Multiple digital data streams forecast COVID-19 activity weeks before traditional epidemiological surveillance.
Abstract
Given still-high levels of coronavirus disease 2019 (COVID-19) susceptibility and inconsistent transmission-containing strategies, outbreaks have continued to emerge across the United States. Until effective vaccines are widely deployed, curbing COVID-19 will require carefully timed nonpharmaceutical interventions (NPIs). A COVID-19 early warning system is vital for this. Here, we evaluate digital data streams as early indicators of state-level COVID-19 activity from 1 March to 30 September 2020. We observe that increases in digital data stream activity anticipate increases in confirmed cases and deaths by 2 to 3 weeks. Confirmed cases and deaths also decrease 2 to 4 weeks after NPI implementation, as measured by anonymized, phone-derived human mobility data. We propose a means of harmonizing these data streams to identify future COVID-19 outbreaks. Our results suggest that combining disparate health and behavioral data may help identify disease activity changes weeks before observation using traditional epidemiological monitoring.
INTRODUCTION
In just over 12 months, coronavirus disease 2019 (COVID-19)—the disease caused by the betacoronavirus SARS-CoV-2—has caused more than 2,200,000 deaths worldwide, 455,000 of which are in the United States (1). In the absence of a widely available vaccine or an effective treatment, authorities have sought to slow epidemic growth by implementing nonpharmaceutical interventions (NPIs), including school and business closures, work-from-home policies, and travel bans. Many U.S. states have progressively reopened their economies, despite estimates of cumulative US COVID-19 incidence, suggesting that most of the U.S. population remains susceptible to SARS-CoV-2 (2). Serological studies also indicate low levels of seroprevalence even in parts of the United States heavily affected by the virus (e.g., 23% in New York City by 29 May 2020) (3, 4). The long development timeline for a vaccine (5) coupled with the possibility that individual immunity to SARS-CoV-2 may decline over time (as is the case with other coronaviruses) portends the emergence of additional epidemic waves (6). The availability of a reliable, robust, real-time indicator of emerging COVID-19 outbreaks would aid in appropriately timing public health interventions.
Despite efforts by the clinical and research community to aggregate and make available data streams that are representative of COVID-19 activity, it is not immediately clear which of these data streams is most dependable for tracking outbreaks in real time. Many metrics for tracking COVID-19, such as confirmed cases, hospitalizations, and deaths, suffer from reporting delays, as well as uncertainties stemming from inefficiencies in data collection, collation, and dissemination processes (7). For example, confirmed cases may be more reflective of testing availability than of disease incidence, and confirmed cases often lag infections by days or weeks (8). Previous work has suggested that clinician-provided reports of the proportion of hospital visits presenting influenza-like illness (ILI), aggregated by the Centers for Disease Control and Prevention (CDC), may be less sensitive to testing availability than confirmed cases, but these reports suffer from reporting lags of 5 to 12 days, are not typically collected during summer months, depend on the thoroughness of clinician reporting, and do not distinguish COVID-19 from other illnesses that may cause similar symptoms (2).
Alternatively, forecasting models can assist in long-term planning, but the accuracy of their predictions is limited by the timeliness of data or parameter updates. Specifically, some models demonstrate predictive skill with respect to hospitalizations and deaths (6, 9), but these predictions are often too late to enable timely NPI implementation. Other models suffer from limited generalizability, with NPI implementation proposed only for a specific city (10). The CDC has launched a state-level forecasting initiative aimed at consolidating predictions from multiple models to estimate future COVID-19–attributable deaths, but the use of these predictions in state-level decision-making is still pending (11).
Over the last decade, innovative methodologies have emerged to track population-level disease spread using data sources not originally conceived for that purpose (12). These approaches have exploited information from internet and clinicians’ search engines (13–19), news reports (20–22), crowd-sourced participatory disease surveillance systems (23, 24), Twitter microblogs (25, 26), electronic health records (27, 28), Wikipedia traffic (29), wearable devices (30), smartphone-connected thermometers (31), and travel websites (32) to estimate disease prevalence in near real time. Several have already been used to track COVID-19 (33, 34). These data sources are liable to bias, however; for example, Google Search activity is sensitive to the intensity of news coverage (15, 35–37). Methodologies to mitigate biases in digital data sources commonly involve combining disease history, mechanistic models, and surveys to produce detrended estimates of disease activity (38, 39).
Our contribution
We propose that several digital data sources may provide earlier indication of epidemic spread than traditional COVID-19 metrics such as confirmed cases or deaths. Six such sources are examined here: (i) Google Trends patterns for a suite of COVID-19–related terms; (ii) COVID-19–related Twitter activity; (iii) COVID-19–related clinician searches from UpToDate; (iv) predictions by the global epidemic and mobility model (GLEAM), a state-of-the-art metapopulation mechanistic model; (v) anonymized and aggregated human mobility data from smartphones; and (vi) Kinsa smart thermometer measurements. We first evaluate each of these “proxies” of COVID-19 activity for their lead or lag relative to traditional measures of COVID-19 activity: confirmed cases, deaths attributed, and ILI. We then propose the use of a metric combining these data sources into a multiproxy estimate of the probability of an impending COVID-19 outbreak. Last, we develop probabilistic estimates of when such a COVID-19 outbreak will occur on the basis of multiproxy variability. These outbreak-timing predictions are made for two separate time periods: the first, a “training” period, from 1 March to 31 May 2020, and the second, a “validation” period, from 1 June to 30 September 2020. Consistent predictive behavior among proxies in both of these subsequent and nonoverlapping time periods would increase the confidence that they may capture future changes in the trajectory of COVID-19 activity.
RESULTS
Visualizing the behavior of COVID-19–tracking data sources: Motivation for designing an early warning system
Figure 1 displays the temporal evolution of all available signals considered in this study for three U.S. states—Massachusetts (MA), New York (NY), and California (CA)—over five time intervals. These states illustrate different early epidemic trajectories within the United States, with NY among the worst affected states initially and CA experiencing a more gradual increase in cases than both MA and NY.
Fig. 1. Visualization of the evolution of each COVID-19 proxy in Massachusetts, New York, and California.
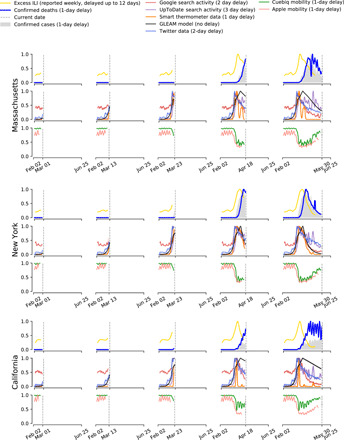
Columns depict progressively increasing time periods over which proxies become available (vertical dashed line indicates the latest date in the time period) to illustrate how the availability of different proxies informs upon the evolution of COVID-19. Time series were normalized between 0 and 1 and smoothed using a simple moving average for purposes of visualization. The legend at the top shows each data stream alongside typical delays between measurement and reporting.
The top row of Fig. 1 for each state displays normalized COVID-19 activity as captured by daily reported confirmed cases, deaths, and “excess ILI” (raw hospitalizations were discounted owing to data sparseness). Excess ILI refers to hospital visits due to ILI in excess of what is expected from a normal flu season (2), which we attribute to COVID-19 in 2020. ILI data were taken from the CDC’s U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet). The middle row for each state displays time series for five proxies of COVID-19 activity. The bottom row for each state displays state-level anonymized and aggregated human mobility data as collected by mobile phones; mobility data are viewed as a proxy for adherence to social distancing recommendations. Similar visualizations for all states are shown in figs. S1 to S17.
Figure 1 demonstrates that for the earliest time period in the pandemic (“time period 1,” 1 March to 31 May 2020) in MA, NY, and CA, COVID-19–related clinicians’ and general population’s internet activity, smart thermometers, and GLEAM model predictions exhibit early increases that led increases of confirmed cases and deaths due to COVID-19. Analogously, decreases in other proxies—especially in mobility—mirror later decreases in COVID-19–attributable confirmed cases and deaths for the three states represented. This is not seen in all states as some, such as Arizona, Florida, and Texas, had not experienced a complete “first wave” of COVID-19 by the end of the first time period. Furthermore, such states did not increase their testing capacity until later into the summer months, making it difficult to observe early changes in COVID-19 activity. Analysis conducted on a subsequent time period (“time period 2,” 1 June to 30 September 2020) allows for a more detailed characterization of these nuances.
Quantifying the timing of growth in proxies of COVID-19 activity
To quantify the relative leads and lags in our collection of disease proxies, we formulated a change of behavior “event” for each proxy and compared it to three “gold standards” of COVID-19 activity: confirmed cases, deaths attributed, and excess ILI. In keeping with classical disease dynamics, we defined an event as any initiation of exponential growth (“uptrend”). Using a Bayesian approach, we obtained a posterior probability distribution for parameter values in a function y(t) ∼ β eγ(t − t0)+ ϵ(t) approximating the behavior of each time series over a time window of 14 days, evaluating β, γ, and the variance of ϵ, denoted σ2, where t0 indicates the first day in the time window. A P value was then calculated for each proxy on each day that represents the posterior probability that γ is less than or equal to zero in the past 14 days. As the P values decrease, we grow more confident that a given time series is exhibiting sustained growth. When the P value decreases below 0.05, we define this as an individual proxy’s “uptrend event.” An additional indicator (discussed further in subsequent paragraphs), built from the combination of the P values of each proxy using the harmonic mean method (see Eq. 5), was calculated for every day of the observed data. We conducted this analysis over both time periods 1 and 2 to assess proxy behavior at different stages of the pandemic.
The sequences of proxy-specific uptrends for an example state, CA, are depicted in Fig. 2. Our choice of this state was motivated by the presence of multiple distinguishable and nonoverlapping growth events over time periods 1 and 2. Upward-pointing triangles denote the date on which a growth event is identifiable by our method for each individual data stream. For CA, Fig. 2 shows that COVID-19–related Twitter posts gave the earliest indication of increasing COVID-19 activity, exhibiting its first uptrend around 2 March. This was closely followed by uptrends in Google searches for “fever,” GLEAM-modeled infections, COVID-19–related searches by clinicians, and fever incidence. Subsequent exponential growth events occurred in most of these proxies.
Fig. 2. Visualization of the event detection procedure applied to COVID-19 proxies.
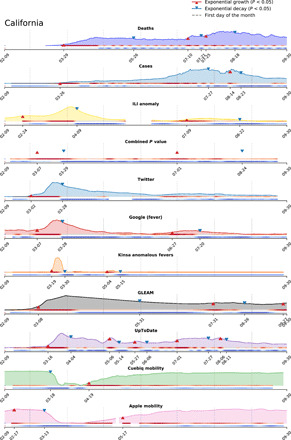
An event is detected by setting a threshold of 0.05 over the P value of the exponential coefficient γ. Under each curve, the P values are shown as colored gradients. Darker red shade signifies increased confidence in the occurrence of an uptrend event, while darker blue shade signifies increased confidence in the occurrence of a downtrend event. Triangular markers are used to signal the date when an uptrend or a downtrend is detected based on the set threshold. The time series are adjusted to account for expected reporting delays in the source of information.
The activation order of COVID-19 early warning indicators in CA is characterized by earlier growth in proxies reflecting public sentiment than in more clinically specific proxies. This ordering is broadly observed across states over time period 1 (Fig. 3A) and, generally, over time period 2 (Fig. 3C). For time period 1, COVID-19–related Twitter posts and Google searches for “fever” were among the earliest proxies to activate, with Twitter activating first for 35 states and Google activating first for 7 states. UpToDate showed the latest activation among proxies, activating last in 37 states albeit still predating an uptrend in confirmed cases (fig. S66). This analysis was conducted for all states with the exception of those where data were unavailable because of reporting delays and/or event information was missing (as was the case for deaths in two states, Kinsa in one state, and excess ILI in five states).
Fig. 3. Event detection results for pairwise comparisons between COVID-19 proxies and gold standards for U.S. states with available data across two time periods.
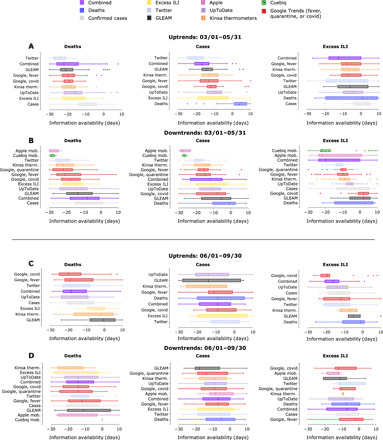
(A) and (B) illustrate the timing of proxy-specific uptrends and downtrends, respectively, relative to deaths, confirmed cases, and excess ILI from 1 March 1 to 31 May 2020. (C) and (D) similarly use box plots to illustrate the lag of COVID-19 gold standards relative to COVID-19 proxies from 1 June to 30 September 2020. Multiple events can be detected per state per time period. Proxy data were unavailable for certain states, which reflects their absence in the plots. Box plots show the median (central vertical lines), interquartile range (vertical lines flanking the median), extrema (whiskers), and outliers (dots); differences between input variable (y axis) and response variable (title) exceeding 30 days are omitted. Negative differences indicate that the input variable event activation preceded the response variable event activation. Deaths, confirmed cases, and excess ILI, as well as a combined measure, are included to intercompare gold standards. Box plots are sorted according to median value and shifted to offset delays in real-time availability.
Later in the pandemic, during time period 2, Twitter posts and Google searches for “covid” showed the earliest median activation relative to deaths, with the latter also showing the earliest median activation relative to ILI, while UpToDate showed the earliest median activation relative to cases (Fig. 3C). For both time periods, we excluded mobility time series data from the uptrend lead-lag analysis as their influence on subsequent disease transmission depends heavily on the underlying disease incidence in a given location at a given point in time. Specifically, if shelter-in-place interventions (captured as reductions in human mobility) were effectively implemented (as in NY earlier in the pandemic), then subsequent increases in human mobility (due to a relaxation of lockdowns) may not lead to increases in disease transmission. In contrast, in regions where shelter-in-place interventions were not observed closely or for long enough, increases in human mobility (due to premature reopenings) may lead to noticeable increased disease transmission. As such, human mobility reductions were only consistently found to influence downtrend events in the epidemiological gold standards.
Although data streams that directly measure public sentiment (i.e., Google and Twitter) are sensitive to the intensity of news reporting, we note that median growth in Google searches for “fever” occurs within 3 days of median growth in fever incidence (as measured by Kinsa) for time period 1, suggesting that many searches may be driven by newly ill people (Fig. 3A). We see a similar closeness in median uptrend activation (i.e., approximately 3 days difference) for Kinsa and Google searches for “fever” during time period 2, specifically with respect to cases. We additionally observed that the median lag between deaths and Google Searches for “fever” was approximately 21 days during both time periods 1 and 2, broadly consistent with previous estimates that the average delay between the onset of symptoms and death is 20 days (40). Detailed time series and event activation dates for the first time period are displayed in figs. S18 to S65.
Consensus among proxies was recorded through a “combined” growth event that acts as a consolidated metric for all proxies’ uptrend events (Figs. 2 and 3). We found that this metric was able to capture changes in behavior across proxies, reflecting a median activation 2 to 3 weeks before uptrend events among deaths, cases, and excess ILI across both time periods. As described in more detail after the next section, this combined event was created via a harmonic mean taken across the P values associated with each of the indicators. The harmonic mean was used because it does not require P values across different proxies to be independent (41). Similar to the case for individual proxies, we defined detection of a growth event to occur when the harmonic mean P value (HMP) decreases below 0.05.
Quantifying the timing of decay in proxies
An examination analogous to that made for the uptrend events was made to identify the timing of exponential decay (“downtrend”) in proxies—now including mobility data—and gold standard time series, for time periods 1 and 2. On each day of the time series and for each proxy, a P value is calculated to represent the posterior probability that γ is greater than or equal to zero. An individual proxy’s downtrend was defined to occur on days when the associated P value crossed a 0.05 threshold, representing the opposite null hypothesis as that adopted for detecting uptrend events. A sample sequence of downtrends is depicted in Fig. 2, where downward-pointing triangles denote the date on which a downtrend event is identifiable. For the example state, Cuebiq and Apple mobility data are the first time series to exhibit downtrends, each activating events around 15 March.
The utility of extending our analysis to include decay events is in possibly characterizing downstream effects of NPIs. Specifically, opportunities to rapidly assess NPI effectiveness may arise if NPI influence on transmission rates is recorded in proxy time series before it is recorded in confirmed case or death time series. We used two smartphone-based metrics of human mobility that are indicators of population-wide propensity to travel within a given U.S. county, as provided by the location analytics company Cuebiq and by Apple. These mobility indicators are used as proxies for adherence to social distancing policies and are described in detail in Materials and Methods. Apple mobility and the Cuebiq mobility index (CMI) are imperfect insofar as they do not account for several important transmission factors, including importing of cases from other states. Although local travel distances are incomplete proxies for the scale at which NPIs are performed, reductions in mobility have been shown to lead to subsequent reduction in fever incidence by an average of 6.5 days during the first time period, an interval approximately equal to the incubation period of COVID-19, across hundreds of U.S. counties (42). Figure 3B supports this observation, with median fever incidence lagging median CMI and Apple mobility activation by 8.5 and 5.5 days, respectively. With the exception of excess ILI, for which median fever incidence lags median Apple mobility activation by roughly 4 days, this relationship is less apparent in time period 2 (Fig. 3D). Our use of two distinct mobility metrics is intended to reduce the influence of systematic biases arising from the methodology of either metric.
During the first time period, the timing of downtrends is consistent between Apple mobility and CMI (maximum difference of 4 days in median activation across all states with available data), with median downtrend activation for CMI preceding median activation of all other proxies and gold standard time series (Fig. 3B). Median decay in these indices predated median decay in deaths and confirmed cases by a median of 3 to 4 weeks; CMI was first to activate in 60% of states (refer to fig. S67). GLEAM, Google searches for “covid,” and UpToDate were among the latest proxies to activate across states. Median downtrend activation for Google searches for “quarantine,” included as a surrogate measure of mobility, lagged Apple mobility and CMI median downtrend activation by an average of 12 and 10.5 days, respectively. Statistically significant decaying events were not detected, or data were not available, for GLEAM in 22 states, excess ILI in 5 states, CMI in 1 state, deaths in 2 states, and confirmed cases in 7 states. For time period 2, we observed that the signals for mobility were less pronounced, only activating earliest relative to excess ILI, with Kinsa thermometer data and GLEAM data instead corresponding to the earliest median activation for deaths and cases, respectively.
Analysis of the event detection approach
As an initial effort to estimate the capacity of the event detection method to identify an impending outbreak, we evaluated whether uptrend or downtrend events detected in proxies were followed within 30 days by the same type of events in confirmed cases, deaths, or excess ILI. This analysis is permissive for estimation of the sensitivity or “true-positive rate” (TPR), defined as the ratio between “successful events” (true positives: when a proxy served as an early warning indicator to an actual gold standard activation) and the total number of observed activation events for the gold standard (the sum of true positives and false negatives). In addition, analysis of “precision,” defined as the ratio between total number of successfully detected events and the total number of times our proxy activated with or without a subsequent gold standard event activating (the sum of true positives and false positives), was carried out. Our results for the test are summarized in Table 1. With the exception of UpToDate, individual proxies do not cross a TPR of 0.6. When combined into a multiproxy estimate, however, a TPR of ∼0.75 was achieved in efforts to predict events in all three gold standard metrics. This result is evidence that a multiproxy approach may permit for more accurate forecasting than using individual data sources (each featuring their own sources of bias and uncertainty). As shown in the high precision scores for each proxy across the task of tracking each gold standard, incorrect events do not originate from a proxy incorrectly activating without any subsequent gold standard events happening but from the fact that the proxy may not activate before an observed gold standard event (low false positives but high false negatives).
Table 1. Sensitivity and precision rates for each proxy as an early indicator for an uptrend in the three different COVID-19 gold standards (confirmed cases, deaths, and excess ILI).
Overall, the combination of the P values achieved the greatest robustness as an early indicator.
| Proxy names | Metric | Excess ILI (n = 86) | Cases (n = 128) | Deaths (n = 127) |
| Combined P value | Sensitivity | 0.78 | 0.75 | 0.76 |
| Precision | 0.9 | 0.96 | 0.98 | |
| UpToDate | Sensitivity | 0.58 | 0.75 | 0.79 |
| Precision | 0.98 | 0.99 | 0.99 | |
| Sensitivity | 0.45 | 0.41 | 0.34 | |
| Precision | 1 | 0.91 | 0.98 | |
| Kinsa | Sensitivity | 0.37 | 0.38 | 0.35 |
| Precision | 1 | 1 | 1 | |
| Google (quarantine) | Sensitivity | 0.39 | 0.46 | 0.48 |
| Precision | 0.97 | 0.97 | 0.98 | |
| Google (fever) | Sensitivity | 0.51 | 0.55 | 0.54 |
| Precision | 1 | 1 | 0.99 | |
| Google (covid) | Sensitivity | 0.44 | 0.52 | 0.55 |
| Precision | 1 | 0.94 | 0.99 | |
| Google (cough) | Sensitivity | 0.58 | 0.5 | 0.54 |
| Precision | 0.98 | 1 | 0.97 | |
| GLEAM | Sensitivity | 0.3 | 0.38 | 0.34 |
| Precision | 1 | 1 | 0.95 |
To complement the event-based identification of anomalous growth in proxies, we also conducted a lagged correlation analysis. We correlated gold standard time series with proxy time series in each of the two time periods, allowing the proxies to lead the gold standards by up to 30 days; this procedure was designed (figs. S68 to S75) to account for the possibility of multiple growth and decay events per time period. The results from this analysis are shown in fig. S76. We found that Twitter and Google searches gave the highest correlations when preceding the gold standards by 2 to 3 weeks during time period 1 and that UpToDate, Google searches, and mobility gave the highest correlation during time period 2. These findings generally correspond to the order and intervals associated with proxy activation in our main analysis.
Using proxy signals to provide early warning of outbreaks
We hypothesize that an early warning system for COVID-19 could be derived from uptrend event dates across a network of proxies. For each state, Δxi, t is defined as the number of days since an uptrend event for proxy i, where t is the current date. A posterior probability distribution of an uptrend in confirmed COVID-19 cases is then estimated for each state conditional on the collection of proxies, p(y ∣ Δx1, t, …, Δxn, t), where each proxy is treated as an independent expert predicting the probability of a COVID-19 event. In this case, n is the number of proxies. See Materials and Methods for a more detailed explanation. A similar analysis is also feasible for downtrends. This method is introduced to better formalize predictions of growth in gold standard indicators using a network of proxies, but further evaluation is required, including relative to subsequent “waves” of COVID-19 cases.
Figure 4A shows uptrend events from proxies (vertical solid lines) and the predicted uptrend probability distribution for confirmed COVID-19 cases (in red) overlaid on confirmed COVID-19 cases (gray). As more proxy-derived uptrend events are observed, the probability mass of the predicted uptrend event becomes increasingly concentrated in the vicinity of identified exponential growth in confirmed COVID-19 cases (long vertical solid line). In the case of NY, exponential growth is identified in confirmed COVID-19 cases in the earlier part of the prediction distribution, although for most states, it occurs near the center as follows from how the prediction is estimated. Figure 4B similarly shows downtrend events in proxies and the estimated downtrend posterior probability distribution for decay in daily reports of confirmed COVID-19 cases. A visualization of the probability distribution for all the states is included in the Supplementary Materials (figs. S66 and S67).
Fig. 4. Illustration of the evolving probability distribution (in red) for the time-to-event estimation as applied to New York.
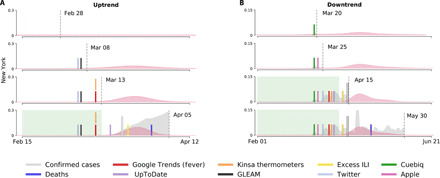
The probability distributions are calculated from information up to a specified time horizon (vertical dashed lines). Events that signaled an exponential increase or decrease more than 7 days before the true event (long vertical solid lines) are contained within a green background. (A) The estimated uptrend posterior probability distribution and uptrend events. (B) The estimated downtrend posterior probability distribution and downtrend events.
We tested our time-to-event estimation approach by training it on data from time period 1 and applying it to data from time period 2. Figure 5 shows the prospective (“out-of-sample”) predictive performance of our approach by time horizon. Further details can be found in Materials and Methods. Performance increases monotonically approaching an event, with 50% of uptrends predicted 2 weeks in advance and 75% predicted 1 week in advance. Skill in predicting downtrends is greater than uptrends by approximately 10%, possibly reflecting that public health interventions are generally implemented over consistent time scales to control emerging outbreaks.
Fig. 5. Predictive performance of our time-to-event estimation approach by time horizon on withheld data.
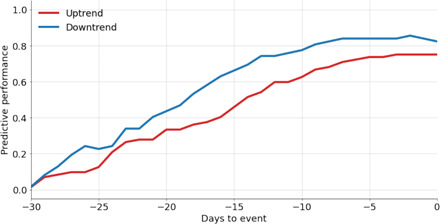
We consider a success to be a high probability corresponding to the date when a confirmed event occurs. Predictive performance is defined as the fraction of confirmed case events our method predicts at a specific time horizon.
DISCUSSION
Here, we have assessed the utility of various digital data streams, individually and collectively, as components of a near–real-time COVID-19 early warning system. Specifically, we focused on identifying early signals of impending outbreak characterized by marked exponential growth and subsiding outbreak characterized by exponential decay. We explored two time periods given that the first submission of this methodology occurred in early June (once outbreaks in the northeastern United States had partially subsided) and our revised version after peer review was prepared 4 months later, giving us the opportunity to evaluate our methods on newly available data. For the first time period, we found that COVID-19–related activity on Twitter and Google Trends showed significant growth 2 to 3 weeks before such growth occurred in confirmed cases and 3 to 4 weeks before such growth occurred in reported deaths; Google Trends displayed a similar pattern for time period 2. We also observed that for exponential decay, as represented by downtrend activation, NPIs—here treated as reductions in human mobility—predated decreases in confirmed cases and deaths by 3 to 4 weeks; for time period 2, mobility data are found to lead excess ILI by roughly 2 weeks (Fig. 3D). Clinicians’ search activity, fever data, estimates from the GLEAM metapopulation mechanistic epidemiological model, and Google searches for COVID-19–related terms were similarly found to anticipate changes in COVID-19 activity. We also developed a consensus indicator of COVID-19 activity using the harmonic mean of all proxies. This combined indicator predated an increase in COVID-19 cases by a median of 19.5 days and an increase in COVID-19 deaths by a median of 29 days, and was synchronous with excess ILI. Such a combined indicator may provide timely information, like a “thermostat” in a heating or cooling system, to guide intermittent activation, intensification, or relaxation of public health interventions as the COVID-19 pandemic evolves.
A challenge that comes with managing multiple data streams with the aim of anticipating emerging infectious diseases is the uncertainty about confounding factors. For example, a recent study has shown that confirmed U.S. cases of COVID-19 may not necessarily track the evolution of the disease considering limited testing frequency at early stages of the pandemic (8). Similarly, some of the signal corresponding to social network data and Google Trends may be driven by media activity about COVID-19. These confounders can lead to increased proxy activity and, therefore, the capturing of false events by our methodology. Nonetheless, the power of our methodology lies in synthesizing digital data streams to reduce the impact of confounding. This aligns with the results of our TPR analysis (Table 1), which shows that the combined P value yielded the highest TPR in predicting changes among the three gold standards. We attribute this to the capacity of the combined P value to remain agnostic to the activation of a single proxy. As the pandemic progressed, we were able to better quantify potential biases in each proxy, suggesting that additional analyses could be done to preprocess the proxies and further improve our methodology.
In extending our approach to two subsequent and nonoverlapping time periods, we observed variability in the behavior of the alternative proxies across states. This may follow from the inherent state-level differences in COVID-19 restrictions and regulations, where states that have been slower to implement COVID-19–countering measures may show more sustained case counts or increased volatility in uptrends and downtrends.
The most reliable metric for tracking the spread of COVID-19 remains unclear, and all metrics discussed in this study feature important limitations. While deaths may seem a more accurate tracker of disease evolution, they are limited in their real-time availability, as they tend to lag cases by nearly 20 days (40). ILI anomalous activity, which may capture COVID-19 (2) activity, similarly suffers from a lag in availability because reports are released with a 5- to 12-day lag; for simplicity, we approximated this lag as 10 days in our analysis. Furthermore, a decrease in ILI reporting is frequently observed in between flu seasons (from early April 2020 and October 2020), rendering ILI-based analyses useful only when surveillance systems are fully operational. Lu et al. (2) support this conjecture, reporting a rapid decrease in the number of patients with ILI reported in late March 2020 despite the number of reporting providers remaining largely unchanged. This decrease may also be attributed to patients forgoing treatment for milder, non–COVID-19–attributed ILI. Hospitalizations, although possibly less biased than confirmed case numbers, were ultimately omitted because of sparseness and poor quality of data.
In contrast to the aforementioned digital data streams, GLEAM assimilates many epidemiological parameters [e.g., literature-derived incubation period and generation time (6, 43, 44)] essential for describing COVID-19. Coupled with internet sources, GLEAM can provide a more robust outbreak detection method because of its different framework and, consequently, at least partially uncorrelated errors with the internet-derived sources. Estimates produced by this model suggest a median increase in cases and deaths of 15 and 5 days later, respectively, for the first time period (Fig. 3A) and 1 and 9 days later, respectively, for the second time period (Fig. 3C). However, some parameters are excluded from the model owing to lack of availability (e.g., age-related COVID-19 susceptibility). These excluded parameters, coupled with the need to regularly update existing parameters, may lead to suboptimal downstream performance by the model.
The analysis we presented focuses largely on temporally analyzing different data streams that are aggregated to the state level. This level of aggregation is able to provide a coarse overview of regional differences within the United States. Smart thermometer, Twitter, and mobility data may help us replicate our analysis at finer spatial resolutions, making them suitable for capturing both regional and local effects. A promising future research avenue is the detection of local COVID-19 clusters (“hotspots”) through more spatially resolved approaches (45). Such an approach would better inform regarding at-risk populations and, therefore, permit for more targeted NPIs. Whereas the data streams that we analyze do not capture important population dynamics, integrated spatial, temporal, and semantic analysis of web data (46) could give a more nuanced understanding of public reaction, such as estimating changes in the public emotional response to epidemics (47).
Using an exponential model to characterize the increase (and decrease) in activity of a COVID-19 proxy offers various advantages in event detection. Our current procedure is capable of estimating the value of γ with a measure on the confidence that γ > 0 or γ < 0. In this work, we provide event dates based on a confidence of 95% (P value <0.05). The degree of confidence can be adjusted to provide earlier event dates (at the cost of less confidence and, consequently, more frequent false positives). P values are combined into a single metric using a harmonic mean, but a more sensitive design may be realized by assuming independence between the proxies and using Fisher’s method. Although this would generally lead to a lower combined P value given decreases in any individual proxy P value (i.e., higher sensitivity), assuming independence would make such an approach prone to false positives (i.e., lower specificity) than the HMP, which does not depend on the assumption of independence. The choice of method for combining proxy indicators requires a trade-off between specificity and sensitivity.
Our assumption that epidemic events exhibit exponential behavior within data stream time series has shown to be especially appropriate at early pandemic stages, particularly when locations have yet to experience their first outbreak or when post-outbreak disease activity has slowed and a pool of susceptible individuals remains. Other models may capture additional data nuances. For example, more linear increases or decreases or inflection points in proxies that, overall, are decreasing in activity are not captured in our current approach but could be included to potentially improve event detection. For example, multimodel event estimates could be produced and combined in an ensemble scheme.
The ability to detect future epidemic changes depends on the stability of historical patterns observed in multiple measures of COVID-19 activity. We posit that using multiple measures in our COVID-19 early warning system leads to improved performance and robustness to measure-specific flaws. The probabilistic framework we developed (Fig. 4) also gives decision-makers the freedom to decide how conservative they want to be in interpreting and consolidating different measures of COVID-19 activity (e.g., by revising the P value required for event detection). Although we can expect COVID-19 activity to increase in the future, given continued opportunities for transmission, the human population in which said transmission occurs may not remain identical in terms of behavior, immune status, or risk. For example, death statistics in the early pandemic have been driven by disease activity in nursing homes and long-term care facilities (48), but the effect of future COVID-19 waves in these settings may be attenuated by better safeguards (e.g., vaccination) implemented after the first wave. Ultimately, to improve the performance of our event detection algorithm in forecasting future waves, our methodology requires the input of additional observed events over a longer time interval. This would also increase the performance and robustness of our analysis at an individual state level, for which there are currently relatively few observations. In the analysis we have presented, we make the assumption that states contribute equally to an ensemble multiproxy forecasting, but we also recognize that each state presents different behavior. It follows that as more state-specific data become available, future research should fine-tune how the changes of COVID-19 activity are preceded by changes in the multiple proxies to improve location-specific predictive performance in methods like ours.
MATERIALS AND METHODS
For our study, we collected the following daily reported data streams: (i) official COVID-19 reports from three different organizations; (ii) ILI cases, as reported weekly by the ILINet; (iii) COVID-19–related search term activity from UpToDate and Google Trends; (iv) Twitter microblogs; (v) fever incidence as recorded by a network of digital thermometers distributed by Kinsa; and (vi) human mobility data, as reported by Cuebiq and Apple.
COVID-19 case reports
Every state in the United States provides daily updates about its COVID-19 situation as a function of testing. Subject to state-to-state and organizational variability in data collection, these reports include information about the daily number of positive, negative, pending, and total COVID-19 tests, hospitalizations, intensive care unit visits, and deaths. Daily efforts in collecting data by research and news organizations have resulted in several data networks, from which official health reports have been made available to the public. Three predominant data networks are the John Hopkins Resource Center, the COVID Tracking Project, and the New York Times Repository (1, 49, 50). We obtained daily COVID-19 testing summaries from all three repositories with the purpose of analyzing the variability and quality in the data networks.
ILINet
ILIs are characterized by fever and either cough or sore throat. An overlap in symptoms of COVID-19 and ILI has been observed, and it has further been shown that ILI signals can be useful in the estimation of COVID-19 incidence when testing data are unavailable or unreliable (2).
ILINet is a sentinel system created and maintained by the U.S. CDC (51, 52) that aggregates information from clinicians’ reports on patients seeking medical attention for ILI symptoms. ILINet provides weekly estimates of ILI activity with a lag of 5 to 12 days; because detailed delay information is unavailable, we arbitrarily apply a lag of 10 days throughout this work. At the national level, ILI activity is estimated via a population-weighted average of state-level ILI data. ILINet data are unavailable for Florida.
The CDC also releases data on laboratory test results for influenza types A and B, shared by laboratories collaborating with the World Health Organization (WHO) and the National Respiratory and Enteric Virus Surveillance System (NREVSS). Both ILI activity and virology data are available from the CDC FluView dashboard (51).
We followed the methodology of Lu et al. (2) to estimate unusual ILI activity, a potential signal of an emerging outbreak such as COVID-19. In particular, we used the divergence-based methods, which treat COVID-19 as an intervention and try to measure the impact of COVID-19 on ILI activity by constructing two control time series representing the counterfactual 2019–2020 influenza season had the COVID-19 outbreak not occurred.
The first control time series is based on an epidemiological model, specifically the Incidence Decay and Exponential Adjustment (IDEA) model (53). IDEA models disease prevalence over time while accounting for factors such as control activities that may dampen the spread of a disease. The model is written as follows
| (1) |
where I(t) represents the incident case count at serial interval time step t. R0 represents the basic reproduction number, and d is a discount factor modeling reductions in the effective reproduction number over time.
In line with the approach of Lu et al. (2), we fit the IDEA model to ILI case counts from the start of the 2019–2020 flu season to the last week of February 2020, where the start of flu season is defined as the first occurrence of two consecutive weeks with an ILI activity above 2%. The serial interval used was half a week, consistent with the influenza serial interval estimates from (54).
The second control time series used the CDC’s virological influenza surveillance data. For any week t, the following was computed
| (2) |
where , Nt, It, and Ft denote positive flu tests, total specimens, ILI visit counts, and the true underlying flu counts, respectively. This can be interpreted as the extrapolation of the positive test percentage to all patients with ILI. Least squares regression (fit on pre–COVID-19 data) is then used to map Ft to an estimate of ILI activity.
The differences between the observed ILI activity time series and these counterfactual control time series can then be used as signals of COVID-19 activity. In particular, we used the difference between observed ILI activity and the virology-based counterfactual control time series to produce excess ILI. The Supplementary Materials show excess ILI computed using both the virology time series and IDEA model–based counterfactuals for all states.
UpToDate trends
UpToDate is a private-access search database, part of Wolters Kluwer, Health, with clinical knowledge about diseases and their treatments. It is used by physicians around the world and most academic medical centers in the United States as a clinical decision support resource given the stringent standards on information within the database (in comparison to Google Trends, information provided within the database is heavily edited and authored by experienced clinicians) (19).
Recently, UpToDate has made available a visualization tool on their website in which they compare their search volumes of COVID-19–related terms to John Hopkins University official health reports (55). The visualization shows that UpToDate trends may have the potential to track confirmed cases of COVID-19. From this tool, we obtained UpToDate’s COVID-19–related search frequencies for every U.S. state. These search frequencies consist only of one time series described as “normalized search intensity” for selected COVID-19–related terms, where normalization is calculated as the total number of COVID-19–related search terms divided by the total number of searches within a location. At the time of analysis, the website visualization appeared to update with a 3-day delay; however, UpToDate is since operationally capable of producing time series with delays of 1 day. More details are available at https://covid19map.uptodate.com/.
Google Trends
Google Search volumes have been shown to track successfully with various diseases such as influenza (18, 56), dengue (57), and Zika (17, 58), among others (59). In recent months, Google has even created a “Coronavirus Search Trends” (60) page that tracks trending pandemic-related searches. We obtained such daily COVID-19–related search trends through the Google Trends Application Programming Interface (API). The original search terms queried using the Google Trends API were composed of the following: (i) official symptoms of COVID-19, as reported by the WHO; (ii) a list of COVID-19–related terms shown to have the potential to track confirmed cases (33); and (iii) a list of search terms previously used to successfully track ILI activity (18). The list of terms can be seen in Box 1. For purposes of the analysis, we narrowed down the list of Google Search terms to those we felt to be most representative of the pandemic to date: “fever,” “covid,” and “quarantine.” Given that lexicon is naturally subject to change as the pandemic progresses, however, other terms, especially more obscure terms that may be less liable to media bias, may become more suitable downstream.
Box 1. Search term list for Google Trends.
anosmia, chest pain, chest tightness, cold, cold symptoms, cold with fever, contagious flu, cough, cough and fever, cough fever, covid, covid nhs, covid symptoms, covid-19, covid-19 who, dry cough, feeling exhausted, feeling tired, fever, fever cough, flu and bronchitis, flu complications, how long are you contagious, how long does covid last, how to get over the flu, how to get rid of flu, how to get rid of the flu, how to reduce fever, influenza, influenza b symptoms, isolation, joints aching, loss of smell, loss smell, loss taste, nose bleed, oseltamivir, painful cough, pneumonia, pneumonia, pregnant and have the flu, quarantine, remedies for the flu, respiratory flu, robitussin, robitussin cf., robitussin cough, rsv, runny nose, sars-cov 2, sars-cov-2, sore throat, stay home, strep, strep throat, symptoms of bronchitis, symptoms of flu, symptoms of influenza, symptoms of influenza b, symptoms of pneumonia, symptoms of rsv, tamiflu dosage, tamiflu dose, tamiflu drug, tamiflu generic, tamiflu side effects, tamiflu suspension, tamiflu while pregnant, tamiflu wiki, and tessalon.
Twitter API
We developed a geocrawler software to collect as much georeferenced social media data as possible in a reasonable time. This software requests data from Twitter’s APIs. Twitter provides two types of APIs to collect tweets: REST (representational state transfer) and streaming (61). The REST API offers various end points to use Twitter functionalities, including the “search/tweets” end point that enables, with limitations, the collection of tweets from the last 7 days. These limitations complicate the collection process, necessitating a complementary strategy to manage the fast-moving time window of the API to harvest all offered tweets with a minimal number of requests. In contrast, the streaming API provides a real-time data stream that can be filtered using multiple parameters. Our software focuses on requesting tweets featuring location information either as a point coordinate from the positioning device of the mobile device used for tweeting or a rectangular outline based on a geocoded place name, using APIs. The combination of APIs makes crawling robust against interruptions or backend issues that would lead to missing data. For example, if data from the streaming API cannot be stored in time, the missing data can be retrieved via the redundant REST API.
All collected tweets are located within the United States. To limit the dataset to COVID-19–relevant tweets, we performed simple keyword-based filtering using the keywords listed in Box 2. This method was chosen for reasons of performance, delivery of results in near real time, and its simplicity. While a machine learning-based semantic clustering method such as guided latent Dirichlet allocation may deliver more comprehensive results [e.g., through identifying co-occurring and unknown terms (62)], controlling the ratio between false positives and false negatives requires extensive experimental work and expert knowledge.
Box 2. Search term list for Twitter.
covid, corona, epidemic, flu, influenza, face mask, spread, virus, infection, fever, panic buying, state of emergency, masks, quarantine, sars, and 2019-ncov.
Kinsa smart thermometer data
County-level estimates of U.S. fever incidence are provided by Kinsa Insights using data from a network of volunteers who have agreed to regularly record and share their temperatures (https://healthweather.us/). Estimates from past years have been shown to correlate closely with reported ILI incidence from the U.S. CDC across regions and age groups (31). Such historical data, coupled with county-specific characteristics (e.g., climate and population size), are used to establish the “expected,” or forecast, number of fevers as a function of time (31, 63). An “excess fevers” time series presumed to represent COVID-19 cases is approximated as the difference between the observed fever incidence and the forecast, with values truncated at zero so that negative excess fevers are not possible. County-level data are aggregated up to the state level by a population-weighted average. A limitation of tracking febrility as a proxy for COVID-19 is that it is not a symptom exclusive to COVID-19, nor is it present in all patients with COVID-19. In a study with more than 5000 patients from NY who were hospitalized with COVID-19, only 30% presented with fever (>100.4°F/38°C) at triage (64).
Cuebiq mobility index
Data are provided by the location analytics company Cuebiq, which collects first-party location information from smartphone users who opted to anonymously provide their data through a General Data Protection Regulation–compliant framework. Cuebiq further anonymizes and then aggregates location data into an index, M, defined as the base-10 logarithm of median distance traveled per user per day; “distance” is measured as the diagonal distance across a bounding box that contains all GPS (Global Positioning System) points recorded for a particular user on a particular day. A county index of 3.0, for example, indicates that a median county user has traveled 1000 m. We were provided with county-level data, derived from these privacy-preserving steps, which we then aggregated up to the state level by a population-weighted average.
Apple mobility
Apple mobility data are generated by counting the number of requests made to Apple Maps for directions in select countries, regions, subregions, and cities. Data that are sent from users’ devices to the Maps service is associated with random, rotating identifiers so Apple does not have a profile of users’ movements and searches. The availability of data in a particular country, region, subregion, or city is based on a number of factors, including minimum thresholds for direction requests per day. Note that some data from late April to early May are unavailable and are represented as a white gap in plots of this data stream. More details are available at www.apple.com/covid19/mobility.
Global epidemic and mobility model
GLEAM is a spatially structured epidemic model that integrates population and mobility data with an individual-based stochastic infectious disease dynamic to track the global spread of a disease (65–68). The model divides the world into more than 3200 subpopulations, with mobility data between subpopulations including air travel and commuting behavior. Air travel data are obtained from origin-destination traffic flows from the Official Aviation Guide and International Air Transport Association databases (69, 70), while short-range mobility flows such as commuting behavior are derived from the analysis and modeling of data collected from the statistics offices for 30 countries on five continents (66). Infectious disease dynamics are modeled within each subpopulation using a compartmental representation of the disease where each individual can occupy a single disease state: susceptible (S), latent (L), infectious (I), and removed (R). The infection process is modeled by using age-stratified contact patterns at the state level (71). These contact patterns incorporate interactions that occur in four settings: school, household, workplace, and the general community. Latent individuals progress to the infectious stage with a rate inversely proportional to the latent period. Infectious individuals progress into the removed stage with a rate inversely proportional to the infectious period. The sum of the average latent and infectious periods defines the generation time. Removed individuals represent those who can no longer infect others, as they were isolated, were hospitalized, have died, or have recovered. To take into account mitigation policies adopted widely across the United States, we incorporated a reduction in contacts in the school, workplace, and community settings (which is reflected in the contact matrices used for each state). Details on the timeline of specific mitigation policies implemented are described in (72). A full discussion of the model for COVID-19 is reported in (65).
Growth and decay event detection
We estimate the probability of exponential growth in a time series that features uncertain error variance using a simple Bayesian method. We model a proxy time series as following an exponential curve
| (3) |
over successive 14-day intervals. Before inference, proxies are normalized, then adjusted to have a common minimum value of 1. The error, ϵ, is assumed Gaussian with zero mean and having an SD of σ. We assess the probability that γ is greater than zero over each successive window. The joint distribution of β and γ, conditional on σ, is proportional to p(y, σ∣β, γ) × p(β) × p(γ). Prior distributions, p(β) and p(γ), are specified as uniform and uninformative, and samples are obtained using the Metropolis-Hastings algorithm (73) with 5 × 103 posterior draws. The first 500 samples are discarded to remove the influence of initial parameter values, and to reduce autocorrelation between draws, only every fifth sample is retained. The conditional posterior distribution for σ is inverse-Gamma and is obtained using Gibbs sampling (74, 75)
| (4) |
where Γ−1 is the inverse-Gamma distribution, y is the vector of observations, and N is the number of observations. Terms α1 and α2 are, respectively, specified to equal 4 and 1. On any given day, a P value for rejecting the null hypothesis of no exponential growth is obtained as one minus the fraction of posterior draws with γ > 0. The procedure is repeated on successive days to obtain a time series of P values.
Our current approach has some important limitations. The mean value in a time series is not inferred, and a highly simplified treatment of errors neglects the possibility of autocorrelation and heteroscedasticity. A more complete treatment might use a more sophisticated sampling strategy and jointly infer (rather than impose) a mean proxy value, nonzero error mean, error autoregressive parameter(s), and heteroscedasticity across each 14-day window.
Multiproxy P value
P values estimated across multiple proxies are combined into a single metric representing the family-wise probability that γ ≤ 0. Because proxies cannot be assumed independent, we use the HMP (41)
| (5) |
where wi are weights that sum to 1 and, for the purposes of our analyses, are treated as equal. Excess ILI, confirmed cases, and deaths were excluded from the harmonic mean because those time series contain the events that the algorithm is attempting to predict. Cuebiq and Apple mobility time series were also excluded because they are not proxies for COVID-19 activity but rather causal effects with long lead times.
As P values of zero may be recorded because of a finite number of samples being drawn in our Monte Carlo approach, we have imposed a lower limit of 0.02 on the possible values that individual proxy P values may take during calculation of the harmonic mean to avoid the harmonic P value event to be triggered by a single proxy.
Predicting false-positive and false-negative rates for uptrend and downtrend events
As a way to quantify the performance of digital data streams as early indicators of COVID-19 activity, we performed a TPR and false-positive rate analysis for each over the following events: (i) If the event detection activates an alarm for a proxy and a subsequent activation of a gold standard is observed within a 30-day time window (given that a proxy may present several dates in which its P value crosses the threshold for an event within a period of time before an opposite event happening, only the first activations were counted for the test), we classified the event as a true positive; (ii) if the event detection mechanism classified an event for a gold standard and no proxy was observed within a 30-day time window in the past, then we considered that event as a false negative for the proxy; and (iii) a false positive is considered as an event when a proxy activates but no subsequent gold standard event is observed within the 30-day time window. TPR and false-positive rate analyses were calculated using the standard formulas for sensitivity (TP/[TP + FN]) and precision (TP/[TP + FP]). We included data from 1 February to 30 September 2020 for this procedure to account for early activations before the first observed outbreak of the pandemic.
Time-to-event estimation
A time–to–outbreak event estimation strategy can be formulated to provide probability estimates for when the next outbreak will occur given early indicators. We propose a strategy based on the timing of detected events among input data sources with respect to the eventual COVID-19 outbreak event in each state, as defined in the preceding section. We first modeled the behavior of the data sources in each state as a function of the state’s underlying COVID-19 case trajectory over the time period studied. Specifically, we modeled the detected events in each data source as conditionally independent given the state-level latent COVID-19 dynamics. This follows from the assumption that exponential behavior in each data source is a causal effect of a COVID-19 outbreak and that other correlations unrelated to COVID-19 are mostly minor in comparison.
The time intervals between each event and the eventual outbreak event were then pooled across states to form an empirical conditional distribution for each dataset. Since observations were sparse relative to the time window, we used kernel density estimation with Gaussian kernels to smooth the empirical distributions, where bandwidth selection for the kernel density estimation was performed using Scott’s rule (76).
Thus, for any given dataset, a detected event implies a distribution of likely timings for the COVID-19 outbreak event. We define Δxit as the number of days since an uptrend event for signal i where t is the current date. Within a state, the relative intervals of the events for each data source Δxit specify a posterior distribution over the probability of a COVID-19 event in y days from current date t
| (6) |
where we decomposed the joint likelihood
| (7) |
using conditional independence.
A uniform prior p(y) over the entire range of possible delays (a period of 180 days) was assumed, and additive smoothing was used when combining the probabilities. Because we modeled y at the daily level, the distributions are discrete, allowing evaluation of the posterior likelihood explicitly using marginalization. This process was repeated for each state. Such an approach can be viewed as pooling predictions from a set of “experts” when they have conditionally independent likelihoods given the truth (77), with each expert corresponding to a data source. We note as a limitation that the assumption of conditionally independent expert likelihoods given the truth is unlikely to hold perfectly as, for example, an increase in measured fevers could be correlated with an increase in fever-related Google searches even when the underlying COVID-19 infection dynamics are similar. Such dependencies may manifest heterogeneously as correlations among locations with similar COVID-19 outbreak timings but are likely to be small since most of our inputs represent disparate data sources. When no event is detected for a data source, that data source’s expert’s prediction is zero across all possible timings, which translates with smoothing to a uniform distribution.
Time-to-event prediction and validation
As the pandemic progressed, the emergence of uptrends and downtrends in COVID-19 gold standards, specifically confirmed cases, meant that we were able to validate our time-to-event estimation approach as well. We trained our approach on data during time period 1 (1 March to 31 May 2020) and tested our approach on data from time period 2 (1 June to 30 September 2020). Figure 5 shows the predictive performance of our approach by time horizon. We consider a success to be a high probability corresponding to the date when a confirmed case event occurred. High probability is defined as a probability exceeding that associated with a uniform distribution over the date range of interest. The threshold set by the uniform distribution is exceeded once probability becomes concentrated over a subset of the dates in the considered interval, and, by extension, probabilities for other dates will grow smaller than the threshold. We note that other choices of thresholds are possible depending on the deployment context. Predictive performance here is defined as the fraction of confirmed cases events our method was able to predict at a specific time horizon. As expected, predictive performance improves as we get closer to the date of an event of interest.
Acknowledgments
We thank Wolters Kluwer Health’s UpToDate team for providing resource-specific data. Funding: M.S. was partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM130668. J.D., M.C., and A.V. acknowledge support from Google Cloud and the Google Cloud Research Credits program, as well as the McGovern Foundation and the Chleck Foundation, for funding this project. B.R., C.H., and A.P. would like to thank the Austrian Science Fund (FWF) for their support through the project “The Scales and Structures of Intra-Urban Spaces” (reference number P 29135-N29) and the Austrian Federal Ministry of Climate Action, Environment, Energy, Mobility, Innovation, and Technology (BMK) through their support of the project “GeoSHARING” (project number 878652). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Author contributions: M.S., N.E.K., L.C., P.L., and J.K. conceptualized the study; M.S. supervised the study. P.L., A.T.N., F.S.L., and P.H. designed and implemented the Bayesian methodology, and L.C., P.L., N.E.K., A.T.N., F.S.L., and J.K. performed all quantitative analyses. B.R., C.H., and A.P. provided Twitter data. J.D., M.C., and A.V. provided the GLEAM data. L.C., N.E.K., and P.L. collected the remaining data sources. N.B.L., F.S.L., P.H., B.M., and W.P.H. provided additional subject matter expertise in shaping the manuscript. N.E.K., L.C., P.L., and M.S. wrote the first draft of the paper. All authors contributed to and approved the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Some data sources may require establishing data sharing agreements with the data providers, which have been established with a diverse set of research teams. Code can be made available upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/10/eabd6989/DC1
REFERENCES AND NOTES
- 1.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu F. S., Nguyen A. T., Link N. B., Lipsitch M., Santillana M., Estimating the early outbreak cumulative incidence of COVID-19 in the United States: Three complementary approaches. medRxiv 2020.04.18.20070821 , (2020). [Google Scholar]
- 3.Rosenberg E. S., Tesoriero J. M., Rosenthal E. M., Chung R., Barranco M. A., Styer L. M., Parker M. M., Leung S.-Y. J., Morne J. E., Greene D., Holtgrave D. R., Hoefer D., Kumar J., Udo T., Hutton B., Zucker H. A., Cumulative incidence and diagnosis of SARS-CoV-2 infection in New York. Ann. Epidemiol. 48, 23–29.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okell L. C., Verity R., Watson O. J., Mishra S., Walker P., Whittaker C., Katzourakis A., Donnelly C. A., Riley S., Ghani A. C., Gandy A., Flaxman S., Ferguson N. M., Bhatt S., Have deaths from COVID-19 in Europe plateaued due to herd immunity? Lancet 395, e110–e111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corey L., Mascola J. R., Fauci A. S., Collins F. S., A strategic approach to COVID-19 vaccine R&D. Science 368, 948–950 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kissler S. M., Tedijanto C., Goldstein E., Grad Y. H., Lipsitch M., Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368, 860–868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipsitch M., Santillana M., Enhancing situational awareness to prevent infectious disease outbreaks from becoming catastrophic. Curr. Top. Microbiol. Immunol. 424, 59–74 (2019). [DOI] [PubMed] [Google Scholar]
- 8.J. Kaashoek, M. Santillana, COVID-19 positive cases, evidence on the time evolution of the epidemic or an indicator of local testing capabilities? A case study in the United States. arXiv:2004.3128874 (2020).
- 9.S. Flaxman, S. Mishra, A. Gandy, H. J. T. Unwin, H. Coupland, T. A. Mellan, H. Zhu, T. Berah, J. W. Eaton, P. N. P. Guzman, N. Schmit, L. Cilloni, K. E. C. Ainslie, M. Baguelin, I. Blake, A. Boonyasiri, O. Boyd, L. Cattarino, C. Ciavarella, L. Cooper, Z. Cucunubá, G. Cuomo-Dannenburg, A. Dighe, B. Djaafara, I. Dorigatti, S. van Elsland, R. F. John, H. Fu, K. Gaythorpe, L. Geidelberg, N. Grassly, W. Green, T. Hallett, A. Hamlet, W. Hinsley, B. Jeffrey, D. Jorgensen, E. Knock, D. Laydon, G. Nedjati-Gilani, P. Nouvellet, K. Parag, I. Siveroni, H. Thompson, R. Verity, E. Volz, C. Walters, H. Wang, Y. Wang, O. Watson, P. Winskill, X. Xi, C. Whittaker, P. G. T. Walker, A. Ghani, C. A. Donnelly, S. Riley, L. C. Okell, M. A. C. Vollmer, N. M. Ferguson, S. Bhatt, Report 13: Estimating the number of infections and the impact of non-pharmaceutical interventions on COVID-19 in 11 European countries (Imperial College London, 2020).
- 10.H. Yang, D. Duque, Ö. Sürer, D. P. Morton, R. Pasco, K. Pierce, S. J. Fox, L. A. Meyers, Staged strategy to avoid hospital surge and preventable mortality, while reducing the economic burden of social distancing measures, in The University of Texas COVID-19 Modeling Consortium (University of Texas at Austin, 2020). [Google Scholar]
- 11.U.S. CDC, Forecasts of total COVID-19 deaths (2020).
- 12.Salathé M., Bengtsson L., Bodnar T. J., Brewer D. D., Brownstein J. S., Buckee C., Campbell E. M., Cattuto C., Khandelwal S., Mabry P. L., Vespignani A., Digital epidemiology. PLOS Comput. Biol. 8, e1002616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsberg J., Mohebbi M. H., Patel R. S., Brammer L., Smolinski M. S., Brilliant L., Detecting influenza epidemics using search engine query data. Nature 457, 1012–1014 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Polgreen P. M., Chen Y., Pennock D. M., Nelson F. D., Weinstein R. A., Using internet searches for influenza surveillance. Clin. Infect. Dis. 47, 1443–1448 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Santillana M., Zhang D. W., Althouse B. M., Ayers J. W., What can digital disease detection learn from (an external revision to) Google Flu Trends? Am. J. Prev. Med. 47, 341–347 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Yang S., Santillana M., Kou S. C., Accurate estimation of influenza epidemics using Google search data via ARGO. Proc. Natl. Acad. Sci. U.S.A. 112, 14473–14478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGough S. F., Brownstein J. S., Hawkins J. B., Santillana M., Forecasting Zika incidence in the 2016 Latin America outbreak combining traditional disease surveillance with search, social media, and news report data. PLOS Negl. Trop. Dis. 11, e0005295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu F. S., Hattab M. W., Clemente C. L., Biggerstaff M., Santillana M., Improved state-level influenza nowcasting in the United States leveraging Internet-based data and network approaches. Nat. Commun. 10, 147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santillana M., Nsoesie E. O., Mekaru S. R., Scales D., Brownstein J. S., Using clinicians’ search query data to monitor influenza epidemics. Clin. Infect. Dis. 59, 1446–1450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownstein J. S., Freifeld C. C., Reis B. Y., Mandl K. D., Surveillance sans frontières: Internet-based emerging infectious disease intelligence and the HealthMap project. PLOS Med. 5, e151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder M. S., Kluberg S., Santillana M., Mekaru S., Brownstein J. S., 2014 Ebola outbreak: Media events track changes in observed reproductive number. PLOS Curr. 7, ecurrents.outbreaks.e6659013c1d7f11bdab6a20705d1e865, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majumder M. S., Santillana M., Mekaru S. R., McGinnis D. P., Khan K., Brownstein J. S., Utilizing nontraditional data sources for near real-time estimation of transmission dynamics during the 2015-2016 Colombian Zika virus disease outbreak. JMIR Public Health Surveill. 2, e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smolinski M. S., Crawley A. W., Baltrusaitis K., Chunara R., Olsen J. M., Wójcik O., Santillana M., Nguyen A., Brownstein J. S., Flu near you: Crowdsourced symptom reporting spanning 2 influenza seasons. Am. J. Public Health 105, 2124–2130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolotti D., Carnahan A., Colizza V., Eames K., Edmunds J., Gomes G., Koppeschaar C., Rehn M., Smallenburg R., Turbelin C., van Noort S., Vespignani A., Web-based participatory surveillance of infectious diseases: The Influenzanet participatory surveillance experience. Clin. Microbiol. Infect. 20, 17–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul M. J., Dredze M., Broniatowski D., Twitter improves influenza forecasting. PLOS Curr. 6, ecurrents.outbreaks.90b9ed0f59bae4ccaa683a39865d9117, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagar R., Yuan Q., Freifeld C. C., Santillana M., Nojima A., Chunara R., Brownstein J. S., A case study of the New York City 2012-2013 influenza season with daily geocoded Twitter data from temporal and spatiotemporal perspectives. J. Med. Internet Res. 16, e236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viboud C., Charu V., Olson D., Ballesteros S., Gog J., Khan F., Grenfell B., Simonsen L., Demonstrating the use of high-volume electronic medical claims data to monitor local and regional influenza activity in the US. PLOS ONE 9, e102429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santillana M., Nguyen A. T., Louie T., Zink A., Gray J., Sung I., Brownstein J. S., Cloud-based electronic health records for real-time, region-specific influenza surveillance. Sci. Rep. 6, 25732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Generous N., Fairchild G., Deshpande A., Del Valle S. Y., Priedhorsky R., Global disease monitoring and forecasting with Wikipedia. PLOS Comput. Biol. 10, e1003892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radin J. M., Wineinger N. E., Topol E. J., Steinhubl S. R., Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: A population-based study. Lancet Digit. Health 2, e85–e93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller A. C., Singh I., Koehler E., Polgreen P. M., A smartphone-driven thermometer application for real-time population- and individual-level influenza surveillance. Clin. Infect. Dis. 67, 388–397 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Kogan N. E., Bolon I., Ray N., Alcoba G., Fernandez-Marquez J. L., Müller M. M., Mohanty S. P., Ruiz de Castañeda R., Wet markets and food safety: TripAdvisor for improved global digital surveillance. JMIR Public Health Surveill. 5, e11477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.V. Lampos, M. S. Majumder, E. Yom-Tov, M. Edelstein, S. Moura, Y. Hamada, M. X. Rangaka, R. A. McKendry, I. J. Cox, Tracking COVID-19 using online search. arXiv:2003.08086 (2020). [DOI] [PMC free article] [PubMed]
- 34.Lu T., Reis B. Y., Internet search patterns reveal clinical course of COVID-19 disease progression and pandemic spread across 32 countries. medRxiv 2020.05.01.20087858, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazer D., Kennedy R., King G., Vespignani A., The parable of Google Flu: Traps in big data analysis. Science 343, 1203–1205 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Bento A. I., Nguyen T., Wing C., Lozano-Rojas F., Ahn Y.-Y., Simon K., Evidence from internet search data shows information-seeking responses to news of local COVID-19 cases. Proc. Natl. Acad. Sci. U.S.A. 117, 11220–11222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiken E. L., McGough S. F., Majumder M. S., Wachtel G., Nguyen A. T., Viboud C., Santillana M., Real-time estimation of disease activity in emerging outbreaks using internet search information. PLOS Comput. Biol. 16, e1008117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santillana M., Nguyen A. T., Dredze M., Paul M. J., Nsoesie E. O., Brownstein J. S., Combining search, social media, and traditional data sources to improve influenza surveillance. PLOS Comput. Biol. 11, e1004513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reich N. G., Brooks L. C., Fox S. J., Kandula S., McGowan C. J., Moore E., Osthus D., Ray E. L., Tushar A., Yamana T. K., Biggerstaff M., Johansson M. A., Rosenfeld R., Shaman J., A collaborative multiyear, multimodel assessment of seasonal influenza forecasting in the United States. Proc. Natl. Acad. Sci. U.S.A. 116, 3146–3154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linton N. M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A. R., Jung S.-m., Yuan B., Kinoshita R., Nishiura H., Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. J. Clin. Med. 9, 538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson D. J., The harmonic mean p-value for combining dependent tests. Proc. Natl. Acad. Sci. U.S.A. 116, 1195–1200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. Liautaud, P. Huybers, M. Santillana, Fever and mobility data indicate social distancing has reduced incidence of communicable disease in the United States. arXiv:2004.09911 (2020).
- 43.Backer J. A., Klinkenberg D., Wallinga J., Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Euro. Surveill. 25, 2000062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verity R., Okell L. C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P. G. T., Fu H., Dighe A., Griffin J. T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C. A., Ghani A. C., Ferguson N. M., Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 20, 669–677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ord J. K., Getis A., Local spatial autocorrelation statistics: Distributional issues and an application. Geogr. Anal. 27, 286–306 (1995). [Google Scholar]
- 46.B. Resch, A. Summa, G. Sagl, P. Zeile, J.-P. Exner, in Progress in Location-Based Services 2014, G. Gartner, H. Huang, Eds. (Springer International Publishing, 2015), pp. 199–212. [Google Scholar]
- 47.Resch B., Summa A., Zeile P., Strube M., Citizen-centric urban planning through extracting emotion information from Twitter in an interdisciplinary space-time-linguistics algorithm. Urban Plan. 1, 114–127 (2016). [Google Scholar]
- 48.C. A. S. P. E. System, Nursing Home COVID-19 Data (2020).
- 49.R. Meyer, E. Kissane, A. Madrigal, The COVID Tracking Project (2020).
- 50.Coronavirus in the US: Latest map and case count; www.nytimes.com/interactive/2020/us/coronavirus-us-cases.html.
- 51.CDC FluView; https://gis.cdc.gov/grasp/fluview/fluportaldashboard.html.
- 52.Centers for Disease Control and Prevention, U.S. Influenza Surveillance System: Purpose and Methods.
- 53.Fisman D. N., Hauck T. S., Tuite A. R., Greer A. L., An IDEA for short term outbreak projection: Nearcasting using the basic reproduction number. PLOS ONE 8, e83622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vink M. A., Bootsma M. C. J., Wallinga J., Serial intervals of respiratory infectious diseases: A systematic review and analysis. Am. J. Epidemiol. 180, 865–875 (2014). [DOI] [PubMed] [Google Scholar]
- 55.UpToDate, COVID-19 search intensity monitoring dashboard (2020); https://covid19map.uptodate.com/.
- 56.Lu F. S., Hou S., Baltrusaitis K., Shah M., Leskovec J., Sosic R., Hawkins J., Brownstein J., Conidi G., Gunn J., Gray J., Zink A., Santillana M., Accurate influenza monitoring and forecasting using novel internet data streams: A case study in the Boston Metropolis. JMIR Public Health Surveill. 4, e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gluskin R. T., Johansson M. A., Santillana M., Brownstein J. S., Evaluation of Internet-based dengue query data: Google Dengue Trends. PLOS Negl. Trop. Dis. 8, e2713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teng Y., Bi D., Xie G., Jin Y., Huang Y., Lin B., An X., Feng D., Tong Y., Dynamic forecasting of Zika epidemics using Google Trends. PLOS ONE 12, e0165085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carneiro H. A., Mylonakis E., Google trends: A web-based tool for real-time surveillance of disease outbreaks. Clin. Infect. Dis. 49, 1557–1564 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Google, Coronavirus Search Trends (2020).
- 61.Twitter, Developer Docs (2020); https://developer.twitter.com/en/docs.
- 62.Resch B., Usländer F., Havas C., Combining machine-learning topic models and spatiotemporal analysis of social media data for disaster footprint and damage assessment. Cartogr. Geogr. Inf. Sci. 45, 362–376 (2018). [Google Scholar]
- 63.Dalziel B. D., Kissler S., Gog J. R., Viboud C., Bjørnstad O. N., Metcalf C. J. E., Grenfell B. T., Urbanization and humidity shape the intensity of influenza epidemics in U.S. cities. Science 362, 75–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson S., Hirsch J. S., Narasimhan M., Crawford J. M., McGinn T., Davidson K. W.; Northwell COVID-19 Research Consortium , Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 323, 2052–2059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chinazzi M., Davis J. T., Ajelli M., Gioannini C., Litvinova M., Merler S., Pastore y Piontti A., Mu K., Rossi L., Sun K., Viboud C., Xiong X., Yu H., Halloran M. E., Longini I. M. Jr., Vespignani A., The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science 368, 395–400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balcan D., Gonçalves B., Hu H., Ramasco J. J., Colizza V., Vespignani A., Modeling the spatial spread of infectious diseases: The GLobal Epidemic and Mobility computational model. J. Comput. Sci. 1, 132–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomes M. F. C., Pastore y Piontti A., Rossi L., Chao D., Longini I., Halloran M. E., Vespignani A., Assessing the international spreading risk associated with the 2014 West African Ebola outbreak. PLOS Curr. 6, ecurrents.outbreaks.cd818f63d40e24aef769dda7df9e0da5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Q., Sun K., Chinazzi M., Pastore y Piontti A., Dean N. E., Rojas D. P., Merler S., Mistry D., Poletti P., Rossi L., Bray M., Halloran M. E., Longini I. M. Jr., Vespignani A., Spread of Zika virus in the Americas. Proc. Natl. Acad. Sci. U.S.A. 114, E4334–E4343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.International Air Transportation Association; www.iata.org/.
- 70.Official Aviation Guide; www.oag.com/.
- 71.D. Mistry, M. Litvinova, A. Pastore y Piontti, M. Chinazzi, L. Fumanelli, M. F. C. Gomes, S. A. Haque, Q.-H. Liu, K. Mu, X. Xiong, M. E. Halloran, I. M. Longini Jr., S. Merler, M. Ajelli, A. Vespignani, Inferring high-resolution human mixing patterns for disease modeling. arXiv:2003.01214 (2020). [DOI] [PMC free article] [PubMed]
- 72.G. Project, COVID-19 Modeling: United States; https://covid19.gleamproject.org/.
- 73.A. Gelman, J. B. Carlin, H. S. Stern, D. B. Dunson, A. Vehtari, D. B. Rubin, Bayesian Data Analysis (CRC Press, 2013). [Google Scholar]
- 74.D. Fink, A compendium of conjugate priors 46 (1997); www.johndcook.com/CompendiumOfConjugatePriors.pdf [accessed 1 July 2020].
- 75.Gelman A., Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 1, 515–534 (2006). [Google Scholar]
- 76.D. W. Scott, Multivariate Density Estimation: Theory, Practice, and Visualization (John Wiley & Sons, 2015). [Google Scholar]
- 77.O. Punska, Bayesian Approaches to Multi-sensor Data Fusion (Cambridge University, 1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/10/eabd6989/DC1


