Abstract
The Coronavirus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) became a pandemic, resulting in an exponentially increased mortality globally and scientists all over the world are struggling to find suitable solutions to combat it. Multiple repurposed drugs have already been in several clinical trials or recently completed. However, none of them shows any promising effect in combating COVID-19. Therefore, developing an effective drug is an unmet global need. RdRp (RNA dependent RNA polymerase) plays a pivotal role in viral replication. Therefore, it is considered as a prime target of drugs that may treat COVID-19. In this study, we have screened a library of compounds, containing approved RdRp inhibitor drugs that were or in use to treat other viruses (favipiravir, sofosbuvir, ribavirin, lopinavir, tenofovir, ritonavir, galidesivir and remdesivir) and their structural analogues, in order to identify potential inhibitors of SARS-CoV-2 RdRp. Extensive screening, molecular docking and molecular dynamics show that five structural analogues have notable inhibitory effects against RdRp of SARS-CoV-2. Importantly, comparative protein-antagonists interaction revealed that these compounds fit well in the pocket of RdRp. ADMET analysis of these compounds suggests their potency as drug candidates. Our identified compounds may serve as potential therapeutics for COVID-19.
Keywords: COVID-19, RNA dependent RNA polymerase, Binding free energy, Molecular docking, Virtual screening, Molecular dynamics simulation
COVID-19, RNA dependent RNA polymerase, Binding free energy, Molecular docking, Virtual screening, Molecular dynamics simulation.
1. Introduction
SARS-CoV-2 is a novel coronavirus that first emerged in late 2019 at Wuhan of China (Ludwig and Zarbock, 2020). This is the 7th coronavirus species discovered, belonging to the order Nidovirales of the Coronaviridae family. Since its emergence, till 16th February 2021, it has reportedly infected 109,706,925 people and caused 2,419,664 deaths in total (Coronavirus Update (Live): 109,706,925 Cases and 2,419,664 Deaths from COVID-19 Virus Pandemic - Worldometer). Ever since the disease broke out in China, the whole world started taking precautionary measures but failed to stop its rapid spread across the globe. Due to its extreme outbreak, the World Health Organization (WHO) declared a state of emergency and called on a global pandemic on March 11, 2020 (Brinks and Ibert, 2020). Initially, the disease was characterized by fever, coughing, sneezing, shortness of breath, or breathing difficulties (Huang et al., 2020); in severe cases the disease can result in pneumonia and multiple organ failure leading to death. Recent cases complaining of a higher rate of renal impairment indicates its possible role in rendering kidney dysfunction (Li et al., 2020). Similar to an influenza outbreak, COVID-19 causes myocardial injury that may be related to increased viscosity, heightened coagulation cascade, pro-inflammatory effects, or endothelial cell dysfunction caused by SARS-CoV-2 virus (JC et al., 2018; Nguyen et al., 2016). The virus attaches to host cells mostly in the upper respiratory tract via an interaction between spike glycoprotein of the virus and ACE-2 receptor of human (Tai et al., 2020).
The molecular biology of this novel coronavirus has already been vividly studied and several salient features of its genome organization have been elucidated in a number of studies. The RNA genome consists of at least six open reading frames (ORF) (Ahmed et al., 2020). One ORF of special importance is the ORF 1a/b which constitutes two-thirds of the genome and synthesizes two very important polypeptides namely polypeptide 1a and polypeptide 1b which eventually produce 16 nonspecific proteins (NSPs) of the virus. The other 5 ORFs constitute nearly one-third of the genome and produce four structural proteins for the virus, which includes spike glycoprotein (S), envelope glycoprotein (E), membrane protein (M) and nucleoprotein (N) (Woo et al., 2010). Eleven cleavage sites have so far been discovered within NSP5 that generates many important proteins, including RdRp. RdRp plays crucial role in the replication of RNA viruses that makes it an ideal drug target for anti-viral drug development against RNA viruses. For treating many RNA virus infections such as Hepatitis C, Zika and several coronaviruses, many anti-viral drugs have already been developed, which target the viral RdRp enzyme (Elfiky, 2020). Some antiviral drugs, including remdesivir and favipiravir, have been briefly tested against SARS-CoV-2 RdRp, though partially successful further validation is required for the clinical approval to use those for treating SARS-CoV-2 infection (Harrison, 2020; Lung et al., 2020; Zhang and Zhou, 2020).
Over the last ten months, scientists have come up with different ways to alleviate the severity of the disease and reduce its contagiousness (Van Kampen et al., 2020). So far, we have only been able to battle the disease by maintaining social distancing, quarantine and hygiene. Very recently, many countries in the world have already started running the vaccination program (D. D. Li and Li, 2021; Rubin and Longo, 2020). Yet, due to production and budget limitation it will take considerable time to vaccinate the entire population of the world. Importantly, this is uncertain whether the currently developed vaccines will give durable immunity against the virus. In addition, acquiring newer mutations by the virus may render the vaccines ineffective or the virus may make its way to infect vaccinated individuals. Therefore, discovery of effective drugs for treating COVID-19 patients is an urgent need. The enzyme, RdRp plays a pivotal role in viral replication, having no host homologues and, therefore, developing therapeutics that inhibit the enzyme would not only inhibit viral replication but also minimize any potential risks in host cells. A number of FDA-approved RdRp inhibitor drugs, which include remdesivir, favipiravir, sofosbuvir, ribavirin, lopinavir, ritonavir, tenofovir and galidesivir, are effective against a broad range of RNA viruses, including past coronaviruses (Li and De Clercq, 2020; Mitjà and Clotet, 2020; Şimşek Yavuz and Ünal, 2020; Sinha and Balayla, 2020). Although remdesivir, an experimental Ebola virus drug, has been approved for emergency use only for hospitalized patients and recent clinical data suggest a moderate clinical improvement by reducing the recovery period (Beigel et al., 2020), however, no impact on mortality; therefore, the potentiality of the drug is not as promising as expected. Thus, identifying novel therapeutics that have the potential to effectively combat the disease is an unmet need. To address this, we, herein, virtually screened FDA approved RdRp inhibitor drugs and their structural analogues (~2400 compounds) based on molecular docking analysis of these compounds in order to identify drug candidates that can potentially inhibit SARS-CoV-2 RdRp. SARS-CoV nsp7 and nsp8 activate and confer the processivity to the nsp12 RNA-synthesizing activity (Subissi et al., 2014). SARS-CoV-2 RNA-dependent RNA polymerase (PDB ID: 6M71) is a complex with those cofactors (nsp7 and nsp8) and nsp12 is a target for nucleotide analog of several antiviral inhibitors and the structure may provide a basis for new antiviral therapeutics (Gao et al., 2020). Based on docking scores, protein-ligand interactions, molecular dynamic simulations and free energy calculation we predicted five RdRp inhibitor drug candidates that may have the potential to combat life-threatening SARS-CoV-2 infection, and therefore, may serve as a cure to treat the disease, addressing the current global need.
2. Methods
2.1. Ligand selection and preparation
An extensive literature assessment was performed to gather information on antiviral drugs that target and inhibit RdRp enzyme. The ability to block RdRp with various extents has been reported in other RNA viruses, which includes H1N11 (Lin et al., 2015), HCV2 (Hofmann et al., 2008), HIV3 (Blair and Cox, 2016), SARS (Fani et al., 2020) and MERS (Fani et al., 2020). Eight antiviral compounds: favipiravir (Allen et al., 2020), sofosbuvir (Buonaguro and Buonaguro, 2020), lopinavir (Chakraborty and Das, 2020), tenofovir (Allen et al., 2020), ribavirin (Gish, 2006), ritonavir (Chakraborty and Das, 2020), galidesivir (Allen et al., 2020) and remdesivir (Fani et al., 2020) have been primarily reported to inhibit RdRp of these viruses. To determine their potential in inhibiting SARS CoV-2 RdRp computationally, their SDF (Spatial Data File) files were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Moreover, structurally similar compounds of the inhibitor drugs were downloaded from Swiss similarity software (Zoete et al., 2016) (http://www.swisssimilarity.ch/) by activating zinc drug-like features. A library of ~2400 compounds has been prepared.
2.2. Macromolecule selection and preparation
The crystal structure of RdRp of SARS-CoV-2 was collected from the RCSB Protein Data Bank (https://www.rcsb.org/search) (PDB ID: 6M71) (Gao et al., 2020) followed by optimization and processing of the structure by Swiss-PDB viewer software packages (Guex and Peitsch, 1997) (https://spdbv.vital-it.ch/) (version 4.1.0) based on their least energy. Some significant factors, such as side-chain geometry, improper bond order, and missing hydrogen, were observed in the crystal structure of the RdRp protein. PyMol (version 1.1) software package (Adams et al., 2010) (https://pymol.org/2/) was used to remove all the water molecules, hetero atoms, and inhibitor existent in the Crystal structure.
2.3. Virtual screening and ligand preparation
As mentioned above the structural analogues of the inhibitor drugs were obtained from the ZINC database (Sterling and Irwin, 2015) for the best optimal hit against the mentioned target (Figure 1). PyRx (Dallakyan and Olson, 2015) from MGLTools (https://ccsb.scripps.edu/mgltools/) was used in the virtual screening executed in the study. For computational drug discovery, PyRx is used as a Virtual Screening software that screens libraries of compounds against potential drug targets according to their target binding affinities.
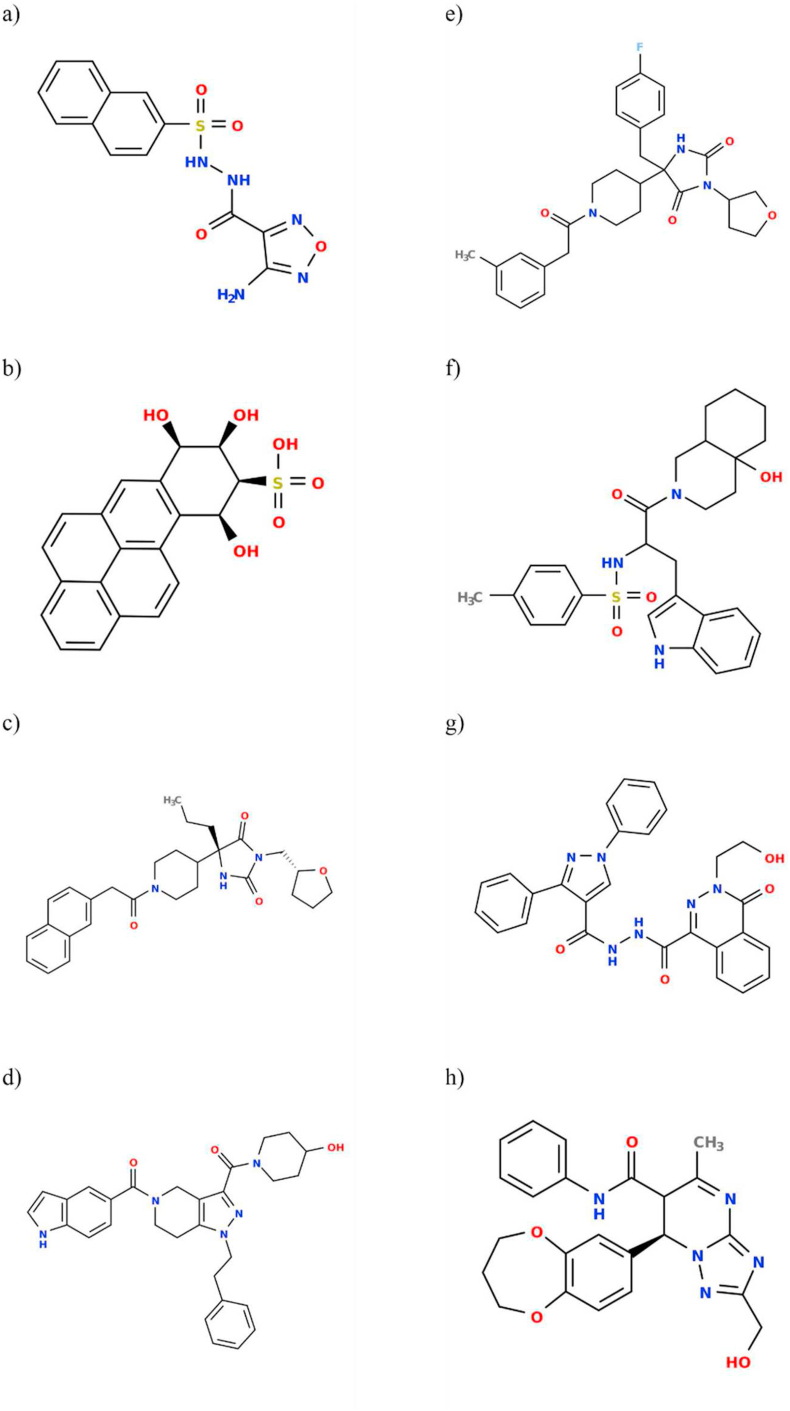
Figure 1.
The chemical structures of ligands that were selected based on the initial virtual screen. a) ZINC000005605139, b) ZINC000006094731, c) ZINC000012161475, d) ZINC000014751834 and e) ZINC000014882040, f) ZINC000012863240, g) ZINC000049581065, h) ZINC000072460420.
2.4. Molecular docking studies
The protein-ligand binding interaction of chosen protein-ligand complexes was performed using Autodock Vina (Trott and Olson, 2010). The docking analyses were performed using a semi-flexible docking approach. Briefly, the protein RdRp was kept rigid and ligands were kept flexible. The allowed degrees of freedom for ligand molecules were considered 10. The steps involving the conversion of molecules into pdbqt format, box type, grid box generation, etc. were specified by AutoDock. Since remdesivir were considered as a potential drug for SARS-CoV-2 based on some initial in vitro findings (Shannon et al., 2020) and the site of interaction of the remdesivir to SARS-CoV-2 RdRp has already been determined (Gao et al., 2020) we have chosen the interaction site for determining the inhibitory potential of the selected compounds in molecular docking. Critical 11 amino acid positions of A chain of RdRp (PDB ID: 6m71) is important for remdesivir binding and were selected for creating remdesivir incorporation model. These include K545, R553, R555, V557, D618, S 623, T680, D682, N691, D760 and D761. The grid box was set to size 22.32 Å × 23.91 Å × 32 .57 Å (x, y and z) parameters and the center 118.660 Å × 117.665 Å × 131.857 Å (x, y and z) parameters. The docking results were analyzed using Discovery studio visualizer (“BIOVIA Discovery Studio — Discngine,”) and Molecular Operating Environment (Vilar et al., 2008) software, and the docking of ligands to RdRp with the least energy was considered to have a significant binding affinity.
2.5. Drug likeness analysis of the potential drug candidates
ADMET profiling is a significant pointer in defining the effectiveness and safety of a drug compound before bringing it into commercialization. Due to cost-effectiveness and proper timing in the experimental evaluation of ADMET properties, computational methods have been developed as a practicable alternative in the high-throughput drug discovery process (Shen et al., 2010). The ADMET properties of the 8 top-ranked ligands were calculated by the online ADMET structure-activity database (admetSAR) (Yang et al., 2019) (http://lmmd.ecust.edu.cn/admetsar2/) and pkCSM (Pires et al., 2015) (http://biosig.unimelb.edu.au/pkcsm/) where SMILES (simplified molecular-input line-entry system) strings were utilized throughout the whole process.
2.6. Molecular dynamics simulation with MMGBSA scoring
Complexes were solvated with TIP3P water model employing a dodecahedron box with a 12 Å packing using tleap available in Amber16 package (Case et al., 2005). The systems were then neutralized with Na+ and Cl− ions until neutrality. The protein, water molecules, and ions were defined by the ff14SB AMBER force field (Duan et al., 2003). Ligands were parameterized by assigning AM1-BCC atomic charges employing the general AMBER force field (GAFF) (Wang et al., 2004). Simulations were run at the NPT ensemble at 310 K via a Langevin thermostat, 1 bar of pressure, and an isotropic Berendsen barostat. Periodic boundary conditions (PBC) were considered with a cutoff of 10 Å for the nonbonded interactions, whereas the electrostatic term was described via the PME method (Darden et al., 1993). A numeric time step of 2 fs and the SHAKE algorithm (Van Gunsteren and Berendsen, 1977) was chosen to constrain bond lengths at individual equilibrium. After constructions and previous MD simulations, a minimization and relaxing protocol was performed comprising of: 1,000 steps of energy minimization by conjugate gradient method; 1000 ps of pre-relaxation with harmonic restraint for protein and ligand; 1000 ps relaxation removing the ligand restrains; 1000 ps of relaxation with no restrictions to the side chains of the residues 5 Å around the ligand; 1000 ps of NPT relaxation without restrictions for the entire residues around the ligand; 50 ns of NPT MD simulations. The RMSD and clustering analysis was performed using the cpptraj tool in Amber16 to check system equilibration and identification of the most populated conformation during the MD simulations. Images were built using PyMOL (“DeLano (2002). The PyMOL Molecular Graphics System. Delano Scientific, San Carlos. - References - Scientific Research Publishing,”).
The molecular mechanics-generalized Born surface area (MMGBSA) (Belouzard et al., 2012) approach was employed to determine the binding free energy (ΔGmmgbsa). This analysis was performed over the last 20 ns of MD simulation, selecting a total of 2000 snapshots at time intervals of 10 ps, using implicit solvent models (Feig et al., 2004), and a salt concentration of 0.10 M. The ΔGmmgbsa values were determined as previously described (Bello, 2018) and embody average values of triplicate MD simulations.
3. Results
3.1. Virtual screening, ligand selection and molecular docking studies
In order to identify potential drug candidates targeting SARS-CoV-2 RdRp we adopted a computational approach. Initially, with extensive literature mining, we selected 8 FDA-approved RdRp inhibitor drugs (favipiravir (Allen et al., 2020), sofosbuvir (Buonaguro and Buonaguro, 2020), lopinavir (Chakraborty and Das, 2020), tenofovir (Allen et al., 2020), ribavirin (Gish, 2006), ritonavir (Chakraborty and Das, 2020), galidesivir (Allen et al., 2020) and remdesivir (Fani et al., 2020) that are effective against broad ranges of RNA viruses, including past coronaviruses. We hypothesized that these inhibitor drugs and/or their structural analogues could have the potential to inhibit SARS-CoV-2 RdRp. Following downloading these drugs and their structural analogues (detailed in method section), we have performed an initial virtual screening using PyRx that eventually yielded 8 best compound hits. The structures of the 8 best compound hits are shown in Figure 1. To further validate the initial screen, we later performed molecular docking with AutoDock Vina (Trott and Olson, 2010) and the drug binding scores for the above mentioned approved drugs and their best hit analogues suggest high affinity binding (Table 1 and Table 2), further strengthening our initial findings. It is important to mention that the structural analogues were predicted to have better binding than the existing FDA-approved drugs tested in this study, as evident from the binding scores (Tables 1 and 2).
Table 1.
Autodock Vina docking results of the selected antiviral drugs.
| Compound | PMCID | Score (KJ/mol) |
|---|---|---|
| Remdesivir | 121304016 | -6.5 |
| Lopinavir | 92727 | -7 |
| Ribavirin | 37542 | -6.2 |
| Galidesivir | 10445549 | -6.8 |
| Sofosbuvir | 45375808 | -7.5 |
| Favipiravir | 492405 | -5.5 |
| Tenofovir | 464205 | -5.9 |
| Ritonavir | 392622 | -7.2 |
Table 2.
Autodock Vina docking results of the top-ranking ligands.
| Compound | IUPAC name | Score (KJ/mol) |
|---|---|---|
| ZINC000005605139 | 4-Amino-N′-naphthalen-2-ylsulfonyl-1,2,5-oxadiazole-3- carbohydrazide | -7.9 |
| ZINC000006094731 | (7R,8R,9S,10S)-7,8,10-Trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene-9-sulfonic acid | -8.1 |
| ZINC000012161475 | 5-[1-(2-Naphthalen-2-ylacetyl)piperidin-4-yl]-3-(oxolan-2-ylmethyl)-5-propylimidazolidine-2,4-dione | -8.9 |
| ZINC000014751834 | [3-(4-Hydroxypiperidine-1-carbonyl)-1-(2-phenylethyl)-6,7-dihydro-4H-pyrazolo[4,3-c]pyridin-5-yl]-(1H-indol-5-yl)methanone | -8.9 |
| ZINC000014882040 | 5-[(4-Fluorophenyl)methyl]-5-[1-[2-(3-methylphenyl) acetyl]piperidin-4-yl]-3-(oxolan-3-yl)imidazolidine-2,4-dione | -9.1 |
| ZINC000012863240 | N-[1-(4a-Hydroxy-1,3,4,5,6,7,8,8a-octahydroisoquinolin-2-yl)-3-(1H-indol-3-yl)-1-oxopropan-2-yl]-4-methylbenzenesulfonamide | -9.1 |
| ZINC000049581065 | N'-(1,3-Diphenylpyrazole-4-carbonyl)-3-(2-hydroxyethyl)-4-oxophthalazine-1-carbohydrazide | -9.1 |
| ZINC000072460420 | 7-(3,4-Dihydro-2H-1,5-benzodioxepin-7-yl)-2-(hydroxymethyl)-5-methyl-N-phenyl-1,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidine-6-carboxamide | -8.6 |
3.2. Docking analyses and drug interactions
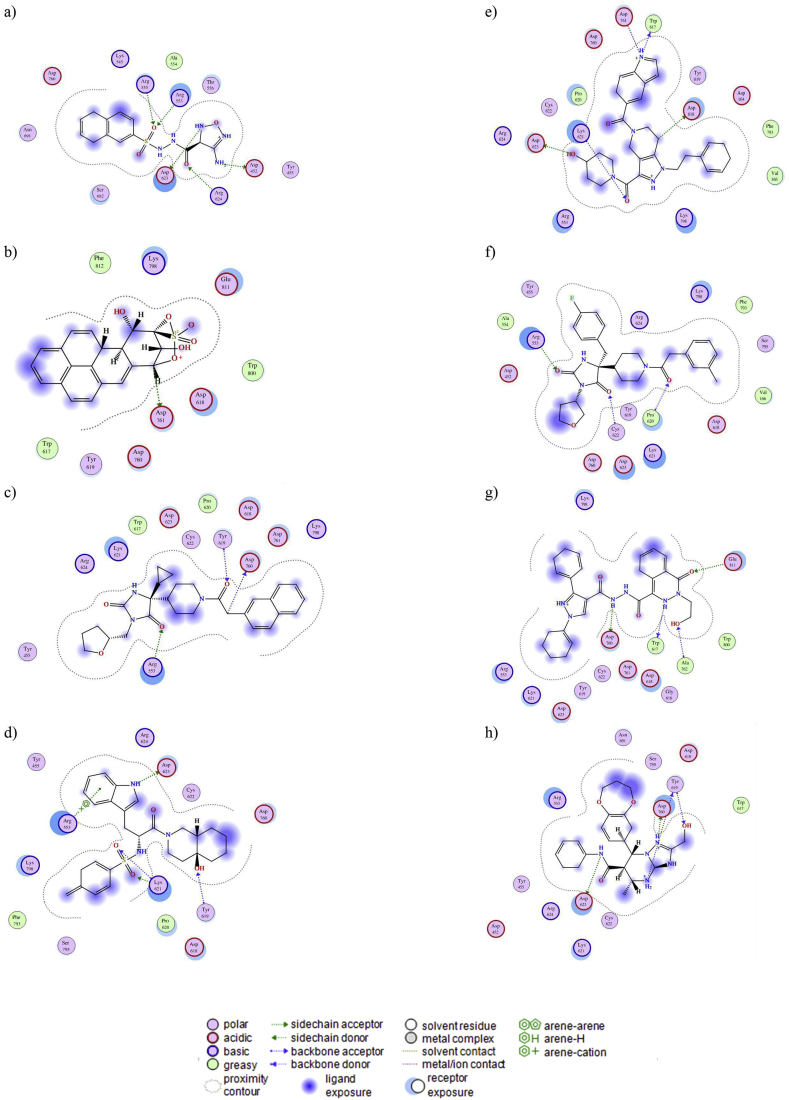
The chosen 8 compounds predicted (by molecular docking) to bind effectively with RdRp were selected for further analysis (the highest-ranked bound compound in all poses made as a cluster at the ligand-binding sites of these target protein). The analyses were performed to check the efficiency of the ligand and the state of interaction using the Discovery studio visualizer (“BIOVIA Discovery Studio — Discngine,”) and show that all the 8 ligands bind efficiently and interact well with RdRp (Figure 2).
Figure 2.
Molecular docking of probable inhibitory molecules onto RdRp. Panels (a−h) show the 2D structures of the binding interaction of the ligands inside the pocket of SARS Cov-2 RdRp a) ZINC000005605139, b) ZINC000006094731, c) ZINC000012161475, d) ZINC000014751834 and e) ZINC000014882040, f) ZINC000012863240, g) ZINC000049581065, h) ZINC000072460420. The types of interaction are shown at the end of the figure.
3.3. Stability of simulated systems
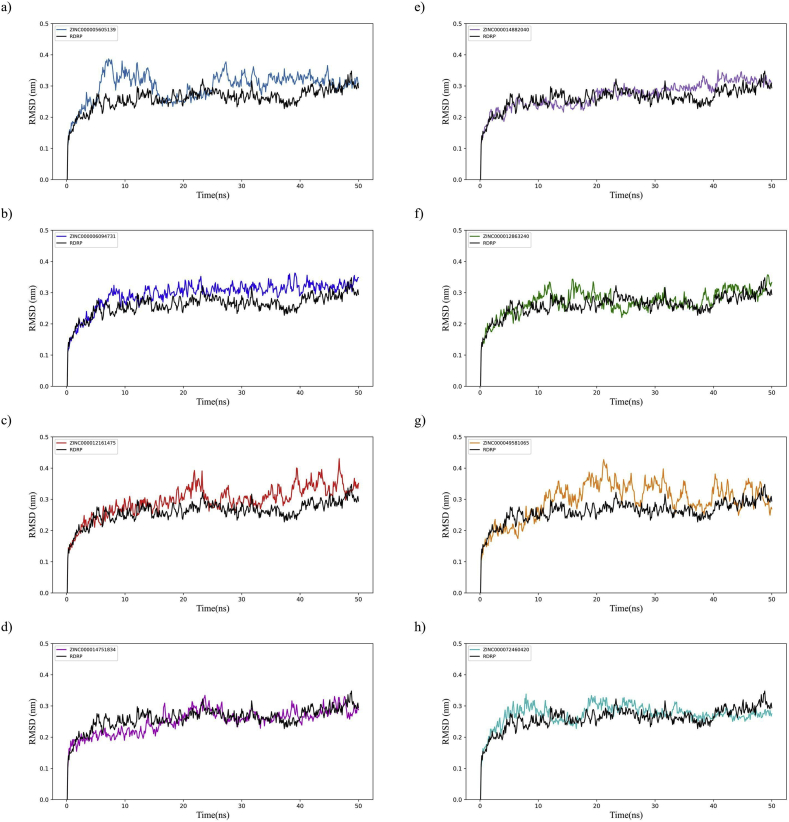
The RMSD analysis plots show the mobility of the receptor-ligand complexes and constant values suggest that the system reached equilibrium (Figure 3). The RMSD values of backbone atoms reached an equilibrium state between 20 to 30 ns with RMSD values that oscillated between 2.7 to 3.2 Å. Based on this, further analyses were performed considering the last 30 ns.
Figure 3.
RMSD pattern of docked complexes compared to Cov-2 RdRp. a) ZINC000005605139, b) ZINC000006094731, c) ZINC000012161475, d) ZINC000014751834 and e) ZINC000014882040, f) ZINC000012863240, g) ZINC000049581065, h) ZINC000072460420. The RMSD pattern show that all the small molecules docked to the RdRp protein remain relatively stable under the simulated environment.
3.4. Structural analysis of receptor-ligand complexes through MD simulation
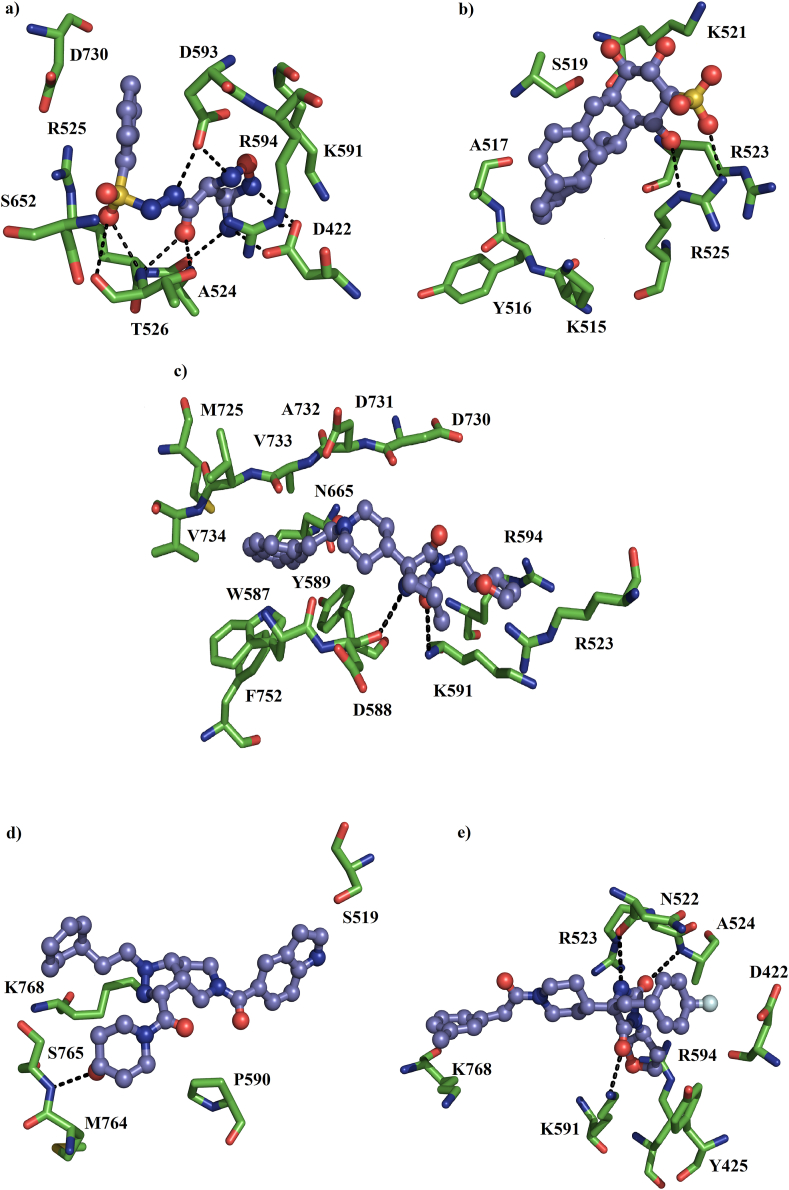
Analysis of the receptor-ligand complexes showed that 3 (ZINC000012863240, ZINC000049581065, and ZINC000072460420) out of 8 ligands lost interactions with the receptor at the beginning of the simulations, therefore, these three compounds were not considered in further analyses. Based on the simulation time in which systems reached equilibrium, a cluster analysis was performed using a cut-off of 2.5 Å to observe the main residues involving in stabilizing the ligand inside the binding site (Figure 2). The ligand orientation at the binding site is shown in Figure 2. ZINC000005605139 compound is coordinated by A524, R525, K591, R594, S652, and D730 through non-polar interactions. Whereas that T526 forms hydrogen bonds with compound through the polar backbone and side-chain atoms, as well as side chains of D422 and D593 (Figure 4A). ZINC000006094731 compound is bound by K515, Y516, A517, S519, K521, and R523 through non-polar interactions, whereas that R525 forms polar interactions through the backbone and side-chain atoms (Figure 4B). ZINC000012161475 compound is coupled through hydrophobic interactions with R523, W587, Y589, R594, N665, M725, D730, D731, A732, V733, V734, and F752, and forms polar interactions with D588 and K591 (Figure 4C). ZINC000014751834 is bound by S519, P590, M764, and K768 by hydrophobic contacts and polar interactions with backbone atoms of S765 (Figure 4D). ZINC000014882040 compound is coordinated through non-polar contacts with D422, Y425, R523, A524, R594, and K768, and polar interaction with side chains of K591 and N522, and with polar backbone atoms of A524 (Figure 4E). Comparative analysis revealed that most of the interactions are hydrophobic, and R523, A524, R525, and R594 residues were present in almost all the complexes, suggesting an important role of these residues in ligand binding stabilization.
Figure 4.
The binding orientation of 5 best ligands inside the receptor binding site of SARS Cov-2 RdRp. a) ZINC000005605139, b) ZINC000006094731, c) ZINC000012161475, d) ZINC000014751834 and e) ZINC000014882040. The binding orientation indicates stable binding for each of the 5 ligands to the RdRp of Cov-2.
3.5. Binding free energy of protein-ligand complexes
To further assess the thermodynamic feasibility of binding of the top-5 ligands to RdRp, MMGBSA method was used to calculate the binding free energy of the protein-ligand complex. Simulation runs were extracted to calculate the MMGBSA free binding energy. All of the five compounds showed negative binding free energy; suggesting successful binding throughout the simulation (Table 3). The molecular mechanic energy showed that Van der Waals energy (ΔEvdw + ΔGnpol, sol) was the main force for stabilizing the ligand at the binding site (Table 3). Despite that all the systems exhibited favorable electrostatic contributions (ΔEele), the higher nonpolar free energy of desolvation (ΔGnpol,sol) cancelled with the favorable electrostatic contribution. Table 3 also shows that the binding affinities are in the order of ZINC000005605139 > ZINC000012161475 > ZINC000014882040 > ZINC000006094731 > ZINC000014751834. Comparative analysis of the binding free energy (DGmmgbsa) values points out that ZINC000005605139 > ZINC000012161475 exhibit the more favourable DGmmgbsa value, suggesting that these two perhaps are better candidates to inhibit RdRp than the other three compounds.
Table 3.
Binding free energy components of protein-ligand systems (in units of kcal/mol).
| System | ΔEvdw | ΔEele | ΔGele,sol | ΔGnpol,sol | DGmmgbsa |
|---|---|---|---|---|---|
| ZINC000005605139 | -31.90 ± 4.0 | -191.27 ± 13.5 | 186.38 ± 12.6 | -4.92 ± 0.40 | -41.72 ± 4.0 |
| ZINC000006094731 | -29.08 ± 3.0 | -18.76 ± 6.5 | 30.18 ± 12.0 | -3.76 ± 0.40 | -21.43 ± 6.0 |
| ZINC000012161475 | -50.73 ± 4.0 | -32.16 ± 7.5 | 52.51 ± 6.4 | -5.99 ± 0.50 | -36.40 ± 5.0 |
| ZINC000014751834 | -29.79 ± 4.0 | -35.13 ± 11.0 | 48.54 ± 8.0 | -4.09 ± 0.70 | -20.47 ± 5.0 |
| ZINC000014882040 | -34.84 ± 6.0 | -33.81 ± 10.7 | 48.09 ± 9.7 | -4.87 ± 0.70 | -25.43 ± 6.0 |
3.6. ADMET properties
Drug bioavailability, metabolism and toxicity are important factors that may limit the efficacy of a drug in clinical settings. Therefore, to determine whether the potential drug candidates are effectively bioavailable and have potentially no carcinogenicity to organs in human, the top-5 ligands were assessed based on their ADMET profile (Table 4) and all of the 5 drug candidates were found to have no potential toxicity (residing below toxicity classes of 4 and 5), supporting their safety for human use.
Table 4.
ADMET results of the top-ranking ligands.
| Property | Model Name | ZINC000005605139 | ZINC000006094731 | ZINC000012161475 | ZINC000014751834 | ZINC000014882040 | Unit |
|---|---|---|---|---|---|---|---|
| Absorption | Water solubility | -3.533 | -2.891 | -5.178 | -4.127 | -4.263 | Numeric (log mol/L) |
| Absorption | Caco2 permeability | 0.492 | 1.237 | 0.846 | 1.307 | 1.159 | Numeric (log Papp in 10−6 cm/s) |
| Absorption | Intestinal absorption (human) | 72.698 | 39.7 | 95.097 | 87.731 | 90.718 | Numeric (% Absorbed) |
| Distribution | BBB permeability | -1.118 | -1.353 | -0.979 | -0.69 | -0.638 | Numeric (log BB) |
| Distribution | CNS permeability | -3.219 | -3.176 | -2.419 | -2.453 | -2.471 | Numeric (log PS) |
| Metabolism | CYP1A2 inhibitor | No | No | No | No | No | Categorical (Yes/No) |
| Metabolism | CYP2C19 inhibitior | No | No | Yes | Yes | Yes | Categorical (Yes/No) |
| Metabolism | CYP2C9 inhibitior | No | No | Yes | Yes | Yes | Categorical (Yes/No) |
| Metabolism | CYP2D6 inhibitior | No | No | No | No | No | Categorical (Yes/No) |
| Metabolism | CYP3A4 inhibitior | No | No | Yes | Yes | Yes | Categorical (Yes/No) |
| Toxicity | AMES toxicity | No | No | No | No | No | Categorical (Yes/No) |
| Toxicity | Hepatotoxicity | Yes | No | Yes | Yes | Yes | Categorical (Yes/No) |
Finally, the top-5 screened ligands ZINC000005605139, ZINC000006094731, ZINC000012161475, ZINC000014751834, and ZINC000014882040 may be considered as potential therapeutic candidates for further exploring their potential in preclinical and clinical settings.
4. Discussion
The COVID-19 caused by rapidly spreading coronavirus, known as SARS-CoV-2 has been declared as a pandemic outbreak by WHO in March 2020. It has made a devastating impact on the public as well as the social health of more than 200 countries around the globe (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Therefore, scientific communities from all over the world have come forward to collaborate and make the best effort for the earliest discovery of effective anti-viral drugs for treating COVID-19. The RdRp of SARS-CoV-2 has been sequenced and the X-ray crystallography 3-D structure is available in the PDB databank (Gao et al., 2020).
Currently, there are no effective therapeutics for treating COVID-19. Therefore, the main focus has been on disease management by prevention of the infection and following other controlling measures. Recently, some anti-malarial drugs like chloroquine, hydroxychloroquine have given positive results in vitro cell culture systems but further clinical tests are needed for them to be approved as effective anti-viral drugs for treating SARS-CoV-2 infection (Enmozhi et al., 2020). A few other anti-viral drugs, including remdesivir, ribavirin etc., used for treating Hepatitis C and Ebola virus disease, and various vaccine candidates are being studied for effectively tackling the COVID-19 (Shoenfeld, 2020). Of particular interest was remdesivir; however, recently published clinical data showed no significant impact on the reduction in mortality, though the drug does reduce the recovery period (Hillaker et al., 2020).
Inhibition of the replication via inhibiting RdRp has been explored in the development of various antiviral drugs such as favipiravir (Allen et al., 2020), sofosbuvir (Buonaguro and Buonaguro, 2020), lopinavir (Chakraborty and Das, 2020), tenofovir (Allen et al., 2020), ribavirin (Gish, 2006), ritonavir (Chakraborty and Das, 2020), galidesivir (Allen et al., 2020), and remdesivir (Fani et al., 2020). Computational studies have also led to the suggestion of using the RdRp protein as a viable drug target against the current threat (Khan et al., 2020b). In order to find effective drug, many computational studies have been conducted, which led to in vitro work and showed promising results (Khan et al., 2020a, 2021) but are not still fully sufficient for mass use. Since there are no effective drugs for treating COVID-19, we adopted a computational approach to identify potential drug candidates. In this study, the inhibitory potential of the candidate drugs targeting SARS-CoV-2 RdRp was predicted via screening from a library of compounds containing existing, approved drugs against RNA viruses and their structural analogues. Careful evaluation of the library by employing various computational screening methods yielded 5 candidate drugs and gave an insight into the potential of these prospective drugs against SARS-CoV-2.
The computational screening approach enables the rapid discovery of promising compounds for developing effective therapeutics against SARS-CoV-2. It has been shown that less binding energy denotes more affinity of a compound for binding to its target (Ortiz et al., 1995). To elucidate the binding affinity to RdRp, the library of compounds was docked against RdRp using the PyRx tool (Dallakyan and Olson, 2015). Since among the approved RdRp drugs the binding site of only remdesivir to SARS-CoV-2 RdRp has already been elucidated as well as the in vitro studies of remdesivir showed some promises we, for our screening, selected 11 critical amino acids in RdRp pocket where the drug interacts. Of note, compounds with a binding affinity of -7.5 kcal/mol or less are evaluated as potent inhibitors of enzymatic mechanisms (R Yunta, 2016). Based on binding affinities, top ranked 8 compounds were selected for further analysis. None of the approved drugs, including remdesivir, showed significant binding, perhaps these best hits are better candidates than the mother compounds.
The binding affinity determined from the docking study indicates the affinity of an individual ligand and the potency by which the ligand interacts and binds with the target protein at a specific pocket. For the best 8 hit compounds, our docking results were successful as they all were found to have the least binding scores and presence of significant hydrogen bonding at the pocket of SARS-CoV-2 RdRp (Table 2; Figure 2).
Molecular dynamics simulation was used to validate the docking study. The overreaching goal of our molecular dynamics (MD) simulation study was to investigate the stability and dynamic activity of the receptor protein in the ligand-protein complex system. Protein flexibility plays an essential role and should be considered when designing inhibitors against a target (Salmaso and Moro, 2018). Molecular docking has been performed implementing a single structure while MD simulation presents an in-depth interpretation of the protein-ligand interactions. The stability and dynamic activity of the interaction of RdRp with the selected compounds was studied by molecular dynamics simulation analysis. The MD simulation showed that most of the interactions between RdRp-ligand complexes are hydrophobic, highlighting R523, A524, R525, and R594 amino acid residues that were present in almost all the complexes, indicating a crucial role of these residues in ligand binding stabilization. Analysis of H-bond based interactions suggests that all of the compounds docked with the RdRp with all the available binding interactions and sustain all these interactions throughout the simulation process. Apart from that the receptor-ligand based interactions, all of the docked ligands on an average formed at least two H-bonds with the nearby water molecules, which bestowed further stability of the ligands docked to RdRp.
MMGBSA analysis gave an insight into the binding energy throughout the simulation. The order of binding affinities of the finally selected 5 drug candidates are: ZINC000005605139 > ZINC000012161475 > ZINC000014882040 > ZINC000006094731 > ZINC000014751834. The other three compounds, ZINC000012863240, ZINC000049581065, and ZINC000072460420, were shown to be unstable inside the binding pocket of the SARS-CoV-2 RdRp. The more negative binding free energy (DGmmgbsa value) indicates the higher favorable binding, perhaps suggesting that ZINC000005605139 and ZINC000012161475 are better candidates to inhibit the enzyme compared to the other three compounds (Table 3).
ZINC000005605139 & ZINC000006094731, ZINC000012161475 & ZINC000014882040, ZINC000014751834 are structural analogues of tenofovir, lopinavir and ritonavir, respectively and this study predicted them to be more potent inhibitors of SARS-CoV-2 RdRp compared to their mother drugs.
Physicochemical properties of the selected compounds were studied using ADMET (Hodgson, 2001) to determine their drug-likeness properties. ADMET evaluates the molecular properties of a drug that are crucial for pharmacokinetics of the drug in human body, which includes absorption, distribution, metabolism, and excretion (ADME). Selected drugs showed satisfactory ADMET profile, which may explain by the fact that these are analogues of available drugs for which safety and efficacy have already been proven. Although 4 of the 5 compounds show hepatotoxicity as predicted from ADMET, however, the level of toxicity to the liver, if any, needs to be addressed in preclinical and clinical settings in future. Our work is limited by the fact that the potential drugs were identified solely by computational approach. Wet lab validation is required which is beyond the scope of our current work, however, is an area that we will try to address in the future.
5. Conclusion
The present in silico investigation explored the potentiality of the analogues of the available drugs in inhibiting SARS-CoV-2 RdRp. The result of the virtual screening yielded top-5 hit ligands. An ideal drug is presumed to not only have a high affinity to a specific target but also should have the drug likeness properties. Taking into account, for better prediction the binding affinities predicted by MMGBSA, binding modes and further 50 ns MD simulation were also determined, and five ligands (ZINC000005605139, ZINC000006094731 ZINC000012161475, ZINC000014751834, and ZINC000014882040) were found to be the most favorable ones. Considering the safety issue of these drugs, all five compounds showed a good ADMET profile. The identified 5 drug candidates may have therapeutic potential in COVID-19, however, our study offers further preclinical and clinical investigation to prove their safety and efficacy against SARS-CoV-2.
Declarations
Author contribution statement
Kamrul Hasan, Mohammad Kamruzzaman: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Omar Hamza Bin Manjur, Araf Mahmud, Martiniano Bello: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Nazmul Hussain, Muhammad Shafiul Alam Mondal: Performed the experiments; Wrote the paper.
Ismail Hosen: Conceived and designed the experiments.
Atiqur Rahman: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors acknowledge Saeed Anwar (University of Alberta) and Sohel Mahmud (Tejgaon College, National University of Bangladesh) for helping with the writing of the manuscript.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S., Mahtarin R., Ahmed S.S., Akter S., Islam M.S., Mamun A. Al, Islam R., Hossain M.N., Ali M.A., Sultana M.U.C., Parves M.R., Ullah M.O., Halim M.A. Investigating the binding affinity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C., Arjona S., Santerre M., Sawaya B.E. Potential use of RNA-dependent RNA polymerase (RdRp) inhibitor against COVID-19 infection. Life. 2020;13(1):608–614. [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R.…Lane H.C. Remdesivir for the treatment of covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello M. Binding mechanism of kinase inhibitors to EGFR and T790M, L858R and L858R/T790M mutants through structural and energetic analysis. Int. J. Biol. Macromol. 2018;118(Pt B):1948–1962. doi: 10.1016/j.ijbiomac.2018.07.042. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA Discovery Studio — Discngine. https://www.discngine.com/discovery-studio
- Blair W., Cox C. Current landscape of antiviral drug discovery. F1000Research. 2016;5 doi: 10.12688/f1000research.7665.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinks V., Ibert O. From corona virus to Corona crisis: the value of an analytical and geographical understanding of Crisis. Tijdschr. Econ. Soc. Geogr. 2020;111(3):275–287. doi: 10.1111/tesg.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L., Buonaguro F.M. Knowledge-based repositioning of the anti-HCV direct antiviral agent Sofosbuvir as SARS-CoV-2 treatment. Infect. Agents Canc. 2020;15(1) doi: 10.1186/s13027-020-00302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D.A., Cheatham T.E., Darden T., Gohlke H., Luo R., Merz K.M., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Das G. 2020. Why re-purposing HIV drugs Lopinavir/ritonavir to inhibit the SARS-Cov2 protease probably wont work - but re-purposing Ribavirin might since it has a very similar binding site within the RNA-polymerase. [Google Scholar]
- Coronavirus Update (live): 109,706,925 cases and 2,419,664 deaths from COVID-19 virus pandemic - Worldometer. https://www.worldometers.info/coronavirus/ Retrieved February 16, 2020, from.
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- DeLano W.L. Delano Scientific - Scientific Research Publishing; San Carlos: 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- Duan Y., Wu C., Chowdhury S., Lee M.C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase Quantum mechanical calculations. J. Comput. Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J. Biomol. Struct. Dynam. 2020:1–10. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani M., Teimoori A., Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol. 2020;15(5):317–323. [Google Scholar]
- Feig M., Onufriev A., Lee M.S., Im W., Case D.A., Brooks C.L. Performance comparison of generalized Born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J. Comput. Chem. 2004;25(2):265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B.…Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish R.G. Treating HCV with ribavirin analogues and ribavirin-like molecules. J. Antimicrob. Chemother. 2006;57(1):8–13. doi: 10.1093/jac/dki405. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38(4):379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- Hillaker E., Belfer J.J., Bondici A., Murad H., Dumkow L.E. Delayed initiation of remdesivir in a COVID-19-positive patient. Pharmacotherapy. 2020;40(6):592–598. doi: 10.1002/phar.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson J. ADMET - turning chemicals into drugs. Rapidly resolving the pharmacokinetic and toxicological properties of drug candidates remains a key challenge for drug developers. Nat. Biotechnol. 2001;19(8):722–726. doi: 10.1038/90761. [DOI] [PubMed] [Google Scholar]
- Hofmann W.P., Herrmann E., Sarrazin C., Zeuzem S. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int. 2008;28(10):1332–1343. doi: 10.1111/j.1478-3231.2008.01896.x. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M.…Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JC K., KL S., MA C. Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 2018;378(26):2538–2541. doi: 10.1056/NEJMc1805679. [DOI] [PubMed] [Google Scholar]
- Khan A., Ali S.S., Khan M.T., Saleem S., Ali A., Suleman M., Babar Z., Shafiq A., Khan M., Wei D.Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Heng W., Wang Y., Qiu J., Wei X., Peng S., Saleem S., Khan M., Ali S.S., Wei D.Q. In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro) Phytother Res. 2021 doi: 10.1002/ptr.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Khan M., Saleem S., Babar Z., Ali A., Khan A.A., Sardar Z., Hamayun F., Ali S.S., Wei D.Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. 2020;12(3):335–348. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.D., Li Q.H. SARS-CoV-2: vaccines in the pandemic era. Milit. Med. Res. 2021;8(1):1. doi: 10.1186/s40779-020-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li Z., Wu M., Yao J., Guo J., Liao X., Song S., Li J., Duan G., Zhou Y., Wu X., Zhou Z., Wang T., Hu M., Chen X., Fu Y., Lei C., Dong H., Xu C., Hu Y.…Yan J. Caution on kidney dysfunctions of COVID-19 patients. SSRN Electr. J. 2020 2020.02.08.20021212. [Google Scholar]
- Lin M.I., Su B.H., Lee C.H., Wang S.T., Wu W.C., Dangate P., Wang S.Y., Huang W.I., Cheng T.J., Lin O.A., Cheng Y.S.E., Tseng Y.J., Sun C.M. Synthesis and inhibitory effects of novel pyrimido-pyrrolo-quinoxalinedione analogues targeting nucleoproteins of influenza A virus H1N1. Eur. J. Med. Chem. 2015;102:477–486. doi: 10.1016/j.ejmech.2015.08.016. [DOI] [PubMed] [Google Scholar]
- Ludwig S., Zarbock A. Coronaviruses and SARS-CoV-2: a brief overview. Anesth. Analg. 2020;93–96 doi: 10.1213/ANE.0000000000004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung J., Lin Y., Yang Y., Chou Y., Shu L., Cheng Y., Liu H. Te, Wu C. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitjà O., Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. The Lancet Glob. Health. 2020;8(5):e639–e640. doi: 10.1016/S2214-109X(20)30114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J.L., Yang W., Ito K., Matte T.D., Shaman J., Kinney P.L. Seasonal influenza infections and cardiovascular disease mortality. JAMA Cardiol. 2016;1(3):274–281. doi: 10.1001/jamacardio.2016.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A.R., Pisabarro M.T., Gago F., Wade R.C. Prediction of drug binding affinities by comparative binding energy analysis. J. Med. Chem. 1995;38(14):2681–2691. doi: 10.1021/jm00014a020. [DOI] [PubMed] [Google Scholar]
- Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Yunta M.J. Docking and ligand binding affinity: uses and pitfalls. Am. J. Model. Optimiz. 2016;4(3):74–114. [Google Scholar]
- Rubin E.J., Longo D.L. SARS-CoV-2 vaccination — an ounce (actually, much less) of prevention. N. Engl. J. Med. 2020;383(27):2677–2678. doi: 10.1056/NEJMe2034717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9(AUG) doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Cheng F., Xu Y., Li W., Tang Y. Estimation of ADME properties with substructure pattern recognition. J. Chem. Inf. Model. 2010;50(6):1034–1041. doi: 10.1021/ci100104j. [DOI] [PubMed] [Google Scholar]
- Shoenfeld Y. Corona (COVID-19) time musings: our involvement in COVID-19 pathogenesis, diagnosis, treatment and vaccine planning. Autoimmun. Rev. 2020;19(6) doi: 10.1016/j.autrev.2020.102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şimşek Yavuz S., Ünal S. Antiviral treatment of covid-19. Turk. J. Med. Sci. 2020;50(SI-1):611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N., Balayla G. Hydroxychloroquine and covid-19. Postgrad. Med. 2020;96(1139):550–555. doi: 10.1136/postgradmedj-2020-137785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T., Irwin J.J. ZINC 15 - ligand discovery for everyone. J. Chem. Inf. Model. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gunsteren W.F., Berendsen H.J.C. Algorithms for macromolecular dynamics and constraintdynamics. Mol. Phys. 1977;34(5):1311–1327. [Google Scholar]
- Van Kampen J.J.A., Van De Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Sc M., Okba N., Van Den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., Van Der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., De Steenwinkel J.E.M., Aron G.I., Van Gorp E.C.M.…Van Der Eijk A.A. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. MedRxiv. 2020 doi: 10.1038/s41467-020-20568-4. 2020.06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8(18):1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhou R. Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. Preprints. 2020 doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- Zoete V., Daina A., Bovigny C., Michielin O. SwissSimilarity: a Web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016;56(8):1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.