor (Fxr), small heterodimer partner (Shp), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The primer sequences (Table 1) for mRNA encoding Ppara, Cpt1, Lfabp1, Apob1, Srebp2, Srebp1c, Scd1, Acc1a, Fas, Hmgcr, Cyp2b10, Fxr, Shp, and Gapdh were used for the SYBR Green assay. The following proprietary TaqMan Gene Expression Assays were also purchased from Applied Biosystems/Life Technologies (Grand Island, NY) and used for real-time quantitative PCR: Adh1 (class I Adh, no. Mm00507711_m1), Adh4 (class II Adh, no. Mm00478838_m1), Aldh2 (no. Mm00477463_m1), Aldh1a1 (no. Mm00657317_m1), Pxr (no. Mm01344139_m1), Car (no. Mm01283978_m1), retinoid X receptor a (Rxra) (no. Mm00441185_m1), uncoupling protein 2 (Ucp2) (no. Mm00627599_m1), Dgat1 (no. Mm00515643_m1), Dgat2 (no. Mm00499536_m1), Akr1b7 (no. Mm00477605_m1), Akr1b8 (no. Mm00484314_m1), and Gapdh (housekeeping gene; no. 99999915_g1). The amplification reactions were carried out in an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) as described previously (Gyamfi et al., 2008). Results are presented as levels of expression relative to that of controls after normalizing with Gapdh mRNA using the comparative threshold cycle method.
TABLE 1 .
Sequences of primers used for real-time quantitative PCR
| Name | Sequence | Accession Number |
|---|---|---|
| PPARa | ||
| Sense | GATTCAGAAGAAGAACCGGAACA | NM011144 |
| Antisense | TGCTTTTTCAGATCTTGGCATTC | |
| CPT1 | ||
| Sense | CGATCATCATGACTATGCGCTACT | NM013495 |
| Antisense | GCCGTGCTCTGCAAACATC | |
| LFABP | ||
| Sense | TGCATGAAGGGAAGAAAATCAAA | NM017399 |
| Antisense | CCCCCAGGGTGAACTCATT | |
| APOB1 | ||
| Sense | GGTGTTGTTTCCATACCACGTTT | XM 137955 |
| Antisense | TCGTAACGCCATGTCACTCTCT | |
| MTP | ||
| Sense | CCGCTGTGCTTGCAGAAGA | NM008642 |
| Antisense | TTTGACACTATTTTTCCTGCTATGGT | |
| SREBP2 | ||
| Sense | ATGATCACCCCGACGTTCAG | NM 033218 |
| Antisense | GCTGCGTTCTGGTATATCAAAGG | |
| SREBP1c | ||
| Sense | CATGCCATGGGCAAGTACAC | NM011480 |
| Antisense | TGTTGCCATGGAGATAGCATCT | |
| SCD1 | ||
| Sense | CGTTCCAGA ATGACGTGTACGA | NM009127 |
| Antisense | AGGGTCGGCGTGTGTTTC | |
| ACC1a | ||
| Sense | ATGTCCGCACTGACTGTAACCA | NM133360 |
| Antisense | TGCTCCGCACAGATTCTTCA | |
| FAS | ||
| Sense | CCCGGAGTCGCTTGAGTATATT | NM007988 |
| Antisense | GGACCGAGTAATGCCATTCAG | |
| HMGCR | ||
| Sense | GGCAGTCAGTGGGAACTATTGC | NM 008255 |
| Antisense | CAGTCTTTCCTCGTCCTTCGA | |
| CYP2B10 | ||
| Sense | TTTCTGCCCTTCTCAACAGGAA | NM 009999 |
| Antisense | ATGGACGTGAAGAAAAGGAACAAC | |
| FXR | ||
| Sense | CACGAAGATCAGATTGCTTTGC | NM 009108 |
| Antisense | CCGCCGAACGAAGAAACAT | |
| SHP | ||
| Sense | CAGGCACCCTTCTGGTAGATCT | NM 011850 |
| Antisense | GTCTTCAAGGAGTTCAGTGATGTCA | |
| GAPDH | ||
| Sense | TGTGTCCGTCGTGGATCTGA | NM001001303 |
| Antisense | CCTGCTTCACCACCTTCTTGA |
Statistical Analysis.
Data are presented as means ± S.E.M. and were analyzed using SigmaPlot 13 software (Systat Software Inc., San Jose, CA). The effect of genotype, binge EtOH, and their interaction was analyzed using a two-way analysis of variance with the Bonferroni post hoc test. A P value < 0.05 was considered statistically significant.
Results
Both WT and Pxr-Null Mice Are Susceptible to Binge EtOH-Induced Steatosis.
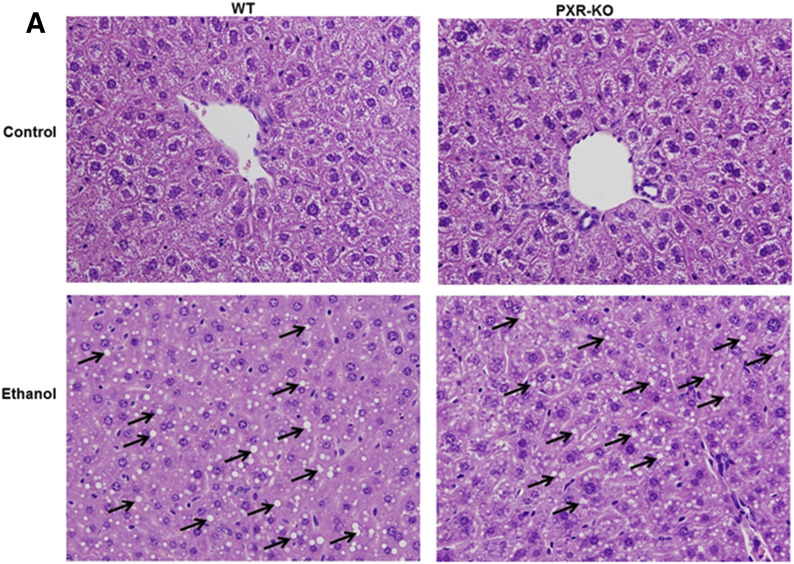
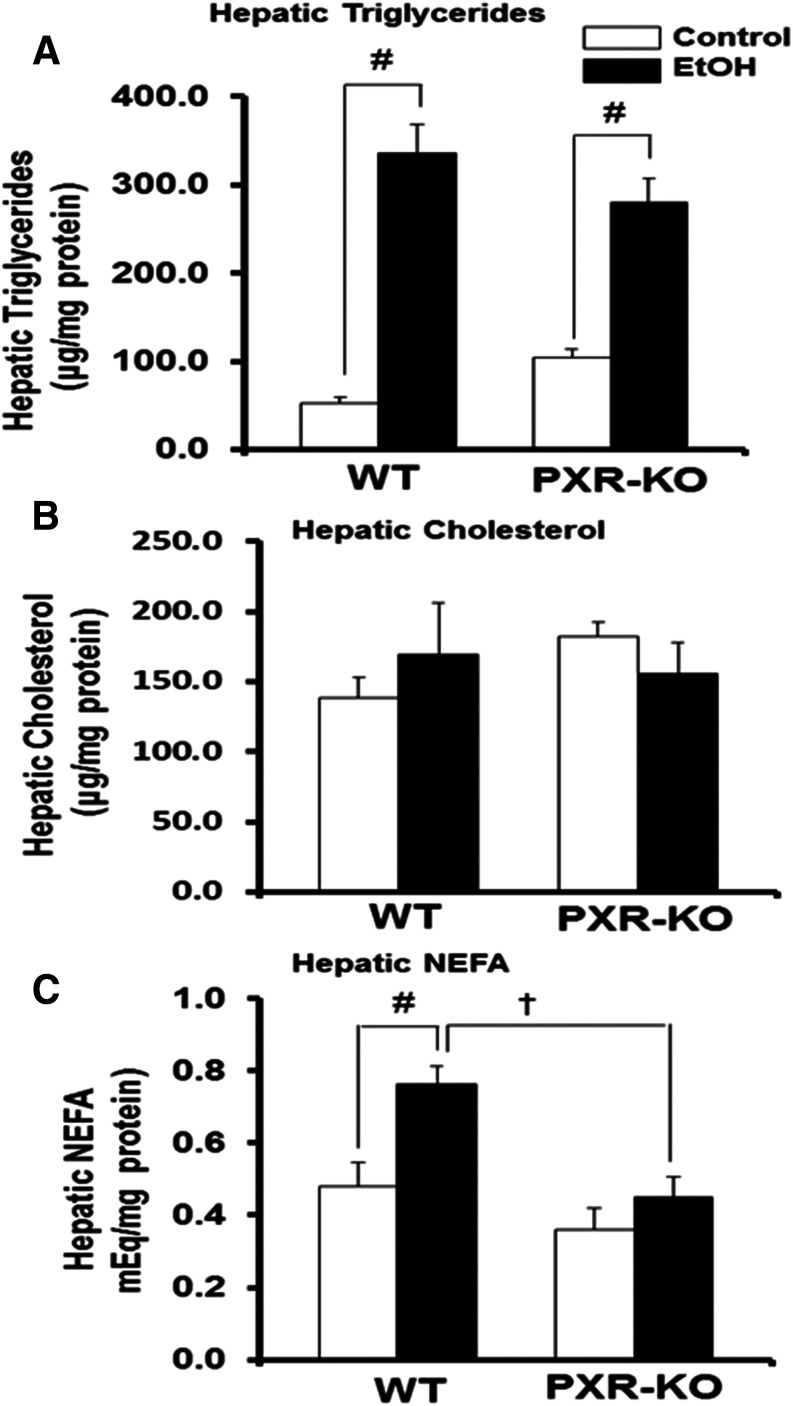
The saline (control) or binge EtOH treatment had no effect on body weight, liver weight, or liver-to-body weight ratios in either genotype (data not shown). Histopathological analysis of liver sections by hematoxylin and eosin staining indicated the presence of many lipid droplets in both genotypes fed EtOH compared with their respective controls (Fig. 1). Similar to our previous report, basal hepatic triglyceride levels were elevated in control-fed Pxr-null mice (1.9-fold) (Spruiell et al., 2014b); however, these differences were not statistically significant (Fig. 2A). Furthermore, the genotype or the interaction between the genotype and EtOH treatment had no effect on hepatic triglyceride levels (P = 0.715). However, binge EtOH ingestion increased hepatic triglyceride levels (P < 0.001) in both WT (5.9-fold) and Pxr-null (3.3-fold) mice but did not increase hepatic cholesterol levels (Fig. 2, A and B). Notably, the genotype influenced hepatic nonesterified fatty acid levels (P = 0.002), which were significantly higher only in EtOH-fed WT mice (Fig. 2C).
Fig. 1.
Representative H&E staining of livers in control and binge EtOH-fed male WT and Pxr-null mice. Male WT and Pxr-null mice were orally administered saline (control) or EtOH (4.5 g/kg) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. H&E stains were assayed as described in the Materials and Methods. Binge EtOH administration produced steatosis (indicated by arrows) in both WT and Pxr-null mice fed EtOH. H&E, hematoxylin and eosin; KO, knockout. Original magnification, 400×.
Fig. 2.
Quantification of hepatic lipids in control and binge EtOH-fed male WT and Pxr-null mice. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–C) Hepatic triglycerides (A), hepatic cholesterol (B), and hepatic NEFAs (C) were assayed as described in the Materials and Methods. Data represent the mean ± S.E.M. (n = 5 to 6). †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout; NEFA, nonesterified fatty acid.
Basal Hepatic mRNA Levels of Srebp1c and Its Target Gene Scd1 Are Upregulated in Pxr-Null Mice.
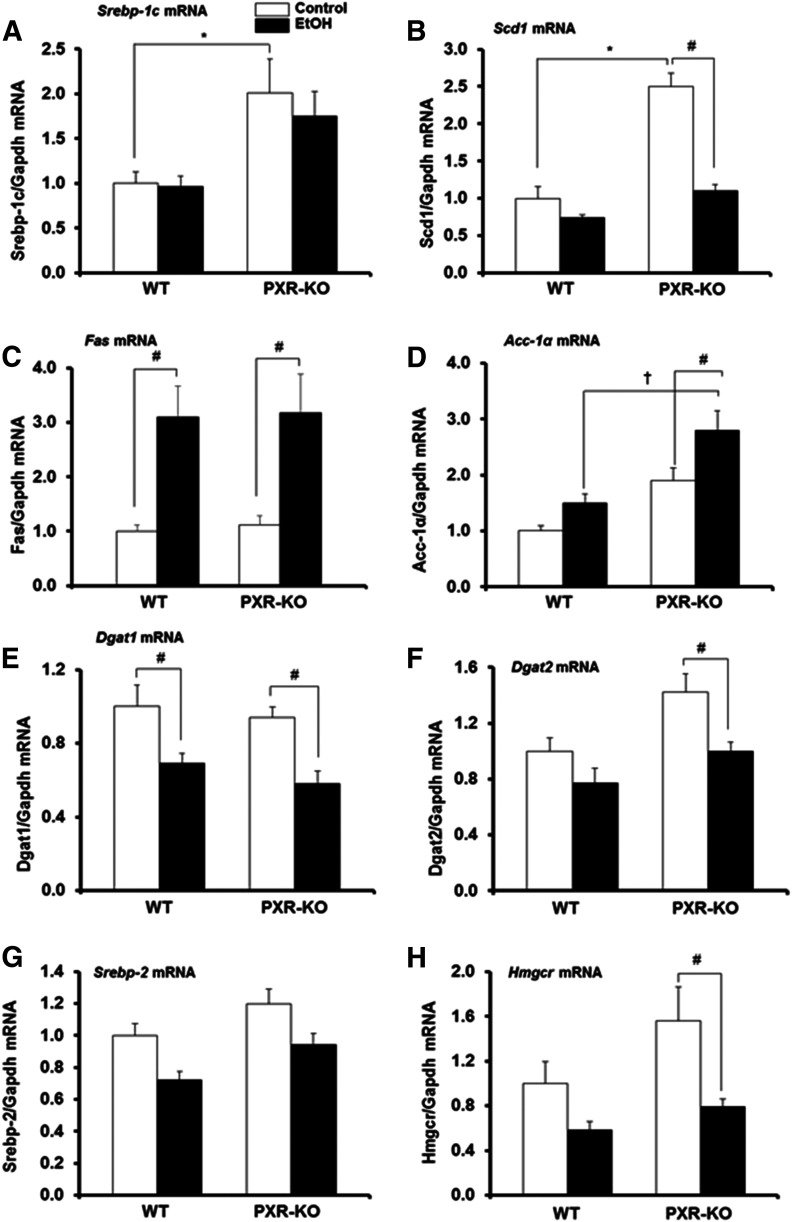
The increased steatosis and hepatic triglyceride content we observed in both genotypes fed EtOH (Figs. 1 and 2A) prompted us to examine the expression of mRNAs encoding Srebp1c and hepatic fatty acid and triglyceride synthesis enzymes. The genotype influenced basal hepatic mRNA levels of both Srebp1c and its target gene Scd1, which were significantly increased 2-fold and 2.5-fold in Pxr-null mice, respectively, compared with saline-treated (control) WT mice (Fig. 3, A and B). In contrast, EtOH administration had no effect on Srebp1c mRNA levels in either genotype but diminished Scd1 mRNA (P < 0.05) in Pxr-null mice (Fig. 3, A and B). Basal hepatic gene expression of other Srebp1c targets, Fas and Acc1a, was not different between the two genotypes (Fig. 3, C and D). EtOH increased Fas mRNA levels in both WT (3.1-fold) and Pxr-null (2.8-fold) mice (Fig. 3C), suggesting a significant effect of treatment (P = 0.001) but not interaction between the genotype and EtOH on Fas mRNA levels. EtOH had no effect on Acc1a mRNA levels in WT mice (Fig. 3D). In contrast, EtOH increased hepatic Acc1a gene expression in Pxr-null mice (1.7-fold) (P = 0.01) (Fig. 3D), leading to significantly higher Acc1α mRNA levels in EtOH-fed Pxr-null mice compared with similarly treated WT mice (Fig. 3D). Pairwise multiple-comparison analysis revealed that both genotype and treatment significantly influenced Acc1a mRNA levels. Constitutive Dgat1 and Dgat2 mRNA levels were not significantly different between the two genotypes (Fig. 3, E and F). Both hepatic Dgat1 and Dgat2 mRNA levels were decreased by EtOH in Pxr-null mice (P = 0.004); however, only Dgat1 mRNA levels were decreased in WT mice (P < 0.001) (Fig. 3, E and F). Basal levels of Srebp2 mRNA and its target gene Hmgcr were similar between control-fed WT and Pxr-null mice; however, EtOH significantly decreased the Hmgcr gene only in Pxr-null mice (Fig. 3, G and H).
Fig. 3.
Gene expression of mouse hepatic enzymes that promote lipogenesis. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–H) Total hepatic mRNA levels of Srebp1c (A), Scd1 (B), Fas (C), Acc1a (D), Dgat1 (E), Dgat2 (F), Srebp2 (G), Hmgcr (H), and Gapdh were quantified by the SensiFast SYBR Hi-ROX Kit (Bioline, Taunton, MA). Data represent the mean ± S.E.M. (n = 5 to 6). *P < 0.05 (significantly different from saline-treated WT control); †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout.
Binge EtOH Ingestion Similarly Affected Hepatic Fatty Acid Oxidation Genes in Both Genotypes.
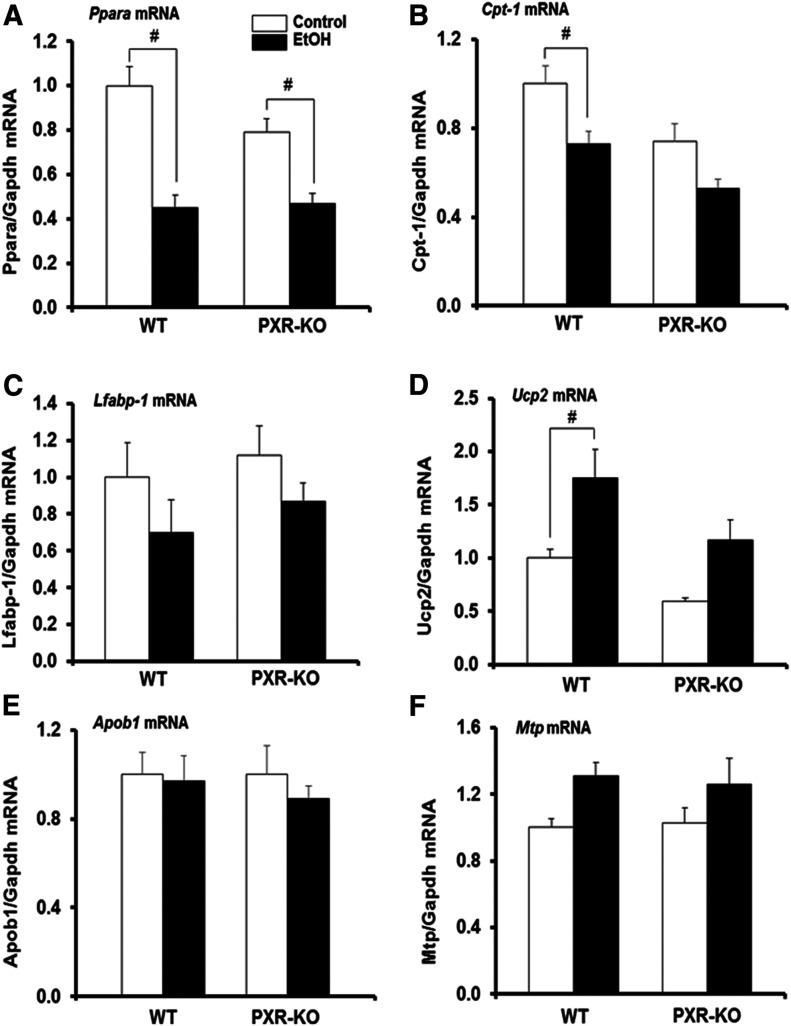
Since alcohol-induced lipid accumulation may also result from inhibition of hepatic fatty acid β-oxidation, we monitored fatty acid uptake and oxidation genes. There was a significant effect of treatment (P = 0.001) but not genotype or interaction between the genotype and EtOH on Ppara mRNA levels. Basal hepatic Ppara mRNA levels were not different between saline-fed WT and similarly treated Pxr-null mice (Fig. 4A). Hepatic Ppara mRNA levels in both WT and Pxr-null mice were significantly decreased by EtOH by 55% and 41%, respectively, compared with their corresponding controls (Fig. 4A). Similarly, basal Cpt1a gene expression did not vary between the two genotypes (Fig. 4B). However, there was a significant effect of both genotype (P = 0.004) and treatment (P = 0.001), in which binge EtOH treatment significantly decreased Cpt1a mRNA levels only in WT mice (Fig. 4B). Constitutive expression levels of Lfabp1 mRNA were similar between both genotypes and EtOH did not significantly affect Lfabp1 gene expression in either genotype (Fig. 4C). Basal hepatic levels of Ucp2 mRNA were similar between both control-fed WT and Pxr-null mice (Fig. 4D). However, there was a significant effect of treatment (P = 0.005) but not genotype on Ucp2 mRNA levels, such that EtOH significantly upregulated Ucp2 mRNA levels only in WT mice (1.8-fold) (Fig. 4D). Constitutive levels of Apob1 and microsomal triglyceride transfer protein Mtp mRNAs in the liver did not vary between the two genotypes and their mRNA levels were not different after EtOH treatment (Fig. 4, E and F).
Fig. 4.
Hepatic gene expression of mouse Pparα and its target genes. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–F) Total hepatic mRNA levels of Ppara (A), Cpt1 (B), Lfabp1 (C), Ucp2 (D), Apob1 (E), Mtp (F), and Gapdh were quantified by the SensiFast SYBR Hi-ROX Kit (Bioline). Data represent the mean ± S.E.M. (n = 5 to 6). #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout, MTP, microsomal triglyceride transfer protein.
Binge EtOH Upregulation of the Cyp2b10 Gene Is PXR Dependent.
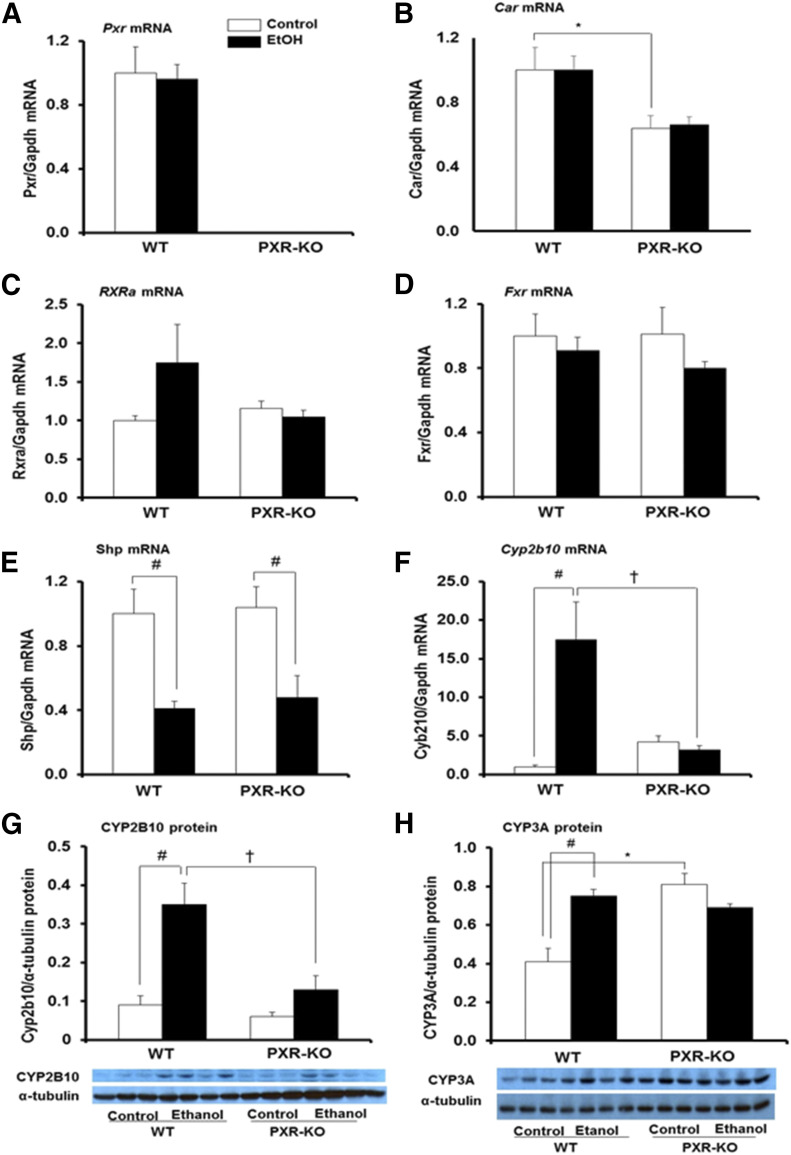
Nuclear receptors including PXR, FXR, CAR, and SHP have also been implicated in lipid homeostasis (Tanaka et al., 2017). Furthermore, due to the crosstalk between PXR and CAR, we evaluated the effect of these nuclear receptors on the development of lipid accumulation by binge EtOH. As expected, Pxr mRNA was absent in the livers of Pxr-null mice (Fig. 5A). Interestingly, EtOH did not alter Pxr gene expression in either genotype (Fig. 5A). There was a significant effect of genotype on basal hepatic Car mRNA levels, which were significantly lower in Pxr-null mice compared with WT controls (Fig. 5B). Interestingly, EtOH did not alter Car gene expression in either genotype (Fig. 5B). Basal mRNA levels of Rxra, Fxr, and Shp were not different between both genotypes and Rxra and Fxr mRNA levels were not affected by EtOH (Fig. 5, C and D). However, there was a significant effect of treatment on Shp mRNA levels (P = 0.001), such that binge EtOH administration significantly reduced Shp mRNA levels in both WT and Pxr-null mice (Fig. 5E). Unexpectedly, binge EtOH induced the CAR target gene Cyp2b10 (17-fold) only in WT mice (Fig. 5F). There was a statistically significant interaction between genotype and treatment on both Cyp2b10 mRNA and CYP2B10 protein levels (P = 0.035). Basal Cyp2b10 mRNA levels were somewhat higher in Pxr-null mice compared with WT mice but were not statistically different (Fig. 5F). Western blot analyses further confirmed that CYP2B10 protein levels were significantly higher in EtOH-fed WT mice (4-fold) compared with WT controls (Fig. 5G). However, EtOH did not alter CYP2B10 protein expression in Pxr-null mice (Fig. 5G). Constitutive hepatic protein expression of CYP3A11 was significantly induced in Pxr-null mice (2.0-fold) compared with saline-fed WT mice as previously reported (Fig. 5H) (Staudinger et al., 2001). Hepatic CYP3A11 protein levels were significantly increased by EtOH in WT mice (1.8-fold) but not in similarly treated Pxr-null mice (Fig. 5H).
Fig. 5.
Hepatic gene expression and immunoblot analysis of mouse type II nuclear receptors and their target genes important for xenobiotic metabolism and lipid homeostasis. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–F) Total hepatic mRNA levels of Pxr (A), Car (B), Rxra (C), Fxr (D), Shp (E), Cyp2b10 (F), and Gapdh were quantified by the SensiFast SYBR Hi-ROX Kit (Bioline). Data represent the mean ± S.E.M. (n = 5 to 6). (G and H) Furthermore, Western blots of liver homogenate (40 μg/lane) were probed with antibodies to CYP2B10 (G) and CYP3A11 (H). Bands were quantified and normalized to α-tubulin. Data represent the mean ± S.E.M. (n = 3 to 4). *P < 0.05 (significantly different from saline-treated WT control); †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout.
Antiapoptotic Protein Bcl-2 Expression Is Decreased in EtOH-Fed WT Mice.
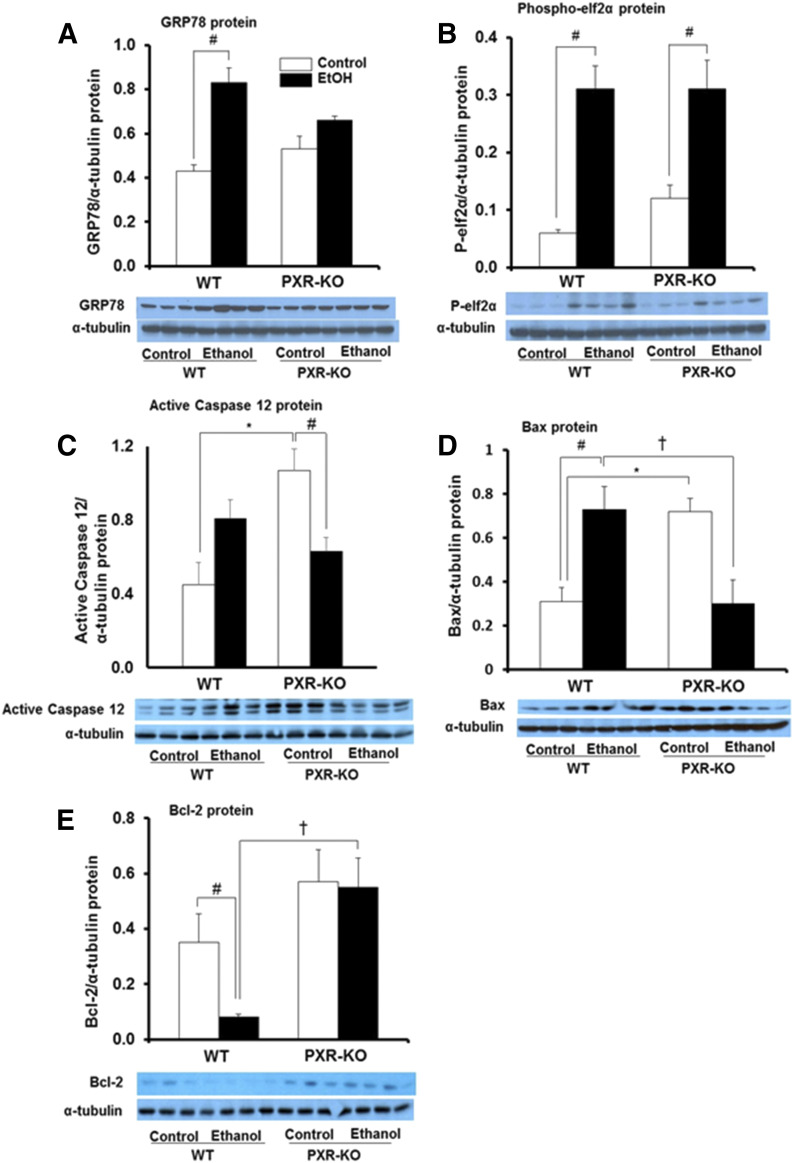
Binge EtOH treatment had a significant effect on protein levels of hepatic endoplasmic reticulum stress markers GRP78 (P = 0.015) and phospho-elF2α (P = 0.001) in WT mice. Hepatic GRP78 and phospho-elF2α protein expression was upregulated 1.9- and 5.0-fold, respectively, in EtOH-treated WT mice versus saline-treated WT mice (Fig. 6, A and B). Phospho-elF2α protein levels were similar in the livers of both genotypes fed saline (Fig. 6B). EtOH exposure increased hepatic phospho-elF2α protein levels (2.6-fold) but not GRP78 in Pxr-null mice (Fig. 6, A and B). Furthermore, basal expression of the endoplasmic-specific apoptotic protein caspase-12 was increased 3.6-fold in saline-fed Pxr-null mice compared with WT controls (P = 0.002) (Fig. 6C). EtOH nonsignificantly increased caspase-12 protein in WT mice (1.8-fold); however, caspase-12 protein was significantly decreased approximately 40% in Pxr-null mice (Fig. 6C). There was a statistically significant interaction between genotype and treatment on Bax protein levels (P = 0.002). Hepatic Bax protein expression levels were significantly higher in saline-fed Pxr-null mice compared with saline-fed WT mice (Fig. 6D). EtOH ingestion decreased Bax protein levels in Pxr-null mice (P = 0.015) but significantly increased Bax protein in WT mice (2.3-fold) to a level higher than that in EtOH-fed Pxr-null mice (P = 0.009) (Fig. 6D). Expression of the antiapoptotic protein Bcl-2 did not differ between the two genotypes fed saline; however, EtOH markedly inhibited the Bcl-2 protein only in WT mice (Fig. 6E). Furthermore, Bcl-2 protein levels were significantly higher in EtOH-fed Pxr-null mice than in similarly treated WT mice (Fig. 6E).
Fig. 6.
Immunoblot analysis of hepatic endoplasmic reticulum stress and apoptotic markers. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–E) Western blots of liver homogenate (40 μg/lane) were probed with antibodies to GRP78 (A), phospho-elF2α (B), active caspase-12 (C), Bax (D), and Bcl-2 (E). Bands were quantified and normalized to α-tubulin. Data represent the mean ± S.E.M. (n = 3 to 4 mice/group). *P < 0.05 (significantly different from saline-treated WT control); †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout; P, phosphorylated.
Basal and EtOH-Exposed Hepatic ADH1 Protein Levels Are Significantly Higher in Pxr-Null Mice.
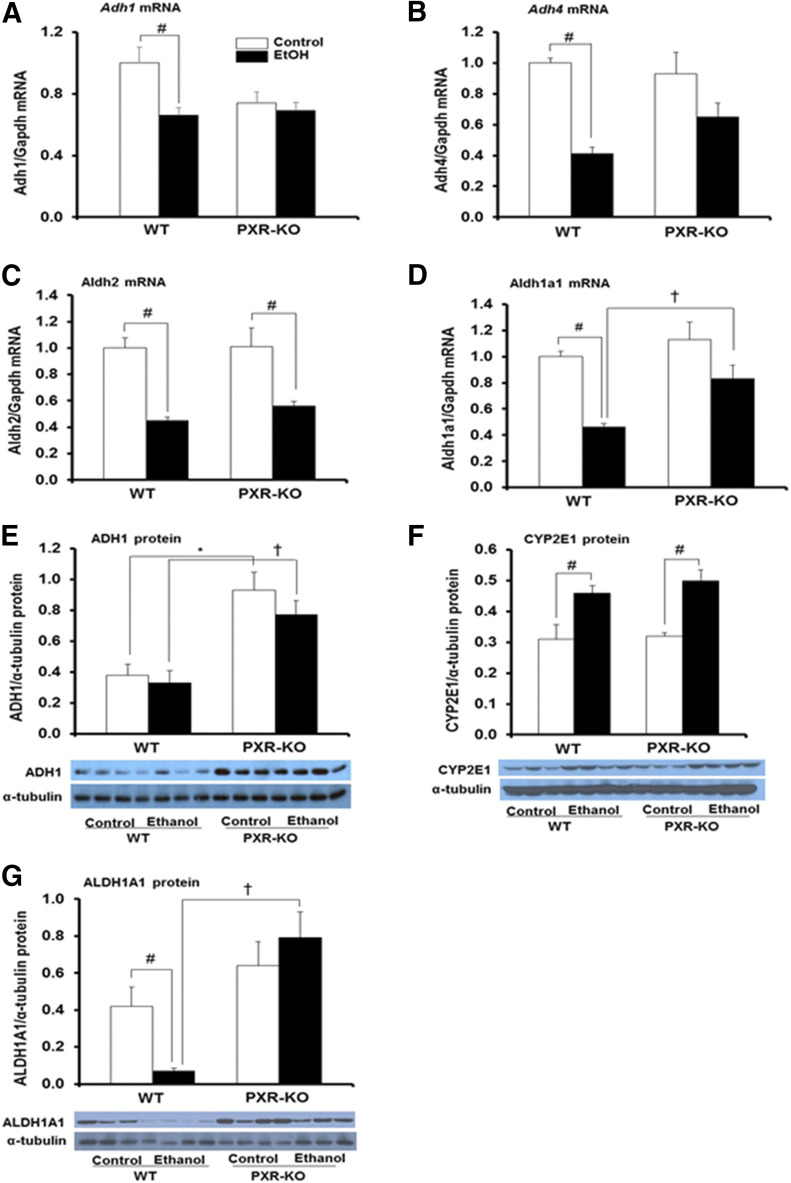
Published reports indicate that nuclear receptors regulate Adh1 expression (Gyamfi et al., 2006; Langhi et al., 2013). Furthermore, EtOH metabolism is known to alter the intracellular redox state due to the generation of increased NADH levels, which promotes excess fatty acids synthesis leading to steatosis (Zakhari and Li, 2007), a phenomenon corroborated in our study (Figs. 1 and 2). Therefore, we examined the effect of PXR deficiency, binge EtOH, and their interaction on mRNAs of Adh1, Adh4, Aldh2, and Aldh1a1 as well as the protein levels of ADH1, CYP2E1, and ALDH1A1, which are known to be involved in EtOH metabolism (Heit et al., 2015). The EtOH treatment had a significant effect on mRNA levels of hepatic Adh1, Adh4, and Aldh2 (P = 0.001) but not the genotype or their interaction. Basal mRNA levels of hepatic Adh1, Adh4, and Aldh2 did not vary significantly between the two genotypes treated with saline (Fig. 7, A–C). EtOH significantly decreased Adh1, Adh4, and Aldh2 mRNA levels by 34%–59% in WT mice but decreased only Aldh2 mRNA (43%) in Pxr-null mice (Fig. 7, A–C). Genotype, treatment, and the interaction between genotype and treatment significantly affected Aldh1a1 mRNA levels. Interestingly, basal Aldh1a1 mRNA levels were not significantly different between the two genotypes (Fig. 7D). However, Aldh1a1 mRNA levels were significantly lower after EtOH ingestion only in WT mice (46% of control) (Fig. 7D). Genotype, but not treatment or the interaction between genotype and EtOH, significantly affected ADH1 protein levels. Contrary to the mRNA data, Western blotting indicated a marked increase in basal hepatic ADH1 protein levels in Pxr-null mice (2.5-fold) compared with WT controls (Fig. 7E). Binge EtOH did not have any significant effect on ADH1 protein levels in WT mice but maintained the higher levels of ADH1 protein in Pxr-null mice (Fig. 7E). There was no significant effect of genotype or interaction between genotype and treatment on hepatic CYP2E1 protein levels; in agreement, basal CYP2E1 protein expression did not vary between WT and Pxr-null mice (Fig. 7F). However, there was a significant effect of EtOH treatment on CYP2E1 protein expression, which was upregulated (1.6-fold) in both genotypes (Fig. 7F). In agreement with the marked inhibition of Aldh1a1 mRNA levels, ALDH1A1 protein expression was also markedly inhibited by EtOH only in WT mice (Fig. 7, D and G). Furthermore, both hepatic Aldh1a1 mRNA and ALDH1A1 protein levels in EtOH-fed Pxr-null mice were higher compared with EtOH-fed WT mice (P < 0.001) (Fig. 7, D and G).
Fig. 7.
Gene expression and immunoblot analysis of mouse hepatic enzymes involved in EtOH metabolism. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–D) Total hepatic mRNA levels of Adh1 (A), Adh4 (B), Aldh2 (C), Aldh1a1 (D), and Gapdh were quantified by the SensiFast SYBR Hi-ROX Kit (Bioline). Data represent the mean ± S.E.M. (n = 5–7). (E–G) Western blots of liver homogenate (40 μg/lane) were probed with antibodies to ADH1 (E), CYP2E1 (F), and ALDH1A1 (G). Data represent the mean ± S.E.M. (n = 3 to 4). *P < 0.05 (significantly different from saline-treated WT control); †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout.
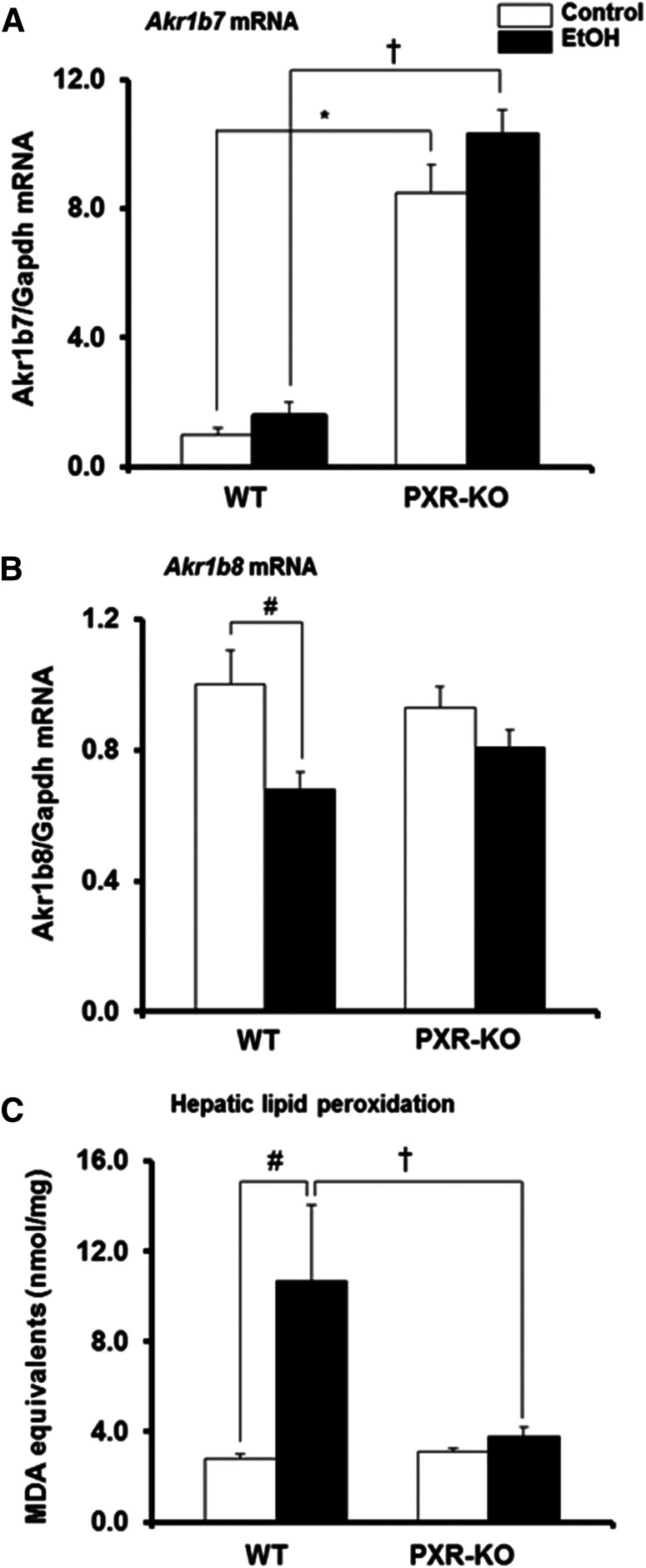
Basal Hepatic Gene Expression of Akr1b7, But Not Akr1b8, Is Elevated in Pxr-Null Mice.
Both Akr1b7 and Akr1b8 (or its human ortholog AKR1B10) have been implicated in lipid metabolism (Tirard et al., 2007; Bitter et al., 2015). Interestingly, Akr1b7 is both a CAR and PXR target gene (Liu et al., 2009). It was therefore of interest to examine the effect of PXR deficiency and binge EtOH exposure on the hepatic expression of Akr1b7 and Akr1b8 genes using quantitative real-time-PCR. Genotype, but not treatment or the interaction between the genotype and treatment, had a major effect on Akr1b7 mRNA levels, such that basal hepatic Akr1b7 mRNA levels were significantly higher in Pxr-null mice (8.4-fold) than in saline-treated WT controls (Fig. 8A). Binge EtOH had no effect on hepatic levels of Akr1b7 gene expression in both genotypes but maintained the higher Akr1b7 mRNA levels in Pxr-null mice, such that Akr1b7 mRNA levels were significantly higher in Pxr-null mice than in WT mice (Fig. 8A). Unlike Akr1b7 mRNA, basal hepatic Akr1b8 gene expression was similar between the two genotypes; however, EtOH significantly decreased hepatic Akr1b8 mRNA levels only in WT mice (33%) (Fig. 8B). Induction of oxidative stress is involved in binge/acute EtOH-induced hepatotoxicity (Meagher et al., 1999). Both Akr1b7 and Akr1b8 can detoxify toxic aldehydes generated during LPO (Spite et al., 2007; Liu et al., 2009). Furthermore, ALDHIA1 is a major cytosolic ALDH involved in cellular defense against oxidative stress in the mouse liver (Makia et al., 2011). Therefore, we measured LPO, an early biochemical event of EtOH hepatotoxicity and a major indicator of oxidative stress, by quantifying the thiobarbituric acid reactive product, MDA. Consistent with reduced hepatic Aldh1a1 and Akr1b8 genes and ALDH1A1 protein expression in EtOH-fed WT mice, hepatic MDA levels were elevated by binge EtOH only in the livers of WT mice (3.8-fold) (Figs. 7, D and G, and 8, B and C). Furthermore, the increase in hepatic MDA levels by EtOH was higher in WT mice compared with Pxr-null mice (P = 0.003) (Fig. 8C).
Fig. 8.
Gene expression of mouse hepatic enzymes involved in the detoxification of lipid peroxides and the extent of lipid peroxidation in liver tissues. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A and B) Total hepatic mRNA levels of Akr1b7 (A), Akr1b8 (B), and Gapdh were quantified by the SensiFast SYBR Hi-ROX Kit (Bioline). Data represent the mean ± S.E.M. (n = 5–7). (C) The extent of lipid peroxidation in liver tissues was quantified by measuring the thiobarbituric acid reactive product, MDA. Data represent the mean ± S.E.M. (n = 5 to 6). *P < 0.05 (significantly different from saline-treated WT control); †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout.
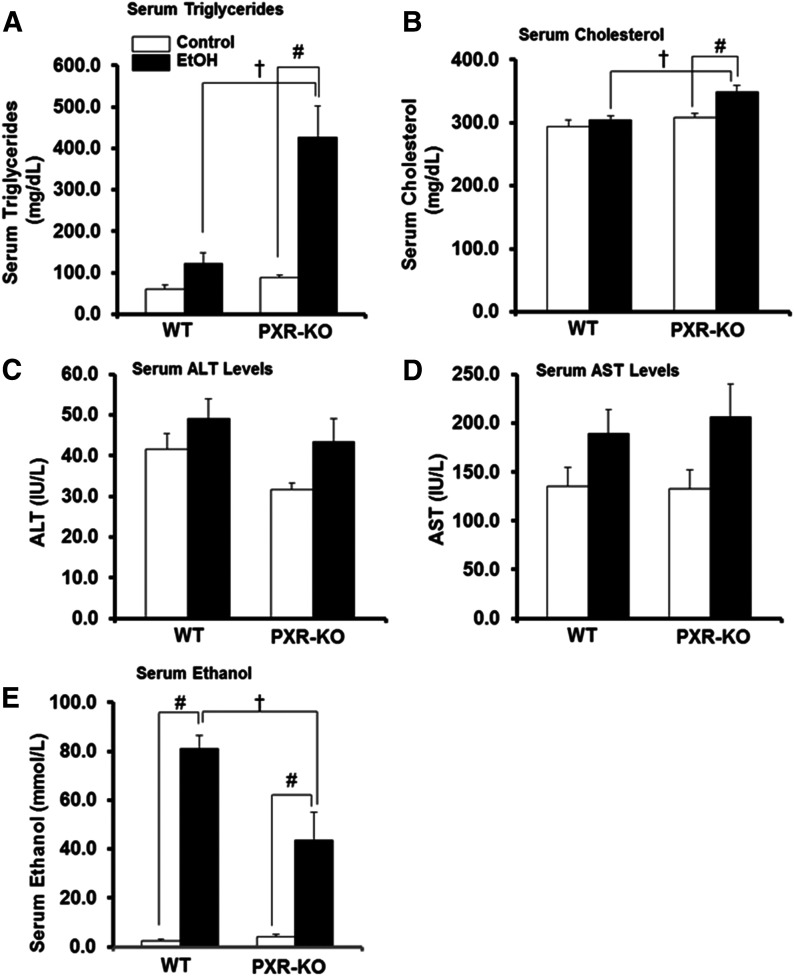
PXR Deficiency Leads to Increased Binge EtOH-Induced Hyperlipidemia.
There was a significant interaction between treatment and genotype on serum triglyceride levels (P = 0.010). Basal serum triglyceride levels tended to increase in Pxr-null mice (1.5-fold) compared with saline-fed WT mice (Fig. 9A). Binge EtOH ingestion tended to increase serum triglyceride levels in WT mice (2.0-fold) (Fig. 9A). By contrast, serum triglyceride levels were markedly increased in EtOH-fed Pxr-null mice (4.8-fold) (Fig. 9A). Similarly, serum cholesterol levels were significantly elevated by EtOH in Pxr-null mice by 13% (Fig. 9B). Furthermore, after EtOH treatment, both serum triglyceride and cholesterol levels in Pxr-null mice were higher (P < 0.05) than in EtOH-fed WT mice (Fig. 9, A and B). Serum ALT and AST activities were measured as indices of hepatocyte/organ injury. Unexpectedly, serum ALT and AST were not significantly induced in either genotype exposed to EtOH (Fig. 9, C and D). Due to the marked decreases in EtOH-metabolizing enzymes (especially hepatic Aldh1a1 expression in EtOH-fed WT mice) but higher ADH1 protein levels in Pxr-null mice, serum EtOH concentrations were measured after administration of three doses of binge EtOH. There was an interaction between EtOH treatment and genotype on the residual serum EtOH concentration (P = 0.014). As expected, the EtOH concentration was higher (P < 0.001) in WT mice compared with similarly treated Pxr-null mice (Fig. 9E).
Fig. 9.
Serum lipid profile, markers of hepatotoxicity, and serum EtOH concentration. Male WT and Pxr-null mice were orally administered saline (control; white bars) or EtOH (4.5 g/kg; black bars) every 12 hours for a total of three doses and were euthanized 4 hours after the final dose. (A–E) Serum was assayed for triglycerides (A), cholesterol (B), ALT (C), AST (D), and serum EtOH concentration (E) as described in the Materials and Methods. Data represent the mean ± S.E.M. (n = 5 to 6). †P < 0.05 (significantly different from treatment-matched WT group); #P < 0.05 (significantly different from genotype-matched saline control). KO, knockout.
Discussion
In this report, we characterized the hepatotoxic responses of WT mice and Pxr-null mice to acute EtOH exposure. CAR and PXR are xenobiotic receptors that share ligands and regulate drug-inducible expression of the same target genes, such as CYP2B and CYP3A, as well as genes unique to each receptor (Wang and Negishi, 2003). CAR preactivation and alcohol administration increases EtOH-induced hepatotoxicity in a CAR-dependent manner (Chen et al., 2011). Although the Car gene was not induced by binge EtOH, our study demonstrates a robust PXR-dependent upregulation of Cyp2b10, a CAR target gene (Ding et al., 2006), in binge EtOH-fed WT mice. In addition, protein expression of CYP3A11, whose expression can be regulated by both PXR and CAR, was also induced by binge EtOH only in WT mice (Wang and Negishi, 2003). Although the molecular basis for Cyp2b10 upregulation by binge EtOH seen here is not fully understood, we confirm that PXR is required for optimal Cyp2b10 upregulation by CAR (Wang and Negishi, 2003; Ding and Staudinger, 2005). Interestingly, in agreement with our findings, a PXR-dependent increase in Cyp2b10 expression by chronic EtOH administration has been reported (Choi et al., 2018). Both PPARγ coactivator 1α and hepatocyte nuclear factor 4α positively regulate Cyp2b10 independent of CAR (Wiwi et al., 2004; Koga et al., 2016). Hepatic hepatocyte nuclear factor 4α and PPARγ coactivator 1α mRNA levels were also examined in this study, but they were not induced by EtOH in both genotypes (data not shown). Together, our results indicate that although Pxr-null mice express CAR, binge EtOH could not induce Cyp2b10 in this model, suggesting that PXR influences EtOH induction of Cyp2b10.
EtOH is metabolized mainly by oxidation to acetaldehyde by multiple enzymes, including ADH, CYP2E1, and catalase (Zakhari and Li, 2007). In this study, EtOH significantly decreased the gene expression of both Adh1, the principal enzyme important for EtOH oxidation, and Adh4, an isoform efficient at high EtOH concentrations (Zakhari and Li, 2007) only in WT mice. Although Adh1 mRNA expression did not differ, basal hepatic protein expression of ADH1 was higher in Pxr-null mice than in WT mice. These findings confirm published reports indicating that hepatic ADH is regulated by a post-transcriptional mechanism (Gyamfi et al., 2006), possibly due to increased serum leptin levels in Pxr-null mice (Mezey et al., 2005; Spruiell et al., 2014b). Acetaldehyde, a toxic product generated from EtOH metabolism, is then oxidized to acetate by three ALDH isoforms (ALDH2, ALDH1B1, and ALDH1A1) (Heit et al., 2015). Among the cytosolic ALDH1 isoforms, ALDH1A1 also plays a major role in the removal of many reactive carbonyl species (Makia et al., 2011). One mechanism identified in the activation of CAR and enhanced EtOH hepatotoxicity involves inhibition of EtOH-metabolizing enzymes including ALDH1A1 expression (Chen et al., 2011). In our study, while Aldh2 mRNA levels were inhibited in both genotypes, PXR deficiency prevented EtOH-induced suppression of both Aldh1a1 gene and ALDH1A1 protein expression in Pxr-null mice, but not in WT mice. In agreement with decreased Adh1, Adh4, and Aldh1a1 mRNAs as well as ADH1 and ALDH1A1 protein expression in EtOH-fed WT mice, the serum EtOH concentration was elevated in WT mice compared with EtOH-fed Pxr-null mice. Thus, our results indicate that PXR ablation results in increases in ADH1 protein expression and prevents binge EtOH-induced suppression of EtOH metabolism, thereby increasing EtOH elimination in Pxr-null mice.
In this study, EtOH-fed mice of both genotypes developed steatosis with higher hepatic triglyceride content. Importantly, expression of hepatic fatty acid β-oxidation genes including Ppara and Cpt1 was suppressed by binge EtOH in WT mice, but only Ppara was suppressed in EtOH-fed Pxr-null mice. Moreover, binge EtOH also upregulated genes involved in fatty acid synthesis, including Acc1a and Fas in Pxr-null mice, whereas it induced only Fas in WT mice. Unexpectedly, binge EtOH treatment resulted in somewhat much larger hepatic triglyceride levels in WT mice (5.9-fold) compared with Pxr-null mice (3.3-fold). In this study, basal hepatic triglyceride levels were somewhat elevated in saline-fed Pxr-null mice (1.9-fold) compared with WT mice. Therefore, despite the fold increase being greater in WT mice, the absolute levels of hepatic triglyceride accumulation were similar between WT and Pxr-null mice. In a previous report, PXR knockdown-induced steatosis in HepG2 cells was associated with elevation of AKR1B10 gene expression (Bitter et al., 2015). However, in this study, basal hepatic mRNA levels of Akr1b8, the mouse ortholog of human AKR1B10, were not different between WT and Pxr-null mice. Taken together, although EtOH-induced inhibition of fatty acid catabolic gene expression is the major mechanism for steatosis in both WT and the Pxr-null mice, marked upregulation of genes involved in fatty acid synthesis may also account for EtOH-induced steatosis in Pxr-null mice.
Acute alcohol consumption can cause hypertriglyceridemia (Zemánková et al., 2015). PPARa activation in rodents and humans markedly reduces hepatic triglyceride synthesis and promotes plasma triglyceride clearance (Kersten, 2008). Although serum triglycerides were not increased in EtOH-fed WT mice, it was unexpected to observe hyperlipidemia in the EtOH-fed Pxr-null mice. It is possible that the concomitant induction of lipogenic genes involved in triglyceride synthesis and suppression of PPARa and its target genes by EtOH contributed to the hypertriglyceridemia seen in Pxr-null mice.
Cyp2b10 upregulation by EtOH in WT mice, as seen in our study, produces reactive oxygen species (ROS) and increased oxidative stress (Dostalek et al., 2007), an important feature of ALD. Interestingly, CYP2E1 only plays a minor role in the mechanism of early alcoholic liver injury, whereas both CYP2B and CYP3A can be major sources of ROS in alcoholic hepatitis (Kono et al., 1999). While EtOH administration caused upregulation of CYP2E1 protein in both genotypes, markers of oxidative stress including thiobarbituric acid reactive substances were elevated only in EtOH-fed WT mice in association with decreased Aldh1a1 and Akr1b8 expression. Unexpectedly, mRNA levels of Akr1b7, another common transcriptional target of both PXR and CAR that is known to detoxify lipid peroxides (Spite et al., 2007; Liu et al., 2009), was massively upregulated in both control and binge EtOH-fed Pxr-null mice compared with EtOH-fed WT mice. It is not clear why basal Akr1b7 mRNA levels are higher in Pxr-null mice. However, in our study, mRNA levels of the liver X receptor, a nuclear receptor known to regulate Akr1b7 (Volle et al., 2004), did not differ between the two genotypes fed saline (data not shown). Furthermore, both Akr1b7 and Akr1b8 are nuclear factor E2–related factor-2 target genes (Beneš et al., 2013; Luo et al., 2015). Whether the elevated Akr1b7 mRNA levels seen in saline-fed Pxr-null mice involve nuclear factor E2–related factor-2 activation remains to be determined. Nevertheless, it is reasonable to propose that elevated Akr1b7 gene expression and resistance to EtOH-induced suppression of both Aldh1a1 and Akr1b8 gene/protein expression may protect against EtOH-induced oxidative stress in Pxr-null mice.
The antiapoptotic protein Bcl-2 can protect cells against ROS (Li et al., 2004). In agreement, Bcl-2 protein was suppressed in EtOH-exposed WT mice, whereas the proapoptotic Bax protein and other oxidative stress or endoplasmic reticulum stress markers including the Ucp2 gene, GRP78, and phospho-elf2α protein (Carmiel-Haggai et al., 2003; Ji and Kaplowitz, 2003) were increased in EtOH-exposed WT mice. While EtOH-induced activation of LPO was noted in EtOH-fed WT mice, unexpectedly, markers of liver damage, ALT and AST were not significantly increased in EtOH-fed WT mice. Similarly, in a previous study, three binge doses of EtOH (6 g/kg, every 12 hours) also failed to induce ALT activity (Abdelmegeed et al., 2013). In contrast with our findings, concomitant with increased LPO, serum ALT activity was significantly increased in male PXR-humanized mice after the same dose of acute EtOH (4.5 g/kg) was administered (Spruiell et al., 2015). Taken together, these findings are consistent with previous studies demonstrating the hepatotoxic effects of EtOH-induced activation of Cyp2b10 due to increased oxidative stress and suppression of EtOH-metabolizing enzymes (Chen et al., 2011; Koga et al., 2016; Wang et al., 2017).
In summary, our findings suggest that PXR ablation prevents binge EtOH-induced upregulation of Cyp2b10 mRNA and CYP2B10 protein expression, suppresses EtOH metabolism, increases BEC, and activates oxidative stress and apoptosis but promotes alcoholic hypertriglyceridemia. However, both PXR and PXR ablation enhance binge EtOH-induced steatosis. Furthermore, our results also suggest that PXR is involved in the transcriptional suppression of Akr1b7 expression and that the PXR-dependent effects of EtOH as determined using Pxr-null mice have clinical relevance to EtOH-induced oxidative stress.
Acknowledgments
We thank Dr. Susan T. Yeyeodu and Dr. William J. Arendshorst for critical reading of the manuscript.
Abbreviations
- ACC
acetyl-CoA carboxylase
- ADH
alcohol dehydrogenase
- AKR
aldo-keto reductase
- ALT
alanine aminotransferase
- ALDH
aldehyde dehydrogenase
- ApoB
apolipoprotein B
- AST
aspartate aminotransferase
- BEC
blood ethanol concentration
- CAR
constitutive androstane receptor
- CPT
carnitine palmitoyltransferase
- DGAT
diacylglycerol acyltransferase
- EtOH
ethanol
- FAS
fatty acid synthase
- FXR
farnesoid X receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GRP
glucose-regulated protein
- HMGCR
3-hydroxy-3-methyl-glutaryl CoA reductase
- LFABP
liver fatty acid binding protein
- LPO
lipid peroxidation
- MDA
malondialdehyde
- PCN
pregnenolone 16α-carbonitrile
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator–activated receptor
- PXR
pregnane X receptor
- ROS
reactive oxygen species
- RXRa
retinoid X receptor a
- SCD
stearoyl-CoA desaturase
- SHP
small heterodimer partner
- SREBP
sterol regulatory element binding protein
- UCP
uncoupling protein
- WT
wild-type
Authorship Contributions
Participated in research design: M. A. Gyamfi.
Conducted experiments: S. Choi, A. A. Gyamfi, P. Neequaye, S. Addo, M. A. Gyamfi.
Contributed new reagents or analytic tools: F. Gonzalez, M. A. Gyamfi.
Performed data analysis: S. Choi, M. A. Gyamfi.
Wrote or contributed to the writing of the manuscript: F. Gonzalez, M. A. Gyamfi.
Footnotes
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grant U54-AA019765] and the National Institutes of Health National Institute on Minority Health and Health Disparities [Grants 1U54-MD012392 and P20-MD000175]. This work was also supported in part by the Intramural Research Program of the National Institutes of Health National Cancer Institute.
References
- Abdelmegeed MA, Banerjee A, Jang S, Yoo SH, Yun JW, Gonzalez FJ, Keshavarzian A, Song BJ (2013) CYP2E1 potentiates binge alcohol-induced gut leakiness, steatohepatitis, and apoptosis. Free Radic Biol Med 65:1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneš H, Vuong MK, Boerma M, McElhanon KE, Siegel ER, Singh SP (2013) Protection from oxidative and electrophilic stress in the Gsta4-null mouse heart. Cardiovasc Toxicol 13:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter A, Rümmele P, Klein K, Kandel BA, Rieger JK, Nüssler AK, Zanger UM, Trauner M, Schwab M, Burk O (2015) Pregnane X receptor activation and silencing promote steatosis of human hepatic cells by distinct lipogenic mechanisms. Arch Toxicol 89:2089–2103. [DOI] [PubMed] [Google Scholar]
- Carmiel-Haggai M, Cederbaum AI, Nieto N (2003) Binge ethanol exposure increases liver injury in obese rats. Gastroenterology 125:1818–1833. [DOI] [PubMed] [Google Scholar]
- Carson EJ, Pruett SB (1996) Development and characterization of a binge drinking model in mice for evaluation of the immunological effects of ethanol. Alcohol Clin Exp Res 20:132–138. [DOI] [PubMed] [Google Scholar]
- Chen X, Meng Z, Wang X, Zeng S, Huang W (2011) The nuclear receptor CAR modulates alcohol-induced liver injury. Lab Invest 91:1136–1145. [DOI] [PubMed] [Google Scholar]
- Choi S, Neequaye P, French SW, Gonzalez FJ, Gyamfi MA (2018) Pregnane X receptor promotes ethanol-induced hepatosteatosis in mice. J Biol Chem 293:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL (2006) Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem 281:26540–26551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Staudinger JL (2005) The ratio of constitutive androstane receptor to pregnane X receptor determines the activity of guggulsterone against the Cyp2b10 promoter. J Pharmacol Exp Ther 314:120–127. [DOI] [PubMed] [Google Scholar]
- Dostalek M, Brooks JD, Hardy KD, Milne GL, Moore MM, Sharma S, Morrow JD, Guengerich FP (2007) In vivo oxidative damage in rats is associated with barbiturate response but not other cytochrome P450 inducers. Mol Pharmacol 72:1419–1424. [DOI] [PubMed] [Google Scholar]
- Gyamfi MA, He L, French SW, Damjanov I, Wan YJ (2008) Hepatocyte retinoid X receptor alpha-dependent regulation of lipid homeostasis and inflammatory cytokine expression contributes to alcohol-induced liver injury. J Pharmacol Exp Ther 324:443–453. [DOI] [PubMed] [Google Scholar]
- Gyamfi MA, Kocsis MG, He L, Dai G, Mendy AJ, Wan YJ (2006) The role of retinoid X receptor alpha in regulating alcohol metabolism. J Pharmacol Exp Ther 319:360–368. [DOI] [PubMed] [Google Scholar]
- Gyamfi MA, Wan YJ (2010) Pathogenesis of alcoholic liver disease: the role of nuclear receptors. Exp Biol Med (Maywood) 235:547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Court MH, Greenblatt DJ, von Moltke LL (2006) Factors influencing midazolam hydroxylation activity in human liver microsomes. Drug Metab Dispos 34:1198–1207. [DOI] [PubMed] [Google Scholar]
- Heit C, Dong H, Chen Y, Shah YM, Thompson DC, Vasiliou V (2015) Transgenic mouse models for alcohol metabolism, toxicity, and cancer. Adv Exp Med Biol 815:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N (2003) Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology 124:1488–1499. [DOI] [PubMed] [Google Scholar]
- Kersten S (2008) Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Res 2008:132960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Yao PL, Goudarzi M, Murray IA, Balandaram G, Gonzalez FJ, Perdew GH, Fornace AJ Jr, Peters JM (2016) Regulation of cytochrome P450 2B10 (CYP2B10) expression in liver by peroxisome proliferator-activated receptor-β/δ modulation of SP1 promoter occupancy. J Biol Chem 291:25255–25263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Bradford BU, Yin M, Sulik KK, Koop DR, Peters JM, Gonzalez FJ, McDonald T, Dikalova A, Kadiiska MB, et al. (1999) CYP2E1 is not involved in early alcohol-induced liver injury. Am J Physiol 277:G1259–G1267. [DOI] [PubMed] [Google Scholar]
- Kostrubsky VE, Strom SC, Wood SG, Wrighton SA, Sinclair PR, Sinclair JF (1995) Ethanol and isopentanol increase CYP3A and CYP2E in primary cultures of human hepatocytes. Arch Biochem Biophys 322:516–520. [DOI] [PubMed] [Google Scholar]
- Langhi C, Pedraz-Cuesta E, Haro D, Marrero PF, Rodríguez JC (2013) Regulation of human class I alcohol dehydrogenases by bile acids. J Lipid Res 54:2475–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Ueta E, Kimura T, Yamamoto T, Osaki T (2004) Reactive oxygen species (ROS) control the expression of Bcl-2 family proteins by regulating their phosphorylation and ubiquitination. Cancer Sci 95:644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Takahashi Y, Wada T, He J, Gao J, Tian Y, Li S, Xie W (2009) The aldo-keto reductase Akr1b7 gene is a common transcriptional target of xenobiotic receptors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol 76:604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Chen Y, Wu D, Shou J, Wang S, Ye J, Tang X, Wang XJ (2015) Butylated hydroxyanisole induces distinct expression patterns of Nrf2 and detoxification enzymes in the liver and small intestine of C57BL/6 mice. Toxicol Appl Pharmacol 288:339–348. [DOI] [PubMed] [Google Scholar]
- Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao DF, Cao D (2008) Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. J Biol Chem 283:3418–3423. [DOI] [PubMed] [Google Scholar]
- Makia NL, Bojang P, Falkner KC, Conklin DJ, Prough RA (2011) Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem Biol Interact 191:278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher EA, Barry OP, Burke A, Lucey MR, Lawson JA, Rokach J, FitzGerald GA (1999) Alcohol-induced generation of lipid peroxidation products in humans. J Clin Invest 104:805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey E, Rennie-Tankersley L, Potter JJ (2005) Effect of leptin on liver alcohol dehydrogenase. Biochem Biophys Res Commun 337:1324–1329. [DOI] [PubMed] [Google Scholar]
- Moreau A, Téruel C, Beylot M, Albalea V, Tamasi V, Umbdenstock T, Parmentier Y, Sa-Cunha A, Suc B, Fabre JM, et al. (2009) A novel pregnane X receptor and S14-mediated lipogenic pathway in human hepatocyte. Hepatology 49:2068–2079. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Moore R, Negishi M, Sueyoshi T (2007) Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem 282:9768–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä O, Parkkila S, Juvonen RO, Viitala K, Gelboin HV, Pasanen M (2000) Cytochromes P450 2A6, 2E1, and 3A and production of protein-aldehyde adducts in the liver of patients with alcoholic and non-alcoholic liver diseases. J Hepatol 33:893–901. [DOI] [PubMed] [Google Scholar]
- Pang M, de la Monte SM, Longato L, Tong M, He J, Chaudhry R, Duan K, Ouh J, Wands JR (2009) PPARdelta agonist attenuates alcohol-induced hepatic insulin resistance and improves liver injury and repair. J Hepatol 50:1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish KM, Dufour MC, Stinson FS, Harford TC (1993) Average daily alcohol consumption during adult life among decedents with and without cirrhosis: the 1986 National Mortality Followback Survey. J Stud Alcohol 54:450–456. [DOI] [PubMed] [Google Scholar]
- Pavek P, Dvorak Z (2008) Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr Drug Metab 9:129–143. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Chang SL (2013) Ethanol concentration-dependent alterations in gene expression during acute binge drinking in the HIV-1 transgenic rat. Alcohol Clin Exp Res 37:1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Pruett SB, Szabo G, Arteel GE (2013) Binge ethanol and liver: new molecular developments. Alcohol Clin Exp Res 37:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spite M, Baba SP, Ahmed Y, Barski OA, Nijhawan K, Petrash JM, Bhatnagar A, Srivastava S (2007) Substrate specificity and catalytic efficiency of aldo-keto reductases with phospholipid aldehydes. Biochem J 405:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruiell K, Gyamfi AA, Yeyeodu ST, Richardson RM, Gonzalez FJ, Gyamfi MA (2015) Pregnane X receptor-humanized mice recapitulate gender differences in ethanol metabolism but not hepatotoxicity. J Pharmacol Exp Ther 354:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruiell K, Jones DZ, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA (2014a) Role of human pregnane X receptor in high fat diet-induced obesity in pre-menopausal female mice. Biochem Pharmacol 89:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruiell K, Richardson RM, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA (2014b) Role of pregnane X receptor in obesity and glucose homeostasis in male mice. J Biol Chem 289:3244–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Aoyama T, Kimura S, Gonzalez FJ (2017) Targeting nuclear receptors for the treatment of fatty liver disease. Pharmacol Ther 179:142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirard J, Gout J, Lefrançois-Martinez AM, Martinez A, Begeot M, Naville D (2007) A novel inhibitory protein in adipose tissue, the aldo-keto reductase AKR1B7: its role in adipogenesis. Endocrinology 148:1996–2005. [DOI] [PubMed] [Google Scholar]
- Tsuzura H, Genda T, Sato S, Murata A, Kanemitsu Y, Narita Y, Ishikawa S, Kikuchi T, Mori M, Hirano K, et al. (2014) Expression of aldo-keto reductase family 1 member b10 in the early stages of human hepatocarcinogenesis. Int J Mol Sci 15:6556–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle DH, Repa JJ, Mazur A, Cummins CL, Val P, Henry-Berger J, Caira F, Veyssiere G, Mangelsdorf DJ, Lobaccaro JM (2004) Regulation of the aldo-keto reductase gene akr1b7 by the nuclear oxysterol receptor LXRalpha (liver X receptor-alpha) in the mouse intestine: putative role of LXRs in lipid detoxification processes. Mol Endocrinol 18:888–898. [DOI] [PubMed] [Google Scholar]
- Wang H, Negishi M (2003) Transcriptional regulation of cytochrome p450 2B genes by nuclear receptors. Curr Drug Metab 4:515–525. [DOI] [PubMed] [Google Scholar]
- Wang X, Zheng L, Wu J, Tang B, Zhang M, Zhu D, Lin X (2017) Constitutive androstane receptor activation promotes bilirubin clearance in a murine model of alcoholic liver disease. Mol Med Rep 15:3459–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwi CA, Gupte M, Waxman DJ (2004) Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4alpha-deficient mice. Mol Endocrinol 18:1975–1987. [DOI] [PubMed] [Google Scholar]
- Wu W, Zhu B, Peng X, Zhou M, Jia D, Gu J (2014) Activation of farnesoid X receptor attenuates hepatic injury in a murine model of alcoholic liver disease. Biochem Biophys Res Commun 443:68–73. [DOI] [PubMed] [Google Scholar]
- Zakhari S, Li TK (2007) Determinants of alcohol use and abuse: impact of quantity and frequency patterns on liver disease. Hepatology 46:2032–2039. [DOI] [PubMed] [Google Scholar]
- Zemánková K, Makoveichuk E, Vlasáková Z, Olivecrona G, Kovář J (2015) Acute alcohol consumption downregulates lipoprotein lipase activity in vivo. Metabolism 64:1592–1596. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W (2006) A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 281:15013–15020. [DOI] [PMC free article] [PubMed] [Google Scholar]