Abstract
Dispersal limitation has been recurrently suggested to shape both macroecological patterns and microevolutionary processes within invertebrates. However, because of potential interactions among biological, environmental, temporal, and spatial variables, causal links among flight-related traits, diversification and spatial patterns of community assembly remain elusive. Integrating genetic variation within species across whole insect assemblages, within a simplified spatial and environmental framework, can be used to reduce the impact of these potentially confounding variables. Here, we used standardized sampling and mitochondrial DNA sequencing for a whole-community characterization of the beetle fauna inhabiting a singular forested habitat (laurel forest) within an oceanic archipelago setting (Canary Islands). The spatial structure of species assemblages together with species-level genetic diversity was compared at the archipelago and island scales for 104 winged and 110 wingless beetle lineages. We found that wingless beetle lineages have: (i) smaller range sizes at the archipelago scale, (ii) lower representation in younger island communities, (iii) stronger population genetic structure, and (iv) greater spatial structuring of species assemblages between and within islands. Our results reveal that dispersal limitation is a fundamental trait driving diversity patterns at multiple hierarchical levels by promoting spatial diversification and affecting the spatial configuration of entire assemblages at both island and archipelago scales.
Keywords: dispersal limitation, wing, population genetics, community structure, arthropod, mtDNA
1. Introduction
Dispersal ability, in particular flight capability, plays a key role in the diversification and evolution of species, as it enables species to disperse in search of mates, food and new habitats, and to escape predators [1–4]. However, flight is also an energetically costly and risky activity [3], and functional wings have been repeatedly lost or reduced across the tree of life, most notably within beetles, due to both spatial and ecological causes [2,4,5]. For many insect lineages, wing loss has an important impact on their dispersal potential, because the loss of flight ability means a dispersal limitation for distances greater than a few hundred metres [3]. This raises the question of how dispersal limitation might shape ecological and evolutionary patterns at both community and intraspecific levels. Within beetles, limited dispersal ability is suggested to constrain species geographical range sizes (e.g. [6,7]), mediate disequilibrium between species distributions and climatic conditions (e.g. [8]), promote stronger spatial community structure (e.g. [9]) and generate steeper latitudinal richness patterns among dispersal-limited lineages (e.g. [10,11]). Similarly, within insect species, it has been shown that dispersal reduction may enhance geographical structuring and diversification (e.g. [12–15]), due to limitations to individual movement and gene flow among isolated populations, potentially leading to higher molecular evolutionary [16] and speciation [14] rates.
There is a suite of traits potentially making up the dispersal potential of a species. Due to the scarcity of empirical data on insect dispersal ability, different biological, ecological and/or morphological traits have been used as surrogates of insect dispersal potential. These include habitat occupancy [17,18], physiological tolerances [19,20], body size [21,22] and presence/morphology of functional wings [23]. Functional wings are a morphological trait essential to flight [3], and thus the presence of functional wings is expected to play a fundamental role in species dispersal potential. To test the hypothesis that flightlessness promotes allopatric speciation and higher species richness, Ikeda et al. [14] applied a population genetic approach comparing flight capable and flightless lineages, finding general support for this hypothesis. However, there is still limited evidence for a causal link among flight-related traits, diversification and spatial patterns of community assembly due to potential interactions among biological, environmental, temporal or spatial variables [5,18]. Thus, studies are required where other variables (mostly niche-based) are controlled for. In this context, islands, and in particular specific insular habitats present across archipelago settings, offer a useful framework for comparative analyses between flighted and flightless lineages. Islands are suggested to have high proportions of flightless species [5], thus facilitating such comparative analyses. They also present discrete geographical entities, which in some circumstances may serve as natural replicates, or ecological and evolutionary time series [24–26].
In the present study, we evaluate the potential effect of dispersal limitation on allopatric diversification, comparing across a large number of winged and wingless lineages within a shared landscape of forest habitat. We examine beetle assemblages from laurel forests of the Canary Islands, which allows us to minimize potential confounding effects associated with differential habitat availability, habitat connectivity or other habitat-specific biological traits. Without considering the limited and degraded remnants of Gran Canaria, laurel forests are found on four of the seven Canary Islands, allowing for comparisons both within and among geographically independent areas. We applied a standardized field survey and molecular identification protocol to sample 25 laurel forest sites across four islands, where samples were sorted to parataxonomic units (PU), and up to four individuals for each PU within each sampling site were sequenced for a region of mitochondrial DNA (mtDNA). To compare dispersal history among lineages across an equivalent time frame, we defined lineages of maternal dispersal history (LMDH) by applying a conservative maximum intraspecific divergence threshold. This approach yielded a total of 104 winged and 110 wingless lineages, which were used to compare diversity patterns at both lineage and assemblage levels in order to test if dispersal limitation acts as a fundamental trait constraining beetle diversity patterns at multiple biological and spatial scales. Specifically, we predict that wingless beetle lineages will: (i) have smaller range sizes, (ii) be less represented in younger islands communities, (iii) have stronger population genetic structures and thus, (iv) present more spatially structured species assemblages at multiple scales.
2. Material and methods
(a). Field sampling
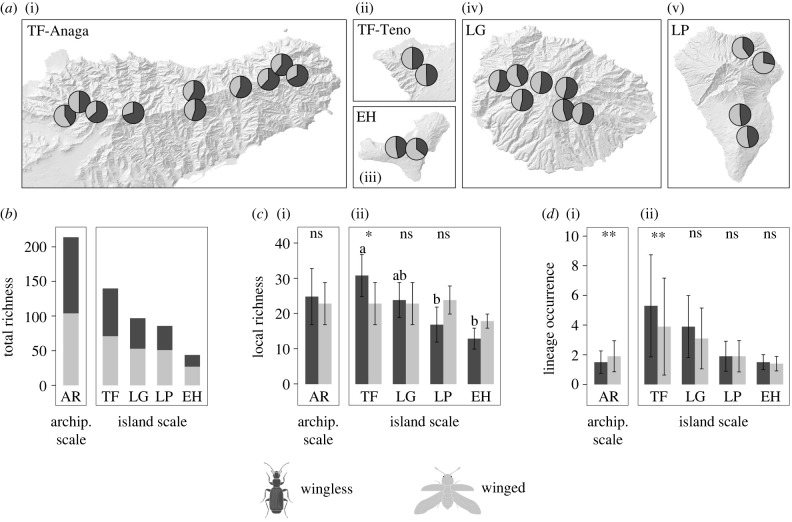
A total of 25 sites of 50 × 50 m were sampled within laurel forests across four of the Canary Islands: Tenerife (12), La Gomera (7), La Palma (4) and El Hierro (2) (figure 1; electronic supplementary material, table S1). Field sites were chosen based on the size and distribution of laurel forest on each island, state of preservation and accessibility. A standardized sampling protocol (modified from COBRA [27]) combining pitfall traps, leaf litter-sifting, foliage beating, vegetation sweeping and active searching (see [28] for details) was applied to each site. Sampling was carried out from 2012 to 2017, sampling adult beetles between the months of November to May, corresponding to optimal insect activity in the laurel forest associated with high humidity.
Figure 1.
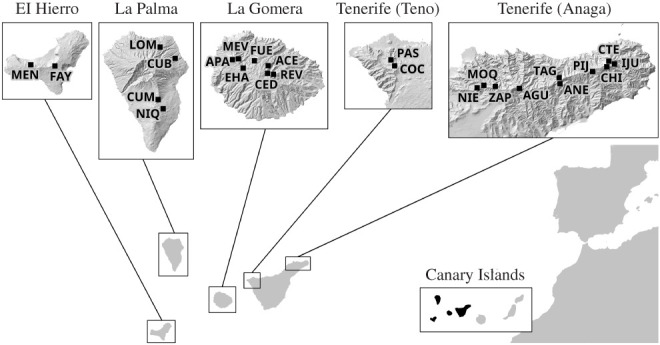
Sampling sites within the laurel forests of the Canary Islands (Tenerife, La Gomera, La Palma and El Hierro). Sampling sites are labelled with three letter codes (see the electronic supplementary material, table S8 for further details).
(b). Selection of specimens and mitochondrial DNA sequencing
Samples were preserved in 100% ethanol at −20°C, and sorted to PUs. PUs are estimations of species as discrete morphological entities based on external morphology only [28]. PUs were defined for each sampling site, and four specimens per PU and site (less if fewer than four were sampled) were selected for DNA extraction and sequencing. PUs were assigned provisional taxonomic identifications to species level where possible, otherwise to genus or family level, based on the authors' entomological knowledge or assisted with the use of taxonomic literature and a reference collection of Canary Island beetles.
DNA extractions were carried out using a chelex protocol [29], using either single legs, several legs, head and pronotum, or the entire individual for extraction, depending upon specimen size. In contrast with Emerson et al. [28], beetle PUs were amplified for an approximately 824 bp region of the mtDNA COI gene using the primers SPatR and Jerry [30]. PCR reaction conditions and sequencing details can be found in the electronic supplementary material (appedinx S1).
(c). Mitochondrial DNA lineages for the estimation of dispersal history
As our interest is to test for relative dispersal-related differences between flight capable and flightless lineages, we defined LMDH using mtDNA sequences. To establish a conservative maximum intraspecific divergence threshold to minimize the probability of a given biological species being assigned to more than one LMDH, we used data from Hendrich et al. [31]. From this dataset of 3514 European beetle species, we used 693 species for which eight or more individuals were sequenced, from which we inferred 8% divergence (Kimura two-parameter, K2P) to be a conservative LDMH threshold, as only 0.6% of all species exceed this value, all of which are suggested to represent morphologically cryptic species [31]. A custom R script was used to (i) produce an unweighted pair group method with arithmetic mean tree from pairwise K2P distances using an alignment of all sequences from all PUs, then (ii) create individual alignments for all lineages exceeding the 8% threshold. LMDHs may thus represent different stages of the speciation process, from single panmictic species, through to geographically structured species, incipient species and species complexes, and ultimately different closely related species, which may or may not be taxonomically diagnosable. Thus, the issue of species-level taxonomic identity is of limited importance within the context of our objectives.
(d). Flight ability assignment
For each LMDH, two categories of flight ability were assigned: (i) wingless, where individuals present atrophied or no wings, and (ii) winged, where well-developed wings are present. Flight polymorphic lineages were considered as winged, as they are able to disperse at the lineage level, even if not at the level of all individuals. Information about the development of wings for each LMDH was obtained by bibliographic searches (electronic supplementary material, table S2). In the case of taxa with no bibliographic reference material, or those LMDHs of uncertain taxonomy, wing morphology was examined under a stereomicroscope.
(e). Lineage richness and proportion of wingless lineages in beetle assemblages
Both the total number (total richness) and the proportions of wingless/winged LMDHs were calculated across the archipelago and at the island scale. Given that estimates of total richness within islands could be impacted by island size, and confounded by the different number of sampling sites within islands, for subsequent analyses, LMDH richness was estimated and compared per sampling site (local richness). Local richness estimates between wingless and winged assemblages were compared with a Wilcoxon signed-rank sum test in R v. 3.6.1 (wilcox.test function with samples paired by sampling site from stats R package), except for El Hierro where only two sites were sampled. Finally, we evaluated the extent to which flightlessness limits long-distance colonization of new areas, by comparing local richness for total (combined wingless and winged), wingless and winged lineages among islands, with a Kruskal–Wallis rank sum test and Dunn's test of multiple comparisons with the Benjamini–Hochberg correction in R v. 3.6.1 (kruskal.test and dunnTest functions from stats and FSA R packages, respectively). The islands of La Palma and El Hierro have maximum subaerial age estimates of 1.7 Myr and 1.1 Myr, respectively, notably younger than the neighbouring islands of Tenerife (11.9 Myr) and La Gomera (9.4 Myr) [32]. In this scenario, dispersal limitation may have conditioned the pool of species colonizing the younger islands.
(f). Occurrence patterns of winged and wingless lineages
To compare occurrence patterns for wingless and winged LMDHs, we evaluated (i) the number of islands, and (ii) the number of sampling sites within each island, where each lineage was found, and significance was tested with a Wilcoxon rank sum test in R v. 3.6.1 (wilcox.test function from stats R package). Additionally, proportions of wingless and winged LMDHs sampled in (i) more than one island at the archipelago scale, and (ii) more than one sampling site within each island, were compared with the Pearson's chi-squared test in R v. 3.6.1 (chisq.test function from stats R package).
(g). Population genetic differentiation within winged and wingless lineages
Two measures of population genetic differentiation were estimated to quantify phylogeographic structure within LMDHs: GST which considers only allelic frequencies, and NST which takes into account genetic distances between alleles, thus incorporating a phylogenetic weight in the measure [33]. LMDHs sampled from a minimum of two populations and comprising more than three overall individuals were selected to estimate GST and NST: (i) within each island, considering sampling sites as populations, (ii) among islands, where islands are considered as a single site, and (iii) pairwise among islands, using Spagedi 1.4b [34] with 10 000 permutations and K2P distances. GST and NST values were compared between winged and wingless lineages both among islands and within islands using Wilcoxon rank sum tests in R v. 3.6.1 (wilcox.test function, stats R package). Phylogeographic structuring within lineages was then evaluated by testing for significant differences from zero for GST and NST, and the proportions of phylogeographically structured wingless and winged lineages were compared both at the archipelago and island scales by applying the Fisher's exact test for count data (fisher.test function from stats R package). Differences in population structure between winged and wingless lineages between pairs of islands were also summarized graphically.
(h). Beta diversity and distance decay of winged and wingless assemblages
Total beta diversity (Sorensen dissimilarity, βsor) and its additive turnover (Simpson dissimilarity, βsim) and nestedness (βsne) components [35,36] were explored at multiple spatial scales for winged and wingless assemblages using the betapart R package [36]. In order to describe major spatial patterns of composition for winged and wingless assemblages across the archipelago: (i) the non-metric multidimensional scaling (NMDS) ordination method (metaMDS function from the vegan R package; [37]) was applied to turnover matrices among sampling sites, creating plots with the ordispider option, and (ii) a permutational multivariate analysis of variance (Permanova) (adonis function from the vegan R package; [37]) was applied with 999 permutations and island as the grouping factor. Additionally, we investigated an alternative beta diversity partition for species replacement and richness difference components [38], using the BAT R package [39].
Distance decay describes how similarity in species composition between communities varies with geographical distance, thus representing the magnitude of spatial structuring [40]. In the absence of environmental gradients, the decay of assemblage similarity with spatial distance represents the probability of species reaching distant localities, which decreases due to their limited dispersal ability [9]. To estimate distance decay for wingless and winged assemblages, we used the function decay.model (betapart R package; [36]), with a negative exponential model (glm(formula = y ∼ x, family = Gaussian(link = log))) and similarity values from the turnover component of beta diversity (1 - pairwise turnover matrices), at both the archipelago and island scales (with the exception of El Hierro, which only has two sampling sites). Matrices were computed as Euclidean distances between the centroids of sampling sites, with a custom R function. The goodness of fit of these decay models was computed as pseudo-R2 = 1 − model deviance/null deviance. Significance was assessed by randomizing spatial distances 1000 times and computing the proportion of times in which model deviance was smaller than the randomized model deviance. Analyses were run independently for wingless and winged assemblages, using R software v. 3.6.1.
Additionally, to understand potential differences in beta diversity between wingless and winged assemblages, abundance–occurrence relationships were explored at both the archipelago and individual island scales. For each LMDH, abundances were calculated as the average across all islands (archipelago scale) or sampling sites (island scale) where the LMDH was sampled [41]. For occurrence, both (i) the number of islands where the LMDH was sampled divided by the total number of islands, and (ii) the number of sampling sites where the LMDH was sampled divided by the total number of sites within each island [41] was used for the archipelago and island scales, respectively. Then, linear regressions were applied, treating occurrence as the response variable and log-transformed abundance as the predictor.
3. Results
(a). Field sampling and sorting into parataxonomic units
A total of 44 331 adult individuals were collected from laurel forests of the Canary Islands (figure 1; electronic supplementary material, table S1). Sampling methods presented marked variation in effectiveness, with 64% of all individuals sampled by pitfall traps, 17% by foliage beating and sweeping, 7% by leaf litter sampling and 12% by active searching (aerial, under decaying trunks, dead wood and stones on the ground, and under bark, lichens and mosses on living trees; for the description of sampling methods see [28]). A total of 685 PUs were established.
(b). Mitochondrial DNA sequencing and lineages of maternal dispersal history
A total of 5328 individuals were selected for mtDNA sequencing, of which 4678 yielded good quality sequences (a sequencing success rate of 87%), resulting in the establishment of 269 LMDHs across all islands. Ten LMDHs were excluded from further analysis as they were identified as introduced species, and a further 45 were excluded as it was not possible to assign wing state (i.e. no available bibliographic information and highly deteriorated material after DNA extraction). The final dataset comprised 214 LMDHs corresponding to 38 Coleoptera families (19 families including only winged LMDHs, 11 only wingless ones and eight including both winged and wingless LMDHs; electronic supplementary material, figure S1). Further lineage details, including taxonomic identification, flight ability assignment, sampling and associated DNA sequences, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.63xsj3v15 [42].
(c). Lineage richness and proportions of wingless lineages in beetle assemblages
At the archipelago scale, local (per sampling site) lineage richness was highest for Tenerife (54 ± 9) followed by La Gomera (48 ± 10), La Palma (41 ± 3) and finally El Hierro (32 ± 1), with differences being significant (Kruskal–Wallis χ23 = 9.980, p = 0.019). The proportions of wingless (51%) and winged (49%) LMDHs were similar, but proportions varied across sampling sites (figure 2a). While the local richness of winged lineages was not significantly different among four islands (Kruskal–Wallis χ23 = 2.366, p = 0.500), the local richness of wingless lineages was significantly different (Kruskal–Wallis χ23 = 13.758, p = 0.003), with higher values in the oldest island of Tenerife (31 ± 6), followed by La Gomera (24 ± 5), and then the youngest islands La Palma (17 ± 5) and El Hierro (13 ± 3) (figure 2c; electronic supplementary material, table S3, table S4). However, only Tenerife showed significant differences between La Palma and El Hierro after multiple comparisons. At the island scale, local winged and wingless lineage richness varied among islands with: (i) higher local richness of wingless lineages in Tenerife, (ii) similar values for both groups in La Gomera, and (iii) lower local richness of wingless lineages in both La Palma and El Hierro, although differences were significant only for Tenerife (figure 2c; electronic supplementary material, table S3).
Figure 2.
Lineage richness and occurrence for wingless (dark grey) and winged (light grey) beetle assemblages, at both the archipelago and island scales. (a) Proportions of lineages across sampling sites. (b) Total richness. (c) Richness per sampling site (local richness). (d) Lineage occurrence measured as the number of islands or sampling sites where each lineage was sampled. TF = Tenerife, LG = La Gomera, LP = La Palma, and EH = El Hierro. **p < 0.01, *p < 0.05 for significant differences between wingless and winged lineages. ns = not significant. Letters represent significant differences of local richness among islands for wingless lineages.
(d). Occurrence patterns of winged and wingless lineages
At the archipelago scale, the number of islands sampled for a given LMDH was significantly lower for wingless lineages (figure 2d; electronic supplementary material, table S5), and the proportion of wingless and winged lineages sampled on two or more islands was significantly different (electronic supplementary material, table S5). At the island scale, the number of sites for which a given wingless LMDH was sampled was higher than that for winged lineages within Tenerife, La Gomera and El Hierro (but only significant in Tenerife), with similar values for both groups in La Palma (figure 2d; electronic supplementary material, table S5). In all islands, higher proportions of wingless lineages were sampled in two or more sampling sites, compared to winged lineages, but differences were only significant in Tenerife (electronic supplementary material, table S5).
(e). Population genetic differentiation within winged and wingless lineages
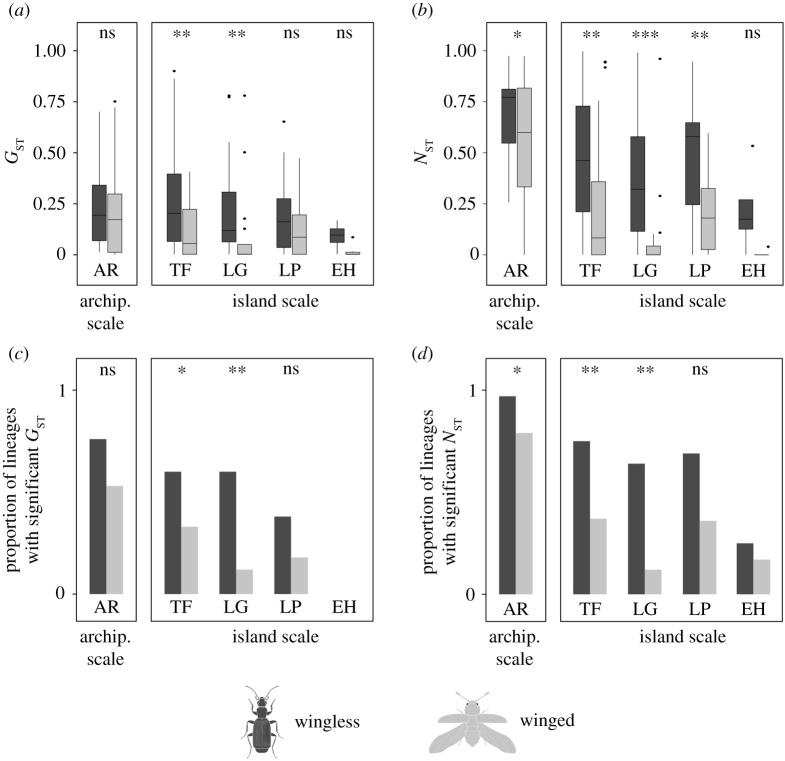
Both GST and NST were higher for wingless compared to winged lineages, with significant differences for NST between winged and wingless lineages at both spatial scales, among islands and within three of the four analysed islands (the difference was marginally non-significant in El Hierro; figure 3b; electronic supplementary material, table S6). Significant differences for GST were only found within the islands of Tenerife and La Gomera (figure 3a; electronic supplementary material, table S6). A significantly higher proportion of wingless lineages were phylogeographically structured (GST and NST significantly higher than zero), compared to winged lineages, among islands and within Tenerife and La Gomera. Proportionately more wingless lineages were also phylogeographically structured within La Palma, but the difference was not significant, and significance could not be tested in El Hierro (figure 3c,d; electronic supplementary material, table S7). Pairwise comparisons of GST and NST between islands (electronic supplementary material, figure S2) indicate lower gene flow for wingless compared to winged lineages, most notably between El Hierro and the other islands.
Figure 3.
Genetic structure for wingless (dark grey) and winged (light grey) beetle assemblages, at both the archipelago and island scales. (a) GST values. (b) NST values. (c) Proportion of lineages with GST significantly different from zero. (d) Proportion of lineages with NST significantly different from zero. AR = archipelago, TF = Tenerife, LG = La Gomera, LP = La Palma and EH = El Hierro. *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant.
(f). Beta diversity and distance decay of winged and wingless assemblages
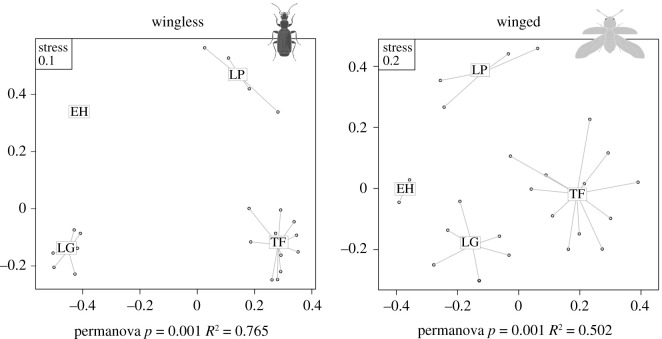
Turnover (βsim) was the largest component of total beta diversity (βsor) both at the archipelago scale and within islands (electronic supplementary material, table S8). At the archipelago scale, multi-site βsim was higher for wingless (0.626) compared to winged lineages (0.462). Both winged and wingless dissimilarity matrices across all sampling sites were highly and significantly explained by the island factor, indicating that variation for both wingless and winged communities is strongly structured by islands (figure 4). However, the explained variation for wingless assemblages across islands (represented by the variance of turnover βsim matrix explained by the island grouping) was higher compared to that for winged lineages (76.5% versus 50.2% of explained variance, respectively). At the island scale, wingless lineages tended to show lower overall turnover among sampling sites (figure 4; electronic supplementary material, table S8). Nestedness (βsne) showed no significant patterns across islands for both winged and wingless lineages (electronic supplementary material, figure S3), although it was higher for wingless lineages within the islands of La Gomera, La Palma and El Hierro (electronic supplementary material, table S8). All these results were consistent when considering the beta diversity partition proposed by Carvalho [38] (electronic supplementary material, figure S4).
Figure 4.
NMDS ordinations of both wingless and winged assemblages according to the turnover component (Simpson dissimilarity, βsim) of beta diversity across all sampling sites, grouping by island (TF = Tenerife, LG = La Gomera, LP = La Palma and EH = El Hierro). R2 and significance (P) from the permutational ANOVAs (permanova) over the community dissimilarity matrices are provided.
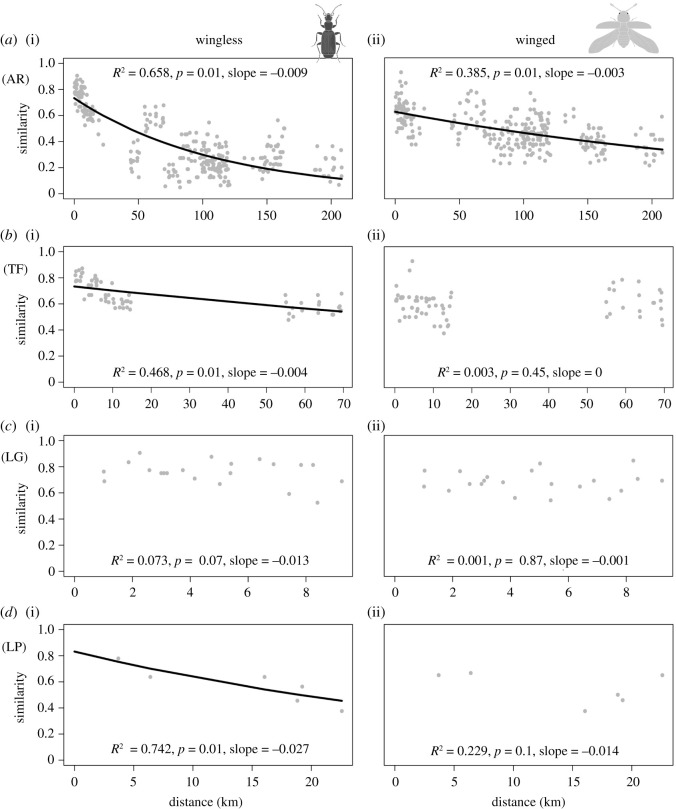
The negative exponential model significantly explained the decay in similarity with spatial distance among islands (p < 0.01), both for winged and wingless assemblages. The fit of the exponential decay model was better for wingless (pseudo-R2 = 0.658) compared to winged assemblages (pseudo-R2 = 0.385), and a steeper slope was also observed for wingless assemblages (slope = −0.009 versus slope = −0.003 for wingless and winged respectively) (figure 5a). At the island scale, the decay in similarity with geographical distance among sampling sites was significantly explained by the negative exponential model only for wingless assemblages within the islands of Tenerife (p < 0.01; figure 5b) and La Palma (p < 0.01; figure 5d). Winged assemblages showed non-significant distance decay at the island scale (figure 5b–d). Positive abundance–occurrence relationships were found for both wingless and winged lineages at the island scale (electronic supplementary material, figure S6). At the archipelago scale, only winged lineages showed a significant relationship.
Figure 5.
Distance decay of wingless and winged assemblage similarities (Simpson index, βsim) at both (a) the archipelago and (b,c,d) island scales. AR = archipelago, TF = Tenerife, LG = La Gomera, LP = La Palma, and EH = El Hierro. Pseudo-R2, significance (P) and slope are provided.
4. Discussion
(a). Dispersal limitation and the geographical structuring of species assemblages and species-level genetic variation
Dispersal limitation may have consequences at the community scale, with the expectation that communities will become increasingly differentiated as lineage exchange among assemblages is reduced. Contrasting macroecological patterns among beetle taxa with different dispersal abilities have previously been reported, indicating dispersal capability may be a key factor determining global patterns of species richness and community assembly (e.g. [9,10]). Our results reveal clearly differentiated spatial patterns of diversity between winged and wingless assemblages. At the archipelago scale, wingless assemblages showed higher beta diversity, primarily mediated by lineage turnover βsim, as wingless lineages tend to occur on fewer islands, compared to winged lineages. Distance decay patterns for wingless lineages were also steeper and less dispersed, compared to winged assemblages, showing that the heterogeneity in assemblage composition was more spatially structured in the former. While there is no significant difference in the local richness of winged lineages across islands, the oldest island of Tenerife presents significantly higher local richness of wingless lineages, pointing to a delayed colonization of younger islands (La Palma and El Hierro) by wingless lineages and/or higher intra-island diversification of wingless lineages over time in the case of Tenerife. Overall, these patterns point to a fundamental role for dispersal limitation constraining the colonization and movement of individuals within a discontinuous landscape. This is broadly consistent with the results of Soininen et al. [22], who examined the effects of dispersal on beta diversity using data from a range of organisms (bacteria, protists, fungi, plants, invertebrates, fishes, amphibians, birds and mammals), and concluded that total beta diversity and turnover were higher for passively dispersed taxa than for flying taxa. While Soininen et al. [22] emphasized the potential interactive effects from factors such as habitat and spatial extent impacting on their findings, our results using whole beetle communities inhabiting the same habitat within the same geographical setting, strengthen the evidence for a primary role of dispersal limitation in structuring community assembly.
At a lower hierarchical level of diversity, analyses of genetic variation across the archipelago also reveal that dispersal limitation constrains movement over evolutionary timescales, facilitating diversification within flightless lineages. Population structure as measured by NST (weighted genetic distance among haplotypes) was significantly higher for wingless lineages at both the archipelago and island scales. A similar pattern, albeit less pronounced, was also found for GST (unweighted genetic distance among haplotypes). This difference between NST and GST is consistent with Pons & Petit [33], who point out that NST will be more informative than GST, as haplotype variation is more efficiently used when ordered alleles are considered, leading to a more powerful test for genetic structure. Significant differences in NST between flightless and flighted lineages were observed both at the archipelago scale and within individual islands. These results indicate that restricted gene flow due to dispersal limitation drives higher genetic divergence in flightless lineages, building upon previous work suggesting that this may be a general rule, at least across arthropods [12–15].
While winged lineages tended to be distributed across more islands and have overall lower beta diversity at the archipelago scale, within islands they tend to be found in a lower number of sampling sites, driving higher intra-island turnover compared to flightless assemblages. This more patchy intra-island distribution of winged lineages could be associated with the lower local abundances of winged lineages found for most of the sampling sites (electronic supplementary material, figure S5, tables S9 and S10). Indeed, a positive local abundance–occupancy relationship of the lineages is found for all islands (electronic supplementary material, figure S6). In general, locally abundant species are expected to be geographically more widespread than those that are locally rare, leading to positive local abundance–occupancy relationships [43–45]. Interestingly, at the archipelago scale, wingless species do not conform to such a relationship, reflecting the greater relevance of dispersal constraint at this scale. In addition to local abundance differences, a potentially higher degree of ecological specialization for winged species, for example to host plants, could also contribute to the more patchy intra-island distribution of these species [46]. An example is provided by the beetle genus Rhopalomesites, which was sampled for both a plant specialist (Rhopalomesites proximus) and generalist (Rhopalomesites persimilis) species [47], being winged and wingless, respectively. Further ecological studies on the differential intra-island occurrence patterns of dispersive and dispersal-limited lineages are needed to clarify the potential mechanisms acting.
Overall, our results comparing beetle assemblages within the same habitat and spatial setting point to a fundamental role for dispersal ability shaping the geographical structure of diversity, from species-level genetic variation to the level of whole communities. Because flightlessness is in many cases a conserved trait, even across complete beetle families, the influence of other biological traits (e.g. trophic level, size or metabolism) could also contribute to some extent to the contrasted patterns found. However, the broad phylogenetic and functional diversity of taxa included within the sampled wingless and winged groups (electronic supplementary material, figure S1), and the consistent patterns found between wingless and winged lineages, even within single families (electronic supplementary material, figure S7), strongly support dispersal limitation, in particular flightlessness, as a key trait shaping spatial patterns of species-level and community-level diversity.
(b). Dispersal limitation and speciation
It has long been recognized that dispersal limitation should positively contribute to isolation by spatial distance, through reduced gene flow between populations [48–51]. Such a relationship may scale up to higher rates of neutral speciation within dispersal-limited lineages, where geographical isolation favours speciation by de novo mutation and random genetic drift [52]. Support for this has come from reductions in dispersal ability being associated with increased rates of speciation [14], although with some controversy as to whether differences in habitat, rather than dispersal ability per se, may be the causal mechanism [18]. By comparing wingless and winged lineages within the same habitat, our community-level approach provides compelling support for a general model where higher genetic differentiation among populations is favoured when dispersal is limited. However, a potential relationship between population-level differentiation and geographical speciation also requires consideration of how species are distributed spatially.
Wingless lineages present higher differentiation among islands, consistent with the extreme physical barrier presented by the continuous ocean between islands. Wingless lineages are unsurprisingly less able to cross the 28–113 km stretches of ocean that separate islands, and this is clearly reflected in our results, which provide support for higher rates of between island speciation for wingless compared to winged lineages. The same trend for higher diversification within wingless lineages is also observed within islands, but may differ in speciation outcomes as it can be explained by either isolation by distance [50,51,53] or isolation by resistance (due to geographical barriers or unfavourable environmental conditions; [54,55]). Within continuously distributed species, higher differentiation among populations will be associated with dispersal limitation, through the action of isolation by distance. This alone is unlikely to translate to higher rates of neutral allopatric speciation, as it does not involve geographical or climatic barriers to gene flow, only intrinsic limitations to gene flow. However, when species are distributed discontinuously due to landscape features or spatial variation in climate, intrinsic limitations to gene flow may exceed geographical distances between populations, thus providing the conditions for allopatric speciation driven by isolation by resistance. Our results demonstrate a relationship between dispersal limitation and the geographical structuring of genetic variation within species consistent with increased rates of speciation between islands for wingless lineages. However, understanding the speciation consequences of population genetic structuring from dispersal limitation within continuous terrestrial habitat will require testing against the null hypothesis of isolation by distance, by applying an approach that explicitly takes into account landscape features and climatic variations (e.g. [56,57]).
5. Conclusion
Causal links between specific dispersal-related traits, diversification and the spatial configuration of assemblages are often difficult to establish. By exploring genetic-level and community-level structure for densely sampled assemblages of Coleoptera within a common and simplified spatio-environmental framework, we provide support for a model where dispersal limitation has a pervasive impact on the diversification history of beetle lineages. This species-level influence in turn scales up to the spatial configuration of entire assemblages at different spatial scales. Despite its importance, estimating dispersal ability for insects is far from trivial, as empirical data on migration rates are extremely scarce. Here, we show how the presence or absence of wings can be used as a proxy for dispersal ability and integrated within a community genetic context to understand how dispersal ability contributes to the spatial structuring of biodiversity. Wing-related dispersal traits in other insect groups, such as wing length or size variation in butterflies [58,59], stoneflies and mayflies [60,61] and damselfies [62], or variation for dispersal-related traits in non-insect arthropods (e.g. ballooning in spiders), may provide similar opportunities to further explore the generality of our findings.
Supplementary Material
Acknowledgements
The authors wish to thank the following for contributing to fieldwork, sample sorting and species identification: Pedro Oromí, Víctor García-Olivares, Nuria Macías-Hernández, Salvador de la Cruz, Benito Pérez Vispo, David Hernández, Rienk Apperloo, Manuel Arechavaleta, Nieves Zurita, Sara Ravagni, Isa Sancibrián Span, Antonio Machado, Daniel Suárez, Irene Santos and Nassam Medina Pérez. We also wish to thank Peter Stüben and Christoph Bayer (CURCULIO Institute e.V.) for providing information on the presence or absence of wings in Curculionidae, based on their recognized taxonomic experience in this family of Coleoptera. Fieldwork was supported by collecting permits AFF-107/17 (sigma no. 2017-00572), 246285, A/EST-034/16 and RE:2349 kindly provided by the Cabildo of Tenerife, Canary Islands Government, Cabildo of La Palma and Cabildo of El Hierro, respectively.
Data accessibility
The information associated with the studied lineages, including taxonomic identification (family level identification), flight ability assignment, sampling and associated DNA sequences, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.63xsj3v15 [42]. The custom R script developed to define lineages of maternal dispersal history (LMDH) by applying a maximum intraspecific divergence threshold is available from GitHub (https://github.com/asalcescastellano/Divergence-threshold.git). All supplementary tables and figures cited in the main text have been uploaded as electronic supplementary material, information.
Authors' contributions
B.C.E. A.S.-C. and P.A. designed and coordinated the study. All authors participated in the fieldwork. A.J.P.-D. and H.L. led sample sorting and flight category classification with input from A.S.-C. A.S.-C. and H.L. generated the mtDNA sequence data. A.S.-C. led the data analyses with input from B.C.E., P.A. and C.A. A.S.-C., B.C.E. and P.A. wrote the manuscript, with contributions from all authors. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
The authors declare no conflict of interest.
Funding
This work was supported by the Spanish Agencia Estatal de Investigación (project code CGL2017-85718-P, awarded to B.C.E.), co-financed by FEDER and by the ERA-Net Net-Biome research framework, financed through a Canary Island Government ACIISI grant (no. SE-12/04, awarded to B.C.E.), co-financed by FEDER. It was also supported by the Spanish Ministerio de Ciencia, Innovación y Universidades (project code EQC2018-004418-P, awarded to B.C.E.), and Spanish grant 1153/2014 awarded to BCE by the Organismo Autonomo Parques Nacionales of the Ministerio de Medio Ambiente y Medio Rural y Marino GRAMA. A.S.-C. was funded by the Spanish Ministerio de Ciencia, Innovación y Universidades through the FPU PhD fellowship (FPU014/02948). C.A. was supported by the Spanish Ministry of Economy and Competitiveness (MINECO, Spain) (CGL2015-74178-JIN MINECO/FEDER, UE).
References
- 1.Engel MS. 2015. Insect evolution. Curr. Biol. 25, R868-R872. ( 10.1016/j.cub.2015.07.059) [DOI] [PubMed] [Google Scholar]
- 2.Grimaldi D, Engel MS. 2005. Evolution of the insects. New York, NY: Cambridge University Press. [Google Scholar]
- 3.Roff DA. 1990. The evolution of flightlessness in insects. Ecol. Monogr. 60, 389-421. ( 10.2307/1943013) [DOI] [Google Scholar]
- 4.Wagner DL, Liebherr JK. 1992. Flightlessness in insects. Trends Ecol. Evol. 7, 216-220. ( 10.1016/0169-5347(92)90047-F) [DOI] [PubMed] [Google Scholar]
- 5.Waters JM, Emerson BC, Arribas P, McCulloch GA. 2020. Dispersal reduction: causes, genomic mechanisms, and evolutionary consequences. Trends Ecol. Evol. 35, 512-522. ( 10.1016/j.tree.2020.01.012) [DOI] [PubMed] [Google Scholar]
- 6.Ribera I, Vogler AP. 2000. Habitat type as a determinant of species range sizes: the example of lotic-lentic differences in aquatic Coleoptera. Biol. J. Linnean Soc. 71, 33-52. ( 10.1006/bijl.1999.0412) [DOI] [Google Scholar]
- 7.Arribas P, Velasco J, Abellán P, Sánchez-Fernández D, Andújar C, Calosi P, Millán A, Ribera I, Bilton DT. 2012. Dispersal ability rather than ecological tolerance drives differences in range size between lentic and lotic water beetles (Coleoptera: Hydrophilidae). J. Biogeogr. 39, 984-994. ( 10.1111/j.1365-2699.2011.02641.x) [DOI] [Google Scholar]
- 8.Sánchez-Fernández D, Lobo JM, Millán A, Ribera I. 2012. Habitat type mediates equilibrium with climatic conditions in the distribution of Iberian diving beetles. Glob. Ecol. Biogeogr. 21, 988-997. ( 10.1111/j.1466-8238.2011.00743.x) [DOI] [Google Scholar]
- 9.Gómez-Rodríguez C, Baselga A. 2018. Variation among European beetle taxa in patterns of distance decay of similarity suggests a major role of dispersal processes. Ecography 41, 1825-1834. ( 10.1111/ecog.03693) [DOI] [Google Scholar]
- 10.Baselga A, Lobo JM, Svenning JC, Aragón P, Araújo MB. 2012. Dispersal ability modulates the strength of the latitudinal richness gradient in European beetles. Glob. Ecol. Biogeogr. 21, 1106-1113. ( 10.1111/j.1466-8238.2011.00753.x) [DOI] [Google Scholar]
- 11.Hortal J, Diniz-Filho JAF, Bini LM, Rodríguez MÁ, Baselga A, Nogués-Bravo D, Rangel TF, Hawkins BA, Lobo JM. 2011. Ice age climate, evolutionary constraints and diversity patterns of European dung beetles. Ecol. Lett. 14, 741-748. ( 10.1111/j.1461-0248.2011.01634.x) [DOI] [PubMed] [Google Scholar]
- 12.Dapporto L, et al. 2019. Integrating three comprehensive data sets shows that mitochondrial DNA variation is linked to species traits and paleogeographic events in European butterflies. Mol. Ecol. Res. 19, 1623-1636. ( 10.1111/1755-0998.13059) [DOI] [PubMed] [Google Scholar]
- 13.Dussex N, Chuah A, Waters JM. 2016. Genome-wide SNPs reveal fine-scale differentiation among wingless alpine stonefly populations and introgression between winged and wingless forms. Evolution 70, 38-47. ( 10.1111/evo.12826) [DOI] [PubMed] [Google Scholar]
- 14.Ikeda H, Nishikawa M, Sota T. 2012. Loss of flight promotes beetle diversification. Nat. Commun. 3, 643. ( 10.1038/ncomms1659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MP, Blinn DW, Keim P. 2002. Correlations between observed dispersal capabilities and patterns of genetic differentiation in populations of four aquatic insect species from the Arizona White Mountains, USA. Freshw. Biol. 47, 1660-1673. ( 10.1046/j.1365-2427.2002.00911.x) [DOI] [Google Scholar]
- 16.Mitterboeck FT, Adamowicz SJ. 2013. Flight loss linked to faster molecular evolution in insects. Proc. R. Soc. B 280, 20131128. ( 10.1098/rspb.2013.1128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulou A, Anastasiou I, Keskin B, Vogler AP. 2009. Comparative phylogeography of tenebrionid beetles in the Aegean archipelago: the effect of dispersal ability and habitat preference. Mol. Ecol. 18, 2503-2517. ( 10.1111/j.1365-294X.2009.04207.x) [DOI] [PubMed] [Google Scholar]
- 18.Vogler AP, Timmermans MJTN. 2012. Speciation: don't fly and diversify? Curr. Biol. 22, 284-286. ( 10.1016/j.cub.2012.03.015) [DOI] [PubMed] [Google Scholar]
- 19.Calosi P, Bilton DT, Spicer JI, Votier SC, Atfield A. 2010. What determines a species' geographical range? Thermal biology and latitudinal range size relationships in European diving beetles (Coleoptera: Dytiscidae). J. Anim. Ecol. 79, 194-204. ( 10.1111/j.1365-2656.2009.01611.x) [DOI] [PubMed] [Google Scholar]
- 20.Arribas P, Abellán P, Velasco J, Bilton DT, Millán A, Sánchez-Fernández D. 2012. Evaluating drivers of vulnerability to climate change: a guide for insect conservation strategies. Glob. Change Biol. 18, 2135-2146. ( 10.1111/j.1365-2486.2012.02691.x) [DOI] [Google Scholar]
- 21.De Bie T, et al. 2012. Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol. Lett. 15, 740-747. ( 10.1111/j.1461-0248.2012.01794.x) [DOI] [PubMed] [Google Scholar]
- 22.Soininen J, Heino J, Wang J. 2018. A meta-analysis of nestedness and turnover components of beta diversity across organisms and ecosystems. Glob. Ecol. Biogeogr. 27, 96-109. ( 10.1111/geb.12660) [DOI] [Google Scholar]
- 23.Sekar S. 2012. A meta-analysis of the traits affecting dispersal ability in butterflies: can wingspan be used as a proxy? J. Anim. Ecol. 81, 174-184. ( 10.1111/j.1365-2656.2011.01909.x) [DOI] [PubMed] [Google Scholar]
- 24.Emerson BC. 2002. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol. Ecol. 11, 951-966. ( 10.1046/j.1365-294X.2002.01507.x) [DOI] [PubMed] [Google Scholar]
- 25.Shaw KL, Gillespie RG. 2016. Comparative phylogeography of oceanic archipelagos: hotspots for inferences of evolutionary process. Proc. Natl Acad. Sci. USA 113, 7986-7993. ( 10.1073/pnas.1601078113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren BH, et al. 2015. Islands as model systems in ecology and evolution: prospects fifty years after MacArthur-Wilson. Ecol. Lett. 18, 200-217. ( 10.1111/ele.12398) [DOI] [PubMed] [Google Scholar]
- 27.Cardoso P. 2009. Standardization and optimization of arthropod inventories: the case of Iberian spiders. Biodivers. Conserv. 18, 3949-3962. ( 10.1007/s10531-009-9690-7) [DOI] [Google Scholar]
- 28.Emerson BC, Casquet J, López H, Cardoso P, Borges PAV, Mollaret N, Oromí P, Strasberg D, Thébaud C. 2017. A combined field survey and molecular identification protocol for comparing forest arthropod biodiversity across spatial scales. Mol. Ecol. Res. 17, 694-707. ( 10.1111/1755-0998.12617) [DOI] [PubMed] [Google Scholar]
- 29.Casquet J, Thebaud C, Gillespie RG. 2012. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Res. 12, 136-141. ( 10.1111/j.1755-0998.2011.03073.x) [DOI] [PubMed] [Google Scholar]
- 30.Timmermans MJTN, Dodsworth S, Culverwell CL, Bocak L, Ahrens D, Littlewood DTJ, Pons J, Vogler AP. 2010. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 38, 1-14. ( 10.1093/nar/gkq807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrich L, Morinière J, Haszprunar G, Hebert PDN, Hausmann A, Köhler F, Balke M. 2015. A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD. Mol. Ecol. Res. 15, 795-818. ( 10.1111/1755-0998.12354) [DOI] [PubMed] [Google Scholar]
- 32.Carracedo JC, Troll VR. 2016. The geology of the Canary Islands. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 33.Pons O, Petit RJ. 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237-1245. ( 10.1093/genetics/144.3.1237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy OJ, Vekemans X. 2002. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618-620. ( 10.1046/j.1471-8286.2002.00305.x) [DOI] [Google Scholar]
- 35.Baselga A. 2010. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19, 134-143. ( 10.1111/j.1466-8238.2009.00490.x) [DOI] [Google Scholar]
- 36.Baselga A, Orme CDL. 2012. Betapart: an R package for the study of beta diversity. Methods Ecol. Evol. 3, 808-812. ( 10.1111/j.2041-210X.2012.00224.x) [DOI] [Google Scholar]
- 37.Oksanen J, et al. 2019. vegan: community ecology package. R package version 2.5-6. See http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 38.Carvalho JC, Cardoso P, Borges PAV, Schmera D, Podani J. 2013. Measuring fractions of beta diversity and their relationships to nestedness: a theoretical and empirical comparison of novel approaches. Oikos 122, 825-834. ( 10.1111/j.1600-0706.2012.20980.x) [DOI] [Google Scholar]
- 39.Cardoso P, Rigal F, Carvalho JC. 2015. BAT - biodiversity assessment tools, an R package for the measurement and estimation of alpha and beta taxon, phylogenetic and functional diversity. Methods Ecol. Evol. 6, 232-236. ( 10.1111/2041-210X.12310) [DOI] [Google Scholar]
- 40.Morlon H, Chuyong G, Condit R, Hubbell S, Kenfack D, Thomas D, Valencia R, Green JL. 2008. A general framework for the distance-decay of similarity in ecological communities. Ecol. Lett. 11, 904-917. ( 10.1111/j.1461-0248.2008.01202.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigal F, Whittaker RJ, Triantis KA, Borges PAV. 2013. Integration of non-indigenous species within the interspecific abundance-occupancy relationship. Acta Oecologica 48, 69-75. ( 10.1016/j.actao.2013.02.003) [DOI] [Google Scholar]
- 42.Salces-Castellano A, Andújar C, López H, Pérez-Delgado AJ, Arribas P, Emerson BC. 2021. Data from: Flightlessness in insects enhances diversification and determines assemblage structure across whole communities. Dryad Digital Repository. ( 10.5061/dryad.63xsj3v15) [DOI] [PMC free article] [PubMed]
- 43.Freckleton RP, Noble D, Webb TJ. 2006. Distributions of habitat suitability and the abundance-occupancy relationship. Am. Nat. 167, 260-275. ( 10.1086/498655) [DOI] [PubMed] [Google Scholar]
- 44.Gaston KJ, Blackburn TIMM, Greenwood JD, Gregory RD, Quinn M, Lawton JH. 2000. Abundance-occupancy relationships. J. Appl. Ecol. 37, 39-59. ( 10.1046/j.1365-2664.2000.00485.x) [DOI] [Google Scholar]
- 45.Gaston KJ, Blackburn TM.. 2000. Pattern and process in macroecology. Oxford, UK: Blackwell Science. [Google Scholar]
- 46.Rádková V, Bojková J, Křoupalová V, Schenková J, Syrovátka V, Horsák M. 2014. The role of dispersal mode and habitat specialisation in metacommunity structuring of aquatic macroinvertebrates in isolated spring fens. Freshw. Biol. 59, 2256-2267. ( 10.1111/fwb.12428) [DOI] [Google Scholar]
- 47.Hernández-Teixidor D, López H, Pons J, Juan C, Oromí P. 2016. Host plant associations and geographical factors in the diversification of the Macaronesian Rhopalomesites beetles (Coleoptera: Curculionidae). J. Biogeogr. 43, 1608-1619. ( 10.1111/jbi.12737) [DOI] [Google Scholar]
- 48.Bohonak AJ. 1999. Dispersal, gene flow, and population structure. Q. Rev. Biol. 74, 21-45. ( 10.1086/392950) [DOI] [PubMed] [Google Scholar]
- 49.Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L.. 2013. Drivers of population genetic differentiation in the wild: Isolation by dispersal limitation, isolation by adaptation and isolation by colonisation. Mol. Ecol. 22, 5983-5999. ( 10.1111/mec.12561) [DOI] [PubMed] [Google Scholar]
- 50.Wright S. 1943. Isolation by distance. Genetics 28, 114-138. ( 10.1093/nq/184.4.114g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright S. 1946. Isolation by distance under diverse systems of mating. Genetics 31, 39-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gavrilets S. 2014. Models of speciation: where are we now? J. Heredity 105, 743-755. ( 10.1093/jhered/esu045) [DOI] [PubMed] [Google Scholar]
- 53.Slatkin M. 1993. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47, 264. ( 10.2307/2410134) [DOI] [PubMed] [Google Scholar]
- 54.McRae BH. 2006. Isolation by resistance. Evolution 60, 1551. ( 10.1554/05-321.1) [DOI] [PubMed] [Google Scholar]
- 55.Peterman WE, Connette GM, Semlitsch RD, Eggert LS. 2014. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 23, 2402-2413. ( 10.1111/mec.12747) [DOI] [PubMed] [Google Scholar]
- 56.García-Olivares V, Patiño J, Overcast I, Salces-Castellano A, de Heredia U López, Mora-Márquez F, Machado A, Hickerson MJ, Emerson BC.. 2019. A topoclimate model for quaternary insular speciation. J. Biogeogr. 46, 2769-2786. ( 10.1111/jbi.13689) [DOI] [Google Scholar]
- 57.Salces-Castellano A, et al. 2020. Climate drives community-wide divergence within species over a limited spatial scale: evidence from an oceanic island. Ecol. Lett. 23, 305-315. ( 10.1111/ele.13433) [DOI] [PubMed] [Google Scholar]
- 58.Dennis RLH, Donato B, Sparks TH, Pollard E. 2000. Ecological correlates of island incidence and geographical range among British butterflies. Biodivers. Conserv. 9, 343-359. ( 10.1023/A:1008924329854) [DOI] [Google Scholar]
- 59.Scalercio S, et al. 2020. How long is 3 km for a butterfly? Ecological constraints and functional traits explain high mitochondrial genetic diversity between Sicily and the Italian Peninsula. J. Anim. Ecol. 89, 2013-2026. ( 10.1111/1365-2656.13196) [DOI] [PubMed] [Google Scholar]
- 60.Malmqvist B. 2000. How does wing length relate to distribution patterns of stoneflies (Plecoptera) and mayflies (Ephemeroptera)? Biol. Conserv. 93, 271-276. ( 10.1016/S0006-3207(99)00139-1) [DOI] [Google Scholar]
- 61.McCulloch GA, Wallis GP, Waters JM. 2017. Does wing size shape insect biogeography? Evidence from a diverse regional stonefly assemblage. Glob. Ecol. Biogeogr. 26, 93-101. ( 10.1111/geb.12529) [DOI] [Google Scholar]
- 62.Rundle SD, Bilton DT, Abbott JC, Foggo A. 2007. Range size in North American Enallagma damselflies correlates with wing size. Freshw. Biol. 52, 471-477. ( 10.1111/j.1365-2427.2006.01712.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Salces-Castellano A, Andújar C, López H, Pérez-Delgado AJ, Arribas P, Emerson BC. 2021. Data from: Flightlessness in insects enhances diversification and determines assemblage structure across whole communities. Dryad Digital Repository. ( 10.5061/dryad.63xsj3v15) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The information associated with the studied lineages, including taxonomic identification (family level identification), flight ability assignment, sampling and associated DNA sequences, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.63xsj3v15 [42]. The custom R script developed to define lineages of maternal dispersal history (LMDH) by applying a maximum intraspecific divergence threshold is available from GitHub (https://github.com/asalcescastellano/Divergence-threshold.git). All supplementary tables and figures cited in the main text have been uploaded as electronic supplementary material, information.