2HG accumulation from IDH1/2 mutation induces synthetic lethality in TET2-mutant cells by reducing TET activity below essentially required. A new TET inhibitor mimics 2HG and selectively restricts clonal evolution of TET2-mutant cells in vitro and in vivo.
Abstract
TET2 is frequently mutated in myeloid neoplasms. Genetic TET2 deficiency leads to skewed myeloid differentiation and clonal expansion, but minimal residual TET activity is critical for survival of neoplastic progenitor and stem cells. Consistent with mutual exclusivity of TET2 and neomorphic IDH1/2 mutations, here we report that IDH1/2 mutant–derived 2-hydroxyglutarate is synthetically lethal to TET dioxygenase–deficient cells. In addition, a TET-selective small-molecule inhibitor decreases cytosine hydroxymethylation and restricted clonal outgrowth of TET2 mutant but not normal hematopoietic precursor cells in vitro and in vivo. Although TET inhibitor phenocopied somatic TET2 mutations, its pharmacologic effects on normal stem cells are, unlike mutations, reversible. Treatment with TET inhibitor suppresses the clonal evolution of TET2-mutant cells in murine models and TET2-mutated human leukemia xenografts. These results suggest that TET inhibitors may constitute a new class of targeted agents in TET2-mutant neoplasia.
Significance:
Loss-of-function somatic TET2 mutations are among the most frequent lesions in myeloid neoplasms and associated disorders. Here we report a strategy for selective targeting of residual TET dioxygenase activity in TET-deficient clones that results in restriction of clonal evolution in vitro and in vivo.
Introduction
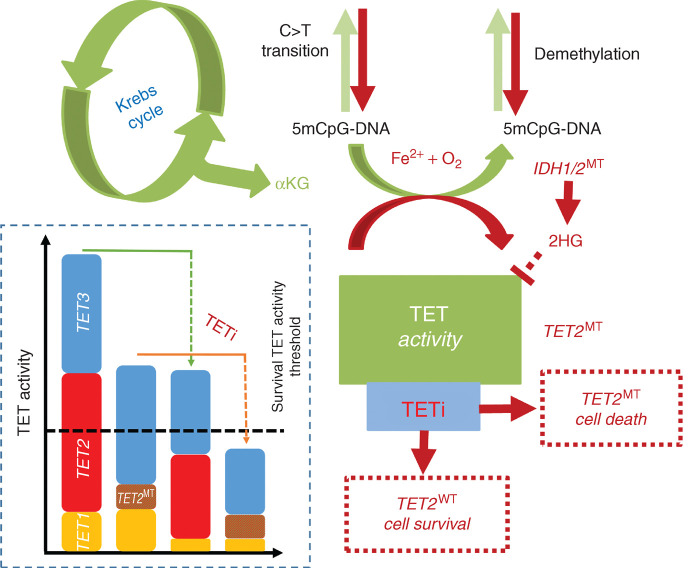
Somatic mutations are essential pathogenic lesions in myeloid leukemias. A subset of these mutations may serve as targets for drug development either directly or through modulation of upstream/downstream pathways and regulatory signaling networks critical for survival and proliferation of malignant cells (1). DNA methylation–demethylation is central to epigenetic gene regulation. In humans, 60% to 80% of CpGs are methylated in somatic cells as a default state (2). Demethylation of enhancer and promoter CpG islands establishes transcription programs that determine cell lineage, survival, and proliferation (3, 4). TET dioxygenases are indirect erasers of methylation from mCpG containing DNA (5) with TET2 accounting for more than 50% of activity in hematopoietic stem and progenitor cells (HSPC). Somatic mutations in the TET2 gene (TET2MT) are among the most common genetic defects in myeloid neoplasia (6, 7). In particular, TET2MT in myelodysplastic syndromes (MDS) increase with age, with >70% of MDS patients 80 years or older having TET2MT (8). Approximately 50% of TET2MT are founder lesions (9). In addition, TET2MT are also detected in blood leukocytes of otherwise healthy older adults, a condition termed clonal hematopoiesis of indeterminate potential (CHIP) associated with a risk for subsequent myeloid neoplasia and cardiovascular disorders (8, 10). The TET2 gene, like TET1/3, is an iron(II) and α-ketoglutarate (αKG)–dependent DNA dioxygenase. TET2MT cause partial loss of dioxygenase catalyzed oxidation of 5-methyl cytosine (5mC)→ 5-hydroxy- methyl cytosine (5hmC)→ 5-formyl cytosine (5fC)→ 5-carboxyl cytosine (5caC). TET-catalyzed reactions require a radical equivalent to abstract a hydrogen from 5mC by cleaving the O–O bond of O2. For this purpose, it uses 2e− gained by decarboxylation of αKG via a Fe2+/Fe3+ redox reaction in two single-electron transfers. Ultimately, 5hmC generated by TET2 passively prevents maintenance methylation due to DNA methyltransferase's inability to recognize 5hmC or causes demethylation as a result of base excision repair of 5fC and 5caC. TET2MT leads to hematopoietic stem cell (HSC) expansion due to perturbation in differentiation programs, resulting in a skewed differentiation toward monocytic predominance (3). In addition, hypermethylation in TET2-mutant cells resulting from 5mC accumulation may increase background C>T mutation rates via mC deamination (11).
The high incidence of TET2MT in MDS and related myeloid neoplasia with a strong age dependence suggests that TET2 is a key pathogenetic factor. Targeting founder TET2MT could disrupt clonal proliferation at its origin and therefore can be a rational target, both for therapeutics in MDS and for strategies preventing CHIP evolution. To date, the only TET2MT-targeted therapeutic compound aimed at restoration of TET2 activity is vitamin C, a cofactor in TET2 catalysis (12, 13). However, several recent reports demonstrated that the effects of ascorbic acid (AA) in myeloid neoplasms are complex and context dependent and often fail to restore TET function in the presence of certain mutations and posttranslational modifications (14, 15, 16). Basal TET function is essential for the expression of several 5mC-sensitive transcription factors, including Myc (17) and Runx1 (18). TET2-deficient leukemic cells rely on the remaining TET activity largely from TET3 and weakly expressed TET1. They must compensate for TET2 loss as evidenced by the persistence of hydroxymethylation in cells with biallelic inactivation of TET2 in human leukemias and in Tet2−/− mice (19). Hence, we hypothesized that TET2MT cells might be more vulnerable to TET inhibition compared with normal HSPCs. The murine model suggests that TET1/3 may play an important compensatory role; a knockout of all three Tet genes in mouse models is embryonically lethal (20) and, in a zebrafish model system (21), results in loss of HSCs. The series of experiments presented in this report indicate that inhibition of the essential residual DNA dioxygenases activity in TET2MT cells may lead to selective synthetic lethality that can be experimentally exploited to study the role of TET enzymes in HSPC biology. Most importantly, our results indicate that TET-specific inhibitors (TETi) may constitute a new class of therapeutics effective in TET2MT-associated myeloid neoplasia.
Results
Lessons from a Natural αKG Antagonist, 2-Hydroxyglutarate
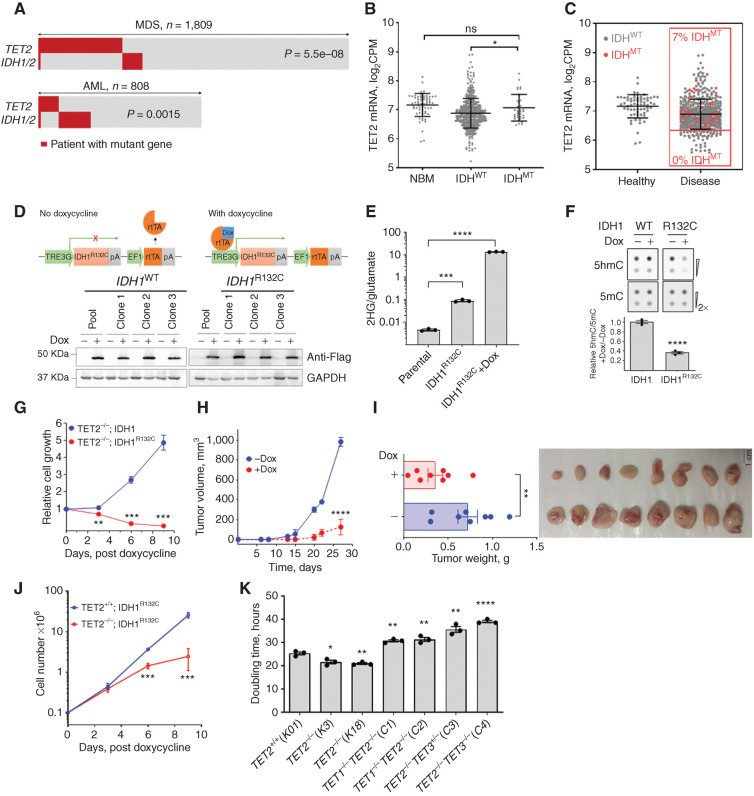
A comprehensive analysis of the configurations of TET2MT in myeloid neoplasia including MDS (n = 1,809) and acute myeloid leukemia (AML; n = 808) showed mutual exclusivity between TET2MT and 2-hydroxyglutarate (2HG)–producing neomorphic IDH1/2MT (Fig. 1A), consistent with earlier reports with much smaller cohorts (22). In a separate MDS cohort (495/1,809 TET2MT cases), only nine double-mutant exceptions were found; they included small TET2MT (or IDH1/2MT) subclones in five cases, missense N-terminal TET2MT in two cases, and non-2HG–producing missense IDH1MT in one case (Supplementary Table S1). Similarly, in an AML cohort, there were 166/808 IDH1/2MT cases, but only eight had co-occurrence of TET2MT. Among these overlapping TET2MT and IDH1/2MT, four of eight had nondeleterious missense TET2 variants, two of eight had low variant allele frequency (VAF) implicating small subclones, and one had a nonneomorphic IDH1 variant (Fig. 1A; Supplementary Table S1). Analysis of TET2 expression in 97 healthy and 909 patients with MDS/myeloproliferative neoplasms or AML from two independent studies (Supplementary Table S2) showed that IDH1/2MT cases have significantly higher TET2 expression (Fig. 1B) and were absent in the lower 25th percentile of TET2-expressing patients (Fig. 1C). These observations suggest that the product of neomorphic IDH1/2MT, 2HG, may further inhibit the residual TET activity (TET1/3) and cause synthetic lethality to cells affected by the loss-of-function TET2MT and decreased expression. As a result, cells with functional TET2 and adequate TET activity may survive and proliferate even after transient and partial TET inhibition, whereas the cells with defective TET2 are eliminated or growth restricted. However, the mutual exclusivity of TET2MT and IDH1/2MT may also arise due to the functional redundancy of these two mutations. To clarify which of these mechanisms is operative, we tested the hypothesis that 2HG, a natural TETi produced by neomorphic IDH1/2MT, may have selective toxicity to TET2MT clones. When we transduced a natural TET2MT cell line, SIG-M5 (TET2 p.I1181fs, p.F1041fs), with IDH1WT or IDH1R132C under doxycycline-inducible tet-on promoter (Fig. 1D), a significant increase in 2HG (∼3,000-fold over baseline) with a concurrent decrease in 5hmC was observed following IDH1R132C induction with doxycycline (Fig. 1D–F). SIG-M5-tet-on-IDH1R132C cells displayed a profound growth inhibition upon doxycycline induction compared with SIG-M5-tet-on-IDH1 cells (Fig. 1G). To further verify the in vivo effects of the neomorphic IDH1R132C in eliminating TET2MT cells, we implanted SIG-M5-tet-on-IDH1R132C cells in the flanks of NSG mice and induced expression of IDH1R132C by treating with doxycycline after the tumor was established. Indeed, doxycycline-induced 2HG production led to a significant inhibition of tumor growth (Fig. 1H and I).
Figure 1.
Effect of 2HG-producing IDH1/2 mutations on TET2MT cells. A, Analysis of TET2 and IDH1/2 mutations in patients with myeloid neoplasia. IDH1/2MT in patients with TET2MT within a CCF cohort of 1,809 MDS patients and TCGA and Beat AML cohorts of 808 patients with AML were analyzed for the co-occurrence of TET2 and IDH1/2 mutations (P values are from Fisher exact test). B, Comparison of TET2 expression among normal healthy donor and myeloid neoplasia patients carrying wild-type or mutant IDH1/2 from different cohorts including Beat AML (www.vizome.org). NBM, normal bone marrow–derived mononuclear cells. C, Distribution of IDH1/2MT and TET2 expression in patients with myeloid neoplasia. Red dots are mutant IDH1/2; no mutations were observed in low TET2-expressing population defined as half of the mean expression of controls. D, Inducible expression of 3XFlag-IDH1 or -IDH1R132C in SIG-M5 cells. Cells were treated with 1 μg/mL doxycycline (Dox) for 3 days. Anti-Flag antibody was used in Western blotfor the detection of induced IDH1 and IDH1R132C. Three independent clones from each cell line along with their pool were analyzed for the expressionanalysis. E, Production of 2HG measured by LC-MS/MS. Cells were treated with or without 1 μg/mL Dox for 3 days, followed by 2HG extraction and analysis. F, Dot blot analysis and quantification of 5hmC and 5mC in the IDH1-inducible SIG-M5 cell line. Cells were treated with or without 1 μg/mL Dox for 3 days. Sodium ascorbate at a final concentration of 100 μmol/L was added 12 hours before harvesting the cells for DNA extraction. G, Effect of 2HG-producing IDH1R132C on the growth of SIG-M5 cells. Cells (105/mL) were treated with 1 μg/mL Dox, and cell proliferation was monitored. The total cell output was plotted as a function of time. H and I, Tumor growth of SIG-M5-IDH1R132C cells in NSG mice (n = 8/group) was monitored upon doxycycline treatment. J, Cell growth curve of K562 TET2+/+ and TET2−/− cells after inducing IDH1R123C expression. K, Doubling time of K562 isogenic TET-mutant cells. Indicated TET dioxygenase genes were knocked out using CRISPR-Cas9, and the genotypes were confirmed by Western blot analysis and Sanger sequencing. The doubling times were determined by exponential growth curve fitting in GraphPad Prism. Three independent clones of each cell line were used in E–G and J. Three biological replicates were used in K. Experiments were performed at least twice for E–G, J, and K. Data, mean ± SEM; statistical significance (P values) from two-tailed t test (except for A) is indicated; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; ns: not significant.
To further understand if the synthetic lethality of neomorphic IDH1/2MT is due to the loss of TET2, we generated isogenic TET2 knockout K562 cells (Supplementary Fig. S1A and S1B; Supplementary Table S3) using CRISPR-Cas9 and transduced with IDH1R132C under a doxycycline-induced promoter (Supplementary Fig. S1C). We did not observe any change in the growth rate of these cells in the absence of doxycycline, most likely caused by some increase of basal 2HG levels due to leakiness of IDHR132C-TET-on expression construct (23) that may have slowed the growth of TET2−/−;IDH1R132C cells while increasing the rate of TET2+/+;IDH1R132C cells (Supplementary Fig. S1D). However, doxycycline induction of neomorphic IDH1R132C led to a significant growth inhibition of K562 TET2−/− versus control K562 TET2+/+ cells (Fig. 1J).
To further support the hypotheses that a threshold level of DNA dioxygenase activity is essential for the growth and survival of leukemia cells, we used genetic approaches to sequentially inactivate TET1 and TET3 in TET2 knockout cells (Supplementary Fig. S1A and S1B; Supplementary Tables S3 and S4). As expected, deletion of TET2 gave proliferative advantage to K562 over the control cells; however, further deletion of either TET1 or TET3 led to significant reduction of global 5hmC correlating with severe growth impairment (Fig. 1K) due to G2–M cell-cycle arrest and increased (2-fold for TET2−/−TET1−/− and 3-fold for TET2−/−TET3MT) basal levels of apoptotic cells (Supplementary Fig. S1E and S1F).
Intrinsically, SIG-M5 cells express significant levels of TET3 and very low levels of TET1 (Supplementary Fig. S1G), and, therefore, they rely heavily on TET3 for their DNA dioxygenase activity. Every attempt to completely inactivate TET3 in SIG-M5 cells failed (Supplementary Table S5). Instead, we were only able to obtain TET3 heterozygous deletion resulting in nearly 2-fold decrease in 5hmC and a significant growth impairment (Supplementary Fig. S1B). The reliance on residual TET3 and TET1 activity was further confirmed by either double TET knockouts or inducible knockdown of TET3 in TET2−/− leukemic cell line SIG-M5 transduced with shTET3 tet-on vector. Doxycycline-induced knockdown of TET3 in SIG-M5 resulted in a significant growth retardation (Supplementary Fig. S1H and S1I).
TETi
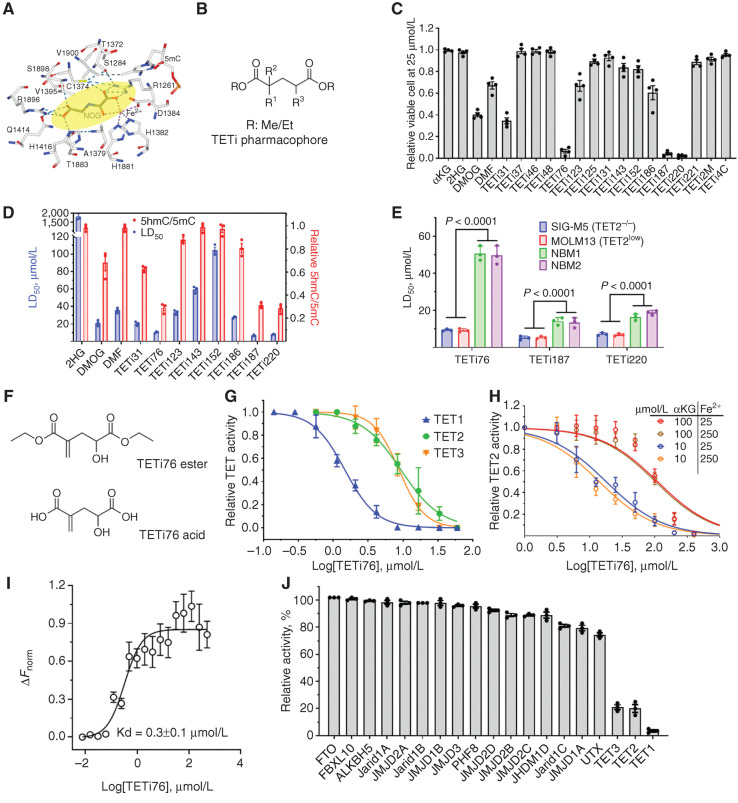
TET dioxygenases not only determine the transcriptional program that guides cell lineage determination, but are also important for efficient transcription of target genes essential for proliferation and survival of malignant cells (4, 17). Although the loss-of-function TET2MT leads to skewing toward myeloid precursors, their survival and proliferation are critically dependent on residual TET activity derived mostly from TET3 and to some extent on TET1. We hypothesized that transient suppression of this residual DNA dioxygenase activity with a small-molecule inhibitor (e.g., designed based on a 2HG scaffold in the αKG binding site of TET2 catalytic domain) may preferentially suppress and eventually eliminate TET-deficient/TET2MT leukemia-initiating clones. 2HG, N-oxalylglycine (NOG), and dimethyl fumarate (DMF) are known to inhibit a variety of αKG-dependent dioxygenases and hence lack specificity, pharmacologic properties, and potency (22, 24). Therefore, we utilized the cocrystal structure of the TET2 catalytic domain (TET2CD) in complex with pseudosubstrate NOG and performed in silico docking to design and synthesize several compounds where the C4 position was substituted with either -keto, -olefin, -methyl, or -cyclopropyl functional groups and the C2 position was single or double substituted with –chloro, -fluoro, -hydroxy, -methyl, or –trifluoromethyl groups (Fig. 2A and B; Supplementary Fig. S2A and S2B). These compounds were tested at 25 μmol/L in SIGM-5 (a TET2−/− leukemia cell line) cells in vitro, and their ability to induce cell death, followed by their ability to inhibit TET dioxygenase activity was used to select the top three compounds (Fig. 2C and D). For further selection of the most potent and pharmacologically active compounds, we calculated the therapeutic index using ratio of LD50 of normal bone marrow–derived mononuclear cells (NBM) and TET2-deficient leukemia cells (SIG-M5 and MOLM13; Fig. 2E; Supplementary Fig. S2C). Interestingly, in this series of compounds, 2-methelene and 4-hydroxy (TETi76) are critical to maintain TET-inhibitory and cell killing activity of these compounds (Fig. 2D and F; Supplementary Fig. S2B). The substitution of the C4 proton in TETi76 either with -CH3 (TETi187) or -CF3 (TETi220) leads to a decrease of the therapeutic index from 5.4 to 2.5, suggesting some off-target effect of these compounds (Fig. 2E). The 2-methelene and 4-hydroxy derivative of αKG binds to TET2CD similar to pseudosubstrate NOG that putatively involved H1801, H1381, and S1898 (Fig. 2A; Supplementary Fig. S2D). These residues are conserved among TET1, TET2, and TET3 (Supplementary Fig. S2E); therefore, we tested the TETi76 against these three dioxygenases in a cell-free assay using recombinant proteins and found that it inhibits all three TETs with 1.5, 9.4, and 8.8 μmol/L IC50, respectively (Fig. 2G; Supplementary Fig. S2F). The effect of TETi76 on TET2CD can be reversed by increasing αKG but not by Fe2+ (Fig. 2H). We performed the direct binding interaction of TET2CD and TETi76 using highly sensitive and label-free MicroScale Thermophoresis (MST) technique. Analysis of thermophoresis binding curves for the association and dissociation of TETi76 with TET2CD using the MO.Affinity software supplied with the instrument demonstrated that TETi76 specifically binds to TET2CD with a dissociation constant of 0.3 μmol/L (Fig. 2I) evaluated with in-built Kd model that utilized the normalized signal with increasing concentration of ligands.
Figure 2.
Lead optimization and efficacy enhancement of TETi. A, Consensus binding site for known pseudosubstrate NOG. B, 2D TETi pharmacophore. C, TET2−/− leukemia cell killing effect of 20 compounds. TET2-deficient SIG-M5 AML cells at 105/mL were treated with 25 μmol/L compounds for 3 days, and viable cells were determined by trypan blue exclusion method. D, LD50 and TET-inhibitory effect of 11 selected compounds. SIG-M5 cells at 105/mL were treated with increasing concentrations of TETi for 3 days, and surviving cells were counted by trypan blue exclusion on automated Vi-CELL counter and LD50 was calculated from viable cell number in GraphPad Prism. TET dioxygenase activity was assessed by measuring 5hmC in a dot blot assay. SIG-M5 cells at 3 × 105/mL were treated with 25 μmol/L compounds for 12 hours in the presence of 100 μmol/L sodium ascorbate and the relative 5hmC/5mC quantification using dot blot was performed on genomic DNA. E, LD50 of TETi against normal bone marrow–derived mononuclear cells (NBM) and TET2-mutant and TET2-deficient leukemia cells (SIG-M5 and MOLM13). Cells were treated with increasing doses of indicated TETi for 3 days, and LD50 was calculated from viable cell number in GraphPad Prism. F, Structure of ester and acid forms of TETi76. Acid forms were used for cell-free assays, whereas diethyl esters were used in cell culture. G and H, Dose-dependent inhibition of TET activity by TETi76 in cell-free condition. Catalytic domains of recombinant TET1, TET2, and TET3 were incubated with TETi76 separately in the presence of different concentrations of cofactors (αKG and Fe2+), and the 5hmC was monitored by ELISA using anti 5hmC antibodies. I, Measurement of direct interaction of TETi76 with TET2CD by MST. Binding interaction of TETi76 with TET2CD was monitored using label-free MST on Monolith NT.LabelFree instrument. The normalized change in MST signal ΔFnorm = (Fbound − Ffree)/Response amplitude at different TETi76 concentrations is plotted. Data were analyzed using MO.Affinity analysis V2.3 software using built-in Kd model for data fitting and calculation of dissociation constant. J, Testing inhibitory effect of TETi76 on αKG- and Fe2+-dependent demethylases. Purified enzymes were used with their substrate in the presence or absence of 15 μmol/L TETi76 in alpha screen (https://bpsbioscience.com), and the relative activity was determined for a vehicle control. Each assay was accompanied by a positive control (see Supplementary Table S6). Data are representative for experiments performed in triplicates at least twice (C–E and G–I). Data, mean ± SEM.
To test if TETi76 can inhibit other αKG/Fe2+-dependent enzymes, we performed in vitro enzyme assays against 16 other known family members including the most closely related RNA dioxygenase FTO. Interestingly, TETi76 does not inhibit any of the enzymes at a concentration of 15 μmol/L, well above the IC50 for TET dioxygenases. Therefore, TETi76 is a TET-specific inhibitor as determined by in vitro enzyme activity assays (Fig. 2J; Supplementary Table S6). We synthesized and purified R- and S-enantiomers of TETi76, performed in vitro TET inhibition assay, and found that there were no significant differences in the IC50 of the two enantiomers under cell-free in vitro conditions (Supplementary Fig. S2G).
Efficacy and Selectivity of TETi76 in a Cell Culture Model
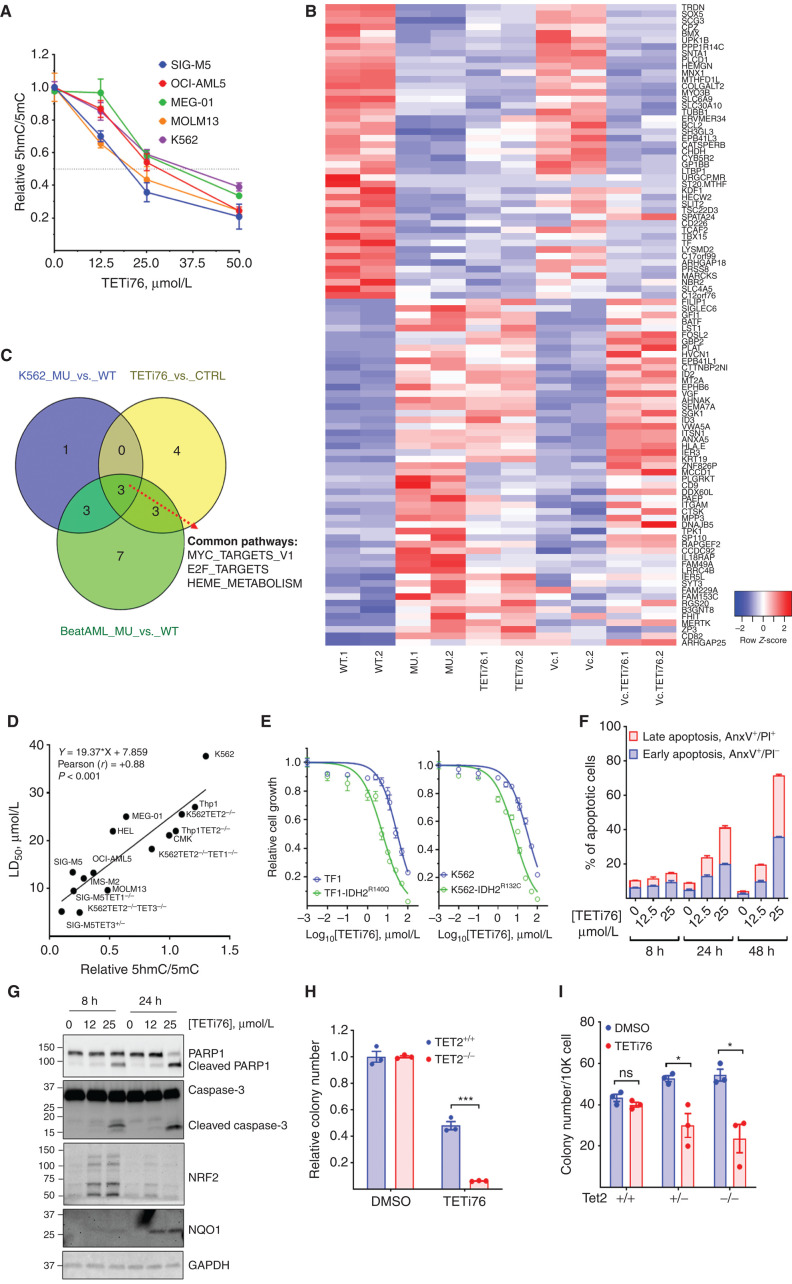
TETi76 preferentially inhibits the catalytic function of TET dioxygenases in a cell-free system; therefore, we further analyze its specificity in cell culture models and examined if it mimics loss of TET function. For this purpose, we generated cell-permeable diethyl ester of TETi76 and treated different human leukemia cell lines (K562, MEG-01, SIG-M5, OCI-AML5, and MOLM13) and measured 5hmC using dot blot assay. Consistent with cell-free data, we observed a dose-dependent decrease in the global 5hmC content in a variety of TET2-proficient and TET2-deficient human leukemia cells (Fig. 3A; Supplementary Figs. S3A and S2C), with 50% inhibition of 5hmC ranging from 20 to 37 μmol/L (Fig. 3A). To characterize target specificity of TETi76, we performed global gene-expression analyses of K562 TET2+/+ and TET2−/− control cells; TETi76 mimicked expression signatures generated by the loss of TET2 in K562. The addition of AA, known to enhance TET activity, counteracted the changes induced by TETi76 (Fig. 3B and C).
Figure 3.
TETi76 mimics loss of TET activity and preferentially restricts the growth of TET dioxygenase–deficient neoplastic cells. A, Dose-dependent inhibition of 5hmC by TETi76 in different leukemia cells. Cells were treated with increasing concentrations of TETi76 in the presence of 100 μmol/L sodium ascorbate for 12 hours. Genomic content of 5hmC and 5mC was detected by dot blot analysis using specific antibodies, and the ratio of 5hmC/5mC is plotted. B, Heat map of significantly upregulated and downregulated genes in K562 TET2+/+ (K01) or K562 TET2−/− (K18) cells treated with 25 μmol/L TETi76 or 100 μmol/L AA for 24 hours, followed by RNA-sequencing (RNA-seq) analysis. C, Venn diagram of pathway analysis of K562 TET2−/− (K18, MU) versus K562 TET2+/+ (K01, WT), K562 TET2+/+ (K01) treated with 25 μmol/L TETi76 for 24 hours versus vehicle (DMSO) control (CTRL), and BeatAML RNA-seq data of TET2 mutant (MU) versus TET2 wild-type (WT; vizome.org/aml/) performed by hallmark gene set enrichment analysis. D, Correlation analysis of LD50 of TETi76 against different leukemia cells with TET activity. The TET activities in different cells were measured by relative ratio of 5hmC/5mC in 15 different leukemia cell lines, including 8 isogenic TET1/2/3 knockout cell lines. Each cell line was treated with increasing concentrations of TETi76 for 72 hours, and the LD50 was calculated from viable cells monitored by methylene blue exclusion on Vi-CELL counter. Pearson correlation coefficient (r) and significance were calculated in GraphPad Prism. E, Effect of TETi76 on cells expressing neomorphic IDH1/2 mutants. Commercially available TF1-IDH2R140Q, house-made K562-IDH1R132C (Fig. 1J), and their parental cells (TF1 and K562) were treated with different concentrations of TETi76 for 3 days. Both K562 and K562-IDH1R132C cells were supplied with 1 μg/mL doxycycline during TETi76 treatment. Relative cell growth was measured by CellTiter-Glo. F, TETi76 induces programmed cell death in TET2-deficient cells. SIG-M5 cells were treated with TETi76, and the apoptotic cell population was determined by Annexin V (AnxV) and propidium iodide (PI) staining using flow cytometer. TETi76 treatment demonstrates a dose- and time-dependent increase in early and late apoptotic cells. G, Western blot analysis of PARP1, caspase-3, NRF2, and NQO1 after TETi76 treatment in SIG-M5 cells. H, Colony-forming abilities of K562 TET2+/+ and TET2−/− cells in the presence and absence of TETi76. I, Bone marrow from Tet2+/+, Tet2+/−, and Tet2−/− mice were harvested and cultured in MethoCult in the presence or absence of TETi76. Data are representative of three independent experiments performed separately. Data, mean or mean ± SEM; statistical significance (P values) from two tailed t test is indicated; *, P < 0.05; ***, P < 0.001; ns, not significant.
Suspension cultures followed by determination of 5hmC indicated that TET2 deficiency was associated with decreased levels of 5hmC and increased proliferation. However, any further suppression of TET dioxygenases by inactivating either TET1 or TET3 induced profound growth suppression in a variety of leukemia cell lines (Fig. 1K). Cell lines with lower TET dioxygenase activity (SIG-M5, MOLM13, and TET-deficient isogenic engineered cell lines) were more sensitive to TETi76, compared with TET dioxygenase–proficient cells like K562 and CMK cells with higher activity, which showed less sensitivity to TETi76-mediated growth inhibition (Fig. 3D). The isogenic TET1/2/3 single- or double-knockout leukemia cells also showed dioxygenase activity-dependent sensitivity to TETi76 (Fig. 3D; Supplementary Fig. S1A and S1B). For example, the parental control K562 cells had an IC50 of 40 μmol/L, whereas TET2 and TET3 double-knockout K562 cells exhibited nearly 4-fold lower IC50. A consistent positive correlation between TET activity and the IC50 was observed in 15 different cell lines, including 8 isogenic TET knockout human leukemia cell lines (Fig. 3D).
It has been shown that neomorphic IDH1/2 mutations phenocopy loss of TET2 in AML and other malignancies (22, 25, 26). Consistent with previous observations, ectopic inducible expression of IDH1R132C in K562 or stably expressing of IDH2R140Q in TF1 demonstrated loss of TET dioxygenase activity, as observed by significant reduction in global 5hmC that can be further suppressed by TETi76 (Supplementary Fig. S3B). To test if TET inhibition in IDH1/2 mutant–expressing cells by TETi76 restricts their growth, we treated IDH1R132C (K562) and IDH2R140Q (TF1) cells with increasing concentrations of TETi76 and observed a 5-fold decrease in the IC50 of IDH1/2 mutant–expressing cells (Fig. 3E).
TETi76 treatment induces apoptotic cell death in TET dioxygenase–deficient leukemia cells in a dose- and time-dependent manner. The treatment of SIG-M5 cells with TETi76 induced early and late stages of apoptotic cell death as probed by the fractions of Annexin V and propidium iodide (PI)–positive cells (Fig. 3F; Supplementary Fig. S3C), a finding further confirmed by PARP1 and caspase-3 cleavage, a hallmark of programmed cell death (Fig. 3G; Supplementary Fig. S3D).
Global gene-expression analyses (RNA sequencing, RNA-seq) of SIG-M5 cells after treatment with TETi76 demonstrated a significant upregulation of TNFα signaling and the downregulation of interferon-α signaling (Supplementary Fig. S3E and S3F). Interestingly, we also observed significant upmodulation of oxidative stress response pathway genes consistent with the inhibition of dioxygenases. In particular, TETi76 treatment induced an 8-fold increase of oxidative stress sensor NQO1 (Fig. 3G; Supplementary Fig. S3D), an NRF2 target gene that has been shown to induce proapoptotic cell death in cancer cells (27).
Consistent with TET dioxygenase deficiency and sensitivity to TETi76 in suspension cells, we also observed that TETi76 preferentially restricted the growth of colony-forming units of TET dioxygenase–deficient cells. The effect of TETi76 on TET2−/− K562 was more profound than on TET2-proficient isogenic cells (Fig. 3H). This observation was striking in murine bone marrow derived from Tet2+/+, Tet2+/−, and Tet2−/− C57BL6 mice. Interestingly, we did not observe any significant growth-inhibitory or anticlonogenic effects of TETi76 on bone marrow–derived mononuclear cells from TET dioxygenase–proficient wild-type mice; on the contrary, it significantly restricted the growth of Tet2−/− and Tet2+/− mouse bone marrow mononuclear cells in a similar assay (Fig. 3I).
Consistent with in vitro specificity of the inhibitory effect, we observed that treatment of cells with TETi76 did not affect the function of αKG-dependent histone dioxygenases that demethylate histone K4, K27, and K36 lysine residues. For example, we did not see any change in H3K4 methylation upon treatment with TETi76, though a cell-permeable αKG psuedosubstrate methyl 2-[(2-methoxy-2-oxoethyl)amino]-2-oxoacetate (DMOG) significantly affected the levels of H3K4 methylation in a dose-dependent manner (Supplementary Fig. S3G and S3H). Interestingly, analysis of Krebs cycle metabolite in TETi76-treated SIG-M5 cells showed a significant increase in the levels of malate, isocitrate, citrate, fumarate, and αKG, as well as a decrease in 2HG and succinate (Supplementary Fig. S3I).
TETi76 Restricts the Growth of Tet2mt Cells in Preventative CHIP Model Systems
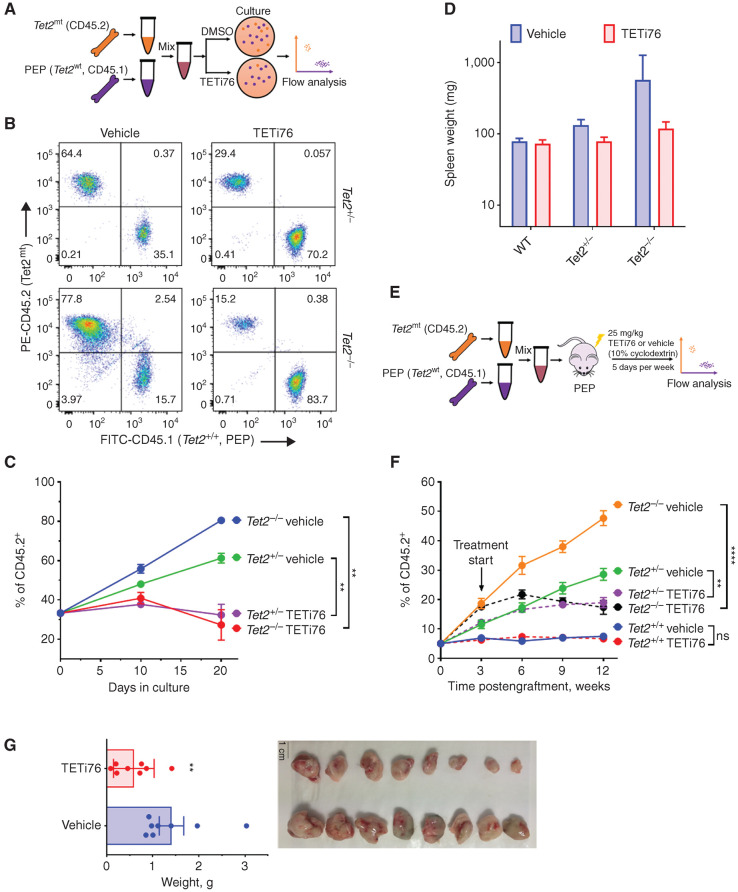
To probe the effects of TETi76 on TET2-deficient cells, Tet2mt/Tet2+/+ bone marrow mononuclear cells were cocultured at fixed ratios to mimic evolving Tet2mt clones, and the differences in surface marker CD45 isoform fractions served as a readout. Marrow from C57BL6 CD45.2 Tet2mt (Tet2+/− or Tet2−/−) versus C57BL6 CD45.1 Pep Boy (Tet2+/+) mice was cocultured (3 mice/group) in a 1:2 ratio with/without TETi76. In culture, TETi76 effectively targeted otherwise dominating Tet2mt cells (Fig. 4A–C). As expected, Tet2mt cells grew at a faster rate in the controls, as reflected in the increased percentage of Tet2mt cells. However, TETi76 treatment selectively restricted the proliferation of Tet2mt cells as determined using CD45.1/CD45.2 surface markers on Tet2+/+ and Tet2mt bone marrow, respectively (Fig. 4B and C).
Figure 4.
TETi selectively restricts the growth of TET2-mutant cells. A, Schematic representation of the mixing experiment of Tet2wt and Tet2mt murine bone marrow in a colony-forming assay. B, Tet2mt bone marrow (CD45.2) cells were mixed in a ratio of 1:2 with Tet2+/+ (CD45.1) and grown in MethoCult for colony formation in the presence or absence of TETi76 (20 μmol/L). On day 10, cells were harvested and the ratio of Tet2mt/Tet2+/+ was measured by flow cytometry using isoform-specific antibodies. C, The ratio was plotted for two consecutive platings. D, Tet2mt or TET2wt mice weretreated with TETi (50 mg/kg, p.o., 5 days/week) for 8 weeks. The spleens were harvested, and weights were plotted compared with vehicle control. E, Schematics of experimental design for in vivo transplant experiment. F, C57BL6 Pep Boy mice expressing CD45.1 surface marker on mononuclear hematopoietic cells were lethally irradiated and transplanted with a mixture of donor mouse bone marrows (5% Tet2−/−, CD45.2; and 95% Tet2+/+, CD45.1). Once mice fully recovered after transplant, the engraftment was accessed by isotype-specific antibodies, and TETi treatment (25 mg/kg, s.c.) 5 days a week at 4 weeks was started. The engraftment of Tet2mt cells in peripheral blood mononuclear cells was monitored and plotted. TETi76 prevented the clonal expansion of Tet2mt cells in vivo. G, Tumor growth of SIG-M5 cells in NSG mice (n = 8/group) was monitored upon TETi76 treatment. Once controlled reached the maximum allowed limit of tumor (<18 mm diameter) burden, tumors were harvested and tumor weight is plotted. TETi76 significantly reduced the tumor size. Data, mean ± SEM; statistical significance (P values) from two-tailed t test is indicated; **, P < 0.01; ****, P < 0.0001; ns, not significant.
To further evaluate the in vivo effects of TETi76 treatment in mice, we used Tet2+/+, Tet2+/−, and Tet2−/− mice and treated them with TETi76 (50 mg/kg, n = 3/group, 5 days/week) for 3 months and observed biweekly body weight and once a month blood count. We did not observe any impact on the overall body weight or any significant change in the overall blood counts of TETi76-treated mice compared with placebo (Supplementary Fig. S4A–S4C). However, TETi76 treatment did decrease spleen sizes in Tet2-deficient mice in a gene dose-dependent manner (Fig. 4D).
To determine the in vivo effects of TETi76 in preventing the clonal evolution of Tet2-deficient cells, for example, on restricting the growth of Tet2+/− or Tet2−/− (Tet2mt) clones, we performed bone marrow competitive reconstitution assays in C57BL6 CD45.1 Pep Boy mice. For this purpose, C57BL6 CD45.1 Pep Boy mice (n = 6 per group) were lethally irradiated, followed by injection of 2 × 106 total bone marrow cells that included 5% of cells from C57BL6 CD45.2 Tet2+/+, Tet2+/−, or Tet2−/− and 95% of CD45.1 Pep Boy Tet2+/+ mice. Proliferation of each genotype was monitored by flow cytometry using surface markers. The CD45.2 versus CD45.1 ratios were plotted as a function of time with treatment and compared with vehicle alone (Fig. 4E). Consistent with several previous reports (19, 28), Tet2mt cells become the dominant fractions in vehicle-treated mice in a gene dose-dependent manner (Fig. 4F). However, treatment with TETi76 preferentially restricted the proliferative advantage of Tet2mt cells compared with vehicle control (Fig. 4F). The ratio of mice receiving Tet2+/+ CD45.2 cells did not change, suggesting that TETi76 restricts the growth of Tet2mt cells only.
TETi76 Restricts the Growth of TET-Deficient Leukemia In Vivo
To test if TET inhibition would inhibit TET2-deficient leukemic cells under in vivo conditions, we used subcutaneous tumor development in immune-compromised mice. Since Tet2mt mice develop a protracted myeloproliferative syndrome rather than overt leukemia, we used a human TET2−/− leukemia cell line xenograft model to test the efficacy of TETi76 for established tumors. SIG-M5, a TET2-deficient cell subcutaneously implanted in NSG mice, grew very aggressive tumors. Once tumors were established, TETi76 was orally (50 mg/kg) administered to the implanted mice. Treatment with TETi76 significantly reduced tumor burden compared with vehicle-treated mice, indicating that TET2MT leukemia may be sensitive to TETi76 in vivo (Fig. 4G).
Discussion
DNA dioxygenases are the key enzymes that catalyze cytosine hydroxymethylation, an essential step for passive DNA demethylation. The high prevalence of somatic loss-of-function mutations in TET2 underscores the important role of this gene in myeloid neoplasia. In hematopoietic cells, TET2 is the most abundantly expressed gene among the TET family of DNA dioxygenases and accounts for nearly 60% of DNA dioxygenase activity, explaining why TET1 and TET3 are rarely mutated in leukemia. However, even in cases with biallelic TET2MT, the residual activity of TET3 and TET1 preserves detectable levels of 5hmC in the genome, suggesting a compensatory role for TET1/TET3 in the survival of TET2MT cells. TET2MT are mutually exclusive with IDH1/2MT, which produce 2HG and inhibit TET dioxygenases (22). It has been proposed previously that the functional redundancy may be responsible for the mutual exclusivity; however, here we demonstrate the elimination of TET2-deficient cells by ectopic expression of neomorphic IDH1R132C mutation.
The results presented here underscore the cellular requirement for preservation of DNA dioxygenase activity and therefore the essential compensatory activity of TET1/TET3 in TET2MT cells. Furthermore, this work suggests a mechanism whereby 2HG is synthetic lethal to TET2-deficient cells and explains the observed mutual exclusivity of TET2 and IDH1/2 mutations.
These observations indicate that chemical dioxygenase inhibitors may be exploited as novel class of agents for TET2MT-associated disorders. The natural Tet2 inhibitor 2HG itself affects a broad array of αKG-utilizing enzymes and hence is not a suitable drug candidate. In addition, its LD50 for TET2MT (in millimolar ranges) cannot be achieved therapeutically. Consequently, using 2HG computer-aided rational drug design and iterative modification, we generated several competitive inhibitors of TET activity. One of these inhibitors, TETi76, proved highly specific and potent and was selected for further studies. We demonstrated, using an inducible IDH1/2MT model that produces 2HG, that treatment with TETi76 further decreases the levels of 5hmC resulting in selective growth inhibition of TET2MT cells, while normal bone marrow cells and leukemic cell lines with robust TET function remain unaffected. In vitro, TETi76 results in selective toxicity (>100-fold that of 2HG) to primary human TET2MT lines, engineered TET2−/− cells, as well as Tet2−/− murine hematopoietic cells. The difference in TET activity in TET2WT and TET2MT cells offers a therapeutic window to selectively eliminate TET2MT clones, either preventatively early on in the pathway toward oncogenesis (e.g., in CHIP; refs. 8, 10) or therapeutically in evolved neoplasms.
There have been two reports of the co-occurrence of IDH2R172 and TET2MT in cases of angioimmunoblastic T-cell lymphoma (AITL; refs. 29, 30). However, the cellular and extracellular levels of 2HG were within the normal range (29). In the other case, a majority of the TET2MT reported have relatively low VAFs, and the exact VAF for IDH2R172 is not clear; therefore, it is hard to access the clonal architecture and clonal/subclonal mosaicism of the AITL patients. Based on our analysis in patients with MDS and AML, most of the co-occurrences were nondamaging or small clone for one or the other variants. In AITL, IDH2 mutations are exclusively restricted to R172, which are commonly mutated to serine or lysine residues (30). It has also been shown that median 2HG level in IDH2R172 AITL patients is significantly lower than the level in IDH1- or IDH2-mutated AML (31). Thus, the coexistence of TET2 with weak neomorphic mutations that produces low levels of 2HG may exist under such conditions.
The effects of TET inhibition observed in TET-deficient malignant HSPCs are consistent with observations in zebrafish models, where loss of all three TET results in loss of HSCs (32). TETi showed potent efficacy in in vivo murine model systems, where the proliferative capacity of Tet2mt bone marrow cells was abrogated. In murine transplant model systems that mimic CHIP, TETi was highly effective in selectively preventing clonal expansion of Tet2−/− hematopoietic cells. Unlike acute leukemia described in genetic mouse models of simultaneous TET inactivation, we did not see evidence in vitro that the pharmacologic manipulation would phenocopy the engineered TET2-deficient leukemia. Moreover, therapeutic intervention with TETi would be applied transiently and discontinued upon elimination of vulnerable TET2-deficient cells (Fig. 5). In general, the effects of TETi in TET2WT cells were minimal with respect to a stable transcriptional profile and preserved cell viability at levels corresponding to a 10-fold therapeutic index. The effect of TETi is relatively specific to DNA dioxygenases, since other αKG-consuming enzymes in humans (33) remained unaffected in vitro. In contrast, 2HG or the αKG pseudosubstrate NOG exhibit “off-target” activity, for example, against histone lysine demethylases.
Figure 5.
Schematic representation of TETi mechanism of action. TET2-mutant and TET dioxygenase–deficient cells are susceptible to further TET inhibition. Loss to TET2 increases C→T transition and mutator phenotype leading to neoplastic evolution. Residual TET dioxygenase activity from TET1 and TET3 in TET2-mutant cells is important for efficient transcription of survival and proliferative genes. Inhibition of the residual TET dioxygenase activity leads to preferential growth restriction and finally elimination of TET2-mutant and TET dioxygenase–deficient clones.
TETi creates transcriptional changes similar to those of the genetically engineered loss of TET2 activity in leukemic cells. The effect of TETi was partially reversed by AA. TETi76 induced apoptosis in TET2MT cells, while no detectable apoptosis was seen in either normal TET2WT bone marrow or untreated TET2MT cells. The analysis of RNA-seq data and metabolic profile further suggested that TET inhibition leads to a major metabolic shift in TET2MT cells, as demonstrated by significant downmodulation of c-MYC target genes, a profile similar to the complete loss of TET2 or the treatment with 2HG (17).
One potential drawback of TETi is that it may phenocopy TET2MT and drive transformation of normal hematopoietic cells. We did not observe disease acceleration or proproliferative effects of TETi administered to Tet2+/+, Tet2+/−, or Tet2−/− mice to support accelerated leukemogenesis. One of the key concerns of TET dioxygenase inhibition stems from a report where combined deletion of Tet2 and Tet3 in early hematopoietic murine cells is suggested to cause an aggressive and transplantable AML (34). To the best of our knowledge, TET3 loss-of-function mutation in human myeloid cancer has not been described, which indicates the importance of TET3 dioxygenase for the survival and the proliferation of hematopoietic cells. Consistent with this observation, Tet2 or Tet3 is required for normal HSC emergence in zebrafish (32).
The reports of Tet2/Tet3 double deletion leading to the development of aggressive myeloid cancer (34) utilized Tet3fl/fl mice crossed with Mx1-Cre or ERT2-Cre recombinase to induce conditional excision of floxed alleles. Polyinosinic–polycytidylic acid (p(rI):p(rC)) or tamoxifen injection, respectively, is essential to induce conditional Tet3 knockout in these models. It has been previously reported that both p(rI):p(rC) and tamoxifen can affect the normal hematopoiesis (35, 36), which in part may explain the effect in mice. P(rI):p(rC) in particular mimics RNA virus infection, activates Toll-like receptor 3 (TLR3), and leads to the immune response, whereas Tet2 is required to resolve inflammation to repress IL6 (37). In our study, we showed that both monoallelic and biallelic TET3 knockout K562 TET2−/− leukemia cells are growth impaired. The triple TET knockout of K562 was not feasible. It will be interesting to characterize Tet2−/−Tet3+/− mice compared with Tet2−/− mice and the effect of TETi on such cells.
Preferential inhibition of the clonal evolution of TET2MT clone can have huge therapeutic implications in CHIP. In the context of CHIP, eliminating and restricting the growth of TET2MT cells by TET inhibition are highly relevant not only to myeloid malignancies and associated disorders but also to the increased risk of cardiovascular disorders (10, 38, 39). Transient TET inhibition by small-molecule inhibitors such as TETi76 would be devoid of any aberration caused by genetic lesion while lethal against Tet2mt and TET dioxygenase deficient. Thus, the therapeutic benefit of TETi in the prevention of cardiovascular disease could be obtained.
The utility of TETi is not limited to CHIP and the myeloid malignancies because a pan TET dioxygenase small-molecule inhibitor (C35) has been reported to achieve somatic cell reprograming with several probable therapeutic utilities (40).
In summary, we demonstrate for the first time that TET inhibition leads to selective synthetic lethality in TET2MT cells. This approach may be developed as a potential targeted therapeutic strategy for TET2MT-associated disorders and myeloid neoplasia associated with TET dioxygenase deficiency and may lead to development of a new class of TET-selective DNA dioxygenase–inhibiting agents.
Methods
Patient Samples
Patient bone marrow samples were obtained from healthy controls or patients with myeloid neoplasia after written informed consent in accordance with Cleveland Clinic Institutional Review Board–approved protocol. Human cord blood was acquired from the Cleveland Cord Blood Center, Cleveland, Ohio, and CD34+ cells were isolated by the human CD34 MicroBead Kit (Miltenyi Biotec).
Cell Lines
All cell lines were purchased recently (2 years or less) and cultured according to the suppliers' guidelines. K562, THP-1, TF1, and TF1-IDH2R140Q cell lines were purchased from ATCC, whereas CMK, MEG-01, MOLM13, HEL, OCI-AML5, and SIG-M5 cell lines were from DSMZ. ISM-M2 cells were a gift from Yogen Saunthararajah of Cleveland Clinic and authenticated by short tandem repeat assay. Normal bone marrow was cultured in IMDM with 10% FBS, 100 U/mL penicillin–streptomycin, and 10 ng/mL each of SCF, FLT3L, IL3, IL6, and TPO. Mouse bone marrow was cultured in IMDM with 10% FBS, 100 U/mL penicillin–streptomycin, 50 ng/mL mSCF, 10 ng/mL mIL3, and 10 ng/mL mIL6. SIG-M5 was cultured in IMDM, whereas all other leukemic cells were grown in RPMI with 10% FBS and 100 U/mL penicillin–streptomycin. The medium was supplied with 2 ng/mL recombinant human GM-CSF. Cells were confirmed Mycoplasma negative by using the MycoAlert Mycoplasma Detection Kit (Lonza, cat. #LT07-118).
Reagents for Cell Experiments
RPMI-1640 and IMDM cell media were purchased from Invitrogen. FBS was purchased from ATLANTA Biologicals (cat. #S11150). Penicillin–streptomycin was purchased from Lerner Research Institute Media Core in Cleveland Clinic. Human and murine recombinant growth factors were purchased from PeproTech. MethoCult H4435 and M3434 were purchased from STEMCELL Technologies. Antibody information is provided in Supplementary Table S7.
IDH1- and IDH1R132C-Inducible Cell Line Generation
Tetracycline-inducible IDH1 and IDH1R132C were generated based on a previously published construct (41, 42). IDH1 was amplified from IDH1 in pDONR221 (Harvard Medical School, Plasmid HsCD00043452) and then R132C mutation was introduced using the In-Fusion HD Cloning Plus CE kit (Takara, 638916). Tet-system– approved FBS (Atlanta Biological) was used for doxycycline-inducible cell lines.
Measurement of Metabolite by LC-MS/MS
Cells (5 × 106) were harvested by centrifugation, washed with PBS, and resuspended in 0.5 mL of chilled 80% methanol (20% ddH2O) by dry ice. The cell suspension was freeze-thawed for two cycles on dry ice and centrifuged twice for 15 minutes at 4°C at 15,000 × g, and then the supernatant was collected, dried by N2, and dissolved in 100 μL HPLC grade water. The solution was filtered by 0.22-μm Eppendorf filter and used for LC-MS/MS. Shimadzu LCMS-8050 with a C18 column (Prodigy, 3 μmol/L, 2 × 150 mm, Phenomenex) was used for LC-MS/MS with the following gradient: 0 to 2 minutes 0% B; 2 to 8 minutes 0% B to 100% B; 8 to 16 minutes 100% B; 16 to 16.1 minutes 100% B to 0% B; 16.1 to 24 minutes 0% B. Mobile phase A was water + 5 mmol/L AmAc. Mobile phase B was methanol + 5 mmol/L ammonium acetate. The flow rate was 0.3 mL/minute. The multiple reaction monitoring (MRM) transition for 2-hydroxyglutaric acid was 147 > 85.
Viable Cell Count and Doubling Time Measurement
Viable cell count was performed by methylene blue exclusion on Vi-CELL counter (Beckman Coulter).
CellTiter-Glo–Based Cell Proliferation Assay for Doubling Time Measurement
Cells were cultured in 96-well plates (Corning; cat. #3610) at 8,000 cells per 100 μL of culture media for each well. Relative cell number was calculated by using CellTiter-Glo assay (Promega). One to one of 1× CellTiter-Glo:1× PBS was mixed, and then 40 μL/well was added. Plates were shaken for 10 minutes to ensure complete lysis before CellTiter-Glo signal reading. Reading was performed every 24 hours for 4 days with new plates for each time point. There were six replicates per condition. Experiments were repeated three times. Doubling time was calculated with exponential growth equation in GraphPad Prism 8.
Generation of TET Dioxygenase Knockout Cells Using CRISPR-Cas9
Zhang lab LentiCRISPR plasmid was used for CRISPR-Cas9–mediated TET2 gene knockout. 5′-caccgGGATAGAACCAACCATGTTG-3′ and 5′-aaacCAACATGGTTGGTTCTATCCc-3′ DNA oligos were ordered from IDT and used for cloning. Vector cloning, virus production, and virus infection were performed according to the reference article (43). Virus-infected K562 cells were seeded in 96-well plates at a density of 0.5 cell per well, and colonies grown from single cells were kept for expansion and analysis. DNA was extracted for sequencing. Sequencing libraries were prepared using Illumina's Nextera Custom Enrichment panel and sequenced with the HiSeq 2000. Variants were extracted according to GATK Best Practices. The sequencing panel covers all the exons of 170 genes, including TET2, commonly mutated in myeloid malignancies as previously described (7). TET1 and TET3 knockout cells were generated using IDT genome editing with the CRISPR-Cas9 system according to the manufacturer's protocol. The gRNA details and sequencing results are summarized in Supplementary Table S4.
RNA-seq and Analysis
RNA was purified by using the NucleoSpin RNA kit (Takara Bio USA, Inc.; cat. #740955) according to the manufacturer's instruction. RNA quality was validated by RNA integrity number (RIN >9) calculated by Agilent 2100 Bioanalyzer. mRNA was enriched using oligo(dT) beads and then was fragmented randomly by adding fragmentation buffer. The cDNA was synthesized using mRNA template and random hexamers primer, after which a custom second-strand synthesis buffer (Illumina, dNTPs, RNase H, and DNA polymerase I) was added to initiate the second-strand synthesis. After a series of terminal repair, a ligation, and sequencing adapter ligation, the double-stranded cDNA library was completed through size selection and PCR enrichment. The qualified library was fed into Illumina sequencer after pooling, and more than 30 million reads were acquired for each sample. The quality of RNA-seq raw reads was checked using FastQC (Galaxy Version 0.72). Raw reads were mapped to the human genome, hg19, using RNA STAR. Differential expression and gene set enrichment analysis were assessed using edgeR 3.24.3 and limma 3.38.3 with R 3.5. Computational analysis was performed using Galaxy server (https://usegalaxy.org/). The RNA-seq data were submitted to the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) archives, with assigned GEO accession number GSE162487.
Computational Docking and Small Molecule
The crystal structure of TET2 catalytic domain (TET2CD) in complex with DNA and αKG pseudosubstrate NOG and DNA oligo (protein data bank ID 4NM6) was used for all docking simulations and structural activity analysis of TETi by Glide running in the Schrodinger Maestro environment. The complex was minimized, and the binding site was analyzed in UCSF Chimera 1.8.
TET2CD Protein Purification
GST-TET2 (1099–1936 Del-insert; ref. 44) expression vector was transformed into Escherichia coli strain BL21(DE3)pLysS. The transformant was grown at 37°C to an OD600 of 0.6 and switched to 16°C for another 2 hours. Ethanol was added to the final concentration of 3% before expression induction by adding isopropyl-b-D-thiogalactopyranoside to the final concentration of 0.05 mmol/L. Cells were cultured for 16 hours at 16°C. Cells from 2 L of culture were harvested and lysed in 50 mL of lysis buffer (20 mmol/L Tris-HCl pH 7.6, 150 mmol/L NaCl, 1× CelLytic B (Sigma C8740), 0.2 mg/mL lysozyme, 50 U/mL benzonase, 2 mmol/L MgCl2, 1 mmol/L DTT, and 1× protease inhibitor (Thermo Scientific A32965) for 30 minutes on ice. Lysate was sonicated by ultrasonic processor (Fisher Scientific FB-505 with ½″ probe) with the setting of 70% amplitude and 18 cycles of 20 seconds on and 40 seconds off. Then the lysate was centrifuged twice at 40,000 × g for 20 minutes each. Supernatant was filtered through the membrane with the pore size of 0.45 μm. Flow-through was diluted four times with the solution of 20 mmol/L Tris-HCl pH 7.6 and 150 mmol/L NaCl. GST-TET2 was purified by GE Healthcare AKTA pure by affinity (GSTPrep FF16/10) and gel filtration (Superdex 200 increase 10/300 GL). For gel filtration, buffer of 10 mmol/L phosphate and 140 mmol/L NaCl, pH 7.4, was used. Protein was dialyzed in 50 mmol/L HEPES, pH 6.5, containing 10% glycerol, 0.1% CHAPS, 1 mmol/L DTT, and 100 mmol/L NaCl. GST tag was removed by TEV enzyme.
Dot Blot Assay for 5hmC and 5mC Detection of Genomic DNA
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega). DNA samples were diluted in ddH2O, denatured in 0.4 M NaOH/10 mmol/L EDTA for 10 minutes at 95°C, neutralized with equal volume of 2 M NH4OAc (pH 7.0), and spotted on a nitrocellulose membrane (prewetted in 1 M NH4OAc, pH 7.0) in 2-fold serial dilutions using a Bio-Dot Apparatus Assembly (Bio-Rad). The blotted membrane was air-dried and cross-linked by Spectrolinker XL-1000 (120 mJ/cm2). Cross-linked membrane was blocked in 5% nonfat milk for 1 hour at room temperature and incubated with anti-5hmC (Active motif, 1:5,000) or anti-5mC (Eurogentec, 1:2,500) antibodies at 4°C overnight. After 3 × 5 minutes washing with TBST, the membrane was incubated with HRP-conjugated anti-rabbit or anti-mouse IgG secondary antibody (Santa Cruz), treated with ECL substrate, and developed using film or captured by ChemiDoc MP Imaging System (Bio-Rad). Membrane was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) to ensure equal loading of input DNA.
ELISA Assay of 5hmC for TET Activity Detection
The 96-well microtiter plate was coated with 10 pmol/L avidin (0.66 μg, SIGMA A8706) suspended in 100 μL 0.1 M NaHCO3, pH 9.6, overnight. Biotin-labeled DNA substrate (10 pmol/L) was added for 2 hours. The 60-bp duplex DNA substrate (forward strand: 5′-ATTACAATATATATATAATTAATTATAATTAACGAAATTATAATTTATAATTAATTAATA-3′ and reverse strand: 5′-Bio-TATTAATTAATTATAAATTATAATTTmCGTTAATTAT AATTAATTATATATATATTGTAAT-3′) was synthesized by IDT. TET2CD, TET1CD (Epigentek; cat. #E12002-1) or TET3CD (BPS Bioscience; cat. #50163) protein (0.1 μmol/L) in 100 μL reaction buffer containing 50 mmol/L HEPES (pH 6.5), 100 mmol/L NaCl, 1 mmol/L DTT, 0.1 mmol/L ascorbate, 25 μmol/L Fe(NH4)2(SO4)2, and 10 μmol/L αKG was added to each well for 2 hours in 37°C. Concentrations of Fe(NH4)2(SO4)2 and αKG are indicated in the relative figure if different than previously stated. Reaction was stopped by incubating with 100 μL of 0.05 M NaOH on a shaking platform for 1.5 hours at room temperature. After washing, the wells were blocked with 2% BSA dissolved in TBST for 30 minutes and incubated with anti-5hmC antibody (Active motif, 39769, 1:3,000) at 4°C overnight. After washing with TBST, the wells were incubated with HRP-conjugated anti-rabbit secondary antibody (Santa Cruz). Signal was developed by adding TMB (SIGMA, T4444). Reaction was stopped by adding 2M H2SO4.
Label-Free Thermophoresis Assay to Measure the Binding of TET2CD and TETi76
Binding interaction of TETi76 with TET2CD was monitored using label-free MST (45). Briefly, TET2CD (0.5 μmol/L) recombinant protein was mixed with different concentrations of TETi76 in modified TET2 reaction buffer without αKG containing 50 mmol/L HEPES (pH 6.5), 100 mmol/L NaCl, 1 mmol/L DTT, 0.1 mmol/L ascorbate, and 25 μmol/L Fe(NH4)2(SO4)2, then was loaded to capillary (NanoTemper, MO-Z022), and measurements were made using the Monolith NT.115 (NanoTemper). Data were analyzed using MO.Affinity analysis V2.3 software using an in-built Kd model for data fitting and calculation of dissociation constant. GraphPad Prism 8.0.2 was used for plotting data.
Dioxygenase Inhibition Screen
The dioxygenase inhibition screen was performed by using alpha screen of BPS Bioscience (https://bpsbioscience.com).
TET3 Dox-Inducible Knockdown by shRNA
Dox-inducible shRNA lentivirus vector EZ-Tet-pLKO-Blast was used for cloning. The vector was a gift from Cindy Miranti (Addgene plasmid # 85973). Primers 5′-CTAGCGAACCTTCTCTTGCGCTATTTTACTAGTAAATAGCGCAAGAGAAGGTTCTTTTTG-3′ and 5′-AATT CAAAAAGAACCTTCTCTTGCGCTATTTACTAGTAAAATAGCGCAAGAGAAGGTTCGA-3′ were purchased from IDT. The procedures in a reference article (46) were followed for cloning, lentivirus production, and stable cell line selection.
Quantitative Real-time PCR
Total RNA was extracted from cells using the NucleoSpin RNA kit (Takara Bio USA, Inc.; cat. #740955). The purity of RNA was confirmed by the 260/280 absorption ratio on Nanodrop. cDNA synthesis was performed by using iScript cDNA Synthesis Kit (Bio-Rad, 1708890). Total RNA (500 ng) was used as a template. qRT-PCR was performed by using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad; cat. 1725270) on a CFX96 real-time PCR detection system (Bio-Rad). Primers 5′-CGATTGCGTCGAACAAATAG-3′ and 5′-CTCCTTCCCCGTGTAGATGA-3′ were used for TET3 detection. Primers 5′-CTCCTCCTGTTCGACAGTCAGC-3′ and 5′-CCATGGAATTTGCCATGGGTGG-3′ were used for GAPDH detection. The values obtained for the target gene expression were normalized to GAPDH.
Chemical Synthesis of TETi
All novel TETi were synthesized in-house and purified using solid phase extraction on silica flash column (all diesters) and C18 Sep-Pak column using water/acetonitrile as mobile phase for all corresponding acid or dilithium salt. The derivatives were synthesized with modified methodology from the previously published literature (47, 48). We used the Barbier reaction method with acetic acid instead of ammonium chloride, which gave better yield. We used the Dess-Martin-Periodinane reagent for the oxidation and Pd/C hydrogenation for the reduction of the respective hydroxyl and methylene group. The details of the chemistry are provided in Supplementary Methods. Briefly, the compounds were characterized by 1HNMR (500 MHz Bruker Asend Avance III HD at room temperature) and 13CNMR (125 MHz for 13C NMR) in the dutrated solvent, and high-resolution mass spectrometry was performed on an Agilent Q-TOF. The purity of compounds was always >95%, based on the combination of 1HNMR and chromatography. Details of the synthesis protocol are provided in Supplementary Methods. TETi diesters stock solutions were dissolved in dry DMSO, and acid or dilithium salts were dissolved in ultrapure nuclease and proteinase-free water for all in vitro cell culture or cell-free assays, respectively. The final DMSO content in all assays was always maintained ≤0.1% v/v.
Annexin V and PI Staining
Cells were incubated with FITC–Annexin V (BD Pharmingen; cat. #556420) in a binding buffer (BD Pharmingen; cat. #556454) containing PI (BD Pharmingen; cat. #556463) and analyzed by flow cytometry (BD FACSVerse).
Mouse Studies
Animal care and procedures were conducted in accordance with institutional guidelines and approved by the Institutional Animal Care and Use Committee, Cleveland Clinic. Tet2-mutant mice (stock# 023359) were procured from The Jackson Laboratory. In cell line–derived tumor xenografts, 1 × 106 doxycycline-inducible IDH1R132C SIG-M5 cells or parental SIG-M5 cells were subcutaneously injected into each flank of NSG mice. For inducible IDH1R132C SIG-M5 cells, mice were intraperitoneally injected with 10 mg/kg doxycycline (Millipore; cat. #198955). For parental SIG-M5 cells, mice were orally administrated with 50 mg/kg TETi76. In the transplantation mouse model, recipient mice received two doses of 480 rad (4.8 Gy) irradiation delivered 3 hours apart; 2 million of a mixture of mononuclear cells from donor mouse bone marrows (5% Tet2−/− cell, CD45.2; and 95% Tet2+/+, PEP, CD45.1) were injected into the tail veins of recipients after the second irradiation. Mice were maintained on antibiotic food for 3 weeks. Blood (50 μL) was drawn from each mouse for hematology profile analysis using DREW HEMAVET 950FS and for ratio of CD45.1/CD45.2 analyses by flow cytometry using FITC-CD45.1 (Invitrogen; cat #11-0453-82) and PE-CD45.2 (Invitrogen; cat. #12-0454-82). Once mice were fully recovered and engraftment was confirmed, TETi treatment (25 mg/kg) was started by intraperitoneal injection. All the treatments were performed once a day, 5 days a week.
Data and Material Availability
Request of any specific raw data or material used in this study but not commercially available can be made to the corresponding authors.
Authors' Disclosures
Y. Guan reports a patent for antitumor tet2 modulating compounds pending. A.D. Tiwari reports a patent for antitumor tet2 modulating compounds (WO2019108796A1) pending and owned by Cleveland Clinic. J.G. Phillips reports grants from R35HL135795-01 during the conduct of the study, as well as a patent for PCT/2018/063069 issued. D.J. Lindner reports grants from NIH/NCI [P30 CA043703-23 (principal investigator, Gerson)] during the conduct of the study. M. Meggendorfer reports personal fees from MLL Munich Leukemia Laboratory during the conduct of the study. M. Abazeed reports grants from Siemens Medical Solutions, USA, and grants and personal fees from Bayer AG outside the submitted work.M.A. Sekeres reports personal fees from Bristol-Myers Squibb/Celgene and Takeda/Millenium outside the submitted work. J.P. Maciejewski reports grants from NIH during the conduct of the study, as well as patents for TET modulators pending. B.K. Jha reports grants from Cleveland Clinic during the conduct of the study, as well as a patent for antitumor tet2 modulating compounds pending and owned by Cleveland Clinic. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This work was supported, in parts, by grants from the NIH/NHLBI (R35HL135795-01), Leukemia & Lymphoma Society (LLS) TRP, and LLS SCOR and Taub Foundation to J.P. Maciejewski and B.K. Jha. The authors thank their colleagues at the Cleveland Clinic: Amy Graham at Flow Core for help with the flow cytometer, Bartlomiej Przychodzen for help with RNA-seq, Dr. Renliang Zhang at Small-Molecule Mass Spectroscopy Core for mass spectrometry experiments, Dr. Smarajit Bandyopadhyay at Molecular Biotechnology Core for help with the thermophoresis experiment, and Phillip Maciejewski for proofreading.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Blood Cancer Discov 2021;2:146–61
Authors' Contributions
Y. Guan: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. A.D. Tiwari: Investigation, methodology, writing–original draft, writing–review and editing. J.G. Phillips: Methodology, writing–review and editing. M. Hasipek: Investigation, writing–review and editing. D.R. Grabowski: Investigation, methodology, writing–review and editing. S. Pagliuca: Data curation, investigation, methodology, writing–review and editing. P. Gopal: Investigation, methodology, writing–review and editing. C.M. Kerr: Data curation, validation, writing–review and editing. V. Adema: Data curation, writing–review and editing. T. Radivoyevitch: Investigation, writing–review and editing. Y. Parker: Investigation, methodology. D.J. Lindner: Investigation, project administration, writing–review and editing. M. Meggendorfer: Resources, data curation, writing–review and editing. M. Abazeed: Methodology, writing–review and editing. M.A. Sekeres: Methodology, writing–review and editing. O.Y. Mian: Investigation, writing–review and editing.T. Haferlach: Resources, writing–review and editing. J.P. Maciejewski: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing. B.K. Jha: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Andre F, Mardis E, Salm M, Soria JC, Siu LL, Swanton C. Prioritizing targets for precision cancer medicine. Ann Oncol 2014;25:2295–303. [DOI] [PubMed] [Google Scholar]
- 2. Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, et al. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res 2002;12:543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 2014;56:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, et al. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv 2018;4:eaau6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schubeler D. Function and information content of DNA methylation. Nature 2015;517:321–6. [DOI] [PubMed] [Google Scholar]
- 6. Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 2009;113:6403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014;28:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirsch CM, Nazha A, Kneen K, Abazeed ME, Meggendorfer M, Przychodzen BP, et al. Consequences of mutant TET2 on clonality and subclonal hierarchy. Leukemia 2018;32:1751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017;49:204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan F, Wingo TS, Zhao Z, Gao R, Makishima H, Qu G, et al. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat Commun 2017;8:15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 2017;170:1079–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 2017;549:476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell stem cell 2018;23:355–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, et al. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell 2017;65:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guan Y, Greenberg EF, Hasipek M, Chen S, Liu X, Kerr CM, et al. Context dependent effects of ascorbic acid treatment in TET2 mutant myeloid neoplasia. Commun Biol 2020;3:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen LL, Lin HP, Zhou WJ, He CX, Zhang ZY, Cheng ZL, et al. SNIP1 recruits TET2 to regulate c-MYC target genes and cellular DNA damage response. Cell Rep 2018;25:1485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu Y, Zhao Z, Sant DW, Zhu G, Greenblatt SM, Liu L, et al. Tet2 regulates osteoclast differentiation by interacting with Runx1 and maintaining genomic 5-hydroxymethylcytosine (5hmC). Genomics Proteomics Bioinformatics 2018;16:172–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 2011;118:4509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dai HQ, Wang BA, Yang L, Chen JJ, Zhu GC, Sun ML, et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 2016;538:528–32. [DOI] [PubMed] [Google Scholar]
- 21. Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 2014;29:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18:553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Das AT, Zhou X, Metz SW, Vink MA, Berkhout B. Selecting the optimal Tet-On system for doxycycline-inducible gene expression in transiently transfected and stably transduced mammalian cells. Biotechnol J 2016;11:71–9. [DOI] [PubMed] [Google Scholar]
- 24. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 2018;172:90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waitkus MS, Diplas BH, Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell 2018;34:186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27:599–608. [DOI] [PubMed] [Google Scholar]
- 27. Huang X, Motea EA, Moore ZR, Yao J, Dong Y, Chakrabarti G, et al. Leveraging an NQO1 bioactivatable drug for tumor-selective use of Poly(ADP-ribose) polymerase inhibitors. Cancer Cell 2016;30:940–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Churchill H, Naina H, Boriack R, Rakheja D, Chen W. Discordant intracellular and plasma D-2-hydroxyglutarate levels in a patient with IDH2 mutated angioimmunoblastic T-cell lymphoma. Int J Clin Exp Pathol 2015;8:11753–9. [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood 2015;126:1741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemonnier F, Cairns RA, Inoue S, Li WY, Dupuy A, Broutin S, et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc Natl Acad Sci U S A 2016;113:15084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li C, Lan Y, Schwartz-Orbach L, Korol E, Tahiliani M, Evans T, et al. Overlapping requirements for Tet2 and Tet3 in normal development and hematopoietic stem cell emergence. Cell Rep 2015;12:1133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem 2018;87:585–620. [DOI] [PubMed] [Google Scholar]
- 34. An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 2015;6:10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velasco-Hernandez T, Sawen P, Bryder D, Cammenga J. Potential pitfalls of the Mx1-Cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Reports 2016;7:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanchez-Aguilera A, Arranz L, Martin-Perez D, Garcia-Garcia A, Stavropoulou V, Kubovcakova L, et al. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell 2014;15:791–804. [DOI] [PubMed] [Google Scholar]
- 37. Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015;525:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaasinen E, Kuismin O, Rajamaki K, Ristolainen H, Aavikko M, Kondelin J, et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat Commun 2019;10:1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh AK, Zhao B, Liu X, Wang X, Li H, Qin H, et al. Selective targeting of TET catalytic domain promotes somatic cell reprogramming. PNAS 2020;117:3621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagata Y, Narumi S, Guan Y, Przychodzen BP, Hirsch CM, Makishima H, et al. Germline loss-of-function SAMD9 and SAMD9L alterations in adult myelodysplastic syndromes. Blood 2018;132:2309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shima H, Koehler K, Nomura Y, Sugimoto K, Satoh A, Ogata T, et al. Two patients with MIRAGE syndrome lacking haematological features: role of somatic second-site reversion SAMD9 mutations. J Med Genet 2018;55:81–5. [DOI] [PubMed] [Google Scholar]
- 43. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 2013;155:1545–55. [DOI] [PubMed] [Google Scholar]
- 45. Sparks RP, Fratti R. Use of microscale thermophoresis (MST) to measure binding affinities of components of the fusion machinery. Methods Mol Biol 2019;1860:191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frank SB, Schulz VV, Miranti CK. A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector. BMC Biotechnol 2017;17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X, Chen H, Laurini E, Wang Y, Dal Col V, Posocco P, et al. 2-Difluoromethylene-4-methylenepentanoic acid, a paradoxical probe able to mimic the signaling role of 2-oxoglutaric acid in cyanobacteria. Org Lett 2011;13:2924–7. [DOI] [PubMed] [Google Scholar]
- 48. Liu X, Wang Y, Laurini E, Posocco P, Chen H, Ziarelli F, et al. Structural requirements of 2-oxoglutaric acid analogues to mimic its signaling function. Org Lett 2013;15:4662–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.