12 Aug 2020
PONE-D-20-17102
Immune gene expression networks in sepsis: A network biology approach
Reviewer #1: Kim et al present a study focus on the expression of ~1000 genes associated with inflammation and immunity in the context of sepsis. They examine publicly available, retrospective gene expression datasets to define transcriptional networks. There are multiple issues with this study that limit its usefulness and originality:
1. The authors focus on only 998 genes. There have been multiple studies over the past nearly 20 years that have examined the full transcriptome after sepsis (with insights into inflammation and immunity, and including detailed network analyses), and so a focus on only inflammation and immunity is anachronistic at best and very incomplete at worst.
--> Thank you for reading and managing this article and the precise comments.
The definition of sepsis was revised in 2016 (SEPSIS-3) that includes dysregulated host response to infection. These response includes cellular, metabolic, circulatory, neurologic, and immune dysregulation. Oxidative stress or mitochondrial dysfunction, insulin resistance, hypoxemia or hypotension, metabolic acidosis, encephalopathy or neurologic dysregulation are known dysfunctions in sepsis. Among these dysregulated host response, the authors intended to study immune dysfunction via 998 immune related gene expression in this study.
Based on the comment, we have performed full transcriptome analysis by differentially expressed genes (DEG) analysis followed by gene set enrichment for pathway analysis (Table 3). Differentially expressed gene (DEG) was performed between control and patient group with equal sample number. After t-test, genes were selected based on false discovery rate of less than 0.01 (FDR<0.01) except GSE54514, which used global P-value. Based on DEG, gene set enrichment for pathway analysis using GAGE package using R software was performed (Supplementary figure S14-S19, Supplementary Table S17).
Among these pathways, the authors intended to study pathway that was related to immune function. Among up regulated or down regulated pathways with statistical significance, pathways associated with immune related pathway was searched. We have found that complement and coagulation cascade pathway and Fc epsilon RI signaling or Fc gamma R mediated phagocytosis pathway was increased in 66% (2/3) and 100% (3/3) datasets from adult and paediatric patients, respectively. T cell receptor signaling pathway was decreased in 100% (3/3) and 100% (3/3) datasets from adult and paediatric patients, respectively. Antigen presentation and processing was decreased in 66% (2/3) and 66% (2/3) datasets from adult and paediatric patients, respectively.
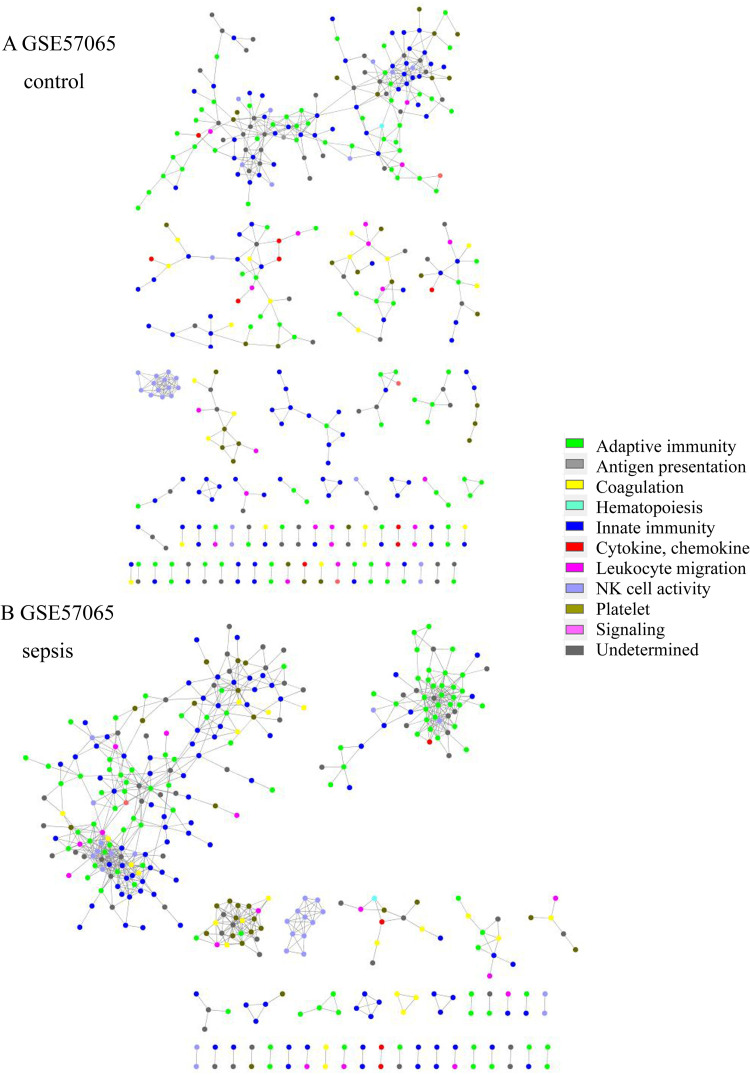
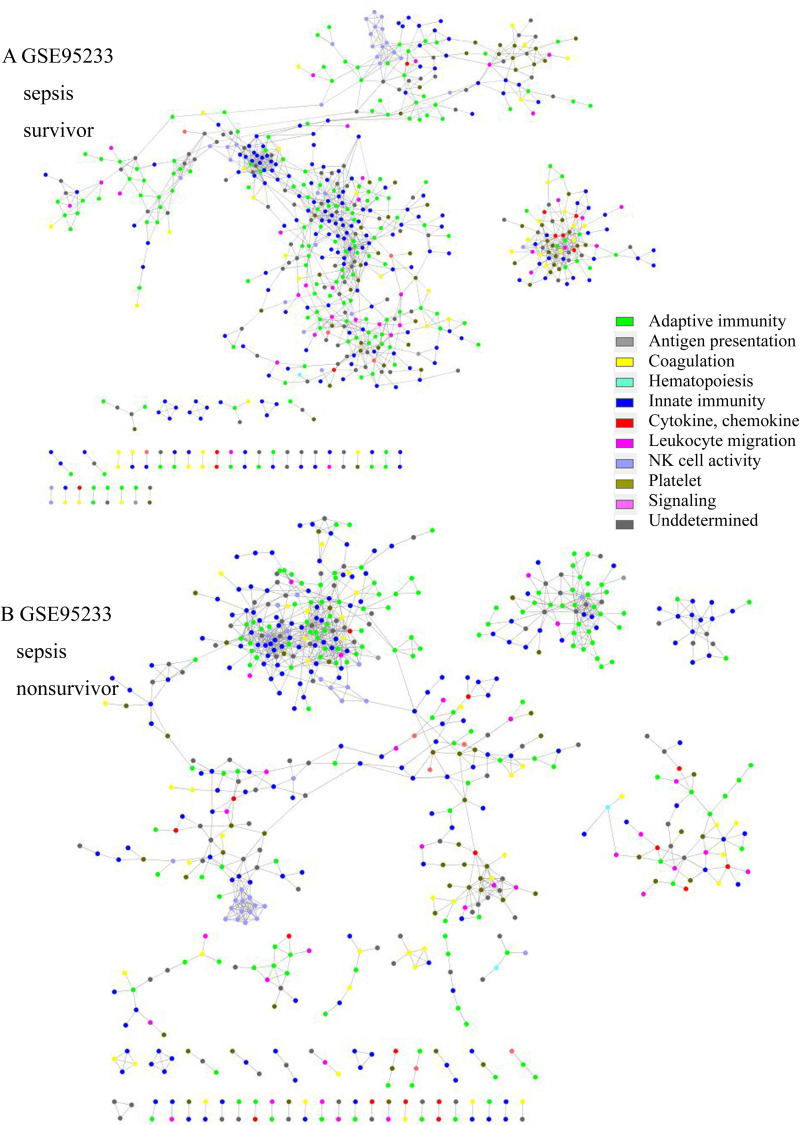
Altogether, these findings could be interpreted along with the network analysis results. In network analysis, adaptive immunity associated with CD3 and CD8 genes were prominent among sepsis patients (Fig. 1). T cell receptor signaling pathway was decreased in 100% (6/6) of the datasets, which implies that T cell function seemed to be decreased or impaired. In network analysis, complement and coagulation cascade was prominent in sepsis survivors of GSE95233 datasets (Fig2). Complement and coagulation cascade was increased among sepsis from 83.3% (5/6) of datasets, which seemed to be a convalescence process.
--> 1.1 Following Table was added in result section
Table 3. Differentially expressed gene analysis between control and sepsis followed by gene set pathway analysis
GSE54514a GSE57065 GSE95233 GSE64456 GSE66099 GSE72829
Patient type Adult Adult Adult Paediatric Paediatric Paediatric
Control cases (n) 18 25 22 19 49 52
Sequential selection
of sepsis cases (n) 18 25 22 19 49 52
Differentially expressed
Genes (n) 2180 7885 6784 3123 8229 7463
Up regulated pathway (n) 4 60 44 13 78 51
Down regulated pathway (n) 1 33 33 16 33 15
Up regulated immune
pathway C, Fc, RIG C, Fc Fc, NK, K, Toll, C K, IgA, Fc, APC, C LK, Fc, NK, B cell, Toll, K, C
Down regulated immune
pathway T cell APC, T cell, H, K APC, IgA, T cell, K, B cell IgA, APC, T cell T cell, H IgA, APC, T cell
aDifferentially expressed gene of GSE54514 was selected based on global P-value (0.05) and other datasets are based on FDR (<0.01).
APC, Antigen processing and presentation; B cell, B cell receptor signaling; C, Complement and coagulation; Fc, Fc gamma R mediated phagocytosis, Fc epsilon RI signaling; H, hematopoietic cell lineage; IgA, intestinal immune network for IgA production; K, chemokine signaling; LM, leukocyte endothelial migration; NK, NK cell mediated cytotoxicity; RIG, RIG-I like receptor signaling; Toll, Toll like receptor signaling; T cell, T cell receptor signaling.
--> 1.2 Following sentence was added in result section
Gene set enrichment for pathway analysis
After differentially expressed gene (DEG) analysis, gene set enrichment for pathway analysis was performed based on KEGG pathway. Among up regulated or down regulated pathways with statistical significance, pathways associated with immune related pathway was searched. We have found that complement and coagulation cascade pathway and Fc epsilon RI signaling or Fc gamma R mediated phagocytosis pathway was increased in 66% (2/3) and 100% (3/3) datasets from adult and paediatric patients, respectively. T cell receptor signaling pathway was decreased in 100% (3/3) and 100% (3/3) datasets from adult and paediatric patients, respectively. Antigen presentation and processing was decreased in 66% (2/3) and 66% (2/3) datasets from adult and paediatric patients, respectively (Table 3). Altogether, these findings could be interpreted along with the network analysis results. In network analysis, adaptive immunity associated with CD3 and CD8 genes were prominent and isolated in sepsis patient group (Fig. 1). T cell receptor signaling pathway was decreased in 100% (6/6) of the datasets, and these data implies that T cell function seemed to be decreased or impaired. In network analysis, complement and coagulation cascade was prominent in sepsis survivors of GSE95233 datasets (Fig2). Complement and coagulation cascade was increased among sepsis in 83.3% (5/6) of datasets, and these data implies that complement and coagulation cascade seemed to be a involved in convalescence process.
--> 1.3 Following supplementary figures were added in supplementary materials
S14 Fig. The gene set analysis result of GSE54514. Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
S15 Fig. The gene set analysis result of GSE57065. Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
S16 Fig. The gene set analysis result of GSE95233. Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
S17 Fig. The gene set analysis result of GSE64456. Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
S18 Fig. The gene set analysis result of GSE66099. Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
S19 Fig. The gene set analysis result of GSE72829 Red cells indicate up regulated gene sets, whereas gene cells indicate down regulated gene sets.
--> 1.4 Following paragraph was added in method section
Gene set enrichment for pathway analysis
Differentially expressed gene (DEG) was performed for total transcriptome between control and patient group with equal sample number. After t-test using basic functions in R software, genes were selected based on false discovery rate of less than 0.01 (FDR<0.01) 53. Based on DEG, gene set enrichment for pathway analysis using GAGE package using R software was performed 54. GAGE package applies input data to KEGG pathway. Significant pathways were plotted for heatmap that was selected based on global P value of less than 0.05.
2. The criteria used for defining sepsis patients are from the 1st consensus definition; we are now at the 3rd consensus definition, and thus the applicability to the clinical setting is possibly limited.
-->Thank you for you kind comment.
Most of the public data are studied based on sepsis-1 criteria.The definition of sepsis-1 was focused on inflammation, whereas sepsis 3 was focused on organ failure. The severe sepsis in sepsis-1 criteria are comparable to sepsis in sepsis-3 criteria. Septic shock in sepsis-1 are comparable to septic shock in sepsis-3. However, this could be limitation of this study and following sentence was added in limitation section.
--> Following sentence was added in the limiation section
" Datasets are based on sepsis-1 criteria for sepsis diagnosis, which was revised to sepsis-3 criteria. This could limit the applicability to the clinical settings."
3. The datasets used involve a mix of adult and pediatric patients. There is extensive literature documenting differences in pediatric vs. adult sepsis patients with regard to multiple inflammatory and immune responses as well as outcomes.
--> Thank you for your kind comments.
The actual analysis was performed based on respective datasets. Adult or paediatric group are not intermingled or analyzed together. The datasets are analyzed independently by each datasets. However, as there was error in Table1, Table 2, we corrected the error in Table 1 and Table 2 as follows. Analysis of the results were corrected according to adult and paediatric datasets, which was as follows.
--> 3.1 Following paragraph was revised in result section
Network parameter analysis
Comparison of topologic parameters revealed that the clustering coefficient in sepsis was increased 66% (2/3) of adult datasets and 100% (3/3) in paediatric datasets (Table 2). Network heterogeneity was increased in 100% (3/3) of adult datasets, but 100% (3/3) were decreased in paediatric datasets, respectively. Modularity value was decreased in 66% (2/3) of adult datasets, whereas modularity value was increased in 100% (3/3) of paediatric datasets in the sepsis group. Average degree and shortest path were increased in the sepsis group among 66% (2/3) of adult and paediatric datasets, respectively. Other parameters did not showed significant results between the healthy control group and the sepsis group (Table 2).
--> 3.2 Table 1 was rearranged as follows
Table 1. Baseline characteristics of recruited datasets for adult and paediatric patient
GSE54514 GSE57065 GSE95233 GSE64456 GSE66099 GSE72829
Patient type Adult Adult Adult Paediatric Paediatric Paediatric
Age, mean (yr) a,b ≥18, 60.3 ≥18, 62 ≥18, 65 ≤2 mo, 29 d ≤10, 3.7 ≤17, 1.9 mo
Sex (male %)b 40 67.9 65 52 58 62
Nonsurvivor (%) 25.7 17.9 (28 d) NA NA 16.3 19.2
Cohort description ICU with sepsis or
septic shock ICU with
septic shock ICU with
septic shock ED with sepsis
(PECARN study) PICU with sepsis or
septic shock Discovery group
(IRIS study)
Severity evaluation APACHEII SOFA SOFA YOSb PRISM NA
Control (n) 18 25 22 19 49 52
Sepsis or septic shock (n) 145 28 102 89 198 94
Randomized selection without
replacement (septic shock) 18 25 - - - -
Randomized selection without
replacement (sepsis) - - 22 18 49 52
Measured time points 5 (1, 2, 3, 4, 5 d) 3 (0, 24, 28 hr) 3 (1-2, 3-4, 7-10 d) 1 1 1
Recruited time pointsc 0-24 hr 0 - 24 hr 1-2 d NA 0 - 24 hr 5 d
Platform Illumina, GPL6947 Affymetrix, GPL570 Affymetrix, GPL570 Illumina, GPL10558 Affymetrix, GPL570 Illumina, GPL10558
Reference [11] [12] [16] [13] [14] [15]
ED, emergency department; ICU, intensive care unit; NA, data not available; hr, hour; d, day.
--> 3.3 Table 2 was rearranged as follows
Table 2. Network topological parameters from gene expression data from GEO dataset for adult and paediatric patient
GSE54514 GSE57065 GSE95233 GSE95233-1 GSE64456 GSE66099 GSE72829
Patient type Adult Adult Adult Adult Paediatirc Paediatirc Paediatirc
Disese type control septic
shock control sepsis control control sepsis control control septic
shock control septic
shock control sepsis
Clustering coefficient 0.319 0.323 0.328 0.280 0.328 0.367 0.390 0.337 0.367 0.390 0.337 0.388 0.381 0.435
Network density 0.013 0.016 0.012 0.010 0.012 0.039 0.026 0.014 0.039 0.026 0.014 0.012 0.030 0.034
Network heterogeneity 1.039 1.142 1.151 1.172 1.151 1.504 1.358 1.193 1.504 1.358 1.193 1.091 1.249 1.129
Connected components 27 30 42 61 42 34 33 47 34 33 47 32 16 25
Network diameter 19 17 14 15 14 12 19 11 12 19 11 14 13 9
Network centralization 0.059 0.079 0.103 0.048 0.103 0.196 0.113 0.075 0.196 0.113 0.075 0.078 0.148 0.121
Shortest path 160514 80598 59638 17222 59638 142640 121152 11730 142640 121152 11730 36902 38092 22532
Characteristic
path length 7.418 5.267 5.276 5.088 5.276 3.862 7.211 3.482 3.862 7.211 3.482 4.705 3.92 2.824
Average degree, <k> 5.93 7.42 5.30 3.64 5.30 17.87 11.71 4.04 17.87 11.71 4.04 4.50 8.43 10.27
Number of nodes, N 464 453 426 367 426 457 448 297 457 448 297 380 284 299
Degree correlation, μ 0.489 0.508 0.422 0.810 0.422 0.403 0.715 0.546 0.403 0.715 0.546 0.210 0.475 0.346
Modularity, Mc 0.743 0.662 0.644 0.806 0.644 0.198 0.581 0.694 0.198 0.581 0.694 0.745 0.434 0.603
--> 3.4 Interpretation of network topology was analyzed by adult and paediatric datasets, respectively. The Following section was corrected as follows:
In that isolated cluster, CD247 (CD3 Zeta chain), CD8A, ITK (tyrosine protein kinase ITK / TSK), LAT (linker for activation of T cells), LCK (leukocyte C-terminal Src kinase) were found in all 6 datasets. CD2, FYN (Src family tyrosine kinase), GATA3 (GATA-binding protein 3), IL7R, RASGRP1 (RAS guanyl-releasing protein 1) were all found in adult datasets, but 66% (2/3) were found in paediatric group. CBLB (E2 ubiquitin protein ligase CBL-B), CD3D, CD3G, ZAP70 (Zeta chain-associated protein kinase70) were found in 66% (2/3) of adult datasets, but all were found in paediatric datasets.
-->3.5 Interpretation of network parameters was analyzed by adult and paediatric datasets, respectively. The Following section was corrected as follows:
Comparison of topologic parameters revealed that the clustering coefficient in sepsis was increased 66% (2/3) of adult datasets and 100% (3/3) in paediatric datasets (Table 2). Network heterogeneity was increased in 100% (3/3) of adult datasets, but 100% (3/3) were decreased in paediatric datasets, respectively. Modularity value was decreased in 66% (2/3) of adult datasets, whereas modularity value was increased in 100% (3/3) of paediatric datasets in the sepsis group. Average degree and shortest path were increased in the sepsis group among 66% (2/3) of adult and paediatric datasets, respectively. Other parameters did not showed significant results between the healthy control group and the sepsis group (Table 2).
4. The source biological material for the transcriptomic analyses is not described. These data are could be from peripheral blood mononuclear cells or whole blood (or were they derived from other tissues?), making interpretation of this study and comparison to other studies difficult.
--> Thank you for your kind comments.
We have added following sentence in the method section
--> All the datasets used transcriptome derived from whole blood.
Reviewer #2: Dear Associate Editor,
The manuscript 'Gene expression networks in sepsis: A network biology approach' by Kim et al analyzes various immune system pathways in patients with sepsis compared to healthy controls using network analysis approach and show that adaptive immune cells are isolated and prominent in that network in patients with sepsis. The strength of this manuscript is that they have used diverse datasets for analysis and obtained comparable results across all the datasets. However, there are some concerns in this manuscript, which the authors have failed to address.
Major comment:
1. In the introduction, the authors nicely summarize the importance of network approach analysis in sepsis as shown by other groups, however two key elements are missing:
a. What is unique or novel about their network approach analysis compared to others and what is the rationale behind that?
b. What was their motivation to perform such an analysis in sepsis?
--> Thank you for your kind comments.
We have added following sentences in introduction.
1-a. These studies constructed network based on measured immune associated molecules that could be regarded as actual observable characteristics of sepsis. Measuring molecules in multiplex methods are relatively unfeasible compared to that of gene expression.
1-b. As molecules could be analyzed independently, analysis of multiple molecules as a system was required. Analysis of these molecules via network analysis including topologic parameters and visualization resulted in informative outcome.
1.1 a, b We have corrected introduction as follows.
--> The immune response between healthy and diseased states is rarely attributed to single molecules but rather to complex inter-related molecules. As sepsis-related immune response molecules are complex, a network approach might enhance our understanding of the equilibrium of the immune response in sepsis18,19. A network approach analysis of a small group of cytokines revealed that the network constructed from sepsis had a lower network diameter and lower shortest path and characteristic path length than the control network20. These topologic features were related to decreased network function or modularity of the network. In addition, compared to that of day 1, the day 4 cytokine network was decreased in sepsis, which was in line with previous literature21. These studies constructed network based on measured immune associated molecules that could be regarded as actual observable characteristics of sepsis. As molecules could be analyzed independently, analysis of multiple molecules as a system was required. Analysis of these molecules via network analysis including topologic parameters and visualization resulted in informative outcome. However, previous studies constructed networks based on small groups of cytokine measurements that resulted in a limited size of recruited molecules. Measuring molecules in multiplex methods are relatively unfeasible compared to that of gene expression.
2. There is a confusion in the two main figures:
a. The dataset identifiers mentioned in the results section of the manuscript are contradictory to those in figures 1 and 2. It looks like the labeling in figure 1 and 2 are swapped.
b. The survivors and non-survivors (mentioned under figure 2 in the results section) have not been labelled in any of the main figures.
--> Thank you for your kind comments.
We have corrected the error in the figures.
3. The discussion section is very long and the authors need to strike a balance between the results and the discussion in the way it is written. There are many elements in the discussion section that actually belongs to the results section.
--> Thank you for your kind comments. We have revised discussion section as follows.
3.1 Deleted sentences in discussion section are as follows.
CD247, CD8A, ITK, LAT, and LCK were found in all 6 datasets, whereas CD2, CD3D, FYN, GATA3, IL7R, RASGRP1, ZAP70, CBLB and CD3G were found in 5 datasets.
3.2 Following discussion section are revised.
most of the genes were annotated to the T cell receptor signalling pathway, which implies that these functions were dysregulated or uncontrolled with other immune networks compared to those in the healthy normal control group. Altogether, adaptive immunity, especially T cell associated gene seemed to be suppressed or dysregulated7.
--> most of the genes were annotated to the T cell receptor signalling pathway, which implies that these functions were dysregulated or uncontrolled with other immune networks compared to those in the healthy normal control group. In addition, gene set enrichment for pathway analysis showed that T cell receptor signalling pathway was down regulated in studies all the datasets. Altogether, adaptive immunity, especially T cell associated gene seemed to be suppressed or dysregulated7.
3.3 Following sentences are deleted.
In the healthy control group, the gene expression network was intermingled with that of innate and adaptive immune genes, whereas genes related to adaptive immunity were isolated and prominent in the sepsis group. In addition, isolated NK cell activity in the control group tended to be integrated into the sepsis group. Isolation or clustering of the network was formed by genes correlating within the cluster and not with the genes outside. Together, these genes might be associated with immune dysregulation or an abnormal host immune response4,5.
3.4 Following sentences are revised as follows
Activation of adaptive immunity is required to resolve sepsis, but the isolated and prominent adaptive immune response in this study seemed to be suppressed or dysregulated. activated without control or harmony with other networks. Complement-coagulation along with platelet-related genes seemed to be activated for convalescence. The gene cluster from nonsurvivors were similar from the genes in isolated network in sepsis. The gene cluster from nonsurvivors shared 4 genes among 5 genes from 6 datasets and 6 genes out of 9 genes from 5 data sets. The gene cluster from nonsurvivors included GATA3, which is known to be the master regulator of CD4 Th2 cell differentiation45.
-> Activation of adaptive immunity is required to resolve sepsis, but the isolated and prominent adaptive immune response in this study seemed to be suppressed or dysregulated.
3.5 Following sentences are moved from discussion to result section
The gene cluster from nonsurvivors were similar from the genes in isolated network in sepsis. shared 4 genes among 5 genes from 6 datasets and 6 genes out of 9 genes from 5 data sets. The gene cluster from nonsurvivors included GATA3, which is known to be the master regulator of CD4 Th2 cell differentiation45
3.6 Following sentence was revised as follows:
Further studies are required to understand the relation between the clustering coefficient and the inflammatory response in the immune network.
--> Network analysis revealed that inflammatory process seemed to be activated in sepsis group and further studies are required to understand the relation between the clustering coefficient and the inflammatory response in the immune network.
3.7 Following sentence was revised as follows:
Shortest path length tended to be decreased in the sepsis group, which was in line with previous literature20
--> Shortest path length was decreased in 66% (2/3) of adult and paediatric datasets, respectively, which was in line with previous literature20
3.8 Following sentence was revised as follows
Modularity of a network represents a subgroup or a cluster within a network28,29. Modularity implies dense connectivity between the nodes within the same cluster but has sparse connection with other nodes outside of the cluster32,33. Modularity is expected to promote evolvability and multifunctionality and function as a driving biological process29,32. In this study, the sepsis group showed increased modularity values in 83.3% of the studied datasets. This result might be due to the prominence or isolation of the adaptive immune network. Although modularity implies the biological process of a network, the isolation and disconnection of a module with a component of a similar functional node might result in adverse reactions in the whole network.
--> Modularity of a network represents a subgroup or a cluster within a network28,29. Modularity implies dense connectivity between the nodes within the same cluster but has sparse connection with other nodes outside of the cluster32,33. Modularity is expected to promote evolvability and multifunctionality and function as a driving biological process29,32.
In this study, the sepsis group in paediatric datasets showed increased modularity values in 100% of the studied datasets. On the contrary, only 33% of sepsis group in adult datasets showed increased modularity. These results are consistent with the data that sepsis group from paediatric datasets showed more significant pathways compared to that of adult group (Table 3). Although modularity implies the biological process of a network, the isolation and disconnection of a module with a component of a similar functional node might result in adverse reactions in the whole network.
3.9 Following sentence was revised as follows
Shortest path length tended to be decreased in the sepsis group, which was in line with previous literature20. This parameter, along with network diameter, is related to the functional and biological processes of a network and increases the probability of the presence of a sub-network or module with biological functions29,30. In this study, these values were decreased in the sepsis group, which might have caused decreased modularity. However, isolated and prominent module formation might have enhanced modularity values in the context of decreasing path length. It has been reported that as a network evolves, complex biological functions are executed through modules, which increases the path length or network diameter34,35. The inconsistency of the decreased shortest path and diameter and increased modularity and clustering coefficient might be an indication of immune dysfunction, which requires further study.
--> Shortest path length was decreased in 66% (2/3) of adult and paediatric datasets, respectively, which was in line with previous literature20. This parameter, along with network diameter, is related to the functional and biological processes of a network and increases the probability of the presence of a sub-network or module with biological functions46,47 . It has been reported that as a network evolves, complex biological functions are executed through modules, which increases the path length or network diameter52,53. The inconsistency of the decreased shortest path and increased clustering coefficient might be an indication of immune dysfunction, which requires further study.
3.10 Following sentence was deleted:
Demographic features, especially sex and age of patients greatly affect prevalence and mortality of the sepsis. There is growing evidence that there are differences in immune system between male and female due to genetic and hormonal causes54,55. Type I interferon activity, T cell numbers and antibody responses are greater in females than in males55. The sex difference in sepsis was supported by the global statistics that the females showed higher incidence of sepsis, whereas males showed higher morality56. The sex difference could not be analyzed in this study due to the small sample sizes and further studies are required to elucidate the sex difference.
3.11 Following sentence was deleted:
CD247, CD8A, ITK, LAT, LCK genes are included in the isolated network in sepsis derived from both age groups. Among them, LCK are intracellular signalling molecule associated with the cytoplasmic tail of CD4 and CD8A molecules.
3.11 Following sentence was deleted:
The sex difference in regard to immune response should be considered, because there is a growing evidence that the genetic and hormonal backgrounds are related with infection54,55. In this study, as the number of samples was limited, the sex difference could not be analyzed.
3.12 Following sentence was revised:
Conclusion
Immune dysfunction might be caused by isolation and prominence of the adaptive immune response from rest of the immune network. The increased modularity and clustering coefficient imply that a modular immune response might be upregulated in the sepsis group compared to that in the normal control group, which seems to be uncontrolled or dysregulated.
--> Conclusion
Immune dysfunction might be caused by isolation and prominence of the adaptive immune response from rest of the immune network. The isolated gene cluster included T cell receptor signaling gene and that pathway was down regulated by gene set enrichment for pathway analysis. T cell signaling or T cell related functions seemed to be impaired or decreased in sepsis cases. Survivors of sepsis showed a prominent cluster of genes that was related to complement and coagulation cascade. As this pathway was up regulated in most of sepsis datasets, this pathway seemed to be related with convalescence process.
4. The current notion is that IL-10 is an anti-inflammatory cytokine, although IL-10 has been shown to be increased amongst non-survivors (ref 28), there is no direct evidence to state that IL-10 is related to unfavorable prognosis. High IL-10 response in non-survivors could be the due to the result of a compensated anti-inflammatory response in sepsis, following the initial pro-inflammation. Therefore, the discussion on high IL-10 is related to unfavorable prognosis in sepsis without a proper evidence is an overstatement and not correct.
--> Thank you for your kind comment.
We have deleted following sentences.
In addition, IL-10 was also included in that cluster; IL-10 is a cytokine that stimulates Th2 cells, and a high concentration of IL-10 along with IL-17, IL-18 and CXCL10 was related to an unfavourable prognosis in sepsis patients28.
5. The discussion about the main computational finding on the role of adaptive immune cells in sepsis identified using their network analysis and their inference in the pathophysiology of SIRS or sepsis is not clear and needs to be elucidated.
--> Thank you for your kind comment.
We have performed additional analysis, which was gene set enrichment for pathway analysis (Supplementary Table S17, Supplmentary Figure S14-S19). Altogether, network analysis showed isolated component or isolated cluster of genes related to adaptive immune response. The genes in the cluster were related to adaptive immune response or T cells and the pathway analysis showed that the T cell signaling pathway was decreased in 100% (3/3) in adult and 100% (3/3) in paedicatric sepsis datasets. From these findings, T cell signaling or T cell related functions seemed to be impaired or decreased in sepsis cases. Among survivor of sepsis, complement and coagulation cascade and platelet related genes was prominent in network analysis. Gene set enrichment analysis showed that complement and coagulation cascade pathway was up regulated in 66% (2/3) in adult and 100% (3/3) in paediatric sepsis cases. From these findings, complement and coagulation pathway seemed to be associated with convalescence process during sepsis process. Clustering coefficient that was related with inflammatory process was increased in 66% (2/3) of adult and 100% (3/3) in paediatric datasets.
5.1 Following sentence was added in discussion section.
--> Altogether, network analysis showed isolated component or isolated cluster of genes related to adaptive immune response. The genes in the cluster were related to adaptive immune response or T cells and the pathway analysis showed that the T cell signaling pathway was decreased in 100% (3/3) in adult and 100% (3/3) in paedicatric sepsis datasets. From these findings, T cell signaling or T cell related functions seemed to be impaired or decreased in sepsis cases. Among survivor of sepsis, complement and coagulation cascade and platelet related genes was prominent in network analysis. Gene set enrichment analysis showed that complement and coagulation cascade pathway was up regulated in 66% (2/3) in adult and 100% (3/3) in paediatric sepsis cases. From these findings, complement and coagulation pathway seemed to be associated with convalescence process during sepsis process. Clustering coefficient that was related with inflammatory process was increased in 66% (2/3) of adult and 100% (3/3) in paediatric datasets.
5.2 Following table below from gene set enrichment for pathway analysis was added as a Supplementary Table S17
Minor comments:
1. The authors should stick to using the abbreviations instead of full form after using it for the first time and have to be consistent throughout the manuscript.
Example: GEO - line 86, KEGG - line 150, GO - line 166
--> Thank you for your kind comment.
We have corrected the errors as commented.
2. Are there any references for the rationale behind using such platforms? Lines 106 and 107.
--> Thank you for your kind comment.
Following articles are cited.
22. Ritchie M, Dunning M, Smith M, Shi W, Lynch A. BeadArray expression analysis using bioconductor. PLoS Comp Biol 2011; 7:e1002276. https://doi.org/10.1371/journal.pcbi.1002276.
23. Jaksik R, Iwanaszko M, Rzeszowska-Wolny R, Kimmel M. Microarray experiments and factors which affect their reliability. Biol Direct 2015; 10:46 10.1186/s13062-015-0077-2.
3. What does '(n)' in the table 1 indicate?
--> Thank you for your kind comment.
We are sorry for confusion . The measured frequency or frequency of blood drawn are described. For example, GSE54514 datasets showed that for eacg patients, as much as 5 time of blood dare drawn at day 1, day 2, day3, day4, and day 5 after admission. Then experimented as much as 5 time per patients. 5 (1, 2, 3, 4, 5 d)
--> We have changed the Table 1 as follows.
Measured freqeuncy, frequency (time points) 5 (1, 2, 3, 4, 5 d) 3 (0, 24, 28 hr) 3 (1-2, 3-4, 7-10 d) 1 1 1
4. Why are the figure legends not listed separately and written along with the text?
--> Thank you for your kind comment.
Submission guideline states as follows.
"Figure captions must be inserted in the text of the manuscript, immediately following the paragraph in which the figure is first cited (read order). Do not include captions as part of the figure files themselves or submit them in a separate document"
5. Maintain a similar font and size throughout the manuscript, line 298 is different.
--> Thank you for your kind comment.
We have corrected the error. Font size was changed to 12 and Times New Roman was used for the font.
Additional revision
Following Supplementary Table S17 was added, which was the result of gene set enrichment for pathway analysis. Gene set enrichment for pathway analysis data was summarized in Table 3.
Supplmentary Table S17.
GSE54514 p.geomean stat.mean p.val q.val set.size
hsa04142 Lysosome 0.025847966 1.269518545 5.09E-08 8.20E-06 114
hsa00970 Aminoacyl-tRNA biosynthesis 0.088028345 0.881659727 0.000130305 0.008555202 40
hsa04141 Protein processing in endoplasmic reticulum 0.108398782 0.852836674 0.000159414 0.008555202 152
hsa00600 Sphingolipid metabolism 0.183824309 0.745837522 0.000888854 0.035776374 33
hsa00500 Starch and sucrose metabolism 0.174616207 0.592419696 0.00683581 0.22011307 32
hsa04260 Cardiac muscle contraction 0.150475266 0.517270132 0.015534859 0.353170066 49
hsa00670 One carbon pool by folate 0.24989082 0.511164476 0.01766595 0.353170066 15
hsa03030 DNA replication 0.140145009 0.502213408 0.019475018 0.353170066 36
hsa00520 Amino sugar and nucleotide sugar metabolism 0.185600438 0.492848731 0.019742426 0.353170066 42
hsa04810 Regulation of actin cytoskeleton 0.142203717 0.470551959 0.023133818 0.372454467 168
hsa04622 RIG-I-like receptor signaling pathway 0.198576053 0.463690703 0.025639176 0.375264301 58
hsa00310 Lysine degradation 0.258921614 0.443101954 0.031635146 0.424438213 37
hsa00561 Glycerolipid metabolism 0.292506444 0.427968945 0.035771277 0.428628393 39
hsa00983 Drug metabolism - other enzymes 0.25490034 0.419779579 0.039240986 0.428628393 27
hsa00564 Glycerophospholipid metabolism 0.255687907 0.414859748 0.040160748 0.428628393 63
hsa04973 Carbohydrate digestion and absorption 0.269226767 0.399200271 0.046898393 0.428628393 31
hsa00630 Glyoxylate and dicarboxylate metabolism 0.258395943 0.407462628 0.047746415 0.428628393 16
hsa02010 ABC transporters 0.255973642 0.394894213 0.049534371 0.428628393 32
hsa03060 Protein export 0.201584586 0.396801811 0.051659627 0.428628393 23
hsa00510 N-Glycan biosynthesis 0.258174577 0.38240521 0.054661249 0.428628393 44
hsa04114 Oocyte meiosis 0.25415967 0.370225054 0.05878054 0.428628393 97
hsa03410 Base excision repair 0.26846419 0.370962959 0.06076491 0.428628393 31
hsa00512 Mucin type O-Glycan biosynthesis 0.297860294 0.371924262 0.061232628 0.428628393 18
hsa04962 Vasopressin-regulated water reabsorption 0.307990132 0.356276752 0.066901165 0.448795313 37
hsa04666 Fc gamma R-mediated phagocytosis 0.149263639 0.326641851 0.083783981 0.484269587 85
hsa04670 Leukocyte transendothelial migration 0.179800404 0.325133026 0.084901015 0.484269587 91
hsa00250 Alanine, aspartate and glutamate metabolism 0.308043361 0.327973968 0.085817892 0.484269587 21
hsa04020 Calcium signaling pathway 0.30905421 0.320665521 0.087228474 0.484269587 121
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.327068995 0.323210599 0.089572826 0.484269587 17
hsa04914 Progesterone-mediated oocyte maturation 0.318424759 0.316995636 0.090236569 0.484269587 74
hsa00531 Glycosaminoglycan degradation 0.28104011 0.312304755 0.097423406 0.495162094 17
hsa00052 Galactose metabolism 0.301508758 0.299776925 0.103657516 0.495162094 23
hsa00100 Steroid biosynthesis 0.309712284 0.303208041 0.10394196 0.495162094 17
hsa04722 Neurotrophin signaling pathway 0.209032778 0.296981402 0.104568392 0.495162094 118
hsa00040 Pentose and glucuronate interconversions 0.339423254 0.297522093 0.107723493 0.495528066 17
hsa04614 Renin-angiotensin system 0.299906414 0.2965293 0.116933139 0.522950984 10
hsa00010 Glycolysis / Gluconeogenesis 0.20023476 0.277225272 0.124573091 0.542061287 53
hsa00790 Folate biosynthesis 0.341498338 0.262181962 0.140744805 0.568424515 10
hsa00640 Propanoate metabolism 0.264798564 0.258017879 0.142572288 0.568424515 29
hsa00770 Pantothenate and CoA biosynthesis 0.290883135 0.2552472 0.146246621 0.568424515 14
hsa04110 Cell cycle 0.217646458 0.248708696 0.147876265 0.568424515 113
hsa04972 Pancreatic secretion 0.338662349 0.247249009 0.148284656 0.568424515 62
hsa00982 Drug metabolism - cytochrome P450 0.367722445 0.238589699 0.157562697 0.58994405 35
hsa04971 Gastric acid secretion 0.3592355 0.233274674 0.162455793 0.594440515 53
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.332875037 0.226637555 0.170044422 0.595901983 38
hsa00140 Steroid hormone biosynthesis 0.354305041 0.22741378 0.170257709 0.595901983 27
hsa00900 Terpenoid backbone biosynthesis 0.332436368 0.218545769 0.184698046 0.630739186 12
hsa00190 Oxidative phosphorylation 0.037330963 0.208137994 0.191092518 0.630739186 98
hsa04910 Insulin signaling pathway 0.245798672 0.205116192 0.1919641 0.630739186 122
hsa04070 Phosphatidylinositol signaling system 0.334222679 0.192350212 0.208023871 0.642372746 63
hsa04977 Vitamin digestion and absorption 0.384343117 0.195255211 0.208279084 0.642372746 14
hsa04912 GnRH signaling pathway 0.342673996 0.189741884 0.211326016 0.642372746 77
hsa00260 Glycine, serine and threonine metabolism 0.321173596 0.19479429 0.21146432 0.642372746 22
hsa04975 Fat digestion and absorption 0.369961393 0.179142864 0.22581784 0.673271708 24
hsa04510 Focal adhesion 0.278160495 0.173626264 0.230738055 0.675433216 152
hsa00562 Inositol phosphate metabolism 0.345858678 0.164064696 0.244301366 0.702366426 49
hsa04120 Ubiquitin mediated proteolysis 0.345414226 0.135754803 0.283040175 0.749230808 124
hsa00565 Ether lipid metabolism 0.393313072 0.13629968 0.283349434 0.749230808 26
hsa00511 Other glycan degradation 0.34406099 0.132503787 0.288935834 0.749230808 15
hsa00360 Phenylalanine metabolism 0.391046096 0.133360583 0.291832263 0.749230808 11
hsa04964 Proximal tubule bicarbonate reclamation 0.37667497 0.132046378 0.293578294 0.749230808 13
hsa04974 Protein digestion and absorption 0.423776406 0.127078542 0.295942314 0.749230808 46
hsa04976 Bile secretion 0.378893273 0.126104602 0.297061262 0.749230808 41
hsa00592 alpha-Linolenic acid metabolism 0.41786042 0.123279378 0.30524373 0.749230808 10
hsa00270 Cysteine and methionine metabolism 0.318554397 0.120569815 0.309732703 0.749230808 29
hsa03320 PPAR signaling pathway 0.386684884 0.116911974 0.310656418 0.749230808 49
hsa00240 Pyrimidine metabolism 0.363458028 0.11620759 0.311791703 0.749230808 86
hsa01040 Biosynthesis of unsaturated fatty acids 0.387275259 0.113252693 0.318749406 0.754686093 17
hsa04210 Apoptosis 0.305235308 0.098878697 0.338606349 0.776437433 82
hsa04610 Complement and coagulation cascades 0.353012098 0.098902363 0.340223888 0.776437433 43
hsa04662 B cell receptor signaling pathway 0.260627497 0.091463676 0.348141754 0.776437433 71
hsa04742 Taste transduction 0.419851888 0.092921726 0.348238904 0.776437433 23
hsa04740 Olfactory transduction 0.352684989 0.088588164 0.35389701 0.776437433 91
hsa00380 Tryptophan metabolism 0.391587333 0.08729509 0.356871863 0.776437433 32
hsa04920 Adipocytokine signaling pathway 0.371053644 0.079635611 0.368659921 0.784034854 60
hsa00053 Ascorbate and aldarate metabolism 0.414632821 0.074706156 0.379728253 0.784034854 12
hsa04146 Peroxisome 0.361172032 0.070587225 0.382781845 0.784034854 61
hsa03450 Non-homologous end-joining 0.435855274 0.071922973 0.384160447 0.784034854 10
hsa04115 p53 signaling pathway 0.391925676 0.069158745 0.384712754 0.784034854 58
hsa00620 Pyruvate metabolism 0.321389717 0.064295199 0.394042236 0.786704121 36
hsa04010 MAPK signaling pathway 0.313311625 0.062364703 0.395795241 0.786704121 206
hsa00290 Valine, leucine and isoleucine biosynthesis 0.334268388 0.061382719 0.401453979 0.788220618 10
hsa04520 Adherens junction 0.324247483 0.048347278 0.416929952 0.793369782 57
hsa04310 Wnt signaling pathway 0.435163792 0.048277858 0.419061166 0.793369782 113
hsa00051 Fructose and mannose metabolism 0.341772085 0.043156679 0.425408167 0.793369782 31
hsa00591 Linoleic acid metabolism 0.446939775 0.042645284 0.429284738 0.793369782 13
hsa00280 Valine, leucine and isoleucine degradation 0.317635293 0.041651145 0.431136109 0.793369782 38
hsa03015 mRNA surveillance pathway 0.431686244 0.039670161 0.433643111 0.793369782 71
hsa03018 RNA degradation 0.346366886 0.032491197 0.446251782 0.807264459 66
hsa04144 Endocytosis 0.333055224 0.026846334 0.454833815 0.808758859 166
hsa00030 Pentose phosphate pathway 0.317824725 0.0257338 0.457249015 0.808758859 24
hsa04970 Salivary secretion 0.416543972 0.022445557 0.46214792 0.808758859 53
hsa03440 Homologous recombination 0.425157786 0.012762907 0.478044595 0.827582578 24
hsa04621 NOD-like receptor signaling pathway 0.277751297 0.010179839 0.48881845 0.832685052 53
hsa04960 Aldosterone-regulated sodium reabsorption 0.437266196 0.00515905 0.4913359 0.832685052 28
hsa00603 Glycosphingolipid biosynthesis - globo series 0.449189859 -0.006807241 0.508352064 0.84747946 10
hsa03050 Proteasome 0.238915289 -0.010484047 0.512024664 0.84747946 41
hsa04340 Hedgehog signaling pathway 0.461833673 -0.009327661 0.515857062 0.84747946 32
hsa00480 Glutathione metabolism 0.384508884 -0.017753272 0.530419193 0.857959851 41
hsa00830 Retinol metabolism 0.474820452 -0.020041995 0.532894318 0.857959851 28
hsa04145 Phagosome 0.306430871 -0.025812974 0.543588662 0.862097576 131
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series 0.431939299 -0.027274556 0.54617362 0.862097576 17
hsa04270 Vascular smooth muscle contraction 0.418991488 -0.032770335 0.554027095 0.865487504 87
hsa04916 Melanogenesis 0.438601359 -0.035310316 0.559072673 0.865487504 72
hsa00330 Arginine and proline metabolism 0.387194909 -0.038035297 0.565743481 0.867473338 35
hsa00350 Tyrosine metabolism 0.451928409 -0.047513912 0.578971415 0.871664378 25
hsa00740 Riboflavin metabolism 0.461009902 -0.051380624 0.582426007 0.871664378 10
hsa00514 Other types of O-glycan biosynthesis 0.46499724 -0.050843399 0.584718961 0.871664378 29
hsa00910 Nitrogen metabolism 0.389061199 -0.056243514 0.590636298 0.872407743 14
hsa00450 Selenocompound metabolism 0.469366125 -0.059659743 0.598334181 0.875743665 14
hsa04664 Fc epsilon RI signaling pathway 0.331403961 -0.067141326 0.606750669 0.878715972 64
hsa04630 Jak-STAT signaling pathway 0.304332486 -0.066561794 0.611280676 0.878715972 112
hsa04512 ECM-receptor interaction 0.311105659 -0.074953004 0.625090399 0.890615525 51
hsa03430 Mismatch repair 0.410418436 -0.081333155 0.633964455 0.895335765 23
hsa04730 Long-term depression 0.448465686 -0.087380224 0.643065335 0.899010592 48
hsa00410 beta-Alanine metabolism 0.429205235 -0.093451249 0.650230831 0.899010592 17
hsa00071 Fatty acid metabolism 0.427905821 -0.097695646 0.658523906 0.899010592 33
hsa04150 mTOR signaling pathway 0.448876505 -0.097928081 0.658902173 0.899010592 45
hsa04320 Dorso-ventral axis formation 0.450974382 -0.112685352 0.680401978 0.920543853 17
hsa00340 Histidine metabolism 0.420885104 -0.124974453 0.699900186 0.939032749 20
hsa04710 Circadian rhythm - mammal 0.517842981 -0.1309111 0.708518549 0.942739557 20
hsa00920 Sulfur metabolism 0.463099405 -0.146929292 0.725639974 0.949930389 10
hsa00650 Butanoate metabolism 0.462646611 -0.14474415 0.725723216 0.949930389 20
hsa00860 Porphyrin and chlorophyll metabolism 0.352093404 -0.166428259 0.75483994 0.963093999 27
hsa00020 Citrate cycle (TCA cycle) 0.345764883 -0.170516025 0.758009742 0.963093999 27
hsa03420 Nucleotide excision repair 0.438925925 -0.170040315 0.762146338 0.963093999 40
hsa04140 Regulation of autophagy 0.501766316 -0.174275298 0.766671141 0.963093999 24
hsa04380 Osteoclast differentiation 0.148156136 -0.189624873 0.786435035 0.963093999 112
hsa00590 Arachidonic acid metabolism 0.503617728 -0.197975009 0.797417779 0.963093999 35
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate 0.532138836 -0.200390682 0.797795062 0.963093999 12
hsa03008 Ribosome biogenesis in eukaryotes 0.333180871 -0.200270704 0.798234973 0.963093999 65
hsa00760 Nicotinate and nicotinamide metabolism 0.528910076 -0.206511288 0.806064252 0.963093999 20
hsa04623 Cytosolic DNA-sensing pathway 0.437963673 -0.20638293 0.806666663 0.963093999 41
hsa04540 Gap junction 0.441976755 -0.206664205 0.807835502 0.963093999 71
hsa04530 Tight junction 0.44193829 -0.208545097 0.809906484 0.963093999 99
hsa04720 Long-term potentiation 0.490372452 -0.21137621 0.813545241 0.963093999 57
hsa04744 Phototransduction 0.55914291 -0.236412336 0.838759444 0.966055827 18
hsa03040 Spliceosome 0.304296201 -0.239609263 0.842039058 0.966055827 122
hsa00230 Purine metabolism 0.46796738 -0.238557953 0.843705602 0.966055827 136
hsa04330 Notch signaling pathway 0.46873001 -0.241488559 0.844589685 0.966055827 42
hsa04130 SNARE interactions in vesicular transport 0.509554977 -0.243339648 0.846048892 0.966055827 31
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.56895764 -0.251407671 0.854575085 0.968919639 21
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.502124633 -0.262852465 0.862913279 0.971531734 19
hsa03013 RNA transport 0.295742887 -0.285477866 0.884918199 0.989387709 131
hsa04360 Axon guidance 0.5175373 -0.304991628 0.900993629 0.992123539 89
hsa00604 Glycosphingolipid biosynthesis - ganglio series 0.593477566 -0.31452579 0.904852663 0.992123539 12
hsa04672 Intestinal immune network for IgA production 0.21337552 -0.31147188 0.905851927 0.992123539 39
hsa04370 VEGF signaling pathway 0.533095949 -0.347139807 0.927802579 1 58
hsa04966 Collecting duct acid secretion 0.54307371 -0.35534608 0.930671803 1 19
hsa04350 TGF-beta signaling pathway 0.566615481 -0.389158586 0.949727553 1 64
hsa03022 Basal transcription factors 0.585770861 -0.395426272 0.951033122 1 30
hsa04012 ErbB signaling pathway 0.554808925 -0.400498237 0.95451798 1 76
hsa04640 Hematopoietic cell lineage 0.399662255 -0.467402264 0.975061345 1 76
hsa04620 Toll-like receptor signaling pathway 0.368806166 -0.475967315 0.976865581 1 82
hsa04062 Chemokine signaling pathway 0.344192063 -0.474163521 0.977260148 1 150
hsa04612 Antigen processing and presentation 0.214081952 -0.481110381 0.977825928 1 69
hsa03020 RNA polymerase 0.620754028 -0.494288871 0.980686154 1 24
hsa04514 Cell adhesion molecules (CAMs) 0.255167259 -0.610800137 0.994744854 1 102
hsa04650 Natural killer cell mediated cytotoxicity 0.472831909 -0.695347858 0.998274891 1 112
hsa04660 T cell receptor signaling pathway 0.667202143 -0.862374107 0.999861365 1 96
hsa03010 Ribosome 0.002095243 -1.890710517 1 1 88
hsa00061 Fatty acid biosynthesis NA NA NA NA 6
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 7
hsa00120 Primary bile acid biosynthesis NA NA NA NA 9
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00300 Lysine biosynthesis NA NA NA NA 3
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 3
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 4
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00730 Thiamine metabolism NA NA NA NA 3
hsa00750 Vitamin B6 metabolism NA NA NA NA 5
hsa00780 Biotin metabolism NA NA NA NA 2
hsa00785 Lipoic acid metabolism NA NA NA NA 3
hsa04122 Sulfur relay system NA NA NA NA 9
hsa04660 T cell receptor signaling pathway 0.136214253 -0.862374107 0.000138635 0.022320274 96
hsa04650 Natural killer cell mediated cytotoxicity 0.116909546 -0.695347858 0.001725109 0.138871256 112
hsa04514 Cell adhesion molecules (CAMs) 0.069023632 -0.610800137 0.005255146 0.282026165 102
hsa03020 RNA polymerase 0.272268632 -0.494288871 0.019313846 0.501890441 24
hsa04612 Antigen processing and presentation 0.081538447 -0.481110381 0.022174072 0.501890441 69
hsa04062 Chemokine signaling pathway 0.135205504 -0.474163521 0.022739852 0.501890441 150
hsa04620 Toll-like receptor signaling pathway 0.13883093 -0.475967315 0.023134419 0.501890441 82
hsa04640 Hematopoietic cell lineage 0.165285289 -0.467402264 0.024938655 0.501890441 76
hsa04012 ErbB signaling pathway 0.277142133 -0.400498237 0.04548202 0.735805808 76
hsa03022 Basal transcription factors 0.298484727 -0.395426272 0.048966878 0.735805808 30
hsa04350 TGF-beta signaling pathway 0.292048704 -0.389158586 0.050272447 0.735805808 64
hsa04966 Collecting duct acid secretion 0.303544751 -0.35534608 0.069328197 0.894137288 19
hsa04370 VEGF signaling pathway 0.286561239 -0.347139807 0.072197421 0.894137288 58
hsa04672 Intestinal immune network for IgA production 0.123902817 -0.31147188 0.094148073 0.99625161 39
hsa00604 Glycosphingolipid biosynthesis - ganglio series 0.358452847 -0.31452579 0.095147337 0.99625161 12
hsa04360 Axon guidance 0.301663998 -0.304991628 0.099006371 0.99625161 89
hsa03013 RNA transport 0.159966018 -0.285477866 0.115081801 1 131
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.323152217 -0.262852465 0.137086721 1 19
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.379562063 -0.251407671 0.145424915 1 21
hsa04130 SNARE interactions in vesicular transport 0.33600731 -0.243339648 0.153951108 1 31
hsa04330 Notch signaling pathway 0.306869511 -0.241488559 0.155410315 1 42
hsa00230 Purine metabolism 0.31234081 -0.238557953 0.156294398 1 136
hsa03040 Spliceosome 0.173578172 -0.239609263 0.157960942 1 122
hsa04744 Phototransduction 0.381424186 -0.236412336 0.161240556 1 18
hsa04720 Long-term potentiation 0.341097583 -0.21137621 0.186454759 1 57
hsa04530 Tight junction 0.294441545 -0.208545097 0.190093516 1 99
hsa04540 Gap junction 0.303891541 -0.206664205 0.192164498 1 71
hsa04623 Cytosolic DNA-sensing pathway 0.304797059 -0.20638293 0.193333337 1 41
hsa00760 Nicotinate and nicotinamide metabolism 0.376359987 -0.206511288 0.193935748 1 20
hsa03008 Ribosome biogenesis in eukaryotes 0.221009065 -0.200270704 0.201765027 1 65
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate 0.386003068 -0.200390682 0.202204938 1 12
hsa00590 Arachidonic acid metabolism 0.361444745 -0.197975009 0.202582221 1 35
hsa04380 Osteoclast differentiation 0.098870485 -0.189624873 0.213564965 1 112
hsa04140 Regulation of autophagy 0.373873992 -0.174275298 0.233328859 1 24
hsa03420 Nucleotide excision repair 0.326468419 -0.170040315 0.237853662 1 40
hsa00020 Citrate cycle (TCA cycle) 0.2497233 -0.170516025 0.241990258 1 27
hsa00860 Porphyrin and chlorophyll metabolism 0.26448502 -0.166428259 0.24516006 1 27
hsa00650 Butanoate metabolism 0.362321227 -0.14474415 0.274276784 1 20
hsa00920 Sulfur metabolism 0.364393515 -0.146929292 0.274360026 1 10
hsa04710 Circadian rhythm - mammal 0.418319238 -0.1309111 0.291481451 1 20
hsa00340 Histidine metabolism 0.342161218 -0.124974453 0.300099814 1 20
hsa04320 Dorso-ventral axis formation 0.373595592 -0.112685352 0.319598022 1 17
hsa04150 mTOR signaling pathway 0.374019224 -0.097928081 0.341097827 1 45
hsa00071 Fatty acid metabolism 0.360393804 -0.097695646 0.341476094 1 33
hsa00410 beta-Alanine metabolism 0.365870176 -0.093451249 0.349769169 1 17
hsa04730 Long-term depression 0.382943389 -0.087380224 0.356934665 1 48
hsa03430 Mismatch repair 0.361918324 -0.081333155 0.366035545 1 23
hsa04512 ECM-receptor interaction 0.277348567 -0.074953004 0.374909601 1 51
hsa04630 Jak-STAT signaling pathway 0.275463443 -0.066561794 0.388719324 1 112
hsa04664 Fc epsilon RI signaling pathway 0.269755422 -0.067141326 0.393249331 1 64
hsa00450 Selenocompound metabolism 0.425995821 -0.059659743 0.401665819 1 14
hsa00910 Nitrogen metabolism 0.354601714 -0.056243514 0.409363702 1 14
hsa00514 Other types of O-glycan biosynthesis 0.427357067 -0.050843399 0.415281039 1 29
hsa00740 Riboflavin metabolism 0.423313094 -0.051380624 0.417573993 1 10
hsa00350 Tyrosine metabolism 0.4176519 -0.047513912 0.421028585 1 25
hsa00330 Arginine and proline metabolism 0.369158011 -0.038035297 0.434256519 1 35
hsa04916 Melanogenesis 0.411020347 -0.035310316 0.440927327 1 72
hsa04270 Vascular smooth muscle contraction 0.385386751 -0.032770335 0.445972905 1 87
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series 0.413805829 -0.027274556 0.45382638 1 17
hsa04145 Phagosome 0.295731261 -0.025812974 0.456411338 1 131
hsa00830 Retinol metabolism 0.457726717 -0.020041995 0.467105682 1 28
hsa00480 Glutathione metabolism 0.375752091 -0.017753272 0.469580807 1 41
hsa04340 Hedgehog signaling pathway 0.455113761 -0.009327661 0.484142938 1 32
hsa03050 Proteasome 0.222295647 -0.010484047 0.487975336 1 41
hsa00603 Glycosphingolipid biosynthesis - globo series 0.441800324 -0.006807241 0.491647936 1 10
hsa04960 Aldosterone-regulated sodium reabsorption 0.440602431 0.00515905 0.5086641 1 28
hsa04621 NOD-like receptor signaling pathway 0.304210931 0.010179839 0.51118155 1 53
hsa03440 Homologous recombination 0.432630488 0.012762907 0.521955405 1 24
hsa04970 Salivary secretion 0.432697161 0.022445557 0.53785208 1 53
hsa00030 Pentose phosphate pathway 0.330594907 0.0257338 0.542750985 1 24
hsa04144 Endocytosis 0.350688271 0.026846334 0.545166185 1 166
hsa03018 RNA degradation 0.371059579 0.032491197 0.553748218 1 66
hsa03015 mRNA surveillance pathway 0.463195069 0.039670161 0.566356889 1 71
hsa00280 Valine, leucine and isoleucine degradation 0.342972261 0.041651145 0.568863891 1 38
hsa00591 Linoleic acid metabolism 0.47793164 0.042645284 0.570715262 1 13
hsa00051 Fructose and mannose metabolism 0.358838557 0.043156679 0.574591833 1 31
hsa04310 Wnt signaling pathway 0.471705527 0.048277858 0.580938834 1 113
hsa04520 Adherens junction 0.339189913 0.048347278 0.583070048 1 57
hsa00290 Valine, leucine and isoleucine biosynthesis 0.372969854 0.061382719 0.598546021 1 10
hsa04010 MAPK signaling pathway 0.34800637 0.062364703 0.604204759 1 206
hsa00620 Pyruvate metabolism 0.359944739 0.064295199 0.605957764 1 36
hsa04115 p53 signaling pathway 0.436600315 0.069158745 0.615287246 1 58
hsa03450 Non-homologous end-joining 0.489929417 0.071922973 0.615839553 1 10
hsa04146 Peroxisome 0.405958937 0.070587225 0.617218155 1 61
hsa00053 Ascorbate and aldarate metabolism 0.468576595 0.074706156 0.620271747 1 12
hsa04920 Adipocytokine signaling pathway 0.426384472 0.079635611 0.631340079 1 60
hsa00380 Tryptophan metabolism 0.452409033 0.08729509 0.643128137 1 32
hsa04740 Olfactory transduction 0.408596481 0.088588164 0.64610299 1 91
hsa04742 Taste transduction 0.486023644 0.092921726 0.651761096 1 23
hsa04662 B cell receptor signaling pathway 0.294646319 0.091463676 0.651858246 1 71
hsa04610 Complement and coagulation cascades 0.424788858 0.098902363 0.659776112 1 43
hsa04210 Apoptosis 0.365273768 0.098878697 0.661393651 1 82
hsa01040 Biosynthesis of unsaturated fatty acids 0.467302308 0.113252693 0.681250594 1 17
hsa00240 Pyrimidine metabolism 0.444599654 0.11620759 0.688208297 1 86
hsa03320 PPAR signaling pathway 0.466931096 0.116911974 0.689343582 1 49
hsa00270 Cysteine and methionine metabolism 0.401737471 0.120569815 0.690267297 1 29
hsa00592 alpha-Linolenic acid metabolism 0.508458368 0.123279378 0.69475627 1 10
hsa04976 Bile secretion 0.464640387 0.126104602 0.702938738 1 41
hsa04974 Protein digestion and absorption 0.522478806 0.127078542 0.704057686 1 46
hsa04964 Proximal tubule bicarbonate reclamation 0.468711064 0.132046378 0.706421706 1 13
hsa00360 Phenylalanine metabolism 0.483639952 0.133360583 0.708167737 1 11
hsa00511 Other glycan degradation 0.42424033 0.132503787 0.711064166 1 15
hsa00565 Ether lipid metabolism 0.490879538 0.13629968 0.716650566 1 26
hsa04120 Ubiquitin mediated proteolysis 0.439426877 0.135754803 0.716959825 1 124
hsa00562 Inositol phosphate metabolism 0.452298465 0.164064696 0.755698634 1 49
hsa04510 Focal adhesion 0.364179241 0.173626264 0.769261945 1 152
hsa04975 Fat digestion and absorption 0.496570773 0.179142864 0.77418216 1 24
hsa00260 Glycine, serine and threonine metabolism 0.45311236 0.19479429 0.78853568 1 22
hsa04912 GnRH signaling pathway 0.47169064 0.189741884 0.788673984 1 77
hsa04977 Vitamin digestion and absorption 0.527591748 0.195255211 0.791720916 1 14
hsa04070 Phosphatidylinositol signaling system 0.457367461 0.192350212 0.791976129 1 63
hsa04910 Insulin signaling pathway 0.334478863 0.205116192 0.8080359 1 122
hsa00190 Oxidative phosphorylation 0.060528215 0.208137994 0.808907445 1 98
hsa00900 Terpenoid backbone biosynthesis 0.478282933 0.218545769 0.815301954 1 12
hsa00140 Steroid hormone biosynthesis 0.518148387 0.22741378 0.829742291 1 27
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.485277778 0.226637555 0.829955578 1 38
hsa04971 Gastric acid secretion 0.530311028 0.233274674 0.837544207 1 53
hsa00982 Drug metabolism - cytochrome P450 0.545327016 0.238589699 0.842437303 1 35
hsa04972 Pancreatic secretion 0.513530861 0.247249009 0.851715344 1 62
hsa04110 Cell cycle 0.361281654 0.248708696 0.852123735 1 113
hsa00770 Pantothenate and CoA biosynthesis 0.444532334 0.2552472 0.853753379 1 14
hsa00640 Propanoate metabolism 0.425501602 0.258017879 0.857427712 1 29
hsa00790 Folate biosynthesis 0.524051286 0.262181962 0.859255195 1 10
hsa00010 Glycolysis / Gluconeogenesis 0.346032518 0.277225272 0.875426909 1 53
hsa04614 Renin-angiotensin system 0.487961985 0.2965293 0.883066861 1 10
hsa00040 Pentose and glucuronate interconversions 0.553628545 0.297522093 0.892276507 1 17
hsa04722 Neurotrophin signaling pathway 0.349423356 0.296981402 0.895431608 1 118
hsa00100 Steroid biosynthesis 0.513321879 0.303208041 0.89605804 1 17
hsa00052 Galactose metabolism 0.488136204 0.299776925 0.896342484 1 23
hsa00531 Glycosaminoglycan degradation 0.474554399 0.312304755 0.902576594 1 17
hsa04914 Progesterone-mediated oocyte maturation 0.542899538 0.316995636 0.909763431 1 74
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.558033864 0.323210599 0.910427174 1 17
hsa04020 Calcium signaling pathway 0.523004003 0.320665521 0.912771526 1 121
hsa00250 Alanine, aspartate and glutamate metabolism 0.534890255 0.327973968 0.914182108 1 21
hsa04670 Leukocyte transendothelial migration 0.317449549 0.325133026 0.915098985 1 91
hsa04666 Fc gamma R-mediated phagocytosis 0.26512417 0.326641851 0.916216019 1 85
hsa04962 Vasopressin-regulated water reabsorption 0.558749396 0.356276752 0.933098835 1 37
hsa00512 Mucin type O-Glycan biosynthesis 0.551808658 0.371924262 0.938767372 1 18
hsa03410 Base excision repair 0.515470076 0.370962959 0.93923509 1 31
hsa04114 Oocyte meiosis 0.479264611 0.370225054 0.94121946 1 97
hsa00510 N-Glycan biosynthesis 0.514702769 0.38240521 0.945338751 1 44
hsa03060 Protein export 0.417899291 0.396801811 0.948340373 1 23
hsa02010 ABC transporters 0.513031406 0.394894213 0.950465629 1 32
hsa00630 Glyoxylate and dicarboxylate metabolism 0.52364952 0.407462628 0.952253585 1 16
hsa04973 Carbohydrate digestion and absorption 0.524658775 0.399200271 0.953101607 1 31
hsa00564 Glycerophospholipid metabolism 0.525247324 0.414859748 0.959839252 1 63
hsa00983 Drug metabolism - other enzymes 0.514204782 0.419779579 0.960759014 1 27
hsa00561 Glycerolipid metabolism 0.596324792 0.427968945 0.964228723 1 39
hsa00310 Lysine degradation 0.558348525 0.443101954 0.968364854 1 37
hsa04622 RIG-I-like receptor signaling pathway 0.456901567 0.463690703 0.974360824 1 58
hsa04810 Regulation of actin cytoskeleton 0.319966449 0.470551959 0.976866182 1 168
hsa00520 Amino sugar and nucleotide sugar metabolism 0.458192385 0.492848731 0.980257574 1 42
hsa03030 DNA replication 0.38849312 0.502213408 0.980524982 1 36
hsa00670 One carbon pool by folate 0.59007797 0.511164476 0.98233405 1 15
hsa04260 Cardiac muscle contraction 0.410814559 0.517270132 0.984465141 1 49
hsa00500 Starch and sucrose metabolism 0.507331557 0.592419696 0.99316419 1 32
hsa00600 Sphingolipid metabolism 0.662377407 0.745837522 0.999111146 1 33
hsa04141 Protein processing in endoplasmic reticulum 0.559852183 0.852836674 0.999840586 1 152
hsa00970 Aminoacyl-tRNA biosynthesis 0.493250441 0.881659727 0.999869695 1 40
hsa04142 Lysosome 0.402794454 1.269518545 0.999999949 1 114
hsa03010 Ribosome 1.40E-06 -1.890710517 1 1 88
hsa00061 Fatty acid biosynthesis NA NA NA NA 6
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 7
hsa00120 Primary bile acid biosynthesis NA NA NA NA 9
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00300 Lysine biosynthesis NA NA NA NA 3
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 3
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 4
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00730 Thiamine metabolism NA NA NA NA 3
hsa00750 Vitamin B6 metabolism NA NA NA NA 5
hsa00780 Biotin metabolism NA NA NA NA 2
hsa00785 Lipoic acid metabolism NA NA NA NA 3
hsa04122 Sulfur relay system NA NA NA NA 9
GSE57065 p.geomean stat.mean p.val q.val set.size
hsa04610 Complement and coagulation cascades 0.006323695 2.557470086 2.86E-34 3.61E-32 22
hsa03320 PPAR signaling pathway 0.009484718 2.423348559 1.51E-31 9.49E-30 23
hsa04512 ECM-receptor interaction 0.033066077 1.752947614 5.78E-18 2.43E-16 32
hsa00190 Oxidative phosphorylation 0.033995879 1.648845137 2.61E-16 8.22E-15 55
hsa04810 Regulation of actin cytoskeleton 0.04412825 1.619092984 4.58E-16 1.16E-14 87
hsa00480 Glutathione metabolism 0.073983063 1.440816773 9.07E-13 1.90E-11 22
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.09832949 1.261096653 4.55E-10 8.19E-09 17
hsa04510 Focal adhesion 0.094310615 1.219373069 6.82E-10 1.07E-08 77
hsa04115 p53 signaling pathway 0.105763393 1.185104354 2.45E-09 3.43E-08 35
hsa00564 Glycerophospholipid metabolism 0.110800687 1.178702969 2.82E-09 3.55E-08 36
hsa04270 Vascular smooth muscle contraction 0.112105143 1.158940797 4.64E-09 5.31E-08 46
hsa04110 Cell cycle 0.084875981 1.156511344 5.05E-09 5.31E-08 73
hsa04114 Oocyte meiosis 0.106659615 1.148173946 6.18E-09 5.99E-08 57
hsa04621 NOD-like receptor signaling pathway 0.117444921 1.131738888 1.34E-08 1.21E-07 24
hsa04666 Fc gamma R-mediated phagocytosis 0.120512095 1.079777444 4.43E-08 3.72E-07 48
hsa00760 Nicotinate and nicotinamide metabolism 0.135047197 1.093036465 6.12E-08 4.82E-07 12
hsa04142 Lysosome 0.122132829 1.062334826 7.07E-08 5.24E-07 51
hsa03050 Proteasome 0.104879832 1.077954357 9.06E-08 6.34E-07 20
hsa04975 Fat digestion and absorption 0.139709561 1.076401023 1.01E-07 6.67E-07 11
hsa00982 Drug metabolism - cytochrome P450 0.132741497 1.047449477 1.63E-07 1.03E-06 18
hsa00512 Mucin type O-Glycan biosynthesis 0.145104296 1.03887001 1.86E-07 1.12E-06 16
hsa00983 Drug metabolism - other enzymes 0.153295604 1.017635843 3.37E-07 1.93E-06 14
hsa00600 Sphingolipid metabolism 0.145321237 0.996496531 5.46E-07 2.99E-06 18
hsa04914 Progesterone-mediated oocyte maturation 0.155146284 0.963241312 8.63E-07 4.53E-06 48
hsa04130 SNARE interactions in vesicular transport 0.157969674 0.927354124 3.10E-06 1.56E-05 19
hsa04620 Toll-like receptor signaling pathway 0.154301705 0.906496796 3.63E-06 1.76E-05 40
hsa00010 Glycolysis / Gluconeogenesis 0.176676377 0.882224723 6.30E-06 2.94E-05 32
hsa04380 Osteoclast differentiation 0.174794935 0.837186175 1.57E-05 7.08E-05 63
hsa04540 Gap junction 0.190927257 0.822060927 2.33E-05 0.000101035 33
hsa04910 Insulin signaling pathway 0.190240471 0.814558066 2.54E-05 0.000106635 61
hsa04740 Olfactory transduction 0.181676931 0.795203478 4.80E-05 0.000194961 21
hsa04920 Adipocytokine signaling pathway 0.208744286 0.77741916 5.86E-05 0.000230558 30
hsa00260 Glycine, serine and threonine metabolism 0.186618432 0.793688228 6.11E-05 0.000233218 13
hsa00565 Ether lipid metabolism 0.210741096 0.772445822 7.57E-05 0.000280648 15
hsa04730 Long-term depression 0.229508887 0.71919779 0.000189212 0.000681163 22
hsa04145 Phagosome 0.201873607 0.700067041 0.000250791 0.00087777 66
hsa04912 GnRH signaling pathway 0.235485716 0.692392594 0.000290031 0.000987672 38
hsa04020 Calcium signaling pathway 0.226863057 0.665419209 0.00046548 0.001543434 60
hsa04622 RIG-I-like receptor signaling pathway 0.226783165 0.66978891 0.000496501 0.00160408 19
hsa04260 Cardiac muscle contraction 0.236662851 0.664122781 0.0005113 0.001610595 26
hsa00052 Galactose metabolism 0.241525952 0.658613474 0.000603231 0.001853831 16
hsa04141 Protein processing in endoplasmic reticulum 0.21008418 0.64753844 0.000641139 0.001923416 73
hsa04976 Bile secretion 0.22367835 0.613242698 0.001270444 0.003722698 24
hsa00520 Amino sugar and nucleotide sugar metabolism 0.238195543 0.606883483 0.001355407 0.003881393 26
hsa04966 Collecting duct acid secretion 0.264173444 0.607748261 0.001447386 0.004052679 12
hsa00051 Fructose and mannose metabolism 0.275016633 0.556020365 0.003126938 0.008565092 15
hsa04370 VEGF signaling pathway 0.276809143 0.535029288 0.003947382 0.010582342 35
hsa04144 Endocytosis 0.269651939 0.495980721 0.006748619 0.017715124 83
hsa04972 Pancreatic secretion 0.297339047 0.483228191 0.008221347 0.021140605 32
hsa00240 Pyrimidine metabolism 0.284675211 0.466165507 0.010305233 0.025969188 41
hsa00380 Tryptophan metabolism 0.298913997 0.450467062 0.013536764 0.033443769 14
hsa04120 Ubiquitin mediated proteolysis 0.266374455 0.430095275 0.016161947 0.039116059 67
hsa00500 Starch and sucrose metabolism 0.242862861 0.438474035 0.016453581 0.039116059 22
hsa00330 Arginine and proline metabolism 0.307542608 0.413545829 0.020406246 0.047614574 25
hsa04623 Cytosolic DNA-sensing pathway 0.306670012 0.411779465 0.021530656 0.049324777 16
hsa04670 Leukocyte transendothelial migration 0.316107805 0.397233084 0.024100342 0.05422577 44
hsa00030 Pentose phosphate pathway 0.333170004 0.372203862 0.034033496 0.075231938 13
hsa00590 Arachidonic acid metabolism 0.323238716 0.351848932 0.040796998 0.08862796 23
hsa03440 Homologous recombination 0.335745663 0.353219158 0.041536054 0.088704116 14
hsa04664 Fc epsilon RI signaling pathway 0.337001288 0.346755923 0.04244902 0.089142941 37
hsa04146 Peroxisome 0.334309761 0.331725449 0.049778762 0.102821704 38
hsa04010 MAPK signaling pathway 0.349075014 0.325789019 0.051986252 0.10564948 115
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.350372309 0.317377344 0.059747911 0.119495823 13
hsa04630 Jak-STAT signaling pathway 0.337957504 0.307136844 0.063172461 0.124370783 58
hsa00670 One carbon pool by folate 0.355395341 0.287184159 0.079626267 0.154352456 12
hsa04210 Apoptosis 0.449418067 0.056735492 0.388846053 0.739940075 46
hsa04722 Neurotrophin signaling pathway 0.431430029 0.054288695 0.393460199 0.739940075 57
hsa03022 Basal transcription factors 0.451147091 0.033928731 0.432168589 0.800782974 18
hsa03060 Protein export 0.425263488 0.024222386 0.449584292 0.820980012 10
hsa04012 ErbB signaling pathway 0.462441166 0.017319915 0.465410076 0.837738138 35
hsa02010 ABC transporters 0.459138946 0.002992497 0.495453384 0.879255301 14
hsa00071 Fatty acid metabolism 0.482006474 -0.012001475 0.523663412 0.916410972 17
hsa04530 Tight junction 0.489833858 -0.019501103 0.538595435 0.929630477 44
hsa04360 Axon guidance 0.460183028 -0.041949749 0.582766049 0.992277327 47
hsa04974 Protein digestion and absorption 0.443277041 -0.055794664 0.607741176 1 22
hsa00350 Tyrosine metabolism 0.47591606 -0.09133216 0.673147094 1 18
hsa00230 Purine metabolism 0.508506455 -0.122534621 0.729318147 1 68
hsa04962 Vasopressin-regulated water reabsorption 0.538837605 -0.160162354 0.785621423 1 17
hsa04720 Long-term potentiation 0.551346292 -0.180977424 0.815553104 1 27
hsa00514 Other types of O-glycan biosynthesis 0.555359733 -0.213419928 0.852991089 1 14
hsa00561 Glycerolipid metabolism 0.554455185 -0.21241531 0.853152495 1 17
hsa04971 Gastric acid secretion 0.560430398 -0.226702517 0.869182106 1 22
hsa04150 mTOR signaling pathway 0.56730353 -0.234823538 0.877524828 1 21
hsa04742 Taste transduction 0.553295103 -0.256586374 0.893726374 1 10
hsa04970 Salivary secretion 0.557646711 -0.253868279 0.89585237 1 31
hsa00830 Retinol metabolism 0.576146507 -0.281975685 0.917646471 1 16
hsa00640 Propanoate metabolism 0.588731209 -0.284453623 0.920097185 1 18
hsa00340 Histidine metabolism 0.556999019 -0.310346216 0.934645239 1 13
hsa00620 Pyruvate metabolism 0.569929301 -0.310559692 0.937656104 1 23
hsa03420 Nucleotide excision repair 0.564507152 -0.323576625 0.944667413 1 25
hsa04350 TGF-beta signaling pathway 0.603247234 -0.336026743 0.95255432 1 35
hsa04520 Adherens junction 0.606326925 -0.373886164 0.967849527 1 24
hsa04330 Notch signaling pathway 0.59429091 -0.379344691 0.969324297 1 24
hsa04960 Aldosterone-regulated sodium reabsorption 0.632741574 -0.416445033 0.978932102 1 10
hsa00310 Lysine degradation 0.630090024 -0.462072457 0.988689747 1 22
hsa04916 Melanogenesis 0.646485588 -0.460283933 0.988879279 1 34
hsa04973 Carbohydrate digestion and absorption 0.563684556 -0.470555615 0.989102108 1 18
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.658066183 -0.503942878 0.993264607 1 13
hsa00270 Cysteine and methionine metabolism 0.679659665 -0.544775497 0.996385101 1 17
hsa04340 Hedgehog signaling pathway 0.678342919 -0.600543156 0.998269544 1 11
hsa03430 Mismatch repair 0.698681355 -0.630980814 0.998908488 1 11
hsa00562 Inositol phosphate metabolism 0.719329022 -0.639779258 0.999231949 1 26
hsa04070 Phosphatidylinositol signaling system 0.752229232 -0.78148155 0.999946204 1 34
hsa00510 N-Glycan biosynthesis 0.742642409 -0.804923087 0.999964123 1 24
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.754347074 -0.827410274 0.99996613 1 10
hsa04062 Chemokine signaling pathway 0.725827791 -0.831323797 0.999982373 1 76
hsa03030 DNA replication 0.72731904 -0.855610501 0.999984305 1 17
hsa04640 Hematopoietic cell lineage 0.732854786 -0.849686685 0.999987777 1 58
hsa04662 B cell receptor signaling pathway 0.760958496 -0.898786315 0.999995839 1 44
hsa00020 Citrate cycle (TCA cycle) 0.742066421 -0.929314515 0.999996686 1 17
hsa03018 RNA degradation 0.746401894 -0.916010176 0.999997028 1 40
hsa00280 Valine, leucine and isoleucine degradation 0.760691731 -0.957904813 0.999998633 1 21
hsa00410 beta-Alanine metabolism 0.769873659 -0.98251582 0.999998816 1 11
hsa00970 Aminoacyl-tRNA biosynthesis 0.830592233 -1.07549933 0.999999915 1 15
hsa03410 Base excision repair 0.809762962 -1.079736532 0.999999921 1 15
hsa04310 Wnt signaling pathway 0.829752605 -1.075144023 0.999999955 1 62
hsa03010 Ribosome 0.600564096 -1.103220314 0.999999967 1 50
hsa03015 mRNA surveillance pathway 0.854753573 -1.17004162 0.999999997 1 43
hsa03040 Spliceosome 0.882692491 -1.608389136 1 1 65
hsa03008 Ribosome biogenesis in eukaryotes 0.933492962 -2.697995688 1 1 44
hsa03013 RNA transport 0.902985199 -2.195454485 1 1 83
hsa04514 Cell adhesion molecules (CAMs) 0.949170919 -2.153334239 1 1 57
hsa04612 Antigen processing and presentation 0.952883458 -3.234270584 1 1 52
hsa04650 Natural killer cell mediated cytotoxicity 0.782493364 -1.926989937 1 1 62
hsa04660 T cell receptor signaling pathway 0.928005785 -1.862981781 1 1 60
hsa04672 Intestinal immune network for IgA production 0.982274511 -2.737004834 1 1 29
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 8
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 6
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 2
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 4
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00140 Steroid hormone biosynthesis NA NA NA NA 8
hsa00232 Caffeine metabolism NA NA NA NA 2
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 5
hsa00300 Lysine biosynthesis NA NA NA NA 1
hsa00360 Phenylalanine metabolism NA NA NA NA 9
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 6
hsa00450 Selenocompound metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 4
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 6
hsa00531 Glycosaminoglycan degradation NA NA NA NA 5
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 8
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 6
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 8
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 7
hsa00650 Butanoate metabolism NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 4
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 9
hsa00780 Biotin metabolism NA NA NA NA 1
hsa00785 Lipoic acid metabolism NA NA NA NA 1
hsa00790 Folate biosynthesis NA NA NA NA 6
hsa00860 Porphyrin and chlorophyll metabolism NA NA NA NA 8
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 6
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 5
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 5
hsa03020 RNA polymerase NA NA NA NA 7
hsa03450 Non-homologous end-joining NA NA NA NA 7
hsa04122 Sulfur relay system NA NA NA NA 4
hsa04140 Regulation of autophagy NA NA NA NA 9
hsa04320 Dorso-ventral axis formation NA NA NA NA 9
hsa04614 Renin-angiotensin system NA NA NA NA 7
hsa04710 Circadian rhythm - mammal NA NA NA NA 8
hsa04744 Phototransduction NA NA NA NA 7
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 8
hsa04977 Vitamin digestion and absorption NA NA NA NA 6
hsa04612 Antigen processing and presentation 0.000523855 -3.234270584 2.90E-55 3.66E-53 52
hsa04672 Intestinal immune network for IgA production 0.003608133 -2.737004834 1.45E-39 9.14E-38 29
hsa03008 Ribosome biogenesis in eukaryotes 0.002848282 -2.697995688 7.18E-39 3.01E-37 44
hsa03013 RNA transport 0.008861591 -2.195454485 1.67E-27 5.26E-26 83
hsa04514 Cell adhesion molecules (CAMs) 0.013269218 -2.153334239 1.64E-26 4.13E-25 57
hsa04650 Natural killer cell mediated cytotoxicity 0.016188012 -1.926989937 1.29E-21 2.71E-20 62
hsa04660 T cell receptor signaling pathway 0.027671058 -1.862981781 1.71E-20 3.08E-19 60
hsa03040 Spliceosome 0.043947735 -1.608389136 1.03E-15 1.62E-14 65
hsa03015 mRNA surveillance pathway 0.115837243 -1.17004162 3.47E-09 4.86E-08 43
hsa03010 Ribosome 0.07335328 -1.103220314 3.33E-08 4.19E-07 50
hsa04310 Wnt signaling pathway 0.1326302 -1.075144023 4.54E-08 5.20E-07 62
hsa03410 Base excision repair 0.131498851 -1.079736532 7.90E-08 8.26E-07 15
hsa00970 Aminoacyl-tRNA biosynthesis 0.139709149 -1.07549933 8.52E-08 8.26E-07 15
hsa00410 beta-Alanine metabolism 0.14997983 -0.98251582 1.18E-06 1.07E-05 11
hsa00280 Valine, leucine and isoleucine degradation 0.149152517 -0.957904813 1.37E-06 1.15E-05 21
hsa03018 RNA degradation 0.153556353 -0.916010176 2.97E-06 2.34E-05 40
hsa00020 Citrate cycle (TCA cycle) 0.152560813 -0.929314515 3.31E-06 2.46E-05 17
hsa04662 B cell receptor signaling pathway 0.164107074 -0.898786315 4.16E-06 2.91E-05 44
hsa04640 Hematopoietic cell lineage 0.170597846 -0.849686685 1.22E-05 8.11E-05 58
hsa03030 DNA replication 0.171875001 -0.855610501 1.57E-05 9.89E-05 17
hsa04062 Chemokine signaling pathway 0.173611772 -0.831323797 1.76E-05 0.000105764 76
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.197394403 -0.827410274 3.39E-05 0.000193981 10
hsa00510 N-Glycan biosynthesis 0.194260047 -0.804923087 3.59E-05 0.000196544 24
hsa04070 Phosphatidylinositol signaling system 0.205451724 -0.78148155 5.38E-05 0.00028243 34
hsa00562 Inositol phosphate metabolism 0.253258602 -0.639779258 0.000768051 0.003870975 26
hsa03430 Mismatch repair 0.252554997 -0.630980814 0.001091512 0.005289635 11
hsa04340 Hedgehog signaling pathway 0.257368442 -0.600543156 0.001730456 0.008075459 11
hsa00270 Cysteine and methionine metabolism 0.280645536 -0.544775497 0.003614899 0.016267044 17
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.291398593 -0.503942878 0.006735393 0.02926412 13
hsa04973 Carbohydrate digestion and absorption 0.254854847 -0.470555615 0.010897892 0.044534123 18
hsa04916 Melanogenesis 0.304475636 -0.460283933 0.011120721 0.044534123 34
hsa00310 Lysine degradation 0.295033806 -0.462072457 0.011310253 0.044534123 22
hsa04960 Aldosterone-regulated sodium reabsorption 0.324334045 -0.416445033 0.021067898 0.080441067 10
hsa04330 Notch signaling pathway 0.317887841 -0.379344691 0.030675703 0.113680548 24
hsa04520 Adherens junction 0.328442497 -0.373886164 0.032150473 0.115741704 24
hsa04350 TGF-beta signaling pathway 0.349250592 -0.336026743 0.04744568 0.166059879 35
hsa03420 Nucleotide excision repair 0.329832375 -0.323576625 0.055332587 0.18842989 25
hsa00620 Pyruvate metabolism 0.342572971 -0.310559692 0.062343896 0.206719234 23
hsa00340 Histidine metabolism 0.333928116 -0.310346216 0.065354761 0.211146149 13
hsa00640 Propanoate metabolism 0.372682622 -0.284453623 0.079902815 0.251693867 18
hsa00830 Retinol metabolism 0.364943768 -0.281975685 0.082353529 0.253086455 16
hsa04970 Salivary secretion 0.367057709 -0.253868279 0.10414763 0.311406438 31
hsa04742 Taste transduction 0.3658902 -0.256586374 0.106273626 0.311406438 10
hsa04150 mTOR signaling pathway 0.389306737 -0.234823538 0.122475172 0.350724356 21
hsa04971 Gastric acid secretion 0.388325221 -0.226702517 0.130817894 0.366290102 22
hsa00561 Glycerolipid metabolism 0.394173247 -0.21241531 0.146847505 0.394108996 17
hsa00514 Other types of O-glycan biosynthesis 0.393926675 -0.213419928 0.147008911 0.394108996 14
hsa04720 Long-term potentiation 0.411840939 -0.180977424 0.184446896 0.484173102 27
hsa04962 Vasopressin-regulated water reabsorption 0.416474537 -0.160162354 0.214378577 0.551259198 17
hsa00230 Purine metabolism 0.416052114 -0.122534621 0.270681853 0.68211827 68
hsa00350 Tyrosine metabolism 0.409470461 -0.09133216 0.326852906 0.807518943 18
hsa04974 Protein digestion and absorption 0.403771361 -0.055794664 0.392258824 0.950473304 22
hsa04360 Axon guidance 0.430196156 -0.041949749 0.417233951 0.991914676 47
hsa04530 Tight junction 0.474244569 -0.019501103 0.461404565 1 44
hsa00071 Fatty acid metabolism 0.472677313 -0.012001475 0.476336588 1 17
hsa02010 ABC transporters 0.46215154 0.002992497 0.504546616 1 14
hsa04012 ErbB signaling pathway 0.474935018 0.017319915 0.534589924 1 35
hsa03060 Protein export 0.443777006 0.024222386 0.550415708 1 10
hsa03022 Basal transcription factors 0.474863816 0.033928731 0.567831411 1 18
hsa04722 Neurotrophin signaling pathway 0.472238096 0.054288695 0.606539801 1 57
hsa04210 Apoptosis 0.493081991 0.056735492 0.611153947 1 46
hsa00670 One carbon pool by folate 0.564890872 0.287184159 0.920373733 1 12
hsa04630 Jak-STAT signaling pathway 0.563463798 0.307136844 0.936827539 1 58
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.585617582 0.317377344 0.940252089 1 13
hsa04010 MAPK signaling pathway 0.596063 0.325789019 0.948013748 1 115
hsa04146 Peroxisome 0.579722145 0.331725449 0.950221238 1 38
hsa04664 Fc epsilon RI signaling pathway 0.595183688 0.346755923 0.95755098 1 37
hsa03440 Homologous recombination 0.595740168 0.353219158 0.958463946 1 14
hsa00590 Arachidonic acid metabolism 0.573712725 0.351848932 0.959203002 1 23
hsa00030 Pentose phosphate pathway 0.608276192 0.372203862 0.965966504 1 13
hsa04670 Leukocyte transendothelial migration 0.607902507 0.397233084 0.975899658 1 44
hsa04623 Cytosolic DNA-sensing pathway 0.602867187 0.411779465 0.978469344 1 16
hsa00330 Arginine and proline metabolism 0.60728957 0.413545829 0.979593754 1 25
hsa00500 Starch and sucrose metabolism 0.531845906 0.438474035 0.983546419 1 22
hsa04120 Ubiquitin mediated proteolysis 0.550911734 0.430095275 0.983838053 1 67
hsa00380 Tryptophan metabolism 0.62017177 0.450467062 0.986463236 1 14
hsa00240 Pyrimidine metabolism 0.619162397 0.466165507 0.989694767 1 41
hsa04972 Pancreatic secretion 0.655654595 0.483228191 0.991778653 1 32
hsa04144 Endocytosis 0.623815554 0.495980721 0.993251381 1 83
hsa04370 VEGF signaling pathway 0.666834121 0.535029288 0.996052618 1 35
hsa00051 Fructose and mannose metabolism 0.678419297 0.556020365 0.996873062 1 15
hsa04966 Collecting duct acid secretion 0.704166055 0.607748261 0.998552614 1 12
hsa00520 Amino sugar and nucleotide sugar metabolism 0.656216144 0.606883483 0.998644593 1 26
hsa04976 Bile secretion 0.628433114 0.613242698 0.998729556 1 24
hsa04141 Protein processing in endoplasmic reticulum 0.644324968 0.64753844 0.999358861 1 73
hsa00052 Galactose metabolism 0.707882858 0.658613474 0.999396769 1 16
hsa04260 Cardiac muscle contraction 0.71065169 0.664122781 0.9994887 1 26
hsa04622 RIG-I-like receptor signaling pathway 0.692420987 0.66978891 0.999503499 1 19
hsa04020 Calcium signaling pathway 0.694595944 0.665419209 0.99953452 1 60
hsa04912 GnRH signaling pathway 0.734398781 0.692392594 0.999709969 1 38
hsa04145 Phagosome 0.675665804 0.700067041 0.999749209 1 66
hsa04730 Long-term depression 0.743032726 0.71919779 0.999810788 1 22
hsa00565 Ether lipid metabolism 0.747723017 0.772445822 0.999924269 1 15
hsa00260 Glycine, serine and threonine metabolism 0.704776342 0.793688228 0.999938919 1 13
hsa04920 Adipocytokine signaling pathway 0.755692937 0.77741916 0.999941446 1 30
hsa04740 Olfactory transduction 0.703626954 0.795203478 0.999952033 1 21
hsa04910 Insulin signaling pathway 0.750087893 0.814558066 0.999974611 1 61
hsa04540 Gap junction 0.756074517 0.822060927 0.999976746 1 33
hsa04380 Osteoclast differentiation 0.732672682 0.837186175 0.99998426 1 63
hsa00010 Glycolysis / Gluconeogenesis 0.776599182 0.882224723 0.999993698 1 32
hsa04620 Toll-like receptor signaling pathway 0.741048659 0.906496796 0.999996367 1 40
hsa04130 SNARE interactions in vesicular transport 0.756803545 0.927354124 0.999996903 1 19
hsa04914 Progesterone-mediated oocyte maturation 0.794016914 0.963241312 0.999999137 1 48
hsa00600 Sphingolipid metabolism 0.784293464 0.996496531 0.999999454 1 18
hsa00983 Drug metabolism - other enzymes 0.822685832 1.017635843 0.999999663 1 14
hsa00512 Mucin type O-Glycan biosynthesis 0.820575761 1.03887001 0.999999814 1 16
hsa00982 Drug metabolism - cytochrome P450 0.789039557 1.047449477 0.999999837 1 18
hsa04975 Fat digestion and absorption 0.826605566 1.076401023 0.999999899 1 11
hsa03050 Proteasome 0.716775376 1.077954357 0.999999909 1 20
hsa04142 Lysosome 0.78760498 1.062334826 0.999999929 1 51
hsa00760 Nicotinate and nicotinamide metabolism 0.827350269 1.093036465 0.999999939 1 12
hsa04666 Fc gamma R-mediated phagocytosis 0.794899718 1.079777444 0.999999956 1 48
hsa04621 NOD-like receptor signaling pathway 0.821443303 1.131738888 0.999999987 1 24
hsa04114 Oocyte meiosis 0.805424104 1.148173946 0.999999994 1 57
hsa04110 Cell cycle 0.737795564 1.156511344 0.999999995 1 73
hsa04270 Vascular smooth muscle contraction 0.834560817 1.158940797 0.999999995 1 46
hsa00564 Glycerophospholipid metabolism 0.845579225 1.178702969 0.999999997 1 36
hsa04115 p53 signaling pathway 0.82909321 1.185104354 0.999999998 1 35
hsa04510 Focal adhesion 0.821324739 1.219373069 0.999999999 1 77
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.849546822 1.261096653 1 1 17
hsa00480 Glutathione metabolism 0.901997925 1.440816773 1 1 22
hsa04810 Regulation of actin cytoskeleton 0.888693247 1.619092984 1 1 87
hsa00190 Oxidative phosphorylation 0.839757082 1.648845137 1 1 55
hsa03320 PPAR signaling pathway 0.985572277 2.423348559 1 1 23
hsa04512 ECM-receptor interaction 0.889855434 1.752947614 1 1 32
hsa04610 Complement and coagulation cascades 0.975002192 2.557470086 1 1 22
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 8
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 6
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 2
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 4
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00140 Steroid hormone biosynthesis NA NA NA NA 8
hsa00232 Caffeine metabolism NA NA NA NA 2
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 5
hsa00300 Lysine biosynthesis NA NA NA NA 1
hsa00360 Phenylalanine metabolism NA NA NA NA 9
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 6
hsa00450 Selenocompound metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 4
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 6
hsa00531 Glycosaminoglycan degradation NA NA NA NA 5
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 8
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 6
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 8
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 7
hsa00650 Butanoate metabolism NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 4
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 9
hsa00780 Biotin metabolism NA NA NA NA 1
hsa00785 Lipoic acid metabolism NA NA NA NA 1
hsa00790 Folate biosynthesis NA NA NA NA 6
hsa00860 Porphyrin and chlorophyll metabolism NA NA NA NA 8
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 6
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 5
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 5
hsa03020 RNA polymerase NA NA NA NA 7
hsa03450 Non-homologous end-joining NA NA NA NA 7
hsa04122 Sulfur relay system NA NA NA NA 4
hsa04140 Regulation of autophagy NA NA NA NA 9
hsa04320 Dorso-ventral axis formation NA NA NA NA 9
hsa04614 Renin-angiotensin system NA NA NA NA 7
hsa04710 Circadian rhythm - mammal NA NA NA NA 8
hsa04744 Phototransduction NA NA NA NA 7
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 8
hsa04977 Vitamin digestion and absorption NA NA NA NA 6
GSE95233 p.geomean stat.mean p.val q.val set.size
hsa00190 Oxidative phosphorylation 0.002405527 2.799196786 5.20E-38 6.45E-36 63
hsa03320 PPAR signaling pathway 0.023874351 2.041525896 3.13E-20 1.94E-18 17
hsa04610 Complement and coagulation cascades 0.027170072 1.925687416 1.70E-18 7.01E-17 20
hsa00600 Sphingolipid metabolism 0.043531155 1.739475885 1.97E-15 6.09E-14 16
hsa04115 p53 signaling pathway 0.059317356 1.532608268 8.32E-13 2.06E-11 30
hsa04260 Cardiac muscle contraction 0.064059077 1.478909164 4.89E-12 1.01E-10 30
hsa00480 Glutathione metabolism 0.074180722 1.419136235 4.36E-11 7.72E-10 20
hsa04621 NOD-like receptor signaling pathway 0.082022662 1.347229837 2.47E-10 3.71E-09 31
hsa04114 Oocyte meiosis 0.0802321 1.336928384 2.69E-10 3.71E-09 49
hsa00770 Pantothenate and CoA biosynthesis 0.09051072 1.367442579 4.36E-10 5.40E-09 10
hsa04110 Cell cycle 0.077352396 1.293514089 9.25E-10 1.04E-08 64
hsa04130 SNARE interactions in vesicular transport 0.120251197 1.166149092 4.17E-08 4.31E-07 23
hsa04966 Collecting duct acid secretion 0.121037232 1.181083 5.34E-08 5.09E-07 10
hsa00564 Glycerophospholipid metabolism 0.11804511 1.137647858 6.87E-08 6.08E-07 31
hsa00512 Mucin type O-Glycan biosynthesis 0.12296791 1.128623959 1.37E-07 1.13E-06 15
hsa04142 Lysosome 0.147791752 0.96042121 3.92E-06 3.04E-05 52
hsa03010 Ribosome 0.101761727 0.952292575 5.58E-06 4.07E-05 57
hsa03050 Proteasome 0.155425607 0.916588863 1.18E-05 8.13E-05 28
hsa04810 Regulation of actin cytoskeleton 0.153795464 0.902471843 1.28E-05 8.34E-05 74
hsa00760 Nicotinate and nicotinamide metabolism 0.172569009 0.918897089 1.65E-05 0.000102243 11
hsa00500 Starch and sucrose metabolism 0.176279294 0.896219269 2.13E-05 0.00012579 13
hsa04622 RIG-I-like receptor signaling pathway 0.18326514 0.832021411 5.99E-05 0.000337554 25
hsa04914 Progesterone-mediated oocyte maturation 0.187167838 0.820907802 6.61E-05 0.000355288 44
hsa04540 Gap junction 0.190249119 0.822086341 6.88E-05 0.000355288 27
hsa04512 ECM-receptor interaction 0.198250317 0.776777836 0.000184832 0.000916769 16
hsa04620 Toll-like receptor signaling pathway 0.16949466 0.757073757 0.000220714 0.001052638 43
hsa00983 Drug metabolism - other enzymes 0.225893716 0.733123885 0.000357773 0.001643104 16
hsa00010 Glycolysis / Gluconeogenesis 0.223996747 0.71951314 0.000418399 0.001852911 28
hsa00565 Ether lipid metabolism 0.229568047 0.699884077 0.000680638 0.002910313 12
hsa04141 Protein processing in endoplasmic reticulum 0.226173816 0.681900788 0.000719969 0.002975871 77
hsa04666 Fc gamma R-mediated phagocytosis 0.229237667 0.65880283 0.001072398 0.004289592 45
hsa03060 Protein export 0.249549271 0.640082208 0.00175819 0.006812988 12
hsa04510 Focal adhesion 0.260932042 0.562041878 0.004324374 0.015891748 65
hsa04910 Insulin signaling pathway 0.24335982 0.561929824 0.004416662 0.015891748 53
hsa04730 Long-term depression 0.271049583 0.564432997 0.004485574 0.015891748 19
hsa00240 Pyrimidine metabolism 0.267918513 0.549812329 0.005175318 0.017826094 46
hsa04912 GnRH signaling pathway 0.280525461 0.546837186 0.005397301 0.018088251 35
hsa00052 Galactose metabolism 0.290773336 0.536632657 0.006730104 0.021509737 12
hsa04120 Ubiquitin mediated proteolysis 0.268014629 0.528831177 0.006765159 0.021509737 62
hsa03022 Basal transcription factors 0.30664421 0.476814175 0.01358191 0.042103921 19
hsa00520 Amino sugar and nucleotide sugar metabolism 0.304550057 0.469809265 0.014674645 0.044381854 20
hsa04270 Vascular smooth muscle contraction 0.292864477 0.459014684 0.016213707 0.04786904 40
hsa04140 Regulation of autophagy 0.304982413 0.463150401 0.01724683 0.049735045 10
hsa04920 Adipocytokine signaling pathway 0.31954882 0.419574248 0.025854521 0.07286274 19
hsa00982 Drug metabolism - cytochrome P450 0.32054652 0.375061263 0.042630916 0.117471858 15
hsa04145 Phagosome 0.305152604 0.355111418 0.048713175 0.131313776 63
hsa04976 Bile secretion 0.361439108 0.292586915 0.087524971 0.230916944 19
hsa00410 beta-Alanine metabolism 0.371970908 0.285064325 0.094677734 0.244584147 10
hsa00670 One carbon pool by folate 0.364183609 0.262694129 0.114236267 0.289087696 10
hsa04623 Cytosolic DNA-sensing pathway 0.366978383 0.254134941 0.119304662 0.293481185 23
hsa00590 Arachidonic acid metabolism 0.378843844 0.25495875 0.120705971 0.293481185 11
hsa00051 Fructose and mannose metabolism 0.408307276 0.181185009 0.200480688 0.46459696 17
hsa00140 Steroid hormone biosynthesis 0.392742352 0.182230976 0.201691477 0.46459696 11
hsa04144 Endocytosis 0.385298326 0.178315479 0.202324483 0.46459696 80
hsa00330 Arginine and proline metabolism 0.400007749 0.174296707 0.209125745 0.471483497 23
hsa04380 Osteoclast differentiation 0.370299683 0.164313504 0.221069318 0.489510633 68
hsa00030 Pentose phosphate pathway 0.432383801 0.144500958 0.252945098 0.550266529 10
hsa04146 Peroxisome 0.429049467 0.130951373 0.270787726 0.578048747 31
hsa04010 MAPK signaling pathway 0.399691392 0.12767921 0.275039323 0.578048747 100
hsa03420 Nucleotide excision repair 0.421770752 0.111723024 0.301650743 0.623411536 26
hsa03430 Mismatch repair 0.425176367 0.105504754 0.314511617 0.639335091 11
hsa04974 Protein digestion and absorption 0.409096433 0.086254184 0.345501396 0.680383033 17
hsa04664 Fc epsilon RI signaling pathway 0.430407946 0.085270771 0.345678477 0.680383033 30
hsa04972 Pancreatic secretion 0.451248965 0.060689464 0.389054944 0.753793954 22
hsa04962 Vasopressin-regulated water reabsorption 0.471824615 0.03268503 0.439642548 0.838702707 19
hsa04740 Olfactory transduction 0.461171799 0.00897602 0.483279626 0.907979902 13
hsa04960 Aldosterone-regulated sodium reabsorption 0.476469895 0.001433239 0.496890405 0.919618063 14
hsa03015 mRNA surveillance pathway 0.483397082 -0.01291453 0.523870469 0.949097087 28
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.46091798 -0.017370575 0.532495006 0.949097087 15
hsa04150 mTOR signaling pathway 0.47292885 -0.019289802 0.535780613 0.949097087 23
hsa04340 Hedgehog signaling pathway 0.462662989 -0.031613012 0.557810197 0.970953798 14
hsa00620 Pyruvate metabolism 0.500093423 -0.037140756 0.568413811 0.970953798 16
hsa00640 Propanoate metabolism 0.493372052 -0.039010701 0.571609897 0.970953798 14
hsa04330 Notch signaling pathway 0.486978011 -0.045831183 0.583663284 0.971335763 16
hsa03018 RNA degradation 0.491614107 -0.047445635 0.587501469 0.971335763 31
hsa00020 Citrate cycle (TCA cycle) 0.505988796 -0.05475885 0.600121557 0.977247166 17
hsa00071 Fatty acid metabolism 0.504199231 -0.058851455 0.606838966 0.977247166 16
hsa04020 Calcium signaling pathway 0.516302166 -0.153175556 0.762847733 1 54
hsa00380 Tryptophan metabolism 0.518422045 -0.175853273 0.788310377 1 12
hsa04360 Axon guidance 0.509780843 -0.18935995 0.811219373 1 41
hsa04210 Apoptosis 0.536704845 -0.190111157 0.812507383 1 49
hsa04973 Carbohydrate digestion and absorption 0.546063843 -0.196097416 0.817571972 1 14
hsa04971 Gastric acid secretion 0.54938446 -0.211472955 0.836938788 1 24
hsa04370 VEGF signaling pathway 0.550486729 -0.212760948 0.839042436 1 31
hsa04720 Long-term potentiation 0.557502193 -0.255018734 0.881460693 1 29
hsa03030 DNA replication 0.539718677 -0.280035079 0.902772456 1 19
hsa03040 Spliceosome 0.565471284 -0.314034859 0.928633923 1 62
hsa00561 Glycerolipid metabolism 0.604228446 -0.34748943 0.945763894 1 16
hsa04670 Leukocyte transendothelial migration 0.594623185 -0.3498243 0.948501202 1 41
hsa00230 Purine metabolism 0.60064389 -0.398751409 0.968850921 1 67
hsa00280 Valine, leucine and isoleucine degradation 0.61161551 -0.405330006 0.969706615 1 22
hsa03020 RNA polymerase 0.627935963 -0.420896642 0.973794645 1 15
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.629197644 -0.425756424 0.973819205 1 10
hsa04916 Melanogenesis 0.62085355 -0.424410724 0.97578663 1 31
hsa00270 Cysteine and methionine metabolism 0.64261013 -0.435500686 0.978174685 1 18
hsa04970 Salivary secretion 0.655957267 -0.481615545 0.987497686 1 29
hsa00514 Other types of O-glycan biosynthesis 0.666225585 -0.517012131 0.991443934 1 14
hsa04662 B cell receptor signaling pathway 0.640081314 -0.517485781 0.991968065 1 41
hsa03410 Base excision repair 0.653636769 -0.526089022 0.992113123 1 14
hsa00310 Lysine degradation 0.66470473 -0.528728928 0.992689874 1 17
hsa04350 TGF-beta signaling pathway 0.695985624 -0.566944806 0.99582273 1 26
hsa00510 N-Glycan biosynthesis 0.668029074 -0.573956449 0.996087483 1 22
hsa00830 Retinol metabolism 0.70030628 -0.612940663 0.997594785 1 13
hsa04710 Circadian rhythm - mammal 0.707529521 -0.62694447 0.997934098 1 10
hsa04520 Adherens junction 0.70653717 -0.627616024 0.998206007 1 22
hsa04722 Neurotrophin signaling pathway 0.677203681 -0.643469671 0.998638926 1 50
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.724090055 -0.693820692 0.999216523 1 10
hsa04012 ErbB signaling pathway 0.713986543 -0.715104655 0.999546754 1 30
hsa00562 Inositol phosphate metabolism 0.757191001 -0.786642798 0.999872327 1 31
hsa04742 Taste transduction 0.802124012 -0.968677766 0.999994491 1 11
hsa04630 Jak-STAT signaling pathway 0.788789658 -0.94752164 0.999995032 1 60
hsa04070 Phosphatidylinositol signaling system 0.811839185 -0.962674659 0.999996316 1 42
hsa04530 Tight junction 0.802053756 -1.008323496 0.99999856 1 34
hsa04310 Wnt signaling pathway 0.816982134 -1.034466898 0.999999277 1 49
hsa00970 Aminoacyl-tRNA biosynthesis 0.825350021 -1.099066804 0.999999584 1 10
hsa04062 Chemokine signaling pathway 0.808304824 -1.110360608 0.999999888 1 76
hsa04640 Hematopoietic cell lineage 0.889669695 -1.499443554 1 1 46
hsa03013 RNA transport 0.899836378 -1.557272524 1 1 61
hsa03008 Ribosome biogenesis in eukaryotes 0.889365877 -1.642285908 1 1 37
hsa04514 Cell adhesion molecules (CAMs) 0.99642481 -3.492975858 1 1 56
hsa04612 Antigen processing and presentation 0.996742098 -4.19632946 1 1 48
hsa04650 Natural killer cell mediated cytotoxicity 0.956887957 -2.589426258 1 1 69
hsa04660 T cell receptor signaling pathway 0.984081578 -2.447563982 1 1 57
hsa04672 Intestinal immune network for IgA production 0.986454584 -2.868405959 1 1 25
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 5
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 4
hsa00061 Fatty acid biosynthesis NA NA NA NA 1
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 3
hsa00100 Steroid biosynthesis NA NA NA NA 8
hsa00120 Primary bile acid biosynthesis NA NA NA NA 2
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 3
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 9
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 3
hsa00300 Lysine biosynthesis NA NA NA NA 1
hsa00340 Histidine metabolism NA NA NA NA 7
hsa00350 Tyrosine metabolism NA NA NA NA 8
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 1
hsa00450 Selenocompound metabolism NA NA NA NA 4
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 7
hsa00531 Glycosaminoglycan degradation NA NA NA NA 7
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate NA NA NA NA 8
hsa00591 Linoleic acid metabolism NA NA NA NA 6
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 4
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 8
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 9
hsa00650 Butanoate metabolism NA NA NA NA 5
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 3
hsa00750 Vitamin B6 metabolism NA NA NA NA 2
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 1
hsa00790 Folate biosynthesis NA NA NA NA 4
hsa00860 Porphyrin and chlorophyll metabolism NA NA NA NA 6
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 7
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 4
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 7
hsa02010 ABC transporters NA NA NA NA 9
hsa03440 Homologous recombination NA NA NA NA 8
hsa03450 Non-homologous end-joining NA NA NA NA 7
hsa04122 Sulfur relay system NA NA NA NA 2
hsa04320 Dorso-ventral axis formation NA NA NA NA 7
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04744 Phototransduction NA NA NA NA 8
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 7
hsa04975 Fat digestion and absorption NA NA NA NA 9
hsa04977 Vitamin digestion and absorption NA NA NA NA 7
hsa04612 Antigen processing and presentation 2.27E-05 -4.19632946 7.01E-78 8.69E-76 48
hsa04514 Cell adhesion molecules (CAMs) 0.000275167 -3.492975858 7.12E-57 4.41E-55 56
hsa04672 Intestinal immune network for IgA production 0.002645237 -2.868405959 1.68E-37 6.93E-36 25
hsa04650 Natural killer cell mediated cytotoxicity 0.003747811 -2.589426258 4.69E-33 1.45E-31 69
hsa04660 T cell receptor signaling pathway 0.007193874 -2.447563982 8.09E-30 2.01E-28 57
hsa03008 Ribosome biogenesis in eukaryotes 0.043118563 -1.642285908 2.12E-14 4.38E-13 37
hsa03013 RNA transport 0.053971972 -1.557272524 2.42E-13 4.30E-12 61
hsa04640 Hematopoietic cell lineage 0.058596705 -1.499443554 1.99E-12 3.08E-11 46
hsa04062 Chemokine signaling pathway 0.114471919 -1.110360608 1.12E-07 1.55E-06 76
hsa00970 Aminoacyl-tRNA biosynthesis 0.136777386 -1.099066804 4.16E-07 5.16E-06 10
hsa04310 Wnt signaling pathway 0.140968001 -1.034466898 7.23E-07 8.15E-06 49
hsa04530 Tight junction 0.144967977 -1.008323496 1.44E-06 1.49E-05 34
hsa04070 Phosphatidylinositol signaling system 0.161567494 -0.962674659 3.68E-06 3.51E-05 42
hsa04630 Jak-STAT signaling pathway 0.156512192 -0.94752164 4.97E-06 4.40E-05 60
hsa04742 Taste transduction 0.164815459 -0.968677766 5.51E-06 4.55E-05 11
hsa00562 Inositol phosphate metabolism 0.20563815 -0.786642798 0.000127673 0.000989466 31
hsa04012 ErbB signaling pathway 0.215333087 -0.715104655 0.000453246 0.003306029 30
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.236119334 -0.693820692 0.000783477 0.005397286 10
hsa04722 Neurotrophin signaling pathway 0.227714622 -0.643469671 0.001361074 0.008882797 50
hsa04520 Adherens junction 0.253445218 -0.627616024 0.001793993 0.011122755 22
hsa04710 Circadian rhythm - mammal 0.258587199 -0.62694447 0.002065902 0.012198657 10
hsa00830 Retinol metabolism 0.259222479 -0.612940663 0.002405215 0.013556667 13
hsa00510 N-Glycan biosynthesis 0.260816553 -0.573956449 0.003912517 0.021093567 22
hsa04350 TGF-beta signaling pathway 0.276808272 -0.566944806 0.00417727 0.021582563 26
hsa00310 Lysine degradation 0.280948854 -0.528728928 0.007310126 0.036258226 17
hsa03410 Base excision repair 0.277480592 -0.526089022 0.007886877 0.036887404 14
hsa04662 B cell receptor signaling pathway 0.267297743 -0.517485781 0.008031935 0.036887404 41
hsa00514 Other types of O-glycan biosynthesis 0.288745698 -0.517012131 0.008556066 0.037891148 14
hsa04970 Salivary secretion 0.298667629 -0.481615545 0.012502314 0.053458172 29
hsa00270 Cysteine and methionine metabolism 0.31779388 -0.435500686 0.021825315 0.090211301 18
hsa04916 Melanogenesis 0.307809028 -0.424410724 0.02421337 0.096853478 31
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.318205481 -0.425756424 0.026180795 0.098468607 10
hsa03020 RNA polymerase 0.318128709 -0.420896642 0.026205355 0.098468607 15
hsa00280 Valine, leucine and isoleucine degradation 0.313740567 -0.405330006 0.030293385 0.110356737 22
hsa00230 Purine metabolism 0.312231752 -0.398751409 0.031149079 0.110356737 67
hsa04670 Leukocyte transendothelial migration 0.333252451 -0.3498243 0.051498798 0.17738475 41
hsa00561 Glycerolipid metabolism 0.344661037 -0.34748943 0.054236106 0.181764247 16
hsa03040 Spliceosome 0.33695989 -0.314034859 0.071366077 0.232878777 62
hsa03030 DNA replication 0.342349169 -0.280035079 0.097227544 0.30913373 19
hsa04720 Long-term potentiation 0.364436535 -0.255018734 0.118539307 0.367471851 29
hsa04370 VEGF signaling pathway 0.388621546 -0.212760948 0.160957564 0.481418815 31
hsa04971 Gastric acid secretion 0.388819729 -0.211472955 0.163061212 0.481418815 24
hsa04973 Carbohydrate digestion and absorption 0.398617215 -0.196097416 0.182428028 0.520195506 14
hsa04210 Apoptosis 0.392313712 -0.190111157 0.187492617 0.520195506 49
hsa04360 Axon guidance 0.372183603 -0.18935995 0.188780627 0.520195506 41
hsa00380 Tryptophan metabolism 0.388325157 -0.175853273 0.211689623 0.570641592 12
hsa04020 Calcium signaling pathway 0.401436606 -0.153175556 0.237152267 0.625678321 54
hsa00071 Fatty acid metabolism 0.458066187 -0.058851455 0.393161034 1 16
hsa00020 Citrate cycle (TCA cycle) 0.463937459 -0.05475885 0.399878443 1 17
hsa03018 RNA degradation 0.455178294 -0.047445635 0.412498531 1 31
hsa04330 Notch signaling pathway 0.452373772 -0.045831183 0.416336716 1 16
hsa00640 Propanoate metabolism 0.46349701 -0.039010701 0.428390103 1 14
hsa00620 Pyruvate metabolism 0.471219028 -0.037140756 0.431586189 1 16
hsa04340 Hedgehog signaling pathway 0.440340955 -0.031613012 0.442189803 1 14
hsa04150 mTOR signaling pathway 0.458495402 -0.019289802 0.464219387 1 23
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.449064886 -0.017370575 0.467504994 1 15
hsa03015 mRNA surveillance pathway 0.473359166 -0.01291453 0.476129531 1 28
hsa04960 Aldosterone-regulated sodium reabsorption 0.477092799 0.001433239 0.503109595 1 14
hsa04740 Olfactory transduction 0.468390194 0.00897602 0.516720374 1 13
hsa04962 Vasopressin-regulated water reabsorption 0.497051442 0.03268503 0.560357452 1 19
hsa04972 Pancreatic secretion 0.498085967 0.060689464 0.610945056 1 22
hsa04664 Fc epsilon RI signaling pathway 0.494742064 0.085270771 0.654321523 1 30
hsa04974 Protein digestion and absorption 0.47183504 0.086254184 0.654498604 1 17
hsa03430 Mismatch repair 0.504461898 0.105504754 0.685488383 1 11
hsa03420 Nucleotide excision repair 0.505818768 0.111723024 0.698349257 1 26
hsa04010 MAPK signaling pathway 0.492553192 0.12767921 0.724960677 1 100
hsa04146 Peroxisome 0.530199964 0.130951373 0.729212274 1 31
hsa00030 Pentose phosphate pathway 0.543688226 0.144500958 0.747054902 1 10
hsa04380 Osteoclast differentiation 0.483918119 0.164313504 0.778930682 1 68
hsa00330 Arginine and proline metabolism 0.530166547 0.174296707 0.790874255 1 23
hsa04144 Endocytosis 0.519600384 0.178315479 0.797675517 1 80
hsa00140 Steroid hormone biosynthesis 0.526475503 0.182230976 0.798308523 1 11
hsa00051 Fructose and mannose metabolism 0.546679988 0.181185009 0.799519312 1 17
hsa00590 Arachidonic acid metabolism 0.570427709 0.25495875 0.879294029 1 11
hsa04623 Cytosolic DNA-sensing pathway 0.557675072 0.254134941 0.880695338 1 23
hsa00670 One carbon pool by folate 0.554795924 0.262694129 0.885763733 1 10
hsa00410 beta-Alanine metabolism 0.585168381 0.285064325 0.905322266 1 10
hsa04976 Bile secretion 0.580590188 0.292586915 0.912475029 1 19
hsa04145 Phagosome 0.555574469 0.355111418 0.951286825 1 63
hsa00982 Drug metabolism - cytochrome P450 0.592457984 0.375061263 0.957369084 1 15
hsa04920 Adipocytokine signaling pathway 0.630891889 0.419574248 0.974145479 1 19
hsa04140 Regulation of autophagy 0.642203171 0.463150401 0.98275317 1 10
hsa04270 Vascular smooth muscle contraction 0.626300305 0.459014684 0.983786293 1 40
hsa00520 Amino sugar and nucleotide sugar metabolism 0.653491632 0.469809265 0.985325355 1 20
hsa03022 Basal transcription factors 0.661041852 0.476814175 0.98641809 1 19
hsa04120 Ubiquitin mediated proteolysis 0.646291348 0.528831177 0.993234841 1 62
hsa00052 Galactose metabolism 0.687462737 0.536632657 0.993269896 1 12
hsa04912 GnRH signaling pathway 0.684718997 0.546837186 0.994602699 1 35
hsa00240 Pyrimidine metabolism 0.668038916 0.549812329 0.994824682 1 46
hsa04730 Long-term depression 0.680593855 0.564432997 0.995514426 1 19
hsa04910 Insulin signaling pathway 0.633543422 0.561929824 0.995583338 1 53
hsa04510 Focal adhesion 0.664757309 0.562041878 0.995675626 1 65
hsa03060 Protein export 0.701516917 0.640082208 0.99824181 1 12
hsa04666 Fc gamma R-mediated phagocytosis 0.691801397 0.65880283 0.998927602 1 45
hsa04141 Protein processing in endoplasmic reticulum 0.707083743 0.681900788 0.999280031 1 77
hsa00565 Ether lipid metabolism 0.718036268 0.699884077 0.999319362 1 12
hsa00010 Glycolysis / Gluconeogenesis 0.735564369 0.71951314 0.999581601 1 28
hsa00983 Drug metabolism - other enzymes 0.747265109 0.733123885 0.999642227 1 16
hsa04620 Toll-like receptor signaling pathway 0.638668924 0.757073757 0.999779286 1 43
hsa04512 ECM-receptor interaction 0.719223739 0.776777836 0.999815168 1 16
hsa04540 Gap junction 0.75049247 0.822086341 0.999931235 1 27
hsa04914 Progesterone-mediated oocyte maturation 0.746126178 0.820907802 0.999933856 1 44
hsa04622 RIG-I-like receptor signaling pathway 0.742904404 0.832021411 0.999940111 1 25
hsa00500 Starch and sucrose metabolism 0.775115118 0.896219269 0.999978697 1 13
hsa00760 Nicotinate and nicotinamide metabolism 0.778282497 0.918897089 0.999983509 1 11
hsa04810 Regulation of actin cytoskeleton 0.739967183 0.902471843 0.999987218 1 74
hsa03050 Proteasome 0.750495362 0.916588863 0.9999882 1 28
hsa03010 Ribosome 0.646488497 0.952292575 0.999994417 1 57
hsa04142 Lysosome 0.776112597 0.96042121 0.999996077 1 52
hsa00512 Mucin type O-Glycan biosynthesis 0.827482899 1.128623959 0.999999863 1 15
hsa00564 Glycerophospholipid metabolism 0.829897708 1.137647858 0.999999931 1 31
hsa04966 Collecting duct acid secretion 0.850884406 1.181083 0.999999947 1 10
hsa04130 SNARE interactions in vesicular transport 0.856609981 1.166149092 0.999999958 1 23
hsa04110 Cell cycle 0.816463858 1.293514089 0.999999999 1 64
hsa00770 Pantothenate and CoA biosynthesis 0.882714031 1.367442579 1 1 10
hsa04114 Oocyte meiosis 0.861120216 1.336928384 1 1 49
hsa04621 NOD-like receptor signaling pathway 0.870466033 1.347229837 1 1 31
hsa00480 Glutathione metabolism 0.883171355 1.419136235 1 1 20
hsa04260 Cardiac muscle contraction 0.890214928 1.478909164 1 1 30
hsa04115 p53 signaling pathway 0.902428216 1.532608268 1 1 30
hsa00600 Sphingolipid metabolism 0.936396717 1.739475885 1 1 16
hsa00190 Oxidative phosphorylation 0.987908268 2.799196786 1 1 63
hsa03320 PPAR signaling pathway 0.962964986 2.041525896 1 1 17
hsa04610 Complement and coagulation cascades 0.937569232 1.925687416 1 1 20
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 5
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 4
hsa00061 Fatty acid biosynthesis NA NA NA NA 1
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 3
hsa00100 Steroid biosynthesis NA NA NA NA 8
hsa00120 Primary bile acid biosynthesis NA NA NA NA 2
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 3
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 9
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 3
hsa00300 Lysine biosynthesis NA NA NA NA 1
hsa00340 Histidine metabolism NA NA NA NA 7
hsa00350 Tyrosine metabolism NA NA NA NA 8
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 1
hsa00450 Selenocompound metabolism NA NA NA NA 4
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 2
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 7
hsa00531 Glycosaminoglycan degradation NA NA NA NA 7
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate NA NA NA NA 8
hsa00591 Linoleic acid metabolism NA NA NA NA 6
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 4
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 8
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 9
hsa00650 Butanoate metabolism NA NA NA NA 5
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 3
hsa00750 Vitamin B6 metabolism NA NA NA NA 2
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 1
hsa00790 Folate biosynthesis NA NA NA NA 4
hsa00860 Porphyrin and chlorophyll metabolism NA NA NA NA 6
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 7
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 4
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 7
hsa02010 ABC transporters NA NA NA NA 9
hsa03440 Homologous recombination NA NA NA NA 8
hsa03450 Non-homologous end-joining NA NA NA NA 7
hsa04122 Sulfur relay system NA NA NA NA 2
hsa04320 Dorso-ventral axis formation NA NA NA NA 7
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04744 Phototransduction NA NA NA NA 8
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 7
hsa04975 Fat digestion and absorption NA NA NA NA 9
hsa04977 Vitamin digestion and absorption NA NA NA NA 7
GSE64456 p.geomean stat.mean p.val q.val set.size
hsa04810 Regulation of actin cytoskeleton 0.024980698 1.900312857 1.38E-16 1.58E-14 71
hsa04380 Osteoclast differentiation 0.023553361 1.784762018 9.44E-15 5.38E-13 65
hsa04670 Leukocyte transendothelial migration 0.035758481 1.760501027 3.10E-14 1.18E-12 40
hsa04666 Fc gamma R-mediated phagocytosis 0.045133485 1.649307533 6.41E-13 1.83E-11 47
hsa04650 Natural killer cell mediated cytotoxicity 0.072225444 1.368939285 2.04E-09 4.64E-08 44
hsa04664 Fc epsilon RI signaling pathway 0.105157532 1.199401081 1.43E-07 2.50E-06 26
hsa04142 Lysosome 0.097778324 1.186075718 1.53E-07 2.50E-06 51
hsa04620 Toll-like receptor signaling pathway 0.111964363 1.141764999 4.56E-07 6.50E-06 36
hsa04510 Focal adhesion 0.125720461 1.109655708 7.59E-07 9.61E-06 64
hsa04010 MAPK signaling pathway 0.12788067 1.078335 1.42E-06 1.62E-05 97
hsa00500 Starch and sucrose metabolism 0.147212645 1.060429956 3.46E-06 3.59E-05 12
hsa04062 Chemokine signaling pathway 0.124300391 1.031280151 4.04E-06 3.84E-05 72
hsa04520 Adherens junction 0.156783987 0.993747894 9.14E-06 7.85E-05 31
hsa00051 Fructose and mannose metabolism 0.154399738 1.004180356 9.63E-06 7.85E-05 15
hsa04920 Adipocytokine signaling pathway 0.153578219 0.980627912 1.23E-05 9.33E-05 27
hsa00030 Pentose phosphate pathway 0.134763581 1.000753343 1.46E-05 0.000104057 13
hsa04910 Insulin signaling pathway 0.17037218 0.922536153 3.23E-05 0.000216303 50
hsa04530 Tight junction 0.177743253 0.889621278 5.93E-05 0.000375845 41
hsa00010 Glycolysis / Gluconeogenesis 0.165722382 0.881968186 7.36E-05 0.000441622 30
hsa04210 Apoptosis 0.180736565 0.849188966 0.000121182 0.00069074 42
hsa04145 Phagosome 0.143549486 0.840558448 0.000139443 0.000756978 72
hsa04722 Neurotrophin signaling pathway 0.200912653 0.798404883 0.000272888 0.001414054 49
hsa04971 Gastric acid secretion 0.217670806 0.761693351 0.000529168 0.002519363 21
hsa04976 Bile secretion 0.215490917 0.762650629 0.000530392 0.002519363 19
hsa03320 PPAR signaling pathway 0.198346671 0.750264896 0.000669556 0.002987829 19
hsa04144 Endocytosis 0.207710515 0.738460837 0.000681435 0.002987829 67
hsa00052 Galactose metabolism 0.230275804 0.731257184 0.000936234 0.003952986 11
hsa04973 Carbohydrate digestion and absorption 0.24191186 0.701405749 0.001407961 0.005732411 11
hsa04610 Complement and coagulation cascades 0.245060211 0.659420134 0.002422761 0.009523956 16
hsa04360 Axon guidance 0.248083663 0.634750607 0.002988767 0.011357316 39
hsa04621 NOD-like receptor signaling pathway 0.227490129 0.624613615 0.003626699 0.013336891 30
hsa04370 VEGF signaling pathway 0.273274253 0.582740232 0.005923901 0.021103896 25
hsa04730 Long-term depression 0.27623823 0.56216917 0.00769666 0.026588463 20
hsa00564 Glycerophospholipid metabolism 0.290202996 0.531137092 0.01098721 0.036839469 27
hsa00520 Amino sugar and nucleotide sugar metabolism 0.294049878 0.518430093 0.012883231 0.041962525 18
hsa00590 Arachidonic acid metabolism 0.283143841 0.510081145 0.014354733 0.045456654 15
hsa04630 Jak-STAT signaling pathway 0.235728391 0.500166708 0.01536124 0.047329226 50
hsa00983 Drug metabolism - other enzymes 0.318271059 0.458430004 0.02518885 0.075333421 11
hsa04115 p53 signaling pathway 0.311111894 0.450529732 0.02577196 0.075333421 28
hsa00600 Sphingolipid metabolism 0.319834909 0.447575713 0.027971255 0.079718078 15
hsa04540 Gap junction 0.305646155 0.433600064 0.030430748 0.084612325 31
hsa00860 Porphyrin and chlorophyll metabolism 0.286189662 0.421130695 0.037565435 0.101963323 11
hsa04150 mTOR signaling pathway 0.340950455 0.397427429 0.04350669 0.113117877 17
hsa04310 Wnt signaling pathway 0.331469051 0.394569796 0.043659531 0.113117877 33
hsa04662 B cell receptor signaling pathway 0.303354669 0.340558377 0.070081134 0.177538873 36
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.368164857 0.324222579 0.083395241 0.206631215 10
hsa04141 Protein processing in endoplasmic reticulum 0.331189279 0.316661248 0.085190062 0.206631215 55
hsa03050 Proteasome 0.359031213 0.268075669 0.127058766 0.30176457 15
hsa04960 Aldosterone-regulated sodium reabsorption 0.390996471 0.265854694 0.130219016 0.302958528 10
hsa04146 Peroxisome 0.389150368 0.23346118 0.156056346 0.355808468 27
hsa04622 RIG-I-like receptor signaling pathway 0.363319151 0.230038908 0.160749313 0.359321993 19
hsa00562 Inositol phosphate metabolism 0.411370149 0.184126116 0.215220712 0.471830023 16
hsa04270 Vascular smooth muscle contraction 0.401505332 0.175943223 0.223055223 0.479779158 32
hsa04974 Protein digestion and absorption 0.389797521 0.148973549 0.261125277 0.551264473 20
hsa04623 Cytosolic DNA-sensing pathway 0.36800736 0.124878059 0.300262436 0.622362141 17
hsa04914 Progesterone-mediated oocyte maturation 0.44240046 0.103737445 0.326500689 0.664662117 37
hsa04512 ECM-receptor interaction 0.436009472 0.061837572 0.392897886 0.777391009 17
hsa04130 SNARE interactions in vesicular transport 0.459979913 0.059400836 0.399168389 0.777391009 17
hsa04320 Dorso-ventral axis formation 0.460532672 0.056790719 0.406367605 0.777391009 10
hsa00480 Glutathione metabolism 0.431579726 0.053231095 0.409153163 0.777391009 18
hsa04912 GnRH signaling pathway 0.479846099 0.033999205 0.441562404 0.824213327 28
hsa00330 Arginine and proline metabolism 0.452189972 0.027380649 0.452115639 0.824213327 17
hsa04640 Hematopoietic cell lineage 0.45550092 0.025942863 0.455486312 0.824213327 49
hsa04975 Fat digestion and absorption 0.483864467 0.013198276 0.477428233 0.850419041 11
hsa04962 Vasopressin-regulated water reabsorption 0.490358214 0.002697565 0.495347886 0.868763984 15
hsa00565 Ether lipid metabolism 0.49525664 -0.006552329 0.510954746 0.872449515 11
hsa04916 Melanogenesis 0.49429824 -0.007438543 0.512755417 0.872449515 25
hsa04972 Pancreatic secretion 0.504725422 -0.025481841 0.543806231 0.908570002 20
hsa00071 Fatty acid metabolism 0.481760323 -0.03007917 0.549923948 0.908570002 14
hsa04012 ErbB signaling pathway 0.509151509 -0.041425859 0.571024586 0.929954325 28
hsa04340 Hedgehog signaling pathway 0.515430457 -0.051318852 0.586236862 0.941281722 11
hsa04970 Salivary secretion 0.52262667 -0.071986586 0.621631808 0.984250362 16
hsa04740 Olfactory transduction 0.507316273 -0.078865103 0.632770843 0.988162687 26
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.528288504 -0.093792889 0.655177429 1 13
hsa00561 Glycerolipid metabolism 0.533062754 -0.099841366 0.667028534 1 21
hsa00640 Propanoate metabolism 0.533575311 -0.125014024 0.704416472 1 12
hsa04070 Phosphatidylinositol signaling system 0.540308736 -0.132137598 0.715115518 1 21
hsa04720 Long-term potentiation 0.546122583 -0.133416835 0.717423883 1 23
hsa03022 Basal transcription factors 0.545041553 -0.159747559 0.751115205 1 13
hsa00310 Lysine degradation 0.557897978 -0.199252793 0.804005476 1 14
hsa04350 TGF-beta signaling pathway 0.56592326 -0.209497793 0.816282057 1 23
hsa00350 Tyrosine metabolism 0.574378348 -0.222558116 0.827279636 1 11
hsa00982 Drug metabolism - cytochrome P450 0.577643382 -0.258736332 0.864660474 1 10
hsa04110 Cell cycle 0.546692757 -0.261309618 0.870768802 1 41
hsa04114 Oocyte meiosis 0.592320415 -0.265090918 0.8745427 1 41
hsa03015 mRNA surveillance pathway 0.595717721 -0.297152628 0.89864673 1 20
hsa04020 Calcium signaling pathway 0.604055818 -0.295711026 0.899774828 1 38
hsa00380 Tryptophan metabolism 0.602509326 -0.314256078 0.909773844 1 11
hsa04330 Notch signaling pathway 0.609621025 -0.385076473 0.950341119 1 13
hsa03440 Homologous recombination 0.642460278 -0.418375221 0.961313798 1 10
hsa00240 Pyrimidine metabolism 0.646417304 -0.421613293 0.966005857 1 35
hsa00340 Histidine metabolism 0.649452139 -0.437050794 0.968744737 1 11
hsa00620 Pyruvate metabolism 0.626413943 -0.437595398 0.970432029 1 22
hsa00280 Valine, leucine and isoleucine degradation 0.651704934 -0.44308936 0.971465917 1 18
hsa00510 N-Glycan biosynthesis 0.655610754 -0.464390398 0.975679095 1 11
hsa04120 Ubiquitin mediated proteolysis 0.662805312 -0.508989698 0.985995059 1 38
hsa04660 T cell receptor signaling pathway 0.618600677 -0.531364514 0.989352139 1 52
hsa03018 RNA degradation 0.663623155 -0.558974754 0.99204445 1 31
hsa00514 Other types of O-glycan biosynthesis 0.699843767 -0.598736591 0.994492248 1 12
hsa03420 Nucleotide excision repair 0.698970853 -0.626920967 0.996132789 1 14
hsa04260 Cardiac muscle contraction 0.630022903 -0.635745402 0.996721052 1 19
hsa00020 Citrate cycle (TCA cycle) 0.703246737 -0.681577417 0.998044929 1 12
hsa03410 Base excision repair 0.738252551 -0.710759384 0.998729203 1 14
hsa03030 DNA replication 0.715694883 -0.722475813 0.998874325 1 13
hsa00970 Aminoacyl-tRNA biosynthesis 0.694849798 -0.765389945 0.999459538 1 16
hsa03008 Ribosome biogenesis in eukaryotes 0.718654093 -0.82675526 0.999797242 1 25
hsa00230 Purine metabolism 0.766696857 -0.926353324 0.99996939 1 53
hsa04514 Cell adhesion molecules (CAMs) 0.783995938 -1.153409151 0.999999634 1 43
hsa03040 Spliceosome 0.818204218 -1.211272899 0.999999911 1 55
hsa00190 Oxidative phosphorylation 0.518124003 -1.286456727 0.99999998 1 50
hsa03013 RNA transport 0.81606492 -1.4303563 1 1 52
hsa04612 Antigen processing and presentation 0.846477013 -1.471357121 1 1 35
hsa04672 Intestinal immune network for IgA production 0.920134709 -1.923000744 1 1 28
hsa03010 Ribosome 0.67474983 -4.82383644 1 1 44
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 6
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 4
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 1
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 2
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00140 Steroid hormone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 0
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 7
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 9
hsa00270 Cysteine and methionine metabolism NA NA NA NA 9
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 7
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 1
hsa00410 beta-Alanine metabolism NA NA NA NA 7
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 1
hsa00450 Selenocompound metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 0
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 2
hsa00512 Mucin type O-Glycan biosynthesis NA NA NA NA 8
hsa00531 Glycosaminoglycan degradation NA NA NA NA 7
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 4
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis NA NA NA NA 5
hsa00591 Linoleic acid metabolism NA NA NA NA 4
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 3
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 4
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 6
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 8
hsa00650 Butanoate metabolism NA NA NA NA 7
hsa00670 One carbon pool by folate NA NA NA NA 5
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 2
hsa00760 Nicotinate and nicotinamide metabolism NA NA NA NA 5
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 8
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 3
hsa00830 Retinol metabolism NA NA NA NA 5
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 4
hsa00910 Nitrogen metabolism NA NA NA NA 5
hsa00920 Sulfur metabolism NA NA NA NA 6
hsa00980 Metabolism of xenobiotics by cytochrome P450 NA NA NA NA 9
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 9
hsa02010 ABC transporters NA NA NA NA 7
hsa03020 RNA polymerase NA NA NA NA 8
hsa03060 Protein export NA NA NA NA 7
hsa03430 Mismatch repair NA NA NA NA 7
hsa03450 Non-homologous end-joining NA NA NA NA 3
hsa04122 Sulfur relay system NA NA NA NA 1
hsa04140 Regulation of autophagy NA NA NA NA 6
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04710 Circadian rhythm - mammal NA NA NA NA 5
hsa04742 Taste transduction NA NA NA NA 8
hsa04744 Phototransduction NA NA NA NA 4
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 4
hsa04966 Collecting duct acid secretion NA NA NA NA 9
hsa04977 Vitamin digestion and absorption NA NA NA NA 4
hsa03010 Ribosome 7.77E-07 -4.82383644 3.43E-78 3.91E-76 44
hsa04672 Intestinal immune network for IgA production 0.025613824 -1.923000744 2.27E-16 1.29E-14 28
hsa04612 Antigen processing and presentation 0.059175179 -1.471357121 1.65E-10 6.27E-09 35
hsa03013 RNA transport 0.062799083 -1.4303563 3.80E-10 1.08E-08 52
hsa00190 Oxidative phosphorylation 0.039003681 -1.286456727 2.00E-08 4.56E-07 50
hsa03040 Spliceosome 0.097913217 -1.211272899 8.89E-08 1.69E-06 55
hsa04514 Cell adhesion molecules (CAMs) 0.101366727 -1.153409151 3.66E-07 5.95E-06 43
hsa00230 Purine metabolism 0.159594793 -0.926353324 3.06E-05 0.000436199 53
hsa03008 Ribosome biogenesis in eukaryotes 0.182118354 -0.82675526 0.000202758 0.002568272 25
hsa00970 Aminoacyl-tRNA biosynthesis 0.199499524 -0.765389945 0.000540462 0.006161265 16
hsa03030 DNA replication 0.22562444 -0.722475813 0.001125675 0.011666083 13
hsa03410 Base excision repair 0.236561864 -0.710759384 0.001270797 0.012072575 14
hsa00020 Citrate cycle (TCA cycle) 0.234045694 -0.681577417 0.001955071 0.017144473 12
hsa04260 Cardiac muscle contraction 0.218658227 -0.635745402 0.003278948 0.026700001 19
hsa03420 Nucleotide excision repair 0.257102696 -0.626920967 0.003867211 0.029390805 14
hsa00514 Other types of O-glycan biosynthesis 0.268746322 -0.598736591 0.005507752 0.039242736 12
hsa03018 RNA degradation 0.264004808 -0.558974754 0.00795555 0.053348984 31
hsa04660 T cell receptor signaling pathway 0.256566763 -0.531364514 0.010647861 0.067436456 52
hsa04120 Ubiquitin mediated proteolysis 0.289484996 -0.508989698 0.014004941 0.084029648 38
hsa00510 N-Glycan biosynthesis 0.314209112 -0.464390398 0.024320905 0.13862916 11
hsa00280 Valine, leucine and isoleucine degradation 0.320869632 -0.44308936 0.028534083 0.153215852 18
hsa00620 Pyruvate metabolism 0.308438214 -0.437595398 0.029567971 0.153215852 22
hsa00340 Histidine metabolism 0.324736621 -0.437050794 0.031255263 0.154917388 11
hsa00240 Pyrimidine metabolism 0.327540915 -0.421613293 0.033994143 0.161472178 35
hsa03440 Homologous recombination 0.333293763 -0.418375221 0.038686202 0.176409082 10
hsa04330 Notch signaling pathway 0.32867722 -0.385076473 0.049658881 0.217735092 13
hsa00380 Tryptophan metabolism 0.366118955 -0.314256078 0.090226156 0.380954879 11
hsa04020 Calcium signaling pathway 0.376159732 -0.295711026 0.100225172 0.398423199 38
hsa03015 mRNA surveillance pathway 0.370820501 -0.297152628 0.10135327 0.398423199 20
hsa04114 Oocyte meiosis 0.387222526 -0.265090918 0.1254573 0.475237309 41
hsa04110 Cell cycle 0.355021303 -0.261309618 0.129231198 0.475237309 41
hsa00982 Drug metabolism - cytochrome P450 0.383153567 -0.258736332 0.135339526 0.48214706 10
hsa00350 Tyrosine metabolism 0.40522162 -0.222558116 0.172720364 0.596670348 11
hsa04350 TGF-beta signaling pathway 0.404982156 -0.209497793 0.183717943 0.615995455 23
hsa00310 Lysine degradation 0.406758356 -0.199252793 0.195994524 0.638382163 14
hsa03022 Basal transcription factors 0.423949328 -0.159747559 0.248884795 0.788135185 13
hsa04720 Long-term potentiation 0.442021952 -0.133416835 0.282576117 0.854653447 23
hsa04070 Phosphatidylinositol signaling system 0.437884934 -0.132137598 0.284884482 0.854653447 21
hsa00640 Propanoate metabolism 0.437630008 -0.125014024 0.295583528 0.864013388 12
hsa00561 Glycerolipid metabolism 0.454933697 -0.099841366 0.332971466 0.948968679 21
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.455986632 -0.093792889 0.344822571 0.958774953 13
hsa04740 Olfactory transduction 0.447156016 -0.078865103 0.367229157 0.996764854 26
hsa04970 Salivary secretion 0.466445299 -0.071986586 0.378368192 1 16
hsa04340 Hedgehog signaling pathway 0.475748551 -0.051318852 0.413763138 1 11
hsa04012 ErbB signaling pathway 0.476830067 -0.041425859 0.428975414 1 28
hsa00071 Fatty acid metabolism 0.459233451 -0.03007917 0.450076052 1 14
hsa04972 Pancreatic secretion 0.484830862 -0.025481841 0.456193769 1 20
hsa04916 Melanogenesis 0.488260643 -0.007438543 0.487244583 1 25
hsa00565 Ether lipid metabolism 0.490277284 -0.006552329 0.489045254 1 11
hsa04962 Vasopressin-regulated water reabsorption 0.492493948 0.002697565 0.504652114 1 15
hsa04975 Fat digestion and absorption 0.494143544 0.013198276 0.522571767 1 11
hsa04640 Hematopoietic cell lineage 0.475761832 0.025942863 0.544513688 1 49
hsa00330 Arginine and proline metabolism 0.472275028 0.027380649 0.547884361 1 17
hsa04912 GnRH signaling pathway 0.506558087 0.033999205 0.558437596 1 28
hsa00480 Glutathione metabolism 0.469997712 0.053231095 0.590846837 1 18
hsa04320 Dorso-ventral axis formation 0.503600092 0.056790719 0.593632395 1 10
hsa04130 SNARE interactions in vesicular transport 0.506161127 0.059400836 0.600831611 1 17
hsa04512 ECM-receptor interaction 0.481232165 0.061837572 0.607102114 1 17
hsa04914 Progesterone-mediated oocyte maturation 0.523878029 0.103737445 0.673499311 1 37
hsa04623 Cytosolic DNA-sensing pathway 0.455529536 0.124878059 0.699737564 1 17
hsa04974 Protein digestion and absorption 0.497807315 0.148973549 0.738874723 1 20
hsa04270 Vascular smooth muscle contraction 0.5332949 0.175943223 0.776944777 1 32
hsa00562 Inositol phosphate metabolism 0.550981155 0.184126116 0.784779288 1 16
hsa04622 RIG-I-like receptor signaling pathway 0.528116453 0.230038908 0.839250687 1 19
hsa04146 Peroxisome 0.567072048 0.23346118 0.843943654 1 27
hsa04960 Aldosterone-regulated sodium reabsorption 0.592768032 0.265854694 0.869780984 1 10
hsa03050 Proteasome 0.557960616 0.268075669 0.872941234 1 15
hsa04141 Protein processing in endoplasmic reticulum 0.564622289 0.316661248 0.914809938 1 55
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.613941996 0.324222579 0.916604759 1 10
hsa04662 B cell receptor signaling pathway 0.524227981 0.340558377 0.929918866 1 36
hsa04310 Wnt signaling pathway 0.627708378 0.394569796 0.956340469 1 33
hsa04150 mTOR signaling pathway 0.643424277 0.397427429 0.95649331 1 17
hsa00860 Porphyrin and chlorophyll metabolism 0.565545027 0.421130695 0.962434565 1 11
hsa04540 Gap junction 0.62150259 0.433600064 0.969569252 1 31
hsa00600 Sphingolipid metabolism 0.652074913 0.447575713 0.972028745 1 15
hsa04115 p53 signaling pathway 0.646496094 0.450529732 0.97422804 1 28
hsa00983 Drug metabolism - other enzymes 0.659874293 0.458430004 0.97481115 1 11
hsa04630 Jak-STAT signaling pathway 0.558502179 0.500166708 0.98463876 1 50
hsa00590 Arachidonic acid metabolism 0.646474592 0.510081145 0.985645267 1 15
hsa00520 Amino sugar and nucleotide sugar metabolism 0.679184835 0.518430093 0.987116769 1 18
hsa00564 Glycerophospholipid metabolism 0.683459054 0.531137092 0.98901279 1 27
hsa04730 Long-term depression 0.686237904 0.56216917 0.99230334 1 20
hsa04370 VEGF signaling pathway 0.702699545 0.582740232 0.994076099 1 25
hsa04621 NOD-like receptor signaling pathway 0.65854571 0.624613615 0.996373301 1 30
hsa04360 Axon guidance 0.704632114 0.634750607 0.997011233 1 39
hsa04610 Complement and coagulation cascades 0.711659505 0.659420134 0.997577239 1 16
hsa04973 Carbohydrate digestion and absorption 0.744492304 0.701405749 0.998592039 1 11
hsa00052 Galactose metabolism 0.74752235 0.731257184 0.999063766 1 11
hsa04144 Endocytosis 0.719766219 0.738460837 0.999318565 1 67
hsa03320 PPAR signaling pathway 0.694428157 0.750264896 0.999330444 1 19
hsa04976 Bile secretion 0.744542264 0.762650629 0.999469608 1 19
hsa04971 Gastric acid secretion 0.752953653 0.761693351 0.999470832 1 21
hsa04722 Neurotrophin signaling pathway 0.75834553 0.798404883 0.999727112 1 49
hsa04145 Phagosome 0.626506377 0.840558448 0.999860557 1 72
hsa04210 Apoptosis 0.752340516 0.849188966 0.999878818 1 42
hsa00010 Glycolysis / Gluconeogenesis 0.742930964 0.881968186 0.999926396 1 30
hsa04530 Tight junction 0.782808536 0.889621278 0.999940656 1 41
hsa04910 Insulin signaling pathway 0.796855098 0.922536153 0.999967744 1 50
hsa00030 Pentose phosphate pathway 0.727141546 1.000753343 0.999985395 1 13
hsa04920 Adipocytokine signaling pathway 0.794788087 0.980627912 0.99998773 1 27
hsa00051 Fructose and mannose metabolism 0.813247994 1.004180356 0.999990366 1 15
hsa04520 Adherens junction 0.819767155 0.993747894 0.999990863 1 31
hsa04062 Chemokine signaling pathway 0.767920201 1.031280151 0.99999596 1 72
hsa00500 Starch and sucrose metabolism 0.837331324 1.060429956 0.999996538 1 12
hsa04010 MAPK signaling pathway 0.81590566 1.078335 0.999998581 1 97
hsa04510 Focal adhesion 0.835612855 1.109655708 0.999999241 1 64
hsa04620 Toll-like receptor signaling pathway 0.81025081 1.141764999 0.999999544 1 36
hsa04142 Lysosome 0.79660643 1.186075718 0.999999847 1 51
hsa04664 Fc epsilon RI signaling pathway 0.814987923 1.199401081 0.999999857 1 26
hsa04650 Natural killer cell mediated cytotoxicity 0.815759204 1.368939285 0.999999998 1 44
hsa04666 Fc gamma R-mediated phagocytosis 0.907783818 1.649307533 1 1 47
hsa04670 Leukocyte transendothelial migration 0.898114304 1.760501027 1 1 40
hsa04380 Osteoclast differentiation 0.777852778 1.784762018 1 1 65
hsa04810 Regulation of actin cytoskeleton 0.916930682 1.900312857 1 1 71
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 6
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 4
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 1
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 2
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00140 Steroid hormone biosynthesis NA NA NA NA 5
hsa00232 Caffeine metabolism NA NA NA NA 0
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 7
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 9
hsa00270 Cysteine and methionine metabolism NA NA NA NA 9
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 7
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 1
hsa00410 beta-Alanine metabolism NA NA NA NA 7
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 1
hsa00450 Selenocompound metabolism NA NA NA NA 6
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 0
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 2
hsa00512 Mucin type O-Glycan biosynthesis NA NA NA NA 8
hsa00531 Glycosaminoglycan degradation NA NA NA NA 7
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 4
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis NA NA NA NA 5
hsa00591 Linoleic acid metabolism NA NA NA NA 4
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 3
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 4
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 6
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 8
hsa00650 Butanoate metabolism NA NA NA NA 7
hsa00670 One carbon pool by folate NA NA NA NA 5
hsa00730 Thiamine metabolism NA NA NA NA 1
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 2
hsa00760 Nicotinate and nicotinamide metabolism NA NA NA NA 5
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 8
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 3
hsa00830 Retinol metabolism NA NA NA NA 5
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 4
hsa00910 Nitrogen metabolism NA NA NA NA 5
hsa00920 Sulfur metabolism NA NA NA NA 6
hsa00980 Metabolism of xenobiotics by cytochrome P450 NA NA NA NA 9
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 9
hsa02010 ABC transporters NA NA NA NA 7
hsa03020 RNA polymerase NA NA NA NA 8
hsa03060 Protein export NA NA NA NA 7
hsa03430 Mismatch repair NA NA NA NA 7
hsa03450 Non-homologous end-joining NA NA NA NA 3
hsa04122 Sulfur relay system NA NA NA NA 1
hsa04140 Regulation of autophagy NA NA NA NA 6
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04710 Circadian rhythm - mammal NA NA NA NA 5
hsa04742 Taste transduction NA NA NA NA 8
hsa04744 Phototransduction NA NA NA NA 4
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 4
hsa04966 Collecting duct acid secretion NA NA NA NA 9
hsa04977 Vitamin digestion and absorption NA NA NA NA 4
GSE66099 p.geomean stat.mean p.val q.val set.size
hsa04610 Complement and coagulation cascades 0.003705049 2.627115289 3.01E-68 3.85E-66 27
hsa04142 Lysosome 0.01266978 2.027149078 2.33E-44 1.49E-42 59
hsa04380 Osteoclast differentiation 0.012269396 1.948024403 3.73E-41 1.59E-39 65
hsa04810 Regulation of actin cytoskeleton 0.017974084 1.886943364 3.68E-39 1.18E-37 77
hsa04666 Fc gamma R-mediated phagocytosis 0.029131543 1.692698376 1.66E-31 4.26E-30 45
hsa03320 PPAR signaling pathway 0.050250178 1.597233745 6.17E-28 1.32E-26 24
hsa04621 NOD-like receptor signaling pathway 0.049312688 1.507315834 5.97E-25 1.07E-23 25
hsa04510 Focal adhesion 0.042514625 1.478517677 6.67E-25 1.07E-23 76
hsa00010 Glycolysis / Gluconeogenesis 0.065143465 1.438619389 2.31E-23 3.28E-22 31
hsa00983 Drug metabolism - other enzymes 0.050625477 1.452537004 8.92E-23 1.14E-21 23
hsa04620 Toll-like receptor signaling pathway 0.054148099 1.404108552 3.23E-22 3.76E-21 35
hsa04910 Insulin signaling pathway 0.060899682 1.37837677 6.64E-22 7.08E-21 61
hsa04145 Phagosome 0.052262049 1.302658365 1.04E-19 1.02E-18 73
hsa04512 ECM-receptor interaction 0.060938568 1.290258349 6.09E-19 5.57E-18 33
hsa04740 Olfactory transduction 0.043112696 1.286221584 2.75E-18 2.29E-17 29
hsa00052 Galactose metabolism 0.093603146 1.272071517 2.87E-18 2.29E-17 16
hsa00531 Glycosaminoglycan degradation 0.098164887 1.257329942 7.38E-17 5.55E-16 10
hsa00051 Fructose and mannose metabolism 0.11236772 1.172167665 6.24E-16 4.44E-15 17
hsa00480 Glutathione metabolism 0.115759676 1.110178902 1.18E-14 7.92E-14 24
hsa04920 Adipocytokine signaling pathway 0.121086301 1.089798802 2.54E-14 1.58E-13 33
hsa00190 Oxidative phosphorylation 0.105760971 1.089125289 2.60E-14 1.58E-13 44
hsa04020 Calcium signaling pathway 0.098336386 1.071985624 5.23E-14 3.04E-13 64
hsa00982 Drug metabolism - cytochrome P450 0.081431459 1.087563285 1.44E-13 7.93E-13 23
hsa04010 MAPK signaling pathway 0.10579043 1.04808766 1.49E-13 7.93E-13 98
hsa04670 Leukocyte transendothelial migration 0.107371889 1.039236046 3.31E-13 1.70E-12 46
hsa04144 Endocytosis 0.115257715 0.971773613 6.94E-12 3.42E-11 81
hsa00564 Glycerophospholipid metabolism 0.129797271 0.976771438 7.98E-12 3.78E-11 31
hsa00140 Steroid hormone biosynthesis 0.108621819 0.999920789 8.37E-12 3.83E-11 18
hsa00860 Porphyrin and chlorophyll metabolism 0.134304384 0.982959373 1.26E-11 5.54E-11 19
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.121269955 0.973182501 1.51E-11 6.46E-11 23
hsa00600 Sphingolipid metabolism 0.143705695 0.958750688 3.00E-11 1.24E-10 17
hsa00030 Pentose phosphate pathway 0.150939894 0.952746064 3.82E-11 1.53E-10 17
hsa04320 Dorso-ventral axis formation 0.160274512 0.935747774 1.42E-10 5.52E-10 11
hsa04540 Gap junction 0.140440959 0.894186191 3.45E-10 1.30E-09 36
hsa04114 Oocyte meiosis 0.133707655 0.888103923 3.92E-10 1.43E-09 50
hsa04130 SNARE interactions in vesicular transport 0.150579419 0.883380558 1.01E-09 3.61E-09 16
hsa04270 Vascular smooth muscle contraction 0.176069445 0.8137631 8.35E-09 2.89E-08 41
hsa04912 GnRH signaling pathway 0.170971979 0.808486958 1.05E-08 3.55E-08 39
hsa04914 Progesterone-mediated oocyte maturation 0.172100632 0.804150142 1.26E-08 4.13E-08 42
hsa00512 Mucin type O-Glycan biosynthesis 0.19059102 0.810585824 1.52E-08 4.85E-08 14
hsa04722 Neurotrophin signaling pathway 0.165526478 0.795151256 1.72E-08 5.37E-08 49
hsa00500 Starch and sucrose metabolism 0.129188063 0.76812641 8.44E-08 2.57E-07 29
hsa00760 Nicotinate and nicotinamide metabolism 0.211669604 0.753077767 1.68E-07 5.02E-07 10
hsa04966 Collecting duct acid secretion 0.207210052 0.740427687 2.33E-07 6.77E-07 12
hsa00330 Arginine and proline metabolism 0.208270356 0.729924788 2.39E-07 6.79E-07 23
hsa04141 Protein processing in endoplasmic reticulum 0.189196825 0.715726473 3.23E-07 8.99E-07 60
hsa04115 p53 signaling pathway 0.212291319 0.695100274 7.24E-07 1.97E-06 35
hsa04370 VEGF signaling pathway 0.200775123 0.695719348 7.84E-07 2.09E-06 29
hsa04622 RIG-I-like receptor signaling pathway 0.224190694 0.683065578 1.29E-06 3.32E-06 18
hsa04976 Bile secretion 0.196657044 0.683424679 1.30E-06 3.32E-06 24
hsa04630 Jak-STAT signaling pathway 0.200846678 0.648508842 3.25E-06 8.15E-06 59
hsa00520 Amino sugar and nucleotide sugar metabolism 0.222656696 0.622511145 8.59E-06 2.11E-05 29
hsa04360 Axon guidance 0.21054589 0.591536051 2.00E-05 4.82E-05 52
hsa04260 Cardiac muscle contraction 0.235501332 0.590618725 2.36E-05 5.59E-05 24
hsa04975 Fat digestion and absorption 0.25565959 0.578036426 4.33E-05 0.000100817 10
hsa04664 Fc epsilon RI signaling pathway 0.234623663 0.560978645 5.14E-05 0.000117457 31
hsa04210 Apoptosis 0.234016116 0.555656094 5.64E-05 0.000126666 46
hsa00830 Retinol metabolism 0.169950294 0.563733787 5.83E-05 0.000128713 26
hsa03050 Proteasome 0.23694637 0.543064357 0.000124335 0.000266461 13
hsa04530 Tight junction 0.263604072 0.526600436 0.000124904 0.000266461 44
hsa00514 Other types of O-glycan biosynthesis 0.237020502 0.4881476 0.000418067 0.000877256 18
hsa04012 ErbB signaling pathway 0.275335156 0.460884513 0.00069433 0.001433455 33
hsa00310 Lysine degradation 0.302109833 0.463259641 0.000740288 0.001504077 13
hsa00380 Tryptophan metabolism 0.307501964 0.448949223 0.001038137 0.002076273 13
hsa04971 Gastric acid secretion 0.280461857 0.439202628 0.001193463 0.002350204 26
hsa04110 Cell cycle 0.174672074 0.438409807 0.001290934 0.00250363 67
hsa00053 Ascorbate and aldarate metabolism 0.244022689 0.446737097 0.001436722 0.002744782 12
hsa04520 Adherens junction 0.280661701 0.415702695 0.001965147 0.0036991 31
hsa04916 Melanogenesis 0.301552839 0.362558429 0.00600789 0.011145072 33
hsa00565 Ether lipid metabolism 0.323048551 0.361186826 0.006930128 0.012672234 11
hsa04962 Vasopressin-regulated water reabsorption 0.334798032 0.341723509 0.009086183 0.016380725 19
hsa00040 Pentose and glucuronate interconversions 0.289787808 0.345056996 0.009290416 0.016516295 15
hsa00561 Glycerolipid metabolism 0.335207685 0.332713947 0.010704146 0.018768913 19
hsa04150 mTOR signaling pathway 0.337606934 0.320797255 0.01321614 0.022860349 23
hsa04742 Taste transduction 0.291028874 0.318718039 0.014402291 0.024579911 16
hsa04730 Long-term depression 0.332568569 0.306091725 0.017434524 0.029363409 21
hsa00590 Arachidonic acid metabolism 0.343245314 0.263932337 0.034058501 0.056616729 20
hsa04623 Cytosolic DNA-sensing pathway 0.354542664 0.24402704 0.04715838 0.077388111 15
hsa04330 Notch signaling pathway 0.35982807 0.192659157 0.091907442 0.148913325 20
hsa03440 Homologous recombination 0.374767582 0.189320952 0.098329508 0.157327212 14
hsa04062 Chemokine signaling pathway 0.292295649 0.15879599 0.133791923 0.211424273 89
hsa04960 Aldosterone-regulated sodium reabsorption 0.390562423 0.147898997 0.1562751 0.243941619 13
hsa04973 Carbohydrate digestion and absorption 0.331954464 0.146179308 0.158477206 0.244398583 21
hsa04146 Peroxisome 0.372717362 0.119390199 0.204114911 0.311032246 38
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.442049424 0.093139652 0.261343285 0.393552241 10
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.437849575 0.066691854 0.322068246 0.479357389 12
hsa04720 Long-term potentiation 0.440235062 0.039588775 0.391737188 0.576348967 24
hsa04972 Pancreatic secretion 0.428445132 0.032656772 0.409181868 0.595173626 33
hsa00071 Fatty acid metabolism 0.413265823 0.022840238 0.437035673 0.628545687 20
hsa04974 Protein digestion and absorption 0.400587787 -0.00438383 0.512075141 0.728284645 19
hsa04310 Wnt signaling pathway 0.437700838 -0.072120803 0.692043725 0.973424141 57
hsa04662 B cell receptor signaling pathway 0.36103007 -0.07710529 0.703307374 0.978514608 39
hsa04970 Salivary secretion 0.512955548 -0.202513164 0.919414566 1 30
hsa00562 Inositol phosphate metabolism 0.545419933 -0.209184316 0.925845588 1 20
hsa00340 Histidine metabolism 0.544360095 -0.232828907 0.943784408 1 11
hsa00350 Tyrosine metabolism 0.592645522 -0.30800538 0.982976014 1 14
hsa00240 Pyrimidine metabolism 0.535418227 -0.304257045 0.983005963 1 45
hsa00640 Propanoate metabolism 0.567389887 -0.32318709 0.9869299 1 15
hsa02010 ABC transporters 0.571371839 -0.33534975 0.98958063 1 19
hsa04120 Ubiquitin mediated proteolysis 0.477484887 -0.33461628 0.990023117 1 60
hsa04350 TGF-beta signaling pathway 0.586336612 -0.347386422 0.99214281 1 38
hsa00620 Pyruvate metabolism 0.543545294 -0.363893801 0.993642594 1 16
hsa00020 Citrate cycle (TCA cycle) 0.548225146 -0.373598925 0.994268151 1 10
hsa00270 Cysteine and methionine metabolism 0.582681441 -0.366283307 0.994290773 1 18
hsa04070 Phosphatidylinositol signaling system 0.621304624 -0.456833384 0.999213644 1 26
hsa04640 Hematopoietic cell lineage 0.560465997 -0.468457781 0.999437017 1 51
hsa00230 Purine metabolism 0.624657536 -0.501315713 0.999763164 1 71
hsa03060 Protein export 0.573804743 -0.547418273 0.999875767 1 12
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.679893069 -0.765756219 0.999999882 1 12
hsa03410 Base excision repair 0.649822419 -0.782178347 0.999999944 1 15
hsa03022 Basal transcription factors 0.694202219 -0.787481501 0.999999947 1 13
hsa03430 Mismatch repair 0.672068333 -0.802861444 0.999999969 1 12
hsa03015 mRNA surveillance pathway 0.738013337 -0.875760598 0.999999999 1 40
hsa00510 N-Glycan biosynthesis 0.735702206 -0.926550133 1 1 20
hsa00280 Valine, leucine and isoleucine degradation 0.684717543 -0.95942875 1 1 20
hsa03420 Nucleotide excision repair 0.637905123 -0.963139829 1 1 22
hsa03030 DNA replication 0.610906963 -0.976498889 1 1 18
hsa00970 Aminoacyl-tRNA biosynthesis 0.708900242 -0.989717566 1 1 13
hsa04514 Cell adhesion molecules (CAMs) 0.664625853 -1.052692299 1 1 59
hsa04660 T cell receptor signaling pathway 0.665854995 -1.108616638 1 1 56
hsa04650 Natural killer cell mediated cytotoxicity 0.654357518 -1.116430068 1 1 49
hsa03008 Ribosome biogenesis in eukaryotes 0.616574079 -1.913281458 1 1 48
hsa03010 Ribosome 0.478525766 -1.58007293 1 1 52
hsa03013 RNA transport 0.628940109 -2.356686231 1 1 78
hsa03018 RNA degradation 0.722605049 -1.260348082 1 1 35
hsa03040 Spliceosome 0.682745561 -1.611086829 1 1 58
hsa04612 Antigen processing and presentation 0.834803569 -2.239990543 1 1 40
hsa04672 Intestinal immune network for IgA production 0.790429155 -1.866945746 1 1 31
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 3
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 3
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 9
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 7
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 3
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 1
hsa00410 beta-Alanine metabolism NA NA NA NA 9
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 4
hsa00450 Selenocompound metabolism NA NA NA NA 9
hsa00460 Cyanoamino acid metabolism NA NA NA NA 2
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 1
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 4
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 6
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 5
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 9
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 6
hsa00650 Butanoate metabolism NA NA NA NA 9
hsa00670 One carbon pool by folate NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 0
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 6
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 5
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 4
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 5
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 8
hsa03020 RNA polymerase NA NA NA NA 9
hsa03450 Non-homologous end-joining NA NA NA NA 5
hsa04122 Sulfur relay system NA NA NA NA 3
hsa04140 Regulation of autophagy NA NA NA NA 9
hsa04340 Hedgehog signaling pathway NA NA NA NA 9
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04710 Circadian rhythm - mammal NA NA NA NA 7
hsa04744 Phototransduction NA NA NA NA 9
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 6
hsa04977 Vitamin digestion and absorption NA NA NA NA 8
hsa03013 RNA transport 0.00224852 -2.356686231 4.45E-58 5.69E-56 78
hsa04612 Antigen processing and presentation 0.007962322 -2.239990543 2.80E-52 1.79E-50 40
hsa03008 Ribosome biogenesis in eukaryotes 0.010169063 -1.913281458 9.27E-39 3.95E-37 48
hsa04672 Intestinal immune network for IgA production 0.019476949 -1.866945746 8.33E-37 2.67E-35 31
hsa03040 Spliceosome 0.023118923 -1.611086829 1.93E-28 4.93E-27 58
hsa03010 Ribosome 0.014619546 -1.58007293 3.80E-26 8.11E-25 52
hsa03018 RNA degradation 0.06964811 -1.260348082 3.16E-18 5.78E-17 35
hsa04650 Natural killer cell mediated cytotoxicity 0.082410013 -1.116430068 7.03E-15 1.12E-13 49
hsa04660 T cell receptor signaling pathway 0.084384899 -1.108616638 8.87E-15 1.26E-13 56
hsa04514 Cell adhesion molecules (CAMs) 0.09348904 -1.052692299 1.62E-13 2.07E-12 59
hsa00970 Aminoacyl-tRNA biosynthesis 0.133072857 -0.989717566 2.16E-11 2.52E-10 13
hsa03030 DNA replication 0.109155318 -0.976498889 2.36E-11 2.52E-10 18
hsa03420 Nucleotide excision repair 0.115046907 -0.963139829 2.97E-11 2.93E-10 22
hsa00280 Valine, leucine and isoleucine degradation 0.128816843 -0.95942875 3.21E-11 2.93E-10 20
hsa00510 N-Glycan biosynthesis 0.152023099 -0.926550133 1.07E-10 9.17E-10 20
hsa03015 mRNA surveillance pathway 0.165892281 -0.875760598 6.44E-10 5.15E-09 40
hsa03430 Mismatch repair 0.175299738 -0.802861444 3.09E-08 2.32E-07 12
hsa03022 Basal transcription factors 0.188589289 -0.787481501 5.30E-08 3.77E-07 13
hsa03410 Base excision repair 0.173489164 -0.782178347 5.62E-08 3.78E-07 15
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.19078658 -0.765756219 1.18E-07 7.55E-07 12
hsa03060 Protein export 0.232311418 -0.547418273 0.000124233 0.000757228 12
hsa00230 Purine metabolism 0.270034866 -0.501315713 0.000236836 0.001377954 71
hsa04640 Hematopoietic cell lineage 0.254676694 -0.468457781 0.000562983 0.003133122 51
hsa04070 Phosphatidylinositol signaling system 0.291470638 -0.456833384 0.000786356 0.004193896 26
hsa00270 Cysteine and methionine metabolism 0.321081727 -0.366283307 0.005709227 0.028218335 18
hsa00020 Citrate cycle (TCA cycle) 0.298123638 -0.373598925 0.005731849 0.028218335 10
hsa00620 Pyruvate metabolism 0.295943726 -0.363893801 0.006357406 0.030138814 16
hsa04350 TGF-beta signaling pathway 0.330278115 -0.347386422 0.00785719 0.035918582 38
hsa04120 Ubiquitin mediated proteolysis 0.268074308 -0.33461628 0.009976883 0.044035897 60
hsa02010 ABC transporters 0.328365142 -0.33534975 0.01041937 0.044455978 19
hsa00640 Propanoate metabolism 0.335237773 -0.32318709 0.0130701 0.053966863 15
hsa00240 Pyrimidine metabolism 0.326085856 -0.304257045 0.016994037 0.066032429 45
hsa00350 Tyrosine metabolism 0.360644035 -0.30800538 0.017023986 0.066032429 14
hsa00340 Histidine metabolism 0.37328606 -0.232828907 0.056215592 0.211635171 11
hsa00562 Inositol phosphate metabolism 0.387769636 -0.209184316 0.074154412 0.271193278 20
hsa04970 Salivary secretion 0.365046672 -0.202513164 0.080585434 0.286525988 30
hsa04662 B cell receptor signaling pathway 0.316557592 -0.07710529 0.296692626 1 39
hsa04310 Wnt signaling pathway 0.386978715 -0.072120803 0.307956275 1 57
hsa04974 Protein digestion and absorption 0.398761984 -0.00438383 0.487924859 1 19
hsa00071 Fatty acid metabolism 0.428667747 0.022840238 0.562964327 1 20
hsa04972 Pancreatic secretion 0.449222421 0.032656772 0.590818132 1 33
hsa04720 Long-term potentiation 0.469431029 0.039588775 0.608262812 1 24
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.486384279 0.066691854 0.677931754 1 12
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.512465415 0.093139652 0.738656715 1 10
hsa04146 Peroxisome 0.457986174 0.119390199 0.795885089 1 38
hsa04973 Carbohydrate digestion and absorption 0.430683001 0.146179308 0.841522794 1 21
hsa04960 Aldosterone-regulated sodium reabsorption 0.497436247 0.147898997 0.8437249 1 13
hsa04062 Chemokine signaling pathway 0.380277652 0.15879599 0.866208077 1 89
hsa03440 Homologous recombination 0.513956383 0.189320952 0.901670492 1 14
hsa04330 Notch signaling pathway 0.494678063 0.192659157 0.908092558 1 20
hsa04623 Cytosolic DNA-sensing pathway 0.529832268 0.24402704 0.95284162 1 15
hsa00590 Arachidonic acid metabolism 0.528832108 0.263932337 0.965941499 1 20
hsa04730 Long-term depression 0.550441643 0.306091725 0.982565476 1 21
hsa04742 Taste transduction 0.495743941 0.318718039 0.985597709 1 16
hsa04150 mTOR signaling pathway 0.570644265 0.320797255 0.98678386 1 23
hsa00561 Glycerolipid metabolism 0.576153301 0.332713947 0.989295854 1 19
hsa00040 Pentose and glucuronate interconversions 0.513728483 0.345056996 0.990709584 1 15
hsa04962 Vasopressin-regulated water reabsorption 0.584312457 0.341723509 0.990913817 1 19
hsa00565 Ether lipid metabolism 0.580006712 0.361186826 0.993069872 1 11
hsa04916 Melanogenesis 0.556507152 0.362558429 0.99399211 1 33
hsa04520 Adherens junction 0.559181787 0.415702695 0.998034853 1 31
hsa00053 Ascorbate and aldarate metabolism 0.513443249 0.446737097 0.998563278 1 12
hsa04110 Cell cycle 0.435995418 0.438409807 0.998709066 1 67
hsa04971 Gastric acid secretion 0.58475622 0.439202628 0.998806537 1 26
hsa00380 Tryptophan metabolism 0.635066091 0.448949223 0.998961863 1 13
hsa00310 Lysine degradation 0.639111986 0.463259641 0.999259712 1 13
hsa04012 ErbB signaling pathway 0.592629829 0.460884513 0.99930567 1 33
hsa00514 Other types of O-glycan biosynthesis 0.540895545 0.4881476 0.999581933 1 18
hsa04530 Tight junction 0.633836273 0.526600436 0.999875096 1 44
hsa03050 Proteasome 0.591292167 0.543064357 0.999875665 1 13
hsa00830 Retinol metabolism 0.468670082 0.563733787 0.999941677 1 26
hsa04210 Apoptosis 0.601301566 0.555656094 0.999943594 1 46
hsa04664 Fc epsilon RI signaling pathway 0.604192424 0.560978645 0.999948613 1 31
hsa04975 Fat digestion and absorption 0.652024693 0.578036426 0.99995668 1 10
hsa04260 Cardiac muscle contraction 0.637269495 0.590618725 0.999976426 1 24
hsa04360 Axon guidance 0.586093277 0.591536051 0.999980033 1 52
hsa00520 Amino sugar and nucleotide sugar metabolism 0.646334324 0.622511145 0.999991412 1 29
hsa04630 Jak-STAT signaling pathway 0.616445633 0.648508842 0.999996753 1 59
hsa04976 Bile secretion 0.635464081 0.683424679 0.999998703 1 24
hsa04622 RIG-I-like receptor signaling pathway 0.693807151 0.683065578 0.999998707 1 18
hsa04370 VEGF signaling pathway 0.656028461 0.695719348 0.999999216 1 29
hsa04115 p53 signaling pathway 0.688345993 0.695100274 0.999999276 1 35
hsa04141 Protein processing in endoplasmic reticulum 0.653812985 0.715726473 0.999999677 1 60
hsa00330 Arginine and proline metabolism 0.707152267 0.729924788 0.999999761 1 23
hsa04966 Collecting duct acid secretion 0.697630823 0.740427687 0.999999767 1 12
hsa00760 Nicotinate and nicotinamide metabolism 0.720020429 0.753077767 0.999999832 1 10
hsa00500 Starch and sucrose metabolism 0.548600828 0.76812641 0.999999916 1 29
hsa04722 Neurotrophin signaling pathway 0.663390448 0.795151256 0.999999983 1 49
hsa00512 Mucin type O-Glycan biosynthesis 0.727336763 0.810585824 0.999999985 1 14
hsa04914 Progesterone-mediated oocyte maturation 0.696081054 0.804150142 0.999999987 1 42
hsa04912 GnRH signaling pathway 0.686409867 0.808486958 0.999999989 1 39
hsa04270 Vascular smooth muscle contraction 0.71107199 0.8137631 0.999999992 1 41
hsa04130 SNARE interactions in vesicular transport 0.686226057 0.883380558 0.999999999 1 16
hsa04114 Oocyte meiosis 0.662166189 0.888103923 1 1 50
hsa04540 Gap junction 0.68400657 0.894186191 1 1 36
hsa04320 Dorso-ventral axis formation 0.750619375 0.935747774 1 1 11
hsa00030 Pentose phosphate pathway 0.753237135 0.952746064 1 1 17
hsa00600 Sphingolipid metabolism 0.734002849 0.958750688 1 1 17
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.675621088 0.973182501 1 1 23
hsa00860 Porphyrin and chlorophyll metabolism 0.728729034 0.982959373 1 1 19
hsa00140 Steroid hormone biosynthesis 0.641876744 0.999920789 1 1 18
hsa00564 Glycerophospholipid metabolism 0.70791611 0.976771438 1 1 31
hsa04144 Endocytosis 0.6613336 0.971773613 1 1 81
hsa04670 Leukocyte transendothelial migration 0.697138615 1.039236046 1 1 46
hsa04010 MAPK signaling pathway 0.707186444 1.04808766 1 1 98
hsa00982 Drug metabolism - cytochrome P450 0.610342455 1.087563285 1 1 23
hsa04020 Calcium signaling pathway 0.695878714 1.071985624 1 1 64
hsa00190 Oxidative phosphorylation 0.737152037 1.089125289 1 1 44
hsa04920 Adipocytokine signaling pathway 0.796193616 1.089798802 1 1 33
hsa00480 Glutathione metabolism 0.786642536 1.110178902 1 1 24
hsa00051 Fructose and mannose metabolism 0.823922855 1.172167665 1 1 17
hsa00531 Glycosaminoglycan degradation 0.803065014 1.257329942 1 1 10
hsa00010 Glycolysis / Gluconeogenesis 0.849968102 1.438619389 1 1 31
hsa00052 Galactose metabolism 0.829285731 1.272071517 1 1 16
hsa00983 Drug metabolism - other enzymes 0.749096419 1.452537004 1 1 23
hsa03320 PPAR signaling pathway 0.886542926 1.597233745 1 1 24
hsa04142 Lysosome 0.818320689 2.027149078 1 1 59
hsa04145 Phagosome 0.668260621 1.302658365 1 1 73
hsa04380 Osteoclast differentiation 0.733913776 1.948024403 1 1 65
hsa04510 Focal adhesion 0.74311781 1.478517677 1 1 76
hsa04512 ECM-receptor interaction 0.69877176 1.290258349 1 1 33
hsa04610 Complement and coagulation cascades 0.899216217 2.627115289 1 1 27
hsa04620 Toll-like receptor signaling pathway 0.75078068 1.404108552 1 1 35
hsa04621 NOD-like receptor signaling pathway 0.790571784 1.507315834 1 1 25
hsa04666 Fc gamma R-mediated phagocytosis 0.781807072 1.692698376 1 1 45
hsa04740 Olfactory transduction 0.577770189 1.286221584 1 1 29
hsa04810 Regulation of actin cytoskeleton 0.811449739 1.886943364 1 1 77
hsa04910 Insulin signaling pathway 0.783235592 1.37837677 1 1 61
hsa00061 Fatty acid biosynthesis NA NA NA NA 2
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 3
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 3
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 9
hsa00260 Glycine, serine and threonine metabolism NA NA NA NA 7
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 3
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 5
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 1
hsa00410 beta-Alanine metabolism NA NA NA NA 9
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 4
hsa00450 Selenocompound metabolism NA NA NA NA 9
hsa00460 Cyanoamino acid metabolism NA NA NA NA 2
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 1
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 0
hsa00511 Other glycan degradation NA NA NA NA 4
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 6
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 5
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series NA NA NA NA 9
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 6
hsa00604 Glycosphingolipid biosynthesis - ganglio series NA NA NA NA 9
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 6
hsa00650 Butanoate metabolism NA NA NA NA 9
hsa00670 One carbon pool by folate NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 0
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00770 Pantothenate and CoA biosynthesis NA NA NA NA 6
hsa00780 Biotin metabolism NA NA NA NA 0
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 5
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 4
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 5
hsa01040 Biosynthesis of unsaturated fatty acids NA NA NA NA 8
hsa03020 RNA polymerase NA NA NA NA 9
hsa03450 Non-homologous end-joining NA NA NA NA 5
hsa04122 Sulfur relay system NA NA NA NA 3
hsa04140 Regulation of autophagy NA NA NA NA 9
hsa04340 Hedgehog signaling pathway NA NA NA NA 9
hsa04614 Renin-angiotensin system NA NA NA NA 5
hsa04710 Circadian rhythm - mammal NA NA NA NA 7
hsa04744 Phototransduction NA NA NA NA 9
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 6
hsa04977 Vitamin digestion and absorption NA NA NA NA 8
GSE72829 p.geomean stat.mean p.val q.val set.size
hsa04670 Leukocyte transendothelial migration 0.05415705 1.300194212 2.01E-20 2.73E-18 51
hsa04810 Regulation of actin cytoskeleton 0.048998803 1.27237948 5.85E-20 3.98E-18 91
hsa04142 Lysosome 0.084005127 1.204835267 3.40E-18 1.54E-16 74
hsa04666 Fc gamma R-mediated phagocytosis 0.061311067 1.166895344 6.26E-17 2.13E-15 51
hsa04380 Osteoclast differentiation 0.061344043 1.083941709 5.54E-15 1.51E-13 76
hsa00030 Pentose phosphate pathway 0.128313338 0.990613986 2.73E-12 6.18E-11 16
hsa04664 Fc epsilon RI signaling pathway 0.135860182 0.913638652 4.95E-11 9.62E-10 32
hsa04145 Phagosome 0.133686986 0.758400951 2.76E-08 4.69E-07 82
hsa00010 Glycolysis / Gluconeogenesis 0.20366167 0.739123308 6.59E-08 9.96E-07 35
hsa00500 Starch and sucrose metabolism 0.230093425 0.705126812 2.63E-07 3.58E-06 20
hsa04966 Collecting duct acid secretion 0.213661309 0.704455122 3.65E-07 4.51E-06 13
hsa04650 Natural killer cell mediated cytotoxicity 0.168056547 0.681314767 5.55E-07 6.29E-06 65
hsa04976 Bile secretion 0.228454012 0.676990106 7.91E-07 8.27E-06 19
hsa00190 Oxidative phosphorylation 0.154362526 0.668569919 9.97E-07 9.69E-06 57
hsa00590 Arachidonic acid metabolism 0.24434838 0.654543094 1.53E-06 1.39E-05 22
hsa04662 B cell receptor signaling pathway 0.154654558 0.651501074 1.75E-06 1.49E-05 48
hsa04510 Focal adhesion 0.188401339 0.641011783 2.16E-06 1.73E-05 77
hsa04722 Neurotrophin signaling pathway 0.21096455 0.636079059 2.56E-06 1.94E-05 62
hsa04520 Adherens junction 0.182161148 0.639337348 2.75E-06 1.97E-05 34
hsa00531 Glycosaminoglycan degradation 0.255402943 0.633352137 3.55E-06 2.42E-05 15
hsa04910 Insulin signaling pathway 0.253927411 0.602859056 7.45E-06 4.83E-05 64
hsa00860 Porphyrin and chlorophyll metabolism 0.232700362 0.62124119 7.93E-06 4.90E-05 12
hsa00520 Amino sugar and nucleotide sugar metabolism 0.269205274 0.598621439 9.98E-06 5.90E-05 24
hsa04620 Toll-like receptor signaling pathway 0.245260934 0.552207624 3.88E-05 0.000219832 46
hsa00480 Glutathione metabolism 0.273746242 0.547138149 5.02E-05 0.000273148 19
hsa04210 Apoptosis 0.253284213 0.523650063 8.82E-05 0.000461596 50
hsa04971 Gastric acid secretion 0.264769995 0.518522828 0.000109677 0.000552449 27
hsa04920 Adipocytokine signaling pathway 0.252463312 0.510432064 0.000131522 0.00063882 37
hsa04270 Vascular smooth muscle contraction 0.286367247 0.505650324 0.000148116 0.000694611 37
hsa04062 Chemokine signaling pathway 0.180036176 0.497523935 0.000179582 0.000814105 93
hsa00770 Pantothenate and CoA biosynthesis 0.29851147 0.489354291 0.000280391 0.001230102 11
hsa00051 Fructose and mannose metabolism 0.311024424 0.465713336 0.000453299 0.001926519 20
hsa04530 Tight junction 0.237591825 0.433256405 0.000986309 0.004064789 49
hsa04730 Long-term depression 0.285485043 0.427601115 0.001197421 0.004789683 19
hsa00052 Galactose metabolism 0.346320092 0.383642286 0.003168516 0.012311946 17
hsa04962 Vasopressin-regulated water reabsorption 0.318861612 0.374360219 0.003867763 0.01461155 20
hsa03320 PPAR signaling pathway 0.344239701 0.370554856 0.0040354 0.014832821 29
hsa04622 RIG-I-like receptor signaling pathway 0.334721092 0.367593145 0.004338377 0.015526822 24
hsa00564 Glycerophospholipid metabolism 0.353347659 0.349951851 0.006080495 0.021203777 40
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.347086045 0.344640139 0.007433866 0.025275145 12
hsa04146 Peroxisome 0.357941218 0.336681878 0.007852141 0.026046126 43
hsa04010 MAPK signaling pathway 0.303773303 0.326024378 0.009486739 0.030718964 116
hsa04540 Gap junction 0.35262357 0.306802879 0.014069983 0.044500412 33
hsa00983 Drug metabolism - other enzymes 0.369557312 0.307722038 0.014480895 0.044759131 16
hsa00330 Arginine and proline metabolism 0.369542922 0.296236364 0.017017578 0.051430903 25
hsa04610 Complement and coagulation cascades 0.332372122 0.293370135 0.018308377 0.054085628 25
hsa00600 Sphingolipid metabolism 0.382277084 0.291911068 0.018691357 0.054085628 23
hsa04144 Endocytosis 0.338752752 0.28779002 0.019277297 0.054619009 83
hsa04260 Cardiac muscle contraction 0.313122423 0.286526928 0.021167507 0.058750631 24
hsa04115 p53 signaling pathway 0.37829683 0.264931243 0.028799736 0.078335282 35
hsa00760 Nicotinate and nicotinamide metabolism 0.380364897 0.252157541 0.037371028 0.099656075 11
hsa00512 Mucin type O-Glycan biosynthesis 0.398577935 0.246671028 0.040446026 0.105781914 11
hsa00982 Drug metabolism - cytochrome P450 0.400001444 0.206047237 0.073351914 0.188223778 10
hsa04370 VEGF signaling pathway 0.386378166 0.192329988 0.083921656 0.20857907 35
hsa04973 Carbohydrate digestion and absorption 0.418162638 0.193062855 0.08435183 0.20857907 18
hsa00511 Other glycan degradation 0.419016152 0.182453464 0.099354195 0.241288759 11
hsa04975 Fat digestion and absorption 0.438299 0.144159942 0.1534701 0.366174274 14
hsa04912 GnRH signaling pathway 0.451066488 0.105164164 0.225725679 0.5292878 34
hsa04360 Axon guidance 0.356112742 0.093695189 0.249742443 0.575677495 58
hsa04630 Jak-STAT signaling pathway 0.369759818 0.077546745 0.287398899 0.638999628 64
hsa04972 Pancreatic secretion 0.460986032 0.078237041 0.28776535 0.638999628 29
hsa04621 NOD-like receptor signaling pathway 0.417932398 0.076897348 0.291308654 0.638999628 33
hsa00071 Fatty acid metabolism 0.462888562 0.068292168 0.312546346 0.674703223 22
hsa02010 ABC transporters 0.472205352 0.053002746 0.353234104 0.750622471 13
hsa00562 Inositol phosphate metabolism 0.464587509 0.044131808 0.376372083 0.773007771 23
hsa04130 SNARE interactions in vesicular transport 0.46712152 0.044084385 0.376543896 0.773007771 22
hsa04974 Protein digestion and absorption 0.476241193 0.042573966 0.380820005 0.773007771 21
hsa04150 mTOR signaling pathway 0.461384546 0.02970994 0.415831959 0.831663918 25
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.475815539 0.022498477 0.436063778 0.859488026 14
hsa04114 Oocyte meiosis 0.452100421 0.014563241 0.458823796 0.891429089 53
hsa04960 Aldosterone-regulated sodium reabsorption 0.4899527 0.006472858 0.481575601 0.915739302 13
hsa00640 Propanoate metabolism 0.470580566 0.001655696 0.495325236 0.915739302 18
hsa01040 Biosynthesis of unsaturated fatty acids 0.49627936 0.000527438 0.498599457 0.915739302 13
hsa00380 Tryptophan metabolism 0.489786822 -0.000565913 0.501391975 0.915739302 17
hsa00450 Selenocompound metabolism 0.497639791 -0.001773829 0.505003291 0.915739302 12
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series 0.501935428 -0.010456592 0.529230933 0.938909975 10
hsa04623 Cytosolic DNA-sensing pathway 0.446327511 -0.011807675 0.53436575 0.938909975 22
hsa04310 Wnt signaling pathway 0.431892011 -0.013720991 0.538492486 0.938909975 70
hsa04320 Dorso-ventral axis formation 0.498647009 -0.023521742 0.566589646 0.965592759 11
hsa00565 Ether lipid metabolism 0.504964205 -0.026166305 0.573383612 0.965592759 15
hsa04914 Progesterone-mediated oocyte maturation 0.498039317 -0.028208371 0.580086656 0.965592759 42
hsa04640 Hematopoietic cell lineage 0.457126988 -0.029081356 0.582195634 0.965592759 56
hsa00604 Glycosphingolipid biosynthesis - ganglio series 0.515383567 -0.055600076 0.651622018 1 11
hsa04110 Cell cycle 0.469470205 -0.067187099 0.68558538 1 70
hsa00310 Lysine degradation 0.518944513 -0.071687324 0.695507903 1 23
hsa00240 Pyrimidine metabolism 0.511212106 -0.077453642 0.710719124 1 50
hsa04710 Circadian rhythm - mammal 0.517044399 -0.082909105 0.719708411 1 13
hsa04512 ECM-receptor interaction 0.520742941 -0.081874779 0.720578404 1 27
hsa00260 Glycine, serine and threonine metabolism 0.521665791 -0.08724246 0.73171996 1 16
hsa00561 Glycerolipid metabolism 0.529267397 -0.091483179 0.743411647 1 27
hsa00830 Retinol metabolism 0.531488923 -0.095501208 0.74959857 1 13
hsa04012 ErbB signaling pathway 0.531334814 -0.108659929 0.781558867 1 36
hsa04916 Melanogenesis 0.536788558 -0.116275365 0.79776179 1 43
hsa04720 Long-term potentiation 0.539812588 -0.122464462 0.808898973 1 25
hsa03022 Basal transcription factors 0.546697669 -0.133308704 0.826859015 1 14
hsa04740 Olfactory transduction 0.546439763 -0.134106761 0.831112177 1 31
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.553232201 -0.143383745 0.844185555 1 13
hsa03050 Proteasome 0.375344809 -0.141609659 0.850481622 1 23
hsa00270 Cysteine and methionine metabolism 0.513154296 -0.151542624 0.859221979 1 18
hsa04070 Phosphatidylinositol signaling system 0.541313547 -0.1573775 0.869816489 1 32
hsa04742 Taste transduction 0.566138028 -0.179815313 0.897661298 1 13
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.565878474 -0.181485207 0.898275033 1 10
hsa00350 Tyrosine metabolism 0.556096966 -0.188749709 0.90935875 1 17
hsa03060 Protein export 0.516360908 -0.198999719 0.920339465 1 14
hsa00650 Butanoate metabolism 0.566745718 -0.207254297 0.927911669 1 13
hsa04141 Protein processing in endoplasmic reticulum 0.531407778 -0.206011985 0.930676734 1 75
hsa04020 Calcium signaling pathway 0.570299867 -0.215597936 0.939172555 1 58
hsa04970 Salivary secretion 0.526780031 -0.225270502 0.946310651 1 23
hsa04340 Hedgehog signaling pathway 0.583940654 -0.228833175 0.947791429 1 17
hsa00280 Valine, leucine and isoleucine degradation 0.579311776 -0.235482498 0.953612291 1 28
hsa00340 Histidine metabolism 0.582973922 -0.243246524 0.95705023 1 13
hsa03020 RNA polymerase 0.580728054 -0.248585184 0.959928142 1 13
hsa03420 Nucleotide excision repair 0.554329955 -0.252428913 0.96352221 1 24
hsa00620 Pyruvate metabolism 0.544684903 -0.263946904 0.970046305 1 22
hsa04120 Ubiquitin mediated proteolysis 0.552684596 -0.272689134 0.974702814 1 62
hsa03410 Base excision repair 0.575019955 -0.275968942 0.974751055 1 19
hsa00020 Citrate cycle (TCA cycle) 0.563113544 -0.283547069 0.977726892 1 16
hsa04330 Notch signaling pathway 0.598547966 -0.285039521 0.978917046 1 23
hsa03440 Homologous recombination 0.609275818 -0.308974306 0.984705069 1 10
hsa00510 N-Glycan biosynthesis 0.604102455 -0.304665233 0.984745201 1 20
hsa03430 Mismatch repair 0.610380943 -0.314208095 0.986416668 1 12
hsa03030 DNA replication 0.595469255 -0.343968193 0.992555544 1 19
hsa03008 Ribosome biogenesis in eukaryotes 0.593444661 -0.388030206 0.99722464 1 38
hsa04350 TGF-beta signaling pathway 0.619756171 -0.392880015 0.997558426 1 41
hsa00970 Aminoacyl-tRNA biosynthesis 0.622543257 -0.422848601 0.998675768 1 24
hsa00230 Purine metabolism 0.632716287 -0.450409799 0.999391342 1 74
hsa00514 Other types of O-glycan biosynthesis 0.665157296 -0.472557863 0.99958388 1 15
hsa03015 mRNA surveillance pathway 0.675444585 -0.545651016 0.999951037 1 33
hsa04660 T cell receptor signaling pathway 0.66128095 -0.605689364 0.999993102 1 68
hsa03018 RNA degradation 0.67967229 -0.646287141 0.999998053 1 35
hsa04514 Cell adhesion molecules (CAMs) 0.639242597 -0.707047442 0.999999778 1 57
hsa03013 RNA transport 0.679393562 -1.044284155 1 1 69
hsa03040 Spliceosome 0.748589683 -1.058781283 1 1 76
hsa03010 Ribosome 0.320420663 -4.234576248 1 1 50
hsa04612 Antigen processing and presentation 0.743070708 -1.247614409 1 1 48
hsa04672 Intestinal immune network for IgA production 0.83971879 -1.555921998 1 1 29
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 8
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 3
hsa00061 Fatty acid biosynthesis NA NA NA NA 4
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 4
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 5
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00140 Steroid hormone biosynthesis NA NA NA NA 7
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 8
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 6
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00410 beta-Alanine metabolism NA NA NA NA 9
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 3
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 1
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 1
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 5
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 3
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 9
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 6
hsa00670 One carbon pool by folate NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 2
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00780 Biotin metabolism NA NA NA NA 1
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 4
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 6
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 8
hsa03450 Non-homologous end-joining NA NA NA NA 5
hsa04122 Sulfur relay system NA NA NA NA 5
hsa04140 Regulation of autophagy NA NA NA NA 8
hsa04614 Renin-angiotensin system NA NA NA NA 7
hsa04744 Phototransduction NA NA NA NA 7
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 4
hsa04977 Vitamin digestion and absorption NA NA NA NA 6
hsa03010 Ribosome 9.17E-07 -4.234576248 5.32E-154 7.24E-152 50
hsa04672 Intestinal immune network for IgA production 0.04881873 -1.555921998 3.72E-28 2.53E-26 29
hsa04612 Antigen processing and presentation 0.073141503 -1.247614409 4.33E-19 1.96E-17 48
hsa03040 Spliceosome 0.118116448 -1.058781283 1.87E-14 6.36E-13 76
hsa03013 RNA transport 0.104722919 -1.044284155 4.33E-14 1.18E-12 69
hsa04514 Cell adhesion molecules (CAMs) 0.181617266 -0.707047442 2.22E-07 5.03E-06 57
hsa03018 RNA degradation 0.232682495 -0.646287141 1.95E-06 3.78E-05 35
hsa04660 T cell receptor signaling pathway 0.239198359 -0.605689364 6.90E-06 0.000117272 68
hsa03015 mRNA surveillance pathway 0.278293382 -0.545651016 4.90E-05 0.000739888 33
hsa00514 Other types of O-glycan biosynthesis 0.313532887 -0.472557863 0.00041612 0.005659234 15
hsa00230 Purine metabolism 0.303521584 -0.450409799 0.000608658 0.007525229 74
hsa00970 Aminoacyl-tRNA biosynthesis 0.315071711 -0.422848601 0.001324232 0.015007958 24
hsa04350 TGF-beta signaling pathway 0.327643526 -0.392880015 0.002441574 0.025542624 41
hsa03008 Ribosome biogenesis in eukaryotes 0.315995839 -0.388030206 0.00277536 0.026960636 38
hsa03030 DNA replication 0.343720855 -0.343968193 0.007444456 0.067496399 19
hsa03430 Mismatch repair 0.372674369 -0.314208095 0.013583332 0.115458325 12
hsa00510 N-Glycan biosynthesis 0.372324425 -0.304665233 0.015254799 0.115561699 20
hsa03440 Homologous recombination 0.376001956 -0.308974306 0.015294931 0.115561699 10
hsa04330 Notch signaling pathway 0.379533174 -0.285039521 0.021082954 0.150909564 23
hsa00020 Citrate cycle (TCA cycle) 0.358399472 -0.283547069 0.022273108 0.151457134 16
hsa03410 Base excision repair 0.370392003 -0.275968942 0.025248945 0.156382607 19
hsa04120 Ubiquitin mediated proteolysis 0.355060165 -0.272689134 0.025297186 0.156382607 62
hsa00620 Pyruvate metabolism 0.35635919 -0.263946904 0.029953695 0.177117501 22
hsa03420 Nucleotide excision repair 0.36979153 -0.252428913 0.03647779 0.206707474 24
hsa03020 RNA polymerase 0.392774662 -0.248585184 0.040071858 0.217990907 13
hsa00340 Histidine metabolism 0.397288327 -0.243246524 0.04294977 0.224660335 13
hsa00280 Valine, leucine and isoleucine degradation 0.398078308 -0.235482498 0.046387709 0.233656608 28
hsa04340 Hedgehog signaling pathway 0.406776113 -0.228833175 0.052208571 0.251784533 17
hsa04970 Salivary secretion 0.366581271 -0.225270502 0.053689349 0.251784533 23
hsa04020 Calcium signaling pathway 0.403703432 -0.215597936 0.060827445 0.275751086 58
hsa04141 Protein processing in endoplasmic reticulum 0.381702908 -0.206011985 0.069323266 0.304127877 75
hsa00650 Butanoate metabolism 0.409377434 -0.207254297 0.072088331 0.306375407 13
hsa03060 Protein export 0.377675947 -0.198999719 0.079660535 0.328297964 14
hsa00350 Tyrosine metabolism 0.412770751 -0.188749709 0.09064125 0.362565001 17
hsa00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis 0.426613556 -0.181485207 0.101724967 0.386612873 10
hsa04742 Taste transduction 0.427173797 -0.179815313 0.102338702 0.386612873 13
hsa04070 Phosphatidylinositol signaling system 0.420503382 -0.1573775 0.130183511 0.478512363 32
hsa00270 Cysteine and methionine metabolism 0.403625067 -0.151542624 0.140778021 0.503837127 18
hsa03050 Proteasome 0.30919273 -0.141609659 0.149518378 0.521397421 23
hsa00534 Glycosaminoglycan biosynthesis - heparan sulfate 0.44199671 -0.143383745 0.155814445 0.529769115 13
hsa04740 Olfactory transduction 0.441576533 -0.134106761 0.168887823 0.560213266 31
hsa03022 Basal transcription factors 0.443549634 -0.133308704 0.173140985 0.560646998 14
hsa04720 Long-term potentiation 0.444469694 -0.122464462 0.191101027 0.604412552 25
hsa04916 Melanogenesis 0.445697524 -0.116275365 0.20223821 0.625099923 43
hsa04012 ErbB signaling pathway 0.446963275 -0.108659929 0.218441133 0.660177645 36
hsa00830 Retinol metabolism 0.457717745 -0.095501208 0.25040143 0.740317271 13
hsa00561 Glycerolipid metabolism 0.4576984 -0.091483179 0.256588353 0.742468426 27
hsa00260 Glycine, serine and threonine metabolism 0.455112992 -0.08724246 0.26828004 0.760126779 16
hsa04512 ECM-receptor interaction 0.457427865 -0.081874779 0.279421596 0.762393123 27
hsa04710 Circadian rhythm - mammal 0.45427317 -0.082909105 0.280291589 0.762393123 13
hsa00240 Pyrimidine metabolism 0.451698641 -0.077453642 0.289280876 0.77141567 50
hsa00310 Lysine degradation 0.463147847 -0.071687324 0.304492097 0.796363946 23
hsa04110 Cell cycle 0.424928972 -0.067187099 0.31441462 0.80679978 70
hsa00604 Glycosphingolipid biosynthesis - ganglio series 0.472727794 -0.055600076 0.348377982 0.8773964 11
hsa04640 Hematopoietic cell lineage 0.432301457 -0.029081356 0.417804366 1 56
hsa04914 Progesterone-mediated oocyte maturation 0.476273347 -0.028208371 0.419913344 1 42
hsa00565 Ether lipid metabolism 0.484641547 -0.026166305 0.426616388 1 15
hsa04320 Dorso-ventral axis formation 0.480447046 -0.023521742 0.433410354 1 11
hsa04310 Wnt signaling pathway 0.419394903 -0.013720991 0.461507514 1 70
hsa04623 Cytosolic DNA-sensing pathway 0.438963293 -0.011807675 0.46563425 1 22
hsa00601 Glycosphingolipid biosynthesis - lacto and neolacto series 0.493861413 -0.010456592 0.470769067 1 10
hsa00450 Selenocompound metabolism 0.49627572 -0.001773829 0.494996709 1 12
hsa00380 Tryptophan metabolism 0.489696095 -0.000565913 0.498608025 1 17
hsa01040 Biosynthesis of unsaturated fatty acids 0.496729691 0.000527438 0.501400543 1 13
hsa00640 Propanoate metabolism 0.473172547 0.001655696 0.504674764 1 18
hsa04960 Aldosterone-regulated sodium reabsorption 0.495021092 0.006472858 0.518424399 1 13
hsa04114 Oocyte meiosis 0.466606892 0.014563241 0.541176204 1 53
hsa00532 Glycosaminoglycan biosynthesis - chondroitin sulfate 0.49316074 0.022498477 0.563936222 1 14
hsa04150 mTOR signaling pathway 0.483898981 0.02970994 0.584168041 1 25
hsa04974 Protein digestion and absorption 0.509542926 0.042573966 0.619179995 1 21
hsa04130 SNARE interactions in vesicular transport 0.501382719 0.044084385 0.623456104 1 22
hsa00562 Inositol phosphate metabolism 0.498536373 0.044131808 0.623627917 1 23
hsa02010 ABC transporters 0.513567334 0.053002746 0.646765896 1 13
hsa00071 Fatty acid metabolism 0.516120141 0.068292168 0.687453654 1 22
hsa04621 NOD-like receptor signaling pathway 0.474619639 0.076897348 0.708691346 1 33
hsa04972 Pancreatic secretion 0.522192854 0.078237041 0.71223465 1 29
hsa04630 Jak-STAT signaling pathway 0.41328336 0.077546745 0.712601101 1 64
hsa04360 Axon guidance 0.413139664 0.093695189 0.750257557 1 58
hsa04912 GnRH signaling pathway 0.533390471 0.105164164 0.774274321 1 34
hsa04975 Fat digestion and absorption 0.550249731 0.144159942 0.8465299 1 14
hsa00511 Other glycan degradation 0.55853026 0.182453464 0.900645805 1 11
hsa04973 Carbohydrate digestion and absorption 0.568165781 0.193062855 0.91564817 1 18
hsa04370 VEGF signaling pathway 0.527348675 0.192329988 0.916078344 1 35
hsa00982 Drug metabolism - cytochrome P450 0.55530716 0.206047237 0.926648086 1 10
hsa00512 Mucin type O-Glycan biosynthesis 0.588221709 0.246671028 0.959553974 1 11
hsa00760 Nicotinate and nicotinamide metabolism 0.567379195 0.252157541 0.962628972 1 11
hsa04115 p53 signaling pathway 0.58016617 0.264931243 0.971200264 1 35
hsa04260 Cardiac muscle contraction 0.508816415 0.286526928 0.978832493 1 24
hsa04144 Endocytosis 0.543355757 0.28779002 0.980722703 1 83
hsa00600 Sphingolipid metabolism 0.608181201 0.291911068 0.981308643 1 23
hsa04610 Complement and coagulation cascades 0.541208764 0.293370135 0.981691623 1 25
hsa00330 Arginine and proline metabolism 0.595473579 0.296236364 0.982982422 1 25
hsa00983 Drug metabolism - other enzymes 0.603809476 0.307722038 0.985519105 1 16
hsa04540 Gap junction 0.579858035 0.306802879 0.985930017 1 33
hsa04010 MAPK signaling pathway 0.520784553 0.326024378 0.990513261 1 116
hsa04146 Peroxisome 0.615680193 0.336681878 0.992147859 1 43
hsa00980 Metabolism of xenobiotics by cytochrome P450 0.603305618 0.344640139 0.992566134 1 12
hsa00564 Glycerophospholipid metabolism 0.620620284 0.349951851 0.993919505 1 40
hsa04622 RIG-I-like receptor signaling pathway 0.606541439 0.367593145 0.995661623 1 24
hsa03320 PPAR signaling pathway 0.624806784 0.370554856 0.9959646 1 29
hsa04962 Vasopressin-regulated water reabsorption 0.584585048 0.374360219 0.996132237 1 20
hsa00052 Galactose metabolism 0.63845918 0.383642286 0.996831484 1 17
hsa04730 Long-term depression 0.573727796 0.427601115 0.998802579 1 19
hsa04530 Tight junction 0.498026244 0.433256405 0.999013691 1 49
hsa00051 Fructose and mannose metabolism 0.658391143 0.465713336 0.999546701 1 20
hsa00770 Pantothenate and CoA biosynthesis 0.653033847 0.489354291 0.999719609 1 11
hsa04062 Chemokine signaling pathway 0.43477825 0.497523935 0.999820418 1 93
hsa04270 Vascular smooth muscle contraction 0.653822166 0.505650324 0.999851884 1 37
hsa04920 Adipocytokine signaling pathway 0.592032085 0.510432064 0.999868478 1 37
hsa04971 Gastric acid secretion 0.618065663 0.518522828 0.999890323 1 27
hsa04210 Apoptosis 0.610866122 0.523650063 0.999911754 1 50
hsa00480 Glutathione metabolism 0.669833063 0.547138149 0.999949789 1 19
hsa04620 Toll-like receptor signaling pathway 0.617618464 0.552207624 0.999961206 1 46
hsa00520 Amino sugar and nucleotide sugar metabolism 0.708634695 0.598621439 0.999990023 1 24
hsa00860 Porphyrin and chlorophyll metabolism 0.631675707 0.62124119 0.999992068 1 12
hsa04910 Insulin signaling pathway 0.687013218 0.602859056 0.999992549 1 64
hsa00531 Glycosaminoglycan degradation 0.711055963 0.633352137 0.999996447 1 15
hsa04520 Adherens junction 0.537875253 0.639337348 0.999997254 1 34
hsa04722 Neurotrophin signaling pathway 0.620863687 0.636079059 0.999997438 1 62
hsa04510 Focal adhesion 0.570032152 0.641011783 0.99999784 1 77
hsa04662 B cell receptor signaling pathway 0.482772868 0.651501074 0.999998246 1 48
hsa00590 Arachidonic acid metabolism 0.71040965 0.654543094 0.999998467 1 22
hsa00190 Oxidative phosphorylation 0.54379846 0.668569919 0.999999003 1 57
hsa04976 Bile secretion 0.691975954 0.676990106 0.999999209 1 19
hsa04650 Natural killer cell mediated cytotoxicity 0.539077201 0.681314767 0.999999445 1 65
hsa04966 Collecting duct acid secretion 0.677815621 0.704455122 0.999999635 1 13
hsa00500 Starch and sucrose metabolism 0.728003334 0.705126812 0.999999737 1 20
hsa00010 Glycolysis / Gluconeogenesis 0.708824854 0.739123308 0.999999934 1 35
hsa04145 Phagosome 0.494910222 0.758400951 0.999999972 1 82
hsa04664 Fc epsilon RI signaling pathway 0.654002099 0.913638652 1 1 32
hsa00030 Pentose phosphate pathway 0.689858521 0.990613986 1 1 16
hsa04380 Osteoclast differentiation 0.480740605 1.083941709 1 1 76
hsa04666 Fc gamma R-mediated phagocytosis 0.559549688 1.166895344 1 1 51
hsa04142 Lysosome 0.745016658 1.204835267 1 1 74
hsa04670 Leukocyte transendothelial migration 0.612557078 1.300194212 1 1 51
hsa04810 Regulation of actin cytoskeleton 0.560717439 1.27237948 1 1 91
hsa00040 Pentose and glucuronate interconversions NA NA NA NA 8
hsa00053 Ascorbate and aldarate metabolism NA NA NA NA 3
hsa00061 Fatty acid biosynthesis NA NA NA NA 4
hsa00072 Synthesis and degradation of ketone bodies NA NA NA NA 4
hsa00100 Steroid biosynthesis NA NA NA NA 6
hsa00120 Primary bile acid biosynthesis NA NA NA NA 5
hsa00130 Ubiquinone and other terpenoid-quinone biosynthesis NA NA NA NA 4
hsa00140 Steroid hormone biosynthesis NA NA NA NA 7
hsa00232 Caffeine metabolism NA NA NA NA 1
hsa00250 Alanine, aspartate and glutamate metabolism NA NA NA NA 8
hsa00290 Valine, leucine and isoleucine biosynthesis NA NA NA NA 8
hsa00300 Lysine biosynthesis NA NA NA NA 0
hsa00360 Phenylalanine metabolism NA NA NA NA 6
hsa00400 Phenylalanine, tyrosine and tryptophan biosynthesis NA NA NA NA 2
hsa00410 beta-Alanine metabolism NA NA NA NA 9
hsa00430 Taurine and hypotaurine metabolism NA NA NA NA 3
hsa00460 Cyanoamino acid metabolism NA NA NA NA 3
hsa00471 D-Glutamine and D-glutamate metabolism NA NA NA NA 1
hsa00472 D-Arginine and D-ornithine metabolism NA NA NA NA 1
hsa00533 Glycosaminoglycan biosynthesis - keratan sulfate NA NA NA NA 6
hsa00591 Linoleic acid metabolism NA NA NA NA 5
hsa00592 alpha-Linolenic acid metabolism NA NA NA NA 3
hsa00603 Glycosphingolipid biosynthesis - globo series NA NA NA NA 9
hsa00630 Glyoxylate and dicarboxylate metabolism NA NA NA NA 6
hsa00670 One carbon pool by folate NA NA NA NA 8
hsa00730 Thiamine metabolism NA NA NA NA 2
hsa00740 Riboflavin metabolism NA NA NA NA 5
hsa00750 Vitamin B6 metabolism NA NA NA NA 4
hsa00780 Biotin metabolism NA NA NA NA 1
hsa00785 Lipoic acid metabolism NA NA NA NA 2
hsa00790 Folate biosynthesis NA NA NA NA 4
hsa00900 Terpenoid backbone biosynthesis NA NA NA NA 6
hsa00910 Nitrogen metabolism NA NA NA NA 7
hsa00920 Sulfur metabolism NA NA NA NA 8
hsa03450 Non-homologous end-joining NA NA NA NA 5
hsa04122 Sulfur relay system NA NA NA NA 5
hsa04140 Regulation of autophagy NA NA NA NA 8
hsa04614 Renin-angiotensin system NA NA NA NA 7
hsa04744 Phototransduction NA NA NA NA 7
hsa04964 Proximal tubule bicarbonate reclamation NA NA NA NA 4
hsa04977 Vitamin digestion and absorption NA NA NA NA 6
AttachmentSubmitted filename: comments.to.reviewer.docx