Abstract
Neuroinflammation plays a crucial role during ageing and various neurological conditions, including Alzheimer's disease, multiple sclerosis and infection. Technical limitations, however, have prevented an integrative analysis of how lymphocyte immune receptor repertoires and their accompanying transcriptional states change with age in the central nervous system. Here, we leveraged single-cell sequencing to simultaneously profile B cell receptor and T cell receptor repertoires and accompanying gene expression profiles in young and old mouse brains. We observed the presence of clonally expanded B and T cells in the central nervous system of aged male mice. Furthermore, many of these B cells were of the IgM and IgD isotypes, and had low levels of somatic hypermutation. Integrating gene expression information additionally revealed distinct transcriptional profiles of these clonally expanded lymphocytes. Our findings implicate that clonally related T and B cells in the CNS of elderly mice may contribute to neuroinflammation accompanying homeostatic ageing.
Keywords: bioinformatics, immune repertoires, central nervous system, single cell
1. Introduction
Ageing of the immune system, commonly referred to as immune senescence, has been shown to hamper adaptive immune responses in the context of vaccination, viral infection and neurological conditions [1–4]. Host-mediated immunopathology is sometimes increased in the elderly during immune challenges [5], further highlighting the importance of studying adaptive immunity in the context of ageing. A number of phenotypic and functional alterations have been linked to dysfunctional adaptive immunity, including the diminished production of new naive B and T cells [6–8], increased host-immunopathology during viral infections and disease [5,9], increased exhaustion and elevated expression levels of dysfunction markers [10,11]. A recent study using high-dimensional mass-cytometry reported an increased percentage of B and T cells located in the elderly murine central nervous system (CNS) [12]. However, the phenotypic analysis of this study was primarily focused on myeloid cells and microglia, thereby leaving the phenotype of adaptive immune cells uncharacterized.
Advances in deep sequencing and microfluidic-based technologies have revolutionized the resolution with which we are now able to extract biologically relevant information from adaptive immune receptor repertoires (immune repertoires) [13–15]. New single-cell sequencing (scSeq) technologies are now enabling a more comprehensive characterization of immune repertoires by capturing both BCR variable light (VL) and heavy (VH) regions and TCR variable alpha (Vα) and beta (Vβ) chains [16,17]. scSeq can also recover transcriptional profiles, which can be linked to immune repertoire clonal identifiers to thus obtain a comprehensive phenotypic characterization of lymphocytes [16].
Here, we performed single-cell immune repertoire and transcriptome sequencing of B and T cells from 3-month-old, 12-month-old and 18-month-old mice in order to phenotypically characterize and determine the extent of clonal expansion of adaptive immune cells in the aged CNS. Our strategy to profile B and T lymphocytes allowed us to relate repertoire parameters such as clonal expansion, germline gene usage and isotype to gene expression profiles at the single-cell resolution. Our findings demonstrated the presence of highly expanded B and T cell clonal lineages in the aged CNS. We could additionally demonstrate that expanded B cells in the aged CNS were predominantly of the IgM isotype and exhibit low levels of somatic hypermutation. While we observed that clonally expanded CNS lymphocytes had distinct transcriptional profiles compared to unexpanded clones, there were few age-associated changes in gene expression. Taken together, our findings demonstrate the accumulation of clonally expanded B and T lymphocytes in the CNS upon ageing that may contribute to age-associated neuroinflammation.
2. Results
(a). Single-cell sequencing of immune repertoires and transcriptomes of lymphocytes from murine CNS
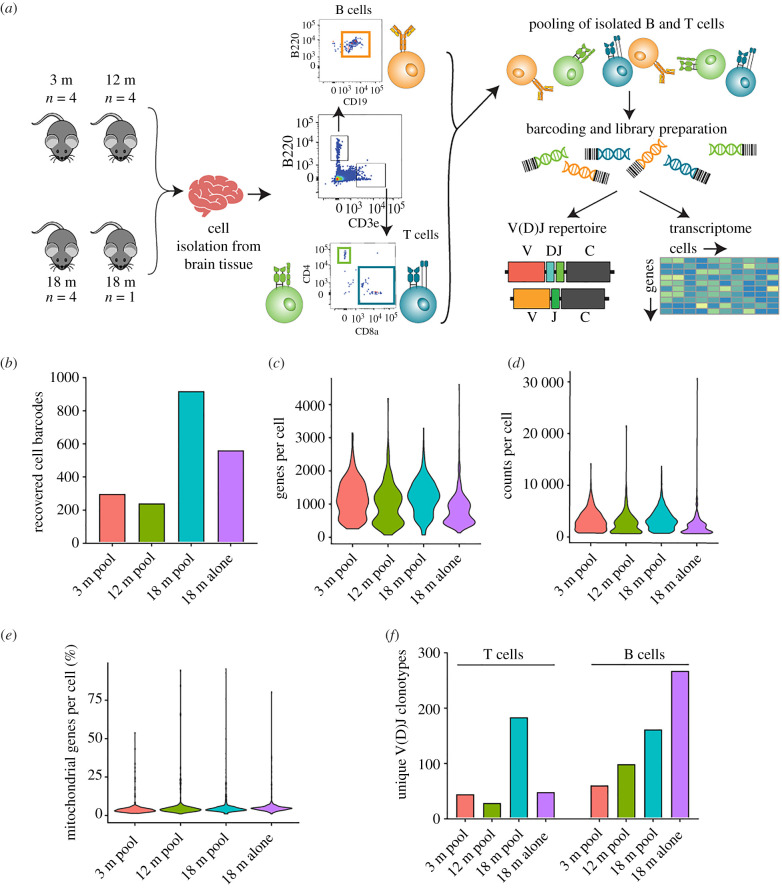
To investigate the lymphocytes in the aged CNS, we performed scSeq on B and T cells isolated from entire brains of pooled 3-month-old (n = 4), 12-month-old (n = 4) and 18-month-old C57/BL6j male mice (n = 4). We additionally processed and sequenced the brain of an individual 18-month-old mouse to ensure both reproducibility and to allow the detection of common immune cell clones shared between mice. B and T lymphocytes were isolated by performing fluorescence-activated cell sorting (FACS) of cell suspensions derived from whole brains of each cohort. B cells were isolated based on CD19+ B220+ surface expression, whereas T cells were sorted based on surface expression of CD3+ and CD4+ or CD8+ (figure 1a). Following sorting, the entirety of the three isolated immune cell populations were pooled and supplied as input for scSeq using the 10× Genomics Chromium pipeline to prepare gene expression libraries (GEX), T cell V(D)J libraries or B cell V(D)J libraries separately. Following scSeq of the three libraries per cohort, we were able to recover transcriptional information for approximately 2000 distinct cells (figure 1b). In accordance with previous reports of increased B and T cells in the aged mouse brain [12], we observed a higher number of cells from both cohorts of 18-month-old animals (figure 1b). We did not observe an increased number of recovered cells in the pooled 18-month-old relative to the individually processed 18-month-old brain, suggesting potential cell loss during pooling and sequencing library preparation. Nevertheless, we detected on average approximately 1000 genes per cell across all cohorts, with an average of approximately 2700 counts per cell (figure 1c,d). We additionally observed that, on average, fewer than 5% of counts per cell mapped to mitochondrial genes, implying adequate sequencing quality (figure 1e).
Figure 1.
Single-cell sequencing recovers repertoire and gene expression information of B and T cells from the aged murine central nervous system. (a) Experimental overview of isolation of B and T cells from entire murine brains. (b) Number of recovered cell barcodes following single-cell sequencing of gene expression libraries for each of the experimental cohorts. (c) Unique genes detect per cell for each of the experimental cohorts. (d) Total number of RNA molecules detected within each cell. (e) Percentage of mitochondrial genes for each cell. (f) Number of unique clones from V(D)J sequencing library for each of the experimental cohorts. Clone is defined as identical, paired amino acid CDR3 sequences (CDRβ3 + CDRα3 for T cells, CDRH3 + CDRL3 for B cells) with exactly one heavy chain and one light chain. (Online version in colour.)
We next analysed full-length paired variable region clones (VH : VL for B cells and Vβ : Vα for T cells); a clone was defined based on the amino acid sequence of complementary determining region 3 (CDR3) of both variable regions (i.e. CDRH3 + CDRL3 for BCR, CDRβ3 + CDRα3 for TCR). We observed increased clonal diversity for both B and T cells in the aged cohorts (figure 1f), as well as for the BCR repertoire from the individual 18-month-old mouse compared to the pooled 18-month-old cohort (figure 1f), potentially suggesting mouse heterogeneity with regard to clonal diversity or that technical biases arose during sample pooling.
(b). Clonally expanded B and T cells are located in the murine CNS of elderly mice
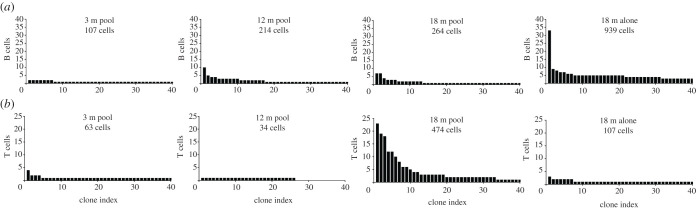
After witnessing the clonal diversity in the immune repertoires of aged mice, we next questioned whether this was due to random infiltration of unrelated, circulating lymphocytes or due to infiltration of clonally related B and T cells. V(D)J recombination generates a diverse panel of BCRs and TCRs, such that they are essentially unique to an individual recombination event [18]. Thus, finding two or more lymphocytes with identical clonal sequences (CDRH3 + CDRL3 or CDRβ3 + CDRα3) strongly supports that clonal expansion occurred. We therefore quantified the number of cell barcodes for the top 40 most expanded B and T cells for the four cohorts (figure 2a,b). In the 3-month-old pooled brain, we observed only 7 BCR clones corresponding to two distinct B cells (figure 2a). Comparing this to the other two cohorts, revealed an increase in B cell clonal expansion for both 12-month-old and 18-month-old mice (figure 2a). Due to our experimental setting, it is not possible to resolve whether this clonal expansion is due to BCR clones recovered in multiple mice (public clones) or clonal expansion within a single mouse. To further investigate this, we performed a similar quantification for the individually processed 18-month-old mouse, which revealed immense clonal expansion relative to the low number of recovered cells (figure 2a and 1b). For example, 33 cells were present in the most expanded clonal family, and additionally there were still 3 cells corresponding to the 40th most expanded clone (figure 2a). The definition of a clonal family here is based on exact CDRH3 + CDRL3 amino acid sequences, which thus excludes BCR sequences that have somatic hypermutation in the CDR3 regions. Thus, we performed an additional clonotyping analysis based on identical VH and VL germline genes and identical CDRH3 and CDRL3 amino acid sequence lengths, which resulted in only a difference for three clones (electronic supplementary material, figure S1), thereby confirming that our initial clonal family definition sufficiently described clonal expansion.
Figure 2.
Clonally expanded B and T cells in the aged CNS. (a,b) The number of recovered cells corresponding to the top 40 most expanded clones for B and T cell repertoires, respectively. Clone is defined as identical, paired amino acid CDR3 sequences (CDRβ3 + CDRα3 for T cells, CDRH3 + CDRL3 for B cells) with exactly one heavy chain and one light chain. Clone index corresponds to the clonal rank determined by frequency.
The presence of clonally expanded B cells in the aged CNS prompted us to question whether similar expansion profiles would be observed for T cells. We therefore performed the identical analysis for the TCR repertoires for each age cohort. Measuring the clonal expansion for the top 40T cell clones demonstrated that we could again recover multiple cells containing the same clonal sequence (figure 2b). In the 3-month-old and 12-month-old pooled brain TCR repertoires, there were very few clonal sequences corresponding to more than 2 cells (figure 2b). However, in the pooled 18-month-old cohort, clonal expansion was nevertheless detected, with the most expanded clone corresponding to 23 cells. While it is again not possible to entirely delineate whether these are arising from a single mouse or public clones, combining these results with the individually sequenced 18-month-old mouse nevertheless supports that clonally expanded lymphocytes are present in the aged CNS (figure 2b).
(c). BCR repertoires in aged CNS have characteristics of naive B cells
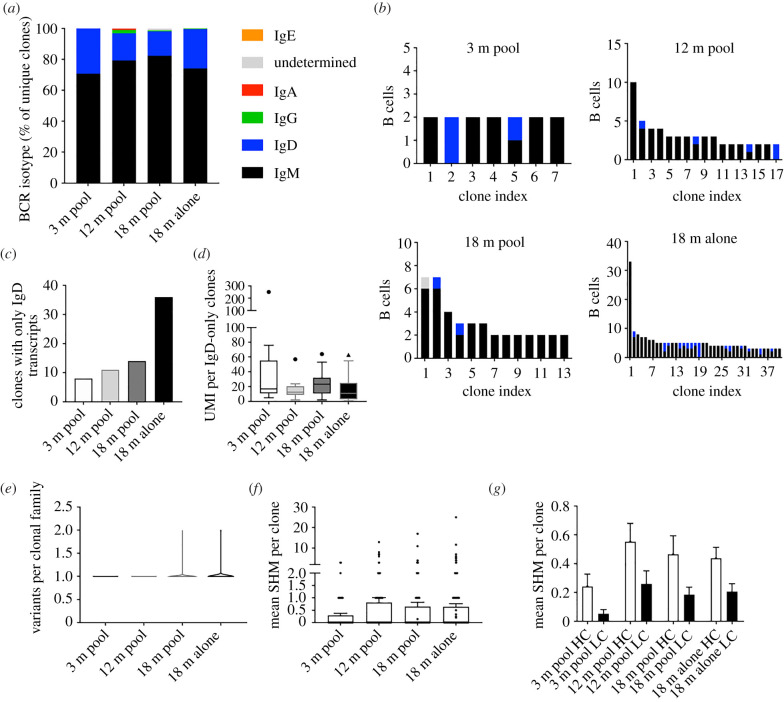
After observing clonal expansion in the brains of aged mice, we next investigated whether these B cells had undergone class-switch recombination, which would be a correlate of a previous interaction with cognate antigen. We, therefore, extracted the heavy chain isotype for each B cell clonal lineage across all four age cohorts. For clonal families containing more than one distinct cell, we used the isotype corresponding to the majority of cells within that family. The majority of the expanded clones were of the IgM isotype for all age cohorts, although surprisingly there were many B cell clones corresponding to the IgD isotype for all four cohorts (figure 3a). We next determined the isotype distribution of clonally expanded B cells (figure 2a), which revealed that the majority of clonally expanded families contained cells of either the IgM, IgD or a combination of the two (figure 3b). The unexpected occurrence of exclusive IgD clones was also observed as trend in many B cells of aged mice (figure 3c). We further assessed whether these BCR sequences were supported by multiple unique molecular identifiers (UMIs), because these isotype results could have been a result of not sampling enough mRNA molecules for each cell, thereby not detecting co-expressed IgM. However, we observed that almost all (greater than 97%) of these IgD clones were inferred using multiple UMIs (figure 3d), suggesting sufficient sequencing and sampling depth and thereby excluding to have missed co-expression of IgM.
Figure 3.
B cells in the CNS are primarily of the IgM isotype with minor somatic hypermutation. (a) Percentage of unique clones with a given BCR. For those clones with multiple cells, the majority isotype was selected. (b) Isotype distribution for those expanded (greater than 1 cell barcode) B cell clones. (c) Number of clones with exclusively IgD BCR transcripts. (d) Tukey box-plot displaying the number of unique molecular identifiers (UMI) per those clones with reads mapping exclusively to the IgD isotype. Horizontal line within the box indicates the median and dot above. (e) The number of somatic variants per expanded B cell clone. Variant is defined as unique, combined VH + VL amino acid sequence. (f) The mean number of nucleotide somatic hypermutations (SHMs) per B cell clone in the full-length V and J regions across both heavy (HC) and light chain (LC). (g) The mean number of nucleotide substitutions per B cell clone detected on either the HC or light chain LC. Both HC and LC sequences correspond to full-length V and J regions. (Online version in colour.)
Our previous analyses demonstrating that relaxing clonotyping stringency did not alter clonality in BCR repertoires suggested that there were not substantial amino acid mutations in the CDR3 regions of B cells in the CNS. This was further implicated by the lack of class-switched B cells. We, therefore, next investigated whether a similar lack of somatic hypermutation was observed in the remaining regions of the BCRs. To accomplish this, we first quantified the number of somatic variants (defined by unique, full-length, paired VH–VL nucleotide sequences) within each clonal family. This demonstrated that the majority of these clonal families containing more than one cell had only one unique antibody sequence (i.e. all cells in a clonal family produced the same antibody) for all age cohorts (figure 3e). We next determined divergence from germline for each clonal family which showed that despite low mutation levels for all BCR repertoires, there was a trend of increased somatic hypermutation in the aged cohort, with certain clones diverging from the reference germline by more than 10 nucleotide mutations (figure 3f). Single-cell sequencing allows us to separately analyse VH and VL regions, thus we additionally determined whether somatic hypermutation was distributed evenly across the two regions. In line with previous findings, the VH region was the dominant target for somatic hypermutation across all cohorts (figure 3g). Taken in concert, these results suggest that the majority of B cells express IgM or IgD BCRs and that there is a slight increase in somatic hypermutation in BCR repertoires of aged mice.
(d). Assessing sequence similarity of immune repertoires
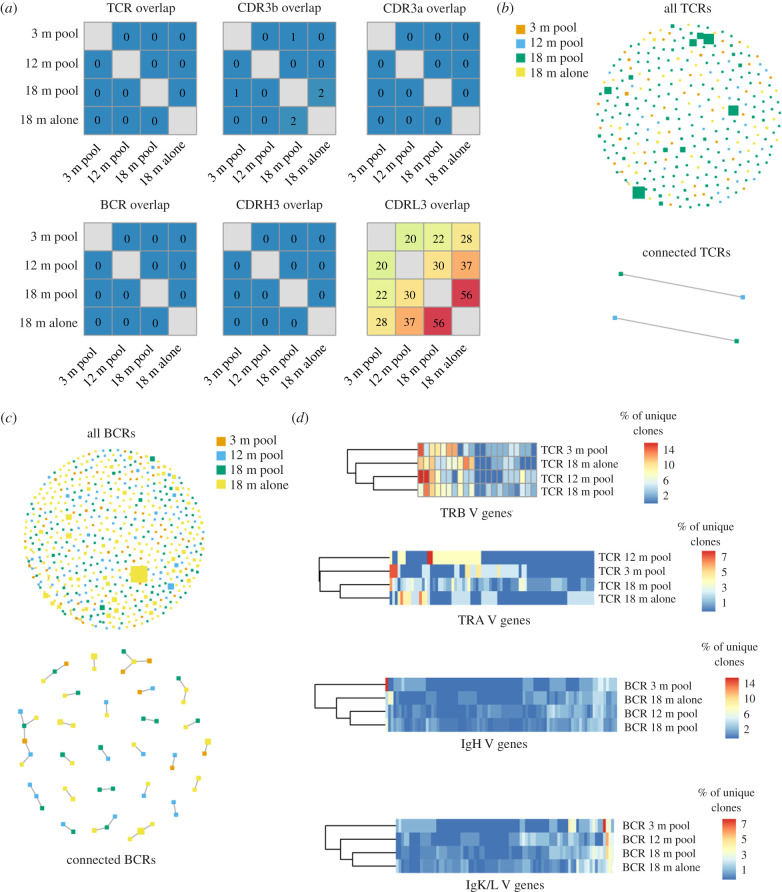
After observing comparable isotype distributions in the BCR repertoires of young and old mice, we next questioned whether we could detect hints of clonal convergence using other repertoire metrics. While multiple bioinformatics analyses exist to quantify the similarity between repertoires, we assessed whether there were identical BCR or TCR clones (defined as identical amino acid sequences of CDRH3 + CDRL3 for BCR and CDRβ3 + CDRα3 for TCR) shared between the different age groups. This analysis revealed that for both BCR and TCR there were no identical clones found between the different age groups (figure 4a). However, when restricting our analysis to a single variable chain, (clone defined as identical amino acid sequence of CDRH3, CDRβ3, CDRL3 or CDRα3), we observed substantial clonal overlap in variable light chains of 18-month-old cohorts (figure 4a). We next determined if relaxing the definition of clonal overlap would alter metrics of repertoire similarity between the aged cohorts. To address this, we employed metrics from graph theory to quantify sequence similarity between clones both within and across repertoires [13]. In sequence similarity networks, clones are represented as nodes and are connected via edges to other clones with high sequence similarity (four or less amino acid mutations across the paired CDR3 s; figure 4b,c). Information corresponding to age and clonal frequency is demonstrated by colour and size of nodes, respectively. We first illustrated the overall connectivity between all repertoires by including each unique clone in our similarity network. We additionally created similarity networks to demonstrate the relationship between the connected nodes, which clearly demonstrated a higher degree of sequence similarity between BCRs relative to TCRs across cohorts (figure 4b,c). There were several clusters in the BCR sequence similarity network in which only clones from the CNS repertoires of aged mice were connected (figure 4c), potentially suggesting these clones may have a similar antigen-specificity and could accumulate with age. Finally, we quantified the percentage of unique clones using each V germline gene for the BCR and TCR variable chains for all cohorts and subsequently performed hierarchical clustering. This revealed that the 18-month-old cohorts clustered together for BCR VH and VL and TCR Vα chains, whereas the pooled repertoires for 12-month-old and 18-month-old mice clustered together for the TCR Vβ chain (figure 4d). Together, these findings suggest that V-gene usage similarity of immune repertoires increases in the elderly CNS.
Figure 4.
Minor clonal convergence between CNS immune repertoires. (a) Heatmap depicting clonal overlap between repertoires. Clonal overlap is defined as the number of identical amino acid CDR3 sequences shared between repertoires. BCR and TCR overlap corresponds to the paired CDRH3 + CDRL3/CDRβ3 + CDRα3 amino acid sequence. (b,c) Similarity networks depicting clonal relatedness and expansion between T cell and B cell repertoires, respectively. Size of nodes corresponds to relative clonal expansion. Colour of node corresponds to experimental cohort. Edges between nodes indicate two clones (paired CDRH3 + CDRL3/CDRβ3 + CDRα3 amino acid sequence) that are separated by four or less amino acid mutations. Lower panel highlights those clones containing edges, whereas upper panel depicts all clones. (d) Percentage of unique clones using each V-gene for each experimental cohort. (Online version in colour.)
(e). Profiling transcriptional landscape in aged CNS lymphocytes
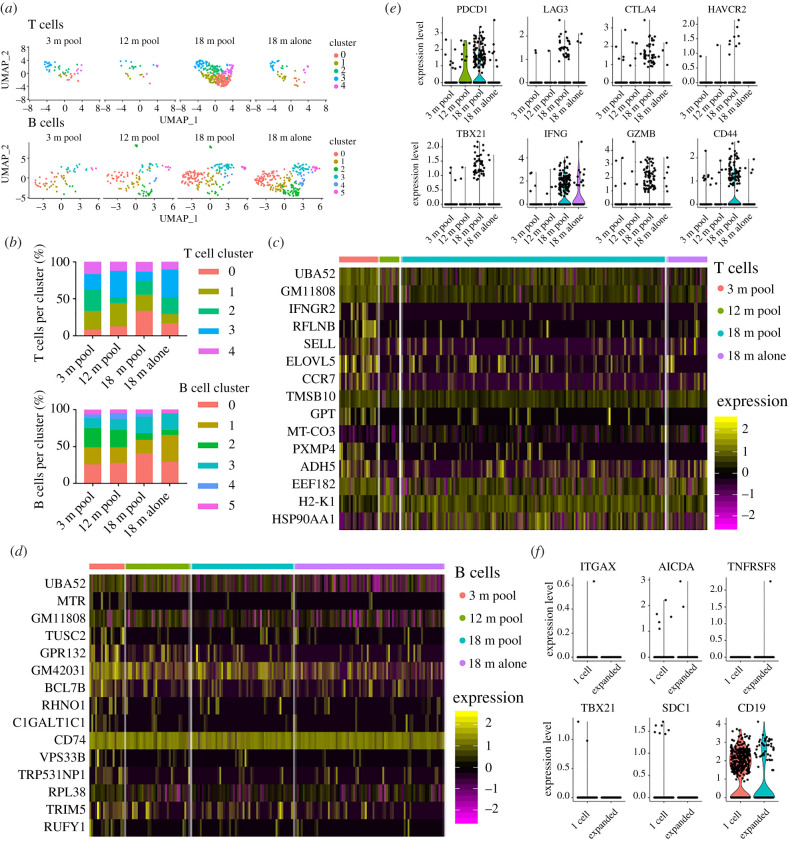
We used transcriptome sequencing data to perform unsupervised clustering and uniform manifold approximation projection (UMAP) to group cells with similar gene expression profiles. This resulted in 11 distinct clusters which were populated by cells from all cohorts (electronic supplementary material, figure S2a,b). We next verified that single-cell transcriptome data was sensitive to B and T cell clusters by visualizing expression levels of CD19, CD3e, CD8a and CD4, which confirmed the expected cell (electronic supplementary material, figure S2c,d). After retaining the cells which were present in both repertoire and transcriptome libraries (n = 1008 cells), we observed that the B and T cells from all age cohorts colocalized based on gene expression information, further highlighting similarities between the aged cohorts when using an unbiased clustering approach (electronic supplementary material, figure S3a). Next we investigated if separating B and T cells would reveal any age-associated transcriptional changes between cohorts. By restricting phenotypic analysis to cells for which we had complete BCR or TCR sequence information (paired CDR3 s, full-length V(D)J sequence) and performing unbiased clustering and UMAP resulted in 5 and 4 transcriptional clusters for the T and B cells, respectively (figure 5a; electronic supplementary material, figure S3b,c). We again observed comparable frequencies of cluster membership across all age cohorts (figure 5b), further suggesting that lymphocytes in the aged CNS maintain comparable transcriptional profiles to those in young mice.
Figure 5.
Transcriptional signatures of clonally expanded lymphocytes. (a) Unsupervised clustering and uniform manifold approximation projection (UMAP) was performed separately on either B or T cells that were also detected in V(D)J libraries. Each dot represents a single cell. Colour corresponds to transcriptional cluster specific to T or B cell analysis. (b) The percentage of T (top) or B (bottom) cells found within each cluster for each of the experimental cohorts. Clusters were determined using either only T cells or only B cells found in V(D)J libraries. (c,d) Heatmap displaying differentially expressed genes between young and 18-month-old T and B cells, respectively. Clusters were calculated separately for B cells and T cells. Displayed cells were randomly sampled from within each cluster proportional to cluster's total number of cells. Intensity corresponds to normalized gene expression. Cells are ordered by p-value after adjusting for multiple hypothesis testing. (e) Normalized gene expression of for T cells for PD-1 (PDCD1), LAG3, CTLA4, TIM3 (HAVCR2), TBET (Tbx21), IFNG, Granzyme B (GZMB) and CD44. (f) Normalized gene expression of for B cells ITGAX, AID (AICDA), CD30 (TNFRSF8), T-bet (TBX21), CD138 (SDC1) and CD19. (Online version in colour.)
We next determined if any age-associated differences in the transcriptional profiles of lymphocytes was observed when cluster membership was not taken into account. To this end we computed the differential expression between the cohorts and found minor differences for both B and T cells (figure 5c,d). However, we observed a trend of upregulation of markers associated with T cell function or exhaustion such as PD-1, Lag3, CTLA4, Tim3 (HAVCR2) in aged but not young mice (figure 5e). Other genes relevant to T cell function, such as expression of granzymes and interferon gamma followed a similar trend of upregulation in the 18-month-old animals, albeit with a low number of cells (figure 5e). With the exception of CD19, characteristic B cell markers, including age-associated genes (CD11c and TBX21) and differentiation genes (AID, CD138, CD30) showed no major differences across cohorts (figure 5f).
(f). Clonally expanded lymphocytes have distinct transcriptional profiles
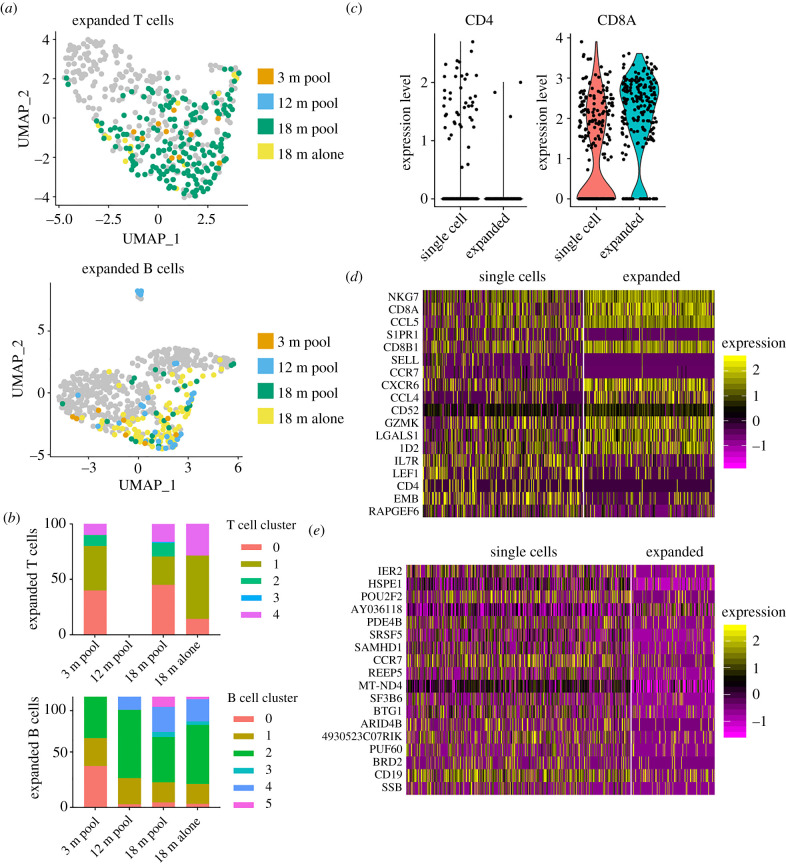
Since we could observe clonal expansion in immune repertoires aged CNS, we investigated the transcriptional landscape of these cells. Highlighting expanded clones (clonal families corresponding to at least two distinct cell barcodes) suggested transcriptional-dependent clustering for both B and T cells (figure 6a,b). While expanded T cells were distributed across all clusters (excluding cluster 3), we observed that the majority were CD8T cells (figure 6c), clarifying our previous clonal expansion findings (figure 2b). In order to detect transcriptional signatures, we explicitly quantified differential gene expression between clonally expanded lineages and those containing only a single cell. This comparison between expanded to unexpanded cells resulted in 26 and 57 genes differentially regulated (p.adj < 0.01) for T and B cells (figure 6d,e) and 247 and 137 genes without multiple hypothesis correcting (p < 0.01) (electronic supplementary material, figure S4a). Strictly taking the top genes based on p-values demonstrated that expanded T cells expressed higher levels of NKG7, CCL5, CXCR6, CCL4, Granzyme K, and ID2 and had downregulated markers associated with a central memory phenotype, such as CD62 L, CCR7, LEF1 and IL7R (figure 6d). The phenotypic profile of expanded B cells was less clear, although some relevant genes such as CCR7 and ones involving proliferation (e.g. BTG1) were detected as downregulated in expanded cells (figure 6e). Quantifying gene expression for a panel of relevant genes demonstrated a trend that clonally expanded T cells expressed more PD-1 (PDCD1), interferon gamma (IFNG), and granzyme B (GZMB) (electronic supplementary material, figure S4b), suggesting recent or persistent activation. Performing a similar analysis using a panel of age- and activation-associated B cell markers resulted in minor differences in gene expression between expanded and unexpanded B cells, although this analysis may have been limited by low count numbers for the selected genes (electronic supplementary material, figure S4c). Extending our analysis to quantify differences between the expanded cells of aged versus young CNS lymphocytes did not reveal major changes in transcription, again consistent with our previous findings (electronic supplementary material, figure S5a–c). Taken together, these results demonstrate distinct transcriptional phenotypes of clonally expanded lymphocytes in the CNS that are robust to age-associated changes following homeostatic ageing.
Figure 6.
Clonally expanded lymphocytes have distinct transcriptional profiles. (a) Location and cluster membership of expanded B and T cells. Expanded B and T cells were defined by those clones containing at least 2 distinct cell barcodes. Clone is defined as identical, paired amino acid CDR3 sequences (CDRβ3 + CDRα3 for T cells, CDRH3 + CDRL3 for B cells) with exactly one heavy chain and one light chain. (b) Cluster membership of clonally expanded T and B cells in V(D)J libraries. Clonally expanded T and B cells correspond to those clones (identical, paired CDR3 amino acid sequence) containing two or more distinct cell barcodes. (c) CD4 and CD8a expression for those clones supported by a single cell or those expanded clones (more than 1 cell). (d,e) Heatmap of top differentially expressed genes between T cell (d) and B cell (e) clones with either one cell or clonally expanded. Cells are ordered by p-value after adjusting for multiple hypothesis testing. All genes displayed correspond corrected p-values less than 0.01. Heatmap intensity depicts normalized gene expression. (Online version in colour.)
3. Discussion
While previous studies have demonstrated increased frequencies of lymphocytes in the aged and diseased CNS [2], it could not be determined whether these cells were clonally related. Our single-cell sequencing approach suggests that lymphocyte CNS infiltration was driven by clonally expanded lineages. Additionally, we were able to profile differences in gene expression between expanded and unexpanded lymphocytes. Further experiments combining immune repertoire sequencing from blood and secondary lymphoid organs could answer whether these expanded clones are specific to the CNS environment or expanded across different physiological compartments.
Investigating whether identical clones would be present in the repertoires across various peripheral organs within the same aged individual would highlight whether the expansion we observed is a CNS-specific phenomenon, or if there is a global accumulation of clonally related lymphocytes in organs typically unassociated with immunity. The discovery of clonally expanded lymphocytes in the aged CNS suggests that determining their cognate antigens would be important in order to elucidate which role, if any, they play under homeostatic and disease states. Relating antigen-specificity information to immune receptor sequence could further inform whether these expanded B and T cell clones originate from cross-reactivity, dysregulation of self-tolerance or simply past environmental exposure (e.g. diet, hygiene, pathogen).
One important consideration is that our study has been restricted to male mice, thereby not excluding that our findings is a sex-specific. Recent work has demonstrated that both male and female mice demonstrate expanded clones across multiple tissues, suggesting that our findings would be comparable in both male and female mice [19]. Another important consideration to our study is that the majority of our analyses were not normalized by the number of cells recovered in each sequencing library in order to use the entirety of the clonal information. While this may have changed the relative clonal expansion, the occurrence of multiple cells expressing identical BCR and TCRs nevertheless suggests a model in which clonally expanded cells populate the aged brain. Indeed, from our data we are not able to conclude that clonally expanded lymphocytes do not populate the young murine CNS, as the number of B and T cells recovered was lower relative to the other aged cohorts. It was recently has demonstrated that clonally related B and T cells do populate various organs and this effect increases with age [19], demonstrating congruencies with our own findings. A further drawback of our study is that we cannot entirely exclude that some of the cells were found in the local vasculature of the brain despite perfusing upon sacrifice. Although further studies comparing circulating repertoires to the lymphocytes found within the CNS would be helpful to exclude this point, visual inspection of the brains after extraction demonstrated only minor traces of blood in CNS-associated capillaries.
Expanded lymphocytes could represent a population of clones that persistently interact with their cognate antigen during homeostatic ageing, and following a disease or infection trigger, such cells may be poised to exacerbate immunopathology. In this case, the high number of predominantly IgM and IgD B cell clones might arise in an antigen-activated manner independent of T cell help. Alternatively, previous reports have demonstrated that an increase in IgD expression coincides with a decrease in IgM levels in the context of autoimmunity and anergy [20,21], providing one possible explanation to the high number of IgD-exclusive B cell clones found in aged CNS. Another hypothesis regarding the role of clonally expanded CNS lymphocytes arises from a recent study that identified virus-reactive T cells in the brains of Alzheimer's patients and even provided an example of a cognate antigen [2]. Future experiments performing a similar validation of clonally expanded B and T cells against pathogen or autoimmune targets could shed light upon age-associated immune dysregulation. In conclusion, our findings demonstrated the presence of clonally expanded lymphocytes in the CNS and furthermore quantitatively describe the immune repertoire and transcriptome landscape accompanying homeostatic ageing.
4. Methods
(a). Murine brain cell isolation
Mouse experiments were performed under the guidelines and protocols approved by the Basel-Stadt cantonal veterinary office (Basel-Stadt Kantonales Veterinäramt Tierversuchsbewilligung #2582). 3-month-old, 12-month-old and 18-month-old male C57/Bl6j mice were purchased (Janvier Laboratories, France) and were housed under pathogen-free conditions and maintained on a standard chow diet. Mice were perfused with cold PBS for 3 min following sacrifice and entire brains were collected in 1 ml of RPMI. Brains were subsequently transferred to a new tube containing 1 ml RPMI with Collagenase I/DNAse I (Roche) and cut to smaller pieces. Following 30 min incubation at 37°C, each brain was mashed through an individual 70 μm cell strainer into a 50 ml falcon tube. Following a 30% Percoll gradient, cells were incubated with fluorescently conjugated antibodies for 30 min at 4°C and subsequently sorted using a FACS Aria. All experiments were performed one a single day and all samples were sorted, captured, and sequenced together. The antibodies used in this study can be found in the electronic supplementary material.
(b). Single-cell immune repertoire sequencing
Integrated repertoire and transcriptome analyses were performed using the functions contained in the R package Platypus following default parameters [22] which heavily relies upon the R package Seurat [23]. Somatic hypermutation was quantified for each heavy and light chain based on the number of nucleotide substitutions in the V and J regions compared to those germline genes with the highest alignment scores determined by MiXCR v. 3.0.1 [24]. Genes defining clusters were determined setting the min.pct argument equal to 0.25 using Seurat's FindMarkers function using the Wilcoxon rank sum test. Heatmaps of cluster defining genes were selected for the top genes ranked by p-value after employing the Bonferroni correction for multiple hypothesis testing. A more detailed description is located in the electronic supplementary material.
Supplementary Material
Acknowledgments
We acknowledge and thank Dr Christian Beisel, Elodie Burcklen, Ina Nissen, and Tobias Schär at the ETH Zurich D-BSSE Genomics Facility Basel for excellent support and assistance. We also thank Mariangela Di Tacchio and Marie-Didiée Hussherr for excellent experimental support.
Contributor Information
Alexander Yermanos, Email: ayermanos@gmail.com.
Sai T. Reddy, Email: sai.reddy@bsse.ethz.ch.
Data accessibility
The VDJ and GEX sequencing files from deep sequencing and code that support the findings of this study are available at 10.5281/zenodo.4264462, 10.5281/zenodo.4264978, and https://github.com/alexyermanos/Platypus. The preprint can be found on bioRxiv [25].
Authors' contributions
A.Y. and D.N. performed experiments and analyses. All authors contributed to the study and manuscript design.
Competing interests
There are no competing interests.
Funding
This work was supported by the European Research Council Starting grant no. 679403 (to S.T.R.) and ETH Zurich Research Grants (to S.T.R. and A.O.).
References
- 1.Doyle KP, et al. 2015. B-lymphocyte-mediated delayed cognitive impairment following stroke. J. Neurosci. 35, 2133-2145. ( 10.1523/JNEUROSCI.4098-14.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gate D, et al. 2020. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer's disease. Nature 577, 399-404. ( 10.1038/s41586-019-1895-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leng J, Goldstein DR. 2010. Impact of aging on viral infections. Microbes Infect. 12, 1120-1124. ( 10.1016/j.micinf.2010.08.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegrist C-A, Aspinall R. 2009. B-cell responses to vaccination at the extremes of age. Nat. Rev. Immunol. 9, 185-194. ( 10.1038/nri2508) [DOI] [PubMed] [Google Scholar]
- 5.Chan Y-H, Ng LFP. 2017. Age has a role in driving host immunopathological response to alphavirus infection. Immunology 152, 545-555. ( 10.1111/imm.12799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britanova OV, et al. 2014. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 192, 2689-2698. ( 10.4049/jimmunol.1302064) [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Zhurbenko N, Quach TD, Hopkins TJ, Rothstein TL, Hernandez AM. 2019. Human B-1 cells and B-1 cell antibodies change with advancing age. Front. Immunol. 10, 483. ( 10.3389/fimmu.2019.00483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zharhary D. 1988. Age-related changes in the capability of the bone marrow to generate B cells. J. Immunol. 141, 1863-1869. [PubMed] [Google Scholar]
- 9.Johnson SA, Cambier JC. 2004. Ageing, autoimmunity and arthritis: senescence of the B cell compartment – implications for humoral immunity. Arthrit. Res. Therapy 6, 131-139. ( 10.1186/ar1180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decman V, Laidlaw BJ, Doering TA, Leng J, Ertl HCJ, Goldstein DR, Wherry EJ. 2012. Defective CD8 T cell responses in aged mice are due to quantitative and qualitative changes in virus-specific precursors. J. Immunol. 188, 1933-1941. ( 10.4049/jimmunol.1101098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KA, Shin KS, Kim GY, Song YC, Bae EA, Kim IK, Koh CH, Kang CY. 2016. Characterization of age-associated exhausted CD8+ T cells defined by increased expression of Tim-3 and PD-1. Aging Cell 15, 291-300. ( 10.1111/acel.12435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrdjen D, et al. 2018. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380-395.e6. ( 10.1016/j.immuni.2018.01.011) [DOI] [PubMed] [Google Scholar]
- 13.Miho E, Yermanos A, Weber CR, Berger CT, Reddy ST, Greiff V. 2018. Computational strategies for dissecting the high-dimensional complexity of adaptive immune repertoires. Front. Immunol. 9, 224. ( 10.3389/fimmu.2018.00224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yermanos AD, Dounas AK, Stadler T, Oxenius A, Reddy ST. 2018. Tracing antibody repertoire evolution by systems phylogeny. Front. Immunol. 9, 2149. ( 10.3389/fimmu.2018.02149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. 2014. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158-168. ( 10.1038/nbt.2782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horns F, Dekker CL, Quake SR. 2020. Memory B cell activation, broad anti-influenza antibodies, and bystander activation revealed by single-cell transcriptomics. Cell Rep. 30, 905-913.e6. ( 10.1016/j.celrep.2019.12.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spindler MJ, et al. 2020. Massively parallel interrogation and mining of natively paired human TCRαβ repertoires. Nat. Biotechnol. 38, 609-619. ( 10.1038/s41587-020-0438-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briney B, Inderbitzin A, Joyce C, Burton DR. 2019. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393-397. ( 10.1038/s41586-019-0879-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almanzar N, et al. 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590-595. ( 10.1038/s41586-020-2496-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutzeit C, Chen K, Cerutti A. 2018. The enigmatic function of IgD: some answers at last. Eur. J. Immunol. 48, 1101-1113. ( 10.1002/eji.201646547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noviski M, Mueller JL, Satterthwaite A, Garrett-Sinha LA, Brombacher F, Zikherman J. 2018. IgM and IgD B cell receptors differentially respond to endogenous antigens and control B cell fate. Elife 7, e35074. ( 10.7554/eLife.35074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yermanos A, et al. 2020. Platypus: an open-access software for integrating lymphocyte single-cell immune repertoires with transcriptomes. bioRxiv ( 10.1101/2020.11.09.374280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. ( 10.1038/nbt.4096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380-381. ( 10.1038/nmeth.3364) [DOI] [PubMed] [Google Scholar]
- 25.Yermanos A, Neumeier D, Sandu I, Borsa M, Waindok AC, Merkler D, Oxenius A, Reddy ST. 2020. Single-cell immune repertoire and transcriptome sequencing reveals that clonally expanded and transcriptionally distinct lymphocytes populate the aged central nervous system in mice. bioRxiv ( 10.1101/2020.05.04.077081) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yermanos A, Neumeier D, Sandu I, Borsa M, Waindok AC, Merkler D, Oxenius A, Reddy ST. 2020. Single-cell immune repertoire and transcriptome sequencing reveals that clonally expanded and transcriptionally distinct lymphocytes populate the aged central nervous system in mice. bioRxiv ( 10.1101/2020.05.04.077081) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The VDJ and GEX sequencing files from deep sequencing and code that support the findings of this study are available at 10.5281/zenodo.4264462, 10.5281/zenodo.4264978, and https://github.com/alexyermanos/Platypus. The preprint can be found on bioRxiv [25].